Abstract
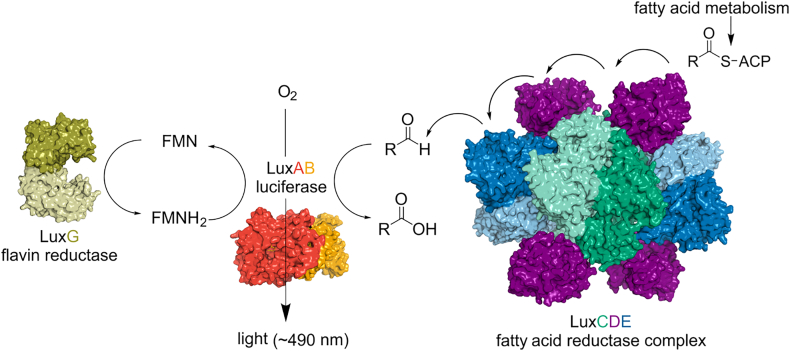
Bioluminescence refers to the production of light by living organisms. Bioluminescent bacteria with a variety of bioluminescence emission characteristics have been identified in Vibrionaceae, Shewanellaceae and Enterobacteriaceae. Bioluminescent bacteria are mainly found in marine habitats and they are either free-floating, sessile or have specialized to live in symbiosis with other marine organisms. On the molecular level, bioluminescence is enabled by a cascade of chemical reactions catalyzed by enzymes encoded by the lux operon with the gene order luxCDABEG. The luxA and luxB genes encode the α- and β- subunits, respectively, of the enzyme luciferase producing the light emitting species. LuxC, luxD and luxE constitute the fatty acid reductase complex, responsible for the synthesis of the long-chain aldehyde substrate and luxG encodes a flavin reductase. In bacteria, the heterodimeric luciferase catalyzes the monooxygenation of long-chain aliphatic aldehydes to the corresponding acids utilizing reduced FMN and molecular oxygen. The energy released as a photon results from an excited state flavin-4a-hydroxide, emitting light centered around 490 nm. Advances in the mechanistic understanding of bacterial bioluminescence have been spurred by the structural characterization of protein encoded by the lux operon. However, the number of available crystal structures is limited to LuxAB (Vibrio harveyi), LuxD (Vibrio harveyi) and LuxF (Photobacterium leiognathi). Based on the crystal structure of LuxD and homology models of LuxC and LuxE, we provide a hypothetical model of the overall structure of the LuxCDE fatty acid reductase complex that is in line with biochemical observations.
Keywords: Bacterial bioluminescence, Luciferase, lux operon, Structure-function relationships, Fatty acid reductase complex, Luciferin, FMN
Graphical Abstract
1. Introduction
Bioluminescence is the phenomenon of light emission that results from an enzyme-catalyzed oxidation reaction in living organisms. Bioluminescence can be found in nearly all kingdoms of life with a variety of luciferases and luciferins, the enzymes and light-emitting molecule involved in light emission, respectively. Consequently, the spectral range of light emission of bioluminescent organisms spans from ca. 400 to 700 nm, i.e. from blue to red light. Varieties of blue are the most common colors in light emission followed by green. Only very few species emit violet, yellow, orange or red light. The reason for this color preference relates to the predominant environment of bioluminescent organisms, mainly living in the seawater, which can best be penetrated by blue light (λmaxca. 475 nm) [1].
One component that nearly all bioluminescent reactions have in common is the dependence on oxygen. While molecular oxygen is used as an oxidizing agent in all cases, different biochemical reactions are catalyzed and various molecules are employed as luciferins. In fact, the enzymes involved in formation of excited state luciferins, the luciferases from evolutionary distant organisms, are neither related at the gene nor the protein sequence level [2]. As the individual mechanisms of the various bioluminescent systems strongly differ from each other, this review is focussed on bacterial bioluminescence.
In principle, the biological functions of bioluminescence can be categorized into four major groups: defense, counterillumination, prey attraction and intraspecific communication [1,3]. However, the purpose of bioluminescence for marine bacteria is still obscure. The possible biological advantages of light emission can be diverse, since bioluminescent bacteria can be free-floating, colonizing the skin of marine animals as saprophytes, living in their intestine as commensal enteric symbionts or in their tissues and body fluids as parasites. One observation revealed that luminous bacteria associated with fecal pellets in the mid-depths of the ocean are involved in nutrition turnover. Various fishes are attracted to the luminous fecal debris, which is then consumed. The bacteria are ingested and transferred to the gut where they find a nutrition rich environment to proliferate. After excretion, the luminous bacterial culture is associated with fecal pellets again, restarting the proliferation-cycle [1,4,5].
Another observation showed that some bioluminescent bacteria form a specific symbiosis with several families of marine fish or squids colonizing extracellular light organs [2]. For luminous bacteria living in symbiosis, the fish has the advantage of using the light emission for its purposes while the bacteria are provided with nutrition and an ideal growth environment [1]. In contrast, only few species were found in freshwater (e.g., V. cholerae) or terrestrial habitats (e.g., Photorhabdus species as symbionts of entomopathogenic nematodes) [2,6]. Although the habitat and the main function of light emission might be different, all luminous bacteria are gram negative, motile, rod-shaped and facultative anaerobic [7]. Beijerinck was the first to categorize them into the genus Photobacterium [8]. With new bioluminescent bacteria being isolated and due to the advancements in analyzing and determining various genera, a new taxonomy and phylogeny was necessary [9]. As a matter of fact, establishment of new clades and reclassification of luminous bacteria is still ongoing.
So far, luminous bacteria have been found among three families, namely Vibrionaceae, Shewanellaceae and Enterobacteriaceae, within five main genera Vibrio, Photobacterium, Aliivibrio, Photorhabdus and Shewanella [2,6,10].
1.1. The lux Operon
Despite classification into three families, all bioluminescent bacteria share the genes coding for the proteins responsible for light emission. The enzymes involved in bacterial bioluminescence are encoded in an operon with a single promoter. The gene order is conserved over various bacterial strains with the core sequence luxCDABE [2]. Due to various species harboring the lux operon and being capable of cold light emission, it was assumed that bioluminescence evolved independently at least 40 times [1,3]. Another hypothesis is that the lux operon arose once in the evolutionary past and that all genes have a common ancestor. Due to vertical gene transfer, gene duplications, losses and new recruitments, the constellations found today have evolved [2,6,9]. A schematic representation of different lux operons summarizes the gene architectures where sequence information is available so far (Fig. 1).
Fig. 1.
Examples of lux gene order of bioluminescent bacterial strains (adapted from Fig. 2 in Dunlap P. Bioluminescence: Fundamentals and Applications in Biotechnology - Volume 1. 2014 [6]). According to Table 1 (supplement) and available gene sequences and orders, the five most divergent bacterial strains (Vibrio harveyi, Aliivibrio fischeri, Photobacterium mandapamensis, Photobacterium leiognathi and Photorhabdus luminescens) were chosen to represent lux operon constellations. The color code of individual genes is also used for the corresponding protein models in Fig. 3, Fig. 4, Fig. 5, Fig. 6, Fig. 7, Fig. 8. The arrow above luxR indicates that its reading frame is oriented in the opposite direction (individual operon not directly linked to the lux operon).
Additionally, Table 1 (supplement) lists all bioluminescent bacterial strains, where DNA and protein sequences of most genes of the lux operon are available. Starting from Photobacterium leiognathi subsp. mandapamensis ATCC 27561, where the gene sequence of the whole lux-rib operon is known, extensive BLAST searches on NCBI were performed to find all obtainable bioluminescent bacterial strains with the core genes luxCDABE. All obtained strains were reanalyzed in order to identify all related sequences and after rigorous evaluation, 49 bioluminescent bacterial strains remained, as listed in Table 1. The bioluminescent bacterial strains given in Table 1 were the basis for sequence alignments to calculate conservation scores with the ConSurf server [11] for all structural models of the luciferase (Fig. 3) and the fatty acid reductase complex (Fig. 4, Fig. 5, Fig. 6, Fig. 7, Fig. 8).
Fig. 3.
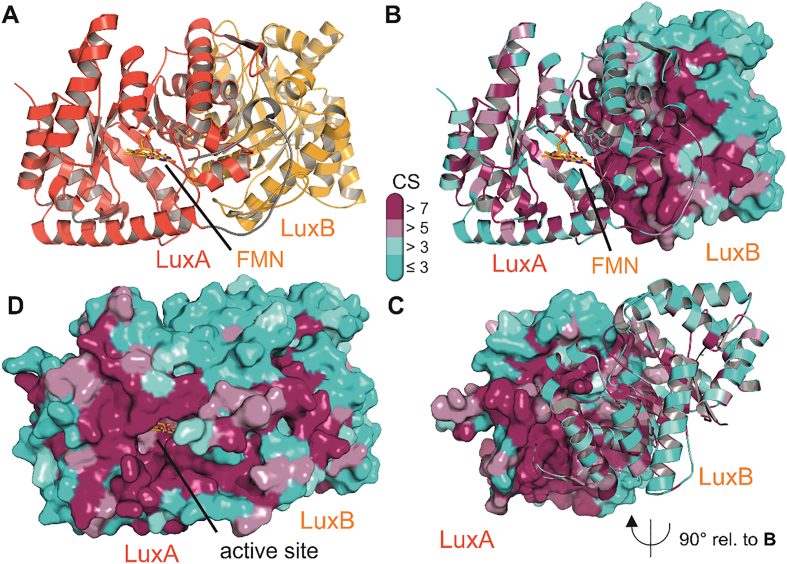
Crystal structure of the bacterial luciferase from Vibrio harveyi (PDB 3FGC). Panel A shows the characteristic heterodimer of LuxA (red) and LuxB (orange) in cartoon representation. The FMN cofactor in the active site is shown as yellow stick model. The characteristic loop region that mediates the contact between the α- and β-subunits is shown in grey [15]. Panels B, C and D feature the same luciferase dimer colored according to the conservation of residues among the members of bioluminescent bacteria (details of which sequences are aligned are shown in supplementary Table 1). Conservation scores from 1 to 9 correspond to an increase in evolutionary conservation and are colored according to the bar legend in the middle of the figure with higher scores (purple) indicating higher conservation. Panels B and C show the high conservation of residues at the heterodimer interface. Either LuxB or LuxA are shown in surface representation in panels B and C, respectively, and panel C features a 90° out of plane rotation of the dimer for better visibility of the highly conserved LuxA interface. Panel D shows both protomers in surface representation and highlights the strict conservation of residues in the open active site as well as its entrance. Conservation scores were computed with the ConSurf server [11].
Fig. 4.
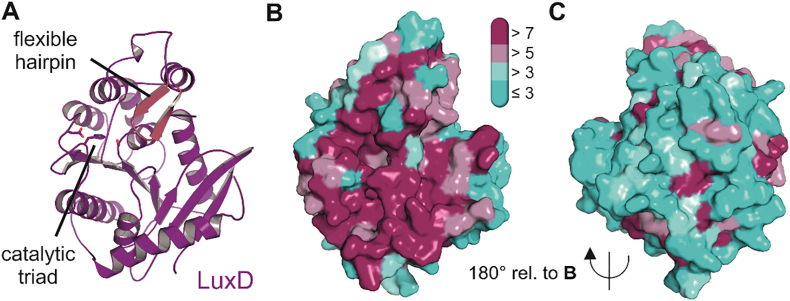
Crystal structure of LuxD, the myristoyl-ACP-specific thioesterase from V. harveyi (PDB 1THT). Panel A shows the cartoon representation of LuxD in purple color with residues of the catalytic triad as stick models. The flexible hairpin element above the catalytic serine residue is colored in light red. Panels B and C feature the same coloration according to evolutionary conservation of residues introduced in Fig. 3. Highly conserved residues of the whole surface region around the fatty acid binding site support its involvement in complex formation with LuxE and/or LuxC (see below).
Fig. 5.
