Abstract
Polycystic ovary syndrome (PCOS) is a common endocrine and metabolic disorder of unclear etiology in women and is characterized by androgen excess, insulin resistance, and mood disorders. The gut microbiome is known to influence conditions closely related with PCOS, and several recent studies have observed changes in the stool microbiome of women with PCOS. The mechanism by which the gut microbiome interacts with PCOS is still unknown.
We used a mouse model to investigate if diet-induced maternal obesity and maternal DHT exposure, mimicking the lean and obese PCOS women, cause lasting changes in the gut microbiome of offspring. Fecal microbiome profiles were assessed using Illumina paired-end sequencing of 16S rRNA gene V4 amplicons.
We found sex-specific effects of maternal and offspring diet, and maternal DHT exposure on fecal bacterial richness and taxonomic composition. Female offspring exposed to maternal obesity and DHT displayed reproductive dysfunction and anxietylike behavior. Fecal microbiota transplantation from DHT and diet-induced obesity exposed female offspring to wild-type mice did not transfer reproductive dysfunction and did not cause the expected increase in anxietylike behavior in recipients.
Maternal obesity and androgen exposure affect the gut microbiome of offspring, but the disrupted estrous cycles and anxietylike behavior are likely not microbiome-mediated.
Keywords: gut microbiota, polycystic ovary syndrome, anxiety, diet-induced obesity, androgens
Polycystic ovary syndrome (PCOS) is one of the most prevalent female endocrine disorders and is characterized by hyperandrogenism, oligo-/anovulation, and polycystic ovarian morphology [1]. Women with PCOS have an increased prevalence of disorders of glucose and lipid metabolism, anxiety, and depression [2–4]. The origins of PCOS are still unclear, with genetic, environmental, and lifestyle-related factors implicated in the development of the disorder [5].
The gut microbiome is known to regulate nutrient uptake, fat storage, and insulin sensitivity [6–8]. In humans, gut microbiome changes have been observed in conditions closely linked to PCOS, such as obesity, type 2 diabetes, anxiety, and depression [9–11]. We recently showed that women with PCOS have reduced fecal bacterial richness and changes in overall community composition compared with non-PCOS women [12]. Two subsequent studies by independent research groups have yielded similar results [13, 14]. Decreased bacterial richness and community changes have also been reported in a mouse model of PCOS [15]. In a rat model of PCOS, an improvement of the reproductive and endocrine PCOS phenotype was achieved through fecal microbiota transplantation (FMT) from healthy rats [16].
In rodents, sheep, and nonhuman primates, a PCOS-like phenotype can be induced by maternal androgen exposure during a critical developmental window in late gestation [17]. Although many maternal androgen exposure models develop glucose intolerance and/or β cell dysfunction, body weight and fat mass are generally not affected [18]. These models are therefore suggested to represent the lean PCOS phenotype [19].
In this study, we used a PCOS mouse model combining maternal androgen exposure [18] with high-fat, high-sugar (HFHS) diet-induced obesity to capture both the reproductive and metabolic aspects of the human PCOS and investigate their individual and combined effects on the gut microbiome [20]. We used DHT as a nonaromatizable androgen to exclude secondary estrogenic effects.
In our phenotypic assessment of the model, we found sex-specific effects of maternal DHT exposure and HFHS diet on anxietylike behavior and estrous cycle disruption in offspring, with female offspring appearing more anxious in response to maternal androgen exposure [21]. Further, we found sex-specific effects of the studied exposures on fecal bacterial richness and taxonomic composition.
To determine whether disturbed estrous cyclicity and increased anxietylike behavior observed in female offspring exposed to maternal androgens was microbiome-mediated, we performed FMT from female maternal DHT-HFHS-exposed offspring to healthy wild-type (WT) mice.
1. Materials and Methods
A. Maternal DHT Exposure Model
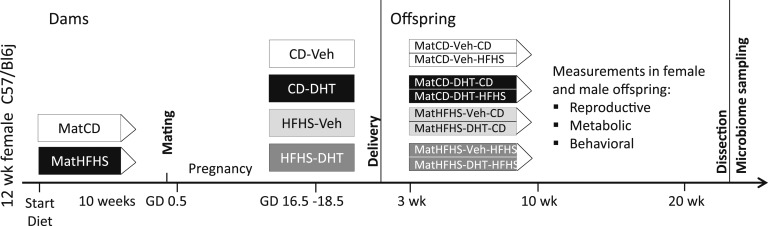
The study design for the prenatal androgenization model is summarized in Fig. 1. Fifty 12-week-old virgin female C57/Bl6j mice were purchased from Janvier Laboratories (Le Genest-Saint-Isle, France). Mice were housed five to a cage under a 12-hour light/dark cycle at a temperature of 21°C to 22°C and 55% to 65% humidity. After 1 week of acclimatization, mice were randomly assigned to either a control diet (CD) (#98052602; Research Diets, Inc., New Brunswick, NJ) containing 10 kcal% fat, 73 kcal% carbohydrates, and 17 kcal% protein, or a HFHS diet consisting of (1) high-fat diet (#D12079B; Research Diets, Inc.) containing 40 kcal% fat, 43 kcal% carbohydrates, and 17 kcal% protein and (2) 20% sucrose solution (S9378, Sigma-Aldrich, St. Louis, MO) supplemented with a vitamin mix (V10001; Research Diets, Inc., 10 g/4000 kcal) and a mineral mix (S10001; Research Diets, Inc., 35 g/4000 kcal). All animals were provided with tap water ad libitum.
Figure 1.
Study design for the maternal DHT-HFHS-exposure mouse model. n = 50 dams at start of experiment and 9 to 12 offspring/group. An additional group of dams was sacrificed at GD 18.5 and fecal material collected for gut microbiome analysis (n = 4 to 6/group). GD, gestational day.
After 10 weeks, females in estrus phase, determined by vaginal smear cytology, were mated overnight with a CD-fed male. The presence of a postcopulation plug was confirmed the following morning, which was defined as gestational day (GD) 0.5. Plug-positive animals were placed in single cages and continued on their respective diets. At GD 16.5, the HFHS and CD groups were subdivided into four groups: CD-Vehicle (Veh), CD-DHT, HFHS-Veh, and HFHS-DHT, testing two factors: diet and DHT exposure.
All mice in the DHT groups were subcutaneously injected with a fixed dose of 250 µg DHT (A8380; Sigma-Aldrich) in a 100-µL Veh solution consisting of 5 µL benzyl benzoate (B6630; Sigma-Aldrich) and 95 µL sesame oil (S3547; Sigma-Aldrich) in the scapular area for 3 days (until GD 18.5). Mice in the Veh groups were injected with an equal volume of sesame oil not containing the hormone. All injections were given between 9 and 10 am.
An additional group of 20 mice underwent the same procedures as dams but was anesthetized with isoflurane and blood was collected from axillar vessels followed by decapitation at GD 18.5, and colonic fecal material was collected, frozen immediately in liquid nitrogen, and stored at −80°C until later analysis.
All offspring were weaned at 21 days, separated by sex, and randomly assigned to continue on the diet of the mother or switch to the opposite diet. Male and female offspring were divided into eight groups: MatCD-Veh-CD, MatCD-DHT-CD, MatHFHS-Veh-CD, MatHFHS-DHT-CD, MatCD-Veh-HFHS, MatCD-DHT-HFHS, MatHFHS-Veh-HFHS, and MatHFHS-DHT-HFHS, testing three factors: maternal diet, maternal DHT exposure, and offspring diet. Offspring were housed two to six to a cage. Body weight and food intake were measured weekly. At 22 weeks of age, offspring were anesthetized with isoflurane and blood was collected from axillar vessels followed by decapitation, and colonic fecal material was collected and frozen immediately in liquid nitrogen. Fecal material was collected from a subset of mice for use in FMT as described below.
B. Behavior Testing
At 16 weeks of age, male and female offspring underwent behavioral testing to assess anxietylike behavior in the elevated plus maze (EPM) and 4 to 5 days later the open field (OF). Animals were tracked for 10 minutes in each arena using the EthoVision XT system (Noldus Information Technology, Wageningen, Netherlands) and the percentage of time spent in predefined areas of the arena was recorded (open and closed arms for EPM, periphery and center for OF). The behavior tests were performed in a soundproof room with no daylight. Prior to the start of the experiment, mice were acclimatized to the behavior room for 20 minutes. The arena was cleaned with 70% ethanol before each mouse to remove bias due to olfactory cues.
