Abstract
MicroRNAs (miRNAs) play important roles in mammalian spermatogenesis, which is highly dependent on Sertoli cells. However, the functions and mechanisms of miRNAs in regulating human Sertoli cells remain largely unknown. Here, we report that hsa-miR-202-3p mediates the proliferation, apoptosis, and synthesis function of human Sertoli cells. miR-202-3p was upregulated in Sertoli cells of Sertoli cell-only syndrome (SCOS) patients compared with obstructive azoospermia (OA) patients with normal spermatogenesis. Overexpression of miR-202-3p induced Sertoli cell apoptosis and inhibited cell proliferation and synthesis, and the effects were opposite when miR-202-3p was knocked down. Lipoprotein receptor-related protein 6 (LRP6) and Cyclin D1 of the Wnt/β-catenin signaling pathway were identified as direct targets of miR-202-3p in Sertoli cells, which were validated by bioinformatics tools and dual-luciferase reporter assay. Differentially expressed LRP6 and Cyclin D1 between OA and SCOS Sertoli cells were also verified. LRP6 small interfering RNA (siRNA) interference not only mimicked the effects of miR-202-3p overexpression, but also antagonized the effects of miR-202-3p inhibition on Sertoli cells. Collectively, miR-202-3p controls the proliferation, apoptosis, and synthesis function of human Sertoli cells via targeting LRP6 and Cyclin D1 of the Wnt/β-catenin signaling pathway. This study thus provides a novel insight into fate determinations of human Sertoli cells and niche of human testis.
Keywords: human Sertoli cells, miR-202-3p, proliferation and apoptosis, synthesis, LRP6, Cyclin D1, Wnt/β-catenin
Introduction
Non-obstructive azoospermia (NOA), which is defined as the absence of sperm in the semen, affects about 10% of infertile men and almost 60% of the azoospermia patients.1, 2 NOA usually results from testicular failure and aberrant spermatogenesis, including maturation arrest and a complete absence of male germ cells in the seminiferous tubules, which is known as Sertoli cell-only syndrome (SCOS).3, 4 Some genetic factors that cause male infertility have been identified and well studied, such as Y chromosome microdeletion and Klinefelter’s syndrome; however, the roles of epigenetic regulation in human spermatogenesis and in the pathogenesis of NOA, especially SCOS, remain largely unknown.5
Spermatogenesis is a complex developmental process whereby highly specialized haploid sperms are generated from spermatogonial stem cells (SSCs) via the self-renew and differentiation of SSCs, meiosis of spermatocytes, and the spermiogenesis.6, 7 Each step of this intricate process is precisely modulated by the microenvironment or the niche of the testis, which is mainly composed of Sertoli cells, growth factors, cytokines, and blood vessels.8, 9 As the only somatic cell type within the seminiferous tubules, Sertoli cells play essential roles in the formation of a functional testis and the regulation of SSC fate by supplying structural, immunological, and nutritional support.10, 11, 12 A precise balance between SSC self-renewal and differentiation is of great significance in maintaining male fertility, which is mainly regulated by various growth factors and cytokines produced by Sertoli cells, e.g., glial cell line-derived neurotrophic factor (GDNF),13, 14 stem cell factor (SCF),15, 16, 17 bone morphogenetic protein 4 (BMP4),18, 19 fibroblast growth factor 2 (FGF2),20, 21 epithelial growth factor (EGF),22, 23 leukemia inhibitory factor (LIF),24, 25 insulin-like growth factor 1 (IGF1),26, 27 and C-X-C motif chemokine ligand 12 (CXCL12).28, 29 Therefore, a better understanding of the roles and mechanisms of Sertoli cells in spermatogenesis is significant for male infertility therapy and male contraceptive. Besides, Sertoli cells derive from mesoderm and can be converted to multipotent neural stem cells and Leydig cells in vitro, suggesting that Sertoli cells have great applications in tissue engineering and cell-based therapy for neural system disorders or testosterone deficiency of the aging male.30, 31
MicroRNAs (miRNAs), a class of endogenous, small (21–25 nt) non-coding RNAs, exert their functions by directly binding to the 3′ UTR of the target mRNAs, causing the degradation of mRNA or translational inhibition of functional proteins.32 As important epigenetic regulators, although the biological functions of most miRNAs have not yet been fully understood, they are believed to play important roles in the regulation of various biological processes, including cell differentiation, proliferation, apoptosis, and cancer development.33, 34, 35, 36 In recent years, miRNAs have attracted increasing attention in the male infertility field, and they are found to be essential for spermatogenesis. However, these studies mainly focused on male germ cells.37, 38, 39 Regulatory functions of miRNAs in human Sertoli cells remain largely unknown. It has been reported that Sertoli cell-specific deletion of Dicer, an indispensable enzyme for miRNA production, severely impairs male fertility and results in testicular degeneration and absence of mature sperms, reflecting the essential roles of miRNAs in Sertoli cells for normal spermatogenesis.40, 41 We have previously compared the miRNA profiles between OA and SCOS Sertoli cells using miRNA microarrays, and we have revealed that miR-133b promotes the proliferation of human Sertoli cells via targeting transcription factor GLI3 (GLI family zinc finger 3) and activating Cyclin B1 and Cyclin D1.42 We also found that miR-202-3p of the let7 family was prominently upregulated in SCOS Sertoli cells compared with that of obstructive azoospermia (OA) patients with normal spermatogenesis.42 miR-202-3p is located within a chromosomal fragile site in 10q26, and it has been reported to regulate the proliferation, invasion, and apoptosis of many types of tumor cells, such as gastric cancer,43 breast cancer,44 cervical squamous cell carcinoma,45, 46 colorectal cancer,47 esophageal squamous cell carcinoma,48, 49 and brain tumors.50 In the male reproductive system, miR-202-3p maintains mouse SSCs by inhibiting cell-cycle regulators and RNA binding proteins.51 However, the effects of miR-202-3p in mediating human Sertoli cells have not been documented.
Low-density lipoprotein receptor-related protein 6 (LRP6) functions as a transmembrane receptor to transduce Wnt signals and activate the canonical Wnt/β-catenin signaling pathway, which is involved in regulation of various developmental processes, including determination of segment polarity during Drosophila larval development52, 53 and differentiation of brain,54 kidney, limb, and reproductive tracts of male and female mice.55, 56 An aberrant LRP6-mediated Wnt/β-catenin pathway has been shown to be involved in many diseases, such as Alzheimer’s disease,57 autosomal-dominant oligodontia,58 and colorectal cancer.59 Proliferation and self-renewal of mouse and human testis cells are also regulated by the Wnt/β-catenin pathway. It has been reported that the Wnt/β-catenin pathway stimulates the proliferation of adult human Sertoli cells via upregulation of c-Myc expression. Mutant mice that expressed constitutively active forms of β-catenin specifically in Sertoli cells developed testicular Sertoli cell tumor at 8 months of age. These results indicated the involvement of abnormal Wnt/β-catenin signaling in impaired Sertoli cell functions and spermatogenesis.60, 61, 62 However, the mechanisms of the Wnt/β-catenin pathway in human Sertoli cells, especially the epigenetic regulations of this signaling pathway, remain unclear.
In this study, we found that miR-202-3p was upregulated in SCOS Sertoli cells. miR-202-3p induced the apoptosis and led to suppression of cell proliferation and synthesis function of Sertoli cells by targeting LRP6 and Cyclin D1 of Wnt/β-catenin signaling pathway. This study could offer new epigenetic mechanisms about the regulation of human Sertoli cell functions and spermatogenesis, and provide new targets for gene therapy of male infertility.
Results
Isolation and Identification of Human Primary Sertoli Cells
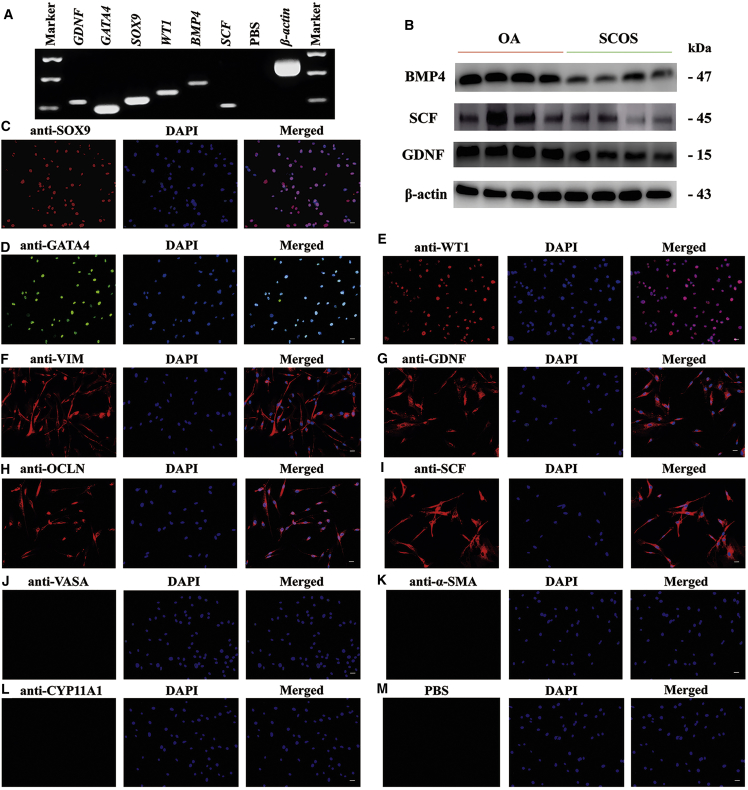
Human Sertoli cells were isolated and purified from 20 OA and 20 SCOS patients using a two-step enzymatic digestion followed by differential plating. Trypan blue exclusion assay was conducted to measure the viability of primary isolated cells, which was over 97% (data not shown). The isolated human cells were identified by detecting various markers for Sertoli cells at both transcriptional and translational levels. RT-PCR showed that GDNF, GATA4, SOX9, WT1, BMP4, and SCF, markers for Sertoli cells, were expressed in the isolated cells. PCR with PBS instead of cDNA served as a negative control, and PCR with β-actin (ACTB) served as a loading control for total RNA (Figure 1A). The protein levels of BMP4, SCF, and GDNF were also detected in both OA and SCOS Sertoli cells, and their expressions were higher in OA Sertoli cells (Figure 1B). The results were consistent with our former finding.63 The purity of isolated human cells was determined by immunofluorescence. More than 98% of the cells were positive for SOX9 (Figure 1C), GATA4 (Figure 1D), wild-type 1 (WT1) (Figure 1E), VIM (Figure 1F), GDNF (Figure 1G), occludin (OCLN) (Figure 1H), and SCF (Figure 1I). Detection of VASA (Figure 1J), α-smooth muscle actin (α-SMA) (Figure 1K), and CYP11A1 (Figure 1L) was also conducted to exclude the potential contamination of germ cells, peritubular myoid cells, and Leydig cells. No fluorescence was observed after substituting PBS for primary antibodies (Figure 1M). Taken together, these data indicated that the primary isolated cells were human Sertoli cells in phenotype.
Figure 1.
Isolation and Identification of Human Sertoli Cells from OA and SCOS Patients
(A) RT-PCR showed the transcripts of GDNF, GATA4, SOX9, WT1, BMP4, and SCF in the isolated cells. PCR with PBS but without cDNA served as a negative control. (B) Western blots showed the protein levels of BMP4, SCF, and GDNF in OA and SCOS Sertoli cells. (C–L) Immunofluorescence demonstrated the expression of SOX9 (C), GATA4 (D), WT1 (E), VIM (F), GDNF (G), OCLN (H), SCF (I), VASA (J), α-SMA (K), and CYP11A1 (L) in the isolated cells. Replacement of primary antibodies with PBS was used as a negative control (M). The cell nuclei were stained with DAPI. Scale bars, 5 μm (C–M).