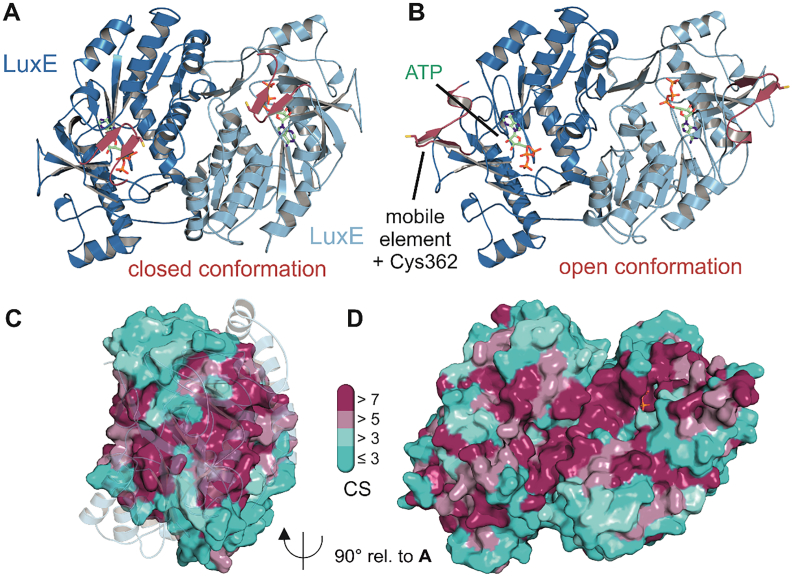
Homology models of LuxE from V. harveyi. Panels A and B correspond to homology models generated with the SWISS-MODEL server [95] based on PDB 2Y4O and PDB 4R1L, respectively, in the closed and open conformations. The mobile element is depicted in red color and the cysteine residue that can be acylated present in this region is shown as stick model. For reasons of clarity individual functional elements and cofactors are only labelled once in both panels A and B. Nevertheless, all red beta hairpins correspond to the same mobile element and all green stick models correspond to ATP. The overall LuxE dimer is shown in cartoon representation with one protomer colored in blue and the other in light blue. The ATP moiety overlaid from the cofactor bound form of the closed LuxE model template (PDB 2Y27) is shown as green stick model and its proximity to Cys362 further supports the relevance of the closed LuxE state. The open conformation is relevant for the LuxE-LuxC interaction as described before. Panels C and D show the evolutionary conservation of residues according to the ConSurf generated conservation scores (CS – bar legend in the middle). Panel C shows the conservation of the dimer interface in a rotated view relative to panel A and with the light blue protomer shown in transparency. Panel D shows the same view as panels A and B to illustrate the high conservation of residues at a specific surface of the dimer, which is therefore likely involved in the interaction with the LuxC reductase component (cf.Fig. 7) [92].
Fig. 6.
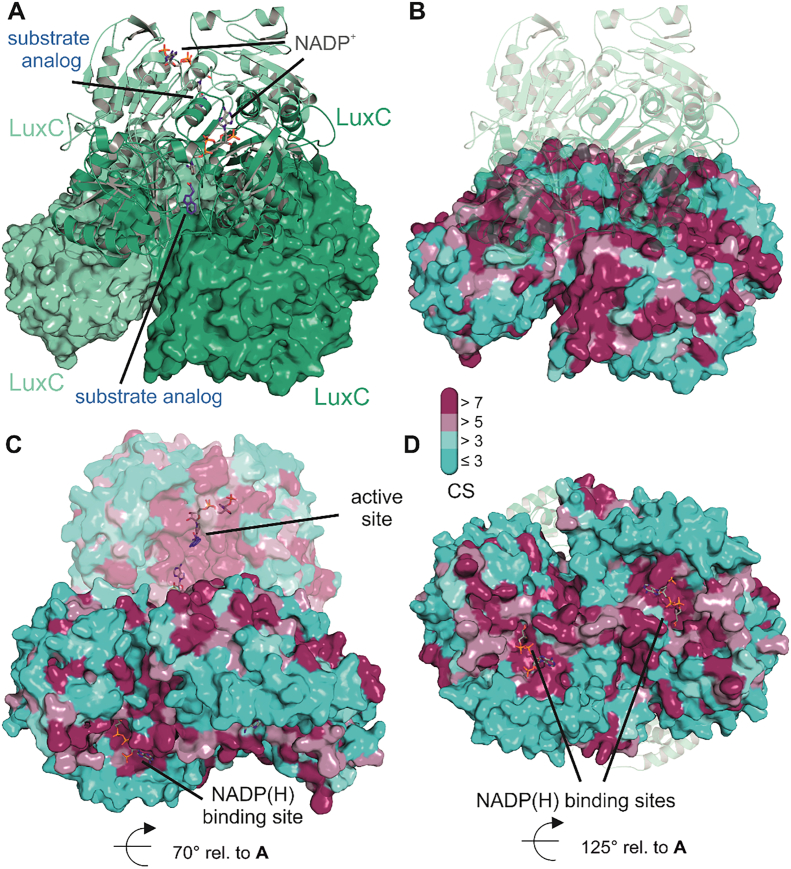
Homology model of LuxC from V. harveyi. Panel A features the tetrameric assembly of LuxC obtained from the SWISS-MODEL server [95] using the crystal structure of methylmalonate semialdehyde dehydrogenase from Bacillus subtilis as template (PDB 1T90). The tetramer corresponds to a dimer of dimers, which are shown once as cartoon representation and once as molecular surface. Individual protomers of each dimer are colored in green and light green. For the dimer in cartoon representation, we also show stick models of the substrate analog (indole-3-acetaldehyde – blue) and the cofactor (NADP+ − grey) in the respective binding sites obtained from the superposition of the LuxC model with indole-3-acetaldehyde dehydrogenase from Pseudomonas syringae (PDB 5IUW) and from the structure of an aldehyde dehydrogenase from Burkholderia multivorans (PDB 5JRY), respectively. Panels B, C and D provide an overview of the evolutionary conservation of residues according to the ConSurf server [11] and computed conservation scores (CS – bar legend). Panel B highlights the conserved residues at the interface of the individual LuxC dimers in the same orientation as panel A. For clarity, the dimer in cartoon representation is shown in transparent mode. Panel C shows a different orientation of the tetrameric assembly, to demonstrate the high degree of conservation in the active site generated at the interface of two subdomains of each LuxC protomer. Importantly, the substrate and the NADPH cofactor approach the active site from opposite sides. Panel D shows another view of one LuxC dimer highlighting the conservation of residues around the NADP+ binding site. For better visibility loop regions covering the NADP+ binding site are not shown in this figure. Similar to the observations for LuxE, patches of strongly conserved residues can be found at surface elements near the active site that are not involved in LuxC oligomerization and are therefore likely involved in complex formation with the LuxE synthetase subunits.
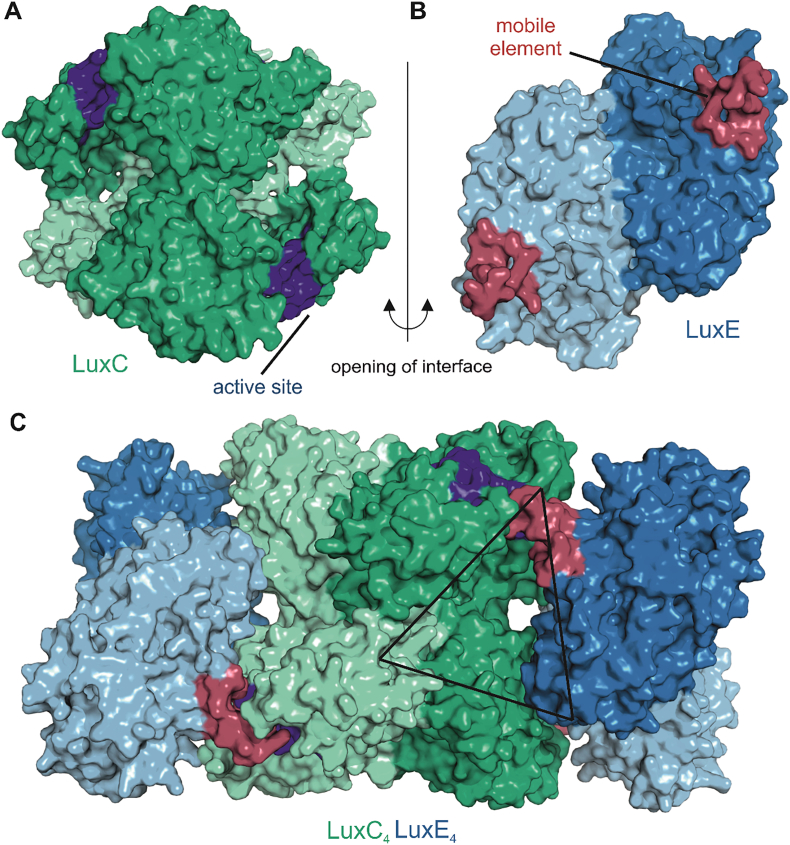
Fig. 7.
Model of the LuxC - LuxE interaction. Panels A and B show surface representations of LuxC and LuxE, respectively, colored according to the details presented in Fig. 5, Fig. 6. The active site region of LuxC is highlighted in dark blue and the mobile element of LuxE containing the acylated residue is colored in red. The central region of complementarity at the LuxC2-LuxE2 interface is lined by many highly conserved residues at specific surfaces of the respective oligomeric structures that have been highlighted in Fig. 5D (LuxE) and Fig. 6B (LuxC). Their complementarity is illustrated by the opening of the interface (right side interface of the complex in panel C) by opposite 90° rotations of LuxC and LuxE. Panel C also shows the second LuxE dimer bound to the opposite side of the LuxC tetramer resulting in an overall LuxC4LuxE4 stoichiometry. The architecture around the active site reveals an interesting triangular complementarity to the LuxD structure (Fig. 4). For generation of the LuxC-LuxE complex we used a different template for homology modeling of LuxC to better reflect the open apo-conformation that might be needed for the initial interaction with LuxE. This model was again generated with the SWISS-MODEL server [95] based on the apo form of the indole-3-acetaldehyde dehydrogenase (PDB 5IUU). The apo form is characterized by a modest opening of the active site accompanied by unstructured loop regions involved in substrate coordination.
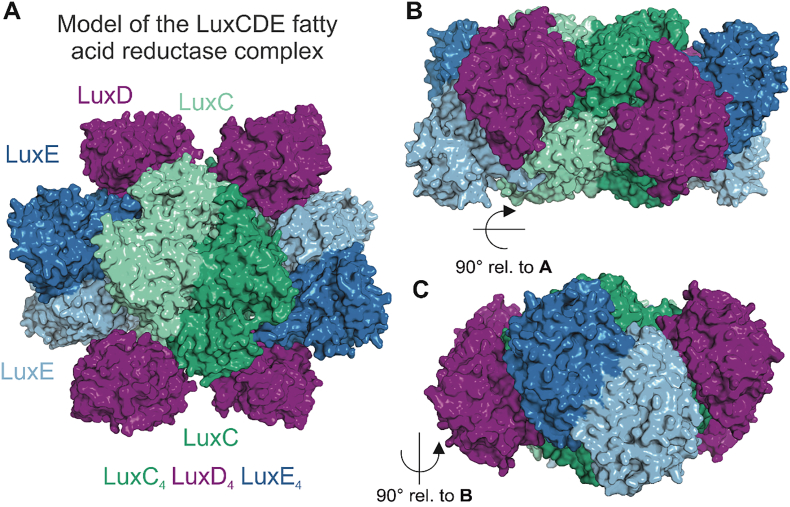
Fig. 8.
Tentative model of the LuxCDE fatty acid reductase complex. Panels A, B and C show different views of the LuxC4LuxD4LuxE4 complex. Individual subunits are colored according to their individual presentations in Fig. 4, Fig. 5, Fig. 6. Dark and light colors correspond to individual protomers of the dimeric LuxC and LuxE subspecies. Monomeric LuxD protomers shield off individual active sites of the complex and can readily dissociate [91] to allow release of the final fatty aldehyde product and reassociate to deliver new fatty acid substrates after unloading them from the ACP.