C. Assessment of Estrous Cyclicity
Vaginal smear cytology was performed on female mice immediately after behavior testing to confirm that results were based on female mice in all cycle stages. At 20 to 21 weeks of age, vaginal smear cytology was performed on female mice to assess estrous cyclicity over 10 consecutive days. Vaginal smears were collected by flushing the vagina with 15 µL sterile 0.9% saline solution, followed by microscopic inspection of the fluid on a glass slide and identification of the cycle phase according to published protocols [22, 23].
D. Preparation of Fecal Inoculate for FMT Experiment
The donors for the FMT experiment were female offspring that remained on the same diet as their mothers (MatCD-Veh-CD, MatCD-DHT-CD, MatHFHS-Veh-HFHS, and MatHFHS-DHT-HFHS). Three cages per group were selected (n = 1 to 4 mice/cage, 6 to 8 mice/group), and the cecal contents were pooled and homogenized in sterile PBS containing 25% glycerol (4 mL per cage). The stool suspension was briefly centrifuged to remove particulate matter and the supernatant was collected and pooled for each group. The absorption of the pooled supernatants was measured on a NanoDrop 2000 spectrophotometer (Thermo Scientific, Waltham, MA) at 620 nm and the dilutions adjusted to the levels of the least concentrated sample. Inoculates were aliquoted and fresh aliquots were thawed on each day of FMT. Immediately prior to gavage, FMT aliquots were diluted 1:5. This dilution approach enabled us to use the same FMT inoculate stocks for all mice in each group for the whole duration of the experiment.
E. FMT Experiment
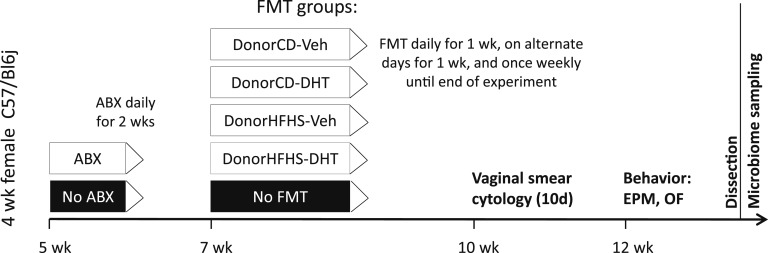
Numerous studies have achieved a successful metabolic and behavior phenotype transfer in mice using conventional housing, frozen FMT material, and antibiotic pretreatment, and these studies served as guidelines for our study [8, 24–27]. We used a cocktail of five antibiotics and one antifungal drug based on previously published protocols for microbiome depletion in mice [25, 26, 28].
An overview of the study design for the FMT experiment is given in Fig. 2. Four-week-old female C57/Bl6j mice (Janvier) were conventionally housed six to a cage and fed standard irradiated chow. Weight gain, food, and water intake were monitored throughout the experiment. Mice were randomly assigned to one of five treatment groups, four receiving FMT and one control group (n = 12/group). Beginning at 5 weeks of age, mice in the FMT groups received a sterile-filtered antibiotic mix consisting of 10 mg/kg vancomycin, 20 mg/kg neomycin, 20 mg/kg metronidazole, 20 mg/kg ampicillin, 10 mg/kg gentamicin, and 1 mg/kg amphotericin B (all Sigma-Aldrich), prepared in sterile water, via once daily oral gavage for 2 weeks. The control group received sterile water without antibiotics. Beginning on the day following the last antibiotic gavage, mice in the FMT groups received 150 µL FMT inoculate via oral gavage. FMT was given once daily for 7 days, every second day for the next 7 days, and then once weekly until the end of the experiment. Mice in the control group received sterile PBS with 25% glycerol following the same schedule. Estrous cyclicity was assessed 3 weeks after FMT at 10 to 11 weeks of age as described earlier. Anxietylike behavior was assessed in the EPM and OF 5 to 6 weeks after FMT at 12 to 13 weeks of age as described earlier. Following behavior testing, mice were anesthetized with isoflurane and blood was collected from axillar vessels followed by decapitation, and cecal contents were collected and frozen immediately in liquid nitrogen.
Figure 2.
Study design for the FMT experiment. n = 12/FMT group. ABX, antibiotic.
F. Next-Generation Sequencing of Fecal Samples and FMT Inoculates
DNA was extracted from fecal samples and FMT inoculates using the Maxwell 16 Tissue DNA Purification Kit (Promega Corporation, Madison, WI) on the Maxwell 16 Instrument according to the manufacturer’s instructions. Up to 50 mg fecal material per sample and 300 µL FMT inoculate were used. DNA was eluted in 300 µL Elution Buffer. The eluted DNA was centrifuged at full speed for 2 minutes and the supernatant collected. The DNA concentration was measured on a NanoDrop 2000 and DNA was diluted to a concentration of 5 ng/µL with diethyl pyrocarbonate-treated water. One negative control was included in each DNA extraction run and negative controls were pooled prior to PCR. Fifty nanograms DNA were used in a PCR reaction to amplify the V4 region of the bacterial 16S rRNA gene using the primers 515fB (GTGYCAGCMGCCGCGGTAA) and 806rB (GGACTACNVGGGTWTCTAAT) (Eurofins Genomics, Ebersberg, Germany) and the Phusion Hot Start II High-Fidelity PCR Master Mix (New England Biolabs, Ipswich, MA). The PCR conditions were initial denaturation at 95°C for 3 minutes followed by 25 cycles of denaturation at 95°C for 30 seconds, annealing at 60°C for 30 seconds, and extension at 72°C for 45 seconds, one final extension cycle at 72°C for 10 minutes, and a final cooling step to 10°C. A sample of diethyl pyrocarbonate-treated water, used to dilute the samples, was included as a PCR negative control. PCR products were visualized on a 1% agarose gel and purified using Agencourt AMPure XP beads (Beckman Coulter, Brea, CA) with a ratio of 1.2× beads to DNA. The purified PCR amplicons were indexed at the Science for Life Laboratory (Stockholm, Sweden) using the Nextera DNA Library Prep Kit (Illumina, San Diego, CA) and sequenced with v3, 600 cycles chemistry on a MiSeq Instrument (Illumina, San Diego, CA). Dam and offspring samples were sequenced in one run, FMT samples in a second run. An additional negative control from the indexing PCR was included. A microbial community standard consisting of eight bacterial species underwent the same DNA isolation, PCR, and sequencing protocol as samples (D6300; Zymo Research, Irvine, CA).
G. Sequencing Data Processing and Analysis
Primers were trimmed from raw reads using cutadapt [29]. Trimmed raw reads were processed in R 3.4.0 using the DADA2 1.4.0 package [30]. Forward and reverse reads were filtered based on their quality score profiles and trimmed at the point where read quality began to drop (F = 230 and R = 180 bases for maternal DHT exposure samples, F = 260 and R = 180 bases for FMT samples). Error rates of forward and reverse reads were calculated using the DADA2 algorithm and error-corrected reads were merged. Chimeric sequences were identified and removed. Taxonomic assignments were made for each ribosomal sequence variant (RSV) using the Silva v123 database (www.arb-silva.de). Subsequent analyses were performed in R using the phyloseq package [31]. Alpha rarefaction analyses were based on the number of observed RSVs and the Shannon diversity index. Beta diversity analyses were based on Bray-Curtis distances and visualized using multidimensional scaling (MDS).
H. Statistical Analysis
Statistical analyses were performed in IBM SPSS Statistics Version 22 or 23 unless otherwise stated. All continuous data were screened for normality and equality of variance prior to statistical testing. Nonnormally distributed data were log-transformed.
Data were analyzed using two-way ANOVA (diet and DHT exposure in dams and FMT mice) or three-way ANOVA (maternal diet, maternal DHT exposure, and offspring diet in DHT-HFHS exposed offspring). ANOVA was followed by posthoc tests with Bonferroni correction. Male and female offspring were analyzed separately. For the microbiome data, α diversity was compared in SPSS using two- or three-way ANOVA with posthoc testing and P-value correction as described above. Bray-Curtis distance matrices were compared using Adonis as part of the phyloseq package. Phylum and genus comparisons were performed using DESeq2 [32].
A P-value < 0.05 was considered statistically significant. The following abbreviations are used to denote statistically significant main effects in two- or three-way ANOVA: (a) [maternal] diet, (b) [maternal] DHT exposure, and (c) offspring diet. The following abbreviations are used to denote statistically significant interaction effects in two- or three-way ANOVA: (1) [maternal] diet and [maternal] DHT exposure, (2) [maternal] DHT exposure and offspring diet, and (3) [maternal] diet and offspring diet.
