Abstract
Despite the amenability of early-stage prostate cancer to surgery and radiation therapy, locally advanced and metastatic prostate cancer is clinically problematic. Chemical castration is often used as a first-line therapy for advanced disease, but progression to the castration-resistant prostate cancer phase occurs with dependable frequency, largely through mutations to the androgen receptor (AR), aberrant AR signaling, and AR-independent mechanisms, among other causes. Semaphorin 3C (SEMA3C) is a secreted signaling protein that is essential for cardiac and neuronal development and has been shown to be regulated by the AR, to drive epithelial-to-mesenchymal transition and stem features in prostate cells, to activate receptor tyrosine kinases, and to promote cancer progression. Given that SEMA3C is linked to several key aspects of prostate cancer progression, we set out to explore SEMA3C inhibition by small molecules as a prospective cancer therapy. A homology-based SEMA3C protein structure was created, and its interaction with the neuropilin (NRP)-1 receptor was modeled to guide the development of the corresponding disrupting compounds. Experimental screening of 146 in silico‒identified molecules from the National Cancer Institute library led to the discovery of four promising candidates that effectively bind to SEMA3C, inhibit its association with NRP1, and attenuate prostate cancer growth. These findings provide proof of concept for the feasibility of inhibiting SEMA3C with small molecules as a therapeutic approach for prostate cancer.
Keywords: semaphorin 3C, prostate cancer, small molecule, neuropilin, plexin
The clinical management of locally advanced and metastatic prostate cancer (PCa) presents a major challenge because of advancement of the disease to treatment-refractory stages. PCa progression is driven largely by the androgen receptor (AR), a nuclear hormone receptor that drives expression of numerous genes responsible for cell growth. Accordingly, currently used therapeutics for patients with advanced PCa include pharmacological inhibitors of the androgen-signaling axis, including treatment with AR antagonists such as enzalutamide. Upregulation of the AR or mutations in its ligand-binding domain invariably render these therapeutic approaches ineffective and limit the extension in life expectancy with enzalutamide treatment to only 4 to 5 months [1]. This reality underscores the urgent need to develop novel treatments to supplement current therapeutic options. The retention of AR activity in treatment-refractory stages of disease suggests that the transcriptional targets of the AR also represent a promising class of therapeutic targets.
The semaphorins constitute a highly conserved family of secreted or membrane-bound signaling proteins that function in embryogenesis and neurogenesis [2–5]. In this context, semaphorins form chemotactic cues that aid in directional neuronal cell migration and axonal outgrowth. Structurally, all semaphorins retain a 500‒amino acid semaphorin domain that folds into a seven-bladed β-propeller. This domain is critical to protein-protein interactions between semaphorins and their cognate receptors, such as neuropilins (NRPs) and plexins (PLXNs). The NRPs have been shown to activate vascular endothelial growth factor receptor signaling, whereas PLXNs are known to crosstalk with the receptor tyrosine kinases human epidermal growth factor receptor 2/V-Erb-B2 avian erythroblastic leukemia viral oncogene homolog 2 (HER2/ErbB2) and MET and can activate MAPK and integrin signaling cascades [6–9]. The eight classes of semaphorins are distinguished by structural features. Classes 1, 2, and 5 are found in invertebrates, whereas classes 3 through 7 are found in vertebrates and the eighth class, V, occurs in viruses [10]. Of the vertebrate semaphorins, class 3 family members are secreted and require NRP coreceptors to bind PLXNs, whereas classes 4 through 7 are membrane-associated semaphorins that directly bind PLXNs.
Semaphorins have been implicated in numerous cancers. For example, semaphorin 3C (SEMA3C) has been shown to partially drive progression of breast, ovarian, lung, gastric, and pancreatic cancers and has exhibited major clinical and prognostic relevance in PCa [8, 11–18]. We recently reported that the AR transcriptionally regulates SEMA3C and established that SEMA3C can drive epithelial-to-mesenchymal transition and stemness [19, 20]. Furthermore, we demonstrated that SEMA3C can activate multiple receptor tyrosine kinases involved in the growth of prostate and other cancers and that antagonizing SEMA3C holds potential utility in the management of PCa [21]. Given marked SEMA3C involvement in PCa growth and treatment resistance and to capitalize on our previously reported inhibition of SEMA3C with biologics, we investigated whether small molecule inhibitors or chemical probes for SEMA3C could also be developed.
The X-ray determination of parts of SEMA3A structure and other biophysical studies have provided a detailed understanding of semaphorin-receptor interactions at the cell surface [22, 23]. Here, a homology model built from SEMA3A-NRP1 crystal structure allowed us to virtually screen small molecules for the potential ability to disrupt SEMA3C-NRP1 interactions. We hypothesized that small molecule inhibitors of SEMA3C would attenuate downstream signaling and suppress invasive cellular phenotypes.
Inhibition of semaphorins was previously explored as a potential anticancer therapeutic strategy, but these studies focused mainly on SEMA3E and SEMA4D [24] proteins. To our knowledge, inhibition of SEMA3C by small molecules has not yet been explored. Here, we identified several chemical probes for SEMA3C that can effectively disrupt its association with NRP1. Furthermore, these molecules demonstrated substantial inhibition of the growth of PCa cell lines and attenuated relevant growth pathways including MAPK. These results demonstrate that inhibition of SEMA3C may hold therapeutic potential and provide a strong rationale for exploring SEMA3C inhibitors as promising PCa drugs.
1. Materials and Methods
A. Preparation of the Protein Structure for Docking
Virtual screening was carried out on the modeled structure of SEMA3C. To prepare the protein structure for docking, bond orders for the ligand and the protein were adjusted. The missing hydrogen atoms were added, and side chains were then energy-minimized using the OPLS-2005 force field with Maestro software (Schrödinger, New York, NY). The ligand-binding region was defined by a 12 Å box centered on amino acids of the NRP1 binding region. No van der Waals scaling factors were applied; the default settings were used for all other adjustable parameters.
B. Ligand Preparation
The National Cancer Institute (NCI) database was used for virtual screening against the SEMA3C/NRP1 binding site. The compounds were imported into a molecular database using Molecular Operating Environment (MOE) version 2012 (Chemical Computing Group Inc., Montreal, QC, Canada). Hydrogen atoms were added after these structures were “washed” (a procedure including salt disconnection, removal of minor components, deprotonation of strong acids, and protonation of strong bases). The following energy minimization was performed with the Merck molecular force field 94×, as implemented by the MOE, and optimized structures were exported into the Maestro suite in .sdf format.
C. Virtual Screening, Consensus Scoring, and Voting
Virtual screening was performed as described previously [25, 26].