The genes luxA and luxB encode the heterodimeric luciferase; luxC, luxD, and luxE are part of the fatty acid reductase complex and luxG encodes a flavin reductase. Next to the core genes luxCDABE(G), additional genes are found within the lux operon (luxF; ribEBHA; luxI) or in a separate operon (luxR) adjacent to the lux operon, where the reading frame is in the oppposite direction (Fig. 1).
1.2. Enzymes and Reaction Mechanisms in Bacterial Bioluminescence
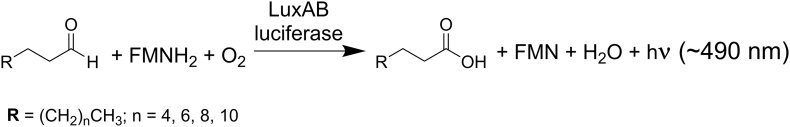
The core genes, luxCDABE(G), code for all enzymes involved in a complex machinery enabling bioluminescence. In bioluminescent bacteria, the heterodimeric enzyme luciferase (LuxAB) catalyzes the monooxygenation of aliphatic aldehydes to the corresponding acids utilizing reduced flavin mononucleotide (FMNH2) as redox cofactor. Tetradecanal is postulated to be the natural substrate of bacterial luciferases, however long-chain aliphatic aldehydes with carbon chain lengths of 8 to 16 are also potential substrates [12]. From an excited state FMN-4a-hydroxide the energy is released as light, thus this intermediate serves as the light emitting luciferin [13,14]. According to the structures of the luciferase, the active site enabling light emission is located in the α-subunit, while the β-subunit is presumably responsible for stability, folding and quantum yield. The two subunits share approximately 30% sequence identity suggesting that luxB arose by gene duplication of luxA [15]. The overal reaction mechanism of bacterial bioluminescence is depicted in Scheme 1.
Scheme 1.
General reaction mechanism of bacterial bioluminescence. Long-chain aldehydes (CH3(CH2)nCHO), reduced flavin mononucleotide (FMNH2) and molecular oxygen (O2) are converted by the enzyme luciferase (LuxAB) to the corresponding long-chain acids (CH3(CH2)nCOOH), oxidized flavin mononucleotide (FMN), water (H2O) and light emission (hν) with an approximate maximum at 490 nm.
The bacterial bioluminescent reaction belongs to the group of two component systems since reduced FMN is necessary for the reaction to proceed and has to be supplied by another enzyme than the luciferase itself [16,17]. Due to its high similarity to Fre, a flavin reductase in E. coli, it was assumed that the protein LuxG has a similar function in bioluminescent bacteria [[18], [19], [20]]. In 2008, Nijvipakul et al. determined the function of LuxG as a NAD(P)H-dependent flavin reductase. LuxG converts free flavin (FMN) to reduced flavin (FMNH2) that can be used by the luciferase for bacterial bioluminescence [21,22]. Because the product FMNH2 is prone to oxidation by molecular oxygen, it is still a matter of debate whether FMNH2 is released by the reductase and freely diffuses to be sequestered by the luciferase or if a LuxG – LuxAB complex is formed in order to directly transfer reduced FMN from the reductase to the luciferase. A separate paragraph is dedicated to provide some more details related to this question.
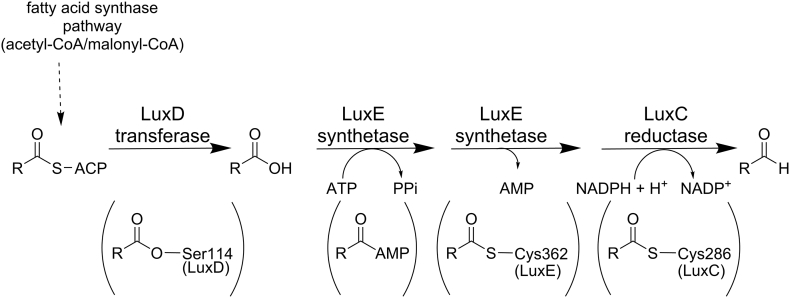
To supply the long-chain aldehyde substrates to the luciferase, the proteins LuxC, LuxD, and LuxE constitute a fatty acid reductase complex [13]. The gene luxC encodes a NADPH-dependent acyl protein reductase (approximately 54 kDa), luxD specifies an acyl-transferase (approximately 33 kDa) and luxE encodes an acyl-protein synthetase (approximately 42 kDa). A schematic overview of the reactions catalyzed by this complex is shown in Scheme 2.
Scheme 2.
Overview of individual reactions catalyzed by the LuxCDE fatty acid reductase complex. Intermediate species covalently linked to individual enzymes are shown in brackets beneath the corresponding reaction steps. The reaction cascade is initiated by the myristoyl transferase LuxD via unloading of myristic acid (R = (CH2)12CH3) bound to the acyl carrier protein (ACP). Covalently linked to Ser114 (Vibrio harveyi numbering) the acyl moiety is transported to the LuxCE complex and released as the free fatty acid that interacts with the LuxE synthetase. At the expense of ATP the fatty acid is activated by LuxE to acyl-AMP and in a second step covalently attached to Cys362. This intermediate is then channeled directly to the active site of the LuxC reductase, where it is initially transferred to Cys286 of LuxC. The latter intermediate is then reduced by NADPH resulting in aldehyde formation and dissociation of the product. Details of the individual processes are provided in the section -The fatty acid reductase complex (luxCDE).
In addition to the canonical luxCDABE(G) gene cluster, a number of bioluminescent Photobacteria carry an additional luxF gene. Natural occurring nonsense mutations in this gene revealed that the strains carrying this inactivated luxF are luminous but to a lower degree. This would support the hypothesis that LuxF is somehow involved in regulating light intensity [23]. The additional gene luxF in the lux operon of photobacterial strains, led to the differentiation of two subgroups of Photobacterium leiognathi, Photobacterium leiognathi subsp. leiognathi (luxF−) and Photobacterium leiognathi subsp. mandapamensis (luxF+). Additional phenotypic characterization led to further discrimination of the two groups. P. leiognathi subsp. leiognathi shows higher light intensities at low salt concentrations and a more intense blue color, while P. leiognathi subsp. mandapamensis shows higher light intensities at high salt concentrations and blue-green color. Ast and Dunlap speculated that luxF arose through a gene duplication event of luxB in an ancestor of Photobacterium, which was lost again in the lineage leading to subsp. leiognathi in accordance with the observation that only Photobacterium leiognathi subsp. mandapamensis and Photobacterium phosphoreum contain luxF [24].
During the structural characterization of the homodimeric protein LuxF, an unusual flavin derivative - 6-(3′-(R)-myristyl)-flavin mononucleotide (myrFMN) - was discovered in keeping with the observed electron density (PDB 1FVP) [[25], [26], [27]]. More recently, it was reported that LuxF binding to myrFMN is important to prevent inhibition of the luciferase by this putative side product of the luciferase reaction. LuxF is thereby acting as a scavenger of myrFMN [28,29]. The chemical nature of this side product of the bioluminescent reaction also provides insights into the mechanism of the luciferase reaction, described in more detail later in this review.
Recently, bacterial bioluminescence has been utilized as a reporter system in plant protoplasts as well as for single-cell imaging [30,31]. Toward this end, Cui et al., have constructed a fusion of luxA and luxB from Photorhabdus luminescens that was further optimized by directed evolution. This resulted in five amino acid exchanges of which four were found in the domain that harbors the active site of the enzyme (corresponding to the α-subunit in the heterodimeric luciferase). However, the amino acid replacements are not in or near the active site and thus their effect on the activity and efficiency of the fused luciferase is unclear [31]. Furthermore, Gregor et al. have subjected the entire lux operon, i.e. including the genes encoding the fatty acid reductase complex as well as the flavin reductase, for optimization using directed evolution. Several beneficial mutations in luxA, luxB, luxC and frp could be identified that led to a seven-fold increase of brightness when expressed in Escherichia coli [30]. As before, the molecular reasons for enhanced light emission remain undetermined and may include several factors such as enhanced gene expression or increased enzyme activity/efficiency. Although, these examples demonstrate the utility of bacterial bioluminescence for further applications, for example in the field of imaging, unfortunately they have failed to provide additional information on critical issues such as the population of the excited state and the involvement of amino acid residues in the active site as well as the dynamic interaction of the α- and β-subunits.
1.3. Genes Associated with the lux Operon
Apart from the core genes luxCDABE(G), various other genes are coregulated, coexpressed or linked to the lux operon. Having a closer look on these additional genes revealed their importance for fine tuning of light emission.
For some luminous bacteria, bioluminescence is regulated via cell density dependent induction or derepression of luciferase-gene expression, so called quorum sensing regulation. Small secondary metabolite levels, e.g. acyl homoserine lactones, reflect the cell density of bioluminescent bacteria. Especially for two bacterial strains, Aliivibrio fischeri and Vibrio harveyi, this regulatory mechanism was investigated in detail. Among many other regulatory factors, the genes luxI, within the lux operon before luxC, and luxR, directly adjacent to the operon with the reading frame in the opposite direction (Fig. 1), are involved in quorum sensing regulation. Furthermore, levels of autoinducers and their synthetases, transcriptional regulators, kinases involved in phosphorylation/dephosphorylation cascades and signal transduction cascades and even small quorum-regulatory RNAs play a role in this complex regulatory mechanism and influence the expression levels of the genes in the lux operon [2,6,32,33].
In strains of the genus Photobacterium, the genes ribEBHA may play a role in bioluminescence. These genes are involved in riboflavin biosynthesis and are an example for gene recruitment, constituting together with the lux genes the lux-rib operon –luxCDAB(F)EG – ribEBHA. These rib genes might facilitate light production as they provide a part of the FMN substrate. Depending on the strain none, one or more of these rib genes are cotranscribed with the lux genes. Interestingly, there is no transcriptional stop or other regulatory terminator between lux and rib genes indicating coexpression [6,34].
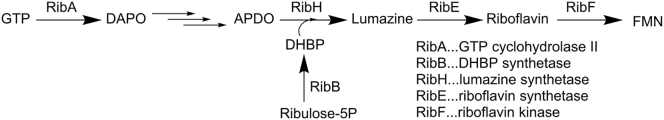
In Scheme 3 the synthesis pathway of riboflavin/FMN is depicted with the involved rib proteins highlighted. DNA sequence analysis and alignment to homologous genes identified the rib genes and the corresponding function of the proteins. The gene ribE encodes a riboflavin synthetase (RibE) converting lumazine to riboflavin; ribB encodes 3,4-dihydroxyl-2-butanone 4-phosphate (DHBP) synthetase (RibB); ribH encodes the lumazine synthetase (RibH) and ribA encodes a GTP cyclohydrolase II (RibA) [34].
Scheme 3.
Schematic representation of the synthesis pathway of riboflavin (adapted from Fig. 2 in Sung, Lee, J. Photosci., [35]). Guanosine triphosphate (GTP) is converted to 2,5-diamino-6-(5′-phosphoribosylamino)-4-pyrimidineone (DAPO) by RibA, which is further converted in three steps to 5-amino-6-(D-ribitylamino)uracil (APDO). Another route starts from ribulose 5-phosphate, which is converted to 3,4-dihydroxy-2-butanone 4-phosphate (DHBP) by RibB. The enzyme RibH produces the product 6,7-dimethyl-8-ribityllumazine (lumazine) from DHBP and APDO. Lumazine is converted to riboflavin by RibE, which is subsequently transformed into flavin mononucleotide (FMN) by a riboflavin kinase.
1.4. Color Change in Light Emission
One of the first descriptions of a bioluminescent bacterial strain emitting light in a different color than blue-green was Photobacterium fischeri Y-1 (later V. fischeri). This strain emits yellow light and was isolated from seawater in 1977 [36]. The yellow fluorescent protein YFP binds FMN and shifts the light emission from around 490 nm to 545 nm. Interestingly, this color change was temperature dependent, where cells grown above 22 °C will emit blue-green light and cells grown below 18 °C will emit yellow light (Fig. 2) [[36], [37], [38]].
Fig. 2.