I. Ethics Approval
All animal experiments were performed at the Department of Physiology and Pharmacology, Karolinska Institutet, Stockholm, Sweden. All animal procedures were approved by the Stockholm Ethical Committee for Animal Research (121-16) in accordance with the legal requirements of the European Community (Decree 86/609/EEC).
J. Sequence Read Archive
The sequencing data sets and metadata files supporting the conclusions of this article are available at the repository [33] with accession numbers SRP136632 https://trace.ddbj.nig.ac.jp/DRASearch/study?acc=SRP136632 and SRP136823 https://trace.ddbj.nig.ac.jp/DRASearch/study?acc=SRP136823. R markdown files containing the details of the microbiome data analysis workflow can be provided upon request. Supplemental files are available at the Dryad Digital Repository [34].
2. Results
A. Phenotypic Characteristics of Female and Male Offspring
The reproductive and behavioral characteristics of female and male offspring have been reported by Manti et al. [21]. To summarize, maternal DHT exposure led to disrupted estrous cycles in female offspring, with a larger percentage of days in diestrus at the expense of proestrus and estrus, indicating reduced fertility. HFHS-diet led to significant weight gain in offspring of both genders compared with CD-fed offspring. Maternal DHT exposure resulted in anxietylike behavior in female offspring, observed in both EPM and OF. This maternal DHT exposure effect was completely absent in male offspring.
B. Fecal Microbiome Composition in Dams and Female and Male Offspring
The predominant phyla and genera across all groups in dams, female offspring, and male offspring are shown in an online repository [34]. Fecal samples were dominated by bacteria from the phylum Bacteroidetes, followed by comparable proportions of Firmicutes and Proteobacteria. Samples from dams showed a high percentage of bacteria from the Verrucomicrobia phylum, which was not seen in offspring samples. This finding was reflected on the genus level, where Akkermansia dominated dam samples, whereas offspring samples were dominated by Desulfovibrio, Bacteroidales family S24-7, and Rikenellaceae group RC9.
C. Differentially Abundant Taxa
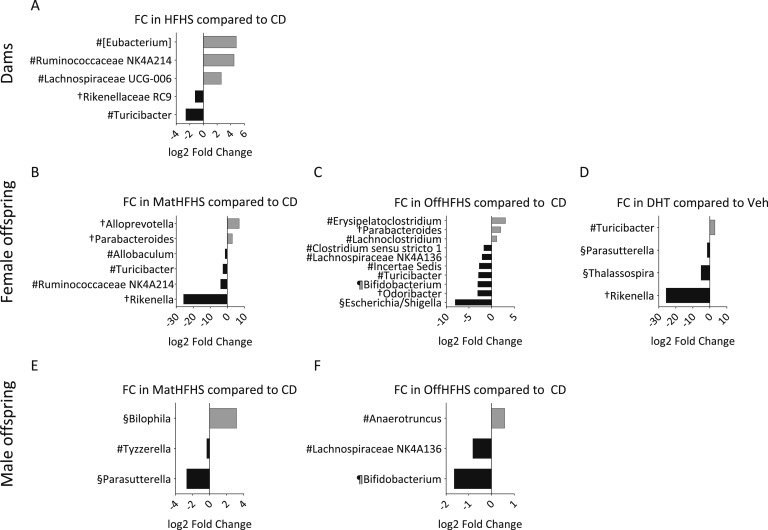
Diet had the strongest overall effect on genus relative abundance in both dams and offspring (Fig. 3). In dams, HFHS-diet caused a downregulation of the genus Turicibacter (Fig. 3A), concordant with that observed in female offspring exposed to either maternal or postnatal HFHS diet (Figs. 3D and 3E).
Figure 3.
Differentially abundant genera in fecal samples from dams and female and male offspring. Log twofold change (FC) of relative abundance for genera, which were found by DESeq2 analysis to be significantly changed in (A) dams, (B–D) female and (E, F) male offspring due to the (A) factors diet, (B, E) maternal diet, (C, F) offspring diet, and (D) maternal DHT exposure. No genera were significantly changed due to DHT exposure in dams or maternal DHT exposure in male offspring. P < 0.05 for all after Benjamini-Hochberg false discovery rate correction. #Firmicutes; †Bacteroidetes; §Proteobacteria; ¶Actinobacteria. Square brackets indicate a Silva suggested taxonomic assignment. n = 9 to 11 animals per tested factor for dams and 34 to 50 animals per tested factor for offspring.
Maternal diet affected genus relative abundance in both female and male offspring, but there were no changes common to both sexes (Figs. 3B and 3E). Ruminococcaceae group NK4A214 was upregulated due to HFHS-diet in dams and downregulated in female offspring (Figs. 3A and 3B).
Offspring HFHS-diet led to a concordant downregulation of two genera, Lachnospiraceae group NK4A136 and Bifidobacterium, in female and male offspring (Figs. 3C and 3F). The response to offspring diet was greater in female than male offspring, with a larger number of up- and downregulated genera (Figs. 3C and 3F).
Maternal DHT exposure significantly affected relative abundance of four genera in female offspring (Fig. 3D), with no significant changes in male offspring or dams. There was no clear pattern of up- or downregulation at the phylum level for any of the three studied factors.
D. Alpha Diversity in Dams and Female and Male Offspring
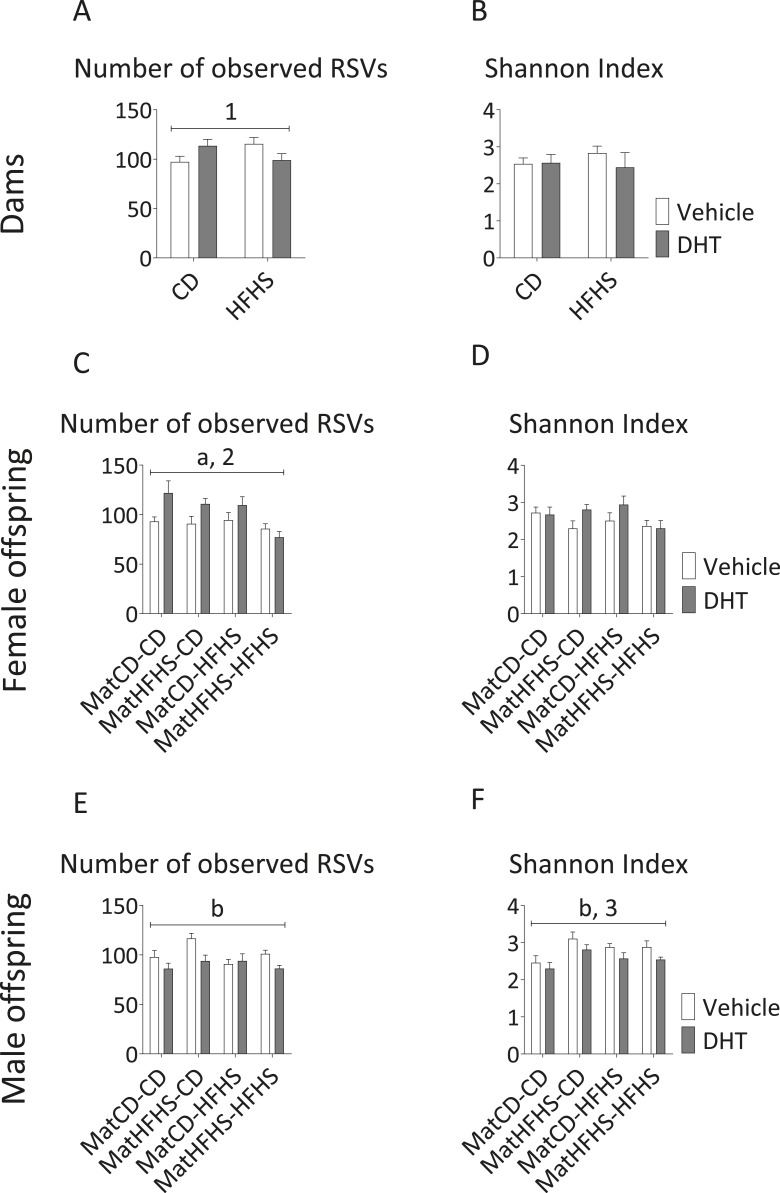
We found sex-specific effects of the studied factors on markers of microbial (α) diversity and richness in fecal samples. In female offspring, maternal DHT exposure increased richness in CD-fed offspring compared with Veh-exposed offspring, but this effect was not seen in HFHS diet-fed offspring (maternal DHT exposure*offspring diet, P < 0.05) (Fig. 4C). This result was mirrored in dams, where richness was increased in CD-fed dams, but not HFHS-fed dams (diet*DHT exposure, P < 0.05) (Fig. 4A). Maternal HFHS-diet led to reduced bacterial richness in female offspring, which was most pronounced when offspring also received HFHS-diet (main effect of maternal diet, P < 0.05) (Fig. 4C).