D. Western Blot
Whole cell extracts were prepared in 50 mM Tris-HCl, 150 mM NaCl, 1% NP40, 10 mM NaF, and 10% glycerol, supplemented with protease inhibitor cocktail (Cat. no. 04693116001; Roche, Basel, Switzerland), and quantitated using a bicinchoninic acid assay approach. Sixty micrograms of protein was run on 10% acrylamide gels and transferred onto a nitrocellulose membrane. Western blots were imaged on a LI-COR Odyssey system. Antibodies were phospho-epidermal growth factor receptor (EGFR) (3777S; Cell Signaling Technology, Danvers, MA [27]), total EGFR (sc-377229; Santa Cruz Biotechnology, Dallas, TX [28]), phospho-ErbB2 (2247; Cell Signaling Technology [29]), total ErbB2 (2165; Cell Signaling Technology [30]), phospho-Src homology 2 domain-containing (SHC) (2434S; Cell Signaling Technology [31]), total SHC (610878; BD Biosciences, Franklin Lakes, NJ [32]), phospho-MAPK (4370S; Cell Signaling Technology [33]), total MAPK (4696S; Cell Signaling Technology [34]), vinculin (V4505; Sigma-Aldrich, St. Louis, MO [35]), anti-rabbit‒700 (A21109; Invitrogen), and anti-mouse‒700 (A21058; Invitrogen, Carlsbad, CA). LNCaP cells were cotreated with SEMA3C at 0.5 µM or epidermal growth factor (EGF) at 10 ng/mL (GF316; Millipore, Burlington, MA) or vehicle (PBS) and increasing concentrations (2.5, 5, 10, 20 µM) of small molecule or vehicle [dimethyl sulfoxide (DMSO)] for 10 minutes. Whole-cell extract was then collected for Western blot analysis. SEMA3C was produced as previously described [21].
E. Displacement Assay
NRP1-FC (sigma, fusion protein with the Fc domain from IgG) was immobilized onto a protein G‒coated plate via incubation at room temperature for 1 hour. Simultaneously, the compounds were incubated at 25 μM with alkaline phosphatase‒conjugated semaphorin 3C (SEMA-AP) at 4°C. SEMA-AP was produced as previously described [21]. The plate was washed with wash buffer (PBS; 0.05% Tween 20, Sigma-Aldrich, P9416) three times. The SEMA-AP compound mixture was added and incubated at 4°C with agitation for 1 hour. The plate was washed with wash buffer three times, followed by the addition of 50 μL of para-Nitrophenylphosphate (Thermo Fisher Scientific; Rockford, IL). Absorbance was measured at 405 nm using a Tecan plate reader (Infinite F500; Tecan, Männedorf, Switzerland) upon sufficient signal development. A hit compound was identified as inhibition >50% of SEMA-AP and NRP1-FC interaction compared with DMSO control.
F. Growth Assay
LNCaP (ATCC, Manassas, VA, CRL-1740), DU 145 cells (ATCC, HTB-81), NIH3T3 (ATCC, CRL-1658), hTERT-HPNE (ATCC, CRL-4023), PANC-1 (ATCC, CRL-1469), and 293T (ATCC, CRL-3216) were cultured as recommended by ATCC. Cell lines were authenticated by IDEXX Laboratories in August 2014. A patient-derived fibroblast cell line [36], which was cultured in RPMI 1640 supplemented with 10% fetal bovine serum, was also tested. Compounds were tested in triplicate. Cells were plated at 4 × 103 cells per well in 96-well plates with 100 μL per well RPMI or DMEM containing 10% v/v fetal bovine serum and 100 μg/mL each of penicillin and streptomycin. Each well was immediately treated during cell plating with one compound at final concentrations of 200 to 0.390625 μM at twofold serial dilutions. Each experiment also included a condition with 0.5% v/v DMSO but no compound. Cell growth was measured using PrestoBlue Cell Viability Reagent (Life Technologies, Carlsbad, CA) following 72 hours of incubation at 37°C in 5% CO2. Eleven microliters of the reagent was added to cells in each well and then incubated 60 minutes at 37°C in 5% CO2. Emission at 612 nm (excitation: 525 nm) was recorded to measure resazurin production. Emission data were normalized so that cells tested against DMSO only (no compound) represent 100% growth.
G. Surface Plasmon Resonance Assay
SEMA3C-FC was expressed in chinese hamster ovary cells as His-tagged proteins and purified by metal-affinity chromatography as previously described [21]. Surface plasmon resonance (SPR) analysis was performed by using Biacore 3000 optical biosensors equipped with carboxylated dextran preimmobilized with nitrilotriacetic acid sensor chips. For the ligand-binding assay, at the beginning of each binding cycle, the sensor chip was preconditioned with a 30-second pulse of regeneration buffer (350 mM EDTA in running buffer), charged for 3 minutes with 500 uM Ni2+ in running buffer, and equilibrated with running buffer (HBS-P buffer supplemented with 1% DMSO). Purified His-tagged SEMA3C-FC (50 µg/mL) was injected at 10 µL/min for 5 minutes across the Ni2+ charged surface to achieve capture levels of >10,000 resonance units. Compounds were first diluted in DMSO to desired concentrations and further diluted 100-fold in running buffer to yield final concentrations of 0, 6.25, 25, and 100 µm (keeping DMSO content constant at 1%). Compound was injected over the SEMA3C-captured surface at a flow rate of 25 µL/min. The sensor chip surface was regenerated between binding cycles with a 30-second injection of regeneration buffer. For data processing and analysis, response from the DMSO control in each concentration series was subtracted to correct for bulk refractive index changes.
G-1. Proximity ligation assay
Proximity ligation assay was performed as previously described [21]. Briefly, cells were seeded on coverslips before treatment with small molecules (or control), fixation, permeabilization, and assaying using the Duolink In-Situ Fluorescence kit (Sigma-Aldrich). Interactions were captured by confocal microscopy and measured using Duolink software. Antibodies were SEMA3C (sc-27796; Santa Cruz Biotechnology [37]), Plexin B1 (sc-25642; Santa Cruz Biotechnology [38]), and NRP1 (TA318985; Origene, Rockville, MD [39]).
2. Results
A. Homology Model of the SEMA3C-NRP1 Interaction
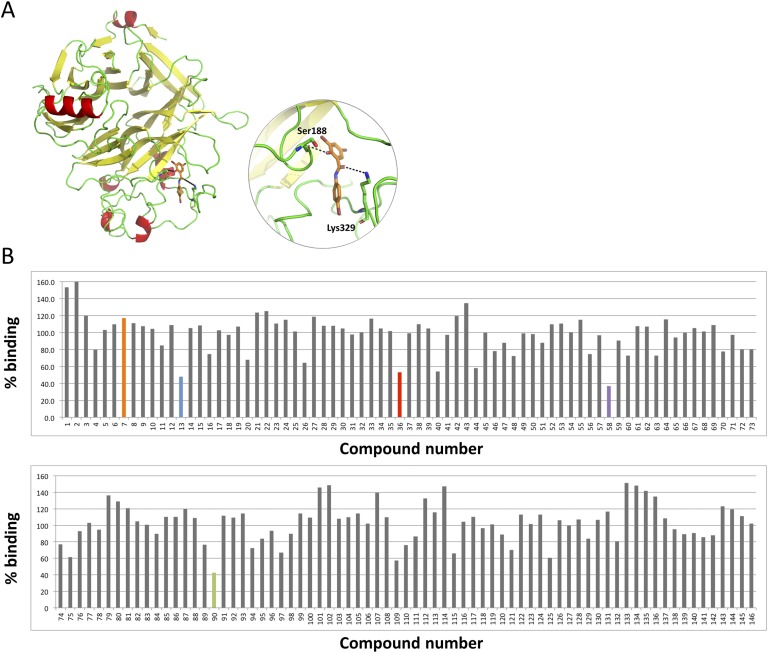
Because no crystal structure is available for human SEMA3C, its primary sequence was retrieved from the UniProt database (accession number Q99985) and aligned with PDB entries. SEMA3A demonstrated high sequence identity (52%) with a Protein Data Bank entry 4GZ8 [40], which was then used to generate the structure of SEMA3C using the Modeler program [41]. Based on the superimposition of experimental- and homology-modeled structures of SEMA3A- and SEMA3C-NRP1 complexes, several potential small molecule‒binding sites were identified at the protein-protein interaction interface and subjected to docking. The positions and residues of SEMA3C that constitute the NRP1-interacting motif are listed in Table 1 alongside the corresponding position in other class 3 semaphorins. The uniqueness of SEMA3C at this position supports specificity for SEMA3C of inhibitors or chemical probes derived from targeting this region.
