Abstract
Background
CC-type glutaredoxins (GRXs) are plant-specific glutaredoxin, play regulatory roles in response of biotic and abiotic stress. However, it is not clear whether the CC-type GRXs are involve in drought response in cassava (Manihot esculenta), an important tropical tuber root crop.
Results
Herein, genome-wide analysis identified 18 CC-type GRXs in the cassava genome, of which six (namely MeGRXC3, C4, C7, C14, C15, and C18) were induced by drought stress in leaves of two cassava cultivars Argentina 7 (Arg7) and South China 124 (SC124). Exogenous abscisic acid (ABA) application induced the expression of all the six CC-type GRXs in leaves of both Arg7 and SC124 plants. Overexpression of MeGRXC15 in Arabidopsis (Col-0) increases tolerance of ABA on the sealed agar plates, but results in drought hypersensitivity in soil-grown plants. The results of microarray assays show that MeGRXC15 overexpression affected the expression of a set of transcription factors which involve in stress response, ABA, and JA/ET signalling pathway. The results of protein interaction analysis show that MeGRXC15 can interact with TGA5 from Arabidopsis and MeTGA074 from cassava.
Conclusions
CC-type glutaredoxins play regulatory roles in cassava response to drought possibly through ABA signalling pathway.
Electronic supplementary material
The online version of this article (10.1186/s12870-018-1528-6) contains supplementary material, which is available to authorized users.
Keywords: Cassava (Manihot esculenta), Drought response, ABA, CC-type glutaredoxin, Transgenic Arabidopsis, TGA factors
Background
As a tropical crop, cassava (Manihot esculenta) evolved different responses to drought stress, such as quick stomata closure, reduction of photosynthetic proteins levels and photosynthetic capacity, induction of senescence in older leaves, and size reduction of leave epidermal cells [1, 2]. The cassava cultivar Argentina 7 (Arg7) display faster senescence in older leaves than the cassava cultivar South China 124 (SC124) [2]. Senescence in cassava is partly controlled by reactive oxygen species (ROS) and ethylene (ET) signaling [3]. Increasing ROS scavenging ability in cassava delays leaf senescence under drought stress [3–5]. It is therefore necessary to analyze genes involved in these pathways for a deeper functional characterization.
Glutaredoxin (GRX) is one of the most important protein modification system in plants [6]. The glutathione/GRX (GSH/GRX) system is essential for redox homeostasis and ROS signalling in plant cells [7]. GRX target proteins are involved in all aspects of plant growth, including basal metabolism, iron/sulfur cluster formation, development, adaptation to the environment, and stress responses [7]. GRX are in particular studied for their involvement in oxidative stress responses [7–9]. GRXs are classified in five subgroups, among which CC-type GRXs are a plant-specific subgroup, also known as the ROXY family in Arabidopsis [10, 11]. CC-type GRXs likely evolved from the CPYC subgroup and expanded during land plant evolution [11]. There are only two CC-type GRXs in the basal land plant Physcomitrella, but between 15 and 24 members in land plants such as rice, Arabidopsis, Vitis and Populus [11]. However, the number of CC-type GRXs in cassava remains unclear.
CC-type GRXs are characterized by the presence of a redox site CC*(C/S/G) as well as their disulfide reductase activity that uses glutathione as cofactor [12]. The first CC-type GRX has been identified as a regulator of petal development [10]. However, CC-type GRXs are also involved in jasmonic acid (JA) / ET mediated biotic stress responses through the interaction with TGA transcription factors in Arabidopsis [13–15]. Moreover, a CC-type GRX GRXS13 is critical in limiting basal and photo-oxidative stress-induced ROS production [16]. Thus, CC-type GRXs may play a key role in the crosstalk between ROS and ethylene. CC-type GRX members are also involved in organ development and biotic stress responses in other plants [13, 17–21]. Since ROS and ethylene signal transduction pathways are involved in cassava drought responses [3], CC-type GRXs might play regulatory roles in these pathways in cassava.
