Abstract
Research on adipobiology has recognized the browning process of white adipocytes as a potential therapeutic strategy for the treatment of obesity and related morbidities. Physical exercise stimulates the secretion of myokines, such as baminoisobutyric acid (BAIBA), which in turn promotes adaptive thermogenesis. White adipocyte conversion to brown cells involves dynamic changes in lipid droplet (LD) dimension and in the transcription of brown-specific marker genes. This study analyzes the effect of different doses of BAIBA and at different days of development on 3T3-L1 cells by evaluating morphological changes in LDs and the expression of browning gene markers. Results suggested that the highest concentration of BAIBA after 4 days of differentiation produced the most significant effects. The number of LDs per cell increased in comparison to control cells, whereas the surface area significantly decreased. Brown adipocyte markers were up-regulated, but the effect of treatment was lost at 10 days of differentiation.
The thermogenic program induced by BAIBA may reflect a rapid adaptation of adipose tissue to physical exercise. This connection stresses the beneficial impact of physical exercise on metabolic health.
Key words: Lipid droplets, 3T3-L1, adipocytes, BAIBA
Introduction
Regular exercise has been shown to have many health benefits, both physically and mentally. Indeed, it increases metabolic rate, helping in weight loss, improves whole-body glucose homeostasis and increases insulin sensitivity. Recently, endocrine factors called myokines, produced by skeletal muscles and released during the contraction of muscle fibers, have been detected and linked to the loss of fat mass through the activation of thermogenesis in adipose tissue.1,2 Thermogenesis is a physiological process typical of uncoupling protein 1 (UCP1)-expressing mature brown adipocytes and recruitable brown-like adipocytes, the so-called beige or brite adipocytes.3 In these cells, UCP1 uncouples the mitochondrial respiration from the production of adenosine triphosphate (ATP) and the energy produced by the catabolic pathways is released in form of heat.4,5 Brown and beige adipocytes are morphologically distinct from white ones for having multiple small lipid droplets (LDs) instead of a unique fat depot that occupies the vast majority of the cytoplasm,6 and for having a high number of mitochondria intimately associated to them.7 It was also demonstrated that the gene expression pattern of the three cell types is very different; despite beige adipocytes may be seen as “intermediate” between white and brown adipocytes, they present a unique gene expression pattern with highly specific molecular markers.8-11
This unique feature of UCP1-expressing cells has raised in recent years a great effort in brown adipobiology and obesity research, as the induction of browning by cold exposure, exercise and dietary/pharmacological factors may represent an effective way to counteract obesity and its related comorbidities.12 In fact, many studies performed on both in vitro and in vivo models, which have been recently reviewed,2 showed that the treatment with browninginducing factors increases energy expenditure by enhancing the expression of Ucp1 gene and other brown adipose tissue (BAT) markers. Canonical BAT gene markers include the elongation of very long chain fatty acids protein 3 (Elovl3), cell deathinducing DNA fragmentation factor A (DFFA)-like effector A (Cidea), PR domain-containing protein 16 (Prdm16) and peroxisome proliferator-activating receptor (PPAR) γ coactivator 1α (Ppargc1a). These last two genes are deeply involved in mitochondrial biogenesis and in early BAT differentiation. 8
The exercise-induced adaptive thermogenesis would not make sense in white adipose tissue (WAT), since exercise per se increases heat production.13 Therefore, myokines, such as irisin, myostatin, meteorin- like 1 (Metrnl) and b-aminoisobutyric acid (BAIBA), which are released during muscle contraction in physical activity, are claimed as browning-promoting factors in WAT.
BAIBA is a non-protein amino acid originating from the catabolism of thymine and valine that plays a critical role in browning and in insulin resistance.14,15 Recently, BAIBA has been shown to protect osteocytes against reactive oxygen species and related apoptosis.16 Despite the role of myokines in WAT browning has been extensively reviewed,2,13 the cellular pathways through which they exert their action are mostly unknown. Roberts and colleagues hypothesized a PPARα-mediated mechanism through which BAIBA positively affects lipid metabolism in both adipocytes and hepatocytes.15 However, the exact molecular pathway of browning, as well as the remodelling of LDs induced by BAIBA needs further investigation.
This paper aims to show the effects of BAIBA treatment on murine 3T3-L1 preadipocytes during their differentiation to mature adipocytes on morphological and numerical changes of LDs and on the expression of a selected gene pattern. The expression of a core set of brown fat-specific genes and mitochondrial biogenesis (Ucp1, Cidea, Elovl3, Ppargc1a, Prdm16 and Cyc1) and a beige marker (Tbx1) was analyzed. Markers of LD formation and dynamics such as perilipin 1 (Plin1) and cell death-inducing DFFA-like effector C (Cidec) were investigated. Finally, the expression of brain-derived neurotrophic factor (Bdnf), a neurotrophin, which has a role in browning on in vivo models and in regulating energy balance and insulin signaling, 17,18 was also assessed.