Differential Expression of miR-202-3p between OA and SCOS Sertoli Cells
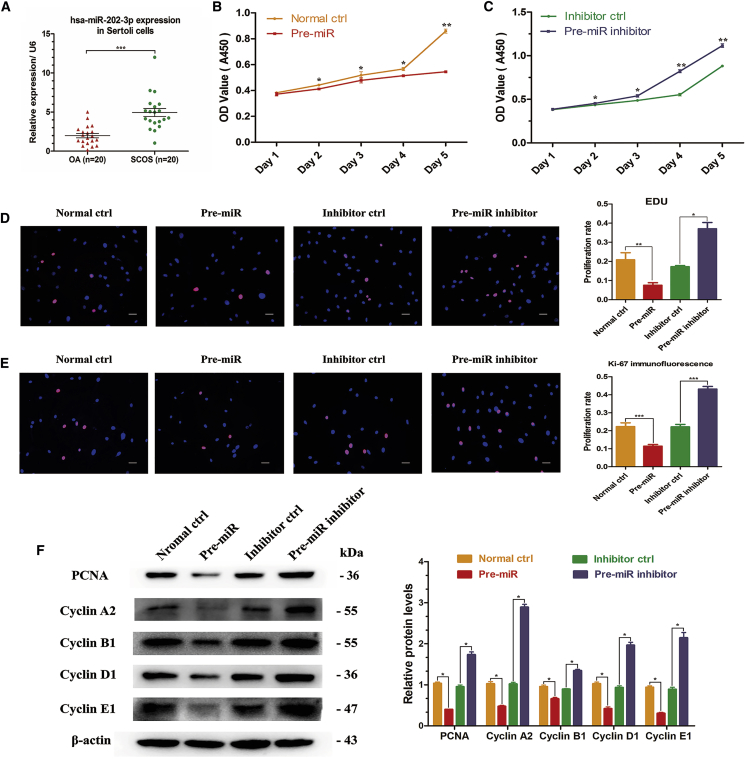
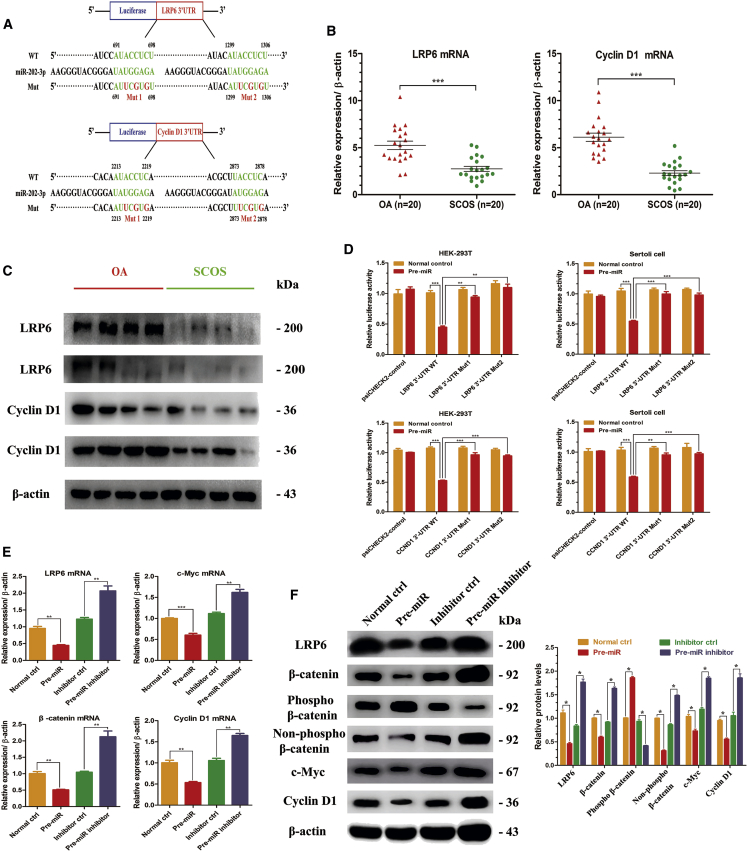
As shown in our previous miRNA microarray data, miR-202-3p was one of the most prominently upregulated miRNAs in SCOS Sertoli cells compared with OA patients with normal spermatogenesis.42 To verify this result, we examined miR-202-3p expression levels in these two types of patients using real-time qPCR. Consistent with the microarray data, expression level of miR-202-3p was significantly upregulated in SCOS Sertoli cells compared with OA Sertoli cells (Figure 2A) (n = 20; p < 0.001).
Figure 2.
Differentially Expressed miR-202-3p Inhibits the Proliferation of Human Sertoli Cells
(A) Real-time qPCR revealed the expression of miR-202-3p in both OA and SCOS Sertoli cells (n = 20). (B and C) CCK-8 assay showed the growth curve of human Sertoli cells for 5 days in the pre-miR group (B) and the pre-miR inhibitor group (C) after virus infection and puromycin screening. (D) EDU incorporation assay showed the EDU-positive cells in human Sertoli cells after virus infection and puromycin screening. Cell nuclei were counterstained with Hoechst 33342. The percentages of EDU-positive cells were counted out of 500 total cells from three independent experiments. (E) Immunofluorescence revealed the ki-67-positive cells in the four cell strains. Cell nuclei were counterstained with DAPI. The percentages of ki-67-positive cells were counted out of 500 total cells from three independent experiments. (F) Western blots demonstrated the expression of PCNA and cell-cycle proteins in human Sertoli cells at 72 hr after virus infection and puromycin screening. β-actin served as a loading control of proteins. Results of the pre-miR group were normalized to the Normal ctrl group, and results of the pre-miR inhibitor group were normalized to the inhibitor ctrl group. *p < 0.05; **p < 0.01; ***p < 0.001. Scale bars, 10 μm (D and E).
Establishment of Stable Cell Strains with Upregulated or Downregulated miR-202-3p
To explore the effects of miR-202-3p on fate determinations of human Sertoli cells, stable cell strains with upregulated or downregulated miR-202-3p and corresponding controls were established using virus infection and puromycin screening. Observation of EGFP fluorescence (Figure S1A) and detection of miR-202-3p expression levels (Figure S1B) indicated that the stable cell strains were successfully established, which were named as normal control (ctrl), pre-miR, inhibitor ctrl, and pre-miR inhibitor, respectively.
miR-202-3p Inhibits the Proliferation of Human Sertoli Cells
To probe whether miR-202-3p influences the proliferation of Sertoli cells, we performed multiple methods to compare the proliferation rates among the four cell strains. Cell counting kit-8 (CCK-8) assay showed that miR-202-3p overexpression significantly reduced the growth rate of Sertoli cells in a time-dependent manner, whereas silencing miR-202-3p expression remarkably promoted the proliferation (Figures 2B and 2C). 5-Ethynyl-2′-deoxyuridine (EDU) incorporation assay was further conducted to examine the influence of miR-202-3p on DNA synthesis of human Sertoli cells. Compared with normal ctrl (20.8% ± 3.7% of EDU-positive cells) and inhibitor ctrl (17.3% ± 0.5% of EDU-positive cells), miR-202-3p downregulation increased the EDU-positive cells up to 37.0% ± 3.3%, whereas miR-202-3p overexpression reduced the EDU-positive cells down to 7.5% ± 1.3%, reflecting that miR-202-3p inhibits DNA synthesis of human Sertoli cells (Figure 2D). PCNA (proliferating cell nuclear antigen) and ki-67 are generally regarded as hallmarks for cellular proliferation. Immunofluorescence showed that the percentage of ki-67-positive cells increased from 22.1% ± 1.4% to 43.1% ± 1.5% after miR-202-3p knockdown, and the change was opposite when miR-202-3p was overexpressed, reducing from 22.3% ± 2.1% to 11.4% ± 0.8% (Figure 2E). Western blots revealed that the expression level of PCNA was obviously decreased by miR-202-3p overexpression (0.401 ± 0.008) compared with normal ctrl (designated as 1.0), whereas its level was significantly increased by miR-202-3p knockdown (1.733 ± 0.068) compared with inhibitor ctrl (designated as 1.0) (Figure 2F). Cell-cycle proteins play essential roles in regulating the entrance of cells to the S phase and cell proliferation. Therefore, we further examined whether miR-202-3p affected the expression levels of cell-cycle regulators. Western blots displayed that expressions of Cyclin A2 (2.891 ± 0.068), Cyclin B1 (1.347 ± 0.029), Cyclin D1 (1.964 ± 0.070), and Cyclin E1 (2.146 ± 0.127) were elevated by miR-202-3p knockdown, whereas the expression levels of these proteins were decreased by miR-202-3p overexpression (Cyclin A2, 0.475 ± 0.025; Cyclin B1, 0.658 ± 0.033; Cyclin D1, 0.420 ± 0.041; Cyclin E1, 0.306 ± 0.020) (Figure 2F).
miR-202-3p Inhibits the Synthesis Function of Human Sertoli Cells
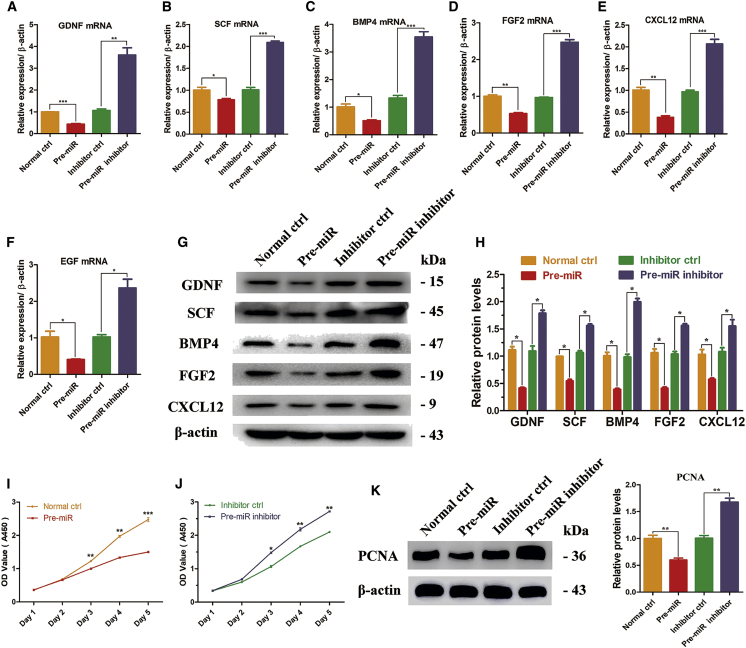
One of the most important roles of the Sertoli cell is to synthesize and secrete essential growth factors that regulate SSC self-renewal and differentiation, such as GDNF, BMP4, SCF, FGF2, CXCL12, and EGF. To figure out whether miR-202-3p influences the synthesis function of Sertoli cells, we conducted real-time qPCR and western blots to examine the transcriptional and translational changes of these growth factors. We observed that miR-202-3p overexpression led to reduction of the mRNA (GDNF, 0.433 ± 0.031; SCF, 0.783 ± 0.029; BMP4, 0.510 ± 0.034; FGF2, 0.529 ± 0.024; CXCL12, 0.379 ± 0.040; EGF, 0.406 ± 0.015) (Figures 3A–3F) and protein (GDNF, 0.415 ± 0.156; SCF, 0.546 ± 0.028; BMP4, 0.392 ± 0.019; FGF2, 0.411 ± 0.026; CXCL12, 0.575 ± 0.023) levels of the growth factors compared with normal ctrl (designated as 1.0) (Figures 3G and 3H), whereas miR-202-3p knockdown enhanced the transcripts (GDNF, 3.604 ± 0.330; SCF, 2.090 ± 0.033; BMP4, 3.546 ± 0.186; FGF2, 2.471 ± 0.073; CXCL12, 2.065 ± 0.110; EGF, 2.364 ± 0.241) (Figures 3A–3F) and protein levels (GDNF, 1.789 ± 0.053; SCF, 1.559 ± 0.031; BMP4, 1.998 ± 0.060; FGF2, 1.564 ± 0.033; CXCL12, 1.552 ± 0.118) compared with inhibitor ctrl (designated as 1.0) (Figures 3G and 3H). However, no significant difference was seen in some genes unrelated to the synthesis function of Sertoli cells among the four cell strains (Figure S2). To confirm the defective synthesis function of Sertoli cells and its effects on spermatogonia, the culture medium of Sertoli cells was collected daily to culture the human SSC line further. This human SSCs cell line was established successfully by our group, which presented unlimited proliferation potentials and no tumor formation. Experiments using this SSCs line were stable and feasible.64 The CCK-8 assay showed that the proliferation of human SSCs was significantly enhanced when cultured with medium collected from the pre-miR inhibitor group, whereas the culture medium from the pre-miR group suppressed the proliferation of SSCs (Figures 3I and 3J). Detection of PCNA protein level at 72 hr showed the same result (Figure 3K). Taken together, these data implicated that miR-202-3p could inhibit the synthesis function of human Sertoli cells, which negatively controlled the proliferation of human SSCs.