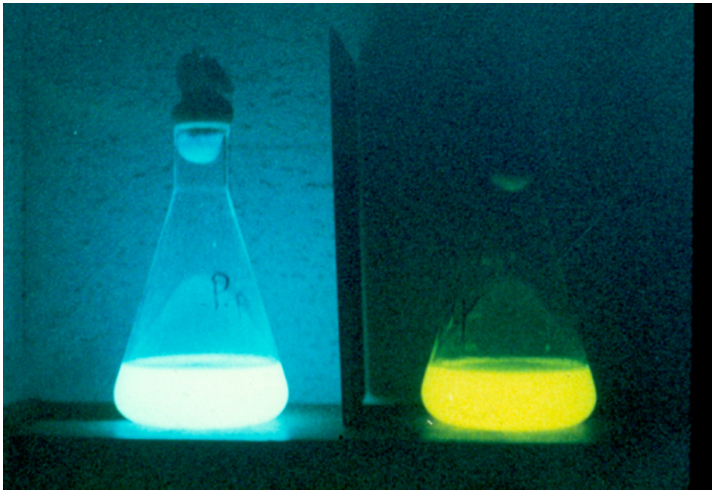
Bacterial cultures of Photobacterium phosphoreum on the left and Vibrio fischeri Y-1 on the right. The light emission in blue and yellow, respectively, shows nicely the effect of LumP and YFP [47].
At the same time as YFP was discovered, another shift in light emission toward blue color was reported. The blue fluorescent protein (BFP, later termed lumazine protein) was isolated from Photobacterium phosphoreum, Photobacterium fischeri and Photobacterium leiognathi (later reclassified as P. mandapamensis) [[39], [40], [41], [42]]. The isolation of BFP from P. phosphoreum and the identification of its ligand as 6,7-dimethyl-8-ribityllumazine (lumazine), a precursor of riboflavin biosynthesis (Scheme 3), led to the renaming to lumazine protein (LumP) [43]. Comparing in vivo and in vitro analysis revealed that in vivo the bioluminescence maximum is at 475 nm, while in vitro the maximum lies at 490 nm. After addition of BFP to the in vitro reaction, the maximum shifts to 475 nm as in the in vivo result. The fluorescence spectrum matches the in vivo bioluminescence spectrum, but the BFP spectrum can be easily altered by temperature, dilution, pH, ionic strength and urea (due to an increase of free lumazine). Both antenna proteins (BFP and YFP) are species specific and can influence the kinetics by enhancing the light intensity and the decay rate [40,41,[44], [45], [46]].
These first analyses led to vigorous discussions about the primary emitter of the bioluminescent reaction and the possible reaction mechanisms behind the energy transfer to the antenna proteins [40,41,44,48,49]. It was speculated that LumP and YFP form a complex with the luciferase enabling weak dipole-dipole Förster-type coupling of the high energy intermediate of the bioluminescent reaction with the antenna transitions. To make FRET (Förster resonance energy transfer) possible, a spectral overlap of the fluorescence spectrum of the donor and the absorption spectrum of the acceptor is needed [48,49]. It was further suggested that the aldehyde substrate holds the two proteins (luciferase and antenna protein) in place. The complex formation has to take place before the excited state intermediate is formed to ensure the energy transfer in time [41,42,45,48,49].
For LumP, X-ray analyses provided further insight in the mechanism. The crystal structure of the L49 N variant of the lumazine protein of P. leiognathi in complex with riboflavin (PDB: 3DDY) gave first structural information on the binding site. The crystal structure has high similarity with the riboflavin synthase of Schizosaccharomyces pombe and Escherichia coli. However, a closer comparison of riboflavin synthase and lumazine protein revealed certain differences, as the latter is a monomer in solution and binds only one molecule of 6,7-dimethyl-8-ribityllumazine (or riboflavin) in the N-terminal domain [50]. The crystal structure of LumP of Photobacterium kishitanii in complex with lumazine, riboflavin and FMN (PDB: 3A3G, 3A35; 3A3B) confirmed one single binding site at the N-terminal region. Computational docking of LumP-Lumazine (PDB: 3A3G) with luciferase of V. harveyi (PDB: 3FGC) predicts complex formation, which is supported by complementary charge distributions on the surfaces of the luciferase and LumP. This suggests a possible direct energy transfer between those two proteins, as the ring system of lumazine is located in close proximity (approximately 10 Å) to the isoalloxazine ring of FMN [48,51]. However, the overlap of the absorption spectrum of the lumazine with the emission spectrum of the luciferase is very small and, moreover, the shift in light emission is to higher energy (for color change toward blue at 475 nm). The question of how energy transfer is achieved remains unanswered [40,44,52]. Recent studies and reviews failed to provide deeper insight into the exact mechanism of the color change of bioluminescent emission [48,53,54] and thus this phenomenon remains puzzling.
Each enzyme presented above contributes to the core machinery of the lux operon enabling light emission, or even modulation of light intensity and color or regulation of bacterial bioluminescence. While the schemes provided above represent the overall reaction, they are simplified representations of complex molecular mechanisms. Using structural information from individual enzymes of the Lux family and addressing evolutionary conserved areas within these structures, allows a better understanding of the underlying core reaction mechanisms. Especially in combination with the comparison of so far not crystallized members of the lux operon with closely related enzyme families and homology modeling, molecular mechanisms and structure function relationships of the enzymes involved in bacterial bioluminescence can be described.
2. Structure-Function Relationships
2.1. The Mechanism of Bacterial Bioluminescence
Bacterial bioluminescence is based on a classical two-component system consisting of an enzyme (termed luciferase) that catalyzes the bioluminescent reaction, and a small molecule that acts as the light-emitting species in the course of the reaction (termed the luciferin, reviewed in [4,55]). In the case of bacterial bioluminescence, the luciferin is derived from flavin mononucleotide (FMN), which undergoes a sequence of reactions as shown in Scheme 4. FMN is widely used in nature as a versatile cofactor in a plethora of biochemical reactions involving the handling of electrons, i.e. oxidation-reduction reactions (EC class 1: oxidoreductases) [56]. The energy required to populate the excited state of the luciferin is derived from the oxidation of a long-chain fatty aldehyde, which is synthesized in bioluminescent bacteria from myristoyl-ACP (see Introduction and The fatty acid reductase complex, below), to the corresponding long-chain fatty acid.
Scheme 4.
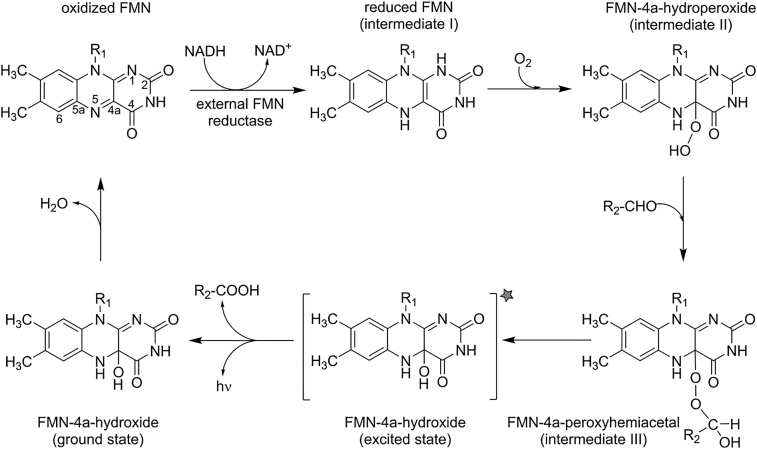
Reaction cycle of FMN during bacterial bioluminescence. Oxidized flavin mononucleotide (FMN; R1: ribityl monophosphate) is reduced by an external FMN reductase employing NAD(P)H as the reducing agent. Reduced FMN (intermediate I) reacts with dioxygen and forms the FMN-4a-hydroperoxide (intermediate II). The addition of long-chain aldehydes (e.g. R2: (CH2)12CH3) leads to the formation of the FMN-4a-peroxyhemiacetal (intermediate III). After monooxygenation of the long-chain aldehyde to the corresponding acid, an excited state FMN-4a-hydroxide is formed, which is the luciferin in the bacterial bioluminescent reaction. As this luciferin relaxes to the ground state, the free energy is released as light with an emission maximum at 490 nm. After release of one water molecule the catalytic cycle is completed and FMN returned to its oxidized ground state.
In addition to the wealth of biochemical experiments addressing mechanistic aspects of bacterial bioluminescence, the elucidation of the crystal structure of a functional luciferase was a major milestone in understanding the involvement of individual amino acids and specific structural elements. Molecular details of the luciferase from Vibrio harveyi have shown that the luxA and luxB gene products form a heterodimer consisting of two (βα)8 barrels [57,58]. The active site of the luciferase that needs to accommodate all substrates, shown in Scheme 1 (Introduction), was speculated to be present only in the LuxA-subunit. Subsequent crystal structures that have the oxidized FMN cofactor bound to the α-subunit confirmed this assignment of the active site and further demonstrated the importance of the β-subunit in stabilizing the active conformation of the α protomer via specific contacts between conserved residues in the β-subunit and an α-subunit loop region close to the active site (Fig. 3 panels A and B) [15]. Thus far, no structural information on the spatial orientation of the FMN and the aldehyde (or intermediates of the reaction) is available, preventing a more detailed interpretation of how the excited state is generated in the course of the overall reaction.
The central question in understanding any bioluminescent process revolves around two issues: (i) what is the structure of the excited state and (ii) how is it populated during the bioluminescent reaction? In order to tackle these questions, research efforts focused initially on the identification of reaction intermediates. A milestone toward a better understanding of the bioluminescent reaction was the identification of the FMN-4a-hydroperoxide (also referred to as intermediate II) that forms upon reaction of luciferase-bound reduced FMN with dioxygen (Scheme 4) [59,60]. This intermediate is fairly stable in the absence of aldehyde substrates, with half-lives of up to 41 min at 4 °C [60,61] and decays to oxidized FMN and hydrogen peroxide. Its stability also enabled characterization by UV–Vis absorption and NMR-spectroscopy lending further support to its chemical structure [60,62]. Interestingly, the same flavin intermediate is involved in the hydroxylation of aromatic substrates, e.g. ρ-hydroxybenzoate hydroxylase and phenol hydroxylase, which also carry out monooxygenation reactions [63]. However, none of these flavin-dependent aromatic hydroxylases produce light during the reaction of their respective substrates suggesting that fundamental mechanistic differences exist between these enzymatic systems. Following the identification of the FMN-4a-hydroperoxide as an intermediate of the bioluminescent reaction, the actual light-emitting species could be identified as the FMN-4a-hydroxide, which forms as a result of the reaction of the FMN-4a-hydroperoxide and the long-chain fatty aldehyde (Scheme 4) [14]. The chemical identity of the FMN-4a-hydroxide as the bioluminophore was also confirmed by a recent theoretical study [64]. These two intermediates are in fact linked by the FMN-4a-peroxyhemiacetal that forms by the reaction of the FMN-4a-hydroperoxide and the long-chain fatty aldehyde (Scheme 4, bottom right). Since the FMN-4a-peroxyhemiacetal is less stable than the hydroperoxide, it could thus far only be detected by absorption spectroscopy [65]. After light emission the FMN-4a-hydroxide splits off water and returns into the oxidized state (Scheme 4, left), thus closing the catalytic cycle. Bacterial luciferases are not capable of reducing the oxidized flavin to the reduced state, in fact oxidized FMN binds rather weakly to the enzyme [66], and therefore reduced FMN needs to be provided by other enzymes, such as NAD(P)H-dependent flavin reductases such as LuxG mentioned above (Scheme 4).