Materials and Methods
Chemicals and culture media
Dulbecco’s modified Eagle medium (DMEM) enriched with 4.5 g/L D-glucose, 110 mg/L sodium pyruvate and 862 mg/L L-alanyl-L-glutamine (GlutaMAX™), DMEM/F-12 (1:1) medium enriched with GlutaMAX™ and 15 mM 4-(2-hydroxyethyl)- 1-piperazineethanesulfonic acid (HEPES), fetal bovine serum (FBS), penicillin/ streptomycin solution and amphotericin B solution were purchased from Gibco by Life Technologies (Thermo Fisher Scientific Inc., Waltham, MA, USA). Rosiglitazone was purchased from Cayman Chemical (Ann Arbor, MI, USA). BAIBA was purchased in a racemic mixture from AdipoGen SA (AdipoGen Life Science Inc., San Diego, CA, USA). Dipyrromethene boron difluoride (BODIPY) 493/503 dye, TRIzol reagent, PureLink™ RNA Mini Kit and Platinum™ SYBR™ Green qPCR SuperMix-UDG kit for real time PCR were purchased from Invitrogen (Thermo Fisher Scientific Inc.). ImProm-II™ Reverse Transcription System was purchased from Promega (Madison, WI, USA). 4,6- diamidin-2-phenylindole (DAPI)-containing mounting medium was purchased from Abcam (Cambridge, MA, USA). All other chemicals used in the experiment and not listed above were purchased from Sigma- Aldrich (Darmstadt, Germany).
Cell culture and treatment
3T3-L1 cells were chosen in the present study for their extensive use in evaluating the effects of compounds or nutrients on adipogenesis and in the potential application of various compounds and nutrients in the treatment of obesity. Moreover, at the best of our knowledge, the browning effect of BAIBA on 3T3-L1 cell model was never investigated. The murine 3T3-L1 preadipocytes (ZenBio Inc., Durham, NC, USA) were grown in high glucose DMEM supplemented with 10% FBS, 1% amphotericin B solution and 1% penicillin/streptomycin solution, at 37 °C with 5% CO2 and 95% relative humidity. Differentiation was induced 48 h after cells reached full confluence with DMEM/F-12 medium supplemented with 10% FBS, 1% amphotericin B solution, 1% penicillin/streptomycin solution, 0.5 μg/mL human insulin, 5 μM dexamethasone, 0.5 mM 3-isobutyl-1-methylxanthine (IBMX), 1 nM triiodothyronine (T3) and 1 μM rosiglitazone. After 3 days, the differentiation medium was switched to a maintenance medium to which 0.5 μg/mL human insulin and 1 nM T3 were added. Maintenance medium was refreshed every 2 days. After the switch from differentiation to maintenance medium, cells were treated with 1, 3 and 5 μM BAIBA until analysis. Cells were analyzed during differentiation at day 4 (4d) and at day 8 (8d), at day 10 (10d); these time points correspond to 2, 6 and 8 days of BAIBA treatment, respectively. A negative vehicle control (CTRL) was established treating cells with sterile milliQ water.
Cell viability assay
Cell viability was determined by 3-(4,5- dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay. Medium was removed from cells plated on a 96-well plate and treated with different concentrations of BAIBA. Prior to incubation with 5 mg/mL MTT solution in Hank’s balanced salt solution (HBSS), cells were rinsed with phosphate buffer saline (PBS) 1x. Incubation with MTT solution was performed at 37°C for 4 h. Accumulated formazan was dissolved in dimethyl sulfoxide (DMSO) and incubated overnight at 37°C. The optical density was used as an indicator of cell viability and was measured at 550 nm.
BODIPY staining
After cell fixation in a 2% formalin solution diluted in PBS 1x at room temperature (RT) for 15 min, the slides were washed three times in PBS 1x and then incubated in a solution of BODIPY493/503 in PBS 1x to fluorescently label lipid droplets. The incubation was performed at RT in dark for 45 min. After the incubation, the slides were washed in PBS 1x three times and then mounted with mounting medium with DAPI. Images were acquired with the fluorescence microscope Axio Observer Z1 equipped with D-PLAN Neofluar objective lenses with N.A. 0.75 and Infinity Color Corrected System (ICS) and with AxioCam and Zen blue software (Carl Zeiss, Jena, Germany). The filters used for BODIPY were 470/525 nm, and 390/460 nm for DAPI.
Morphology of LDs
MRI_Lipid Droplets tool (http://dev.mri.cnrs.fr/projects/imagejmacros/wiki/LipidDroplets_Tool), a macro of ImageJ1.50b software (http://rsb.info.nih.gov/ij/), was used to measure LD area.19 The images were analyzed as already described.20 For each LD area, perimeter, maximum Feret diameter (MFD) and integrated optical density (IOD) were measured. MFD is defined as the maximum distance between the two parallel planes restricting the object perpendicular to that direction and is a measure of an object size along a specified direction. It is generally used in optical microcopy to measure the size of particles.21,22 The IOD reports the average intensity/density of each object and increases linearly with the triglycerides content in 3T3-L1 adipocytes.23
Total area occupied by LDs in each field at different days of differentiation was calculated by summing up the area of each LD. The count of cells in each field was used to calculate the area and the number of LDs per cell. The average area of a single LD was calculated by dividing the area of LDs/cells by the number of LDs/cells. The area (μm2) and number of LDs were measured and counted on 15 fields for each time point.
