ABSTRACT
Background
Humans and mice absorb bovine milk exosomes and their RNA cargos.
Objectives
The objectives of this study were to determine whether milk exosome– and RNA-depleted (ERD) and exosome- and RNA-sufficient (ERS) diets alter the concentrations of purine metabolites in mouse livers, and to determine whether diets depleted of bovine milk alter the plasma concentration and urine excretion of purine metabolites in adults and infants, respectively.
Methods
C57BL/6 mice were fed ERD (providing 2% of the microRNA cargos compared with ERS) and ERS diets starting at age 3 wk; livers were collected at age 7 wk. Plasma and 24-h urine samples were collected from healthy adults who consumed (DCs) or avoided (DAs) dairy products. Spot urine samples were collected from healthy infants fed human milk (HM), milk formula (MF), or soy formula (SF) at age 3 mo. Purine metabolites were analyzed in liver, plasma, and urine; mRNAs and microRNAs were analyzed in the livers of female mice.
Results
We found that 9 hepatic purine metabolites in ERD-fed mice were 1.76 ± 0.43 times the concentrations in ERS-fed mice (P < 0.05). Plasma concentrations and urine excretion of purine metabolites in DAs was ≤1.62 ± 0.45 times the concentrations in DCs (P < 0.05). The excretion of 13 purine metabolites in urine from SF infants was ≤175 ± 39 times the excretion in HM and MF infants (P < 0.05). mRNA expression of 5′-nucleotidase, cytosolic IIIB, and adenosine deaminase in mice fed ERD was 0.64 ± 0.52 and 0.60 ± 0.28 times the expression in mice fed ERS, respectively.
Conclusion
Diets depleted of bovine-milk exosomes and RNA cargos caused increases in hepatic purine metabolites in mice, and in plasma and urine from human adults and infants, compared with exosome-sufficient controls. These findings are important, because purines play a role in intermediary metabolism and cell signaling.
Keywords: bovine milk, exosomes, metabolomics, purines, RNA
Introduction
Virtually every living cell produces and secretes a class of nanoparticles called exosomes (1, 2). Exosomes deliver cargos such as lipids, proteins, and various species of RNA to adjacent or distant recipient cells. Exosome uptake by recipient cells is mediated by endocytosis (3). The delivery of exosome cargos elicits changes in gene expression and metabolism in the recipient cells (1, 2). Among exosome cargos, microRNAs are of particular interest, because >60% of human genes have conserved microRNA-binding sites and loss of microRNA maturation is embryonically lethal (4, 5). MicroRNAs have been implicated in most physiologic and pathologic conditions (6, 7).
We have provided strong evidence that exosomes and their RNA cargos do not originate exclusively in endogenous synthesis, but may also be obtained from dietary sources such as bovine milk. The following lines of evidence were presented in support of this paradigm-shifting theory. First, the plasma concentrations of milk-borne microRNAs increased following a milk meal in humans, whereas the concentration of a microRNA not detectable in milk (miR-1) did not increase (8, 9). Recently, we confirmed these findings by the use of a nucleotide-specific PCR protocol, RNase H2 PCR, which distinguishes microRNAs derived from endogenous synthesis and those obtained from bovine milk (10). Second, human and rodent intestinal cells, human immune cells, and human vascular endothelial cells accumulate bovine milk exosomes by endocytosis and secrete microRNA across the basolateral membrane (11–13). These observations are consistent with reports suggesting that orally administered milk exosomes accumulate in peripheral tissues and that only about half of the extracellular RNAs in human plasma are of human origin (14, 15). Third, in vitro evidence suggests that microRNAs are protected against degradation by RNases and low pH through encapsulation in exosomes, and partially protected against degradation in the TNO intestinal model (16, 17). Some voices of concern remain, casting doubt on the absorption of dietary microRNAs being quantitatively meaningful [reviewed by Zempleni et al. (18) and discussed below]. However, evidence is accumulating that RNAs from dietary sources other than milk are also bioavailable [reviewed by Zempleni et al. (18); additional recent evidence provided by Luo et al. (19)].
Of note, when mice were fed an AIN-93G-based diet, modified to be defined by its content of bovine milk exosomes for 4 wk, plasma microRNA levels were ∼60% lower in mice fed an exosome- and RNA-depleted (ERD) diet compared with mice fed the exosome- and RNA-sufficient (ERS) diet (8). The observation that endogenous microRNA synthesis cannot compensate for dietary depletion raises the important question as to what the phenotypes of dietary RNA depletion might be. Our first attempts at identifying phenotypes of milk RNA depletion included an unbiased liver metabolomics screen of mice fed ERD and ERS diets, which suggested that hepatic adenosine and inosine were among the metabolites for which the concentrations were higher in ERD mice than in ERS mice. Here, we have followed up on this initial observation and conducted a series of studies, specifically targeting purine metabolites with the following objectives: 1) to conduct a comprehensive analysis of hepatic purine metabolites in mice fed ERD and ERS diets; 2) to assess the effects of ERD and ERS diets on the expression of enzymes in purine metabolism in murine livers; and 3) to assess the effects of milk intake on purine metabolites in human adults and infants. This study is significant because it provides a new insight into hepatic purine metabolism pathways impacted by dietary milk–derived microRNA in mice and shows, for the first time, that marked purine metabolic shifts also take place in infants fed milk-free formula. Purines play a role in numerous pathways in energy metabolism and cell signaling (20).
Methods
Mouse feeding studies
C57BL/6 mice (Jackson Laboratories), aged 3 wk, were randomly assigned to 2 diet groups at weaning (21) and housed in groups that were kept at ∼22°C, with a 12:12 light cycle. The absence of sex effects on liver metabolites was demonstrated by referencing against 5 male C57BL/6 mice, determined by hydrophilic-interaction chromatography-multiple reaction monitoring-tandem mass spectrometry (HILIC-MRM-MS/MS, Supplemental Table 1). The diets were based on the AIN-93G formula, and modified by their content of exosomes and their RNA cargos from bovine milk (8, 22). One group was fed an ERD diet and the other group was fed an ERS diet, both for 4 wk. In the diets, lyophilized milk powder (and soy protein) is substituted for milk casein in the AIN-93G formulation to eliminate the dairy exosomes present in the AIN-93G formulation. The milk added to the diets provides the equivalent of 0.5 L of milk consumed by a human adult per day, adjusted by body weight for mice. The milk used to prepare the powder for the ERD diet was ultrasonicated for 1.5 h and incubated for 1 h at 37°C prior to lyophilization; the milk used to prepare the powder for the ERS was not ultrasonicated.