Figure 3.
miR-202-3p Inhibits the Synthesis Function of Human Sertoli Cells
The mRNA levels of GDNF (A), SCF (B), BMP4 (C), FGF2 (D), CXCL12 (E), and EGF (F) at 72 hr after virus infection and puromycin screening. (G) Western blots demonstrated GDNF, SCF, BMP4, FGF2, and CXCL12 proteins in human Sertoli cells at 72 hr after virus infection and puromycin screening. Results of three independent assays were concluded in (H). β-actin served as loading control of proteins. (I and J) CCK-8 assay showed the growth curve of a human spermatogonial stem cell line that was cultured with culture medium collected daily from the four Sertoli cell strains with upregulation of miR-202-3p (I) and downregulation of miR-202-3p (J). (K) Western blot demonstrated the expression of PCNA in human spermatogonial stem cell line after 72 hr of culture with culture medium collected daily from the four Sertoli cell strains. Results of the pre-miR group were normalized to the normal ctrl group, and results of the pre-miR inhibitor group were normalized to the inhibitor ctrl group. *p < 0.05; **p < 0.01; ***p < 0.001.
miR-202-3p Promotes the Apoptosis of Human Sertoli Cells
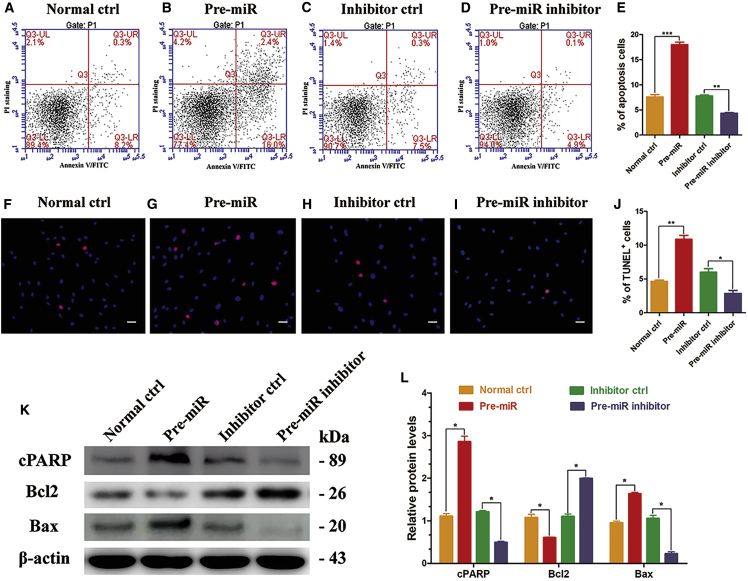
We also examined the influence of miR-202-3p on the apoptosis of human Sertoli cells. Annexin V and propidium iodide (PI) staining and flow cytometry showed that the percentage of apoptosis in human Sertoli cells increased from 7.6% ± 0.5% to 18.0% ± 0.5% after miR-202-3p upregulation and decreased from 7.8% ± 0.2% to 4.3% ± 0.1% when miR-202-3p was knocked down (Figures 4A–4E). Also, the percentage of terminal deoxynucleotidyl transferase-mediated deoxyuridine triphosphate-biotin nick end-labeling-positive (TUNEL+) cells significantly increased when miR-202-3p was overexpressed, and reduced after miR-202-3p inhibition (Figures 4F–4J). Furthermore, miR-202-3p overexpression resulted in an increase in protein levels of apoptosis indicator cleaved-PARP (cPARP) (2.857 ± 0.127) and Bax (1.634 ± 0.033) compared with normal ctrl (designated as 1.0), whereas downregulation of miR-202-3p decreased cPARP (0.494 ± 0.026) and Bax (0.225 ± 0.044) expression levels compared with inhibitor ctrl (Figures 4K and 4L). Meanwhile, the expression of Bcl2, which suppresses apoptosis, significantly decreased in the pre-miR group (0.605 ± 0.016) and increased in the pre-miR inhibitor group (1.996 ± 0.013) (Figures 4K and 4L).
Figure 4.
miR-202-3p Promotes the Apoptosis of Human Sertoli Cells
(A–D) Annexin V-APC/PI and flow cytometry analysis revealed apoptosis in human Sertoli cells at 72 hr in the normal ctrl group (A), pre-miR group (B), inhibitor ctrl group (C), and pre-miR inhibitor group (D) after virus infection and puromycin screening. Results of three independent assays were concluded in (E). A total of 10,000 cells were analyzed. (F–I) TUNEL assay revealed the percentages of TUNEL+ cells in the normal ctrl group (F), pre-miR group (G), inhibitor ctrl group (H), and pre-miR inhibitor group (I) in human Sertoli cells after virus infection and puromycin screening. Results of three independent assays were concluded in (J). (K) Western blots demonstrated cPARP, Bcl2, and Bax proteins in human Sertoli cells at 72 hr after virus infection and puromycin screening. Results of three independent assays were concluded in (L). β-actin served as loading control of proteins. Results of the pre-miR group were normalized to the normal ctrl group, and results of the pre-miR inhibitor group were normalized to the inhibitor ctrl group. *p < 0.05; **p < 0.01; ***p < 0.001. Scale bars, 10 μm (F–I).
LRP6 and Cyclin D1 of the Wnt/β-Catenin Signaling Pathway Are Direct Targets of miR-202-3p in Human Sertoli Cells
To gain novel insights into molecular mechanisms underlying the functions of miR-202-3p in human Sertoli cells, we identified the targets of miR-202-3p using miRNA predict databases, namely TargetScan (http://www.targetscan.org), PicTar (https://pictar.mdc-berlin.de/), and microRNA.org (http://www.microrna.org) (Figure 5A). Among hundreds of genes that were predicted to be the potential targets, we selected LRP6 and Cyclin D1 for the following reasons: (1) both LRP6 and Cyclin D1 mRNAs have two potential binding sites for miR-202-3p at their 3′ UTR (Figure 5A); (2) both LRP6 and Cyclin D1 are important components of the Wnt/β-catenin signaling pathway; and (3) the Wnt/β-catenin signaling pathway is closely associated with cell proliferation, invasion, and differentiation.
Figure 5.
Identification of Direct Targets of miR-202-3p in Human Sertoli Cells
(A) There were two potential binding sites of miR-02-3p at the 3′ UTR region of LRP6 and Cyclin D1 mRNA based on the bioinformatic analysis. 3′ UTR of LRP6 and Cyclin D1 mRNA containing wild-type (WT), mutant-1 (Mut1), and mutant-2 (Mut2) was cloned into dual-luciferase plasmids. (B) The mRNA levels of LRP6 and Cyclin D1 were detected using real-time qPCR in 20 paired OA and SCOS Sertoli cells. (C) Western blots revealed the different expression patterns of LRP6 and Cyclin D1 between OA and SCOS Sertoli cells (n = 8). β-actin served as a loading control. (D) Empty plasmids (psiCHECK2 control) and dual-luciferase plasmids containing WT, Mut1, or Mut2 of LRP6 or Cyclin D1 3′ UTR were transfected into HEK293T cells and OA Sertoli cells with miR-202-3p overexpression plasmids or control plasmids, respectively. Firefly and Renilla luciferase signals were performed for luciferase activity after 36 hr of transfection. (E) mRNA levels of LRP6, c-Myc, Cyclin D1, and β-catenin, as measured by real-time qPCR in the four cell strains. (F) Western blots showed the LRP6, c-Myc, Cyclin D1, β-catenin, phospho-β-catenin, and non-phospho-β-catenin protein levels at 72 hr after virus infection and puromycin screening. β-actin served as a loading control. Results of the pre-miR group were normalized to the normal ctrl group, and results of the pre-miR inhibitor group were normalized to the inhibitor ctrl group. *p < 0.05; **p < 0.01; ***p < 0.001.
To validate the hypothesis mentioned above, the mRNA levels of LRP6 and Cyclin D1 were measured by real-time qPCR in 20 paired OA and SCOS Sertoli cells. The differential expressions of LRP6 and Cyclin D1 proteins were also examined using western blots. Notably, we found that both transcriptional and translational levels of LRP6 and Cyclin D1 were significantly higher in OA Sertoli cells compared with SCOS patients (Figures 5B and 5C). Further, the recombinant plasmids (psiCHECK2) with the 3′ UTR sequences of these two genes containing the predicted binding site sequence (wild-type [WT]) of miR-202-3p were constructed for luciferase assays, and recombinant plasmids containing the LRP6 and Cyclin D1 3′ UTR sequence with mutant nucleotides (mutant type [Mut]) were also constructed to serve as controls (Figure 5A). The WT and Mut vectors were transfected with miR-202-3p overexpression plasmid or control plasmid into both HEK293T cells and OA Sertoli cells, and the level of luciferase enzyme activity was measured according to the manufacturer’s instructions. We revealed that overexpression of miR-202-3p suppressed the luciferase activity of the reporter gene, whereas mutation of three nucleotides within the two miRNA binding sites abolished this repression of luciferase activity, confirming the specificity of the action (Figure 5D). In concordance with these results, LRP6 and Cyclin D1 mRNA and protein levels significantly decreased in miR-202-3p-overexpressed Sertoli cells and enhanced in miR-202-3p-depleted Sertoli cells. Furthermore, in Sertoli cells with upregulated miR-202-3p, the protein levels of c-Myc, β-catenin, and non-phospho β-catenin (active β-catenin) reduced significantly, whereas the expression of phospho-β-catenin increased significantly. The results were opposite when miR-202-3p was downregulated in Sertoli cells (Figures 5E and 5F). Considered together, these results implicate that miR-202-3p inhibits the activation of the Wnt/β-catenin signaling pathway by targeting LRP6 and Cyclin D1 in human Sertoli cells.
LRP6 Knockdown Inhibits the Proliferation and Synthesis Function of Human Sertoli Cells
Then we utilized small RNAs to elucidate the regulating effects of miR-202-3p targets on human Sertoli cells. Because Cyclin D1 was the downstream factor of the Wnt/β-catenin signaling pathway, and it may be involved in some other signaling pathways, we mainly focused on the function of LRP6. To improve the efficiency of LRP6 knockdown, we assessed three pairs of LRP6 small interfering RNAs (siRNAs) targeting different regions of LRP6 mRNA, including LRP6 siRNA-1, siRNA-2, and siRNA-3. At 6 hr after transfection, the transfection efficiency of LRP6 siRNAs in OA Sertoli cells was over 80%, as evidenced by the transfection of FAM-labeled green fluorescent oligo (Figure S3A). Trypan blue exclusion assay revealed that the viability of human Sertoli cells at 6 hr of transfection was about 98% (data not shown), when we changed the culture medium containing transfection reagent to normal DMEM/F12 with 10% FBS. Compared with negative control (only Lipofectamine 3000 was transfected) and siRNA control (designated as 1.0), real-time qPCR showed that LRP6 siRNA-1, -2, and -3 reduced LRP6 mRNA level in Sertoli cells after 24 hr of transfection (siRNA-1, 0.693 ± 0.017; siRNA-2, 0.546 ± 0.013; siRNA-3, 0.364 ± 0.014), and the interfering effect of LRP6 siRNA-3 was the most prominent (Figure S3B). Western blots revealed that LRP6 siRNA-3 significantly reduced the expression level of LRP6 protein at 48 hr after transfection (0.456 ± 0.023). Protein levels of Cyclin D1 (0.596 ± 0.024), c-Myc (0.393 ± 0.010), β-catenin (0.623 ± 0.013), phospho-β-catenin (1.817 ± 0.025), and non-phospho-β-catenin (0.372 ± 0.022) also changed significantly by LRP6 knockdown compared with siRNA control (designated as 1.0), reflecting the inhibition of the Wnt/β-catenin signaling pathway by LRP6 knockdown (Figures S3C and S3D). Therefore, LRP6 siRNA-3 was chosen to further examine the effects of LRP6 on human Sertoli cells.