A nonparametric representation of the LDs size probability density function (kernel) was chosen to avoid making assumptions about the distribution of the data; the kernel size distribution was evaluated with a routine written in Matlab 7.1 (Mathworks, MA, USA).
Of note, 2D examination of the LDs, which are 3D objects, leads to an underestimation of the size of bigger LDs with respect to smaller LDs, increasing the probability to detect the first ones. Despite this approach could affect the absolute kernel density distribution of the LDs size, it is used here in order to obtain a relative comparison only between different populations.
RNA extraction and RT-PCR
After culture medium removal from 6- well plates, 1 mL/10 cm2 of TRIzol reagent was added in each well and repeatedly pipetted to induce a severe breakdown of cell structures. These samples were immediately processed with the PureLink™ RNA Mini Kit following the manufacturer’s instructions.
The concentration of the extracted total RNA was quantified using a spectrophotometer (NanoDrop 1000 Spectropho-tometer; ThermoScientific, Wilmington, DE, USA) and the purity of RNA samples ranged between 1.8 and 1.9. RNA integrity was evaluated through the observation of 18S and 28S ribosomal bands after electrophoresis on 1% agarose gel, in the presence of GelRed. Primer3 Input software was used to design primers.24 GenBank accession, primer sequences, product lengths and relative annealing temperatures for each gene are listed in Table 1, according to the HUGO Gene Nomenclature Committee.
Table 1.
Oligonucleotide primer sequences for RT-PCR.
| Gene | GenBank accession | Primer sets | Product length (bp) | Tm (°C) |
|---|---|---|---|---|
| Tbp | NM_013684.3 | Forward: 5’-CCAATGACTCCTATGACCCCTA-3’ Reverse: 5’-CAGCCAAGATTCACGGTAGAT-3’ |
104 | 58.5 |
| Gapdh | NM_008084 | Forward: 5’-AATGTGTCCGTCGTGGATCTGA-3’ Reverse: 5’-AGTGTAGCCCAAGATGCCCTTC-3’ |
117 | 60 |
| Ucp1 | NM_009463.3 | Forward: 5'-CTTTGCCTCACTCAGGATTGG-3’ Reverse: 5'-ACTGCCACACCTCCAGTCATT-3’ |
123 | 59.8 |
| Ppargc1a | NM_008904.2 | Forward: 5'-TATGGAGTGACATAGAGTGTGCT-3’ Reverse: 5'-CTGGGCAAAGAGGCTGGTC-3’ |
191 | 60.0 |
| Cidea | NM_007702.2 | Forward: 5-ATCACAACTGGCCTGGTTACG-3’ Reverse: 5’-TACTACCCGGTGTCCATTTCT-3’ |
136 | 58.9 |
| Cidec | NM_178373.4 | Forward: 5’-ACCTTCGACCTGTACAAGCT-3’ Reverse: 5’-GTGCAGGTCATAGGAAAGCG-3’ |
99 | 58.4 |
| Elovl3 | NM_007703.2 | Forward: 5'-TTCTCACGCGGGTTAAAAATGG-3’ Reverse: 5'-GGCCAACAACGATGAGCAAC-3’ |
139 | 58.9 |
| Cyc1 | NM_025567.3 | Forward: 5’-GGCATCAGAACCAGAGCATG-3’ Reverse: 5’-CTGACCACTTATGCCGCTTC-3’ |
110 | 59.4 |
| Plin1 | NM_175640.2 | Forward: 5'-TGGACCACCTGGAGGAAAAG-3’ Reverse: 5'-CTTCGAAGGCGGGTAGAGATG-3’ |
94 | 60.6 |
| Prdm16 | NM_027504.3 | Forward: 5'-CCACCAGCGAGGACTTCAC-3’ Reverse: 5'-GGAGGACTCTCGTAGCTCGAA-3’ |
107 | 61.4 |
| Ppara | NM_011144.6 | Forward: 5 -TCTGTCGGGATGTCACACAA-3 Reverse: 5 -CGGGCTTTGACCTTGTTCAT-3 |
191 | 57.3 |
| Bdnf | NM_007540.4 | Forward: 5’-ATTACCTGGATGCCGCAAAC-3’ Reverse: 5’-CCTTCCTTGGTGTAACCCAT-3’ |
241 | 58.5 |
| Tbx1 | NM_011532.2 | Forward: 5’-AGGCGGAAGGAAGTGGTATT-3’ Reverse: 5’-TACCAGTATCTACACCGCCC-3’ |
118 | 58.4 |
Tm, annealing temperature; Tbp; TATA box binding protein; Gapdh, glyceraldehyde-3-phosphate dehydrogenase; Ucp1, uncoupling protein 1; Ppargc1a, peroxisome proliferator-activated receptor coactivator 1 α; Cidea, cell death-inducing DFFA-like effector A; Cidec, cell death inducing DFFA like effector C; Elovl3, elongation of very long chain fatty acids protein 3; Cyc1, cytochrome C1; Plin1, perilipin 1; Prdm16, proline rich domain containing 16; Ppara, peroxisome proliferator-activated receptor α; Bdnf, brain derived neurotrophic factor; Tbx1, T-box 1.