Ultrasonication leads to a transient disruption of exosome membranes and a >98% depletion of RNA cargos in exosomes, a 20% decrease in exosome count (9.1 × 1012 ± 7.1 × 1011 exosomes/mL in ERS milk compared with 7.3 × 1012 ± 3.5 × 1011 exosomes/L in ERD milk; P < 0.05, n = 3), and a >60% decrease in intestinal exosome transport rates (23). Dietary ingredients other than milk are not ultrasonicated, i.e., nutrients other than exosomes and their RNA cargos, including purines, are the same in both sonicated milk and nonsonicated milk, used for ERD and ERS diets, respectively (see below). Differences in the purine contents of the 2 diets were tested for by HPLC, with 3 samples analyzed from separate batches of both ERD and ERS milk. Mice had free access to the diets and water for the entire study period, but no differences in food and water intake were noted between the 2 groups (not shown). At age 7 wk, the mice were killed by the use of carbon dioxide when in a postabsorptive state, and their livers were flash frozen in liquid nitrogen and immediately stored at –80°C until analysis. Body composition was assessed by dual-energy X-ray absorptiometry at the time of the study termination but prior to the liver extraction. Studies were approved by the University of Nebraska-Lincoln Institutional Animal Care and Use Committee (protocol 1229).
Human feeding studies
Cross-sectional studies were conducted in infants and adults. Spot urine samples were collected from infants fed human milk (HM, n = 45), milk formula (MF, n = 41), or soy formula (SF, n = 46) at age 3 mo. The key to group assignments was broken after the purine and creatinine analyses had been completed. The infant population and the formulas fed have been described elsewhere [(24); The Beginnings Study, Clinical Trial # NCT00616395]. In addition, 2 cohorts of apparently healthy adults were recruited (Supplemental Table 2). Dairy consumers (DCs) were persons who regularly consumed a self-chosen diet providing ≥0.5 L milk/d and perhaps other dairy products, whereas dairy avoiders (DAs) were persons who regularly consumed diets free of animal milk and <2 weekly servings of dairy products (1 serving = 0.25 L) other than milk. Dairy consumption was assessed by questionnaire. DAs were instructed to avoid dairy products other than milk altogether for ≥2 wk before sample collection. Exclusion criteria shared by both groups included history of metabolic disease, including lactose intolerance, body weight of <55 kg, history of intestinal surgery, smoking, and use of prescription and over-the-counter medications <4 wk before sample collection. Blood samples and 24-h urine samples were collected after an overnight fast. Ethylenediaminetetraacetic acid tubes were used to avoid inhibition of PCR reactions (10). Plasma was collected by centrifugation and stored at –80°C until analysis. Urine samples were briefly centrifuged to remove debris and stored at –80°C until analysis. Studies were approved by the Institutional Review Board at the University of Nebraska-Lincoln (protocol NU ID 14,778). We estimated the power of human studies to detect changes in purine metabolism based on the differential concentrations of 7 purine metabolites between ERD and ERS diets, including adenosine, guanosine, inosine, xanthine, adenine, hypoxanthine, and ADP. A cohort of 14 infants allowed to detect a 40% change in urinary purine metabolites, and 6 adults allowed us to detect a 60% change of purine metabolites in urine and plasma at 1 – β = 0.8 and α = 0.05. There were no apparent sex effects on purine metabolism in either of these human cohorts.
Analyses of purine metabolites
Polar and nonpolar metabolites were extracted from mouse livers with a mixture of chloroform, methanol, and water, and analyzed by LC-MS as described previously (25) for nontargeted metabolomics. Metabolites were identified with the use of the METLIN database (26). Multivariate analyses were performed with the use of Metaboanalyst 3.0 (27). Mouse livers and human plasma and urine metabolites were extracted with the use of methanol, dried, and stored at –80°C until analyzed as previously described (28), differing only by the use of a 5-µm Luna NH2 column (150 × 2 mm) at a flow rate of 0.4 mL/min. Prior to mass spectrometry, samples were resuspended in 40 µL of 85% acetonitrile and a 20 µL aliquot was injected. Targeted metabolite quantification was performed with the use of a QTRAP 6500 mass spectrometer (AB Sciex LLC) coupled to a Shimadzu Nexera X2 HPLC system (Shimadzu Co.). Peak analysis was performed with the use of Analyst Software (AB Sciex LLC). Multiple reaction monitoring transition data were collected for each purine metabolite (28). Select purines in livers, plasma, and urine samples in the different diets were also analyzed by HPLC (inosine and hypoxanthine) as described previously with minor modifications (29), with the use of the Amplex Red Xanthine Assay Kit (Life Technologies) and the BioVision Uric Acid Colorimetric/Fluorometric Assay Kit (Biovision Inc.), following the manufacturers’ instructions. The concentrations of purine metabolites in urine were normalized with the use of creatinine, which was measured with the use of a colorimetric assay (30). A uric acid test was performed on samples of adult urine collected in a 24-h period, whereas purines from infants were analyzed in spot urine samples, which require normalization by creatinine.
RNA sequencing analysis
Total RNA and microRNAs were extracted from mouse livers with the use of spin columns (Qiagen, Inc.) and shipped to BGI, Inc. for sequencing with the use of the Illumina HiSeq 2500 platform, with protocols of 125-bp read length, paired end and 50-bp read length, single end, respectively. Data quality control was performed with the use of FastQC (31). For RNA, adaptor sequences and reads containing ambiguous bases or having an average quality score of <30 were removed. The remaining high-quality reads were aligned to the reference sequences in murine RNA (GRCm38, mm10) by the use of RSEM (32) and Tophat (33). Cufflinks and cuffdiff were used to obtain the transcripts and to quantify them according to the sum of normalized reads units, displayed as fragments per kilobase million (34). DESeq2 R was used to determine differentially expressed genes, based mainly on fold change, P value, and Q value (false discovery rate) (35). For microRNA, expression was quantified with the use of CAP-miSeq, and miRBase (Version 21) was used as a reference library for annotation (36, 37). We carefully filtered out the low-quality reads and strictly mapped the qualified reads to all known mature sequences, precursor sequences, and genomes of the mouse. Raw sequence data were deposited in the NCBI-BioProject database under Accession No. PRJNA428903 for RNA sequencing (RNA-Seq) data and PRJNA428904 for microRNA-Seq data.
Spin columns for RNA purification may be contaminated with microbial RNAs and produce false positive results in microRNA analysis (38). We confirmed hepatic microRNA-Seq data by real-time qPCR, with the use of microRNAs isolated by spin columns that were purified by treatment with 0.5% sodium hypochlorite (38). We selected 3 microRNAs that represented the top (miR-340-5p), middle (miR-99b-5p), and bottom (miR-362-3p) tertiles of microRNA abundance in microRNA-Seq analysis. In addition, miR-340-5p has a nucleotide sequence complementary to the mRNAs implicated in purine metabolism. mRNA-microRNA interactions were predicted by the use of miRWalk (39).