Table 1.
NRP1 Binding Site Position on SEMA3C and Respective Amino Acid Residues in Class 3 Semaphorins
|
|
Position in the Chain
|
|||||
|---|---|---|---|---|---|---|
| Semaphorin | 189 | 221 | 222 | 329 | 336 | 379 |
| SEMA3C | K | L | T | K | L | L |
| SEMA3A | R | A | I | R | W | A |
| SEMA3B | R | A | V | K | W | L |
| SEMA3D | Y | S | Q | K | W | V |
| SEMA3E | R | A | L | K | W | L |
| SEMA3F | R | S | A | K | W | L |
| SEMA3G | S | V | P | R | W | L |
The amino acid residues and positions on SEMA3C that interact with NRP1 are listed along with residues at the corresponding position in other class 3 semaphorins.
B. Virtual Screening of Small Molecules That Bind to SEMA3C and Disrupt Its Interaction With NRP1
Using our in-house developed computational drug discovery pipeline, we conducted a virtual screening of ~260,000 compounds from the NCI database [42] to identify potential SEMA3C binders. Our in silico pipeline included large-scale docking, in-site rescoring, and consensus voting procedures [25, 26]. First, all chemical structures were collected, washed, and docked into the SEMA3C model using the Glide SP program [43] (no constraints applied). A set of ~50,000 compounds that received a docking score <5.0 were then redocked into the protein using the eHiTS docking protocol [44] with the corresponding docking score threshold set to 3.0. This step allowed for the reduction of the molecular data set to ~20,000 entries. After that, to identify the most consistently predicted binding orientations of the compounds, the root-mean-square deviation was calculated between the docking poses generated by Glide and eHiTS. Only molecules with docking poses with root-mean-square deviation values below 2.0 Å were subjected to further analysis. Next, selected docked ligands were subjected to additional on-site scoring using the Ligand Explorer program and the pKi predicting module of the MOE (Chemical Computing Group Inc.).
With this information, a cumulative scoring of five different predicted parameters (Glide score, eHiTS score, and pKi predicted by the MOE) was generated with each molecule, receiving a binary 1.0 score for every “top 10% appearance.” The final cumulative vote was then used to select ~1000 compounds that consistently demonstrated high predicted binding affinity toward the SEMA3C binding site. These compounds were then visually inspected, and a list of 146 promising chemicals was determined for purchasing and testing. Homology-modeled SEMA3C in association with compound 20526 is shown as an example (Fig. 1A).
Figure 1.
Screening of small molecules for displacement of SEMA3C from NRP1. (A) Homology-modeled structure of human SEMA3C (left); binding mode of compound 20526 (referred to as D13 throughout this report) at the binding region of NRP1 (in circle). This compound forms H-bond interactions with Ser188 and Lys329. Moreover, the compound makes strong van der Waals interactions with Leu379, Ala327, and Leu221. H-bonds are shown as black dotted lines. (B) The y-axis represents percent binding where vehicle (DMSO) treatment (no binding inhibition) was set to 100%. A hit was defined as a compound that inhibited SEMA3C binding to NRP1 >50% compared with DMSO control. Hits are shown in color along with a compound with no inhibitory activity (D7) shown in orange, which served as a negative control for subsequent experiments.
C. Blocking SEMA3C-NRP1 Interactions
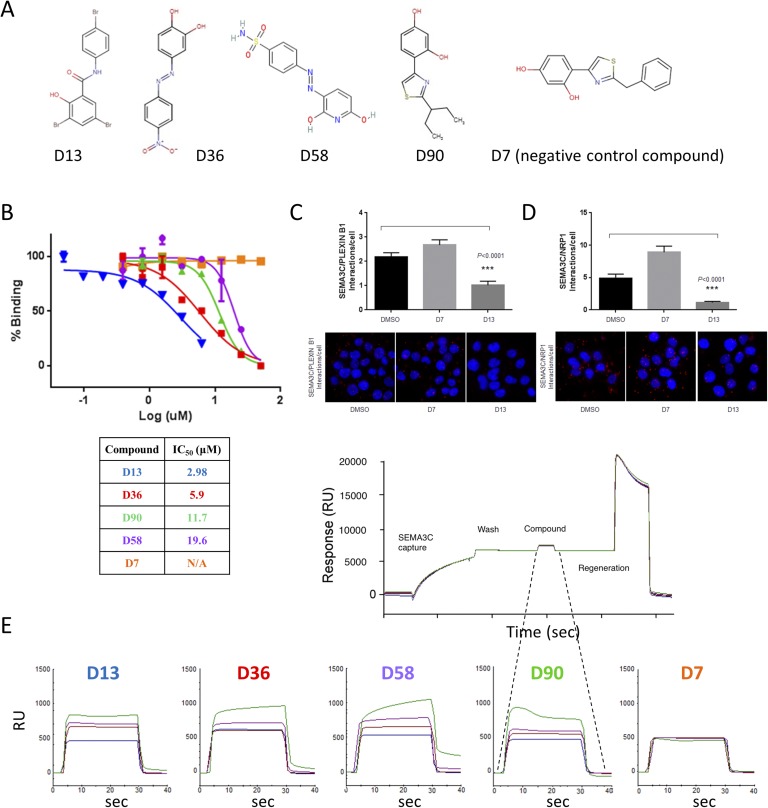
The promising candidates that emerged from the in silico screen were evaluated for their ability to prevent SEMA3C binding to its biological receptor, NRP1. We used an immunosorbent assay‒like approach to measure the displacement of SEMA3C from NRP1 using purified recombinant proteins (Fig. 1B). NRP1 was immobilized on 96-well plates followed by the addition of SEMA3C-AP with or without small molecules; para-Nitrophenylphosphate substrate was then added to measure the presence or absence of SEMA3C, reflecting no blocking (nonhit) or blocking activity (hit), respectively. A compound was identified as a “hit” when it disrupted SEMA3C-NRP1 interaction by >50% compared with DMSO control (Fig. 1B). From this set of 146 compounds originally selected from the in silico screen, four hits (D90, D13, D36, and D58; Fig. 2A) were identified at 25-μM concentration and furthermore inhibited the SEMA3C-NRP1 interaction in a dose-dependent manner (Fig. 2B). The resulting IC50 values ranged from 2.98 to 19.6 μM, with compound D13 identified as the most potent inhibitor of protein-protein interactions. Compound D7 was used as a negative control, which did not block SEMA3C-NRP1 interactions even at the highest concentration tested (100 μM). Of note, we confirmed the ability of the lead compound (D13), but not the negative control (D7), to disrupt the interactions between SEMA3C and either PLXNB1 or NRP1 using proximity ligation assay in DU145 cells, a system that endogenously produces all three proteins (Fig. 2C and 2D). Taken together, these data collectively demonstrate that the hit compounds could disrupt interaction between SEMA3C and its binding partners in vitro using purified components and also in live cells.
