Abstract
Background
Primary glioblastoma cell (GC) cultures have emerged as a key model in brain tumor research, with the potential to uncover patient-specific differences in therapy response. However, there is limited quantitative information about the stability of such cells during the initial 20–30 passages of culture.
Methods
We interrogated 3 patient-derived GC cultures at dense time intervals during the first 30 passages of culture. Combining state-of-the-art signal processing methods with a mathematical model of growth, we estimated clonal composition, rates of change, affected pathways, and correlations between altered gene dosage and transcription.
Results
We demonstrate that GC cultures undergo sequential clonal takeovers, observed through variable proportions of specific subchromosomal lesions, variations in aneuploid cell content, and variations in subpopulation cell cycling times. The GC cultures also show significant transcriptional drift in several metabolic and signaling pathways, including ribosomal synthesis, telomere packaging and signaling via the mammalian target of rapamycin, Wnt, and interferon pathways, to a high degree explained by changes in gene dosage. In addition to these adaptations, the cultured GCs showed signs of shifting transcriptional subtype. Compared with chromosomal aberrations and gene expression, DNA methylations remained comparatively stable during passaging, and may be favorable as a biomarker.
Conclusion
Taken together, GC cultures undergo significant genomic and transcriptional changes that need to be considered in functional experiments and biomarker studies that involve primary glioblastoma cells.
Keywords: aneuploidy, clones, GBM DNA methylation, GBM subtype, glioma stem cell cultures, patient derived GBM cell cultures, systems biology
Importance of the study
Patient-derived cancer cell cultures are an increasingly important tool for oncological research. Such cell cultures are an improvement over classical cancer cell lines, but their properties are not fully scrutinized. We analyzed the stability of patient-derived cell cultures from the brain tumor glioblastoma. Using molecular and mathematical analyses, our results show that the cells undergo both systematic adaptations and sequential clonal takeovers. Such changes tend to affect a broad spectrum of pathways. A systematic analysis of cell culture stability will therefore be essential to make use of primary cells for translational oncology.
Glioblastoma (GBM) is the most frequent grade IV primary brain cancer in adults. The standard-of-care therapy for GBM combines surgery, radiation, and chemotherapy but is not effective due to relapse of the tumor.1 One of the key approaches taken to study new interventions against GBM is the use of patient-derived GBM cell (GC) cultures, a technique based on propagation of tumor cells in a defined medium, supplemented with growth factors.2 Compared with traditional tissue culture models, GC cultures retain cells transcriptionally similar to the tumor of origin, and retain properties important for the study of tumor regeneration.3 One promising application of GCs is based on their potential to facilitate functional studies on cells across a population of patients4,5 (eg, to determine biomarkers of drug response). For such studies to be efficient, however, it is necessary to consider the stability of the GC cultures, and the possible impact of time-dependent fluctuations in clonal substructure.6 Both GBMs and GC cultures have been shown to exhibit intra- and intergenomic heterogeneity,4,7 with observable consequences at the level of drug responses, in vivo tumor initiation capacity, and in vitro cell culture properties.8,9 Given the importance of GC cultures as a model for GBM, it is therefore necessary to understand how clonal composition develops over time, and to determine to what degree such clonal changes impact the transcriptional or epigenetic state of the cells. It is also important to investigate which pathways are affected by such changes and which are not. As part of a larger characterization effort of patient-derived GC cultures in our laboratory,4 we therefore studied the clonal stability of 3 patient-derived GC cultures over time, from early (<10) to late (<30) passages, sampled at regular intervals. High-resolution profiling of DNA copy number aberrations, DNA methylation, targeted exome sequencing, and transcriptomes were combined with mathematical modeling to characterize the rate and impact of genomic changes of the GC cultures.
Materials and Methods
GBM Cell Culture Establishment and Passaging
All surgical samples and records used in this study were obtained from Uppsala University Hospital in accordance with protocols approved by the regional ethical review board and after obtaining written consent from all patients. Passage 6 GC cultures were established and stored at −150°C using the procedures of our Human Glioma Cell Culture (HGCC) biobank.4 One million cell frozen vials of U3021MG, U3028MG, and U3088MG GC cultures from this collection were grown in Neurobasal and Dulbecco’s modified Eagle’s medium/F12 (1:1 mix) supplemented with N2, B27 (ThermoFisher Scientific), human recombinant fibroblast growth factor 2 (10 ng/mL, Peprotech), and epidermal growth factor (10 ng/mL, Peprotech), maintained at 37°C in 5% CO2. As in previous work by us4 and others,2 thawing results in only a limited loss of approximately 10% of cells, unlikely to affect the temporal evolution of cultures. During passages 7–29 the GC cultures were grown in mouse laminin coated 6-well BD Primaria plates. At each passage, once the cells reached 80%–90% confluence, we pooled all cells from 6 wells into one tube and counted the cells using the trypan blue method (Countess, Invitrogen). For the next passage, we seeded 100,000 cells per well into a new 6-well plate and the remaining cells were frozen and stored at −150°C. At regular intervals, an aliquot of cells was used for preparation of DNA and RNA.
Longitudinal Genomic Profiling of GC Cultures
DNA and RNA samples were obtained from the longitudinally passaged GC cultures, as specified in Supplementary Table S1. DNA was isolated from the GCs and formalin-fixed paraffin-embedded (FFPE) GBM tissues by using the DNeasy Blood & Tissue Kit (Qiagen) and the QIAamp DNA FFPE Tissue Kit (Qiagen) according to manufacturer’s protocol. Total RNA was extracted from GCs by the miRNeasy Mini Kit (Qiagen). DNA copy number profiles were measured using Affymetrix CytoScan (DNA from cells) and OncoScan (DNA from FFPE tissues) in accordance with the manufacturer’s instructions. Raw intensities were processed into copy number estimates with the R packages Rawcopy and TAPS.10,11 Copy number data (log ratio and single nucleotide polymorphism allele ratio) were studied for signs of segments with non-integer copy number caused by underlying genetic heterogeneity, as previously described.10–13 Where such heterogeneity was detected, the relative abundance of cells with each copy number composition was then estimated from the observed allele ratio or log ratio of the segment and their expected value(s) if they had been homogeneous. Exome sequencing for 597 selected genes was performed using the Illumina sequencing platform InView assay (GATC Biotech). Single nucleotide variants, insertions, and deletions were detected by using LoFreq.14 Variants detected were screened for known clinical significance by using the ClinVar database.15 Methylomes were profiled using Illumina HumMeth450K assays following the manufacturer’s instructions. Bisulfite conversion was performed using the EZ-96 DNA Methylation Gold Kit (Zymo D5007). The data were processed using the R package ChAMP16 to obtain DNA methylation beta values. RNA profiles were measured using the Affymetrix GeneChip HTA 2.0 arrays, and robust multi-array averages were normalized employing the Affymetrix Expression Console.
