Abstract
Peripheral Artery Disease (PAD) is a chronic, activity-limiting disease that is caused by atherosclerotic occlusion of blood vessels outside the heart. Type 1 Diabetes (T1D) not only increases an individual’s likelihood of developing PAD, but also contributes to poor clinical outcomes after PAD manifestation. Although there is some evidence suggesting that hyperglycemia might alter expression of genes involved in regulating PAD severity or outcomes, our knowledge about the specific genes and pathways involved remains incomplete.
We induced experimental PAD or hind limb ischemia in T1D and non-diabetic mice and subjected the ischemic gastrocnemius muscle tissues to genome-wide mRNA transcriptome and pathway analysis. We identified 513 probe sets that represented 443 different genes with highly significant expression differences (p < 0.005) between the ischemic diabetic and ischemic non-diabetic muscle tissues. Moreover, pathway analysis of the differentially expressed genes identified pathways involved in essential biological processes such as “cell cycle,” “DNA replication,” “metabolic pathways,” “focal adhesion,” “regulation of actin cytoskeleton,” and “nucleotide excision repair”. Taken together, our data offer the opportunity to test hypotheses on the roles played by the altered genes/molecular pathways in poor PAD outcomes in diabetes. Such studies may lead to the development of specific therapies to improve PAD outcomes in patients with comorbid diabetes.
Keywords: Peripheral artery disease, Type 1 diabetes mellitus, Genome-wide gene expression profiling, Molecular pathway analysis
Introduction
Peripheral artery disease (PAD) is a chronic, activity-limiting disease that is caused by atherosclerotic occlusion of blood vessels outside the heart and the arteries of the lower extremity are the most common site of disease [1], [2]. PAD affects approximately 12 million people in the United States and over 200 million people worldwide [3], [4]. There are two classic clinical presentations of PAD, intermittent claudication (IC, lower extremity pain with ambulation relieved by rest) and critical limb ischemia (CLI, pain at rest that may be associated with ulceration or gangrene) [1], [5].
The risk factors for developing PAD include smoking, advanced age, hypercholesterolemia and diabetes [6]. However, smoking and diabetes accounts for about 80% of the risk of developing PAD. Both Type 1 and Type 2 diabetic mellitus (T1D and T2D) patients are at an increased risk of developing PAD [5], [7]. Moreover, diabetic patients with PAD have poorer clinical outcomes than nondiabetic patients with PAD [8], [9];for example, diabetics with concurrent PAD have a seven-fold higher risk of developing CLI compared to PAD patients without diabetes [10], [11]. How diabetes contributes to poorer PAD outcomes is poorly understood, but hyperglycemia in diabetes is thought to contribute to inflammation, endothelial cell dysfunction, and hypercoagulability [7].
Using a mouse model of experimental PAD, i.e. the mouse hind limb ischemia or HLI model (detailed in Materials and Methods), we previously showed that Ins2Akita (Akita) mice, a genetic non-obese T1D model, have impaired perfusion recovery and lower capillary density in its ischemic hind limbs compared to non-diabetic C57BL/6 (B6) controls [12]. Moreover our lab and others have shown some data suggesting that hyperglycemia might alter expression of genes involved in regulating PAD severity or outcomes [12], [13], [14], [15]. However, our knowledge about the specific genes and pathways involved in poor PAD outcomes in diabetes remains incomplete.
In this study, we explored the effect of hyperglycemia on ischemia-induced gene expression following experimental PAD by comparing the gene expression profile in ischemic hind limbs of T1D to that of non-diabetic mice. Given that diabetes contributes to poorer PAD outcomes in both humans and mice, we hypothesized that the gene expression profile of diabetic and non-diabetic mice post-HLI will differ considerably. Therefore, the goal of our study was to elucidate genes and pathways that influence PAD severity and outcomes in diabetes.
Materials and Methods
Mice
All mice (male C57BL/6 [n = 3] and male C57BL/6J-Ins2Akita [n = 3]) were obtained from the Jackson Laboratory (Bar Harbor, ME, USA) either directly or bred internally from parental strains obtained from the Jackson Laboratory. The C57BL/6J-Ins2Akita is the Ins2Akita strain on a C57BL/6 background. The Ins2Akita is a previously described mouse model of Type 1 diabetes [16]. Hyperglycemia exposure was assessed by HbA1c measurement (A1cNow kit, Bayer Health Care, Sunnyvale, CA) in Akita mice at 8 weeks and then monthly afterwards. All TID mice used in this study had Hba1c > 8 (9.6 ± 0.6%). Strain, age and sex matched non-diabetic mice were used as controls. All non-diabetic control mice had Hba1c < 6 (4.4 ± 0.2%).
Hindlimb ischemia
Hindlimb ischemia (HLI) was achieved by unilateral femoral artery ligation and excision, as described previously [17]. Blood flow in the ischemic and contralateral non-ischemic limbs was measured by laser Doppler perfusion imaging, as described previously [18], [19]. Controls were strain, age, and sex matched.