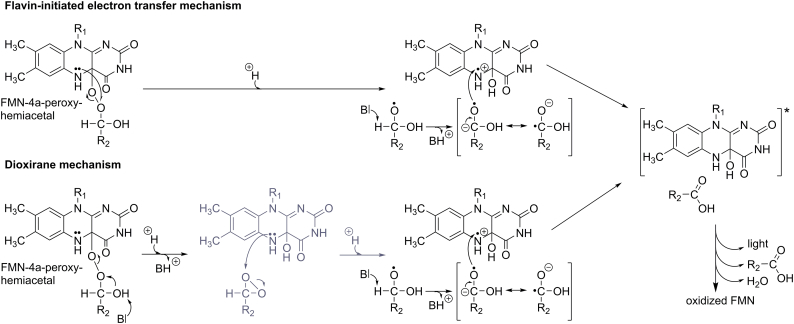
Although the sequence of chemical events shown in Scheme 4 is generally agreed upon, the generation of the excited FMN-4a-hydroxide from the FMN-4a-peroxyhemiacetal remains a matter of debate [67,68]. Based on the overall bioluminescent reaction, Eberhard and Hastings proposed a Baeyer-Villiger mechanism for the bacterial luciferase reaction [69]. Although in keeping with the stoichiometry of the overall reaction, it does not per se explain how the excited state of the FMN-4a-hydroxide is populated. In fact, enzymes that reportedly follow a Baeyer-Villiger reaction mechanism, such as cyclohexanone monooxygenase, lack the production of light entirely [63]. Moreover, cyclohexanone monooxygenase converts boronic to boric acids in a Baeyer-Villiger reaction mechanism, whereas bacterial luciferase fails to catalyzes these oxidation reactions [61,70]. This inconsistency led to the development of alternative mechanistic models that better explain the generation of the excited state of the luciferin [[71], [72], [73]]. Based on kinetic studies using flavin analogs bearing different substituents at the 8-position of the isoalloxazine ring, it was proposed that a single electron transfer from the isoalloxazine moiety of the flavin to the distal oxygen atom of the peroxide initiates bond cleavage and the concomitant formation of a radical pair consisting of the FMN-4a-hydroxide radical cation and the substrate-derived carboxylic acid radical anion, as shown in Scheme 5 [71]. The transfer of an electron from the carboxylic acid radical anion to the FMN-4a-hydroxide radical cation eventually leads to the generation of the excited state of the FMN-4a-hydroxide. This mechanism, reminiscent of the so-called chemically initiated electron exchange luminescence (CIEEL) mechanism [74], has received additional support from the observation that the electron transfer from the carboxylic acid radical anion to the FMN-4a-hydroxide radical cation provides sufficient energy (ca. 90 kcal/mol) to populate the excited state [72]. In a variation of this mechanism, the FMN-4a-peroxyhemiacetal directly decays to the FMN-4a-hydroxide and a dioxirane intermediate (Scheme 5, lower reaction path). This step is followed by an electron transfer, in this case from the FMN-4a-hydroxide rather than the FMN-4a-peroxyhemiacetal, yielding the same radical pair as before (Scheme 5). However, the dioxirane mechanism suffers from a major drawback: the FMN-4a-hydroxide appears twice in the reaction sequence, before and after the rate-limiting step that populates the excited state. This is in contrast to experimental findings that have demonstrated its occurrence only as a result of the relaxation of the excited state FMN-4a-hydroxide, i.e. after the rate-limiting step [14,75].
Scheme 5.
Mechanistic details of the flavin-initiated electron transfer and the dioxirane mechanism. In the former mechanism (upper reaction path), electron transfer from N5 of the isoalloxazine ring to the distal oxygen atom of the flavin-4a-peroxyhemiacetal (R1: ribityl monophosphate) leads to the formation of a substrate-derived alkoxy radical (e.g. R2: (CH2)12CH3) and the flavin-4a-hydroxy radical cation. Deprotonation of the alkoxy radical generates a resonance stabilized anion radical, which transfers an electron back to the flavin-4a-hydroxy radical cation thus leading to the population of the excited state of the flavin-4a-hydroxide. In an alternative route to this mechanism, the dioxirane mechanism (lower reaction path), the flavin-4a-peroxyhemiacetal forms a dioxirane intermediate, which then receives an electron from the flavin-4a-hydroxide (depicted in dark grey). As before, this leads to the flavin-4a-hydroxide radical cation and the subsequent generation of the excited state similar to the mechanism in the flavin-initiated electron transfer mechanism. In both reactions the rate limiting step is the electron donation from the reduced flavin moiety to the substrate moiety.
As mentioned above, the flavin-initiated electron transfer mechanism is conceptually derived from the CIEEL mechanism originally proposed by Schuster and was applied to rationalize the generation of a high energy intermediate in firefly bioluminescence [74,76]. However, redetermination of chemiexcitation efficiencies yielded much lower values than originally reported and have led to a critical reevaluation of the validity of the CIEEL mechanism [77]. Since the classical CIEEL mechanism involves the transfer of an electron from the donor to the peroxide followed by back-transfer of an electron to generate the excited state of the fluorophore (or in the case of bioluminescence the excitated state of the luciferin), it has been argued that a charge transfer, i.e. not a “full” electron, may be sufficient to trigger the decomposition of the peroxide and subsequently leads to the generation of an excited state [[77], [78], [79], [80]]. Although most theoretical studies in the field of bioluminescence have focused on the firefly [[78], [79], [80]] and Cypridina bioluminescence [81,82], it is conceivable that an equivalent charge-transfer process occurs in the bacterial bioluminescent reaction. Taking this into consideration, the mechanism depicted in Scheme 5 (top) represents the extreme case of a “full” electron transfer but may in fact only involve the transfer of charge from the flavin to the peroxide bond to trigger the events leading to the generation of the excited state FMN-4a-hydroxide.
In mechanistic terms it is also important to consider in which step the carbon-hydrogen bond at C1 of the aldehyde substrate is broken. As shown in Scheme 5, electron transfer is presumably followed by carbon-hydrogen bond cleavage. In order to assess whether this step is rate-limiting, kinetic isotope effects, using deuterated [1−2H]-aldehyde substrates, were determined. The moderate kinetic isotope effects of 1.4–1.9 [73,83] suggested that carbon-hydrogen bond breakage is only partially rate-limiting and thus supports the electron transfer mechanism depicted in Scheme 5. More recently, a computational study suggested that the excited state is populated by a charge-transfer rather than an electron transfer mechanism [67]. In the proposed mechanism, carbon-hydrogen bond cleavage is directly connected to the generation of the excited state and thus a strong kinetic isotope effect should be observed, which is arguably not the case. On the other hand, it is also unclear how charge-transfer coupled to carbon-hydrogen bond cleavage results in the population of the excited state of the FMN-4a-hydroxide.
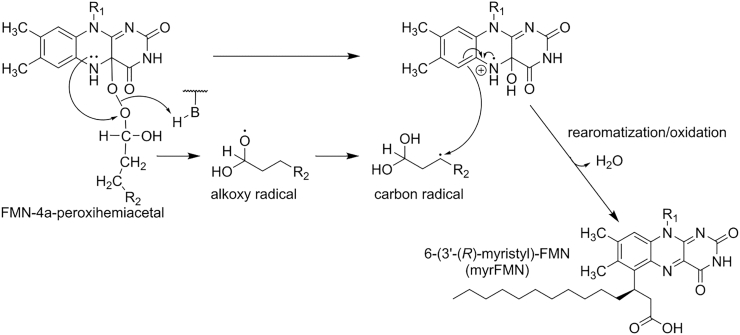
The transient occurrence of radicals during the bioluminescent reaction received further support by studies aiming to understand the origin of a mysterious flavin adduct, the so-called myristylated FMN derivative, i.e. 6-(3′-(R)-myristyl)-flavin mononucleotide (myrFMN) first discovered in bioluminescent Photobacteria [26,28,84]. In addition, it was shown that some bioluminescent marine bacteria have an additional gene, luxF, which encodes a protein (LuxF) that specifically binds four molecules of myrFMN per dimer [26]. LuxF presumably evolved by gene duplication of luxB and while LuxF retained parts of the overall fold, specific elements were removed and replaced by myrFMN specific binding elements [28]. This led to the hypothesis that LuxF acts as a scavenger of myrFMN to prevent the observed inhibitory effect on the luciferase reaction. However, the origin of myrFMN remained unclear until it was shown recently that myrFMN is in fact a side product of the luciferase reaction [29]. Based on this discovery, a mechanism was postulated that rationalizes the formation of myrFMN (Scheme 6). This mechanism invokes the rearrangement of the transient substrate-derived radical anion to a carbon radical that subsequently recombines with the FMN-4a-hydroxide radical cation to form the covalent myrFMN adduct (Scheme 6).
Scheme 6.
Proposed mechanism for myrFMN formation (adapted from Scheme 3 in Tabib, Brodl, Macheroux, Mol. Microbiol. 2017 [29]). As shown in Scheme 5, an electron is transferred from the N5 of the flavin to the distal oxygen atom of the peroxy moiety. A hydrogen rearrangement of the alkoxy radical (R2: (CH2)10CH3) intermediate leads to a C3 carbon radical. This combines with the flavin-4a-hydroxide radical cation forming a covalent bond between the C6 of the isoalloxazine ring and the C3 carbon of the myristyl aldehyde. After rearomatization and the oxidation of the aldehyde to the acid followed by release of water, 6-(3′-(R)-myristyl)-FMN (myrFMN) is formed.
In our view, the formation of a carbon-carbon bond between the C6 position of the isoalloxazine ring and the C3 carbon of the aliphatic substrate molecule requires radical chemistry thus providing indirect support for an electron transfer mechanism being in operation in bacterial luciferase. In conclusion, the available data indicate the involvement of electron transfer as a central step on the reaction path to the excited state of the FMN-4a-hydroxide as postulated in flavin-initiated electron transfer mechanism (Scheme 5, top line).
2.2. Generation of Reduced FMN
As mentioned above and indicated in Scheme 4, bacterial luciferase is a flavin-dependent monooxygenase and thus requires the supply of reduced FMN in order to initiate the bioluminescent reaction. This task is carried out by flavin reductases, which may use NADH and/or NADPH as reducing agents [16,17]. Flavin reductases are either flavin-dependent (class 1) or independent (class 2) [68] and thus exhibit different properties in terms of the formation of binary and ternary enzyme substrate complexes. A central point in this context revolves around the question whether the reduced flavin (FMNH2) is directly transferred from the flavin reductase to the luciferase or simply released and subsequently sequestered by luciferase. Since free reduced flavin may rapidly react with dioxygen to yield oxidized flavin and hydrogen peroxide, the latter mechanism is potentially wasteful and thus the direct transfer of reduced FMN to the luciferase presents an attractive alternative mechanism that avoids this complication. Indeed, in vitro studies have provided evidence for such a direct transfer of reduced FMN between the flavin reductase and luciferase from V. fischeri [85]. In contrast, a recent study using LuxG and the luciferase from P. leiognathi TH1 indicated that the acquisition of reduced FMN by luciferase occurs by free diffusion [86]. Clearly, direct transfer of reduced flavin from the reductase to the luciferase requires the formation of a stable protein complex in which the reduced flavin is “handed over”. However, attempts to establish the formation of such a reductase-luciferase complex have been unsuccessful thus far. Similarly, it was shown that light emission by V. harveyi luciferase in transgenic Escherichia coli is supported by Fre, which is an oxidoreductase similar to LuxG from P. leiognathi [87]. Although it cannot be ruled out entirely, it appears unlikely that Fre (from E. coli) forms a protein complex with a luciferase from V. harveyi thus adding further support to the free diffusion mechanism. In agreement with this assessment, Campbell and Baldwin found no evidence for the existence of a Fre-luciferase complex [87]. From a structural perspective it should be noted that Fre from E. coli and LuxG from V. harveyi share about 40% sequence identity. Therefore, their molecular architecture is expected to be very similar and the mechanistic details involving substrate specificity and NAD(P)H binding can be inferred from the crystal structure of E. coli Fre [18]. With respect to the potential direct interaction of LuxG with the luciferase, the structural information cannot provide a direct answer. It should, however, be emphasized that during evolution the same flavin reductase fold has frequently been used to increase specificity for certain biochemical pathways by altering side chain functionalities in loop regions or by directly fusing the reductase domain with other functionalities such as ferredoxin units, FMN-flavodoxin modules or heme domains. Therefore, the inclusion of luxG in the lux operon might indicate some specialization of LuxG for enhanced light production, even though reduced FMN provided by other flavin reducing enzymes is also accepted by the luciferase. In conclusion, LuxG is an important NAD(P)H-dependent flavin reductase providing the luciferase with reduced FMN via free diffusion.