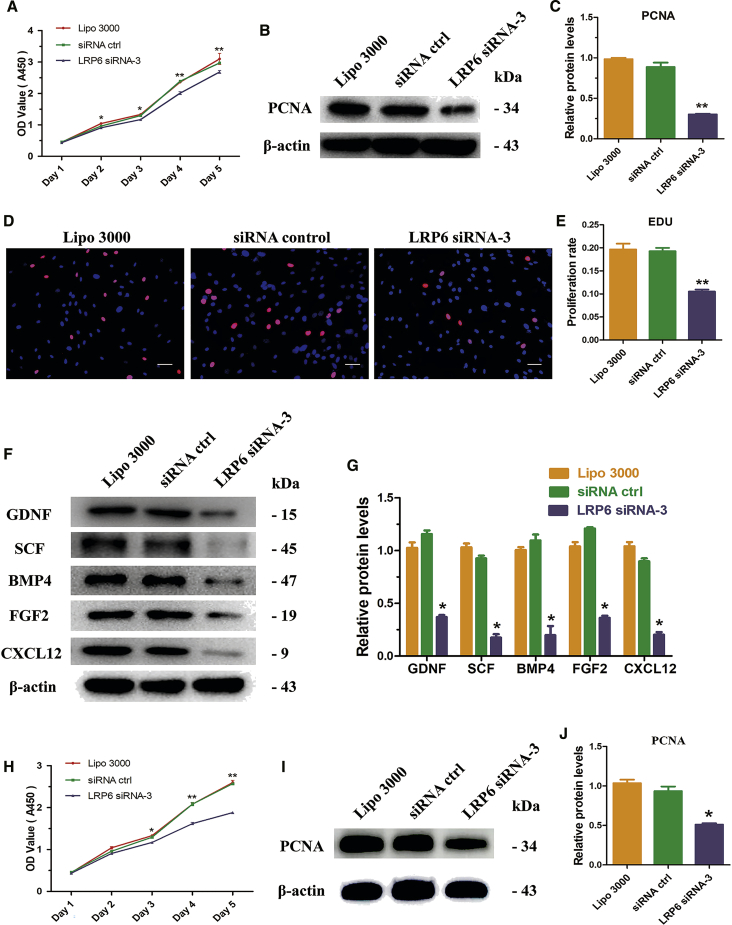
We asked whether LRP6 influences Sertoli cell proliferation. CCK-8 assay was conducted from 24 to 120 hr after transfection of LRP6 siRNA-3 in OA Sertoli cells, and LRP6 siRNA-3 significantly reduced the proliferation from 48 to 120 hr (Figure 6A). The protein level of PCNA was also remarkably reduced by LRP6 siRNA-3 (Figures 6B and 6C). Correspondingly, the percentage of EDU-positive cells reduced significantly in OA Sertoli cells (10.5% ± 2.4%) with LRP6 knockdown compared with the negative control (20.0% ± 1.2%) and siRNA control (19.3% ± 2.9%) at 48 hr after transfection (Figures 6D and 6E). The synthesis function of Sertoli cells was also suppressed by LRP6 knockdown, as evidenced by the reduction of GDNF (0.370 ± 0.018), BMP4 (0.279 ± 0.025), SCF (0.175 ± 0.132), FGF2 (0.362 ± 0.020), and CXCL12 (0.202 ± 0.025) protein levels after LRP6 siRNA-3 treatment, compared with negative control and siRNA control (designated as 1.0) (Figures 6F and 6G). To confirm the defective synthesis function of Sertoli cells and its effects on spermatogonia, the culture medium of Sertoli cells was collected daily to culture the human SSCs line further. The CCK-8 assay showed that the proliferation of human SSCs was significantly reduced when cultured with medium collected from the LRP6 knockdown group (Figure 6H). Detection of PCNA protein level at 72 hr showed the same result (Figures 6I and 6J). Collectively, these data indicate that LRP6 downregulation inactivates the Wnt/β-catenin signaling pathway and inhibits the proliferation and synthesis function of human Sertoli cells.
Figure 6.
LRP6 Knockdown Inhibits the Proliferation and Synthesis Function of Human Sertoli Cells
(A) CCK-8 assay showed the growth curve of human Sertoli cells after transfection of control siRNA or LRP6 siRNA-3 for 1–5 days. (B) Western blots demonstrated the expression of PCNA after 48 hr of transfection of control siRNA or LRP6 siRNA-3. Results of three independent assays were concluded in (C). β-actin served as a loading control of proteins. (D) EDU incorporation assay showed the EDU-positive cells in human Sertoli cells at 48 hr after transfection of control siRNA or LRP6 siRNA-3. Results of three independent assays were concluded in (E). Cell nuclei were counterstained with Hoechst 33342. The percentages of EDU-positive cells were counted out of 500 total cells from three independent experiments. (F) Western blots demonstrated GDNF, SCF, BMP4, FGF2, and CXCL12 proteins in human Sertoli cells at 48 hr after transfection of control siRNA or LRP6 siRNA-3. Results of three independent assays were concluded in (G). β-actin served as a loading control of proteins. (H) CCK-8 assay showed the growth curve of human spermatogonial stem cell line that cultured with culture medium collected daily from Sertoli cells transfected with control siRNA or LRP6 siRNA-3. (I) Western blots demonstrated PCNA protein in human spermatogonial stem cell line after 72 hr of culture with culture medium collected daily from Sertoli cells transfected with control siRNA or LRP6 siRNA-3. Results of three independent assays were concluded in (J). β-actin served as a loading control of proteins. *p < 0.05; **p < 0.01. Scale bars, 20 μm (D).
LRP6 Knockdown Induces Apoptosis of Human Sertoli Cells
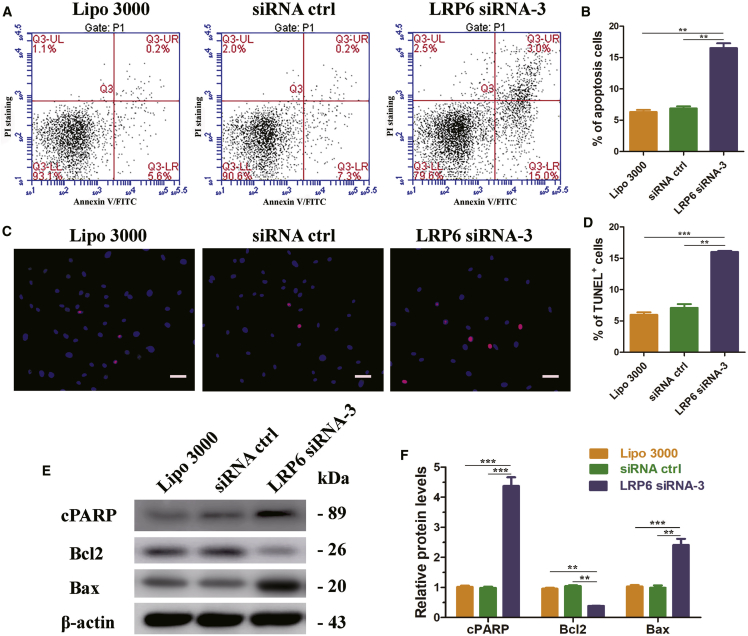
Then we examined the influence of LRP6 knockdown on the apoptosis of human Sertoli cells. Annexin V and PI staining and flow cytometry showed that the percentage of apoptosis in human Sertoli cells increased from 6.3% ± 0.3% to 16.5% ± 0.7% after LRP6 knockdown (Figures 7A and 7B). The percentage of TUNEL+ cells also significantly increased when LRP6 was downregulated (Figures 7C and 7D). Furthermore, LRP6 knockdown resulted in an increase in protein levels of apoptosis indicators cPARP (4.373 ± 0.291) and Bax (2.411 ± 0.207) compared with siRNA ctrl (designated as 1.0). Meanwhile, the expression of Bcl2, which suppresses apoptosis, significantly decreased after LRP6 knockdown (0.384 ± 0.010) (Figures 7E and 7F).
Figure 7.
LRP6 Knockdown Promoted Apoptosis of Human Sertoli Cells
(A) Annexin-V and propidium iodide (PI) staining and flow cytometry showed the percentage of apoptosis in human Sertoli cells after LRP6 knockdown. Results of three independent assays were concluded in (B). A total of 10,000 cells were analyzed. (C) TUNEL assay revealed the percentages of TUNEL+ cells in human Sertoli cells transfected with control siRNA or LRP6 siRNA-3. Results of three independent assays were concluded in (D). (E) Western blots demonstrated cPARP, Bcl2, and Bax proteins in human Sertoli cells at 48 hr after transfection with control siRNA or LRP6 siRNA-3. Results of three independent assays were concluded in (F). β-actin served as loading control of proteins. **p < 0.01; ***p < 0.001. Scale bars, 10 μm (C).
LRP6 Knockdown Attenuated the Effects of miR-202-3p Inhibition on Sertoli Cells
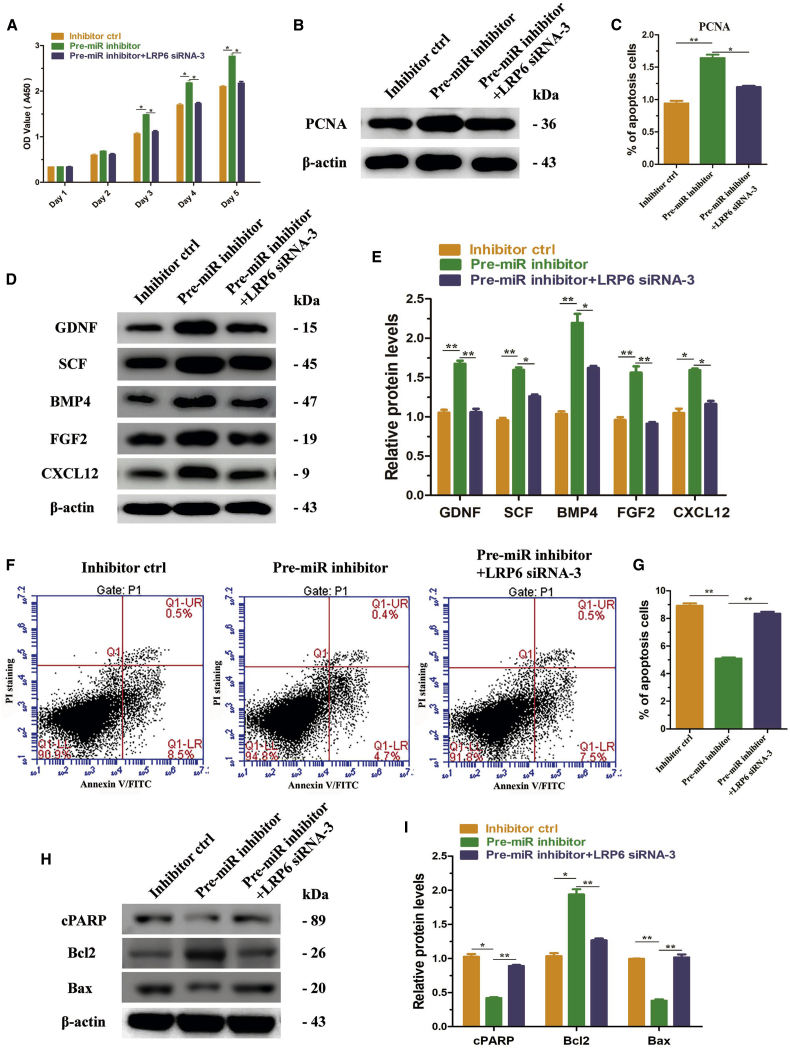
The results above showed that knockdown of LRP6 can mimic the effects of overexpression of miR-202-3p. Then we explored whether LRP6 knockdown could mediate the effects of miR-202-3p. We transfected LRP6 siRNA into the Sertoli cell strain with low expression of miR-202-3p, then compared the Sertoli cell functions among three groups, including inhibitor ctrl, pre-miR inhibitor, and pre-miR inhibitor+LRP6 siRNA-3. We found that LRP6 knockdown could antagonize the effects of miR-202-3p inhibition. CCK-8 assay showed that growth of Sertoli cells with low expression of miR-202-3p was significantly restrained after transfection with LRP6 siRNA (Figure 8A). Detection of PCNA showed the same result (Figures 8B and 8C). The expressions of GDNF, SCF, BMP4, FGF2, and CXCL12 were also significantly reduced at 48 hr of transfection with LRP6 siRNA (Figures 8D and 8E). Annexin V and PI staining and flow cytometry demonstrated that the apoptosis rate increased from 5.1% ± 0.1% to 8.4% ± 0.1% at 48 hr of transfection (Figures 8F and 8G); the protein levels of cPARP and Bax also increased significantly, whereas the expression of Bcl2 decreased (Figures 8H and 8I).
Figure 8.
LRP6 Knockdown Attenuated the Effects of miR-202-3p Inhibition on Sertoli Cells
(A) CCK-8 assay displayed the proliferation of human Sertoli cells treated with miRNA inhibitor control, miR-202-3p inhibitor, and miR-202-3p inhibitor+LRP6 siRNA-3 for 5 days. (B) Western blot showed the expression of PCNA in Sertoli cells treated with miRNA inhibitor control, miR-202-3p inhibitor, and miR-202-3p inhibitor+LRP6 siRNA-3 at 48 hr. Results of three independent assays were concluded in (C). (D) Western blot showed the expression of GDNF, SCF, BMP4, FGF2, and CXCL12 in Sertoli cells treated with miRNA inhibitor control, miR-202-3p inhibitor, and miR-202-3p inhibitor+LRP6 siRNA-3 at 48 hr. Results of three independent assays were concluded in (E). (F) Annexin V and propidium iodide (PI) staining and flow cytometry showed the percentage of apoptosis in human Sertoli cells treated with miRNA inhibitor control, miR-202-3p inhibitor, and miR-202-3p inhibitor+LRP6 siRNA-3 at 48 hr. Results of three independent assays were concluded in (G). A total of 15,000 cells were analyzed. (H) Western blots demonstrated cPARP, Bcl2, and Bax proteins in human Sertoli cells treated with miRNA inhibitor control, miR-202-3p inhibitor, and miR-202-3p inhibitor+LRP6 siRNA-3 at 48 hr. Results of three independent assays were concluded in (I). *p < 0.05; **p < 0.01.