Statistical analysis
Multivariate statistical analysis, pathway enrichment analysis, and topologic analysis approaches were used to compare high-throughput data between ERD and ERS with the use of Metaboanalyst 3.0 (27). Homogeneity of variances was confirmed with the use of the F-test. The Kolmogorov-Smirnov test was used to confirm normal distribution. Differences between 2 groups were determined by unpaired t-tests. If variances were heterogeneous, we used Welch's test for pairwise comparisons. One-way ANOVA and Tukey's post hoc test were used for comparisons between >2 groups. Homogeneity of variances was confirmed with the use of Barlett's test. Effects of diets were considered statistically significant if P < 0.05. Data in figures and tables are reported as means ± SEMs and means ± SDs, respectively. Prism 6 (Graph-Pad), R Studio, and SPSS were used to perform statistical analyses.
Results
ERD and ERS feeding in mice
The ERD and ERS diets contained the same concentrations of the purine metabolites hypoxanthine and inosine (Supplemental Table 3). Physical activity, food consumption, feeding frequency, water consumption, and respiratory exchange rate were the same in mice fed the ERD and ERS diets (23). Body weight, lean body weight, and fat mass were the same in the 2 diet groups at age 7 wk (Supplemental Figure 1).
Hepatic purine metabolites
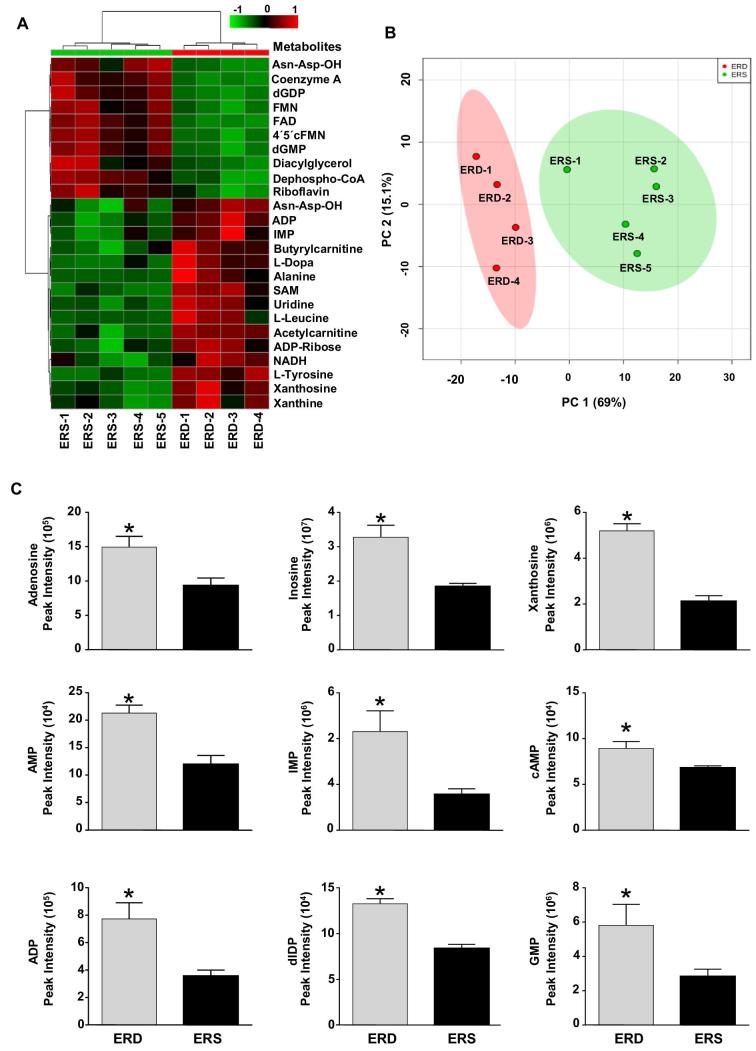
When the metabolome in mouse livers was analyzed with the use of nontargeted mass spectrometry, 7 purine metabolites were among the 25 metabolites (purines and nonpurines) affected the most by ERD or ERS diets (Figure 1A). In principal component analysis, mouse livers were clustered separately by dietary treatment (Figure 1B), which was primarily driven by the differential abundance of purine metabolites. When nontargeted metabolite analysis was followed up with a targeted analysis of purine metabolites by LC-MS, the concentrations of 9 measured hepatic purine metabolites were significantly higher in ERD mice than in ERS mice (Figure 1C). Metabolite–set enrichment analysis of hepatic metabolites suggested that purine metabolism was one of the pathways most affected by ERD diet in mice (Supplemental Figure 2). When individual purines were analyzed by the use of colorimetric assays, HPLC, and HILIC-MRM-MS/MS, the hepatic concentrations of xanthine and guanosine triphosphate were significantly higher in mice fed ERD than in mice fed ERS (Supplemental Figure 3), whereas the concentrations of 7 other purine metabolites in ERD mice were not significantly different from ERS mice (0.07 < P < 0.46).
FIGURE 1.
Hepatic metabolome in female mice fed ERD and ERS diets for 4 wk. (A) Heat map with the top 25 differential liver metabolites between the ERD and ERS diets. (B) PCA leveraging 191 polar metabolites indicates clear separation of mice by their diet treatment group. PC1 and PC2 explain 69% and 15%, respectively, of the variance. Clusters defined by the ERD and ERS diets are shown, with each symbol representing the scores plot value for each mouse. Ovals depict the 95% CIs. (C) Diet-dependent purine metabolites in mouse livers. Values are means ± SEMs. *P < 0.05 vs. ERS (n = 4–5 mice/group in all panels). Asn-Asp-OH, l-asparaginyl-l-aspartic acid; dGDP, 2'-deoxyguanosine-5'-diphosphate; dGMP, 2'-deoxyguanosine-5'-monophosphate; dIDP, 2'-deoxyinosine-5'-diphosphate; ERD, exosome- and RNA-depleted; ERS, exosome- and RNA-sufficient; FMN, flavin mononucleotide; IMP, inosine 5'-monophosphate; PC, principal component; PCA, principal component analysis; SAM, S-adenosyl methionine.