Total RNA (500 ng) from each sample was reverse-transcribed and amplified using an MJ thermal cycler (PT-100; MJ Research, Inc., Waltham, MA, USA). For each gene, an aliquot of cDNA samples were pooled and standard curves with serial dilution of pool were used to optimize PCR conditions and to calculate the efficiency, fluorescence baseline and threshold. Real time PCRs were performed for each sample in triplicate form using Platinum® SYBR® Green qPCR SuperMix-UDG. cDNA concentrations and primers molarities were different for each gene and determined with standard curves analyses. PCR amplification was conduced applying 45 cycles (1 sec at 95.C, 30 sec at the specific annealing temperature, 30 sec at 72.C) in a 96-well spectrofluorometric thermal cycler (CFX, Biorad, Milan Italy). The melting curve analysis of amplification products was performed at the end of each PCR reaction to confirm that a single PCR product was detected.
The expression of target genes was normalized using the TATA box-binding protein (Tbp) mRNA levels or geometric means between Tbp and glyceraldehyde 3- phosphate dehydrogenase (Gapdh) genes and analyzed using DDCt method.25 For all the cell culture experiments, the results are generated from biological triplicates and represent similar results from at least three independent experiments.
Statistical analysis
Results are presented as relative values (means ± SEM). All experiments were performed at least three times. Two-way ANOVA was used for statistical analysis for MTT test by SPSS ver. 20.0 software. The measurements of the area of LDs/cell, number of LDs/cell and the area of a single LD were analyzed by SPSS ver. 20.0 software. The results, obtained from ten biological replicates, were compared using Kruskal- Wallis statistical test followed by pairwise comparisons using the Mann-Whitney approach.
Results
Cell viability
The percentage of cell viability after exposure of BAIBA at 4d, 8d and 10d was always around 100% (data not shown). No significant differences were observed for time point and for concentration x time point interaction.
Lipid droplet distribution and analysis
Figure 1 illustrated LDs formation in 3T3-L1 cells at 4d, 8d and 10d in presence of different BAIBA doses. Interestingly, at 4d with 3 and 5 mM of BAIBA, cells showed very small LDs (Figure 1, panels g and l, respectively) in comparison to CTRL cells (Figure 1a).
Figure 1.
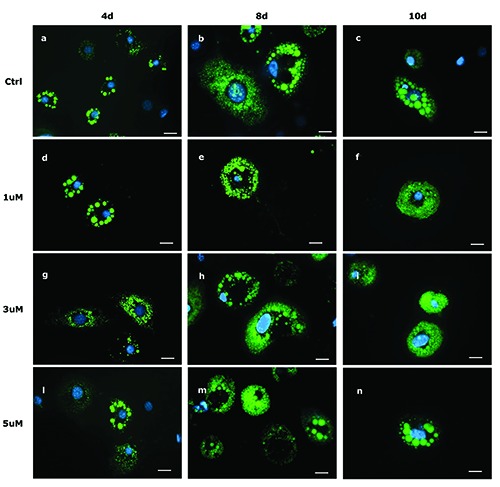
Lipid droplet pattern (stained with BODIPY 493/503) in fixed 3T3-L1 cells at 4d (a,d,g,l), 8d (b,e,h,m), and 10d (c,f,i,n). Control cells (a,b,c) and cells treated with 1 mM (d,e,f ), 3 mM (g,h,i) and 5 mM (l,m,n) of BAIBA are shown. Nuclear staining DAPI. Images are representative of n. 5 biological replicates. Scale bars: 20 mm.
Indeed, kernel densiy distribution suggested an high number of cells with a small Feret diameter when incubated with 5 mM of BAIBA after 4 and 10 days of differentiation (Figure 2a). The kernel distributions of MFD at 4d, 8d and 10d are shown in Figure 2a. For all the three time points, LD size distribution of the treated cells is similar to that of control cells. In particular, using the least squares method evaluated on 100 equally spaced Feret dimensions, the differences at 4d between control and the treatments (1 mM, 3 mM and 5 mM) are respectively 0.0045, 0.0047 and 0.0538; at 8d are 0.0106, 0.0075 and 0.0062; at 10d are 0.0367, 0.0807 and 0.0736. Figure 2b evidenced statistically significant differences of IOD, number of LDs/cell and area of a single LD/cell at 4d in presence of different BAIBA doses. To avoid any technical or biological variance due to cell number among slides, the total LD area was normalized to the number of cells.
Figure 2.