Microarray procedures and Real-time PCR
Total RNA was extracted from the ischemic gastrocnemius muscle on day 3 post-HLI, as described previously [13], [20]. Day 3 was chosen because this is a time point that we previously showed there was no significant difference in perfusion recovery between the non-diabetic C57bL/6 and the diabetic C57BL/6J-Ins2Akita strain. RNA was processed and hybridized onto Affymetrix Mouse430 expression arrays according to the manufacturer’s protocols. Expression values were normalized and provided expression analysis data on a total of 45,101 probe sets. Microarray was performed at the DNA Science Core, Department of Microbiology, Immunology and Cancer Biology, University of Virginia. These probe set expression values were then subjected to statistical analyses using previously described approaches [21]. Microarray results were validated by quantitative Real-time PCR of representative genes from affected molecular pathways. Expression values from ischemic C57BL/6 and C57BL/6J-Ins2Akita tissues were normalized to non-ischemic B6 tissues.
Statistical analysis
Statistical analysis of these gene expression profiles was performed on GeneSpring software (version 14.9, Silicon Genetics, Redwood, CA). The complete dataset was first filtered on expression values which excluded transcript IDs in which 10% of the samples did not express a value above a set cut-off out of the 45,101 probe sets present on the AffyMetrix GeneChip. Next, the dataset was filtered again on a fold change (FC) of 1.2. T test was performed between C57BL/6J-Ins2Akita and C57BL/6 mice to give differentially expressed transcript IDs without multiple test correction. The list of transcript IDs with a p-value < 0.005 was selected for data mining. GeneSpring was also used to produce a heat map of differentially expressed genes. Hierarchical clustering was chosen for the gene list. The list was clustered on expression values and conditions using normalized intensity values, Euclidean distance metric and centroid linkage rule parameters.
Data mining analysis
The lists of differentially expressed genes were subjected to data mining in Web-based Gene Set Analysis Toolkit (WebGestalt, www.webgestalt.org). From this, enriched KEGG pathways and gene ontology categories (GO) were obtained. Ingenuity Pathway Analysis (IPA, www.ingenuity.com, version 44691306) was used as described previously [21] to generate a merged molecular network from the gene list.
Results
Genes differentially expressed between non-diabetic and T1D mice
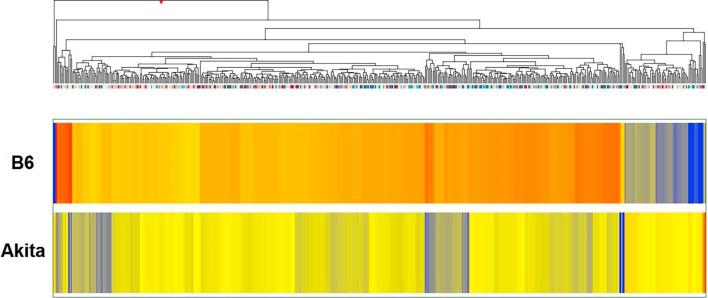
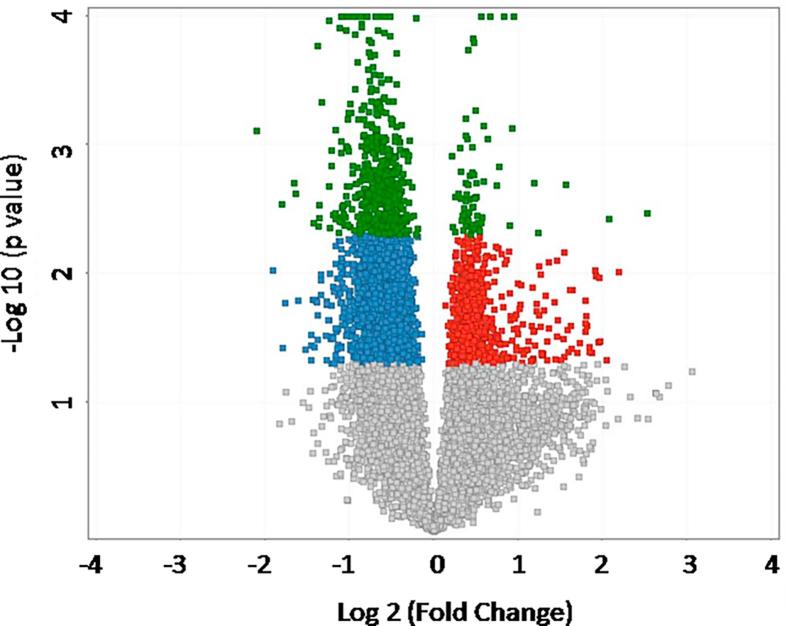
We compared the whole genome mRNA expression from ischemic gastrocnemius muscle tissue from male C57BL/6J-Ins2Akita (diabetic) mice to that of age, and sex matched ischemic gastrocnemius muscle tissue from C57BL/6 (non-diabetic) controls. We identified 513 probe sets with highly significant expression differences between controls and experimental mice (p < 0.005, no multiple test correction). The 513 probe sets represented 443 different genes. Of these 443 differentially regulated genes, 387 genes had lower expression and 56 genes had higher expression in the T1D mice compared to non-diabetic mice (Fig. 1). A volcano plot of the array data showing the distribution of the probe set IDs is also provide (Fig. 2).