Liver transcriptomics
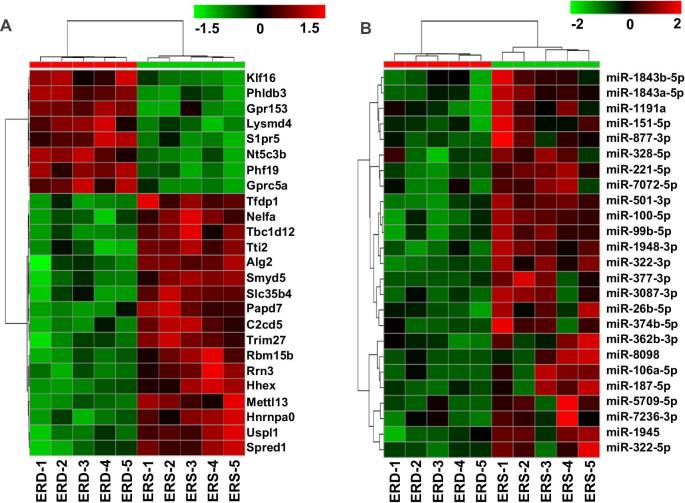
A total of 272 mRNAs were differentially expressed, with criteria of ≥1.5-fold difference, a P value of <0.05, and a Q value of <0.05 (|log2FC| ≥ 0.8, P < 0.05, Q < 0.05), in livers from mice fed ERD or ERS diets (Figure 2A). The mRNAs that were differentially expressed included adenylate kinase 1 and cAMP-specific 3′,5′-cyclic phosphodiesterase 4D, which were 6216% and 175% more abundant, respectively, in ERD than in ERS. The unbiased RNA-Seq analysis was supplemented with a targeted in-depth analysis of mRNA, which detected 9 enzymes in purine metabolism for which the expression of mRNA was significantly different between mice fed ERD and ERS diets (Table 1). A total of 49 microRNAs were expressed at a lower level in livers from mice fed the ERD diet compared with mice fed the ERS diet (|log2FC| ≥ 1.0, P < 0.05) (Figure 2B). Of these differentially expressed microRNAs, 5 (miR-338-5p, miR-340-5p, miR-17-5p, miR-362-3p, and miR-3087-3p) have putative binding sites in mRNAs implicated in purine metabolism, including Polr3k (coding for DNA-directed RNA polymerase III subunit RPC11), Polr3f (DNA-directed RNA polymerase III subunit RPC6), Dck (deoxycytidine kinase), Pde4b (cAMP-specific phosphodiesterase 4), Pde7b (cAMP-specific phosphodiesterase 7), Pfas (phosphoribosylformylglycinamidine synthase), and Prps2 (phosphoribosyl pyrophosphate synthetase 2) (Supplemental Table 4).
FIGURE 2.
Transcriptomic analysis in female mice fed ERD and ERS diets for 4 wk. Panels depict mRNA. (A) Heat map with the top 25 differentially expressed transcripts in livers between the ERD and ERS diets. (B) Differentially abundant microRNAs in mouse livers between the ERD and ERS diets. Alg2, α-1,3/1,6-mannosyltransferase; C2cd5, C2 calcium dependent domain containing 5; ERD, exosome- and RNA-depleted; ERS, exosome- and RNA-sufficient; Gprc5a, G protein-coupled receptor class C group 5 member A; Gpr153, G protein-coupled receptor 15; Hhex, hematopoietically expressed homeobox; Hnrnpa0, heterogeneous nuclear ribonucleoprotein A0; Lysmd4, LysM domain containing 4; Mettl13, methyltransferase like 13; miR, microRNA; Nelfa, negative elongation factor complex member A; Nt5c3b, 5'-nucleotidase, cytosolic IIIB; Papd7, terminal nucleotidyltransferase 4A; Phf19, PHD finger protein 19; Phldb3, pleckstrin homology like domain family B member 3; Rbm15b, RNA binding motif protein 15B; Rrn3, RRN3 homolog, RNA polymerase I transcription factor; Slc35b4, solute carrier family 35 member B4; Smyd5, SMYD family member 5; Spred1, sprouty related EVH1 domain containing 1; S1pr5, sphingosine-1-phosphate receptor 5; Tbc1d12, TBC1 domain family member 12; Tfdp1, transcription factor Dp-1; Trim27, tripartite motif containing 27; Tti2, TELO2 interacting protein 2; Uspl1, ubiquitin specific peptidase like 1; 3Klf16, kruppel like factor 16.
TABLE 1.
Differential expression of mRNAs involved in purine metabolism in livers from mice fed ERD or ERS diets for 4 wk1
| mRNA | ERD | ERS |
|---|---|---|
| Ak1 | 9.0 ± 19.6 | 0.1 ± 0.1 |
| Pde4d | 1.2 ± 0.8 | 0.7 ± 0.1 |
| Nt5c3b | 2.4 ± 0.3* | 1.5 ± 0.1 |
| Ada | 1.0 ± 0.2* | 0.6 ± 0.2 |
| Pole | 0.3 ± 0.1* | 0.2 ± 0.1 |
| Nudt16l1 | 12.1 ± 0.8* | 10.3 ± 1.4 |
| Nme6 | 19.3 ± 3.3* | 25.4 ± 3.2 |
| Entpd7 | 1.1 ± 0.3* | 1.8 ± 0.3 |
| Enpp3 | 9.0 ± 1.4* | 12.5 ± 2.8 |
| Polr1b | 5.0 ± 0.4* | 6.1 ± 0.5 |
| Ak4 | 5.5 ± 0.4* | 7.6 ± 1.9 |
1Values are means ± SD fragments per kilobase million (n = 5 mice/group).
*Significantly different from ERS, P < 0.05. Ada, adenosine deaminase; Ak, adenylate kinase; Enpp3, ectonucleotide pyrophosphatase/phosphodiesterase family member 1/3; Entpd7, ectonucleoside triphosphate diphosphohydrolase 7; ERD, exosome- and RNA-depleted; ERS, exosome– and RNA-sufficient; Nme6, nucleoside diphosphate kinase 6; Nt5c3b, 5′-nucleotidase, cytosolic IIIB; Nudt16l1, (nucleoside diphosphate linked moiety X)-type motif 16-like 1; Pde4d, cAMP-specific phosphodiesterase 4D; Pole, DNA polymerase epsilon subunit 1; Polr1b, DNA-directed RNA polymerase I subunit RPA2.
MicroRNA analysis by real-time qPCR revealed the same expression patterns that were observed in microRNA-Seq analysis. The expression of miR-340-5p was higher in livers from mice fed ERS compared with ERD [cycle threshold (Ct) values 26.7 ± 0.3 compared with 27.8 ± 0.6]. The expression of miR-99b-5p was not different between the 2 diet groups (Ct values 20.3 ± 0.2 compared wtih 20.3 ± 0.4), and miR-362-3p was not detectable (Ct > 30.0) (Supplemental Table 5).
Human purine metabolites
Concentrations of purine metabolites followed the same pattern in the urine of DA human adults as the liver patterns observed in the ERD mice. When purines were analyzed by colorimetric assays, HPLC, and HILIC-MRM-MS/MS, urinary concentrations of AMP and plasma concentrations of guanosine 5′-monophosphate were significantly higher in DAs than in DCs (Figure 3). Although the excretion rates and/or concentrations of 21 purine metabolites were also elevated in DAs, these differences were not significantly different from those in DCs (Supplemental Figure 4). The excretion of creatinine was not statistically different in DAs and DCs (P > 0.10).
FIGURE 3.