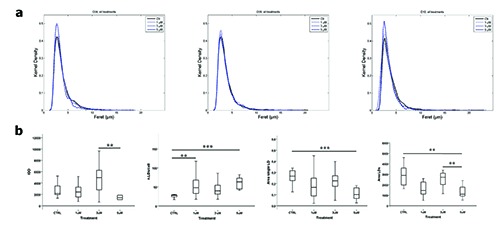
a) Kernel distribution of the Feret of the lipid droplets at 4, 8 and 10 days of differentiation in control and BAIBA-treated cells. b) Lipid droplet measurements on 3T3-L1 in presence of different BAIBA doses: Integrated optical density (IOD), number of LDs per cell, area of single LD after 4 days of differentiation and total area of LD and after 10 days of differentiation. Box plots show median (horizontal lines), first-to-third quartile (box), and the extreme values with the interquartile range (vertical lines). The area measures are expressed in mm2 and are the average of 10 fields of each concentration. ***P<0.001, **P<0.05.
The IOD significantly (P<0.05) decreased between 3 mM and 5 mM, the number of LDs/cell increased between CTRL and 1 mM (P<0.05) and between CTRL and 5 mM (P<0.01). The area of a single LD/cell was significantly different between CTRL and 5 mM (P<0.001). Figure 2b evidenced statistically significant differences in the area of LDs at 10d. In particular, the area was higher (P<0.05) in CTRL and 3 mM in comparison to 5 mM. No statistical differences were found at 8d.
The decrease in IOD, which is inversely related with the amount of triglycerides in LDs, the lowest area of a single LD and the increase of the number of LDs/cell observed with 5 mM BAIBA at 4d can account for a LD generation through the lipolytic release of fatty acids.
Transcription data analysis
Figure 3 indicated the relationship between the different concentrations of BAIBA and the significant differentially expressed genes at 4d and 8d, respectively. At 10d, gene expression did not display any significant variation. Only significant differentially expressed genes were showed. At 4d the expression of Tbx1 was significantly (P<0.05) different between 5 mM and lower doses. No significant differences were found with CTRL, maybe due to the high variability at the 5 mM dose. Ppargc1a (P<0.05) Prdm16, and Bdnf were up-regulated (P<0.002) at the same dose, while Ppara displayed significant differences for 1 mM and 5 mM in comparison to CTRL and 3 mM (Figure 3a). Cidea was significantly upregulated (P<0.05) at 5 mM, whereas Plin1 showed the significantly (P<0.05) lowest expression at 5 mM in comparison to CTRL (Figure 3b).The expression of Ppargc1a, Ucp1 and Ppara (P<0.001) was at its highest with 1 and 3 mM BAIBA doses in comparison to CTRL and 5 mM (Figure 3c).
Figure 3.
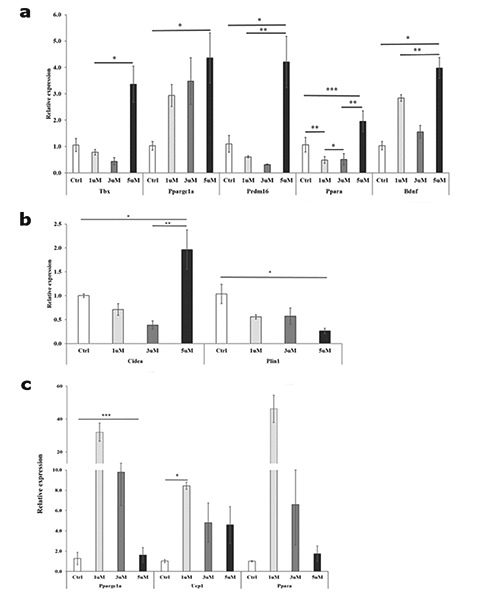
Differentially expressed genes between BAIBA doses after 4 and 8 days of differentiation. a) Genes involved in browning (three biological replicates) at 4d. Tbx1, T-box 1; Ppargc1a, peroxisome proliferator-activated receptor γ coactivator 1 α; Ppara, peroxisome proliferator-activated receptor α; Prdm16, proline rich domain containing 16; Bdnf, brain derived neurotrophic factor. b) Lipid droplets related genes at 4d. Cidea, cell death-inducing DFFA-like effector A; Plin1, perilipin 1. c) Genes involved in browning (three biological replicates) at 8d. Ucp1, uncoupling protein 1. Bars represent the mean plus/minus standard error of the mean (SEM). *P<0.05; **P<0.01; ***P<0.001.
Discussion
The present study reports the browning effect of BAIBA at different concentrations on 3T3-L1 cells at 4, 8 and 10 days of differentiation. It provides evidence showing the dynamics of LD morphology together with the expression of a panel of genes related to adaptive thermogenesis and LD formation. Although it is well established that this molecule, secreted by myocytes during exercise and circulating in blood, triggers the browning of WAT, no studies were conducted after its description by Roberts et al. in 2014.15 This study tested for the first time on 3T3-L1 cells known as reliable cellular model used in obesity researches, the browning effect of BAIBA by measuring variations in morphology of LDs that have recently attracted great interest as dynamic structures at the hub of lipid and energy metabolism.