Fig. 1.
Hierarchical clustering of genes whose expression was different in non-diabetic and T1D mice. The list of genes was identified with a T test unpaired unequal variance (p < 0.005, no multiple test correction) of mRNA expression data. The color of the rectangles representing averaged expression values for each gene in each set of samples (n = 3 for B6 and n = 3 for Akita) indicates the degree of increase (red) or decrease (blue) of each gene expression signal relative to the mean signal intensity (yellow).
Fig. 2.
Volcano plot representing microarray probe set IDs differentially expressed in an analysis of Akita versus B6 mice by a Student’s t-test. Dots highlighted in red represent probe set IDs with a fold change greater than 1.2 in the Akita mice compared to the B6. Conversely, dots highlighted in blue are probe set IDs with a fold change greater than −1.2 in the Akita compared to B6. Dots highlighted in green represent probe set IDs with both a p-value of < 0.005 and a fold change greater than ±1.2. Here, the 513 probe set IDs highlighted in green correspond to the 443 individual genes that were analyzed in this study.
Kyoto encyclopedia of genes and genomes (KEGG) pathways of altered genes between non-diabetic and T1D mice
To better understand the molecular pathways and biological processes associated with the genes altered in T1D and non-diabetic mice, we performed KEGG pathway analyses. Top 10 enriched KEGG pathways generated at a high stringency level (p < 0.005, no multiple test correction) are shown in Table 1. The KEGG pathways that are enriched are highly significant and include important biological pathways such as “cell cycle,” “DNA replication,” “metabolic pathways,” “focal adhesion,” “regulation of actin cytoskeleton,” and “nucleotide excision repair”. Interestingly, almost all of the genes involved in these pathways had lower expression in T1D mice compared to non-diabetic mice.
Table 1.
Enriched KEGG pathways for the list of altered genes between non-diabetic and T1D mice.
| KEGG Pathway | Adjusted p-value | No. of genes | Genes in the pathway |
|---|---|---|---|
| Cell cycle | 4.20e−13 | 16 | Mad2l1(−), Ccne2(−), PIk1(−), Rbl1(−), E2f1(−), Skp2(−), Ccnb2(−), Cdk6(−), Smc1a(−), Aurkb(−), Mcm7(−), Mcm5(−), Cdk1(−), Cdc25a(−), Espl1(−), Dbf4(−) |
| DNA replication | 4.32e−07 | 7 | Lig1(−), Pole3(−), Mcm7(−), Pole(−), Mcm5(−), Pole2(−), Pola1(−) |
| Metabolic pathways | 7.03e−06 | 28 | Rrm1(−), Tk1(−), Galnt1(−), Etnk1(−), Acaa1a(−), Atp6v1b2(−), Pole2(−), Pola1(−), Hmgcs1(−), Hsd17b7(−), Ahcyl1(−), Dnmt1(−), Pole3(−), Man1c1(−), Pole(−), Ugcg(−), Lpcat1(−), Ak5(+), Nanp(−), Umps(−), Pck2(−), Galns(−), B4galt6(−), Lta4h(−), Enpp1(−), Ada(−), B3gnt2(−), Sephs1(−) |
| Pathways in cancer | 7.45e−06 | 14 | Ccne2(−), Itga6(−), Fzd5(−), Prkcb(−), Rad51(−), Ctnnb1(−), Akt3(−), Flt3l(−), E2f1(−), Skp2(−), Cdk6(−), Hsp90aa1(−), Birc5(−), Pdgfrb(−) |
| Progesterone-mediated oocyte maturation | 8.76e−06 | 8 | Ccna2(−), Mad2l1(−), PIk1(−), Cdk1(−), Akt3(−), Ccnb2(−), Cdc25a(−), Hsp90aa1(−) |
| Oocyte meiosis | 4.92e−05 | 8 | Mad2l1(−), Ccne2(−), PIk1(−), Cdk1(−), Ccnb2(−), Fbxo5(−), Espl1(−), Smc1a(−) |
| Focal adhesion | 5.79e−05 | 10 | Ppp1r12a(−), Itga6(−), Prkcb(−), Fyn(−), Akt3(−), Ctnnb1(−), Rock1(−), Col1a1(−), Pdgfrb(−), Src(+) |
| Regulation of actin cytoskeleton | 9.98e−05 | 10 | Ppp1r12a(−), Pip4k2a(−), Diaph3(−), Itga6(−), Wasf2(−), Iqgap2(−), Gna13(−), Rock1(−), Enah(−), Pdgfrb(−) |
| Pyrimidine metabolism | 1.00e−04 | 7 | Pole3(−), Rrm1(−), Tk1(−), Umps(−), Pole(−), Pole2(−), Pola1(−) |
| Nucleotide excision repair | 2.00e−04 | 5 | Lig1(−), Pole3(−), Cul4b(−), Pole(−), Pole2(−) |
The lists of differentially expressed genes were analyzed in WebGestalt (www.webgestalt.org) for enriched KEGG pathways. Best 10 pathways sorted by adjusted p-values are shown. Pathways indicated in bold font might play a role in post-ischemic recovery. “−” signifies that the gene has lower expression in T1D compared to non-diabetic mice, while “+” indicates that the gene has higher expression in T1D mice compared to non-diabetic mice (see Table 1).