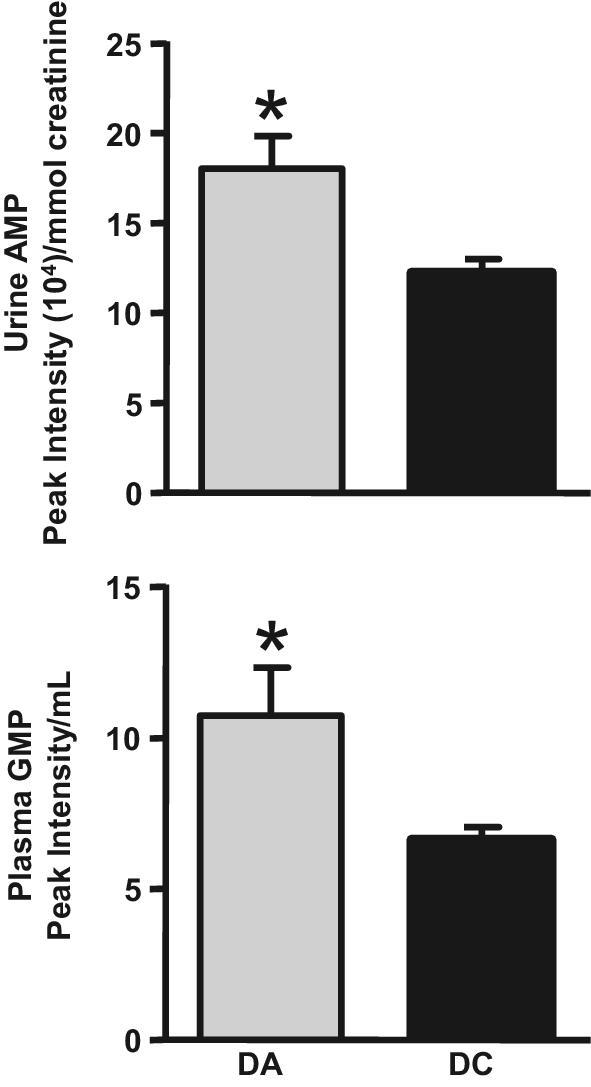
Urinary concentration of AMP and plasma concentration of GMP in adult human DAs and DCs. Values are means ± SEMs, corrected for creatinine. *P < 0.05 vs. DC (n = 6 adults/group). DA, dairy avoider; DC, dairy consumer.
When infant urine samples were analyzed by HPLC and colorimetric assays, uric acid and inosine were significantly higher in the SF group and lower in the HM and MF groups (Figure 4). The concentrations of 12 out of 21 purine metabolites analyzed in infant urine samples by the use of HILIC-MRM-MS/MS were significantly higher in SF compared with HM and MF (Table 2). The excretion of creatinine was not statistically different among the SF, HM, and MF groups (P > 0.10).
FIGURE 4.
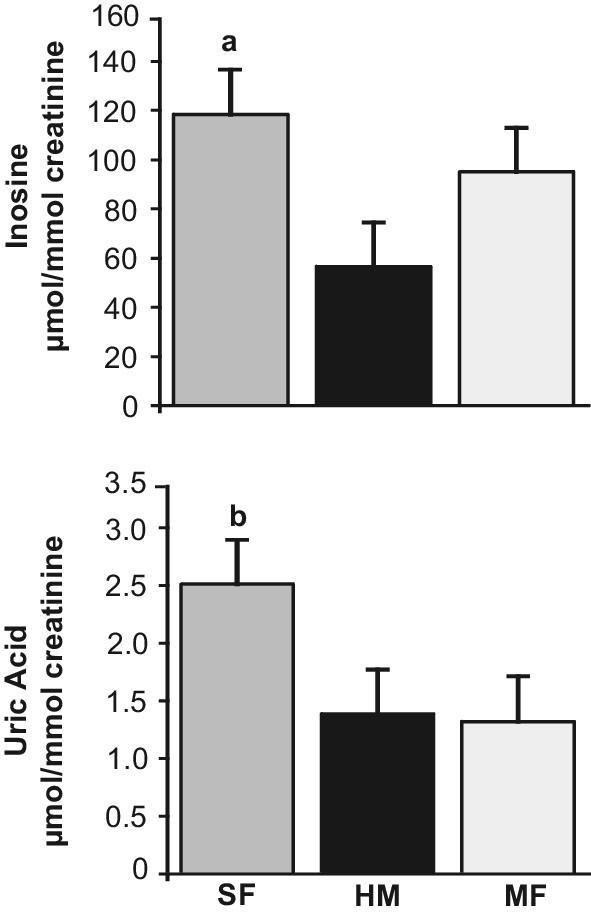
Purine metabolites in urine samples from infants, aged 3 mo, fed SF, MF, or HM. Values are means ± SEMs. aP < 0.05 vs. the HM and MF groups, bP < 0.05 vs. the HM and MF groups (n = 46 for SF, 40 for HM, and 40 for MF). HM, human milk; MF, milk formula; SF, soy formula.
TABLE 2.
Purine metabolites in infants fed SF, HM, or MF1
| Metabolite | Multiplier2 | SF | HM | MF |
|---|---|---|---|---|
| Adenosine | 1000,000 | 242 ± 129b | 135 ± 71 | 167.3 ± 78.2 |
| ADP | 1000 | 34 ± 27b | 7 ± 5 | 10.6 ± 6.6 |
| AICAR | 10,000 | 92 ± 293 | 3 ± 2 | 10.2 ± 10.0 |
| Allantoin | 100,000 | 13 ± 14 | 9 ± 10 | 7.7 ± 6.3 |
| AMP | 100,000 | 21 ± 39b | 1 ± 1 | 1.6 ± 1.5 |
| ATP | 100,000 | 12 ± 26 | 0.1 ± 0.1 | 0.0 ± 0.0 |
| cAMP | 100,000 | 88 ± 79b | 35 ± 25 | 51 ± 33 |
| GDP | 1000 | 57 ± 52b | 32 ± 22 | 31 ± 26 |
| GMP | 1000 | 60 ± 52b | 10 ± 9 | 22 ± 26 |
| GTP | 1000 | 69 ± 51b | 9 ± 9 | 34 ± 33 |
| Guanine | 1000,000 | 68 ± 38b | 35 ± 20 | 45 ± 21 |
| Guanosine | 10,000 | 29 ± 22b | 14 ± 8 | 20 ± 9 |
| Hypoxanthine | 1000,000 | 84 ± 73a | 75 ± 51 | 91 ± 47 |
| IMP | 10,000 | 21 ± 25b | 3 ± 3 | 6 ± 6 |
| Inosine | 1000,000 | 14 ± 9b | 12 ± 7 | 14 ± 10 |
| PRPP | 100,000 | 32 ± 21b | 14 ± 10 | 28 ± 16 |
| Uric acid | 1000,000 | 166 ± 179b | 40 ± 35 | 63 ± 52 |
| Xanthine | 1000,000 | 83 ± 50a | 77 ± 48 | 81 ± 67 |
| Xanthosine | 1000,000 | 17 ± 23b | 5 ± 3 | 7 ± 8 |
| XMP | 100 | 77 ± 79a | 12 ± 13 | 31 ± 19 |
1Values are mean ± SD peak intensity (n = 13 for SF, 14 for HM, and 13 for MF) normalized by the total number of peaks and per millimole of creatinine.
a P < 0.05 vs. HM group.
b P < 0.05 vs. HM and MF groups. AICAR, 5-aminoimidazole-4-carboxamide ribonucleotide; HM, human milk; IMP, inosine monophosphate; MF, milk formula; PRPP, 5-phospho-α-d-ribosyl 1-pyrophosphate; SF, soy formula; XMP, xanthosine monophosphate.