Validation of Ploidy Status by Fluorescence In Situ Hybridization and Flow Cytometry
The fluorescence in situ hybridization (FISH) probes CEp4 (cat no: 06J54-004), C5p (cat no:05J03-005), and C5q (cat no:05J04-005) were purchased from Abbott. The procedure was followed according to the manufacturer’s instruction. The individual cells were quantified for signals under the fluorescence microscope. For each cell line, at least 200 cells were counted for fluorescent signals, and graph was plotted in terms of percentage. GCs were analyzed for DNA content and ploidy status by flow cytometry (FC) as previously described.17 The raw data from flow cytometry were analyzed by using ModFit LT software.
Mathematical Model of Subpopulation Differential Growth Rate
To quantify cell growth rates, we used the exponential model , in which n(t) is the number of cells as a given time point, Δt is a time interval (in days), and r is the growth rate parameter (in doublings per day). From this basic assumption, we first derived equations to (i) estimate growth rates in the unit doublings/day, and (ii) estimate corresponding of doubling times (tD) in the unit hours/doubling, described in the Supplementary Methods. Next, we derived a technique to estimate the difference in growth rate from alteration fraction data as follows. Consider a mixture of 2 populations of cells, where n1(t) is the number of cells of type 1 at time t and n2(t) is the number of cells of type 2 at time t. The proportion of cells of type 2 at an arbitrary starting time point t0 is defined as
| (1) |
ie, the fraction of type 2 cells of the total pool of cells. Assume that cells grow exponentially from time The number of cells of types 1 and 2 are given by
| (2) |
where r is the doubling rate in 1/hours and and Δr is the difference in doubling rate between population 2 and population 1. At time the proportion of cells in population 2 is given by
| (3) |
Substitution of equations 1 and 2 into equation 3, followed by simplification and solving for Δr (details in Supplementary equations S1) gives the result in equation 4 (Results, below).
Solving equations 2 and 3 for Δr gives the result in equation 1. Note that the time unit t is arbitrary, where passage is used, implying that Δr calculated using equation 1 is the differential cell growth per passage is exp(Δr).
Analysis of Pathways, Gene Dosage Effects, and Glioblastoma Subtypes
To detect consistently altered pathways between early and late passages, we used the Gene Set Enrichment Analysis (GSEA)–PreRanked algorithm, as described in the Supplementary Methods. To quantify the effect of gene dosage on transcription between early and late passages, we used linear regression in which the log fold change of each transcript, was modeled as a linear function of the corresponding change in gene dosage, as the predictor variable. We applied this approach both to the full set of human genes in our data, as well as to each of the pathways available in the Kyoto Encyclopedia of Genes and Genomes database. Permutation tests and P-value false discovery rate (FDR) correction were used to find significant associations, as described in the Supplementary Methods.
Comparison of Stability for Different Genomic Data Types
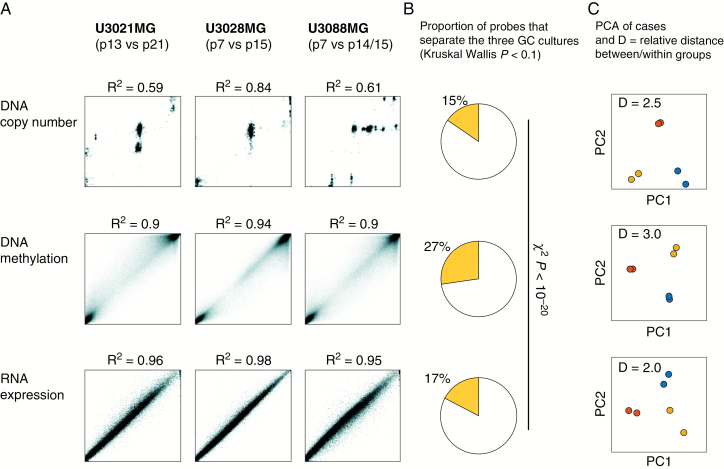
To assess and compare the stability of DNA copy number, DNA methylation, and RNA expression data, we used linear regression with each data type (Fig. 5A), in which X = the earlier time point sample and Y = the later time point sample. To compare the different assays (Fig. 5B), we used a nonparametric assessment. We thus used Kruskal–Wallis test statistic H, with cell cultures as 3 groups and the early and late time points as replicates within each group. We then used 1000 permutations to obtain simulated P-values and kept probes as P < 0.1 as differential between cases. (We are aware that 0.1 is too high a cutoff to call individual probes, but the present design with 3 groups does not permit a stricter calling; what matters here is the differential percentage of probes between the technologies, for which a 0.1 cutoff is adequate.) For Fig. 5C, standard principal component analysis (PCA) was applied to row-centered data for each data type separately.
Fig. 5.