Discussion
Sertoli cells are indispensable for maintaining normal spermatogenesis because they provide a suitable microenvironment or niche with nutritional and immunological support for male germ cell development. Abnormal number and/or functions of Sertoli cells can lead to impaired spermatogenesis and male infertility ultimately.10 Therefore, it is essential to explore the biology of Sertoli cells, which would gain novel insights into the etiology of sterility or infertility and offer new targets for gene therapy of male infertility. It will also facilitate the development of new approaches for male contraception, because it is feasible to modify the functions at epigenetic level, instead of modifying DNA sequences, which is difficulty to do and is irreversible.
Unlike germ cells, human Sertoli cells can be cultured for a long term in vitro, while maintaining their primary morphology, phenotype, and global gene expression pattern.65, 66 Studies on Sertoli cells are mainly focused on their proliferation, apoptosis, secretion, and immunoprotection functions, as well as the effect of aberrant Sertoli cell function on SSC self-renewal and differentiation. Adult Sertoli cells have previously been considered to be unable to divide or proliferate because they are the terminally differentiated cells. Nevertheless, this concept has been challenged, because Sertoli cells from adult hamster and human could regain proliferation in vitro.65 We have revealed that BMP4, BMP6, and NODAL can promote human Sertoli cell proliferation.67, 68, 69 BMP4 knockdown inhibits the synthesis of FGF2, SCF, zonula occludens 1, and claudin 11,67 whereas BMP6 suppresses the apoptosis of Sertoli cells via the Smad2/3 and Cyclin D1 pathway.68 NODAL enhances the secretion of Sertoli cells, especially GDNF, SCF, and BMP4.69 Distinct characteristics of morphology and biochemical phenotype in human Sertoli cells between NOA patients and OA patients are identified, as evidenced by the fact that Sertoli cells isolated from NOA patients have a series of abnormal ultrastructural features and their transcript and protein levels of SCF, BMP4, and GDNF are significantly lower compared with OA Sertoli cells.63 PRPS2, relaxin, and retinoic acid have also been reported to affect Sertoli cell function.70, 71, 72 These studies illustrate the involvement of aberrant Sertoli cell function in the pathogenesis of NOA. However, other regulating factors, especially epigenetic regulators of human Sertoli cells, remain to be clarified.
As important epigenetic regulators, miRNAs can interact with the targeting genes through specific base-pairing with the key domain of mature miRNAs, particularly at bases 2–8 of the 5′ end known as the “seed” region. The mRNA is cleaved under the condition of perfect base-pairing between miRNAs and miRNA regulatory elements (MREs) in the 3′ UTR of targeting mRNAs, whereas imperfect interaction between the miRNA and MREs leads to translational repression.32 miRNAs have been suggested to modulate more than 60% of human protein coding genes and participate in many cell processes, including development, virus defense, hematopoiesis, organ formation, apoptosis, and cell proliferation. Accumulating evidence has indicated that miRNAs play critical roles in regulating male germ cell development and are essential for epigenetic regulation of the mitosis, meiosis, and spermiogenesis.37, 39 Conditional knockout of dicer in either primordial germ cells or undifferentiated spermatogonia results in infertility.41 It has been reported that miR-21 promotes the self-renewal of Thy1+ mouse SSCs by the regulation of transcription factor EVT5.38 We have also demonstrated that miR-20 and miR-106a play essential roles in regulating the proliferation and maintenance of mouse SSCs via targeting Stat3.73 Other miRNAs involved in spermatogenesis and spermiogenesis have been identified. Nevertheless, it remains largely unclear about the function and targets of miRNAs in human Sertoli cells. We have previously compared the miRNA profiles between OA and SCOS Sertoli cells using miRNA microarrays, and among the 174 distinctly expressed miRNAs, miR-202-3p is one of the most significantly upregulated miRNAs in SCOS Sertoli cells compared with OA patients.42
miR-202-3p is highly conserved across animal species, and it belongs to a member of the let-7 family. Consistent with the role of let-7 family members, miR-202-3p has been reported to function as a novel tumor suppressor, inducing apoptosis and inhibiting the proliferation and invasion of various tumor cells, such as gastric cancer, neuroblastoma, lung cancer, and colorectal cancer. However, the function of miR-202-3p in male Sertoli cells has not been elucidated. Our study revealed that miR-202-3p is upregulated in SCOS Sertoli cells compared with OA patients with normal spermatogenesis, indicating a potential function of miR-202-3p in NOA progression. We found that ectopic expression of miR-202-3p suppressed the proliferation and synthesis function of human Sertoli cells, whereas inhibition of miR-202-3p promoted Sertoli cell proliferation and synthesis. Moreover, Sertoli cell apoptosis increased by miR-202-3p overexpression and reduced by miR-202-3p inhibition.
Using bioinformatics tools, we predicted that LRP6 and Cyclin D1 of the Wnt/β-catenin signaling pathway were direct binding targets of miR-202-3p in human Sertoli cells. The prediction was validated using a series of assays, including dual-luciferase assay. Because Cyclin D1 was a downstream factor, and its function was not exclusive to the Wnt/β-catenin signaling pathway, we mainly focused on the functions of LRP6. As an indispensable coreceptor for the Wnt signaling pathway, LRP6 has been reported to function as an onco-protein. Overexpression of LRP6 contributes to the hyperactivation of the Wnt/β-catenin signaling pathway, which could promote the progression and invasion of various tumors, including hepatocellular carcinomas, colorectal cancer, prostate cancer, breast cancer, and pancreatic cancer.59, 74, 75 Aberrant function of LRP6 is also related to many other diseases, such as autosomal-dominant oligodontia, Alzheimer’s disease, atherosclerosis, and diabetic retinopathy. In the male reproductive system, Wnt/β-catenin signaling plays important roles in the differentiation of Müllerian duct, organization of testicular cords, development of primordial germ cells, and proliferation and differentiation of spermatogonia.61 With regard to Sertoli cells, Boyer et al.76, 77 have reported that expression of a constitutively activated form of β-catenin in post-natal Sertoli cells causes male infertility via progressive deterioration of seminiferous tubules, germ cell loss, and testicular atrophy. The Wnt/β-catenin signaling pathway stimulates the proliferation of adult human Sertoli cells via upregulation of c-Myc expression.78 It has been shown that testicular Sertoli cell tumor was formed by overactive Wnt/β-catenin signaling in mice.79 In this study, we found that overexpression of miR-202-3p significantly inhibited the activation of the LRP6-mediated Wnt/β-catenin signaling pathway. We also demonstrated that the expression levels of LRP6 and Cyclin D1 were downregulated in SCOS Sertoli cells compared with OA patients with normal spermatogenesis. LRP6 knockdown inactivated the Wnt/β-catenin signaling pathway, increased apoptosis, and suppressed the proliferation and synthesis function of Sertoli cells. Furthermore, we found that LRP6 could mediate the functions of miR-202-3p. LRP6 knockdown significantly attenuated the effects exerted on Sertoli cells that were induced by miR-202-3p inhibition. However, as we all know, various signaling pathways interact with each other to regulate cellular activities. For example, Akt can promote Wnt/β-catenin signaling by inactivating GSK3β in different cells. Cyclin D1 is also involved in many other signaling pathways, such as mitogen-activated protein kinase (MAPK) and JAK-STAT signaling pathways. Whether they are related to miR-202-3p and regulation of Sertoli cell functions still need further study.
In summary, we have revealed that miR-202-3p is upregulated in SCOS Sertoli cells compared with OA patients with normal spermatogenesis. We have demonstrated that miR-202-3p induces the apoptosis and inhibits the proliferation and synthesis function of Sertoli cells by targeting both LRP6 and Cyclin D1 of the Wnt/β-catenin signaling pathway. Because Sertoli cells play key roles in regulating spermatogenesis and they have significant applications in regenerative medicine, this study could offer new insights into the etiology for azoospermia and might offer new targets for treating male reproductive disorders and other human diseases.
Materials and Methods
Ethics Statement
The work was approved by the Institutional Review Board of Shanghai General Hospital (license number of ethics statement: 2016KY196). All experimental protocols were performed in accordance with relevant guidelines and regulation of the Institutional Review Board of Shanghai General Hospital. Written informed consents for testicular biopsies were obtained from the donors for the research only.
Acquisition of Testicular Tissues from OA and SCOS Patients
Testicular tissues were obtained from 20 OA patients and 20 SCOS patients who underwent micro-dissection testicular sperm extraction from June 2016 to August 2017 at Shanghai General Hospital, affiliated to Shanghai Jiao Tong University. All OA patients were caused by the vasoligation or inflammation, and normal spermatogenesis was observed in these patients. The diagnosis of SCOS was dependent on histological analysis, which showed that only Sertoli cells were present within the seminiferous tubules.
Isolation, Identification, and Culture of Human Sertoli Cells from OA and SCOS Patients
Testicular tissues from OA and SCOS patients were washed three times with aseptic DMEM Nutrient Mixture F-12 (DMEM/F-12) (GIBCO, Grand Island, NY, USA) with 2% antibiotics containing penicillin and streptomycin (GIBCO). Two-step enzymatic digestion and differential plating was utilized to isolate Sertoli cells from human testicular tissues. The testicular tissues were cut into pieces and incubated with 10 mL of DMEM containing 2 mg/mL type IV collagenase (GIBCO) and 10 μg/mL DNase I (Sigma, St. Louis, MO, USA) in water bath at 34°C for 15 min. Seminiferous tubules were washed extensively with DMEM to remove Leydig cells and peritubular myoid cells. Sertoli cells and human male germ cells were obtained from seminiferous tubules using a second enzymatic digestion comprising 4 mg/mL collagenase IV, 2.5 mg/mL hyaluronidase (Sigma), 2 mg/mL trypsin (Sigma), and 10 μg/mL DNase I in water bath at 34°C for 10–15 min. The suspension was centrifuged at 300 × g for 5 min, and the supernatant was removed. The precipitant was resuspended in DMEM/F-12 with 10% fetal bovine serum (FBS) (GIBCO) and then filtered by a 40-μm cell strainer and seeded into a 10-cm Matrigel (BD Biosciences)-coated dish. The cells were incubated in DMEM/F-12 (GIBCO) supplemented with 10% FBS at 34°C in 5% CO2 for 1 day. The suspending male germ cells and other types of cells were removed, and Sertoli cells were attached to culture dishes. The viability of freshly isolated human Sertoli cells was assessed by the trypan blue staining. Freshly isolated human Sertoli cells were identified by RT-PCR and immunostaining with antibodies against GATA4, WT1, SOX9, GDNF, SCF, VIM, and OCLN. Immunostaining with antibodies against α-SMA, CYP11A1, and VASA was also conducted to rule out the possibility of contamination by peritubular myoid cells, Leydig cells, and germ cells.
Trypan Blue Staining
The viability of freshly isolated human Sertoli cells and the cells after 6 hr of LRP6 siRNA transfection was assessed by trypan blue staining. The cells were incubated with 0.4% trypan blue, and viability of these cells was calculated by percentage of the cells excluding trypan blue staining.
Immunocytochemistry
For immunocytochemistry staining, freshly isolated and cultured human Sertoli cells were fixed with 4% paraformaldehyde (PFA) for 30 min, washed three times with cold PBS (Medicago, Uppsala, Sweden), and permeabilized with 0.4% Triton X-100 (Sigma) for 5 min. After extensive wash with PBS, the cells were blocked in 5% bovine serum albumin (BSA) (Sigma) for an hour at room temperature. The cells were then incubated with primary antibodies overnight at 4°C, including GATA4 (1:200; Santa Cruz, Dallas, TX, USA), WT1 (1:200; Santa Cruz, Dallas, TX, USA), SOX9 (1:500; Millipore, Bedford, MA, USA), GDNF (1:300; Santa Cruz, Dallas, TX, USA), SCF (1:300; Santa Cruz, Dallas, TX, USA), VIM (1:100; Cell Signaling Technology [CST], Danvers, MA, USA), OCLN (1:200; Abcam, Cambridge, UK), α-SMA (1:200; Abcam, Cambridge, UK), CYP11A1 (1:200; Abcam, Cambridge, UK), VASA (1:100; Santa Cruz, Dallas, TX, USA), and ki-67 (1:200; Santa Cruz, Dallas, TX, USA). After extensive washes with PBS, the cells were incubated with the secondary antibody, namely immunoglobulin G (IgG) conjugated with fluorescein isothiocyanate (FITC) (Sigma) or rhodamine-conjugated IgG (Sigma), at a 1:200 dilution for 1 hr at room temperature. DAPI was used to label the nuclei. Replacement of primary antibodies with PBS was used as a negative control, and images were captured with a Nikon microscope.