Gene ontology categories of altered genes between non-diabetic and T1D mice
In addition to KEGG pathways, we performed Gene ontology analyses (p < 0.005, no multiple test correction) to better categorize genes found to be differentially expressed between non-diabetic and T1D mice. The best categories for these genes sorted by p-value are shown in Table 2. Gene ontology categories most strongly represented are “cell cycle,” “primary metabolic processes,” and “organelle organization” within the “Biological Processes” category; “protein binding” and “ATP binding” within the “Molecular Function” category; and “organelle” and “nucleus” within the “Cellular Component” category.
Table 2.
Enriched Gene ontology categories for the list of altered genes between non-diabetic and T1D mice.
| Gene Ontology | No. of genes in category | p-value |
|---|---|---|
| Biological process | ||
| Cell cycle | 92 | 5.33E−33 |
| Primary metabolic process | 248 | 9.83E−29 |
| Organelle organization | 109 | 1.35E−27 |
| Molecular function | ||
| Protein | 198 | 1.49E−19 |
| ATP | 71 | 3.31E−14 |
| Purine nucleoside | 80 | 3.32E−14 |
| Cellular compartment | ||
| Cell | 373 | 1.09E−41 |
| Organelle | 309 | 1.95E−36 |
| Nucleus | 211 | 2.40E−33 |
| Chromosome | 58 | 3.92E−25 |
The lists of differentially expressed genes were analyzed in Web Gestalt (www.webgestalt.org). Best categories sorted by p-values are shown.
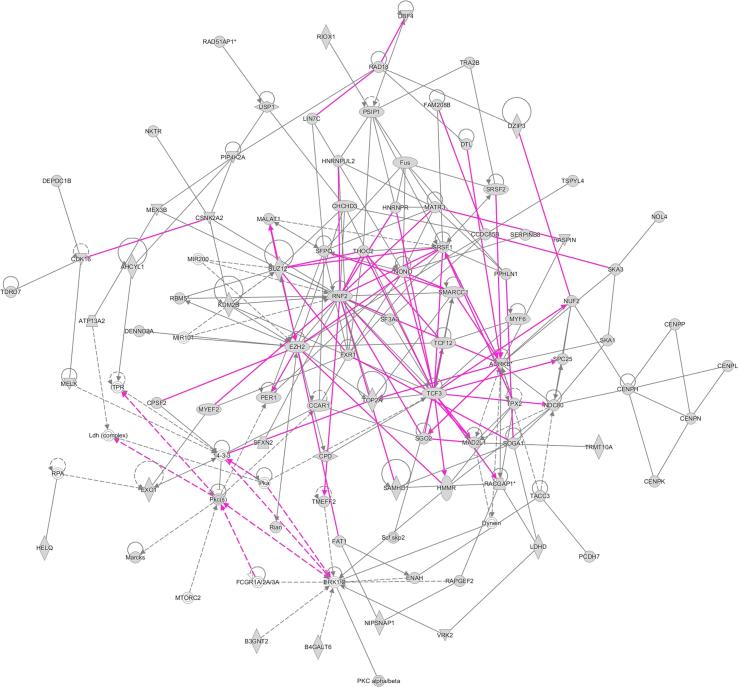
Ingenuity pathway networks of altered genes between non-diabetic and T1D mice
Ingenuity Pathway Analysis (IPA) generated molecular networks are shown in Fig. 3. This network map gives us insight into genes/molecules that might be related to those found in our gene lists, but not directly differentially expressed in our analysis. This serves to help us determine additional pathways and processes that the genes from our analysis (focus genes) might be involved in. Among the genes/molecules related to our focus genes, ERK1/2 stand out as having some of the most numerous connections to the focus genes. ERK1/2 are MAP kinases that participate in the Ras-Raf-MEK-ERK signal transduction pathway. These are involved in cell cycle regulation, proliferation and differentiation among other things [22]. Additionally, within our focus genes, AURKB, MAD2L1, TCF3, EZH2, RNF2 also appear to have numerous connections.
Fig. 3.
Molecular network generated using Ingenuity Pathway Analysis (IPA) software. The merged network was generated from the top three networks expressed in a list of 513 probe sets (representing 443 different genes) differentially expressed in non-diabetic and T1D mice. The list of genes was generated from T test unpaired unequal variance (p < 0.005, no multiple test correction) of mRNA expression data whose expression was altered in T1D mice compared to non-diabetic mice. Genes from our uploaded gene list (focus genes) are shown as gray icons while genes or endogenous chemicals derived from the IPA database that could be algorithmically connected to these focus genes are shown as white icons. The shapes of the icons represent classes of genes/molecules e.g. squares = cytokines/growth factors, ovals = transcription factors, etc.