It is largely known that brown adipocytes are occupied by multilocular LDs to facilitate the efficiency of lipases and subsequent uncoupling in response to cold or other stimuli.26 Recently, changes in LD dimension and in their associated proteins, such as CIDEC, CIDEA and PLIN1, have been demonstrated after the stimulation of browning.6 In the present study, statistical analysis evidenced that at 4 days of differentiation and at the highest BAIBA concentration, the number of LDs per cell increased in comparison to control cells, whereas the surface area of a single LD significantly decreased. The decrease of IOD observed at the same time and concentration suggested a smaller triglyceride accumulation compared to controls. Interestingly these results occurred at 4 days of differentiation, that correspond to the peak of lipogenesis.27 The results on LD morphology at 4d were corroborated by gene expression analysis. The browning adaptation included the up-regulation of Ppargc1a, Tbx1, Ucp1, Cidea, Elovl3 and Prdm16. In particular, 5 mM BAIBA at 4d influenced the expression of Tbx1 (Figure 3a), while the expression of Ucp1 was induced after 8 days of differentiation (Figure 3c). This is in agreement with the results obtained on human primary subcutaneous adipocytes treated with irisin, a myokine with dubious potential role in inducing browning of white adipose tissue. 28,29 Moreover, a significant increase of Cidea with 5 mM BAIBA treatment (Figure 3b), but not of Cidec, was observed, as reported in cells obtained by primary stromal vascular fraction of inguinal WAT of mice.15 Cidea is known as a brown adipocyte specific gene, related to multilocular phenotype,8 and it is induced in WAT following cold exposure or β-adrenergic signaling.6,30 In fact, the highest surface/volume ratio of smaller LDs increases the contact area of the LD with lipases and the subsequent facilitation of free fatty acids transport to mitochondria. CIDEA and CIDEC proteins are involved together in the fusion and enlargement of LDs, but while CIDEC is crucial in the formation of large LD in WAT, CIDEA alone is not sufficient.27 Recently, Nishimoto et al demonstrated that, in BAT, an isoform of CIDEC inhibits the homo dimerization of CIDEA and suppresses the formation of large LDs.31 As further evidence of browning effect induced by BAIBA at 4d, Plin1 gene was significantly down-regulated by all treatments in comparison to control (Figure 3b). In fact, perilipin 1 cooperates with CIDEC, but not with CIDEA, in inducing a super-sized, unilocular LD.32
After 4 days of differentiation, the expression of Ppargc1a and Prdm16 was also significantly increased by 5 mM BAIBA. This is in line with in vivo experiments that showed that exercise training has marked effects on mitochondrial gene expression and activity in subcutaneous WAT in mice and human.33 Ppargc1a controls mitochondrial biogenesis and heat production through induction of expression of Ucp1 and other respiratory factors.34 The higher expression level of this gene and Prdm16, a classic marker of brown fat, at 4d and later at 8d is coherent with the followed higher transcription of Ucp1 (Figure 3c).
PPARα, a nuclear receptor, stimulates genes involved in mitochondrial fatty acid oxidation and increases the expression of Ucp1 by directly interacting with Ppargc1a in primary murine brown adipocytes.35 Roberts et al. demonstrated that browning process induced by BAIBA involved a specific Ppara-dependent mechanism,15 without clarifying the upstream mechanism of action. Very recently, it has been shown that the loss of Ppara affected mature brown adipocytes by reducing the expression of brown markers in vitro but not in vivo, suggesting as well an important role of glycerol kinase in maintaining the activity of PPAR proteins.36 Moreover, it should be considered that PPARα regulates Cidea in mouse liver without altering the expression of Ucp1.37 In the present paper, the expression of Ppara is induced by 5 mM BAIBA at 4d and its expression was further increased at 8d with the lowest BAIBA concentration, as well as the expression of Ppargc1a and Ucp1 (Figure 3c). However, at 8d no significant variations in LD morphology and number were appreciated, as observed in PPARγ2-programmed cells treated with BAIBA.15 At 10d, BAIBA treatments did not significantly affect gene expression, although a decrease in the area of LDs was detected at the highest dose (Figure 3d).
BDNF is a neurotrophin expressed in brain, which regulates neuronal development and plasticity. However, it was also found in non-neuronal tissues, such as WAT and BAT in rodents and cows,38,39 where it could be involved in fat metabolism and in the activation of the sympathetic response.39,40 Moreover, mRNA and protein BDNF expression increases in human skeletal muscle after exercise, suggesting a role, as a myokine, on skeletal muscle metabolism,41 by enhancing local and peripheral fatty acid oxidation.42 The action of BAIBA at 4d on 3T3-L1 cells increased the Bdnf expression (Figure 3a), which in turn activates AMPK,43 thus facilitating the browning conversion.
This paper demonstrated that BAIBA promotes browning phenotype on 3T3-L1 cells during first 4-8 days of differentiation. In agreement with Roberts et al.15 the highest concentration during the early days of differentiation produced the strongest effect on cells in terms of LD dynamics and expression of beiging gene markers. After 8 days of differentiation, no changes were identified on LD morphology, although a browning effect was achieved as observed by brown gene marker expression. This browning effect appeared to be lost in cells after 10 days of differentiation.
The action of BAIBA significantly affects the early stage of adipocyte differentiation and can reflect a rapid adaptation of adipose tissue to physical exercise. This connection stresses the beneficial impact of physical exercise on metabolic health, providing more evidence of the preventive and therapeutic role of physical activity against obesity.