DNA methylation is stable in glioblastoma cell cultures. We analyzed the effect of clonal takeover events on each of (i) DNA copy number, (ii) DNA methylation and (iii) RNA transcription. (A) Scatter plots of early vs late time points; while there was a numerical change in DNA copy number, DNA methylation and RNA transcription remained more stable. (B) Each measured gene locus, transcript, and methylation event was scored by Kruskal‒Wallis test to identify observations that were significantly different between U3021MG, U3028MG, and U3088MG, viewing the early and late passage time points as within-group replicates. DNA methylation had a higher fraction of observed variables with Kruskal‒Wallis P-value < 0.1. The significance of the altered fraction was confirmed by the chi-square test (P < 10−20). (C) PCA, indicating separation of the 3 GC cultures, showing the 2 time points of each GC in the same color. While the PCA plot is different for each platform, the relative average distance between the GC cultures compared with the average distance within the GC cultures was higher for DNA methylation profiles (3.0 vs 2.0–2.5).
Results
Genetically Different Cells Appear and Expand in GC Cultures
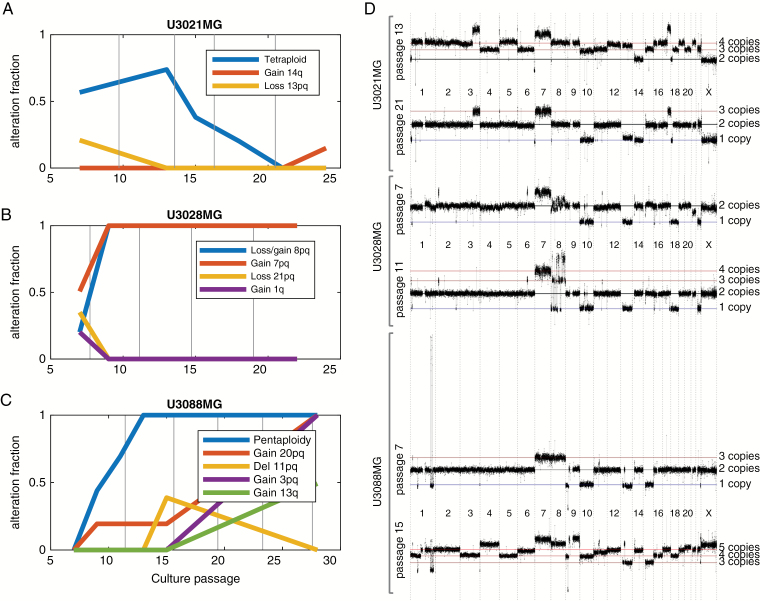
We selected 3 GC cultures (U3021MG, U3028MG, and U3088MG) from our HGCC biobank of GC cultures from Swedish patients.4 The 3 cultures were propagated from passage 6 to 30 in defined, serum-free medium. We subsequently collected samples for profiling, at regular intervals of ~2–8 passages (Supplementary Table S1). We used high-resolution arrays (Affymetrix CytoScanHD) to probe the genome with approximately one probe every 1000 base pairs. We first visualized the results using Rawcopy10 and TAPS,11 which were used to determine the genome-wide absolute copy number at each time point. When there was evidence of genetic heterogeneity, the fraction of cells and their associated copy number were estimated as previously described.12,13 In all 3 GC cultures, we found evidence that subpopulations expanded and retracted. This was observed both as a change in the estimated population fraction of specific genetic alterations over time and as the appearance of specific alterations (Fig. 1A–C). Situations in which a genetic alteration increased from a moderate fraction to 100% of the culture were designated as clonal takeovers. The data also indicated that while the cultures were heterogeneous at the copy number level, many or most copy number alteration (CNA) events were shared between subpopulations of the same cell line, proving a common ancestry in the primary tumor (Fig. 1D). This was also supported by a direct genetic comparison between the parental tumor and the cell cultures by both CNA profiling and targeted exome sequencing (597 genes, Supplementary Tables S2 and S3, Supplementary Fig. S1).
Fig. 1.
Genetically different cells appear and expand in GC cultures. (A–C) Emergence, expansion, and diminishing of clones over time in 3 GCs. (D) Absolute genome-wide copy number of each set of matched subclone genomes at one earlier and one later passage. Note the detected overall change in ploidy in U3021MG and U3088MG.
The GC culture U3021MG, derived from a 50-year-old male, first displayed a complex copy number profile at passage 7, interpreted as a mixture of near-tetraploid and near-diploid cells. At passage 13, however, U3021MG was 74% near-tetraploid, with 3–4 copies per cell of most of the genome. The tetraploid subpopulation was gradually reduced after passage 13 and was no longer detected at passage 21, when all cells appeared near diploid (Fig. 1A).
U3028MG, derived from a 72-year-old female patient, was near-diploid at passage 7 but contained a duplication of chromosome 7 (in 50% of the cells) and a complex chromosome 8 rearrangement (in 20% of cells). At passage 9, the events on chromosomes 7–8 were fully clonal, indicating a rapid clonal takeover. From passage 9 the copy number profile remained homogeneous and unchanged until at least passage 15 (Fig. 1B).
The GC culture U3088MG, derived from a 67-year-old female patient, started as a near-diploid, apparently homogeneous cell population at passage 7. It carried a deletion on chromosome 1q where a few segments of otherwise deleted material had formed a high-level amplification, likely in the form of a circular double-minute chromosome.18 Such a 1q amplification is recurrent in GBM and present in about 10% of primary tumors.19 At passage 9, the copy number profile had become more complex and contained a higher-ploidy subpopulation with a median copy number of 5 (near pentaploid). The pentaploid subpopulation constituted the majority of cells at passage 11 and virtually all cells at passages 13 and 15. Meanwhile, the cell fraction of the 1q amplification fell steadily from passage 7 through 15 and was absent at passage 28. About 60% of the cells also had gain of 13q, which means that at least 2 subpopulations were present at passage 28. Thus, near-pentaploid subpopulations have taken over the culture (Fig. 1C).