2.3. The Fatty Acid Reductase Complex (luxCDE)
Three enzymes are required for the production of the fatty aldehyde substrate of luciferase, a transferase, a synthetase and a reductase. Initially, the transferase (encoded by luxD) takes over the acyl moiety from acyl-ACP or acyl-CoA using the alcohol group of a serine residue to form an ester derivative, which is eventually hydrolyzed to the free acid [88], a step that is greatly enhanced by interaction with the LuxE synthetase [89]. Subsequent elucidation of the crystal structure of LuxD from Vibrio harveyi and biochemical experiments showed that Ser114, which is part of a classical catalytic triad, is the acylated residue (Fig. 4) [90]. In addition, a flexible β-hairpin element is observed close to the catalytic serine that might control activation and/or release of the fatty acid in the appropriate environment of the LuxCDE reductase complex. Strikingly, the relatively flat triangular surface of LuxD featuring the catalytic residues is lined mostly by evolutionary conserved residues, suggesting that this part of the molecule is involved in formation of a specific complex with the remaining subunits of the fatty acid reductase complex [91].
The synthetase subunit (encoded by luxE) activates the free acid at the expense of ATP, leading to the formation of acyl-AMP. This reacts with an invariant C-terminal cysteine residue to the corresponding thioester [92]. No structural information is available for LuxE, however, the close relationship with acyl-CoA synthetases provides an opportunity to generate homology models of LuxE. Even though sequence identities to the closest homologs are below 18%, the core regions of the synthetase units feature highly conserved residues with structural implications indicating the conservation of the overall protein fold. Also, the ATP binding site features full conservation of residues involved in base recognition and phosphate binding as well as the magnesium coordination shell close to the triphosphate of ATP. Interesting differences can be observed in the C-terminal part of the proteins where a flexible subdomain is involved in coenzyme A coordination and formation of the CoA derivatives [93]. For LuxE this subdomain is missing and replaced by a hairpin element that contains the acylated C-terminal cysteine residue (Cys362, [92]). Based upon open and closed structures of acyl-CoA synthetases, we generated models of LuxE in the two states using the structures of acyl-CoA ligase from Bacteroides thetaiotaomicron (PDB 4R1L) and phenylacetate-CoA ligase from Burkholderia cenocepacia [94], respectively (Fig. 5). Importantly, while biochemical data suggested LuxE to be monomeric [91], the modeling approach strongly supports a dimeric LuxE species. This is supported by the observation of untypical elution behavior during gel filtration considering monomeric LuxE and the fact that the monomer was assigned based on the absence of lysine specific crosslinking experiments [91] – Lysines that are absent near the interface of the dimeric models. The dimeric assembly is further supported by the high degree of evolutionary conservation of residues at the corresponding molecular interface (Fig. 5 C).
The flexibility and surface exposure of the catalytically important cysteine residue located at the C-terminus of LuxE allows it to reach into the active site of the reductase (encoded by luxC) where the thioester is reduced by NADPH to the aldehyde with concomitant release of the free fatty aldehyde. Several studies have confirmed the interaction between LuxC and LuxE and it has been shown that their interaction greatly stimulates their respective activities [89,91,92,[96], [97], [98]]. In order to rationalize the interaction on a molecular level, we employed a similar homology modeling approach as for LuxE. Although the sequence identity of LuxC to aldehyde dehydrogenases is rather low (around 18%), we could generate homology models that nicely confirm the tetrameric assembly of LuxC, as indicated by biochemical experiments [91,98] (Fig. 6). In addition, an overlay of the model with a substrate bound indole-3-acetaldehyde dehydrogenase (PDB 5IUW) [99] supports the biochemical observation of Cys286 being the active site residue that is acylated prior to reduction by the NADPH cofactor [97] (Fig. 6 A). Also, the high evolutionary conservation of residues at the tetrameric interfaces as well as the substrate and cofactor binding sites further demonstrate the validity of the overall LuxC model (Fig. 6 B-D).
It is well established that LuxD, LuxE and LuxC form a large multifunctional complex consisting of four reductase, four synthetase and two to four transferase subunits with an overall molecular mass of ca.500 kDa [91]. It was demonstrated that the LuxD transferase subunit is readily lost during purification of the complex, but that the maximum stimulation of activity was observed for equimolar concentrations of all components [91]. Therefore, we have attempted the reconstruction of a LuxC4LuxD4LuxE4 complex based on our homology models in combination with available functional data and conservation of residues among LuxCDE containing organisms. The complex is highly specific for the processing of myristoyl-ACP in agreement with reports that myristyl aldehyde (tetradecanal) is the main substrate in the luciferase reaction [12,88]. In this context, it should be noted that other acyl-CoA derivatives are accepted by the reductase and thus aldehydes with different chain length may be used by the luciferase for light production. Considering the overall good agreement of the homology models of LuxC and LuxE with biochemical data, we had a closer look at the complementarity of individual interfaces. Based on the evolutionary conservation of residues and distance restraints between individual active sites in the high affinity complex of LuxC and LuxE, we observed a tantalizing molecular interface for their direct interaction. As shown in Fig. 7, the distance between two active sites present in LuxC and their relative orientation perfectly matches the distance and orientation of the mobile LuxE element that harbors the acylated cysteine residue. It is known that the interaction between LuxC and LuxE is important for stimulating the acylation of LuxE [97] and even though molecular details of the presented structural models should be considered with caution, the observed interaction of the open LuxE conformation with LuxC might correspond to a functional state where the transfer of the acyl group between LuxE and LuxC can be realized. Considering, however, the importance of protein dynamics in forming open and closed conformations in the synthetase family [93] as well as in the reductase family [100], it should be stressed that the manually placed model of the LuxC-LuxE complex presented here is an approximation and will require experimental support from crystallographic data or potentially reconstructions from electron micrographs.
Based on biochemical data and crosslinking experiments also LuxD is implicated to be involved in complex formation with LuxE and LuxC [89,91]. Considering the relatively open active site and the triangular shape needed to shield the active site from bulk solvent (Fig. 7 C) we noticed that the outline of LuxD (Fig. 4) perfectly matches this substrate access route. Since the active site serine of LuxD carrying the acyl moiety is positioned centrally at the triangular interacting surface and the LuxCE access route is relatively open, the exact rotational position of LuxD is difficult to address based on our LuxC4E4 model. Nevertheless, the elongated triangular shape of LuxD and its surface complementarity to the complex model (Fig. 7) allowed a manual placement of four LuxD protomers to obtain a symmetric LuxC4D4E4 complex (Fig. 8) that is in line with biochemical observations and the evolutionary conservation of residues at the molecular surfaces of individual subunits. In fact, this model also supports the idea that the active site cavity formed at the interface of a LuxCDE complex is used for substrate channeling from LuxD to LuxE and further to LuxC, preventing unwanted dissociation of free fatty acids or unwanted side reactions of the acyl-AMP moiety. The final product of the reductase subunit, the aldehyde, can eventually be transferred to the luciferase for initiating light production.
3. Conclusions
The mesmerizing phenomenon of light emission by living organisms has attracted many scientists of various fields for many decades. As described in this review and summarized in Fig. 9, bacterial bioluminescence is a wonderful showcase for the tantalizing complexity of bioluminescence. Although the enzyme luciferase catalyzes a rather trivial oxidation of an aldehyde to the corresponding acid, the mechanism that leads to the population of an excited state remains a major challenge for mechanistic and structural biochemists in particular as sound high-resolution data on reaction intermediates is still lacking. On the organismic level, many questions revolving around the putative functions of bioluminescence for bacteria are still unanswered. Despite the progress achieved over the last decades, the fascination with bioluminescence will surely prompt many more research efforts to challenge the current frontiers of our knowledge.
Fig. 9.
Bacterial bioluminescence in a nut shell. The central player in bacterial bioluminescence is the heterodimeric luciferase (red/orange; see Fig. 3), which carries out the oxidation of long-chain fatty aldehydes to the corresponding acid accompanied by light emission (see Scheme 1). The required reduced FMN is provided by a NAD(P)H-dependent FMN reductase (LuxG, on the left side the structure of the closely related enzyme Fre of E. coli is shown in olive; PDB 1QFJ [18]) and the fatty aldehyde is synthesized through the multifunctional complex consisting of LuxCDE (green, violet and blue model on the right; see Fig. 8).
Conflict of Interest
The authors declare no conflict of interest.
Acknowledgements
We are grateful to the Austrian Fonds zur Förderung der Wissenschaftlichen Forschung (FWF) for financial support through the Ph.D. program Molecular Enzymology to P.M. (W901).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2018.11.003.