Figure 2.
Lead molecules from displacement assay screening. (A) Chemical structures of hit compounds. (B) Dose-dependent inhibition of SEMA3C-NRP1 interaction by the hit compounds revealed IC50s. (C and D) Lead compound D13 disrupted SEMA3C-receptor interactions in the proximity ligation assay (PLA), where DMSO and D7 served as inhibitory-null controls. D13 inhibited the interaction (C) between SEMA3C and PLXNB1 and (D) between SEMA3C and NRP1 in DU145 relative to vehicle and compound negative controls, DMSO and D7, respectively. Corresponding photomicrographs of in situ PLA are shown below. Punctate red fluorescence indicates protein interactions between the indicated molecules on the y-axis of the graphs from cells representative from five fields of view. PLA analysis was done by seeding 40,000 DU145 cells on 1-cm coverslips. Cells were then treated with either DMSO as control or compound D13 in HBHA-binding buffer (20mM HEPES, 150mM NaCl, 5mM CaCl2, 1mM MgCl2) containing 5% BSA on ice for 1 h. The coverslips were then washed three times in HBHA buffer before PLA probe binding, ligation, and amplification steps. PLA was carried out under the manufacturer’s protocol and analyzed using Duolink Image tool software. Bars represent mean and SEM interactions per cell. The data were statistically significant using the nonparametric Mann-Whitney statistical test as calculated with GraphPad Prism software. The results are representative of independent repeated experiments. Cells were treated with compound D7 or compound D13 at 5 µM. Cells were treated with SEMA3C at 0.5 µM. (E) Biochemical analysis of SEMA3C‒small molecule interaction by surface plasmon resonance. Association curves are shown for the four hit compounds and negative control (compound D7). Various concentrations (0, 6.25, 25, and 100 µM) of each compound were injected over the SEMA3C-capture surface. Refractive indices were normalized to DMSO vehicle. A representative SEMA3C-compound regeneration cycle on the Biacore is shown below; SEMA3C capture, wash, compound addition, and regeneration steps of a Biacore cycle are indicated. N/A, data not available; RU, response unit; sec, seconds.
D. Measuring the Direct Interaction Between Compounds and SEMA3C
To test for a direct interaction between the hit compounds and SEMA3C, we conducted SPR assays (Biacore 3000; GE Healthcare Life Sciences) that were sensitive enough to directly detect protein:compound binding. These experiments were enabled by the overexpression of His-tagged SEMA3C recombinant protein in chinese hamster ovary cells and metal affinity protein purification (see Materials and Methods). SPR data collection proceeded with preconditioning of the carboxylated dextran preimmobilized with nitrilotriacetic acid sensor chip, capture of His-SEMA3C, compound association/dissociation, and removal of protein by application of a regeneration buffer. Analysis of the resulting SPR curves revealed that all four hit compounds bound SEMA3C reversibly and in a dose-dependent manner (Fig. 2E). Compound D7 (negative control) showed no interaction with SEMA3C, consistent with earlier displacement assay results with NRP1. Collectively, results from SPR indicate that D13 and D58 demonstrated the highest affinities for SEMA3C, whereas D36 and D90 bound only weakly to moderately at lower concentrations (6.25 and 25 µM). When comparing these findings with our displacement assay results, we noted that D13 remained the most potent molecule across both assays, D36 and D90 performed moderately in both displacement and in SPR, and D58 performed poorly in displacement but well in SPR. D58 binding to SEMA3C, although strong, may occur in a fashion that does not effectively obstruct NRP1 binding.
E. Effects of Inhibition of SEMA3C on the Growth of PCa Cell Lines
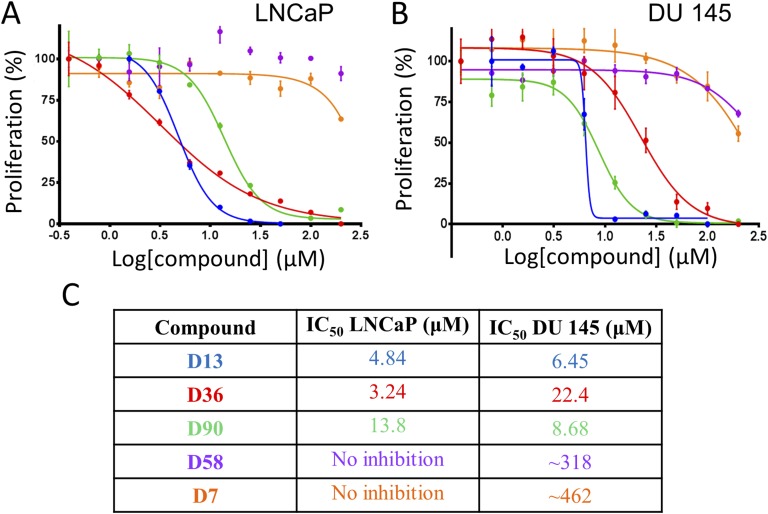
Inhibitory effects of the hit compounds were monitored in two different PCa cell lines, LNCaP (AR positive) and DU145 (AR negative). A dose-dependent growth inhibition of both LNCaP (Fig. 3A) and DU145 (Fig. 3B) cell lines was observed following 72-hour treatment with compounds D13, D36, and D90. In LNCaP cells, compound D36 showed the greatest growth inhibition with an IC50 of 3.24 µM (Fig. 3A; red curve; Fig. 3C). By contrast, in DU145 cells, compound D13 was most effective, with an IC50 of 6.45 µM (Fig. 3B; blue curve, Fig. 3C). These growth inhibition profiles are consistent with the displacement assay results, where compounds D13 and D36 were the top two hits (lowest IC50). As expected, compound D7 (negative control) showed no inhibition in the displacement assay and in the growth inhibition assays. The growth inhibitory activity of D13, D36, and D90 was then examined in additional cell types, including NIH3T3, patient-derived fibroblasts, HPNE, PANC-1, and 293T cell lines. IC50s for D13 were lowest among PCa cell lines, but D13 also showed activity against 293T cells, pointing to possible selectivity deficiencies. D36 and D90 showed a range of potencies against the other cell lines tested, indicating the potential application of these molecules in other cancer types but simultaneously illustrating the need for improved specificity [45]. IC50s are summarized in Table 2. Taken together, the strong potency of D13 was seen across each assay used, whereas D36 was moderate in displacement and binding (SPR) but generally strong in growth inhibition. D90 was moderate in each of the following: displacement, binding, and growth inhibition. D58 bound strongly in SPR but performed poorly in displacement and growth inhibition assays. In summary, D13 demonstrated the most robust and consistent activity in our assays; however, improvements to specificity will likely be required. D36, D90, and D58 will require medicinal chemistry to improve both activity and selectivity.