Near-Diploid Cells May Harbor Pseudo-Subclones with Doubled Genomes
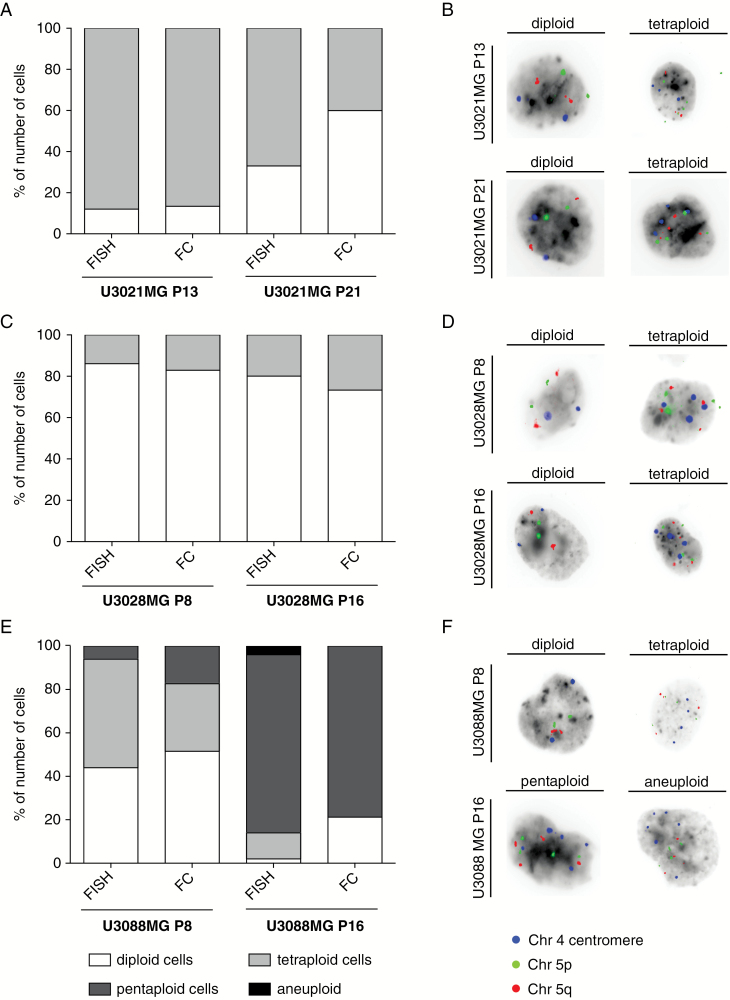
We used 2 independent experimental methods, FISH and FC, to confirm the observed differences in ploidy and clonal heterogeneity. The results confirmed the detection of aneuploidy populations in U3021MG, but at higher proportions than originally observed on the array copy number data. For instance, FISH showed that U3021MG still contained >40% tetraploid cells at passage 21 (Fig. 2A, B) and that U3028MG contained approximately 20% tetraploid cells at passage 16 (Fig. 2C, D). The dramatic increase of pentaploid cells in U3088MG at passage 16 was confirmed, but a tetraploid population was also detected (Fig. 2E, F). Consistent results were obtained using FC (Supplementary Fig. S2). The quantitative difference to the array data likely reflects a technical limitation; with array technology, the DNA of a genome-doubled cell is indistinguishable from the DNA of 2 nondoubled cells. As FISH data consistently showed evidence of the tetraploids, and FC data (Fig. 2, Supplementary Fig. S2) confirmed their existence, presence of the near-tetraploid cells is unquestionable. But the observation of such tetraploid cells that are similar to the near-diploid population in all 3 cultures casts doubt over whether they constitute actual subclones. The data are likely best explained by a process in which tetraploid cells indeed arise frequently through whole genome doublings during cell culture. Thus, the near-tetraploid cells can be viewed as a whole genome doubled version of the near-diploid cells in the culture at that time. Subsequent segregation errors in near-tetraploid cells could then form a source of new aneuploid cells. The implication of such tetraploid cells for functional experiments, if any, remains to be investigated.
Fig. 2 .
Confirmation of shifting ploidy status by FISH and FC. (A) U3021MG passage (P)13 displayed the same amount of diploid and tetraploid population, whereas U3021MG P21 contained 27% more diploid cells when analyzed by FC. (B) Representative FISH images of U3021MG. (C) U3028MG P8 and P16 contained the almost same percentage of diploid and tetraploid population between both methods. (D) FISH of U3028MG. (E) FISH and FC data confirmed the presence of diploid, tetraploid, and pentaploid cells in U3088MG P8 with slight variation in the percentage of population between both techniques. In U3088MG P16, the tetraploid population was not found by FC, whereas by FISH analysis 12% of tetraploid cells were identified. (B, D, F) Representative FISH images of U3021MG, U3028MG, and U3088MG. (F) Representative FISH images U3088MG.
High Rates of Clonal Takeover in GC Cultures
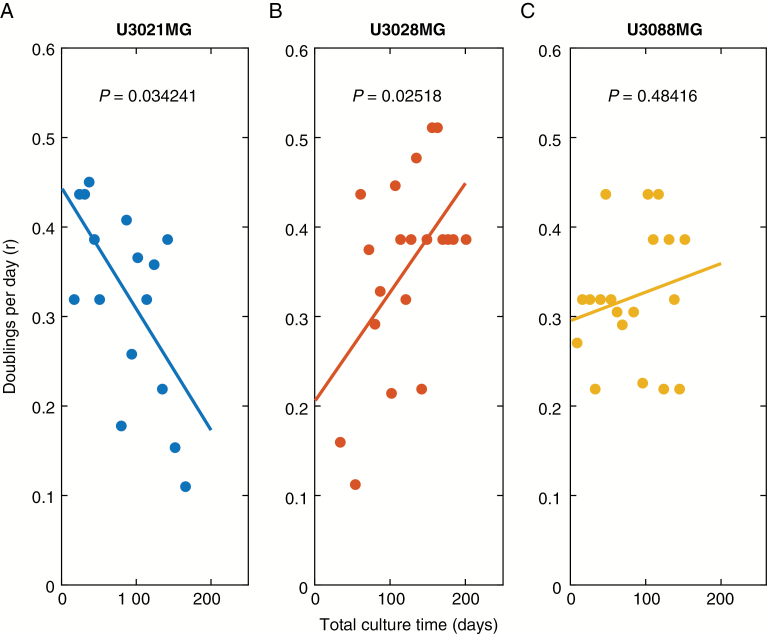
In order to further analyze cell growth and clonal takeover, we interpreted our array data in the context of a mathematical model of exponential cell growth. An exponential model of cell growth states that at any interval in time, the cell population expands by a factor of , where Δt is the number of days in culture, and r is the rate parameter r (in the unit doublings per day). Using data from 29 passages (209 days) of culture, we found that r varied in all 3 cultures between about 0.1 and 0.5 doublings per day, corresponding to approximately 48–240 hours per doubling. There was some evidence that r changes over time: U3021MG reduced its growth rate (Fig. 3A, P = 0.034), whereas U3028MG showed signs of accelerated growth (Fig. 3B, P = 0.025).