RNA Extraction, RT-PCR, and Real-Time qPCR
Total RNA was extracted from human Sertoli cells using TRIzol (Takara, Kusatsu, Japan), and the quality and concentrations of total RNA were measured by NanoDrop (Thermo Scientific). The ratio of A260/A280 of total RNA was set as 1.9–2.0 to ensure quality. Reverse transcription (RT) of total RNA was conducted using the First Strand cDNA Synthesis Kit (Thermo Scientific, USA), and PCR of the cDNA was carried out according to the protocol as described previously.80 The primer sequences of chosen genes were designed and listed in Table S1. The PCR started at 94°C for 2 min and was performed in terms of the following conditions: denaturation at 94°C for 30 s, annealing at 55°C–60°C for 45 s, and elongation at 72°C for 45 s, for 35 cycles. The samples were incubated for an additional 5 min at 72°C. PCR with PBS but without cDNA served as a negative control. PCR products were separated by electrophoresis on 2% agarose gel and visualized with ethidium bromide. Images were recorded and band intensities were analyzed using chemiluminescence (Chemi-Doc XRS; Bio-Rad).
Real-time qPCR was performed using Power SYBR Green PCR Master Mix (Applied Biosystems, Woolston Warrington, UK) in a Veriti 96-Well Thermal Cycler (Applied Biosystems, Carlsbad, CA, USA). To quantify the PCR products, we used the comparative Ct (threshold cycle) method as described previously.67 The Ct values of genes were normalized against the threshold value of human housekeeping gene β-actin [ΔCt = Ct (target gene) − Ct (β-actin)], and the relative expression of target genes in the treatment group to the controls was calculated by formula 2−ΔΔCt [ΔΔCt = ΔCt(treatment) − ΔCt(control)]. The primers of detected genes were listed in Table S1.
Real-Time qPCR Analysis for miRNA
For miRNA real-time qPCR, RT reaction was performed using miScript II RT Kit (Qiagen, Germany). Each RT reaction was in a total volume of 20 μL, comprising 0.1 μg of total RNA, 4 μL of miScript HiSpec Buffer, 2 μL of Nucleics Mix, and 2 μL of miScript Reverse Transcriptase Mix. Reaction was performed in a Veriti 96-Well Thermal Cycler (Applied Biosystems, Carlsbad, CA, USA) for 60 min at 37°C and followed by heat inactivation of RT at 95°C for 5 min. RT reaction mix was diluted by five times with nuclease-free water and preserved at −20°C. Primer sequence of miR-202-3p used for real-time qPCR was 5′-AGAGGTATAGGGCATGGGAA-3′. Real-time qPCR was performed using 7500 Fast Real-Time PCR System (Applied Biosystems, Woolston Warrington, UK) with 25 μL of PCR mixture containing 2 μL of cDNA, 12.5 μL of QuantiTect SYBR Green PCR Master Mix (Qiagen, Germany), 2.5 μL of universal primer (Qiagen, German), 2.5 μL of miRNA-specific primer, and 5.5 μL of nuclease-free water. Reactions were incubated in a 96-well optical plate (Applied Biosystems, Woolston Warrington, UK) at 95°C for 10 min and followed by 40 cycles of 95°C for 10 s, 60°C for 30 s. Each sample was run in triplicate. In the end of the PCR cycles, melting curve analysis was performed to validate the specific generation of the expected PCR products. The expression levels of miRNA were normalized to U6 and calculated using the 2−ΔΔCt method described above.
Lentivirus Production and Transduction
The pGMLV-MA2, pGMLV-MA2-miR-202-3p, pGMLV-SC5, and pGMLV-SC5-miR-202-3p inhibitor plasmids were cotransfected into HEK293T cells along with the packaging plasmid pCMV-Δ8.9 and the envelope plasmid VSVG using Lipofectamine 3000 (Invitrogen). Forty-eight hours after cotransfection, virus particles were harvested and filtered by a 0.2-μm cell strainer. The titer of the lentivirus was 1 × 108 transducing units (TU)/mL. The particles were individually used to infect OA Sertoli cells with 6 mg/mL polybrene (Sigma). Culture medium was changed to DMEM/F-12 with 10% FBS 24 hr later, and EGFP fluorescence was observed under the fluorescent microscope after 96 hr to ensure successful infection. Once green fluorescence was seen, 2 μg/mL puromycin was added into the culture medium for screening. About 5 days later, only Sertoli cells that were infected successfully survived, which were cultured for further experiments.
Western Blots
Human Sertoli cells from OA and SCOS patients with or without infection or siRNA transfection were lysed with RIPA buffer (Biotech Well, Guangzhou, Shanghai, China) for 30 min on ice. Cell lysates were centrifugated at 12,000 × g for 20 min at 4°C, and the protein concentration was measured using BCA kit (Dingguo Changsheng Biotech, Beijing, China). Twenty micrograms of cell lysate from each sample was used for SDS-PAGE (Bio-Rad Laboratories), and western blots were performed according to the protocol as described previously.81 In brief, samples were resolved in the XCell Sure Lock Novex Mini-Cell apparatus (Invitrogen, Carlsbad, CA, USA) and transferred to nitrocellulose membranes for 1.5 hr on ice. The membranes were washed with TBS containing 0.1% Tween (TBST) (Dingguo Changsheng Biotech, Beijing, China) and blocked with 5% nonfat dry milk in TBST for 1 hr at room temperature. After extensive wash with TBST, the membranes were incubated with the chosen primary antibodies overnight at 4°C, including PCNA (1:200; Santa Cruz, Dallas, TX, USA), Cyclin A2 (1:2,000; CST, Danvers, MA, USA), Cyclin B1 (1:1,000; CST, Danvers, MA, USA), Cyclin D1 (1:1,000; CST, Danvers, MA, USA), Cyclin E1 (1:1,000; CST, Danvers, MA, USA), β-actin (1:5,000; Proteintech, Chicago, MI, USA), cPARP (1:1,000; CST, Danvers, MA, USA), Bcl2 (1:500; Santa Cruz, Dallas, TX, USA), Bax (1:500; Santa Cruz, Dallas, TX, USA), GDNF (1:300; Santa Cruz, Dallas, TX, USA), SCF (1:300; Santa Cruz, Dallas, TX, USA), BMP4 (1:1,000, CST, Danvers, MA, USA), FGF2 (1:1,000; CST, Danvers, MA, USA), CXCL12 (1:1,000; CST, Danvers, MA, USA), LRP6 (1:1,000; CST, Danvers, MA, USA), c-Myc (1:1,000; CST, Danvers, MA, USA), phospho-β-catenin (1:1,000; CST, Danvers, MA, USA), β-catenin (1:300; Santa Cruz, Dallas, TX, USA), and non-phospho-β-catenin (1:1,000; CST, Danvers, MA, USA). The next day, the membranes were washed three times with TBST and then incubated with horseradish peroxidase-conjugated anti-rabbit IgG or anti-mouse IgG (Santa Cruz Biotechnology, CA, USA) at a 1:2,000 dilution for 1 hr at room temperature. The blots were detected by chemiluminescence (Chemi-Doc XRS; Bio-Rad, Hercules, CA, USA) after extensive washes with TBST. Optical density analyses were processed with Adobe Photoshop CC. The relative levels of proteins were normalized to the expression of β-actin.
Cell Proliferation Assay
Human Sertoli cells from the stable cell strains were seeded at a density of 1,000 cells/well in 96-well microtiter plates in DMEM/F-12 supplemented with 10% FBS overnight. The medium was changed every day. The proliferation potential of human Sertoli cells was detected by CCK-8 assay (Dojin Laboratories, Kumamoto, Japan) according to the manufacturer’s instructions. Proliferation of human SSCs and Sertoli cells treated with or without LRP6 siRNA transfection was detected the same way.
For EDU incorporation assay, human Sertoli cells from the stable cell strains were seeded in 96-well plates with 1,000 cells/well in DMEM/F-12 supplemented with 10% FBS overnight to allow the cells to attach. Then, 20 μΜ EDU (RiboBio, Guangzhou, China) was added to the medium and incubated for 16 hr. Afterward, the cells were washed twice with PBS and fixed with 4% PFA at room temperature, and 50 μL of 2 mg/mL glycine (DingGuoChangSheng Biotech) was added to each well to neutralize the PFA. The cells were washed with 0.5% Tritonx-100 in PBS and exposed to 100 μL of Apollo-Fluor (RiboBio, Guangzhou, China) for 30 min in the dark at room temperature. Cell nuclei were stained with Hoechst 33342 for 30 min. The percentage of EDU-positive cells was counted from 500 cells, and three independent experiments were performed. Proliferation of Sertoli cells treated with or without LRP6 siRNA transfection was detected by the same method.
ki-67 immunofluorescence and western blot of PCNA were also performed to show the proliferation of Sertoli cells and human SSCs, which was described in detail above.
Annexin-V and PI Staining and Flow Cytometry
Human Sertoli cells from the stable cell strains were seeded at a density of 1 × 105 cells/well in six-well plates in DMEM/F-12 supplemented with 10% FBS overnight. Forty-eight hours later, cells were harvested and washed with cold PBS twice, and apoptosis percentages of human Sertoli cells were detected using the Annexin V-FITC/PI kit by flow cytometry according to the manufacturer’s instructions (BioLegend, London, UK). It was feasible to identify and quantify apoptotic cells by conjugating FITC to Annexin V using flow cytometry. Staining cells simultaneously with Annexin V-FITC (green fluorescence) and the non-vital dye PI (red fluorescence) allowed the discrimination of intact cells (FITC−PI−) and early apoptotic (FITC+PI−) and late apoptotic or necrotic cells (FITC+PI+). Apoptosis of Sertoli cells treated with or without LRP6 siRNA transfection was detected by the same method.
TUNEL Assay
Further analysis of apoptotic cells was conducted by TUNEL Apoptosis Detection Kit (Yeasen, Shanghai, China). Human Sertoli cells were treated according to the methods mentioned above. The cells were fixed with 4% PFA for 25 min at 4°C. After several washes, the cells were incubated with proteinase K (20 mg/mL) and 1× DNase I buffer for 5 min at room temperature. Cells were then treated with 10 U/mL DNase I for 10 min at room temperature and followed by washes in deionized water. These cells were incubated with 1× equilibration buffer for 30 min at room temperature and labeled by Alexa Fluor in buffer premixed with terminal-deoxynucleoitidyl transferase (TdT) enzyme for 60 min at 37°C. After being washed with PBS, the cells were finally stained with DAPI and analyzed under a fluorescence microscope (Nikon, Tokyo, Japan).
Dual-Luciferase Assay
HEK293T cells and Sertoli cells were transfected with 500 ng of psiCHECK-2 vectors in which wild-type or mutant form of 3′ UTR of LRP6 or Cyclin D1 was cloned (contained firefly luciferase as the reporter gene controlled by SV40 promoter gene and Renilla luciferase as the tracking gene controlled by CMV promoter) with 2 μg of miR-202-3p normal control or overexpression plasmids in reduced serum Opti-MEM, using 3.75 μL of Lipofectamine 3000 and 5 μL of P3000. Cell extracts were prepared 24 hr after transfection, and luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega, USA) pursuant to the manufacturer’s protocol. Firefly luciferase activity was used for normalization and an internal control for transfection efficiency.
RNAi of LRP6
The siRNA sequences targeting LRP6 mRNA were purchased from GenePharma (Suzhou, China), and the sequences were listed in Table S3. To optimize the effect of LRP6 RNAi, we constructed three siRNAs targeting different regions of LRP6. Human Sertoli cells were seeded at 2 × 105/cm2 density and cultured in DMEM/F-12 supplemented with 10% FBS overnight. The medium was changed to DMEM/F-12 supplemented with 1% FBS. FAM-labeled control siRNA and LRP6 siRNAs were transfected using Lipofectamine 3000 according to the manufacturer’s manual. The viability of the cells after 6 hr of transfection was evaluated by trypan blue staining. The transfection efficiency was detected using a Nikon fluorescence microscope. After 48 or 72 hr, the cells were harvested to examine the expression changes of various genes and proteins accordingly.
Statistical Analysis
All data were presented as mean ± SEM. The data obtained in experiments with multiple treatments were subjected to one-way ANOVA followed by Newman-Keuls test of significance using GraphPad Prism (version 5, GraphPad Software). Student’s t test was employed to study statistical significance in experiments with only two treatments, and p < 0.05 was considered statistically significant.
Author Contributions
Conception and Design: C. Yang, C. Yao, Z.L.; Acquisition of Data: C. Yang, C. Yao, L.Z.; Analysis and Interpretation of Data: C. Yang, C. Yao, Z.Z.; Writing – Review and/or Revision of the Manuscript: C. Yang, C. Yao, Z.L., Z.H.; Administrative, Technical, or Material Support: P.L., R.T., H.C., E.Z., Y.H., Y.G., Y.X., H.W., Q.Y.; Study Supervision: Z.L., Z.H.
Conflicts of Interest
The authors declare no conflict of interest.