2Multiplication factor to be used to calculate the peak counts.
Discussion
Evidence is accumulating that exosomes and their RNA cargos in milk are bioavailable (8–13, 40). In a previous study, consumption of the ERD diet caused an ∼60% decrease in miR-29b and miR-200c concentrations in murine plasma compared with mice fed the ERS diet (8). Phenotypes associated with the dietary depletion of milk exosomes and their RNA cargos remain elusive. To the best of our knowledge, this is the first report linking the dietary depletion of milk exosomes with a physiologically important metabolic phenotype, namely: 1) a shift in hepatic purine biochemical pathways, and 2) an increase in the concentrations of purine metabolites in tissues and body fluids compared with controls fed milk exosome–sufficient diets.
Changes in purine metabolism have exceptional biological significance because of the essential roles of purines in major pathways in intermediary metabolism and in cell signaling (20). The roles of ATP and adenosine in purinergic receptor signaling in cognitive performance is of particular interest, and ongoing research in our laboratory suggests that spatial learning and memory are impaired in mice fed ERD compared with ERS controls (41). It is widely acknowledged that purinergic receptor signaling plays important roles in spatial learning and memory (42, 43). Herein, we made the novel observation of significantly higher concentrations of urinary purine metabolites in soy-fed infants. Further studies are needed to fully determine if there are direct links between limited postnatal milk intake, purine biochemistry, and brain development.
It is particularly noteworthy that the same purine metabolic phenotype was consistently observed in 3 independent experimental situations, i.e., mice fed ERD or ERS diets, DA compared with DC human adults, and infants fed SF, HM, or MF. Although it can be challenging to model human disease and nutrition on animal studies (44, 45), this study established cause-and-effect in animal feeding studies and translated the findings in animal studies to human adults and infants.
The diets used in mouse feeding studies are based on the AIN-93G formula, and are defined by their RNA content in the milk exosomes added to the diet (8, 22). We formally excluded differences in dietary purines as confounders in this study. Recently, we demonstrated that ERD and ERS differ by their content of milk exosomes, and that the exosomes in the ERD diet are depleted of RNA compared with the exosomes in the ERS diet (S Sukreet and J Zempleni, unpublished observation). The amount of bovine milk exosomes added to the diets is the equivalent of exosomes in 0.5 L of milk consumed by a human adult. The diets consumed by the 2 human cohorts in this study were less stringently controlled than the murine diets, yet the same patterns in purine metabolism were evident in the humans.
Five independent laboratories, including ours, have demonstrated that milk exosomes are transported by intestinal cells, immune cells, and vascular endothelial cells, and are bioavailable in mice (11–14, 40, 46, 47). These observations have gone undisputed. In contrast, concerns were raised by Laubier et al. (48), Auerbach et al. (49), Title et al. (50), and Kang et al. (51) as to whether the low concentrations of microRNAs in bovine milk exosomes can elicit phenotypes in nonbovine species. Laubier et al. (48) fostered wild-type pups to transgenic mice that overexpressed miR-30b and failed to see a substantial increase in tissue levels of miR-30b in pups. The failure to observe an increase in miR-30b in pup tissues was probably owing to the fact that the miR-30b in overexpression dams was not encapsulated in milk exosomes, thereby compromising the miR-30b's stability and bioavailability (11, 16–18). Auerbach et al. (49) reported a failure to detect bovine miR-29b and miR-200c in human plasma following a milk meal. Subsequent studies suggest that the integrity of the samples used in that study was compromised and the RNA was degraded (10). Title et al. (50) detected only trace amounts of miR-375 in the plasma of miR-375 knockout mouse pups fostered to wild-type dams. Our studies suggest that miR-375 in milk, unlike many other microRNAs, is subject to “first passage elimination” in intestinal mucosa and liver, and therefore its concentrations in circulation and peripheral tissues are low [(52, 53); S Manca and J Zempleni, unpublished data). Kang et al. (51) conducted a meta-analysis of published RNA-Seq datasets and concluded that the abundance of dietary microRNAs in body fluids is very low, and is possibly caused by assay artifacts. Their analysis was biased by applying considerably lower levels of stringency when mapping human microRNAs (3 mismatches allowed) compared with dietary microRNAs (1 mismatch), by disregarding the abundance of microRNAs in foods, by withholding details of data normalization protocols across datasets, and by dismissing the possibility that local concentrations of dietary microRNAs at the site of absorption might be high.
Some uncertainties remain. For example, we do not know how exosomes and their cargos alter purine metabolism. The binding of microRNAs to binding sites in mRNAs implicated in purine metabolism is a plausible mechanism, but the evidence in support of this theory is circumstantial and based on in silico predictions. Alternative scenarios include the docking of exosomes to the cell surface triggering cell signaling cascades and exosome-dependent changes in the gut microbiome and microbial metabolites (54–56). Another limitation of our study is that the expression of mRNAs and microRNAs was assessed only in female, and not in male, livers. Note that we did not observe any effects of sex on the hepatic concentrations of purine metabolites in mice. This observation is consistent with a previous report suggesting that sex does not affect the hepatic expression of mRNAs encoding enzymes in purine metabolism in mice (57).
Ongoing and future work in our laboratory is focused on the bioavailability and distribution of microRNAs implicated in brain function, and the roles of milk exosomes and their RNA cargos in spatial learning and memory. For example, we have observed that spatial memory is impaired in young female mice born to parents fed the ERD diet and continued on the parental diet compared with ERS controls. We speculate that altered purinergic receptor signaling plays a role in loss of spatial learning and memory in mice fed the ERD diet compared with ERS controls because adenosine and ATP play crucial roles in purinergic receptor signaling (42). Urine patterns of purine-relevant metabolites in low milk-consuming adults and infants were consistent with those predicted from basic research studies in mice fed ERD. This strongly supports the idea that delivery of RNA cargos from milk exosomes is a fundamental mammalian process, with potentially profound impacts on physiology and development.