Figure 4.
Alpha diversity of fecal samples from dams and female and male offspring. Number of observed RSVs (left panel) and Shannon Index (right panel) are presented for (A, B) dams, (C, D) female offspring, and (E, F) male offspring. Two-/three-way ANOVA main effect of (a) maternal diet and (b) maternal DHT exposure (P < 0.05). Two-/three-way ANOVA interaction between (1) diet and DHT exposure, (2) maternal DHT exposure and offspring diet, and (3) maternal diet and offspring diet (P < 0.05). Mean and SEM are shown. n = 4 to 6 animals per group for dams and 7 to 10 animals per group for offspring.
In male offspring, DHT exposure led to reductions in both studied diversity measures (main effect of maternal DHT exposure, P < 0.05) (Figs. 4E and 4F). The combination of maternal HFHS diet and offspring CD in male offspring resulted in a higher Shannon index compared with the other groups (maternal diet*offspring diet, P < 0.05).
E. β Diversity in Dams and Female and Male Offspring
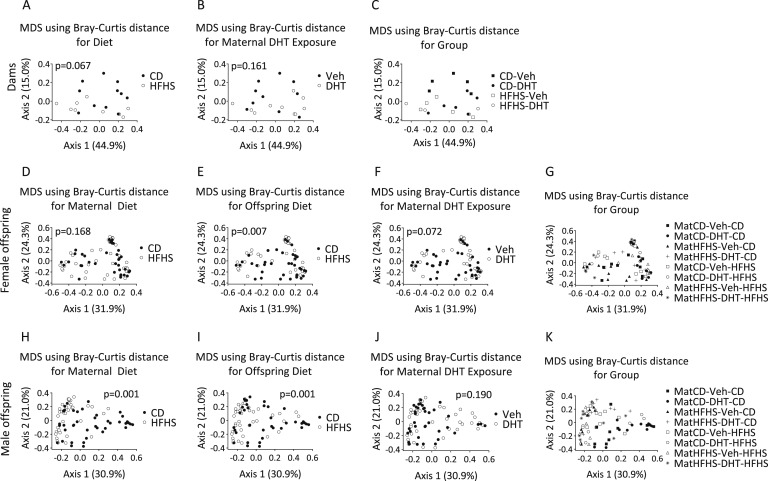
Bray-Curtis distances were calculated as a nonphylogenetic measure of bacterial community diversity and plotted using MDS to compare treatment groups.
Neither diet nor DHT exposure affected clustering in dams (Figs. 5A and 5B). Offspring diet had a significant effect on clustering in the MDS plot in both female and male offspring exposed to maternal DHT (P = 0.001 for both factors) (Figs. 5E and 5I). Maternal diet had a significant effect on clustering only in male offspring (P = 0.001) (Fig. 5H). DHT exposure did not affect clustering in either male or female offspring (Figs. 5F and 5J).
Figure 5.
Beta diversity of fecal samples from dams and female and male offspring. MDS plots of Bray-Curtis distances for the (A) factors diet, (B) DHT exposure, and (C) group in dams and (D, H) maternal diet, (E, I) offspring diet, (F, J) maternal DHT exposure, and (G, K) group in offspring. Each dot represents the total bacterial community composition of one sample. The amount of variation explained by each MDS coordinate is indicated in square brackets. Factors were compared using Adonis.
F. Phenotype Transfer by FMT
As PCOS is a female disorder and we observed greater microbiome and behavioral changes in female offspring exposed to maternal DHT than in male offspring, we investigated whether FMT could transfer these characteristics to unexposed mice. Healthy female WT mice were depleted of their gut microbiome via an antibiotic mix administered via oral gavage and subsequently recolonized with fecal material from female donors belonging to four of the previously described offspring groups: MatCD-Veh-CD, MatCD-DHT-CD, MatHFHS-Veh-HFHS, and MatHFHS-DHT-HFHS. To simplify, because maternal and offspring diet in the donors were the same, these groups will be referred to as DonorCD-Veh, DonorCD-DHT, DonorHFHS-Veh, and DonorHFHS-DHT. A control group received neither antibiotics nor FMT, but instead was gavaged with sterile water/PBS.
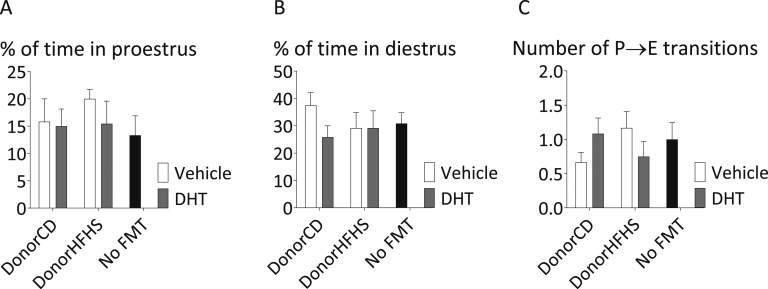
Estrous cyclicity appeared unaffected by FMT, with no significant difference in the percentage of time spent in proestrus or diestrus or in the number of proestrus to estrus transitions in FMT recipients (Fig. 6).
Figure 6.
Estrous cyclicity in FMT recipients. The percent of time spent in (A) proestrus, (B) diestrus, and (C) the number of proestrus to estrus transitions over 10 consecutive d. Mean and SEM are shown. n = 12 mice per group.
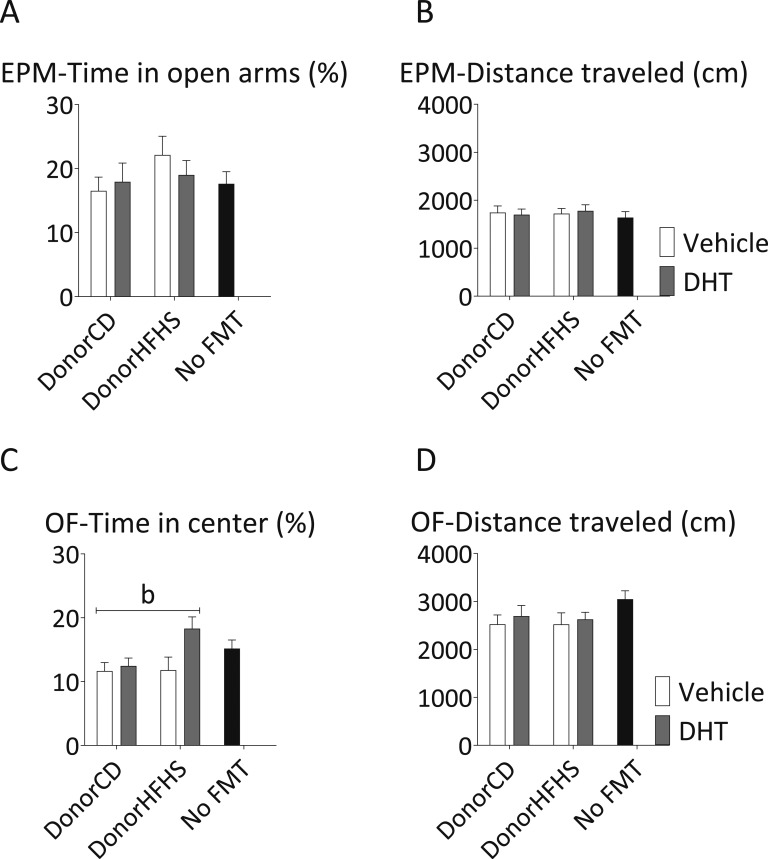
FMT did not induce the anxietylike behavior observed in maternal DHT-exposed donor mice. There was no significant difference between the groups in the time spent in the open arms of the EPM (Fig. 7A). In the OF, three of the four FMT recipient groups showed no significant difference compared with the control mice (Fig. 7C). Mice that received FMT from maternal HFHS-DHT donors spent significantly more time in the center of the arena than mice from the other three groups of FMT recipients (main effect of donor maternal DHT exposure, P < 0.05), indicating reduced anxietylike behavior in these mice (Fig. 7C). There was no difference in the total distance traveled in either arena (Figs. 7B and 7D).