Validation of microarray data by quantitative real-time PCR
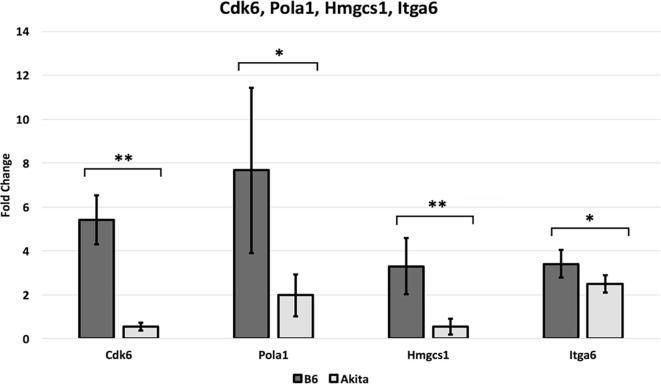
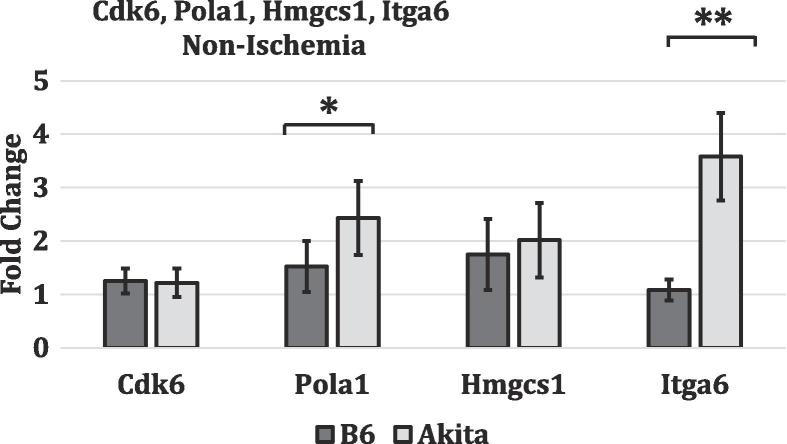
To validate the microarray data, we chose one gene from each of the top 4 pathways identified in our KEGG pathway analysis for validation by qRT-PCR. Cdk6 from “Cell Cycle”, Pola1 from “DNA Replication”, Hmgcs1 from “Metabolic Pathways”, and Itga6 from “Pathways in Cancer”. In all genes tested, the gene expression differs between the ischemic non-diabetic C57BL/6 and the ischemic diabetic hind limb tissues (Fig. 4). This is consistent with our gene array findings (Fig. 2, all four genes had lower expression in T1D mice compared to non-diabetic mice). Additionally, to test whether the differential expression of these 4 genes reflect impaired ischemia induced gene expression or existed at baseline in non-ischemic tissues, we analyzed their level of expression in non-ischemic hind limb tissues from non-diabetic and T1D mice. We found at baseline there was no difference in CDk6 and Hmgcs mRNA expression between non-diabetic and T1DM tissues. (Fig. 5). However, Pola1, and Itga6 are expressed at a higher level in T1D compared to non-diabetic tissues (Fig. 5). Despite the higher expression of Pola1 and Itga6 in TID non ischemic tissues the expression in ischemic tissues was lower than that observed in non-diabetic tissues. Therefore, the result is consistent with impaired ischemia induced upregulation of all 4 genes.
Fig. 4.
Validation of microarray data by quantitative real-time PCR. Quantitative real-time PCR was performed for 4 genes (Cdk6, Pola1, Hmgcs1, and Itga6) from different enriched molecular pathways. Fold change values represent expression levels of genes in ischemic C57BL/6 (non-diabetic) and C57BL/6J-Ins2Akita (diabetic) tissues normalized to non-ischemic C57BL/6 tissues. (n = 3–4/grp and * represents p < 0.05, while ** signifies p < 0.01). The results are consistent with the microarray data results.
Fig. 5.
Validation of microarray data by quantitative real-time PCR. Quantitative real-time PCR was performed for 4 genes (Cdk6, Pola1, Hmgcs1, and Itga6) from different enriched molecular pathways. Fold change values represent expression levels of genes in non ischemic C57BL/6J-Ins2Akita (diabetic) tissues normalized to non-ischemic C57BL/6 (non-diabetic tissues) tissues. (n = 3–4/grp and * represents p < 0.05, while ** signifies p < 0.01). The results are consistent with the microarray data results.
Discussion
Normal adaptation to ischemia involves a variety of time-dependent biological mechanisms and genomic regulation. Whereas inflammation, angiogenesis, and stress-related genes are upregulated early in the time-course (e.g. day 1) of post-ischemic injury, cell cycle and cytoskeletal genes are upregulated later in the process (e.g. day 3) [23]. In fact, it has been previously shown that upregulation of cell cycle genes peaks at day 3 post-HLI, and these genes play an important role in the skeletal muscle regenerative process that involves satellite cell proliferation and differentiation [24]. Myogenesis appears essential in post-ischemic recovery as muscle function that is depressed at day 3 post-HLI returns to normal by day 7 post-HLI [24]. Clearly, skeletal muscle repair and regeneration are essential adaptive processes to minimize complications from PAD.