Supplementary Material
Acknowledgments
We acknowledge the use of the Biomedical and Obesity Research Core in the Nebraska Center for the Prevention of Obesity Disease through Dietary Molecules (NIH 1P20GM104320). We also acknowledge Sean H. Adams for collaborating with editorial comments. The authors’ responsibilities were as follows—AA–L and JZ: wrote the paper; AA–L, SB, RG, JS, DG, JC, KEM, TMB, AA, AL, and JA: conducted the research; AA–L, JS, RG, and JA: analyzed the data; JZ: had primary responsibility for final content; and all authors: read and approved the final manuscript.
Notes
This material is based upon work that is supported by the National Institute of Food and Agriculture (NIFA), USDA, under award numbers 2015-67017-23181 and 2016-67001-25301, NIH grant 1P20GM104320, Egg Nutrition Center, the Gerber Foundation, the University of Nebraska Agricultural Research Division (Hatch Act), and USDA multistate group W3002 (all to JZ), and a Food for Health grant from the University of Nebraska President's Office (to JC and JZ). Funding for the clinical study was from the USDA-Agricultural Research Service Project 6026-51000-010-05S (to AA and TB). JZ serves as a consultant for PureTech Health, Inc. The granting agencies had no influence on the study design; the collection, analysis, and interpretation of the data; the writing of the manuscript; or the decision to submit the manuscript for publication.
Supplemental Tables 1–5 and Supplemental Figures 1–4 are available from the “Supplementary data” link in the online posting of the article and from the same link in the online table of contents at https://academic.oup.com/jn/.
Author disclosures: AA-L, SB, RG, JS, DG, AL, JA, JC, KEM, TMB, AA, and JZ, no conflicts of interest.
Abbreviations used:
- Ct
cycle threshold
- DA
dairy avoider
- DC
dairy consumer
- ERD
exosome- and RNA-depleted
- ERS
exosome- and RNA-sufficient
- HILIC-MRM-MS/MS
hydrophilic-interaction chromatography-multiple reaction monitoring-tandem mass spectrometry
- HM
human milk
- MF
milk formula
- RNA-Seq
RNA-sequencing
- SF
soy formula.
References
- 1. Yanez-Mo M Siljander PR Andreu Z Zavec AB Borras FE Buzas EI Buzas K Casal E Cappello F Carvalho J et al.. Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles 2015;4:27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Abels ER, Breakefield XO. Introduction to extracellular vesicles: biogenesis, RNA cargo selection, content, release, and uptake. Cell Mol Neurobiol 2016;36:301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Svensson KJ, Christianson HC, Wittrup A, Bourseau-Guilmain E, Lindqvist E, Svensson LM, Morgelin M, Belting M. Exosome uptake depends on ERK1/2-heat shock protein 27 signaling and lipid Raft-mediated endocytosis negatively regulated by caveolin-1. J Biol Chem 2013;288:17713–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ. Dicer is essential for mouse development. Nat Genet 2003;35:215–7. [DOI] [PubMed] [Google Scholar]
- 5. Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 2009;19:92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Vojtech L Woo S Hughes S Levy C Ballweber L Sauteraud RP Strobl J Westerberg K Gottardo R Tewari M et al.. Exosomes in human semen carry a distinctive repertoire of small noncoding RNAs with potential regulatory functions. Nucleic Acids Res 2014;42:7290–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Danielson KM, Das S. Extracellular vesicles in heart disease: excitement for the future? Exosomes Microvesicles 2014;2:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Baier SR, Nguyen C, Xie F, Wood JR, Zempleni J. MicroRNAs are absorbed in biologically meaningful amounts from nutritionally relevant doses of cow's milk and affect gene expression in peripheral blood mononuclear cells, HEK-293 kidney cell cultures, and mouse livers. J Nutr 2014;144:1495–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Shu J, Chiang K, Zempleni J, Cui J. Computational characterization of exogenous microRNAs that can be transferred into human circulation. PLoS One 2015;10:e0140587. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10. Wang L, Sadri M, Giraud D, Zempleni J. RNase H2-dependent polymerase chain reaction and elimination of confounders in sample collection, storage, and analysis strengthen evidence that microRNAs in bovine milk are bioavailable in humans. J Nutr 2018;148:153–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wolf T, Baier SR, Zempleni J. The intestinal transport of bovine milk exosomes is mediated by endocytosis in human colon carcinoma caco-2 cells and rat small intestinal IEC-6 cells. J Nutr 2015;145:2201–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Izumi H, Tsuda M, Sato Y, Kosaka N, Ochiya T, Iwamoto H, Namba K, Takeda Y. Bovine milk exosomes contain microRNA and mRNA and are taken up by human macrophages. J Dairy Sci 2015;98:2920–33. [DOI] [PubMed] [Google Scholar]
- 13. Kusuma Jati R, Manca S, Friemel T, Sukreet S, Nguyen C, Zempleni J. Human vascular endothelial cells transport foreign exosomes from cow's milk by endocytosis. Am J Physiol Cell Physiol 2016;310:C800–C7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Munagala R, Aqil F, Jeyabalan J, Gupta RC. Bovine milk-derived exosomes for drug delivery. Cancer Lett 2016;371:48–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang K, Li H, Yuan Y, Etheridge A, Zhou Y, Huang D, Wilmes P, Galas D. The complex exogenous RNA spectra in human plasma: an interface with human gut biota? PLoS One 2012;7:e51009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Izumi H, Kosaka N, Shimizu T, Sekine K, Ochiya T, Takase M. Bovine milk contains microRNA and messenger RNA that are stable under degradative conditions. J Dairy Sci 2012;95:4831–41. [DOI] [PubMed] [Google Scholar]
- 17. Benmoussa A, Lee CH, Laffont B, Savard P, Laugier J, Boilard E, Gilbert C, Fliss I, Provost P. Commercial dairy cow milk microRNAs resist digestion under simulated gastrointestinal tract conditions. J Nutr 2016;146:2206–15. [DOI] [PubMed] [Google Scholar]
- 18. Zempleni J, Aguilar-Lozano A, Sadri M, Sukreet S, Manca S, Wu D, Zhou F, Mutai E. Biological activities of extracellular vesicles and their cargos from bovine and human milk in humans and implications for infants. J Nutr 2017;147:3–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Luo Y Wang P Wang X Wang Y Mu Z Li Q Fu Y Xiao J Li G Ma Y et al.. Detection of dietetically absorbed maize-derived microRNAs in pigs. Sci Rep 2017;7:645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Garrett RH, Grisham CM. Biochemistry. Fort Worth (TX): Saunders College Publishing; 1995. [Google Scholar]
- 21. Couzin-Frankel J. When mice mislead. Science 2013;342:922–3, 5. [DOI] [PubMed] [Google Scholar]