Figure 3.
Inhibition of prostate cancer cell growth by lead compounds. Inhibition of growth of prostate cancer cells by the hit compounds. Compounds D13, D36, and D90 dose dependently inhibited growth of (A) LNCaP and (B) DU145 cells. Cells were incubated with various concentrations of compound (starting concentration at 200 µM, 1:2 serial dilution) for 3 d at which point cell viability was assessed using PrestoBlue. Data represent mean ± SEM. (C) IC50 values of the hit compounds in LNCaP and DU145.
Table 2.
IC50 Summary of Small Molecule Inhibitor Leads
| Compound | Displacement Assay IC50 (µM) | LNCaP IC50 (µM) | DU145 IC50 (µM) | NIH3T3 IC50 (µM) | Patient-Derived Fibroblast IC50 (µM) | HPNE IC50 (µM) | PANC-1 IC50 (µM) | 293T IC50 (µM) |
|---|---|---|---|---|---|---|---|---|
| D13 | 2.98 | 4.84 | 6.45 | 11.48 | 9.1 | 9.145 | 10.19 | 5.7 |
| D36 | 5.90 | 3.24 | 22.4 | 2.77 | 9.9 | 2.65 | 2.52 | 4.3 |
| D90 | 11.7 | 13.8 | 8.68 | 3.80 | 14.4 | 6.54 | 8.36 | 14.7 |
| D58 | 19.6 | N/A | ∼318 | N/A | N/A | N/A | N/A | N/A |
| D7 | N/A | N/A | N/A | 13.91 | 67.1 | 15 | 32.77 | 49.9 |
IC50s of lead compounds in the displacement assay (second column) and growth inhibition assays are summarized. Compounds (first column) were tested in prostate cancer cell lines (LNCaP, DU145), fibroblasts (NIH3T3, patient-derived), and pancreatic cancer cell lines (HPNE, PANC-1) and in 293T. Some IC50s were indeterminably high (displacement assay and growth inhibition assays for LNCaP and DU145); D58 was not examined in NIH3T3, patient-derived fibroblasts, HPNE, PANC-1, and 293T. N/A, data not available.
F. Small Molecule Inhibitors of SEMA3C Attenuate Cell Growth Pathways in PCa Cell Lines
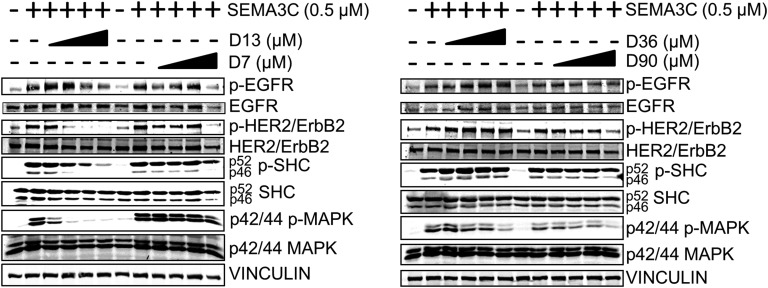
The compounds were then tested for their ability to inhibit SEMA3C-induced cell growth pathways in LNCaP cells. We previously showed that SEMA3C can initiate phosphorylation of EGFR, HER2/ErbB2, SHC, and MAPK [21], which comprise key players in the overall signaling pathway that stimulates cell division. Of all the compounds, D13 had the greatest effect in inhibiting SEMA3C-induced phosphorylation of EGFR, HER2/ErbB2, SHC, and MAPK, whereas negative-control compound D7 inhibited only at the highest concentration tested (Fig. 4). By comparison, compound D36 treatment resulted only in inhibition of SEMA3C-induced phosphorylation of MAPK, whereas compound D90 elicited a somewhat broader effect to block phosphorylation of HER2/ErbB2, SHC, and MAPK (Fig. 4). These results illustrate the capacity of the compounds to inhibit key signaling pathways related to cancer cell growth.
Figure 4.
Inhibition of cell growth signaling by small molecule inhibitor leads. LNCaP cells were treated with recombinant SEMA3C (0.5 µM) alone or in combination with increasing concentrations of small molecules (0, 2.5, 5, 10, and 20 µM) followed by detection of phospho-EGFR, HER2/ErbB2, SHC, and MAPK. D13 attenuated SEMA3C-induced phosphorylation of EGFR, HER2/ErbB2, SHC, and MAPK. D36 inhibited SEMA3C-induced phosphorylation of MAPK only. D90 attenuated SEMA3C-induced phosphorylation of HER2/ErbB2, p46 SHC, and MAPK. Negative control small molecule D7 inhibited phosphorylation of EGFR, HER2/ErbB2, and p46 SHC but only at high concentrations.
To probe the selectivity of these molecules for SEMA3C-induced signaling, we conducted the parallel study in EGF-stimulated LNCaP rather than SEMA3C-stimulated cells. EGF triggered phosphorylation of EGFR, HER2/ErbB2, SHC, and MAPK; however, small molecule compounds did not largely affect EGF-induced phosphorylation. Notable exceptions were the attenuation of phospho-MAPK by D13 at all concentrations and of phospho-EGFR at higher concentrations of D7 and D90; however, total EGFR also decreased correspondingly in these two treatments [46]. Apart from these selected exceptions, the findings suggest that our compounds are indeed selective for SEMA3C-induced signaling.
3. Discussion
Using a combination of in silico and in vitro screening approaches, we identified several small molecule inhibitors and/or chemical probes of SEMA3C and characterized their mechanism of action. These compounds suppress the growth of PCa cell lines with the corresponding IC50s established in the micromolar range. Although SPR showed that these chemicals bind reversibly to SEMA3C, the selectivity of their actions in interrupting SEMA3C-NRP1 interaction‒driven cancer progression needs to be addressed by additional experimentation and medicinal chemistry. We further established that identified SEMA3C small molecule inhibitors diminish phosphorylation of EGFR, HER2/ErbB2, SHC, and MAPK, collectively providing evidence that SEMA3C inhibition may present a prospective avenue for targeted therapy in PCa. Previously, we demonstrated that a decoy protein termed B1SP inhibited cell growth, cell signaling, and castration-resistant tumor growth in vivo [21]. The biologic B1SP is a recombinant protein constituting the extracellular sema domain of PLXNB1, the signal-transducing receptor to SEMA3C. This decoy disrupts binding of SEMA3C to PLXNB1, thereby attenuating downstream SEMA3C signaling and its associated activities. These studies illustrate the importance of the semaphorin-NRP association, which is thought to be a requisite event for class 3 semaphorin-PLXN signaling [23]. Here, our small molecules acted similarly to B1SP by inhibiting binding of SEMA3C to NRP1 and preventing their interactions with Plexin B1. Such disruption of critical protein-protein interactions leads to inactivation of several downstream pathways, including EGFR, HER2/ErbB2, SHC, and MAPK.