Figure 7.
Anxietylike behavior in FMT recipients. Percentage of time spent in the open arms of the (A) EPM and (C) center of the OF and (B, D) the total distance traveled. Two-way ANOVA main effect of (b) donor maternal DHT exposure (P < 0.05). Mean and SEM are shown. n = 12 mice per group.
Antibiotic treatment was tolerated well and did not cause a drop in body weight or food intake. Body weight and food intake remained constant between the FMT groups and the control group throughout the experiment (data not shown).
G. Efficiency of the FMT Protocol
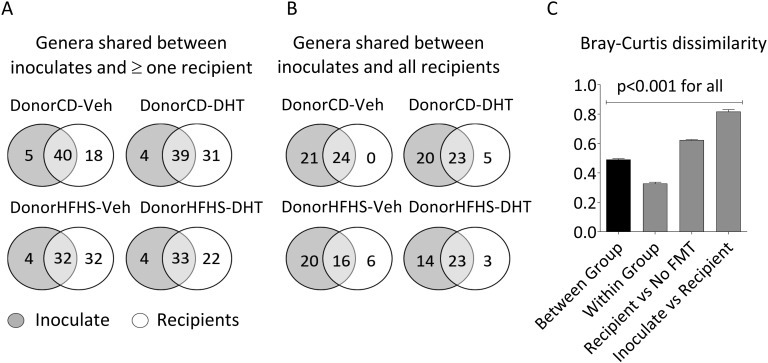
We compared microbiome profiles of FMT inoculates with fecal samples from FMT recipients to assess the efficiency of the procedure. Approximately 90% of genera in the inoculate were transferred to at least one recipient (Fig. 8A). Forty-four percent to 60% of genera in the inoculate were transferred to all recipients (Fig. 8B). Genera in the inoculate that were transferred to all recipients made up over 70% of “core” genera that were present in all recipients (Fig. 8B). Of the remaining “noncore” genera, mice collectively adopted ~90% of the inoculate, but also acquired individual-specific bacteria, which were not derived from the inoculate and not found in all animals from the same group (Fig. 8A).
Figure 8.
Efficiency of the FMT based on the number of genera shared between FMT inoculate and recipient samples and Bray-Curtis dissimilarity. (A) Genera that were shared between FMT inoculate and at least one recipient sample. (B) Genera that were shared between FMT inoculate and all recipient samples. (C) Bray-Curtis dissimilarity. Samples from all groups were pooled together to evaluate dissimilarities of microbiome profiles between groups, within groups, between FMT recipients and No FMT controls, and between FMT recipients and the inoculate. Mean and SEM are shown. n = 12 animals per group.
When comparing Bray-Curtis dissimilarity measures, the greatest level of similarity was found within mice from the same treatment group, whereas a high level of dissimilarity was observed between mice that received FMT and no FMT control mice (Fig. 8C). The greatest dissimilarity was observed between FMT inoculate and recipient samples, indicating that the processing of inoculates, which involved centrifugation and dilution, significantly impacted bacterial profiles.
H. Fecal Microbiome Analysis of FMT Recipients
The most prevalent phyla and genera in fecal material from FMT recipients and in FMT inoculates are summarized in an online repository [34]. The most abundant phylum in FMT recipient samples was Bacteroidetes, followed by Firmicutes [34]. The most abundant genus was from Bacteroidales S24-7 group, followed by Lachnospiraceae NK4A136 and Desulfovibrio. The dominant phyla and genera were the same in FMT recipients as those found in the dams and offspring exposed to maternal DHT, but differed in their relative abundances across samples.
FMT recipient samples did not contain the Akkermansia overgrowth that was observed in dams. FMT recipient samples contained a markedly lower proportion of Rikenellaceae RC9 and a higher proportion of Lachnospiraceae NK4A136 than maternal DHT exposed samples.
FMT inoculates contained a greater proportion of bacteria from the phylum Proteobacteria and a lower proportion of Bacteroidetes than FMT recipient and maternal DHT exposed samples [34]. The dominant genus in FMT inoculates was Desulfovibrio, followed by Akkermansia. The sterile PBS/glycerol solution used to inoculate no-FMT control mice was largely devoid of bacteria and composed almost exclusively of the phylum Proteobacteria, presumably due to reagent contamination, which was amplified in the absence of other bacteria in the solution.
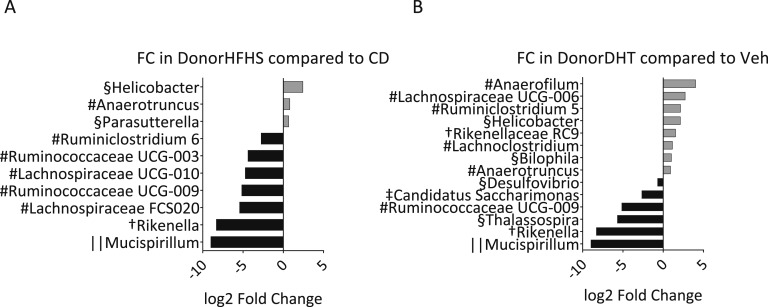
DESeq2 analysis identified several differentially abundant bacterial genera due to donor diet and maternal DHT exposure (Fig. 9). Donor HFHS diet led to a lower relative abundance of bacteria from the families Ruminococcaceae and Lachnospiraceae and the genus Rikenella (Fig. 9A). These same groups were also found to be less abundant in the corresponding mice exposed to maternal DHT (Figs. 3B and 3C). Donor DHT exposure led to a lower relative abundance of the genera Thalassospira and Rikenella, which were also less abundant in the corresponding maternal DHT exposed mice (Figs. 9B and 3D).
Figure 9.
Differentially abundant genera in FMT recipient. Log twofold change (FC) of relative abundance for genera that were found by DESeq 2 analysis to be significantly changed due to (A) donor diet and (B) DHT exposure. P < 0.05 for all after Benjamini-Hochberg false discovery rate correction. #Firmicutes; †Bacteroidetes; §Proteobacteria; ||Deferribacteres; ‡Saccharibacteria. n = 24 animals per tested factor.
I. α Diversity in FMT Recipients
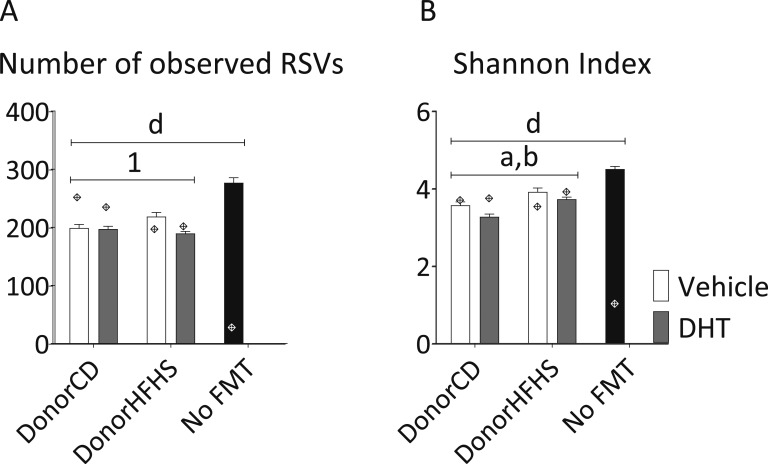
Mice receiving FMT from HFHS-diet fed donors showed increased bacterial richness, which was attenuated by DHT exposure (donor diet*donor maternal DHT exposure, P < 0.05) (Fig. 10A). Donor DHT exposure led to a decreased Shannon index in FMT recipients, whereas donor HFHS-diet led to an increase (main effect of donor diet and donor maternal DHT exposure, P < 0.05) (Fig. 10B).
Figure 10.
Alpha diversity of fecal samples from FMT recipients and inoculates. The number of observed (A) RSVs and (B) Shannon index. Diamonds show the diversity of the respective FMT inoculate. Two-way ANOVA main effect of (a) donor diet and (b) donor maternal DHT exposure (P < 0.05). Two-way ANOVA interaction between (1) donor diet and donor maternal DHT exposure (P < 0.05). P < 0.05 for (d) all groups compared with No FMT. Mean and SEM are shown. n = 12 animals per group.