Type 1 Diabetes is a complex, polygenic disease that not only increases a patient’s likelihood of developing PAD, but also promotes poor clinical outcomes after PAD manifestation [8], [9], [10], [25]. There are many mouse models of T1D each with its limitations as it relates to how well it models human disease [26]. Hence, one limitation of the C57BL/6J-Ins2Akita type 1 DM model is that the hyperglycemia seen is not due to beta cell loss because of autoimmune islet inflammation as seen in humans, instead its beta cell loss is due to accumulation of misfolded proteins within the beta cells resulting in beta cell loss [16]. Nevertheless, this model has one of the key metabolic abnormalities in T1D, that is, hyperglycemia and therefore allows for testing hypothesis related to effects of hyperglycemia in vivo. Previously, our lab and others have shown that hyperglycemia alters expression of genes involved in regulating PAD severity or outcomes [12], [13], [15]. For example, we have shown that hyperglycemia in T1D reduces VEGFR2 protein expression in ischemic hind limbs, which in effect impairs perfusion recovery [13]. Moreover, we have shown ADAM12 is a gene that plays a critical role in perfusion recovery following experimental PAD and we find impaired upregulation of ADAM12 through altered miR29a expression in ischemic diabetic hind limbs [13], [19]. Although these targeted studies allowed us to dissect the underlying mechanisms at the individual gene level, they limited our ability to attain a global understanding of the effects of hyperglycemia on ischemia-induced gene expression. Therefore, we speculated that a comparison of overall gene expression (e.g. non-targeted approach) between ischemic tissues from non-diabetic mice and T1D mice would help elucidate which pathways and groups of genes are altered. We hypothesized that the altered pathways/genes might contribute to the poor post-ischemic adaptation and outcomes seen in PAD patients with comorbid T1D.
Our results confirmed that hyperglycemia alters gene expression and advanced our knowledge about the pathways that may modulate PAD severity and outcomes in T1D. We identified 513 probe sets that represented 443 different genes with highly significant expression differences (p < 0.005) between non-diabetic and T1D mice on day 3 post-HLI. Of these 443 differentially regulated genes, 387 (∼87%) genes had lower expression and 56 genes had higher expression in T1D mice compared to non-diabetic mice. One limitation of our study is that we did not use multiple test corrections in our analysis and this raises the possibility that each single gene may not be differentially expressed if multiple test corrections were applied. Therefore, we have laid more emphasis on the pathway analysis. Even if individual genes in themselves are not significant (with a multiple test correction) the highly significant enrichment of specific pathways would not occur from a list of random genes. Hence, the pathway analysis of genes ensures that the conclusions are sound. KEGG pathway analysis on the altered list of genes between ischemic, non-diabetic and T1D tissues enriched pathways such as “cell cycle,” “DNA replication,” “metabolic pathways,” “focal adhesion,” “regulation of actin cytoskeleton,” and “nucleotide excision repair.” Interestingly some of the genes within the pathways we identified have been implicated as playing a role in post-ischemic recovery [19], [24], [27], [28], [29], [30]. For example, DNA replication proteins (e.g. lig1), cell cycle proteins (e.g. cdk6 and cdk1), cytoskeletal proteins (e.g. iqgap2) and focal adhesion proteins (e.g. col1a1) all play an important role in skeletal regeneration, an important mid-phase (day 3 post-HLI) adaptive mechanism to ischemic injury [24], [31], [32]. Interestingly, all the genes encoding these proteins and many others from these pathways have lower expression in T1D mice compared to non-diabetic mice, thereby suggesting that impairment of these pathways/genes might explain why PAD patients with comorbid diabetes have poorer clinical outcomes compared to PAD patients without diabetes [25]. Additionally, both the KEGG pathway analysis and the Gene ontology analysis identified the metabolic pathway (Table 1, Table 2) as a pathway affected in impaired ischemia induced gene expression. To our knowledge, this is the first time these pathways have been implicated in regulating post-ischemic recovery in hyperglycemia. Although it is not yet clear how hyperglycemia regulates expression of the genes in these pathways, our data offers, insight that can be exploited in future studies.
Moreover, we generated molecular networks using Ingenuity Pathway Analysis (IPA) in order to visualize connections among the genes from our analysis. For instance, AURKB and MAD2L1 are two genes that have numerous connections with other genes from our analysis and outside our analysis. While AURKB is a protein that is a component of the chromosomal passenger complex which ensures proper chromosome alignment and separation, MAD2L1 is a component of the spindle-assembly checkpoint of mitosis [33], [34]. Therefore, it is conceivable that impaired expression of these genes in ischemic T1D mice (Table 1) may affect post-ischemic cell proliferation and recover from ischemic injury. Importantly, IPA also gives us insight into genes/molecules that might be related to those found in our gene lists, but not directly differentially expressed in our analysis. Among the genes/molecules related to our focus genes, ERK1/2 have the most numerous connections. ERK1/2 are MAP kinases that participate in the Ras-Raf-MEK-ERK signal transduction pathway and are involved in cell cycle regulation, proliferation and differentiation, among other things [22]. The numerous connections that ERK1/2 have with genes in our analysis suggest that the Ras-Raf-MEK-ERK pathway could potentially interact upstream or downstream to other genes/pathways in our analysis.