Therapeutic endeavors targeting semaphorins and their receptors have been explored and are summarized elsewhere [24]. Indeed, attenuation of various semaphorins by small molecules and by other approaches has been tested for anticancer effects. The class 3 semaphorin SEMA3E, which shares a common partner-receptor Plexin D1 with SEMA3C, has been shown to promote metastasis [6, 47]. In addition, these studies reported that the proteolytically cleaved forms of SEMA3E promoted a metastatic phenotype, whereas the full-length (uncleaved) form suppressed metastasis. Accordingly, the authors demonstrated that treatment with a recombinant SEMA3E, which is resistant to proteolytic cleavage by furin, reduced metastasis in mice [6]. Other work on SEMA3E-Plexin D1 signaling in metastatic breast cancer indicates that administration of the sema domain of the SEMA3C receptor, Plexin D1, a ligand trap for SEMA3E, inhibited primary and metastatic cancer growth [48]. Moreover, SEMA4D is upregulated in numerous cancers [17] and participates in angiogenesis, cell migration, and cell growth [49–53]. These actions are thought to be mediated by the PLXN receptor Plexin B1 and crosstalk with MET and Rho [6, 53, 54]. Inhibition of SEMA4D has been effective in deterring angiogenesis and tumor formation [52]. Finally, SEMA3A is regarded as a tumor suppressor and is downregulated in several cancers [55, 56]; in addition, its overexpression decreases angiogenesis and metastasis in vitro and in vivo [56–58], opening up the possibility that administration of SEMA3A could counteract the acquisition of tumor vasculature and metastasis. Until now, and in contrast to the previously mentioned studies, investigation of SEMA3C as a therapeutic target has remained largely unexplored despite its well-documented roles in PCa.
The drawbacks in targeting semaphorins include possible redundancies when one semaphorin member is blockaded. In addition, the activity of a semaphorin depends on the predominant modified form in which that semaphorin exists. For example, the full-length form of semaphorins is thought to have different cellular effects than the truncated form, therein implicating the relative abundance of proteolytic enzymes in different microenvironmental contexts. Consequently, although SEMA3C inhibition is effective in the context of certain tumors, it may potentiate cancer in tissues in which the population of proteolytic enzymes is different. On the other hand, semaphorins have a strong appeal as drug targets because of their extracellular location. Furthermore, semaphorins function primarily during development, potentially resulting in lower cytotoxicity for patients when inhibited by a therapeutic agent.
Castration, or hormone therapy, remains the first-line therapy for patients with locally advanced and metastatic forms of PCa. Unfortunately, disease progression invariably occurs through a variety of AR- and non-AR‒dependent mechanisms. We have demonstrated that SEMA3C may be implicated in such resistance to castration, and future work should explore the effects of our prototypical SEMA3C inhibitors on various PCa cell lines, castration-resistant ones in particular. Given the implication of SEMA3C in other cancers, including breast, bladder and kidney, the identified small molecules may prove to be effective in targeting them as well. Of note, further in vivo studies are needed to determine tolerance, efficacy, pharmacokinetics, and stability of the identified small molecules in animal models. Overall, the findings presented here represent a proof of principle for targeting SEMA3C with small molecules as potential therapeutic agents and/or chemical probes. This lays a foundation for further detailed examination and better characterization of inhibition of SEMA3C as a viable therapeutic avenue in cancer.
Acknowledgments
The authors acknowledge Andy Un for his assistance with compound chemistry.
Financial Support: This research was funded by the Michael Smith Foundation for Health Research Grant 17318 (to C.J.O.); Prostate Cancer Canada Grants TAG2014-06 (to C.J.O.) and GS2015-06 (to K.J.T.); the Terry Fox Foundation Grant TFF-116129 (to C.J.O.); the Canadian Cancer Society Research Institute Grant 700347 (to C.J.O.); the Cancer Research Society Grant F09-60564 (to C.J.O.); the National Institutes of Health Pacific Northwest Prostate Cancer SPORE Grant NCI P50 CA097186 (to C.J.O.); Networks of Centres of Excellence of Canada (CECR PC-TRIADD; to C.J.O.); and the Prostate Cancer Foundation BC (to K.J.T.).
Disclosure Summary: The authors have nothing to disclose.
Glossary
Abbreviations:
- AR
androgen receptor
- DMSO
dimethyl sulfoxide
- EGF
epidermal growth factor
- EGFR
epidermal growth factor receptor
- ErbB2
V-Erb-B2 Avian Erythroblastic Leukemia Viral Oncogene Homolog 2
- HER2
human epidermal growth factor receptor 2
- MOE
Molecular Operating Environment
- NCI
National Cancer Institute
- NRP
neuropilin
- PCa
prostate cancer
- PLXN
plexin
- SEMA3C
semaphorin 3C
- SEMA-AP
alkaline phosphatase‒conjugated semaphorin 3C
- SHC
Src homology 2 domain-containing
- SPR
surface plasmon resonance
References and Notes
- 1. Scher HI, Fizazi K, Saad F, Taplin ME, Sternberg CN, Miller K, de Wit R, Mulders P, Chi KN, Shore ND, Armstrong AJ, Flaig TW, Fléchon A, Mainwaring P, Fleming M, Hainsworth JD, Hirmand M, Selby B, Seely L, de Bono JS; AFFIRM Investigators . Increased survival with enzalutamide in prostate cancer after chemotherapy. N Engl J Med. 2012;367(13):1187–1197. [DOI] [PubMed] [Google Scholar]
- 2. Raper JA, Kapfhammer JP. The enrichment of a neuronal growth cone collapsing activity from embryonic chick brain. Neuron. 1990;4(1):21–29. [DOI] [PubMed] [Google Scholar]
- 3. Kolodkin AL, Matthes DJ, O’Connor TP, Patel NH, Admon A, Bentley D, Goodman CS. Fasciclin IV: sequence, expression, and function during growth cone guidance in the grasshopper embryo. Neuron. 1992;9(5):831–845. [DOI] [PubMed] [Google Scholar]
- 4. Luo Y, Raible D, Raper JA. Collapsin: a protein in brain that induces the collapse and paralysis of neuronal growth cones. Cell. 1993;75(2):217–227. [DOI] [PubMed] [Google Scholar]