Both the number of observed RSVs and the Shannon index were significantly lower in all FMT recipients compared with no FMT controls, reflecting the antibiotic effect (Fig. 10). FMT recipients showed comparable diversity values to their respective inoculates, indicating that the FMT successfully restored bacterial diversity after antibiotic-induced depletion.
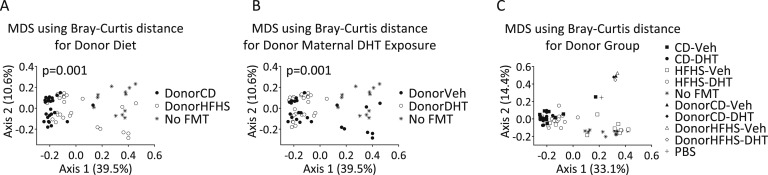
J. β Diversity in FMT Recipients
In β diversity analyses, both donor diet and donor DHT exposure significantly affected clustering in the MDS plot based on Bray-Curtis distance matrices (Figs. 11A and 11B). All fecal samples from mice clustered separately from all FMT inoculate samples, highlighting the bacterial community differences between these two sample types (Fig. 11C).
Figure 11.
Beta diversity of fecal samples from FMT recipients and inoculates. MDS plots of Bray-Curtis distances for the (A) factors donor diet, (B) donor maternal DHT exposure, and (C) donor group. No FMT animals received PBS instead of fecal inoculate. Each dot represents the total bacterial community composition of one sample. The amount of variation explained by each MDS coordinate is indicated in brackets. Groups were compared using Adonis. PBS, phosphate-buffered saline.
3. Discussion
This study describes the gut microbiome of a mouse model of PCOS induced by maternal DHT exposure combined with maternal and offspring HFHS-diet induced obesity. We demonstrate sex-specific effects on the gut microbiome of offspring exposed to maternal androgens. In male offspring, we found a DHT-dependent reduction in bacterial richness and diversity. This result agrees with the findings of our previous study of women with PCOS, and with a recent study of a letrozole-induced mouse model of PCOS [12, 15]. On the contrary, DHT exposure tended to increase richness in female offspring, with the exception of the MatHFHS-HFHS groups, where richness was decreased and lowest in mice exposed to maternal DHT. Although high diversity is usually considered a sign of a stable microbial community, an unexpected increase in diversity compared with the control group might also be indicative of a dysfunctional microbiome [35].
Both maternal and offspring diet significantly affected microbiome parameters. Female offspring were more susceptible than males to α diversity changes due to maternal diet, whereas in males the maternal diet had a greater effect on overall community composition. Female mice appeared to be more susceptible to both DHT exposure and offspring HFHS-diet than males, with a larger number of differentially abundant genera identified by DESeq2. The majority of genera identified by DESeq2 were unique for males and females; however, Lachnospiraceae NK4A136 and Bifidobacterium showed a concordant decrease in relative abundance due to offspring HFHS diet in both male and female offspring. Sex-specific responses of the gut microbiome to diet in mice have been reported previously. Gonadectomy has been reported to cause changes in the microbiome profiles of both male and female mice and this effect was more prevalent in males on a CD and in females on a high-fat diet [36]. These results fit well with our observations, where a DHT-induced diversity reduction was stronger in males on a CD and females on a HFHS-diet.
Contradictory data exist on the effect of diet-induced obesity on bacterial richness and diversity in mice, with different studies reporting a decrease [37], an increase [38, 39], or no change [40]. High-fat diet-induced gut microbiome changes may depend on diet composition rather than diet-induced obesity, as isocaloric diets containing different fat sources were shown to induce specific gut microbiome changes in mice [41]. The gut microbiome is highly variable depending on genetic, environmental, and individual factors, with “individual” and “time” described as two of the strongest contributors to variation in the mouse gut microbiome [42, 43]. Our experiment involved both a large number of individual mice and a long duration; therefore, changes induced by diet or androgen exposure may have been affected by these factors.
The overall bacterial composition in male and female samples exposed to maternal DHT agreed with previously published data from the same mouse strain [44]. In our study, samples from dams showed an unexpected bloom of Akkermansia. Such blooms of single species can occur due to a loss of stability in the intestinal ecosystem as a result of stress, disease, or antibiotic treatment, among other factors [45]. Pregnancy may represent a stressful situation in itself and blooms of single taxa, including Akkermansia, have been reported [46, 47]. The bloom appeared not to have been carried over to the offspring or FMT mice.
We performed FMT to test the hypothesis that the gut microbiome contributes to reproductive and behavioral dysfunction in this mouse model. Few data exist on the effect of the gut microbiome on reproductive function. It has been reported that male germ-free (GF) mice have lower testosterone levels than specific pathogen-free mice, and transfer of male-type microbiota to female GF mice resulted in increased testosterone levels [48, 49]. In a rat model of PCOS, FMT from healthy donors was able to reverse hyperandrogenemia, disrupted estrous cyclicity, and polycystic ovarian morphology after just 2 weeks [16]. Although these data suggest that the gut microbiome may have the potential to affect reproductive function, no such effect was discernible in our study. WT FMT recipients did not show changes in estrous cyclicity compared with the untreated control group. Variation within the groups of mice was high, and a larger number of mice or a longer observation period may be necessary to observe subtle changes. In our study setup, the maternal DHT-HFHS exposure preceded the gut microbiome changes. This effect could be one-directional without a reciprocal effect on reproductive function in the FMT recipients, or it can be that DHT-HFHS exposure causes both but they are not related. Thus, we cannot completely exclude that lack of an effect of the microbiome changes seen in offspring under these specific experimental conditions are not causal.
Various gut microbiome manipulations have been shown to affect anxietylike behavior in mice, including GF breeding, antibiotic and prebiotic treatment, and FMT from humans suffering from depression [11, 50]. Though FMT led to changes in anxietylike behavior in our study, these changes did not mirror the donor phenotype. Instead of the expected increase in anxietylike behavior in mice receiving FMT from DHT-exposed donors, we found no significant difference in three of the four recipient groups. In the DonorHFHS-DHT group, we observed a pronounced anxiolytic response in the OF test. Due to these contradictory results, we cannot conclude that the gut microbiome was involved in mediating anxiety-related effects.
Antibiotic treatment has been reported to affect anxietylike behavior in mice, even if nonabsorbable antibiotics are used [24, 51]. We detected no negative effects of the antibiotic treatment on body weight or food intake, and antibiotic-treated animals did not perform differently to no-FMT control mice in the EPM or OF. However, we observed a nonsignificant decrease in three of the four FMT groups in the time spent in the center of the OF compared with no FMT controls, indicating that the antibiotic administration may have the potential to promote anxietylike behavior in this setting.
All mice receiving FMT had significantly lower α diversity measures than non-FMT controls, which was incompletely restored by FMT. As the inoculates had been diluted in preparation for oral gavage and to enable the use of the same inoculate stocks throughout the whole experiment, the lower α diversity observed in FMT recipients may be due to the fact that the inoculates themselves were not more diverse. Had undiluted material been used, diversity levels after FMT may have risen to those of non-FMT controls.
The overall community composition between inoculate and recipient samples differed substantially, as seen in β diversity plots and comparisons of Bray-Curtis dissimilarity. This was expected, as inoculates were processed prior to FMT and mice were exposed to bacteria from external sources. We did not aim to achieve perfect colonization efficiency with our protocol, but rather to seed the recipient gut with dominant donor microbes and allow the bacterial community to organize and stabilize around these species. Booster FMT doses were given once weekly until the end of the experiment to reinforce the donor microbiome profile. We found comparable colonization efficiency, measured in the number of shared genera between inoculate and recipient, in all FMT groups. A large percentage of the core microbial community, found in all FMT recipients, was derived from the FMT inoculate. Several genera that showed altered relative abundances in FMT donors showed concordant changes in FMT recipients, including a downregulation of bacteria from the families Lachnospiraceae and Ruminococcaceae in response to a HFHS diet and a downregulation of Thalassospira and Rikenella in response to DHT exposure. We conclude that our FMT protocol was successful in transferring various aspects of the donor microbiome, including dominant taxa, discriminant taxa for the donor exposures, and α diversity characteristics.