- 22. Reeves PG, Nielsen FH, Fahey GC., Jr AIN-93 purified diets for laboratory rodents: final report of the American Institute of Nutrition ad hoc writing committee on the reformulation of the AIN-76A rodent diet. J Nutr 1993;123:1939–51. [DOI] [PubMed] [Google Scholar]
- 23. Leiferman A, Shu J, Grove R, Cui J, Adamec J, Zempleni J. A diet defined by its content of bovine milk exosomes and their RNA cargos has moderate effects on gene expression, amino acid profiles and grip strength in skeletal muscle in C57BL/6 mice. J Nutr Biochem 2018;59:123–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Andres A, Casey PH, Cleves MA, Badger TM. Body fat and bone mineral content of infants fed breast milk, cow's milk formula, or soy formula during the first year of life. J Pediat 2013;163:49–54. [DOI] [PubMed] [Google Scholar]
- 25. Nygren H, Seppanen-Laakso T, Castillo S, Hyotylainen T, Oresic M. Liquid chromatography-mass spectrometry (LC-MS)-based lipidomics for studies of body fluids and tissues. Methods Mol Biol 2011;708:247–57. [DOI] [PubMed] [Google Scholar]
- 26. Smith CA, O'Maille G, Want EJ, Qin C, Trauger SA, Brandon TR, Custodio DE, Abagyan R, Siuzdak G. METLIN: a metabolite mass spectral database. Ther Drug Monit 2005;27:747–51. [DOI] [PubMed] [Google Scholar]
- 27. Xia J, Wishart DS. Using MetaboAnalyst 3.0 for comprehensive metabolomics data analysis. Curr Protoc Bioinformatics 2016;55:14.01–0 91. [DOI] [PubMed] [Google Scholar]
- 28. Yuan M, Breitkopf SB, Yang X, Asara JM. A positive/negative ion-switching, targeted mass spectrometry-based metabolomics platform for bodily fluids, cells, and fresh and fixed tissue. Nat Protoc 2012;7:872–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Burdett TC, Desjardins CA, Logan R, McFarland NR, Chen X, Schwarzschild MA. Efficient determination of purine metabolites in brain tissue and serum by high-performance liquid chromatography with electrochemical and UV detection. Biomed Chromatogr 2013;27:122–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Husdan H, Rapoport A. Estimation of creatinine by the Jaffe reaction. A comparison of three methods. Clin Chem 1968;14:222–38. [PubMed] [Google Scholar]
- 31. FastQC [Internet]. 2017 [cited 24 August, 2017]. Available from: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- 32. Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 2011;12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 2009;25:1105–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol 2013;31:46–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sun Z, Evans J, Bhagwate A, Middha S, Bockol M, Yan H, Kocher JP. CAP-miRSeq: a comprehensive analysis pipeline for microRNA sequencing data. BMC Genomics 2014;15:423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 2014;42:D68–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Heintz-Buschart A Yusuf D Kaysen A Etheridge A Fritz JV May P de Beaufort C Upadhyaya BB Ghosal A Galas DJ et al.. Isolation of nucleic acids from low biomass samples: detection and removal of sRNA contaminants. BMC Biol 2018;16:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dweep H, Gretz N. miRWalk2.0: a comprehensive atlas of microRNA-target interactions. Nat Methods 2015;12:697. [DOI] [PubMed] [Google Scholar]
- 40. Chen T Xie MY Sun JJ Ye RS Cheng X Sun RP Wei LM Li M Lin DL Jiang QY et al.. Porcine milk-derived exosomes promote proliferation of intestinal epithelial cells. Sci Rep 2016;6:33862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Mutai E, Zhou F, Zempleni J. Depletion of dietary bovine milk exosomes impairs sensorimotor gating and spatial learning in C57BL/6 mice. FASEB J 2017;31:150.4 [peer-reviewed meting abstract]. [Google Scholar]
- 42. Duster R, Prickaerts J, Blokland A. Purinergic signaling and hippocampal long-term potentiation. Curr Neuropharmacol 2014;12:37–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Burnstock G, Krugel U, Abbracchio MP, Illes P. Purinergic signalling: from normal behaviour to pathological brain function. Prog Neurobiol 2011;95:229–74. [DOI] [PubMed] [Google Scholar]
- 44. McGonigle P, Ruggeri B. Animal models of human disease: challenges in enabling translation. Biochem Pharmacol 2014;87:162–71. [DOI] [PubMed] [Google Scholar]
- 45. Baker DH. Animal models in nutrition research. J Nutr 2008;138:391–6. [DOI] [PubMed] [Google Scholar]
- 46. Agrawal AK Aqil F Jeyabalan J Spencer WA Beck J Gachuki BW Alhakeem SS Oben K Munagala R Bondada S et al.. Milk-derived exosomes for oral delivery of paclitaxel. Nanomedicine 2017;13:1627–36. [DOI] [PubMed] [Google Scholar]
- 47. Liao Y, Du X, Li J, Lonnerdal B. Human milk exosomes and their microRNAs survive digestion in vitro and are taken up by human intestinal cells. Mol Nutr Food Res 2017;61:11. [DOI] [PubMed] [Google Scholar]
- 48. Laubier J, Castille J, Le Guillou S, Le Provost F. No effect of an elevated miR-30b level in mouse milk on its level in pup tissues. RNA Biol 2015;12:26–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Auerbach A, Vyas G, Li A, Halushka M, Witwer K. Uptake of dietary milk miRNAs by adult humans: a validation study. F1000Res 2016;5:721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Title AC, Denzler R, Stoffel M. Uptake and function studies of maternal milk-derived microRNAs. J Biol Chem 2015;290:23680–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kang W, Bang-Berthelsen CH, Holm A, Houben AJ, Muller AH, Thymann T, Pociot F, Estivill X, Friedlander MR. Survey of 800+ data sets from human tissue and body fluid reveals xenomiRs are likely artifacts. RNA 2017;23:433–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pond SM, Tozer TN. First-pass elimination. Basic concepts and clinical consequences. Clin Pharmacokinet 1984;9:1–25. [DOI] [PubMed] [Google Scholar]
- 53. Zempleni J, Baier SR, Hirschi K. Diet-responsive microRNAs are likely exogenous. J Biol Chem 2015;290:25197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Purushothaman A, Bandari SK, Liu J, Mobley JA, Brown EE, Sanderson RD. Fibronectin on the surface of myeloma cell-derived exosomes mediates exosome-cell interactions. J Biol Chem 2016;291:1652–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Muller L, Mitsuhashi M, Simms P, Gooding WE, Whiteside TL. Tumor-derived exosomes regulate expression of immune function-related genes in human T cell subsets. Sci Rep 2016;6:20254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhou F, Paz AH, Sadri M, Fernando CS, Zempleni J. A diet defined by its content of bovine milk exosomes alters the composition of the intestinal microbiome in C57BL/6 mice. FASEB J 2017;31:965.27920150 [Google Scholar]
- 57. Gatti DM, Zhao N, Chesler EJ, Bradford BU, Shabalin AA, Yordanova R, Lu L, Rusyn I. Sex-specific gene expression in the BXD mouse liver. Physiol Genomics 2010;42:456–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.