- 5. Kolodkin AL, Matthes DJ, Goodman CS. The semaphorin genes encode a family of transmembrane and secreted growth cone guidance molecules. Cell. 1993;75(7):1389–1399. [DOI] [PubMed] [Google Scholar]
- 6. Casazza A, Finisguerra V, Capparuccia L, Camperi A, Swiercz JM, Rizzolio S, Rolny C, Christensen C, Bertotti A, Sarotto I, Risio M, Trusolino L, Weitz J, Schneider M, Mazzone M, Comoglio PM, Tamagnone L. Sema3E-plexin D1 signaling drives human cancer cell invasiveness and metastatic spreading in mice [published correction appears in J Clin Invest. 2011;121(7):2945] J Clin Invest. 2010;120(8):2684–2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Giordano S, Corso S, Conrotto P, Artigiani S, Gilestro G, Barberis D, Tamagnone L, Comoglio PM. The semaphorin 4D receptor controls invasive growth by coupling with Met. Nat Cell Biol. 2002;4(9):720–724. [DOI] [PubMed] [Google Scholar]
- 8. Tseng CH, Murray KD, Jou MF, Hsu SM, Cheng HJ, Huang PH. Sema3E/plexin-D1 mediated epithelial-to-mesenchymal transition in ovarian endometrioid cancer. PLoS One. 2011;6(4):e19396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sakurai A, Gavard J, Annas-Linhares Y, Basile JR, Amornphimoltham P, Palmby TR, Yagi H, Zhang F, Randazzo PA, Li X, Weigert R, Gutkind JS. Semaphorin 3E initiates antiangiogenic signaling through plexin D1 by regulating Arf6 and R-Ras. Mol Cell Biol. 2010;30(12):3086–3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Goodman CS, Kolodkin AL, Luo Y, Puschel AW, Raper JA, Comm SN; Semaphorin Nomenclature Committee . Unified nomenclature for the semaphorins/collapsins. Cell. 1999;97(5):551–552. [DOI] [PubMed] [Google Scholar]
- 11. Blanc V, Nariculam J, Munson P, Freeman A, Klocker H, Masters J, Williamson M. A role for class 3 semaphorins in prostate cancer. Prostate. 2011;71(6):649–658. [DOI] [PubMed] [Google Scholar]
- 12. Herman JG, Meadows GG. Increased class 3 semaphorin expression modulates the invasive and adhesive properties of prostate cancer cells. Int J Oncol. 2007;30(5):1231–1238. [PubMed] [Google Scholar]
- 13. Martín-Satué M, Blanco J. Identification of semaphorin E gene expression in metastatic human lung adenocarcinoma cells by mRNA differential display. J Surg Oncol. 1999;72(1):18–23. [DOI] [PubMed] [Google Scholar]
- 14. Miyato H, Tsuno NH, Kitayama J. Semaphorin 3C is involved in the progression of gastric cancer. Cancer Sci. 2012;103(11):1961–1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yamada T, Endo R, Gotoh M, Hirohashi S. Identification of semaphorin E as a non-MDR drug resistance gene of human cancers. Proc Natl Acad Sci USA. 1997;94(26):14713–14718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Esselens C, Malapeira J, Colomé N, Casal C, Rodríguez-Manzaneque JC, Canals F, Arribas J. The cleavage of semaphorin 3C induced by ADAMTS1 promotes cell migration. J Biol Chem. 2010;285(4):2463–2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rehman M, Tamagnone L. Semaphorins in cancer: biological mechanisms and therapeutic approaches. Semin Cell Dev Biol. 2013;24(3):179–189. [DOI] [PubMed] [Google Scholar]
- 18. Xu X, Zhao Z, Guo S, Li J, Liu S, You Y, Ni B, Wang H, Bie P. Increased semaphorin 3c expression promotes tumor growth and metastasis in pancreatic ductal adenocarcinoma by activating the ERK1/2 signaling pathway. Cancer Lett. 2017;397:12–22. [DOI] [PubMed] [Google Scholar]
- 19. Tam KJ, Dalal K, Hsing M, Cheng CW, Khosravi S, Yenki P, Tse C, Peacock JW, Sharma A, Chiang YT, Wang Y, Cherkasov A, Rennie PS, Gleave ME, Ong CJ. Androgen receptor transcriptionally regulates semaphorin 3C in a GATA2-dependent manner. Oncotarget. 2017;8(6):9617–9633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tam KJ, Hui DHF, Lee WW, Dong M, Tombe T, Jiao IZF, Khosravi S, Takeuchi A, Peacock JW, Ivanova L, Moskalev I, Gleave ME, Buttyan R, Cox ME, Ong CJ. Semaphorin 3 C drives epithelial-to-mesenchymal transition, invasiveness, and stem-like characteristics in prostate cells. Sci Rep. 2017;7(1):11501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Peacock JW, Takeuchi A, Hayashi N, Liu L, Tam KJ, Al Nakouzi N, Khazamipour N, Tombe T, Dejima T, Lee KC, Shiota M, Thaper D, Lee WC, Hui DH, Kuruma H, Ivanova L, Yenki P, Jiao IZ, Khosravi S, Mui AL, Fazli L, Zoubeidi A, Daugaard M, Gleave ME, Ong CJ. SEMA3C drives cancer growth by transactivating multiple receptor tyrosine kinases via Plexin B1. EMBO Mol Med. 2018;10(2):219–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Antipenko A, Himanen JP, van Leyen K, Nardi-Dei V, Lesniak J, Barton WA, Rajashankar KR, Lu M, Hoemme C, Püschel AW, Nikolov DB. Structure of the semaphorin-3A receptor binding module. Neuron. 2003;39(4):589–598. [DOI] [PubMed] [Google Scholar]
- 23. Janssen BJC, Malinauskas T, Weir GA, Cader MZ, Siebold C, Jones EY. Neuropilins lock secreted semaphorins onto plexins in a ternary signaling complex. Nat Struct Mol Biol. 2012;19(12):1293–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Worzfeld T, Offermanns S. Semaphorins and plexins as therapeutic targets. Nat Rev Drug Discov. 2014;13(8):603–621. [DOI] [PubMed] [Google Scholar]
- 25. Singh K, Munuganti RSN, Leblanc E, Lin YL, Leung E, Lallous N, Butler M, Cherkasov A, Rennie PS. In silico discovery and validation of potent small-molecule inhibitors targeting the activation function 2 site of human oestrogen receptor α. Breast Cancer Res. 2015;17(1):27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Munuganti RSN, Leblanc E, Axerio-Cilies P, Labriere C, Frewin K, Singh K, Hassona MDH, Lack NA, Li H, Ban F, Tomlinson Guns E, Young R, Rennie PS, Cherkasov A. Targeting the binding function 3 (BF3) site of the androgen receptor through virtual screening: 2. development of 2-((2-phenoxyethyl) thio)-1H-benzimidazole derivatives. J Med Chem. 2013;56(3):1136–1148. [DOI] [PubMed] [Google Scholar]
- 27.RRID:AB_2096270. http://antibodyregistry.org/search?q=3777S.
- 28.RRID:AB_2732851. http://antibodyregistry.org/search.php?q=AB_2732851.
- 29.RRID:AB_331725. http://antibodyregistry.org/search.php?q=AB_331725.
- 30.RRID:AB_10692490. http://antibodyregistry.org/search.php?q=AB_10692490.
- 31.RRID:AB_2188167. http://antibodyregistry.org/search?q=2434S.