Fig. 3.
Estimation of growth rates during long-term cell culturing. The r values for each cell culture passage for the cultures (A) U3021MG, (B) U3028MG, and (C) U3088MG lines are fitted with robust linear regression and the P-value refer to the rejection of null model that the slope is zero (no change in r over time).
To estimate the rate of clonal takeover, we next considered a model with 2 cell populations, called population 1 (the background population) and population 2 (carrying a distinct genetic aberration compared with the background population). In this model, we assume that while population 1 grows at a default rate r, population 2 grows at a modified rate r + Δr. In other words, a genetically altered subpopulation for which Δr > 0 will tend to take over the whole population, whereas one with Δr < 0 will tend to disappear. A useful feature of the model is that it implies (proof in the Supplementary Methods) that Δr can be directly estimated from knowing the fraction of population 2 at 2 different time points:
| (4) |
where p0 is the alteration fraction at the starting time point, and p is the corresponding alteration fraction after an observation time of Δt. Using this result, we obtained relative growth rate of cells with different alterations (Table 1). The results were consistent with differential rates of subpopulation growth. For instance, in U3028MG, one subpopulation carrying chr 8 alterations had a doubling time that was 40 hours faster, whereas a second subpopulation with chr 21 loss had a doubling time of more than 120 hours slower (Supplementary Table S4). While this analytical model is clearly an approximation (eg, it assumes exponential growth and analyses one single alteration at a time), our results do provide a first estimate of rates of subclonal genetic drift in primary GBM cultures; genetically different cancer cells may coexist in the culture and rapidly change their proportions.
Table 1.
Relative growth rate of the different observed alterations
| Culture | Event | Between Passages | Delta r (doublings/day) | Population Doubling Time (h) | Altered Fraction Doubling Time (h) | Difference (h) |
|---|---|---|---|---|---|---|
| U3021MG | gain14q | 21–24 | >0.096 | 61 | <49 | <−12 |
| tetraploid | 7–13 | 0.027 | 57 | −4 | ||
| 13–15 | −0.076 | 76 | 15 | |||
| 15–18 | −0.058 | 72 | 11 | |||
| 18–21 | −0.103 | 83 | 22 | |||
| loss13pq | 7–13 | −0.057 | 72 | 10 | ||
| U3028MG | loss_gain8pq | 7–9 | >0.346 | 78 | <37 | <−41 |
| gain7pq | >0.234 | <44 | <−34 | |||
| loss21pq | <−0.187 | >200 | >121 | |||
| gain1q | <−0.126 | >132 | >54 | |||
| U3088MG | pentaploid | 7–9 | 0.23 | 82 | 46 | −36 |
| 9–11 | 0.153 | 54 | −28 | |||
| 11–13 | >0.219 | <47 | <−35 | |||
| gain20pq | 7–9 | >0.129 | <57 | <−25 | ||
| 15–28 | >0.064 | <67 | <−15 | |||
| del11pq | 13–15 | >0.256 | <44 | <−38 | ||
| 15–28 | <−0.037 | >93 | >12 | |||
| gain3pq | 15–28 | >0.087 | <63 | <−19 | ||
| gain13q | 15–28 | >0.043 | <71 | <−10 |
Note. The Δr is estimated as the difference in growth rate for cells harboring a particular genetic alteration. Model does not assume alterations to be mutually exclusive; it is therefore possible that an individual subclone carries more than one alteration.
The precise mechanism of the clonal fluctuations would require further study. For instance, the fluctuations of the tetraploid subpopulation in U3021MG and chr 11 deletion in U3088MG suggest that the relative growth of genetically distinct subpopulations (Δr) may depend on functional interactions with other subpopulations, or effects due to long-term culture, like senescence. The one cell line (U3028MG) that increased its growth rate (Fig. 3) did so after an initial takeover of a subpopulation with chr 7 gains (Fig. 1B), both known as recurrent events in GBM.19,20 The increased rate may thus be a result of acquisition of additional oncogenic changes.