Acknowledgments
This study was supported by grants from the National Key Research and Development Program (2017YFC1002003), the National Nature Science Foundation of China (81671512, 31671550, and 8170060493), and the Ministry of Science and Technology of the People's Republic of China (grants 2016YFC1000606 and 2014CB943101).
Footnotes
Supplemental Information includes three figures and three tables and can be found with this article online at https://doi.org/10.1016/j.omtn.2018.10.012.
Contributor Information
Zuping He, Email: zupinghe@sjtu.edu.cn.
Zheng Li, Email: lizhengboshi@163.com.
Supplemental Information
References
- 1.De Kretser D.M., Baker H.W. Infertility in men: recent advances and continuing controversies. J. Clin. Endocrinol. Metab. 1999;84:3443–3450. doi: 10.1210/jcem.84.10.6101. [DOI] [PubMed] [Google Scholar]
- 2.Matsumiya K., Namiki M., Takahara S., Kondoh N., Takada S., Kiyohara H., Okuyama A. Clinical study of azoospermia. Int. J. Androl. 1994;17:140–142. doi: 10.1111/j.1365-2605.1994.tb01233.x. [DOI] [PubMed] [Google Scholar]
- 3.Ezeh U.I. Beyond the clinical classification of azoospermia: opinion. Hum. Reprod. 2000;15:2356–2359. doi: 10.1093/humrep/15.11.2356. [DOI] [PubMed] [Google Scholar]
- 4.Hu Z., Li Z., Yu J., Tong C., Lin Y., Guo X., Lu F., Dong J., Xia Y., Wen Y. Association analysis identifies new risk loci for non-obstructive azoospermia in Chinese men. Nat. Commun. 2014;5:3857. doi: 10.1038/ncomms4857. [DOI] [PubMed] [Google Scholar]
- 5.Raman J.D., Schlegel P.N. Testicular sperm extraction with intracytoplasmic sperm injection is successful for the treatment of nonobstructive azoospermia associated with cryptorchidism. J. Urol. 2003;170:1287–1290. doi: 10.1097/01.ju.0000080707.75753.ec. [DOI] [PubMed] [Google Scholar]
- 6.Dym M. Spermatogonial stem cells of the testis. Proc. Natl. Acad. Sci. USA. 1994;91:11287–11289. doi: 10.1073/pnas.91.24.11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feng L.X., Chen Y., Dettin L., Pera R.A., Herr J.C., Goldberg E., Dym M. Generation and in vitro differentiation of a spermatogonial cell line. Science. 2002;297:392–395. doi: 10.1126/science.1073162. [DOI] [PubMed] [Google Scholar]
- 8.Oatley J.M., Brinster R.L. The germline stem cell niche unit in mammalian testes. Physiol. Rev. 2012;92:577–595. doi: 10.1152/physrev.00025.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jan S.Z., Hamer G., Repping S., de Rooij D.G., van Pelt A.M., Vormer T.L. Molecular control of rodent spermatogenesis. Biochim. Biophys. Acta. 2012;1822:1838–1850. doi: 10.1016/j.bbadis.2012.02.008. [DOI] [PubMed] [Google Scholar]
- 10.Griswold M.D. The central role of Sertoli cells in spermatogenesis. Semin. Cell Dev. Biol. 1998;9:411–416. doi: 10.1006/scdb.1998.0203. [DOI] [PubMed] [Google Scholar]
- 11.Elftman H. Sertoli cells and testis structure. Am. J. Anat. 1963;113:25–33. doi: 10.1002/aja.1001130104. [DOI] [PubMed] [Google Scholar]
- 12.Hai Y., Hou J., Liu Y., Liu Y., Yang H., Li Z., He Z. The roles and regulation of Sertoli cells in fate determinations of spermatogonial stem cells and spermatogenesis. Semin. Cell Dev. Biol. 2014;29:66–75. doi: 10.1016/j.semcdb.2014.04.007. [DOI] [PubMed] [Google Scholar]
- 13.Meng X., Lindahl M., Hyvönen M.E., Parvinen M., de Rooij D.G., Hess M.W., Raatikainen-Ahokas A., Sainio K., Rauvala H., Lakso M. Regulation of cell fate decision of undifferentiated spermatogonia by GDNF. Science. 2000;287:1489–1493. doi: 10.1126/science.287.5457.1489. [DOI] [PubMed] [Google Scholar]
- 14.He Z., Jiang J., Kokkinaki M., Golestaneh N., Hofmann M.C., Dym M. Gdnf upregulates c-Fos transcription via the Ras/Erk1/2 pathway to promote mouse spermatogonial stem cell proliferation. Stem Cells. 2008;26:266–278. doi: 10.1634/stemcells.2007-0436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng L.X., Ravindranath N., Dym M. Stem cell factor/c-kit up-regulates cyclin D3 and promotes cell cycle progression via the phosphoinositide 3-kinase/p70 S6 kinase pathway in spermatogonia. J. Biol. Chem. 2000;275:25572–25576. doi: 10.1074/jbc.M002218200. [DOI] [PubMed] [Google Scholar]
- 16.Ohta H., Yomogida K., Dohmae K., Nishimune Y. Regulation of proliferation and differentiation in spermatogonial stem cells: the role of c-kit and its ligand SCF. Development. 2000;127:2125–2131. doi: 10.1242/dev.127.10.2125. [DOI] [PubMed] [Google Scholar]
- 17.Blume-Jensen P., Jiang G., Hyman R., Lee K.F., O’Gorman S., Hunter T. Kit/stem cell factor receptor-induced activation of phosphatidylinositol 3′-kinase is essential for male fertility. Nat. Genet. 2000;24:157–162. doi: 10.1038/72814. [DOI] [PubMed] [Google Scholar]
- 18.Hu J., Chen Y.X., Wang D., Qi X., Li T.G., Hao J., Mishina Y., Garbers D.L., Zhao G.Q. Developmental expression and function of Bmp4 in spermatogenesis and in maintaining epididymal integrity. Dev. Biol. 2004;276:158–171. doi: 10.1016/j.ydbio.2004.08.034. [DOI] [PubMed] [Google Scholar]
- 19.Carlomagno G., van Bragt M.P., Korver C.M., Repping S., de Rooij D.G., van Pelt A.M. BMP4-induced differentiation of a rat spermatogonial stem cell line causes changes in its cell adhesion properties. Biol. Reprod. 2010;83:742–749. doi: 10.1095/biolreprod.110.085456. [DOI] [PubMed] [Google Scholar]
- 20.Lenhard T., Schober A., Suter-Crazzolara C., Unsicker K. Fibroblast growth factor-2 requires glial-cell-line-derived neurotrophic factor for exerting its neuroprotective actions on glutamate-lesioned hippocampal neurons. Mol. Cell. Neurosci. 2002;20:181–197. doi: 10.1006/mcne.2002.1134. [DOI] [PubMed] [Google Scholar]
- 21.Zhang Y., Wang S., Wang X., Liao S., Wu Y., Han C. Endogenously produced FGF2 is essential for the survival and proliferation of cultured mouse spermatogonial stem cells. Cell Res. 2012;22:773–776. doi: 10.1038/cr.2012.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yan Y.C., Sun Y.P., Zhang M.L. Testis epidermal growth factor and spermatogenesis. Arch. Androl. 1998;40:133–146. doi: 10.3109/01485019808987936. [DOI] [PubMed] [Google Scholar]
- 23.Kurokawa S., Kojima Y., Mizuno K., Nakane A., Hayashi Y., Kohri K. Effect of epidermal growth factor on spermatogenesis in the cryptorchid rat. J. Urol. 2005;174:2415–2419. doi: 10.1097/01.ju.0000180414.81767.68. [DOI] [PubMed] [Google Scholar]
- 24.Kanatsu-Shinohara M., Inoue K., Ogonuki N., Miki H., Yoshida S., Toyokuni S., Lee J., Ogura A., Shinohara T. Leukemia inhibitory factor enhances formation of germ cell colonies in neonatal mouse testis culture. Biol. Reprod. 2007;76:55–62. doi: 10.1095/biolreprod.106.055863. [DOI] [PubMed] [Google Scholar]
- 25.Huleihel M., Lunenfeld E. Regulation of spermatogenesis by paracrine/autocrine testicular factors. Asian J. Androl. 2004;6:259–268. [PubMed] [Google Scholar]
- 26.Wang S., Wang X., Wu Y., Han C. IGF-1R signaling is essential for the proliferation of cultured mouse spermatogonial stem cells by promoting the G2/M progression of the cell cycle. Stem Cells Dev. 2015;24:471–483. doi: 10.1089/scd.2014.0376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nakayama Y., Yamamoto T., Abé S.I. IGF-I, IGF-II and insulin promote differentiation of spermatogonia to primary spermatocytes in organ culture of newt testes. Int. J. Dev. Biol. 1999;43:343–347. [PubMed] [Google Scholar]
- 28.Yang Q.E., Kim D., Kaucher A., Oatley M.J., Oatley J.M. CXCL12-CXCR4 signaling is required for the maintenance of mouse spermatogonial stem cells. J. Cell Sci. 2013;126:1009–1020. doi: 10.1242/jcs.119826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yoon K.A., Chae Y.M., Cho J.Y. FGF2 stimulates SDF-1 expression through the Erm transcription factor in Sertoli cells. J. Cell. Physiol. 2009;220:245–256. doi: 10.1002/jcp.21759. [DOI] [PubMed] [Google Scholar]
- 30.Sheng C., Zheng Q., Wu J., Xu Z., Wang L., Li W., Zhang H., Zhao X.Y., Liu L., Wang Z. Direct reprogramming of Sertoli cells into multipotent neural stem cells by defined factors. Cell Res. 2012;22:208–218. doi: 10.1038/cr.2011.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang L., Chen M., Wen Q., Li Y., Wang Y., Wang Y., Qin Y., Cui X., Yang L., Huff V., Gao F. Reprogramming of Sertoli cells to fetal-like Leydig cells by Wt1 ablation. Proc. Natl. Acad. Sci. USA. 2015;112:4003–4008. doi: 10.1073/pnas.1422371112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 33.Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 34.Yao C., Liu Y., Sun M., Niu M., Yuan Q., Hai Y., Guo Y., Chen Z., Hou J., Liu Y., He Z. MicroRNAs and DNA methylation as epigenetic regulators of mitosis, meiosis and spermiogenesis. Reproduction. 2015;150:R25–R34. doi: 10.1530/REP-14-0643. [DOI] [PubMed] [Google Scholar]
- 35.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 36.Zhou H., Hu H., Lai M. Non-coding RNAs and their epigenetic regulatory mechanisms. Biol. Cell. 2010;102:645–655. doi: 10.1042/BC20100029. [DOI] [PubMed] [Google Scholar]
- 37.Song R., Ro S., Michaels J.D., Park C., McCarrey J.R., Yan W. Many X-linked microRNAs escape meiotic sex chromosome inactivation. Nat. Genet. 2009;41:488–493. doi: 10.1038/ng.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Niu Z., Goodyear S.M., Rao S., Wu X., Tobias J.W., Avarbock M.R., Brinster R.L. MicroRNA-21 regulates the self-renewal of mouse spermatogonial stem cells. Proc. Natl. Acad. Sci. USA. 2011;108:12740–12745. doi: 10.1073/pnas.1109987108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hayashi K., Chuva de Sousa Lopes S.M., Kaneda M., Tang F., Hajkova P., Lao K., O’Carroll D., Das P.P., Tarakhovsky A., Miska E.A., Surani M.A. MicroRNA biogenesis is required for mouse primordial germ cell development and spermatogenesis. PLoS ONE. 2008;3:e1738. doi: 10.1371/journal.pone.0001738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Papaioannou M.D., Pitetti J.L., Ro S., Park C., Aubry F., Schaad O., Vejnar C.E., Kühne F., Descombes P., Zdobnov E.M. Sertoli cell Dicer is essential for spermatogenesis in mice. Dev. Biol. 2009;326:250–259. doi: 10.1016/j.ydbio.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Papaioannou M.D., Lagarrigue M., Vejnar C.E., Rolland A.D., Kühne F., Aubry F., Schaad O., Fort A., Descombes P., Neerman-Arbez M. Loss of Dicer in Sertoli cells has a major impact on the testicular proteome of mice. Mol. Cell. Proteomics. 2011;10 doi: 10.1074/mcp.M900587-MCP200. M900587MCP900200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yao C., Sun M., Yuan Q., Niu M., Chen Z., Hou J., Wang H., Wen L., Liu Y., Li Z., He Z. MiRNA-133b promotes the proliferation of human Sertoli cells through targeting GLI3. Oncotarget. 2016;7:2201–2219. doi: 10.18632/oncotarget.6876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao Y., Li C., Wang M., Su L., Qu Y., Li J., Yu B., Yan M., Yu Y., Liu B., Zhu Z. Decrease of miR-202-3p expression, a novel tumor suppressor, in gastric cancer. PLoS ONE. 2013;8:e69756. doi: 10.1371/journal.pone.0069756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iorio M.V., Ferracin M., Liu C.G., Veronese A., Spizzo R., Sabbioni S., Magri E., Pedriali M., Fabbri M., Campiglio M. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 45.Zhang Y., Dai Y., Huang Y., Ma L., Yin Y., Tang M., Hu C. Microarray profile of micro-ribonucleic acid in tumor tissue from cervical squamous cell carcinoma without human papillomavirus. J. Obstet. Gynaecol. Res. 2009;35:842–849. doi: 10.1111/j.1447-0756.2009.01055.x. [DOI] [PubMed] [Google Scholar]