During evolution, CC-type GRXs might have gained new functions in higher land plants [6, 11, 13]. Although several CC-type GRXs have been characterized in Arabidopsis and rice [13, 21], no previous work has profiled them in cassava. Moreover, protein interactions and enzymatic activities during abiotic stress responses in cassava are equally important [2], but currently not well understood. With the recent release and annotation of the cassava genome [22, 23], it is now possible to identify drought responsive CC-type GRX genes and to better characterize their functions and interactors during drought response.
Based on our previously reported transcriptomic data of cassava cultivars [24], we identified six CC-type GRXs (MeGRXC3, C4, C7, C14, C15, and C18) that responded to drought using qPCR analysis in cassava leaves. Expression of the six drought-responsive CC-type GRXs is also induced in cassava leaves by application of exogenous ABA. Our results showed that CC-type GRXs may function as a component in drought stress in an ABA-dependent pathway in both Arg7 and SC124 plants. Furthermore, we found that MeGRXC15 is specifically expressed by exposure to drought in different tissues including leaf, petiole, and abscission zone. Overexpression of MeGRXC15 in Arabidopsis induces insensitivity to ABA on sealed agar plates, and confers drought susceptibility in soil-grown conditions. In addition, gene expression analysis reveals that MeGRXC15 overexpression in Arabidopsis altered the expression of a set of genes which involve in multiple stress responses, ABA, and JA/ET signalling pathways. Protein-protein interaction analysis reveals that MeGRXC15 could interact with TGA transcription factor from both Arabidopsis and cassava. Together, our study could increase the understanding of cassava gene regulation under the condition of drought stress and expand the knowledge of land plant specific CC-type GRXs.
Results
Phylogenetic and protein sequence analysis of cassava CC-type GRXs
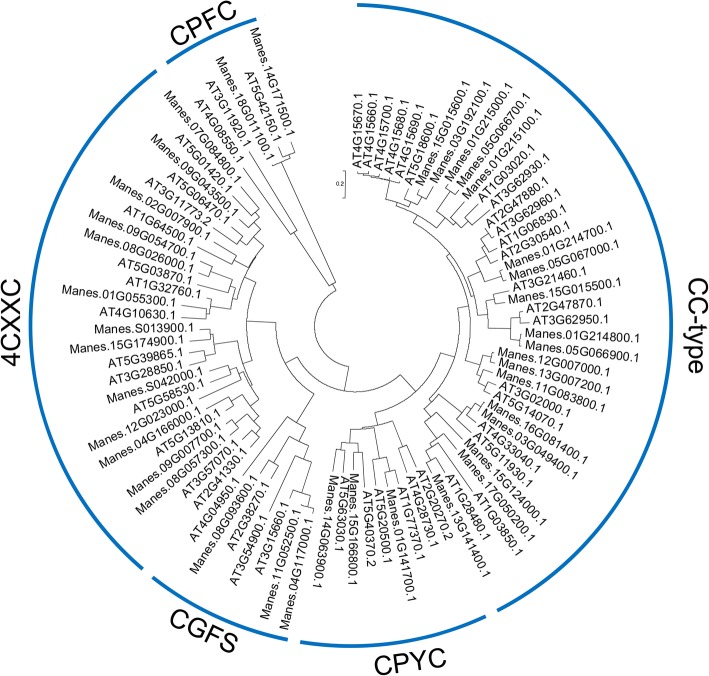