For our next-generation sequencing approach, we sequenced the V4 segment of the 16S rRNA gene to achieve a full overlap of forward and reverse reads, leading to more accurate sequence inference [52]. We opted to use the DADA2 algorithm rather than operational taxonomic unit (OTU)-picking. DADA2 does not perform clustering according to a predefined similarity threshold and thereby avoids the widespread problem of artificial OTUs as a result of sequencing errors [30]. DADA2 calculates sequencing error rates and infers sequences ad hoc and has been shown to outperform other OTU-picking methods on various mock community and biological data sets [30].
To conclude, we showed that maternal DHT exposure and maternal and offspring diet affect gut microbiome profiles in mice in a sex-specific manner. We showed that FMT did not transfer the disrupted estrous cycles and anxietylike behavior observed in DHT-exposed female offspring, suggesting that these effects were likely not microbiome-mediated.
Acknowledgments
The authors thank Rodrigo Marcondes, Manuel Maliqueo, and Qi Xiaojuan for assistance with animal procedures, Stefano Gastaldello for providing laboratory workspace, and members of the Center for Translational Microbiome Research at the Science for Life Laboratory in Stockholm, Sweden, for assistance with next-generation sequencing.
Financial Support: This work is part of the PhD thesis of Lisa Lindheim, who received funding from the Austrian Science Fund FWF (W1241) and the Medical University of Graz through the PhD program Molecular Fundamentals of Inflammation (DK-MOLIN). This work was further funded by the Swedish Medical Research Council (Project No. 2014-2775); Jane and Dan Ohlsson Foundation; the Adlerbert Research Foundation; the Novo Nordisk Foundation (NNF16OC0020744 and NNF17OC0026724); the Strategic Research Programme (SRP) in Diabetes at Karolinska Institutet; the Swedish federal government under the LUA/ALF agreement ALFGBG-429501, the Regional agreement on medical training and clinical research (ALF) between Stockholm County Council and Karolinska Institutet, and Vetenskapsrådet (2014-2775) (all E.S.V.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Disclosure Summary: The authors have nothing to disclose.
Glossary
Abbreviations:
- CD
control diet
- EPM
elevated plus maze
- FMT
fecal microbiota transplantation
- GD
gestational day
- GF
germ-free
- HFHS
high-fat, high-sugar
- MDS
multidimensional scaling
- OF
open field
- OTU
operational taxonomic unit
- PCOS
polycystic ovary syndrome
- RSV
ribosomal sequence variant
- Veh
vehicle
- WT
wild-type
References and Notes
- 1. Rotterdam ESHRE/ASRM-Sponsored PCOS Consensus Workshop Group Revised 2003 consensus on diagnostic criteria and long-term health risks related to polycystic ovary syndrome (PCOS). Hum Reprod. 2004;19:41–47. [DOI] [PubMed] [Google Scholar]
- 2. Spritzer PM, Lecke SB, Satler F, Morsch DM. Adipose tissue dysfunction, adipokines, and low-grade chronic inflammation in polycystic ovary syndrome. Reproduction. 2015;149(5):R219–R227. [DOI] [PubMed] [Google Scholar]
- 3. Cooney LG, Lee I, Sammel MD, Dokras A. High prevalence of moderate and severe depressive and anxiety symptoms in polycystic ovary syndrome: a systematic review and meta-analysis. Hum Reprod. 2017;32(5):1075–1091. [DOI] [PubMed] [Google Scholar]
- 4. Chan JL, Kar S, Vanky E, Morin-Papunen L, Piltonen T, Puurunen J, Tapanainen JS, Maciel GAR, Hayashida SAY, Soares JM Jr, Baracat EC, Mellembakken JR, Dokras A. Racial and ethnic differences in the prevalence of metabolic syndrome and its components of metabolic syndrome in women with polycystic ovary syndrome: a regional cross-sectional study. Am J Obstet Gynecol. 2017;217(2):189.e1–189.e8. [DOI] [PubMed] [Google Scholar]
- 5. Azziz R, Carmina E, Chen Z, Dunaif A, Laven JSE, Legro RS, Lizneva D, Natterson-Horowtiz B, Teede HJ, Yildiz BO. Polycystic ovary syndrome. Nat Rev Dis Primers. 2016;2:16057. [DOI] [PubMed] [Google Scholar]
- 6. Bäckhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, Semenkovich CF, Gordon JI. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci USA. 2004;101(44):15718–15723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444(7122):1027–1031. [DOI] [PubMed] [Google Scholar]
- 8. Ellekilde M, Selfjord E, Larsen CS, Jakesevic M, Rune I, Tranberg B, Vogensen FK, Nielsen DS, Bahl MI, Licht TR, Hansen AK, Hansen CHF. Transfer of gut microbiota from lean and obese mice to antibiotic-treated mice. Sci Rep. 2014;4(1):5922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Le Chatelier E, Nielsen T, Qin J, Prifti E, Hildebrand F, Falony G, Almeida M, Arumugam M, Batto JM, Kennedy S, Leonard P, Li J, Burgdorf K, Grarup N, Jørgensen T, Brandslund I, Nielsen HB, Juncker AS, Bertalan M, Levenez F, Pons N, Rasmussen S, Sunagawa S, Tap J, Tims S, Zoetendal EG, Brunak S, Clément K, Doré J, Kleerebezem M, Kristiansen K, Renault P, Sicheritz-Ponten T, de Vos WM, Zucker JD, Raes J, Hansen T, Bork P, Wang J, Ehrlich SD, Pedersen O; MetaHIT Consortium . Richness of human gut microbiome correlates with metabolic markers. Nature. 2013;500(7464):541–546. [DOI] [PubMed] [Google Scholar]
- 10. Forslund K, Hildebrand F, Nielsen T, Falony G, Le Chatelier E, Sunagawa S, Prifti E, Vieira-Silva S, Gudmundsdottir V, Pedersen HK, Arumugam M, Kristiansen K, Voigt AY, Vestergaard H, Hercog R, Costea PI, Kultima JR, Li J, Jørgensen T, Levenez F, Dore J, Nielsen HB, Brunak S, Raes J, Hansen T, Wang J, Ehrlich SD, Bork P, Pedersen O, Pedersen O; MetaHIT Consortium . Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature. 2015;528(7581):262–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zheng P, Zeng B, Zhou C, Liu M, Fang Z, Xu X, Zeng L, Chen J, Fan S, Du X, Zhang X, Yang D, Yang Y, Meng H, Li W, Melgiri ND, Licinio J, Wei H, Xie P. Gut microbiome remodeling induces depressive-like behaviors through a pathway mediated by the host’s metabolism. Mol Psychiatry. 2016;21(6):786–796. [DOI] [PubMed] [Google Scholar]
- 12. Lindheim L, Bashir M, Münzker J, Trummer C, Zachhuber V, Leber B, Horvath A, Pieber TR, Gorkiewicz G, Stadlbauer V, Obermayer-Pietsch B. Alterations in gut microbiome composition and barrier function are associated with reproductive and metabolic defects in women with polycystic ovary syndrome (PCOS): a pilot study. PLoS One. 2017;12(1):e0168390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Liu R, Zhang C, Shi Y, Zhang F, Li L, Wang X, Ling Y, Fu H, Dong W, Shen J, Reeves A, Greenberg AS, Zhao L, Peng Y, Ding X. Dysbiosis of gut microbiota associated with clinical parameters in polycystic ovary syndrome. Front Microbiol. 2017;8:324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Torres PJ, Siakowska M, Banaszewska B, Pawelczyk L, Duleba AJ, Kelley ST, Thackray VG. Gut microbial diversity in women with polycystic ovary syndrome correlates with hyperandrogenism. J Clin Endocrinol Metab. 2018;103(4):1502–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kelley ST, Skarra DV, Rivera AJ, Thackray VG. The gut microbiome is altered in a letrozole-induced mouse model of polycystic ovary syndrome. PLoS One. 2016;11(1):e0146509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Guo Y, Qi Y, Yang X, Zhao L, Wen S, Liu Y, Tang L. Association between polycystic ovary syndrome and gut microbiota. PLoS One. 2016;11(4):e0153196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Maliqueo M, Benrick A, Stener-Victorin E. Rodent models of polycystic ovary syndrome: phenotypic presentation, pathophysiology, and the effects of different interventions. Semin Reprod Med. 2014;32(3):183–193. [DOI] [PubMed] [Google Scholar]
- 18. Caldwell ASL, Middleton LJ, Jimenez M, Desai R, McMahon AC, Allan CM, Handelsman DJ, Walters KA. Characterization of reproductive, metabolic, and endocrine features of polycystic ovary syndrome in female hyperandrogenic mouse models. Endocrinology. 2014;155(8):3146–3159. [DOI] [PubMed] [Google Scholar]