References
- 1.Bostrom P, Wu J, Jedrychowski MP, Korde A, Ye L, Lo JC, et al. A PGC-1α- dependent myokine that drives brownfat- like development of white fat and thermogenesis. Nature 2012;481:463-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Montanari T, Pošćić N, Colitti M. Factors involved in white-to-brown adipose tissue conversion and in thermogenesis: a review. Obes Rev 2017;18:495-513. [DOI] [PubMed] [Google Scholar]
- 3.Wu J, Bostrom P, Sparks LM, Ye L, Choi JH, Giang AH, et al. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell 2012;150:366-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shabalina IG, Petrovic N, de Jong JMA, Kalinovich AV, Cannon B, Nedergaard J. UCP1 in brite/beige adipose tissue mitochondria is functionally thermogenic. Cell Rep 2013;5:1196-203. [DOI] [PubMed] [Google Scholar]
- 5.Ricquier D. UCP1, the mitochondrial uncoupling protein of brown adipocyte: a personal contribution and a historical perspective. Biochimie 2017;134:3-8. [DOI] [PubMed] [Google Scholar]
- 6.Barneda D, Frontini A, Cinti S, Christian M. Dynamic changes in lipid droplet-associated proteins in the “browning” of white adipose tissues. Biochim Biophys Acta 2013;1831:924-33. [DOI] [PubMed] [Google Scholar]
- 7.Yu J, Zhang S, Cui L, Wang W, Na H, Xhu X, et al. Lipid droplet remodeling and interaction with mitochondria in mouse brown adipose tissue during cold treatment. Biochim Biophys Acta 2015;1853:918-28. [DOI] [PubMed] [Google Scholar]
- 8.Harms M, Seale P. Brown and beige fat: development, function and therapeutic potential. Nat Med 2013;19:1252-63. [DOI] [PubMed] [Google Scholar]
- 9.Rosenwald M, Wolfrum C. The origin and definition of brite versus white and classical brown adipocytes. Adipocyte 2014;3:4-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia RA, Roemmich JN, Claycombe KJ. Evaluation of markers of beige adipocytes in white adipose tissue of the mouse. Nutr Metab 2016;13:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ikeda K, Maretich P, Kajimura S. The common and distinct features of brown and beige adipocytes. Trends Endocrinol Metab 2018;29:191-200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ruiz JR, Martinez-Tellez B, Sanchez- Delgado G, Osuna-Prieto FJ, Rensen PCN, Boon MR. Role of human brown fat in obesity, metabolism and cardiovascular disease: strategies to turn up the heat. Prog Cardiovasc Dis 2018;61:232-45. [DOI] [PubMed] [Google Scholar]
- 13.Lehnig AC, Stanford KI. Exerciseinduced adaptations to white and brown adipose tissue. J Exp Biol 2018;221:jeb161570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Kuilenburg ABP, Stroomer AEM, van Lenthe H, Abeling NG, Van Gennip AH. New insights in dihydropyrimidine dehydrogenase deficiency: a pivotal role for beta-aminoisobutyric acid? Biochem J 2004;379:119-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roberts LD, Bostrom P, O’Sullivan JF, Schinzel RT, Lewis GD, Dejam A, et al. β-aminoisobutyric acid induces browning of white fat and hepatic β-oxidation and is inversely correlated with cardiometabolic risk factors. Cell Metab 2014;19:96-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kitase Y, Vallejo JA, Gutheil W, Vemula H, Jahn K, Yi J, et al. β-aminoisobutyric acid, l -BAIBA, is a muscle-derived osteocyte survival factor. Cell Rep 2018;22:1531-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cao L, Choi EY, Liu X, Martin A, Wang C, Xu X, et al. White to brown fat phenotypic switch induced by genetic and environmental activation of a hypothalamic- adipocyte axis. Cell Metab 2011;14:324-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamanaka M, Tsuchida A, Nakagawa T, Nonomura T, Ono-Kishino M, Sugaru E, et al. Brain-derived neurotrophic factor enhances glucose utilization in peripheral tissues of diabetic mice. Diabetes Obes Metab 2007;9:59-64. [DOI] [PubMed] [Google Scholar]
- 19.Backer V. ImageJ macro tool sets for biological image analysis. In: ImageJ user developer conference. 2012. [Google Scholar]
- 20.Montanari T, Colitti M. Simpson- Golabi-Behmel syndrome human adipocytes reveal a changing phenotype throughout differentiation. Histochem Cell Biol 2018;149:593-605. [DOI] [PubMed] [Google Scholar]
- 21.Rizzatti V, Boschi F, Pedrotti M, et al. Lipid droplets characterization in adipocyte differentiated 3T3-L1 cells: size and optical density distribution. Eur J Histochem 2013;57:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Costanzo M, Boschi F, Carton F, Conti G, Covi V, Tabaracci G, et al. Low ozone concentrations promote adipogenesis in human adipose-derived adult stem cells. Eur J Histochem 2018; 62:2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramirez-Zacarias JL, Castro-Munozledo F, Kuri-Harcuch W. Quantitation of adipose conversion and triglycerides by staining intracytoplasmic lipids with Oil red O. Histochemistry 1992;97:493-7. [DOI] [PubMed] [Google Scholar]