- 32.RRID:AB_398195. http://antibodyregistry.org/search?q=610878.
- 33.RRID:AB_2315112. http://antibodyregistry.org/search?q=4370S.
- 34.RRID:AB_390780. http://antibodyregistry.org/search?q=4696S.
- 35.RRID:AB_477617. http://antibodyregistry.org/search?q=V4505.
- 36. Frame FM, Pellacani D, Collins AT, Maitland NJ. Harvesting human prostate tissue material and culturing primary prostate epithelial cells. Methods Mol Biol. 2016;1443:181–201. [DOI] [PubMed] [Google Scholar]
- 37.RRID:AB_2185540. http://antibodyregistry.org/search.php?q=AB_2185540.
- 38.RRID:AB_2166415. http://antibodyregistry.org/search.php?q=AB_2166415.
- 39.RRID;AB_2734705. http://antibodyregistry.org/search.php?q=AB_2734705.
- 40. Lee CCW, Munuganti RSN, Peacock JW, Dalal K, Jiao IZF, Shepherd A, Liu L, Tam KJ, Sedgwick CG, Bhasin S, Lee KCK, Gooding L, Vanderkruk B, Tombe T, Gong Y, Gleave ME, Ong CJ. Data from: Targeting semaphorin 3C in prostate cancer with small molecules. figshare 2018. Deposited 18 September 2018 https://figshare.com/s/fd82ca11028ac7a4d551. [DOI] [PMC free article] [PubMed]
- 41. Fiser A, Sali A. Modeller: generation and refinement of homology-based protein structure models. Methods Enzymol. 2003;374:461–491. [DOI] [PubMed] [Google Scholar]
- 42. Ihlenfeldt WD, Voigt JH, Bienfait B, Oellien F, Nicklaus MC. Enhanced CACTVS browser of the open NCI database. J Chem Inf Comput Sci. 2002;42(1):46–57. [DOI] [PubMed] [Google Scholar]
- 43. Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, Shaw DE, Francis P, Shenkin PS. Glide: a new approach for rapid, accurate docking and scoring: 1. Method and assessment of docking accuracy. J Med Chem. 2004;47(7):1739–1749. [DOI] [PubMed] [Google Scholar]
- 44. Zsoldos Z, Reid D, Simon A, Sadjad SB, Johnson AP. eHiTS: a new fast, exhaustive flexible ligand docking system. J Mol Graph Model. 2007;26(1):198–212. [DOI] [PubMed] [Google Scholar]
- 45. Lee CCW, Munuganti RSN, Peacock JW, Dalal K, Jiao IZF, Shepherd A, Liu L, Tam KJ, Sedgwick CG, Bhasin S, Lee KCK, Gooding L, Vanderkruk B, Tombe T, Gong Y, Gleave ME, Ong CJ. Data from: Targeting semaphorin 3C in prostate cancer with small molecules. figshare 2018. Deposited 18 September 2018 https://figshare.com/s/b34917854392a915482e. [DOI] [PMC free article] [PubMed]
- 46. Lee CCW, Munuganti RSN, Peacock JW, Dalal K, Jiao IZF, Shepherd A, Liu L, Tam KJ, Sedgwick CG, Bhasin S, Lee KCK, Gooding L, Vanderkruk B, Tombe T, Gong Y, Gleave ME, Ong CJ. Data from: Targeting semaphorin 3C in prostate cancer with small molecules. figshare 2018. Deposited 18 September 2018 https://figshare.com/s/1b2c561a34f0b8456fa9. [DOI] [PMC free article] [PubMed]
- 47. Christensen C, Ambartsumian N, Gilestro G, Thomsen B, Comoglio P, Tamagnone L, Guldberg P, Lukanidin E. Proteolytic processing converts the repelling signal Sema3E into an inducer of invasive growth and lung metastasis. Cancer Res. 2005;65(14):6167–6177. [DOI] [PubMed] [Google Scholar]
- 48. Luchino J, Hocine M, Amoureux MC, Gibert B, Bernet A, Royet A, Treilleux I, Lécine P, Borg JP, Mehlen P, Chauvet S, Mann F. Semaphorin 3E suppresses tumor cell death triggered by the plexin D1 dependence receptor in metastatic breast cancers. Cancer Cell. 2013;24(5):673–685. [DOI] [PubMed] [Google Scholar]
- 49. Soong J, Chen Y, Shustef EM, Scott GA. Sema4D, the ligand for Plexin B1, suppresses c-Met activation and migration and promotes melanocyte survival and growth. J Invest Dermatol. 2012;132(4):1230–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Basile JR, Castilho RM, Williams VP, Gutkind JS. Semaphorin 4D provides a link between axon guidance processes and tumor-induced angiogenesis. Proc Natl Acad Sci USA. 2006;103(24):9017–9022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sun Q, Zhou H, Binmadi NO, Basile JR. Hypoxia-inducible factor-1-mediated regulation of semaphorin 4D affects tumor growth and vascularity. J Biol Chem. 2009;284(46):32066–32074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Zhou H, Binmadi NO, Yang YH, Proia P, Basile JR. Semaphorin 4D cooperates with VEGF to promote angiogenesis and tumor progression. Angiogenesis. 2012;15(3):391–407. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 53. Conrotto P, Valdembri D, Corso S, Serini G, Tamagnone L, Comoglio PM, Bussolino F, Giordano S. Sema4D induces angiogenesis through Met recruitment by Plexin B1. Blood. 2005;105(11):4321–4329. [DOI] [PubMed] [Google Scholar]
- 54. Driessens MHE, Olivo C, Nagata K, Inagaki M, Collard JG. B plexins activate Rho through PDZ-RhoGEF. FEBS Lett. 2002;529(2-3):168–172. [DOI] [PubMed] [Google Scholar]
- 55. Barresi V, Vitarelli E, Cerasoli S. Semaphorin3A immunohistochemical expression in human meningiomas: correlation with the microvessel density. Virchows Arch. 2009;454(5):563–571. [DOI] [PubMed] [Google Scholar]
- 56. Maione F, Molla F, Meda C, Latini R, Zentilin L, Giacca M, Seano G, Serini G, Bussolino F, Giraudo E. Semaphorin 3A is an endogenous angiogenesis inhibitor that blocks tumor growth and normalizes tumor vasculature in transgenic mouse models. J Clin Invest. 2009;119(11):3356–3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Chakraborty G, Kumar S, Mishra R, Patil TV, Kundu GC. Semaphorin 3A suppresses tumor growth and metastasis in mice melanoma model. PLoS One. 2012;7(3):e33633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Casazza A, Fu X, Johansson I, Capparuccia L, Andersson F, Giustacchini A, Squadrito ML, Venneri MA, Mazzone M, Larsson E, Carmeliet P, De Palma M, Naldini L, Tamagnone L, Rolny C. Systemic and targeted delivery of semaphorin 3A inhibits tumor angiogenesis and progression in mouse tumor models. Arterioscler Thromb Vasc Biol. 2011;31(4):741–749. [DOI] [PubMed] [Google Scholar]