- 19. Kauffman AS, Thackray VG, Ryan GE, Tolson KP, Glidewell-Kenney CA, Semaan SJ, Poling MC, Iwata N, Breen KM, Duleba AJ, Stener-Victorin E, Shimasaki S, Webster NJ, Mellon PL. A novel letrozole model recapitulates both the reproductive and metabolic phenotypes of polycystic ovary syndrome in female mice. Biol Reprod. 2015;93(3):69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rosario FJ, Kanai Y, Powell TL, Jansson T. Increased placental nutrient transport in a novel mouse model of maternal obesity with fetal overgrowth. Obesity (Silver Spring). 2015;23(8):1663–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Manti M, Fornes R, Qi X, Folmerz E, Hirschberg AL, de Castro Barbosa T, Maliqueo M, Benrick A, Stener-Victorin E. Maternal androgen excess and obesity induce sexually dimorphic anxiety-like behavior in the offspring. FASEB J. 2018;32(8):4158–4171. [DOI] [PubMed] [Google Scholar]
- 22. Byers SL, Wiles MV, Dunn SL, Taft RA. Mouse estrous cycle identification tool and images. PLoS One. 2012;7(4):e35538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Caligioni CS. Assessing reproductive status/stages in mice. Curr Protoc Neurosci. 2009; 48:A.4I.1–A.4I.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bercik P, Denou E, Collins J, Jackson W, Lu J, Jury J, Deng Y, Blennerhassett P, Macri J, McCoy KD, Verdu EF, Collins SM. The intestinal microbiota affect central levels of brain-derived neurotropic factor and behavior in mice. Gastroenterology. 2011;141(2):599–609, 609.e1–609.e3. [DOI] [PubMed] [Google Scholar]
- 25. Bruce-Keller AJ, Salbaum JM, Luo M, Blanchard E IV, Taylor CM, Welsh DA, Berthoud H-R. Obese-type gut microbiota induce neurobehavioral changes in the absence of obesity. Biol Psychiatry. 2015;77(7):607–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gacias M, Gaspari S, Santos P-MG, Tamburini S, Andrade M, Zhang F, Shen N, Tolstikov V, Kiebish MA, Dupree JL, Zachariou V, Clemente JC, Casaccia P. Microbiota-driven transcriptional changes in prefrontal cortex override genetic differences in social behavior. eLife. 2016;5:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. He B, Nohara K, Ajami NJ, Michalek RD, Tian X, Wong M, Losee-Olson SH, Petrosino JF, Yoo S-H, Shimomura K, Chen Z. Transmissible microbial and metabolomic remodeling by soluble dietary fiber improves metabolic homeostasis. Sci Rep. 2015;5(1):10604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hill DA, Hoffmann C, Abt MC, Du Y, Kobuley D, Kirn TJ, Bushman FD, Artis D. Metagenomic analyses reveal antibiotic-induced temporal and spatial changes in intestinal microbiota with associated alterations in immune cell homeostasis. Mucosal Immunol. 2010;3(2):148–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011;17:10. [Google Scholar]
- 30. Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13(7):581–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8(4):e61217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Leinonen R, Sugawara H, Shumway M, International Nucleotide Sequence Database Collaboration . The sequence read archive. Nucleic Acids Res. 2010;39:D19–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lindheim L, Manti M, Fornes R, Bashir M, Czarnewski P, Diaz OE, Seifert M, Engstrand L, Villablanca EJ, Obermayer-Pietsch B, Stener-Victorin E. Data from: Reproductive and behavior dysfunction induced by maternal androgen exposure and obesity is likely not gut microbiome-mediated. Dryad 2018. Deposited 4 October 2018. https://datadryad.org/resource/doi:10.5061/dryad.7h4338m. [DOI] [PMC free article] [PubMed]
- 35. Shade A. Diversity is the question, not the answer. ISME J. 2017;11(1):1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Org E, Mehrabian M, Parks BW, Shipkova P, Liu X, Drake TA, Lusis AJ. Sex differences and hormonal effects on gut microbiota composition in mice. Gut Microbes. 2016;7(4):313–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Turnbaugh PJ, Bäckhed F, Fulton L, Gordon JI, Bäckhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe. 2008;3(4):213–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Evans CC, LePard KJ, Kwak JW, Stancukas MC, Laskowski S, Dougherty J, Moulton L, Glawe A, Wang Y, Leone V, Antonopoulos DA, Smith D, Chang EB, Ciancio MJ. Exercise prevents weight gain and alters the gut microbiota in a mouse model of high fat diet-induced obesity. PLoS One. 2014;9(3):e92193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Shang Y, Khafipour E, Derakhshani H, Sarna LK, Woo CW, Siow YL, O K. Short term high fat diet induces obesity-enhancing changes in mouse gut microbiota that are partially reversed by cessation of the high fat diet. Lipids. 2017;52(6):499–511. [DOI] [PubMed] [Google Scholar]
- 40. Kim K-A, Gu W, Lee I-A, Joh E-H, Kim D-H. High fat diet-induced gut microbiota exacerbates inflammation and obesity in mice via the TLR4 signaling pathway. PLoS One. 2012;7(10):e47713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Huang EY, Leone VA, Devkota S, Wang Y, Brady MJ, Chang EB. Composition of dietary fat source shapes gut microbiota architecture and alters host inflammatory mediators in mouse adipose tissue. JPEN J Parenter Enteral Nutr. 2013;37(6):746–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hildebrand F, Nguyen TL, Brinkman B, Yunta RG, Cauwe B, Vandenabeele P, Liston A, Raes J. Inflammation-associated enterotypes, host genotype, cage and inter-individual effects drive gut microbiota variation in common laboratory mice. Genome Biol. 2013;14(1):R4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hoy YE, Bik EM, Lawley TD, Holmes SP, Monack DM, Theriot JA, Relman DA. Variation in taxonomic composition of the fecal microbiota in an inbred mouse strain across individuals and time. PLoS One. 2015;10(11):e0142825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gu S, Chen D, Zhang J-N, Lv X, Wang K, Duan L-P, Nie Y, Wu X-L. Bacterial community mapping of the mouse gastrointestinal tract. PLoS One. 2013;8(10):e74957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Stecher B, Maier L, Hardt W-D. ‘Blooming’ in the gut: how dysbiosis might contribute to pathogen evolution. Nat Rev Microbiol. 2013;11(4):277–284. [DOI] [PubMed] [Google Scholar]
- 46. Jašarević E, Howard CD, Misic AM, Beiting DP, Bale TL. Stress during pregnancy alters temporal and spatial dynamics of the maternal and offspring microbiome in a sex-specific manner. Sci Rep. 2017;7(1):44182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gohir W, Whelan FJ, Surette MG, Moore C, Schertzer JD, Sloboda DM. Pregnancy-related changes in the maternal gut microbiota are dependent upon the mother’s periconceptional diet. Gut Microbes. 2015;6(5):310–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Markle JG, Frank DN, Mortin-Toth S, Robertson CE, Feazel LM, Rolle-Kampczyk U, von Bergen M, McCoy KD, Macpherson AJ, Danska JS. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science. 2013;339(6123):1084–1088. [DOI] [PubMed] [Google Scholar]
- 49. Yurkovetskiy L, Burrows M, Khan AA, Graham L, Volchkov P, Becker L, Antonopoulos D, Umesaki Y, Chervonsky AV. Gender bias in autoimmunity is influenced by microbiota. Immunity. 2013;39(2):400–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Luczynski P, McVey Neufeld K-A, Oriach CS, Clarke G, Dinan TG, Cryan JF. Growing up in a bubble: using germ-free animals to assess the influence of the gut microbiota on brain and behavior. Int J Neuropsychopharmacol. 2016;19(8):pyw020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tochitani S, Ikeno T, Ito T, Sakurai A, Yamauchi T, Matsuzaki H. Administration of non-absorbable antibiotics to pregnant mice to perturb the maternal gut microbiota is associated with alterations in offspring behavior. PLoS One. 2016;11(1):e0138293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kozich JJ, Westcott SL, Baxter NT, Highlander SK, Schloss PD. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl Environ Microbiol. 2013;79(17):5112–5120. [DOI] [PMC free article] [PubMed] [Google Scholar]