- 24.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 2000;132:365-86. [DOI] [PubMed] [Google Scholar]
- 25.Rao X, Huang X, Zhou Z, Lin X. An improvement of the 2ˆ(-delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostat Bioinforma Biomath 2013;3:71-85. [PMC free article] [PubMed] [Google Scholar]
- 26.Cinti S. The adipose organ at a glance. Dis Model Mech 2012;5:588-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barneda D, Christian M. Lipid droplet growth: regulation of a dynamic organelle. Curr Opin Cell Biol 2017;47:9-15. [DOI] [PubMed] [Google Scholar]
- 28.Kristof E, Doan-Xuan Q-M, Bai P, Bacso Z, Fesus L. Laser-scanning cytometry can quantify human adipocyte browning and proves effectiveness of irisin. Sci Rep 2015;5:12540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hofmann T, Elbelt U, Stengel A. Irisin as a muscle-derived hormone stimulating thermogenesis—a critical update. Peptides 2014;54:89-100. [DOI] [PubMed] [Google Scholar]
- 30.Rosell M, Kaforou M, Frontini A, Okolo A, Chan YW, Nikolopoulou E, et al. Brown and white adipose tissues: intrinsic differences in gene expression and response to cold exposure in mice. Am J Physiol Endocrinol Metab 2014;306:E945-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nishimoto Y, Nakajima S, Tateya S, Saito M, Ogawa W, Tamori Y. Cell death-inducing DNA fragmentation factor A-like effector A and fat-specific protein 27β coordinately control lipid droplet size in brown adipocytes. J Biol Chem 2017;292:10824-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grahn THM, Zhang Y, Lee M-J, Sommer AG, Mostoslavsky G, Fried SK, et al. FSP27 and PLIN1 interaction promotes the formation of large lipid droplets in human adipocytes. Biochem Biophys Res Commun 2013;432:296-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dewal RS, Stanford KI. Effects of exercise on brown and beige adipocytes. Biochim Biophys Acta Mol Cell Biol Lipids 2019;1864(1):71-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell 1999;98:115-24. [DOI] [PubMed] [Google Scholar]
- 35.Barbera MJ, Schluter A, Pedraza N, Iglesias R, Villarroya F, Giralt M. Peroxisome proliferator-activated receptor alpha activates transcription of the brown fat uncoupling protein-1 gene. A link between regulation of the thermogenic and lipid oxidation pathways in the brown fat cell. J Biol Chem 2001;276:1486-93. [DOI] [PubMed] [Google Scholar]
- 36.Lasar D, Rosenwald M, Kiehlmann E, Balaz M, Tall B, Opitz L, et al. Peroxisome proliferator activated receptor gamma controls mature brown adipocyte inducibility through glycerol kinase. Cell Rep 2018;22:760-73. [DOI] [PubMed] [Google Scholar]
- 37.Viswakarma N, Yu S, Naik S, Kashireddy P, Matsumoto K, Sarkar J, et al. Transcriptional regulation of Cidea, mitochondrial cell death-inducing DNA fragmentation factor alphalike effector A, in mouse liver by peroxisome proliferator-activated receptor alpha and gamma. J Biol Chem 2007;282:18613-24. [DOI] [PubMed] [Google Scholar]
- 38.Sornelli F, Fiore M, Chaldakov GN, Aloe L. Adipose tissue-derived nerve growth factor and brain-derived neurotrophic factor: results from experimental stress and diabetes. Gen Physiol Biophys 2009;28:179-83. [PubMed] [Google Scholar]
- 39.Colitti M, Loor JJ, Stefanon B. Expression of NGF, BDNF and their receptors in subcutaneous adipose tissue of lactating cows. Res Vet Sci 2015;102:196-9. [DOI] [PubMed] [Google Scholar]
- 40.Chaldakov GN, Fiore M, Tonchev AB, Aloe L. Neuroadipology: a novel component of neuroendocrinology. Cell Biol Int 2010;34:1051-3. [DOI] [PubMed] [Google Scholar]
- 41.Matthews VB, Astrom M-B, Chan MHS, Bruce CR, Krabbe KS, Prelovsek O, et al. Brain-derived neurotrophic factor is produced by skeletal muscle cells in response to contraction and enhances fat oxidation via activation of AMPactivated protein kinase. Diabetologia 2009;52:1409-18. [DOI] [PubMed] [Google Scholar]
- 42.Pedersen BK, Pedersen M, Krabbe KS, Bruunsgaard H, Matthews VB, Febbraio MA. Role of exercise-induced brain-derived neurotrophic factor production in the regulation of energy homeostasis in mammals. Exp Physiol 2009;94:1153-60. [DOI] [PubMed] [Google Scholar]
- 43.Pedersen BK, Febbraio MA. Muscles, exercise and obesity: skeletal muscle as a secretory organ. Nat Rev Endocrinol 2012;8:457-65. [DOI] [PubMed] [Google Scholar]