Appendix A. Supplementary Data
Supplementary material
References
- 1.Widder E.A. Bioluminescence in the ocean: origins of biological, chemical, and ecological diversity. Science. 2010;328:704–708. doi: 10.1126/science.1174269. 80- [DOI] [PubMed] [Google Scholar]
- 2.Dunlap P. In: Bioluminescence, microbial. Schaechter M., editor. Elsevier; Oxford: 2009. pp. 45–61. Encycl Microbiol. [Google Scholar]
- 3.Haddock S.H.D., Moline M.A., Case J.F. Bioluminescence in the sea. Ann Rev Mar Sci. 2010;2:443–493. doi: 10.1146/annurev-marine-120308-081028. [DOI] [PubMed] [Google Scholar]
- 4.Hastings J.W., Potrikus C.J., Gupta S.C., Kurfürst M., Makemson J.C. Biochemistry and physiology of bioluminescent bacteria. Adv Microb Physiol. 1985;26:235–291. doi: 10.1016/s0065-2911(08)60398-7. [DOI] [PubMed] [Google Scholar]
- 5.Zarubin M., Belkin S., Ionescu M., Genin A. Bacterial bioluminescence as a lure for marine zooplankton and fish. Proc Natl Acad Sci. 2012;109:853–857. doi: 10.1073/pnas.1116683109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dunlap P. Biochemistry and genetics of bacterial bioluminescence. In: Thouand G., Marks R., editors. Biolumin. fundam. appl. biotechnol. - vol. 1. vol. 144. Springer Berlin Heidelberg; Berlin, Heidelberg: 2014. pp. 37–64. [Google Scholar]
- 7.Meighen E.A. Molecular biology of bacterial bioluminescence. Microbiol Rev. 1991;55:123–142. doi: 10.1128/mr.55.1.123-142.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robertson L.A., Figge M.J., Dunlap P.V. Beijerinck and the bioluminescent bacteria: Microbiological experiments in the late 19th and early 20th centuries. FEMS Microbiol Ecol. 2011;75:185–194. doi: 10.1111/j.1574-6941.2010.01004.x. [DOI] [PubMed] [Google Scholar]
- 9.Urbanczyk H., Ast J.C., Dunlap P.V. Phylogeny, genomics, and symbiosis of Photobacterium. FEMS Microbiol Rev. 2011;35:324–342. doi: 10.1111/j.1574-6976.2010.00250.x. [DOI] [PubMed] [Google Scholar]
- 10.Urbanczyk H., Ast J.C., Kaeding A.J., Oliver J.D., Dunlap P.V. Phylogenetic analysis of the incidence of lux gene horizontal transfer in Vibrionaceae. J Bacteriol. 2008;190:3494–3504. doi: 10.1128/JB.00101-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ashkenazy H., Abadi S., Martz E., Chay O., Mayrose I., Pupko T. ConSurf 2016: an improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016;44:344–350. doi: 10.1093/nar/gkw408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ulitzur S., Hastings J.W. Evidence for tetradecanal as the natural aldehyde in bacterial bioluminescence. Proc Natl Acad Sci U S A. 1979;76:265–267. doi: 10.1073/pnas.76.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meighen E.A. Bacterial bioluminescence: organization, regulation, and application of the lux genes. FASEB J. 1993;7:1016–1022. doi: 10.1096/fasebj.7.11.8370470. [DOI] [PubMed] [Google Scholar]
- 14.Kurfürst M., Ghisla S., Hastings J.W. Characterization and postulated structure of the primary emitter in the bacterial luciferase reaction. Proc Natl Acad Sci U S A. 1984;81:2990–2994. doi: 10.1073/pnas.81.10.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Campbell Z.T., Weichsel A., Montfort W.R., Baldwin T.O. Crystal structure of the bacterial luciferase/flavin complex provides insight into the function of the β subunit. Biochemistry. 2009;48:6085–6094. doi: 10.1021/bi900003t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ellis H.R. The FMN-dependent two-component monooxygenase systems. Arch Biochem Biophys. 2010;497:1–12. doi: 10.1016/j.abb.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 17.van Berkel W.J.H., Kamerbeek N.M., Fraaije M.W. Flavoprotein monooxygenases, a diverse class of oxidative biocatalysts. J Biotechnol. 2006;124:670–689. doi: 10.1016/j.jbiotec.2006.03.044. [DOI] [PubMed] [Google Scholar]
- 18.Ingelman M., Ramaswamy S., Nivière V., Fontecave M., Eklund H. Crystal structure of NAD(P)H:flavin oxidoreductase from Escherichia coli. Biochemistry. 1999;38:7040–7049. doi: 10.1021/bi982849m. [DOI] [PubMed] [Google Scholar]
- 19.Zenno S., Saigo K. Identification of the genes encoding NAD(P)H-flavin oxidoreductases that are similar in sequence to Escherichia coli Fre in four species of luminous bacteria: Photorhabdus luminescens, Vibrio fischeri, Vibrio harveyi, and Vibrio orientalis. J Bacteriol. 1994;176:3544–3551. doi: 10.1128/jb.176.12.3544-3551.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin J.W., Chao Y.F., Weng S.F. Characteristic analysis of the luxG gene encoding the probable flavin reductase that resides in the lux operon of Photobacterium leiognathi. Biochem Biophys Res Commun. 1998;246:446–452. doi: 10.1006/bbrc.1998.8641. [DOI] [PubMed] [Google Scholar]
- 21.Nijvipakul S., Wongratana J., Suadee C., Entsch B., Ballou D.P., Chaiyen P. LuxG is a functioning flavin reductase for bacterial luminescence. J Bacteriol. 2008;190:1531–1538. doi: 10.1128/JB.01660-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nijvipakul S., Ballou D.P., Chaiyen P. Reduction kinetics of a flavin oxidoreductase LuxG from Photobacterium leiognathi (TH1): half-sites reactivity. Biochemistry. 2010;49:9241–9248. doi: 10.1021/bi1009985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaeding A.J., Ast J.C., Pearce M.M., Urbanczyk H., Kimura S., Endo H. Phylogenetic diversity and cosymbiosis in the bioluminescent symbioses of “Photobacterium mandapamensis”. Appl Environ Microbiol. 2007;73:3173–3182. doi: 10.1128/AEM.02212-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ast J.C., Dunlap P.V. Phylogenetic analysis of the lux operon distinguishes two evolutionarily distinct clades of Photobacterium leiognathi. Arch Microbiol. 2004;181:352–361. doi: 10.1007/s00203-004-0663-7. [DOI] [PubMed] [Google Scholar]
- 25.Moore S.A., James M.N.G., O'Kane D.J., Lee J. Crystallization of Photobacterium leiognathi non-fluorescent flavoprotein with limited sequence identity to bacterial luciferase. J Mol Biol. 1992;224:523–526. doi: 10.1016/0022-2836(92)91015-h. [DOI] [PubMed] [Google Scholar]
- 26.Moore S.A., James M.N.G., O'Kane D.J., Lee J. Crystal structure of a flavoprotein related to the subunits of bacterial luciferase. EMBO J. 1993;12:1767–1774. doi: 10.1002/j.1460-2075.1993.tb05824.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moore S.A., James M.N.G. Structural refinement of the non-fluorescent flavoprotein from Photobacterium leiognathi at 1.60 Å resolution. J Mol Biol. 1995;249:195–214. doi: 10.1006/jmbi.1995.0289. [DOI] [PubMed] [Google Scholar]
- 28.Bergner T., Tabib C.R., Winkler A., Stipsits S., Kayer H., Lee J. Structural and biochemical properties of LuxF from Photobacterium leiognathi. Biochim Biophys Acta Proteins Proteom. 2015;1854:1466–1475. doi: 10.1016/j.bbapap.2015.07.008. [DOI] [PubMed] [Google Scholar]
- 29.Tabib C.R., Brodl E., Macheroux P. Evidence for the generation of myristylated FMN by bacterial luciferase. Mol Microbiol. 2017;104:1027–1036. doi: 10.1111/mmi.13676. [DOI] [PubMed] [Google Scholar]
- 30.Gregor C., Gwosch K.C., Sahl S.J., Hell S.W. Strongly enhanced bacterial bioluminescence with the ilux operon for single-cell imaging. Proc Natl Acad Sci. 2018 doi: 10.1073/pnas.1715946115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cui B., Zhang L., Song Y., Wei J., Li C., Wang T. Engineering an enhanced, thermostable, monomeric bacterial luciferase gene as a reporter in plant protoplasts. PLoS One. 2014 doi: 10.1371/journal.pone.0107885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miyashiro T., Ruby E.G. Shedding light on bioluminescence regulation in Vibrio fischeri. Mol Microbiol. 2012;84:795–806. doi: 10.1111/j.1365-2958.2012.08065.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Defoirdt T., Boon N., Sorgeloos P., Verstraete W., Bossier P. Quorum sensing and quorum quenching in Vibrio harveyi: lessons learned from in vivo work. ISME J. 2008;2:19–26. doi: 10.1038/ismej.2007.92. [DOI] [PubMed] [Google Scholar]
- 34.Lin J.W., Chao Y.F., Weng S.F. Riboflavin synthesis genes ribE, ribB, ribH, ribA reside in the lux operon of Photobacterium leiognathi. Biochem Biophys Res Commun. 2001;284:587–595. doi: 10.1006/bbrc.2001.5013. [DOI] [PubMed] [Google Scholar]
- 35.Sung N.-D., Lee C.Y. The lux genes and riboflavin genes in bioluminescent system of Photobacterium leiognathi are under common regulation. J Photosci. 2004;11:41–45. [Google Scholar]
- 36.Ruby E.G., Nealson K.H. A luminous bacterium that emits yellow light. Science. 1977;80:432–434. doi: 10.1126/science.850787. 196. [DOI] [PubMed] [Google Scholar]
- 37.Daubner S.C., Astorga A.M., Leisman G.B., Baldwin T.O. Yellow light emission of Vibrio fischeri strain Y-1: purification and characterization of the energy-accepting yellow fluorescent protein. Proc Natl Acad Sci U S A. 1987;84:8912–8916. doi: 10.1073/pnas.84.24.8912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Macheroux P., Schmidt K.U., Steinerstauch P., Ghisla S., Colepicolo P., Buntic R. Purification of the yellow fluorescent protein from Vibrio fischeri and identity of the flavin chromophore. Biochem Biophys Res Commun. 1987;146:101–106. doi: 10.1016/0006-291x(87)90696-6. [DOI] [PubMed] [Google Scholar]
- 39.Gast R., Neering I.R., Lee J. Separation of a blue fluorescence protein from bacterial luciferase. Biochem Biophys Res Commun. 1978;80:14–21. doi: 10.1016/0006-291x(78)91097-5. [DOI] [PubMed] [Google Scholar]
- 40.O'Kane D.J., Karle V.A., Lee J. Purification of lumazine proteins from Photobacterium leiognathi and Photobacterium phosphoreum: bioluminescence properties. Biochemistry. 1985;24:1461–1467. doi: 10.1021/bi00327a026. [DOI] [PubMed] [Google Scholar]
- 41.Lee J., O'Kane D.J., Visser A.J.W.G. Spectral properties and function of two lumazine proteins from Photobacterium. Biochemistry. 1985;24:1476–1483. doi: 10.1021/bi00327a028. [DOI] [PubMed] [Google Scholar]
- 42.Small E.D., Koka P., Lee J. Lumazine protein from the bioluminescent bacterium Photobacterium phosphoreum. J Biol Chem. 1980;255:8804–8810. [PubMed] [Google Scholar]
- 43.Koka P., Lee J. Separation and structure of the prosthetic group of the blue fluorescence protein from the bioluminescent bacterium Photobacterium phosphoreum. Proc Natl Acad Sci U S A. 1979;76:3068–3072. doi: 10.1073/pnas.76.7.3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gast R., Lee J. Isolation of the in vivo emitter in bacterial bioluminescence. Proc Natl Acad Sci U S A. 1978;75:833–837. doi: 10.1073/pnas.75.2.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Petushkov V.N., Ketelaars M., Gibson B.G., Lee J. Interaction of Photobacterium leiognathi and Vibrio fischeri Y1 luciferases with fluorescent (antenna) proteins: Bioluminescence effects of the aliphatic additive. Biochemistry. 1996 doi: 10.1021/bi9608931. [DOI] [PubMed] [Google Scholar]
- 46.Eckstein J.W., Cho K.W., Colepicolo P., Ghisla S., Hastings J.W., Wilson T. A time-dependent bacterial bioluminescence emission spectrum in an in vitro single turnover system: energy transfer alone cannot account for the yellow emission of Vibrio fischeri Y-1. Proc Natl Acad Sci U S A. 1990 doi: 10.1073/pnas.87.4.1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Macheroux P. University of Konstanz; 1986. Untersuchungen zum Reaktionsmechanismus der bakteriellen Biolumineszenz. [Google Scholar]
- 48.Titushin M.S., Feng Y., Lee J., Vysotski E.S., Liu Z.J. Protein-protein complexation in bioluminescence. Protein Cell. 2011;2:957–972. doi: 10.1007/s13238-011-1118-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Petushkov V.N., Gibson B.G., Lee J. Direct measurement of excitation transfer in the protein complex of bacterial luciferase hydroxyflavin and the associated yellow fluorescence proteins from Vibrio fischeri Y1. Biochemistry. 1996 doi: 10.1021/bi952691v. [DOI] [PubMed] [Google Scholar]