Hyperglycemia has profound effects on gene expression, and these effects might contribute to impaired adaptive mechanisms (e.g. skeletal muscle regeneration) and poor outcomes in PAD patients with comorbid diabetes. There is still much to be learned, but we believe our study provides a foundation for future studies. Taken together, our data offer the opportunity to test hypotheses on the roles played by the altered genes/molecular pathways in poor PAD outcomes in diabetes. Such studies may lead to the development of specific therapies to improve PAD outcomes in diabetes.
Funding
This work was supported by the National Heart, Lung, and Blood Institute (R01 HL130399 to AO Dokun). RP & LG were supported by a Short-Term Research Training Grant (T32-5T35DK113964-02) from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK). Software licenses were supported by UC4 DK104155.
Conflict of interest
None.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jcte.2018.11.003.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Aronow W.S. Peripheral arterial disease of the lower extremities. Arch Med Sci. 2012;8(2):375–388. doi: 10.5114/aoms.2012.28568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thiruvoipati T., Kielhorn C.E., Armstrong E.J. Peripheral artery disease in patients with diabetes: Epidemiology, mechanisms, and outcomes. World J Diabetes. 2015;6(7):961–969. doi: 10.4239/wjd.v6.i7.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fowkes F.G. Comparison of global estimates of prevalence and risk factors for peripheral artery disease in 2000 and 2010: a systematic review and analysis. Lancet. 2013;382(9901):1329–1340. doi: 10.1016/S0140-6736(13)61249-0. [DOI] [PubMed] [Google Scholar]
- 4.Criqui M.H., Aboyans V. Epidemiology of peripheral artery disease. Circ Res. 2015;116(9):1509. doi: 10.1161/CIRCRESAHA.116.303849. [DOI] [PubMed] [Google Scholar]
- 5.Norgren L. Inter-society consensus for the management of peripheral arterial disease (TASC II) Eur J Vasc Endovasc Surg. 2007;33:S1–S75. doi: 10.1016/j.ejvs.2006.09.024. [DOI] [PubMed] [Google Scholar]
- 6.Shammas N.W. Epidemiology, classification, and modifiable risk factors of peripheral arterial disease. Vasc Health Risk Manage. 2007;3(2):229–234. doi: 10.2147/vhrm.2007.3.2.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Association A.D. Peripheral arterial disease in people with diabetes. Diabetes Care. 2003;26(1):3333–3341. doi: 10.2337/diacare.26.12.3333. [DOI] [PubMed] [Google Scholar]
- 8.Marso S.P., Hiatt W.R. Peripheral arterial disease in patients with diabetes. J Am Coll Cardiol. 2006;47(5):921–929. doi: 10.1016/j.jacc.2005.09.065. [DOI] [PubMed] [Google Scholar]
- 9.Zander E., Heinke H.P., Rendel J., Kohnert K.D., Kairies U., Braun J. Peripheral arterial disease in diabetes mellitus type 1 and type 2: are the risk factors different? Eur J Vasc Med. 2002;31:249–254. doi: 10.1024/0301-1526.31.4.249. [DOI] [PubMed] [Google Scholar]
- 10.Selvin E. HbA1c and peripheral arterial disease in diabetes. Diabetes Care. 2006;29(4):877–882. doi: 10.2337/diacare.29.04.06.dc05-2018. [DOI] [PubMed] [Google Scholar]
- 11.Jude E.B., Eleftheriadou I., Tentolouris N. Peripheral arterial disease in diabetes—a review. Diabet Med. 2010;27(1):4–14. doi: 10.1111/j.1464-5491.2009.02866.x. [DOI] [PubMed] [Google Scholar]
- 12.Dokun A.O. Glycaemic control improves perfusion recovery and VEGFR2 protein expression in diabetic mice following experimental PAD. Cardiovasc Res. 2014;101(3):364–372. doi: 10.1093/cvr/cvt342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen L. Modulation of miR29a improves impaired post-ischemic angiogenesis in hyperglycemia. Exp Biol Med (Maywood) 2017;242(14):1432–1443. doi: 10.1177/1535370217716424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caporali A., Meloni M., Vollenkle C., Bonci D., Sala-Newby G.B., Addis R. Deregulation of microRNA-503 contributes to diabetes mellitus – induced impairment of endothelial function and reparative angiogenesis after limb ischemia. Circulation. 2011;123:282–291. doi: 10.1161/CIRCULATIONAHA.110.952325. [DOI] [PubMed] [Google Scholar]