Transcriptional Stability of GC Cultures: Adaptations, Gene Dosage, and Subtype
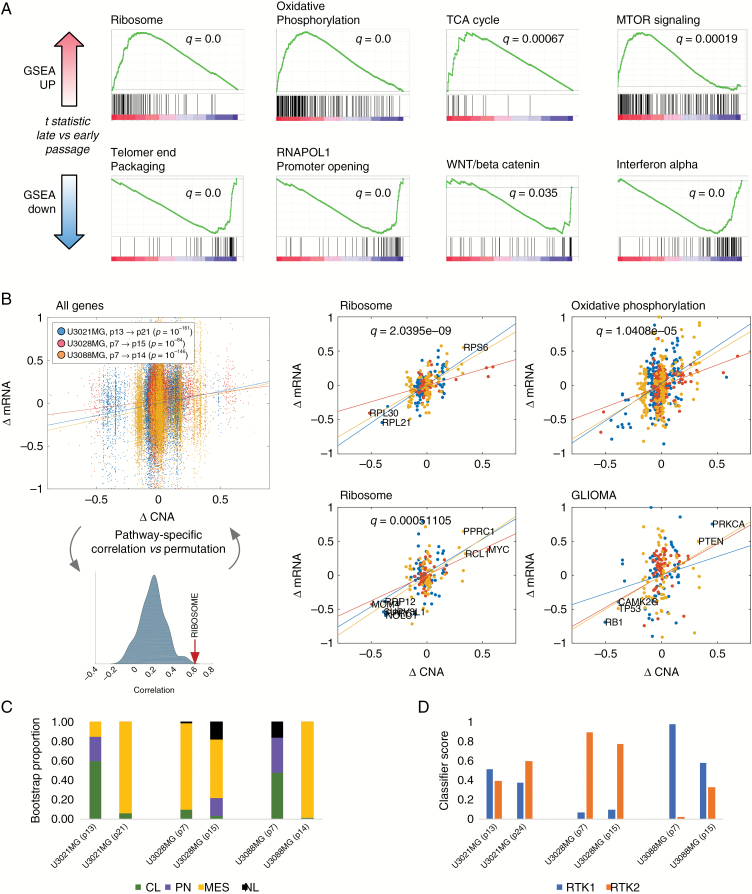
What were the transcriptional effects of clonal takeover? We analyzed global mRNA profiles from pairs of samples, selected to represent time points before and after clonal takeover. We first identified the number of genes that were detected with at least 2-fold differences in expression. Identified were 1592 genes in U3021MG, 517 genes in U3028MG, and 2148 genes in U3088MG (Supplementary Table S5). Thus, the genomic changes are in all 3 cases accompanied by profound changes in gene expression that affect large numbers of genes. Secondly, we tested the hypothesis that cells would show consistent transcriptional changes between earlier and later passages. For this, we scored each individual transcript by a moderated t statistic, which had a positive sign for transcripts that were consistently upregulated during passaging and a negative sign for transcripts that were downregulated. We then scored pathways in the Broad Institute Molecular Signatures Database (MSigDB) for consistent positive or negative bias of t scores using GSEA (Fig. 4A). The 3 GC cultures showed strongly significant (FDR q-value < 0.001) induction of pathways associated with ribosomal biogenesis, oxidative phosphorylation, tricarboxylic acid cycle, mammalian target of rapamycin signaling, and hypoxia (q = 0), which is consistent with broad metabolic adaptations to culture conditions.21 Downregulated pathways included telomere end packaging (q = 0.0), opening of RNA pol 1 promoter (q = 0.0), as well as downregulation of Wnt/beta catenin (q = 0.035) and alpha-interferon pathways (q = 0) (Fig. 4A). A full set of results is provided in a browsable format in (Supplementary File S1).
Fig. 4.
Transcriptional consequences of culturing and altered gene dosage. (A) Detection of pathways that were consistently up- or downregulated in U3021MG, U3028MG, and U3088MG cells, between 7–8 passages of culture, as measured by a moderated t score and the preranked GSEA statistic. (B) Correlation between changes in gene dosage (ΔCNA) and change in gene expression (ΔRNA) for all genes (left) and selected pathways (right; Supplementary Table S4); a permutation based simulation was used to obtain an FDR corrected q-value for each pathway (schematic). (C) Scoring of subtype signatures using bootstrapped K nearest neighbor. The relative proportion of each color indicates the fraction of bootstrap runs in which the cell culture was assigned to the corresponding subtype. (D) Scoring of DNA methylation signatures RTK1 and RTK2 using the DKFZ classifier.
A similar application of GSEA to DNA copy number aberrations indicated consistent gain and loss of multiple chromosomal bands (Supplementary Fig. S3), suggesting that some of the pathway alterations may be due to altered gene dosage. To quantify the gene dosage effect, we considered a regression model in which the change of DNA copy number for each gene locus (ΔCNA) was correlated with the corresponding change of gene expression for that locus (ΔRNA) First considering all transcripts, there was clear evidence that drift in gene dosage affects transcript levels (r > 0.11, P < 10‒20, Fig. 4B). Based on this observation, we next tested the idea that particular pathways may be more strongly affected by CNA-driven gene dosage changes. To quantify this, we computed the correlation between ΔCNA and ΔRNA in each pathway in the MSigDB to obtain a pathway-specific P-value by simulation (Fig. 4B). Using a pooled statistical test of correlation, we identified 21 pathways for which ΔCNA and ΔRNA correlated more than expected at random (Supplementary Table S4). The most extreme example of this was the MSigDB subset of ribosomal genes, MYC transcriptional targets, oxidative phosphorylation genes, and ubiquitin pathway components (Fig. 4B). These observations would suggest that the observed clonal takeover is associated with changes in metabolism and protein turnover driven by CNA changes. For these pathways, high R-square between ΔCNA and ΔRNA in combination with a slope near 1:1 also seems to suggest that in some cases few other factors than CNA changes are at play. A more detailed functional investigation would be required to explore this effect.
It has previously been suggested that GC culturing in mice has resulted in shifts of the observable transcriptional subtype of the culture.4 We investigated whether a similar shift occurred during clonal takeover by applying the transcriptional subtype calling algorithm (bootstrapped k-nearest neighbor4) to assign subtype status (classical, proneural, mesenchymal22). As a result, we observed 2 GCs shifted status to mesenchymal from classical (Fig. 4C). This observation further illustrates that subtype status is not a constant property of a GBM cell culture, which raises a question of relevance of subtype scoring in therapeutic aspects.23,24 By comparison, classification by the German Cancer Research Center (DKFZ) brain tumor methylation classifier25 was relatively stable; all samples were classified as isocitrate dehydrogenase wild-type GBM, and either receptor tyrosine kinase (RTK)1 or RTK2 subtype at relatively constant scores (Fig. 4D).