- 50.Chatwell L., Illarionova V., Illarionov B., Eisenreich W., Huber R., Skerra A. Structure of lumazine protein, an optical transponder of luminescent bacteria. J Mol Biol. 2008;382:44–55. doi: 10.1016/j.jmb.2008.06.052. [DOI] [PubMed] [Google Scholar]
- 51.Sato Y., Shimizu S., Ohtaki A., Noguchi K., Miyatake H., Dohmae N. Crystal structures of the lumazine protein from Photobacterium kishitanii in complexes with the authentic chromophore, 6,7-dimethyl-8-(1′-D-ribityl) lumazine, and its analogues, riboflavin and flavin mononucleotide, at high resolution. J Bacteriol. 2010;192:127–133. doi: 10.1128/JB.01015-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee J. Lumazine protein and the excitation mechanism in bacterial bioluminescence. Biophys Chem. 1993;48:149–158. doi: 10.1016/0301-4622(93)85006-4. [DOI] [PubMed] [Google Scholar]
- 53.Shimomura O. World Scientific Publishing Co. Pte. Ltd.; 2006. Bioluminescence - chemical principles and methods. [Google Scholar]
- 54.Wilson T., Hastings J.W. Harvard University Press; 2013. Bioluminescence living lights, lights for living. [Google Scholar]
- 55.Tu S.C. Reduced flavin: donor and acceptor enzymes and mechanisms of channeling. Antioxid Redox Signal. 2001;3:881–897. doi: 10.1089/15230860152665046. [DOI] [PubMed] [Google Scholar]
- 56.Macheroux P., Kappes B., Ealick S.E. Flavogenomics - a genomic and structural view of flavin-dependent proteins. FEBS J. 2011;278:2625–2634. doi: 10.1111/j.1742-4658.2011.08202.x. [DOI] [PubMed] [Google Scholar]
- 57.Fisher A.J., Rayment I., Raushel F.M., Baldwin T.O. Three-dimensional structure of bacterial luciferase from Vibrio harveyi at 2.4 Å resolution. Biochemistry. 1995;34:6581–6586. doi: 10.1021/bi00020a002. [DOI] [PubMed] [Google Scholar]
- 58.Fisher A.J., Thompson T.B., Thoden J.B., Baldwin T.O., Rayment I. The 1.5 Å resolution crystal structure of bacterial luciferase in low salt conditions. J Biol Chem. 1996;271:21956–21968. doi: 10.1074/jbc.271.36.21956. [DOI] [PubMed] [Google Scholar]
- 59.Hastings J.W., Balny C. The oxygenated bacterial luciferase-flavin intermediate. Reaction products via the light and dark pathways. J Biol Chem. 1975;250:7288–7293. [PubMed] [Google Scholar]
- 60.Hastings J.W., Balny C., Peuch C.L., Douzou P. Spectral properties of an oxygenated luciferase-flavin intermediate isolated by low-temperature chromatography. Proc Natl Acad Sci U S A. 1973;70:3468–3472. doi: 10.1073/pnas.70.12.3468. [doi:066/4] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ahrens M., Macheroux P., Eberhard A., Ghisla S., Branchaud B.P., Hastings J.W. Boronic acids as mechanistic probes for the bacterial luciferase reaction. Photochem Photobiol. 1991;54:295–299. [Google Scholar]
- 62.Vervoort J., Muller F., van den Berg W.A.M., Moonen C.T.W., Lee J. Identifications of the true carbon-13 nuclear magnetic resonance spectrum of the stable intermediate II in bacterial luciferase. Biochemistry. 1986;25:8062–8067. [Google Scholar]
- 63.Palfey B.A., McDonald C.A. Control of catalysis in flavin-dependent monooxygenases. Arch Biochem Biophys. 2010;493:26–36. doi: 10.1016/j.abb.2009.11.028. [DOI] [PubMed] [Google Scholar]
- 64.Luo Y., Liu Y.J. Bioluminophore and flavin mononucleotide fluorescence quenching of bacterial bioluminescence—a theoretical study. Chem A Eur J. 2016 doi: 10.1002/chem.201603314. [DOI] [PubMed] [Google Scholar]
- 65.Macheroux P., Ghisla S., Hastings J.W. Spectral detection of an intermediate preceding the excited state in the bacterial luciferase reaction. Biochemistry. 1993;32:14183–14186. doi: 10.1021/bi00214a017. [DOI] [PubMed] [Google Scholar]
- 66.Baldwin T.O., Nicoli M.Z., Becvar J.E., Hastings J.W. Bacterial luciferase. Binding of oxidized flavin mononucleotide. J Biol Chem. 1975;250:2763–2768. [PubMed] [Google Scholar]
- 67.Hou C., Liu Y.J., Ferré N., Fang W.H. Understanding bacterial bioluminescence: a theoretical study of the entire process, from reduced flavin to light emission. Chem A Eur J. 2014;20:7979–7986. doi: 10.1002/chem.201400253. [DOI] [PubMed] [Google Scholar]
- 68.Tu S.C. Mechanisms of bacterial luciferase and related flavin reductases. In: Hille R., Miller S.M., Palfey B.A., editors. Handb. Flavoproteins. De Gruyter; Berlin: 2013. pp. 101–118. [Google Scholar]
- 69.Eberhard A., Hastings J.W. A postulated mechanism for the bioluminescent oxidation of reduced flavin mononucleotide. Biochem Biophys Res Commun. 1972;47:348–353. doi: 10.1016/0006-291x(72)90719-x. [DOI] [PubMed] [Google Scholar]
- 70.Branchaud B.P., Walsh C.T. Functional group diversity in enzymatic oxygenation reactions catalyzed by bacterial flavin-containing cyclohexanone oxygenase. J Am Chem Soc. 1985;107:2153–2161. [Google Scholar]
- 71.Eckstein J.W., Hastings J.W., Ghisla S. Mechanism of bacterial bioluminescence: 4a,5-dihydroflavin analogs as models for luciferase hydroperoxide intermediates and the effect of substituents at the 8-position of flavin on luciferase kinetics. Biochemistry. 1993;32:404–411. doi: 10.1021/bi00053a004. [DOI] [PubMed] [Google Scholar]
- 72.Merényi G., Lind J., Mager H.I.X., Tu S.C. Properties of 4a-hydroxy-4a,5-dihydroflavin radicals in relation to bacterial bioluminescence. J Phys Chem. 1992;96:10528–10533. [Google Scholar]
- 73.Francisco W.A., Abu-Soud H.M., Delmonte A.J., Singleton D.A., Baldwin TO, Raushel F.M. Deuterium kinetic isotope effects and the mechanism of the bacterial luciferase reaction. Biochemistry. 1998;37:2596–2606. doi: 10.1021/bi972266x. [DOI] [PubMed] [Google Scholar]
- 74.Schuster G.B. Chemiluminescence of organic peroxides. Conversion of ground-state reactants to excited-state products by the chemically initiated electron-exchange luminescence mechanism. Acc Chem Res. 1979 [Google Scholar]
- 75.Matheson I.B., Lee J., Müller F. Bacterial bioluminescence: spectral study of the emitters in the in vitro reaction. Proc Natl Acad Sci U S A. 1981;78:948–952. doi: 10.1073/pnas.78.2.948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Koo J.A., Schmidt S.P., Schuster G.B. Bioluminescence of the firefly: key steps in the formation of the electronically excited state for model systems. Proc Natl Acad Sci. 1978 doi: 10.1073/pnas.75.1.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Catalani L.H., Wilson T. Electron transfer and chemiluminescence. Two inefficient systems: 1,4-dimethoxy-9,10-diphenylanthracene peroxide and diphenoyl peroxide. J Am Chem Soc. 1989 [Google Scholar]
- 78.Ding B.W., Liu Y.J. Bioluminescence of firefly squid via mechanism of single electron-transfer oxygenation and charge-transfer-induced luminescence. J Am Chem Soc. 2017 doi: 10.1021/jacs.6b09119. [DOI] [PubMed] [Google Scholar]
- 79.Yue L., Liu Y.J., Fang W.H. Mechanistic insight into the chemiluminescent decomposition of firefly dioxetanone. J Am Chem Soc. 2012 doi: 10.1021/ja302979t. [DOI] [PubMed] [Google Scholar]
- 80.Yue L., Lan Z., Liu Y.J. The theoretical estimation of the bioluminescent efficiency of the firefly via a nonadiabatic molecular dynamics simulation. J Phys Chem Lett. 2015 doi: 10.1021/jz502305g. [DOI] [PubMed] [Google Scholar]
- 81.Pinto Da Silva L., Pereira R.F.J., Magalhães C.M., Esteves Da Silva J.C.G. Mechanistic insight into Cypridina bioluminescence with a combined experimental and theoretical chemiluminescent approach. J Phys Chem B. 2017 doi: 10.1021/acs.jpcb.7b06295. [DOI] [PubMed] [Google Scholar]
- 82.Min C.G., Ferreira P.J.O., Pinto da Silva L. Theoretically obtained insight into the mechanism and dioxetanone species responsible for the singlet chemiexcitation of Coelenterazine. J Photochem Photobiol B Biol. 2017 doi: 10.1016/j.jphotobiol.2017.07.012. [DOI] [PubMed] [Google Scholar]
- 83.Shannon P., Presswood R.B., Spencer R., Becvar J.E., Hastings J.W., Walsh C.T. A study of deuterium isotope effects on the bacterial bioluminescence reaction. In: Singer T.P., Ondarza R.N., editors. Mech Oxid Enzym. Elsevier; North Holland, Amsterdam: 1978. pp. 69–78. [Google Scholar]
- 84.Kasai S., Matsui K., Nakamura T. Purification and some properties of FP390 from P. phosphoreum. In: Edmondson D.E., McCormick D.B., editors. Flavins and flavoproteins, berlin. Walter de Gruyter; New York: 1987. pp. 647–650. [Google Scholar]
- 85.Jeffers C.E., Tu S.C. Differential transfers of reduced flavin cofactor and product by bacterial flavin reductase to luciferase. Biochemistry. 2001;40:1749–1754. doi: 10.1021/bi0024310. [DOI] [PubMed] [Google Scholar]
- 86.Tinikul R., Pitsawong W., Sucharitakul J., Nijvipakul S., Ballou D.P., Chaiyen P. The transfer of reduced flavin mononucleotide from LuxG oxidoreductase to luciferase occurs via free diffusion. Biochemistry. 2013;52:6834–6843. doi: 10.1021/bi4006545. [DOI] [PubMed] [Google Scholar]
- 87.Campbell Z.T., Baldwin T.O. Fre is the major flavin reductase supporting bioluminescence from Vibrio harveyi luciferase in Escherichia coli. J Biol Chem. 2009;284:8322–8328. doi: 10.1074/jbc.M808977200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ferri S.R., Meighen E.A. A lux-specific myristoyl transferase in luminescent bacteria related to eukaryotic serine esterases. J Biol Chem. 1991;266:12852–12857. [PubMed] [Google Scholar]
- 89.Carey L.M., Rodriguez A., Meighen E. Generation of fatty acids by an acyl esterase in the bioluminescent system of Photobacterium phosphoreum. J Biol Chem. 1984;259:10216–10221. [PubMed] [Google Scholar]
- 90.Lawson D.M., Derewenda U., Serre L., Wei Y., Derewend Z.S., Ferri S. Structure of a myristoyl-ACP-specific thioesterase from Vibrio harveyi. Biochemistry. 1994;33:9382–9388. doi: 10.1021/bi00198a003. [DOI] [PubMed] [Google Scholar]
- 91.Wall L., Meighen E.A. Subunit structure of the fatty acid reductase complex from Photobacterium phosphoreum. Biochemistry. 1986;25:4315–4321. [Google Scholar]
- 92.Soly R.R., Meighen E.A. Identification of the acyl transfer site of fatty acyl-protein synthetase from bioluminescent bacteria. J Mol Biol. 1991;219:69–77. doi: 10.1016/0022-2836(91)90858-4. [DOI] [PubMed] [Google Scholar]
- 93.Li Z., Nair S.K. Structural basis for specificity and flexibility in a plant 4-coumarate:CoA ligase. Structure. 2015;23:2032–2042. doi: 10.1016/j.str.2015.08.012. [DOI] [PubMed] [Google Scholar]
- 94.Law A., Boulanger M.J. Defining a structural and kinetic rationale for paralogous copies of phenylacetate-CoA ligases from the cystic fibrosis pathogen Burkholderia cenocepacia J2315. J Biol Chem. 2011;286:15577–15585. doi: 10.1074/jbc.M111.219683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Biasini M., Bienert S., Waterhouse A., Arnold K., Studer G., Schmidt T. SWISS-MODEL: modeling protein tertiary and quaternary structure using evolutionary information. Nucleic Acids Res. 2014;42 doi: 10.1093/nar/gku340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wall L., Rodriguez A., Meighen E. Intersubunit transfer of fatty acyl groups during fatty acid reduction. J Biol Chem. 1986;261:15981–15988. [PubMed] [Google Scholar]
- 97.Lee C.Y., Meighen E.A. Cysteine-286 as the site of acylation of the Lux-specific fatty acyl-CoA reductase. Biochim Biophys Acta Protein Struct Mol Enzymol. 1997;1338:215–222. doi: 10.1016/s0167-4838(96)00203-8. [DOI] [PubMed] [Google Scholar]
- 98.Rodriguez A., Riendeau D., Meighen E. Purification of the acyl coenzyme A reductase component from a complex responsible for the reduction of fatty acids in bioluminescent bacteria. J Biol Chem. 1983;258:5233–5237. [PubMed] [Google Scholar]
- 99.McClerklin S.A., Lee S.G., Harper C.P., Nwumeh R., Jez J.M., Kunkel B.N. Indole-3-acetaldehyde dehydrogenase-dependent auxin synthesis contributes to virulence of Pseudomonas syringae strain DC3000. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1006811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Luo M., Tanner J.J. Structural basis of substrate recognition by aldehyde dehydrogenase 7A1. Biochemistry. 2015;54:5513–5522. doi: 10.1021/acs.biochem.5b00754. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material