- 15.Leeper N., Cooke J.P. MicroRNA and mechanisms of impaired angiogenesis in diabetes mellitus. Circulation. 2011;123:236–238. doi: 10.1161/CIRCULATIONAHA.110.003855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang J. A mutation in the insulin 2 gene induces diabetes with severe pancreatic beta-cell dysfunction in the Mody mouse. J Clin Invest. 1999;103(1):27–37. doi: 10.1172/JCI4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Couffinhal T. Mouse model of angiogenesis. Am J Pathol. 1998;152(6):1667–1679. [PMC free article] [PubMed] [Google Scholar]
- 18.Hazarika S. Impaired angiogenesis after hindlimb ischemia in type 2 diabetes mellitus: differential regulation of vascular endothelial growth factor receptor 1 and soluble vascular endothelial growth factor receptor 1. Circ Res. 2007;101(9):948–956. doi: 10.1161/CIRCRESAHA.107.160630. [DOI] [PubMed] [Google Scholar]
- 19.Dokun A.O. ADAM12: a genetic modifier of preclinical peripheral arterial disease. Am J Physiol Heart Circ Physiol. 2015;309(5):H790–H803. doi: 10.1152/ajpheart.00803.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hazarika S. MicroRNA-93 controls perfusion recovery after hindlimb ischemia by modulating expression of multiple genes in the cell cycle pathway. Circulation. 2013;127(17):1818–1828. doi: 10.1161/CIRCULATIONAHA.112.000860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu J., Kakoola D.N., Lenchik N.I., Desiderio D.M., Marshall D.R., Gerling I.C. Molecular phenotyping of immune cells from young NOD mice reveals abnormal metabolic pathways in the early induction phase of autoimmune diabetes. PLoS ONE. 2012;7(10) doi: 10.1371/journal.pone.0046941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roskoski R. ERK1/2 MAP kinases: structure, function, and regulation. Pharmacol Res. 2012;66(2):105–143. doi: 10.1016/j.phrs.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 23.Lee C.W. Temporal patterns of gene expression after acute hindlimb ischemia in mice: insights into the genomic program for collateral vessel development. J Am Coll Cardiol. 2004;43(3):474–482. doi: 10.1016/j.jacc.2003.09.033. [DOI] [PubMed] [Google Scholar]
- 24.Paoni N.F. Time course of skeletal muscle repair and gene expression following acute hind limb ischemia in mice. Physiol Genomics. 2002;11(3):263–272. doi: 10.1152/physiolgenomics.00110.2002. [DOI] [PubMed] [Google Scholar]
- 25.Jude E.B. Peripheral arterial disease in diabetic and nondiabetic patients. Diabetes Care. 2001;24(8):1433–1437. doi: 10.2337/diacare.24.8.1433. [DOI] [PubMed] [Google Scholar]
- 26.Van Belle T.L., Taylor P., von Herrath M.G. Mouse models for Type 1 diabetes. Drug Discovery Today Dis Models. 2009;6(2):41–45. doi: 10.1016/j.ddmod.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dai Q. Alterations in endothelial cell proliferation and apoptosis contribute to vascular remodeling following hind-limb ischemia in rabbits. Vasc Med. 2002;7(2):87–91. doi: 10.1191/1358863x02vm430oa. [DOI] [PubMed] [Google Scholar]
- 28.Cazes G.A., Galaup A., Chomel C. Extracellular matrix-bound angiopoietin-like 4 inhibits endothelial cell adhesion, migration, and sprouting and alters actin cytoskeleton. Circ Res. 2006;99(11):1207–1215. doi: 10.1161/01.RES.0000250758.63358.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peng X., Ueda H., Zhou H., Stokol T., Shen T.L., Alcaraz A. Overexpression of focal adhesion kinase in vascular endothelial cells promotes angiogenesis in transgenic mice. Cardiovasc Res. 2004;64(3):421–430. doi: 10.1016/j.cardiores.2004.07.012. [DOI] [PubMed] [Google Scholar]
- 30.Ingber D.E., Prusty D., Betensky H., Sunz H., Wang N. Cell shape, cytoskeletal mechanics, and cell cycle control in angiogenesis. J Biochem. 1995;28(12):1471–1484. doi: 10.1016/0021-9290(95)00095-x. [DOI] [PubMed] [Google Scholar]
- 31.Ml C. Cytoskeleton and adhesion in myogenesis. ISRN Mol Bol. 2014;1(15) [Google Scholar]
- 32.Chargé S.B., Rudnicki M.A. Cellular and molecular regulation of muscle regeneration. Physiol Rev. 2004;84:209–238. doi: 10.1152/physrev.00019.2003. [DOI] [PubMed] [Google Scholar]
- 33.Nguyen A.L., Schindler K. Specialize and Divide (Twice): functions of three aurora kinase homologs in mammalian oocyte meiotic maturation. Trends Genet (Cell) 2017;33(5):349–363. doi: 10.1016/j.tig.2017.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sotillo R., Hernando E., Diaz-Rodriguez E., Teruya-Feldstein J., Cordon-Cardo C., Lowe S.W. Mad2 overexpression promotes aneuploidy and tumorigenesis in mice. Cancer Cell. 2007;11(1):9–23. doi: 10.1016/j.ccr.2006.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.