- 46.Yi Y., Li H., Lv Q., Wu K., Zhang W., Zhang J., Zhu D., Liu Q., Zhang W. miR-202 inhibits the progression of human cervical cancer through inhibition of cyclin D1. Oncotarget. 2016;7:72067–72075. doi: 10.18632/oncotarget.12499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Q., Huang Z., Guo W., Ni S., Xiao X., Wang L., Huang D., Tan C., Xu Q., Zha R. microRNA-202-3p inhibits cell proliferation by targeting ADP-ribosylation factor-like 5A in human colorectal carcinoma. Clin. Cancer Res. 2014;20:1146–1157. doi: 10.1158/1078-0432.CCR-13-1023. [DOI] [PubMed] [Google Scholar]
- 48.Ma G., Zhang F., Dong X., Wang X., Ren Y. Low expression of microRNA-202 is associated with the metastasis of esophageal squamous cell carcinoma. Exp. Ther. Med. 2016;11:951–956. doi: 10.3892/etm.2016.3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meng X., Chen X., Lu P., Ma W., Yue D., Song L., Fan Q. MicroRNA-202 inhibits tumor progression by targeting LAMA1 in esophageal squamous cell carcinoma. Biochem. Biophys. Res. Commun. 2016;473:821–827. doi: 10.1016/j.bbrc.2016.03.130. [DOI] [PubMed] [Google Scholar]
- 50.Li Y.G., He J.H., Yu L., Hang Z.P., Li W., Shun W.H., Huang G.X. microRNA-202 suppresses MYCN expression under the control of E2F1 in the neuroblastoma cell line LAN-5. Mol. Med. Rep. 2014;9:541–546. doi: 10.3892/mmr.2013.1845. [DOI] [PubMed] [Google Scholar]
- 51.Chen J., Cai T., Zheng C., Lin X., Wang G., Liao S., Wang X., Gan H., Zhang D., Hu X. MicroRNA-202 maintains spermatogonial stem cells by inhibiting cell cycle regulators and RNA binding proteins. Nucleic Acids Res. 2017;45:4142–4157. doi: 10.1093/nar/gkw1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bhanot P., Brink M., Samos C.H., Hsieh J.C., Wang Y., Macke J.P., Andrew D., Nathans J., Nusse R. A new member of the frizzled family from Drosophila functions as a Wingless receptor. Nature. 1996;382:225–230. doi: 10.1038/382225a0. [DOI] [PubMed] [Google Scholar]
- 53.Noordermeer J., Klingensmith J., Perrimon N., Nusse R. dishevelled and armadillo act in the wingless signalling pathway in Drosophila. Nature. 1994;367:80–83. doi: 10.1038/367080a0. [DOI] [PubMed] [Google Scholar]
- 54.Clevers H. Wnt/beta-catenin signaling in development and disease. Cell. 2006;127:469–480. doi: 10.1016/j.cell.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 55.He X., Semenov M., Tamai K., Zeng X. LDL receptor-related proteins 5 and 6 in Wnt/beta-catenin signaling: arrows point the way. Development. 2004;131:1663–1677. doi: 10.1242/dev.01117. [DOI] [PubMed] [Google Scholar]
- 56.Wehrli M., Dougan S.T., Caldwell K., O’Keefe L., Schwartz S., Vaizel-Ohayon D., Schejter E., Tomlinson A., DiNardo S. arrow encodes an LDL-receptor-related protein essential for Wingless signalling. Nature. 2000;407:527–530. doi: 10.1038/35035110. [DOI] [PubMed] [Google Scholar]
- 57.Liu C.C., Tsai C.W., Deak F., Rogers J., Penuliar M., Sung Y.M., Maher J.N., Fu Y., Li X., Xu H. Deficiency in LRP6-mediated Wnt signaling contributes to synaptic abnormalities and amyloid pathology in Alzheimer’s disease. Neuron. 2014;84:63–77. doi: 10.1016/j.neuron.2014.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Massink M.P., Créton M.A., Spanevello F., Fennis W.M., Cune M.S., Savelberg S.M., Nijman I.J., Maurice M.M., van den Boogaard M.J., van Haaften G. Loss-of-function mutations in the WNT co-receptor LRP6 cause autosomal-dominant oligodontia. Am. J. Hum. Genet. 2015;97:621–626. doi: 10.1016/j.ajhg.2015.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lemieux E., Cagnol S., Beaudry K., Carrier J., Rivard N. Oncogenic KRAS signalling promotes the Wnt/β-catenin pathway through LRP6 in colorectal cancer. Oncogene. 2015;34:4914–4927. doi: 10.1038/onc.2014.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tanwar P.S., Kaneko-Tarui T., Zhang L., Rani P., Taketo M.M., Teixeira J. Constitutive WNT/beta-catenin signaling in murine Sertoli cells disrupts their differentiation and ability to support spermatogenesis. Biol. Reprod. 2010;82:422–432. doi: 10.1095/biolreprod.109.079335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Takase H.M., Nusse R. Paracrine Wnt/β-catenin signaling mediates proliferation of undifferentiated spermatogonia in the adult mouse testis. Proc. Natl. Acad. Sci. USA. 2016;113:E1489–E1497. doi: 10.1073/pnas.1601461113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lombardi A.P., Royer C., Pisolato R., Cavalcanti F.N., Lucas T.F., Lazari M.F., Porto C.S. Physiopathological aspects of the Wnt/β-catenin signaling pathway in the male reproductive system. Spermatogenesis. 2013;3:e23181. doi: 10.4161/spmg.23181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ma M., Yang S., Zhang Z., Li P., Gong Y., Liu L., Zhu Y., Tian R., Liu Y., Wang X. Sertoli cells from non-obstructive azoospermia and obstructive azoospermia patients show distinct morphology, Raman spectrum and biochemical phenotype. Hum. Reprod. 2013;28:1863–1873. doi: 10.1093/humrep/det068. [DOI] [PubMed] [Google Scholar]
- 64.Hou J., Niu M., Liu L., Zhu Z., Wang X., Sun M., Yuan Q., Yang S., Zeng W., Liu Y. Establishment and characterization of human germline stem cell line with unlimited proliferation potentials and no tumor formation. Sci. Rep. 2015;5:16922. doi: 10.1038/srep16922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Guo Y., Hai Y., Yao C., Chen Z., Hou J., Li Z., He Z. Long-term culture and significant expansion of human Sertoli cells whilst maintaining stable global phenotype and AKT and SMAD1/5 activation. Cell Commun. Signal. 2015;13:20. doi: 10.1186/s12964-015-0101-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Su H., Luo F., Bao J., Wu S., Zhang X., Zhang Y., Duo S., Wu Y. Long-term culture and analysis of cashmere goat Sertoli cells. In Vitro Cell. Dev. Biol. Anim. 2014;50:918–925. doi: 10.1007/s11626-013-9648-7. [DOI] [PubMed] [Google Scholar]
- 67.Hai Y., Sun M., Niu M., Yuan Q., Guo Y., Li Z., He Z. BMP4 promotes human Sertoli cell proliferation via Smad1/5 and ID2/3 pathway and its abnormality is associated with azoospermia. Discov. Med. 2015;19:311–325. [PubMed] [Google Scholar]
- 68.Wang H., Yuan Q., Sun M., Niu M., Wen L., Fu H., Zhou F., Chen Z., Yao C., Hou J. BMP6 regulates proliferation and apoptosis of human Sertoli cells via Smad2/3 and Cyclin D1 pathway and DACH1 and TFAP2A activation. Sci. Rep. 2017;7:45298. doi: 10.1038/srep45298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tian R.H., Yang S., Zhu Z.J., Wang J.L., Liu Y., Yao C., Ma M., Guo Y., Yuan Q., Hai Y. NODAL secreted by male germ cells regulates the proliferation and function of human Sertoli cells from obstructive azoospermia and nonobstructive azoospermia patients. Asian J. Androl. 2015;17:996–1005. doi: 10.4103/1008-682X.159722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lei B., Wan B., Peng J., Yang Y., Lv D., Zhou X., Shu F., Li F., Zhong L., Wu H., Mao X. PRPS2 expression correlates with Sertoli-cell only syndrome and inhibits the apoptosis of TM4 Sertoli cells. J. Urol. 2015;194:1491–1497. doi: 10.1016/j.juro.2015.04.116. [DOI] [PubMed] [Google Scholar]
- 71.Nicholls P.K., Harrison C.A., Rainczuk K.E., Wayne Vogl A., Stanton P.G. Retinoic acid promotes Sertoli cell differentiation and antagonises activin-induced proliferation. Mol. Cell. Endocrinol. 2013;377:33–43. doi: 10.1016/j.mce.2013.06.034. [DOI] [PubMed] [Google Scholar]
- 72.Nascimento A.R., Pimenta M.T., Lucas T.F., Porto C.S., Lazari M.F. Relaxin and Sertoli cell proliferation. Ital. J. Anat. Embryol. 2013;118(Suppl 1):26–28. [PubMed] [Google Scholar]
- 73.He Z., Jiang J., Kokkinaki M., Tang L., Zeng W., Gallicano I., Dobrinski I., Dym M. MiRNA-20 and mirna-106a regulate spermatogonial stem cell renewal at the post-transcriptional level via targeting STAT3 and Ccnd1. Stem Cells. 2013;31:2205–2217. doi: 10.1002/stem.1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lu W., Lin C., Li Y. Rottlerin induces Wnt co-receptor LRP6 degradation and suppresses both Wnt/β-catenin and mTORC1 signaling in prostate and breast cancer cells. Cell. Signal. 2014;26:1303–1309. doi: 10.1016/j.cellsig.2014.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Arensman M.D., Nguyen P., Kershaw K.M., Lay A.R., Ostertag-Hill C.A., Sherman M.H., Downes M., Liddle C., Evans R.M., Dawson D.W. Calcipotriol targets LRP6 to inhibit Wnt signaling in pancreatic cancer. Mol. Cancer Res. 2015;13:1509–1519. doi: 10.1158/1541-7786.MCR-15-0204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Boyer A., Hermo L., Paquet M., Robaire B., Boerboom D. Seminiferous tubule degeneration and infertility in mice with sustained activation of WNT/CTNNB1 signaling in sertoli cells. Biol. Reprod. 2008;79:475–485. doi: 10.1095/biolreprod.108.068627. [DOI] [PubMed] [Google Scholar]
- 77.Boyer A., Yeh J.R., Zhang X., Paquet M., Gaudin A., Nagano M.C., Boerboom D. CTNNB1 signaling in sertoli cells downregulates spermatogonial stem cell activity via WNT4. PLoS ONE. 2012;7:e29764. doi: 10.1371/journal.pone.0029764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li Y., Gao Q., Yin G., Ding X., Hao J. WNT/β-catenin-signaling pathway stimulates the proliferation of cultured adult human Sertoli cells via upregulation of C-myc expression. Reprod. Sci. 2012;19:1232–1240. doi: 10.1177/1933719112447126. [DOI] [PubMed] [Google Scholar]
- 79.Chang H., Guillou F., Taketo M.M., Behringer R.R. Overactive beta-catenin signaling causes testicular sertoli cell tumor development in the mouse. Biol. Reprod. 2009;81:842–849. doi: 10.1095/biolreprod.109.077446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yang S., Ping P., Ma M., Li P., Tian R., Yang H., Liu Y., Gong Y., Zhang Z., Li Z., He Z. Generation of haploid spermatids with fertilization and development capacity from human spermatogonial stem cells of cryptorchid patients. Stem Cell Reports. 2014;3:663–675. doi: 10.1016/j.stemcr.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhao L., Zhu Z., Yao C., Huang Y., Zhi E., Chen H., Tian R., Li P., Yuan Q., Xue Y. VEGFC/VEGFR3 signaling regulates mouse spermatogonial cell proliferation via the activation of AKT/MAPK and Cyclin D1 pathway and mediates the apoptosis by affecting Caspase 3/9 and Bcl-2. Cell Cycle. 2018;17:225–239. doi: 10.1080/15384101.2017.1407891. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.