Glioblastoma Cell DNA Methylation Patterns Are Robust to Clonal Takeover
We went on to analyze the impact of clonal takeovers on DNA methylation of the GC cultures. Passages nearly matching those analyzed for gene expression (U3021MG passage [P]13; P24, U3028MG P7; P15 and U3088MG P7; P15) were thus analyzed for genome-wide methylation status using Illumina 450k arrays. The DNA methylation patterns remain comparatively stable in samples from the same culture that had undergone CNAs and showed large differences in gene expression and doubling times (Fig. 5). Firstly, the correlation of the methylome profile between earlier and later passage was high (R2 > 0.9) in all 3 GC cultures. This suggested that DNA methylation profiles were more preserved than CNA profiles (R2 0.59 to 0.84) but not necessarily mRNA profiles (R2 > 0.95) (Fig. 5A). This motivated us to analyze how the gradual change in genomic patterns would affect our ability to use each type of measurement as a biomarker. To quantify this, we applied a Kruskal–Wallis H statistic to relate the variation across the 3 cell cultures to the variation within each cell culture between passages. Applied to our data, this nonparametric test should thus identify mRNAs, gene loci, or methylation probes that contain robust signals useful to separate individual cases. Among the 3 data types tested, DNA methylation had approximately twice as many variables with a positive H test (Fig. 5B). This significantly higher proportion (chi-square test, P < 10−20) indicates that DNA methylation is a comparatively robust choice for biomarker analyses in GC cultures. The robustness of DNA methylation to separate cases (in terms of relative distances) was also evident in a PCA of each GC (Fig. 5C). Overall, no regions of differential methylation could be detected except in regions that had undergone unbalanced allele-specific CNA. Thus, the observed DNA methylation differences are largely a result of different composition of chromosomes with similar DNA methylation. We thus concluded that DNA methylation was relatively unaffected by multiple passages, and may in that particular sense be a stable biomarker, with differences in methylation between different tumor cell cultures. Those are likely to be caused by differences in methylation in the progenitor cells.
Discussion
A fundamental challenge for cancer research is to establish models that are, on the one hand, accessible for large-scale experimentation and, on the other hand, retain key aspects of patient-specific biology. Ongoing efforts such as the HGCC program aim to establish patient-specific GC cultures4 and xenograft mouse models26 across multiple molecular variants of GBM. In our own HGCC biobank, which is a public resource with widespread international distribution, there is currently a high demand for GC cultures that (i) harbor particular genomic lesions, (ii) show high or low activation of particular pathways, or (iii) cover different GBM subtypes.4 In brain tumor research, these primary GC cultures have emerged as a standard model to study drug sensitivity and identify new molecular targets.5,27–30 Considerable effort has been invested in supporting the representivity of primary GC cultures and comparing cell cultures isolated from the same tumor.2–4 By comparison, we have relatively little information on the temporal evolution of GC cultures. In one study, Garcia-Romero et al demonstrated that long-term passaging of glioma sphere cultures can affect the drug response of cells,6 warranting a systematic longitudinal investigation across several layers of genomic observation.
Our analysis of 3 of the GC cultures from HGCC illustrates clearly that each of these factors (mutation status, pathway activation, and subtype) is subject to time-dependent changes. The genomic copy number changes are often rapid and can be sequential, as illustrated by the succession of different dominant subpopulations. The genomic changes are in all cases accompanied by profound changes in mRNA expression. Some of these changes in specific pathways are likely to be caused directly by gene dosage effects. Some of the pathways with correlation between gene dosage and expression were recurrently affected in all 3 GC cultures. This suggests that the culturing of GC cells in itself may lead to changes in particular pathways or characteristics. However, further experimentation is required to more closely describe how often specific pathways are affected. Our analysis of growth rates shows that the drift in the cell cultures is likely associated with changes in cell phenotype, including phenotypes of key interest for GBM drug development. For instance, further work will be needed to elucidate the stability of in vitro drug responses, tumor-initiating capability in mice, or migratory capacity in GC cultures grown from both primary lesions and recurrent lesions, respectively. It will also be of considerable interest to understand how the pathway-specific temporal changes of GC cultures detected in our analysis relate to intratumor heterogeneity of GBM.7–9,31–33 This would require an extended experimental design, in which multiple tumor regions are sampled and analyzed over time to identify common and divergent time trends; such extensions are reserved for future work.
Using a simple mathematical technique (equation 4), clonal fluctuations can be used to obtain rates of relative growth of a clone or subpopulation. Future development of this model is needed to understand the mechanisms of clonal drift, which may involve functional interactions (eg, cooperative growth) between multiple subpopulations or non-exponential growth modes. In addition to the clonal turnover, we suggest that pseudo-subclones of tetraploid cells are continuously generated. The functional significance of such polyploid GBM cells warrants further study. While polyploidy per se does not cause changes in relative gene dosage, it can potentially affect other cellular phenotypes, like size, increased tolerance to chromosomal instability, and stress tolerance.34,35
It is important to emphasize that while there are time-dependent changes, patient-specific information is clearly retained to a high degree. This bodes well for future studies that use GC cultures to extract biomarkers. But our results underline that such studies must be designed with an awareness of the effect of passaging, particularly when pathways prone to drift—as defined in this work—are under study. Possible solutions will be to involve DNA methylation as a biomarker (since it is stable) and to evaluate if the genomic composition of the cultured cells has changed during the experiment.
Supplementary material
Supplementary material is available at Neuro-Oncology online.
Funding
This study was funded by the Swedish Research Council (2014-03314, SN), the Swedish Cancer Society (CAN 2017/628, SN) and the Swedish Foundation fro Strategic Research (BD15-088, SN).
Conflict of interest statement
There are no conflicts of interest.
Supplementary Material
Acknowledgments
We thank the SciLifeLab SNP-SEQ platform and Clinical and Experimental Pathology Program at Uppsala University for help with the experiments conducted.
References
- 1. Stupp R, Mason W, van den Bent MJ, et al. . Radiotherapy plus concomitant\and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. [DOI] [PubMed] [Google Scholar]
- 2. Pollard SM, Yoshikawa K, Clarke ID, et al. . Glioma stem cell lines expanded in adherent culture have tumor-specific phenotypes and are suitable for chemical and genetic screens. Cell Stem Cell. 2009;4(6):568–580. [DOI] [PubMed] [Google Scholar]
- 3. Lee J, Kotliarova S, Kotliarov Y, et al. . Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9(5):391–403. [DOI] [PubMed] [Google Scholar]
- 4. Xie Y, Bergström T, Jiang Y, et al. . The human glioblastoma cell culture resource: validated cell models representing all molecular subtypes. EBioMedicine. 2015;2(10):1351–1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schmidt L, Baskaran S, Johansson P, et al. . Case-specific potentiation of glioblastoma drugs by pterostilbene. Oncotarget. 2016;7(45):73200–73215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. García-Romero N, González-Tejedo C, Carrión-Navarro J, et al. . Cancer stem cells from human glioblastoma resemble but do not mimic original tumors after in vitro passaging in serum-free media. Oncotarget. 2016;7(40):65888–65901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sottoriva A, Spiteri I, Piccirillo SGM, et al. . Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci U S A. 2013;110(10):4009–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Soeda A. The evidence of glioblastoma heterogeneity. Sci Rep. 2015;5:7979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Meyer M, Reimand J, Lan X, et al. . Single cell-derived clonal analysis of human glioblastoma links functional and genomic heterogeneity. Proc Natl Acad Sci U S A. 2015;112(3):851–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mayrhofer M, Viklund B, Isaksson A. Rawcopy: improved copy number analysis with Affymetrix arrays. Sci Rep. 2016;6:36158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rasmussen M, Sundström M, Göransson Kultima H, et al. . Allele-specific copy number analysis of tumor samples with aneuploidy and tumor heterogeneity. Genome Biol. 2011;12(10):R108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mengelbier LH, Karlsson J, Lindgren D, et al. . Intratumoral genome diversity parallels progression and predicts outcome in pediatric cancer. Nat Commun. 2015;6:6125. [DOI] [PubMed] [Google Scholar]
- 13. Walther C, Mayrhofer M, Nilsson J, et al. . Genetic heterogeneity in rhabdomyosarcoma revealed by SNP array analysis. Genes Chromosomes Cancer. 2016;55(1):3–15. [DOI] [PubMed] [Google Scholar]
- 14. Wilm A, Aw PP, Bertrand D, et al. . LoFreq: a sequence-quality aware, ultra-sensitive variant caller for uncovering cell-population heterogeneity from high-throughput sequencing datasets. Nucleic Acids Res. 2012;40(22):11189–11201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Landrum MJ, Lee JM, Benson M, et al. . ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016;44(D1):D862–D868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Morris TJ, Butcher LM, Feber A, et al. . ChAMP: 450k chip analysis methylation pipeline. Bioinformatics. 2014;30(3):428–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tribukait B, Gustafson H, Esposti PL. The significance of ploidy and proliferation in the clinical and biological evaluation of bladder tumours: a study of 100 untreated cases. Br J Urol. 1982;54(2):130–135. [DOI] [PubMed] [Google Scholar]
- 18. Carroll SM, DeRose ML, Gaudray P, et al. . Double minute chromosomes can be produced from precursors derived from a chromosomal deletion. Mol Cell Biol. 1988;8(4):1525–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Paugh BS, Qu C, Jones C, et al. . Integrated molecular genetic profiling of pediatric high-grade gliomas reveals key differences with the adult disease. J Clin Oncol. 2010;28(18):3061–3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Brennan CW, Verhaak RG, McKenna A, et al. ; TCGA Research Network The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vlashi E, Lagadec C, Vergnes L, et al. . Metabolic state of glioma stem cells and nontumorigenic cells. Proc Natl Acad Sci U S A. 2011;108(38):16062–16067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Verhaak RG, Hoadley KA, Purdom E, et al. ; Cancer Genome Atlas Research Network Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bhat KPL, Balasubramaniyan V, Vaillant B, et al. . Mesenchymal differentiation mediated by NF-κB promotes radiation resistance in glioblastoma. Cancer Cell. 2013;24(3):331–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ozawa T, Riester M, Cheng YK, et al. . Most human non-GCIMP glioblastoma subtypes evolve from a common proneural-like precursor glioma. Cancer Cell. 2014;26(2):288–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sturm D, Witt H, Hovestadt V, et al. . Hotspot mutations in H3F3A and IDH1 define distinct epigenetic and biological subgroups of glioblastoma. Cancer Cell. 2012;22(4):425–437. [DOI] [PubMed] [Google Scholar]
- 26. Joo KM, Kim J, Jin J, et al. . Patient-specific orthotopic glioblastoma xenograft models recapitulate the histopathology and biology of human glioblastomas in situ. Cell Rep. 2013;3(1):260–273. [DOI] [PubMed] [Google Scholar]
- 27. Hubert CG, Bradley RK, Ding Y, et al. . Genome-wide RNAi screens in human brain tumor isolates reveal a novel viability requirement for PHF5A. Genes Dev. 2013;27(9):1032–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schmidt L, Kling T, Monsefi N, et al. . Comparative drug pair screening across multiple glioblastoma cell lines reveals novel drug-drug interactions. Neuro Oncol. 2013;15(11):1469–1478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Barretina J, Caponigro G, Stransky N, et al. . The cancer cell line encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483(7391):603–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Garnett MJ, Edelman EJ, Heidorn SJ, et al. . Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483(7391):570–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Segerman A, Niklasson M, Haglund C, et al. . Clonal variation in drug and radiation response among glioma-initiating cells is linked to proneural-mesenchymal transition. Cell Rep. 2016;17(11):2994–3009. [DOI] [PubMed] [Google Scholar]
- 32. Lee JK, Wang J, Sa JK, et al. . Spatiotemporal genomic architecture informs precision oncology in glioblastoma. Nat Genet. 2017;49(4):594–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Patel AP, Tirosh I, Trombetta JJ, et al. . Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344(6190):1396–1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Coward J, Harding A. Size does matter: why polyploid tumor cells are critical drug targets in the war on cancer. Front Oncol. 2014;4:123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Storchova Z, Kuffer C. The consequences of tetraploidy and aneuploidy. J Cell Sci. 2008;121(Pt 23):3859–3866. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.