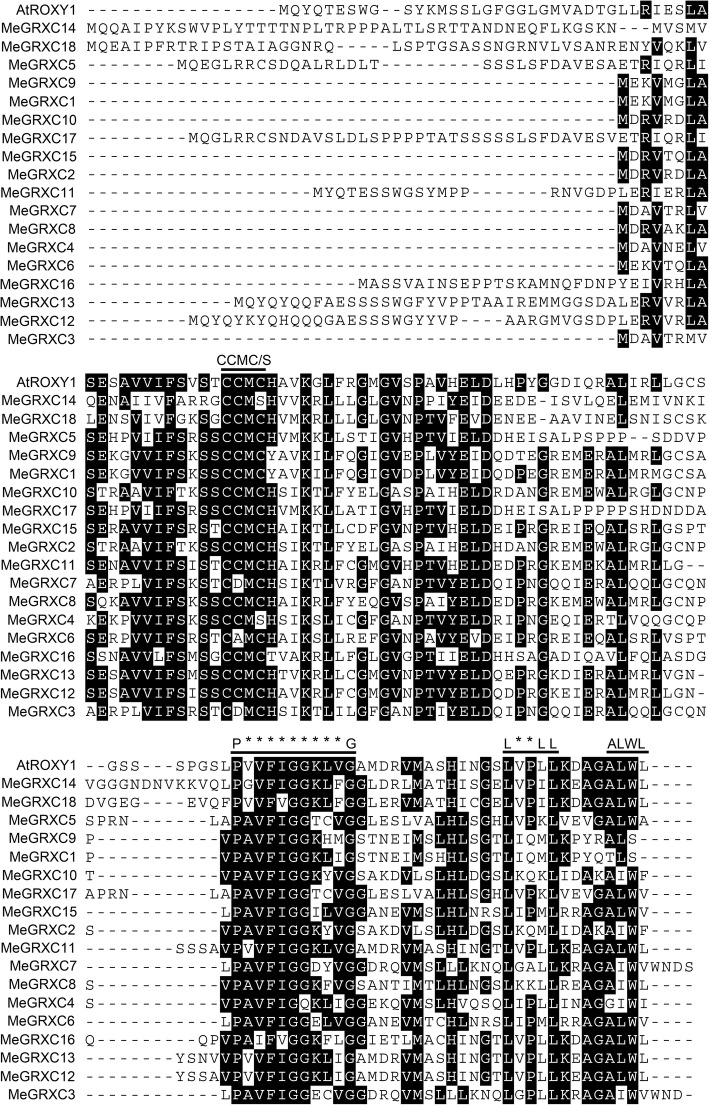
We predicted a total of 39 putative GRX proteins in the cassava genome using the Arabidopsis ROXYs in a BLAST search against the genome of the cassava cultivar AM560 (https://phytozome.jgi.doe.gov/pz/portal.html, Manihot esculenta v6.1). To understand the relationship between GRX proteins in cassava and Arabidopsis, we built a neighbor-joining phylogenetic tree using MEGA5.0 on the basis of the protein sequences in Additional file 1: Table S1. The results show that many cassava GRXs are highly similar to their Arabidopsis counterparts (Fig. 1). We found that the CC-type subgroup had the most members among five GRX subgroups in cassava. Our analysis predicted 18 full-length CC-type GRX members of cassava (Table 1), less than the 21 of Arabidopsis [25]. Cassava CC-type GRX genes are represented on nine chromosomes (Table 1). 16 of cassava CC-type GRXs share an ALWL motif at the C-terminus and extend to A(G)L(I)WL(A/F/V/I) (Fig. 2, Table 1). However, two members, MeGRXC1 and MeGRXC9, are not present ALWL motif at the C-terminus (Fig. 2, Table 1). CC-type GRXs have a distinctive conserved CC(M/L)(C/S) redox site motif in Arabidopsis, whereas this motif extends to C(C/G/F/Y/P)(M/L)(C/S/I/A) in rice [6, 11, 13, 25]. Most cassava CC-type GRXs shares a distinctive CCM(C/S) redox site (Fig. 2, Table 1). However, CDMC is extended in two CC-type GRXs (MeGRXC3 and MeGRXC7) and CAMC is extended in MeGRXC6 in cassava (Fig. 2, Table 1).
Fig. 1.
Phylogenetic tree of glutaredoxins (GRXs) from cassava and Arabidopsis. Members of GRXs were classified by their redox activate site
Table 1.
CC-type GRXs in cassava genome
| JGI identifier V4.1 | JGI identifier V6.1 | Chromosome location | Gene Symbol | Redox Site | ALWL-motif | AA |
|---|---|---|---|---|---|---|
| cassava4.1_019956m | Manes.01G214700.1 | Chr01:30392540..30393324 | MeGRXC1 | CCMC | – | 102 |
| cassava4.1_024597m | Manes.01G214800.1 | Chr01:30394902..30395210 | MeGRXC2 | CCMC | AIWF | 103 |
| cassava4.1_033785m | Manes.01G215000.1 | Chr01:30421882..30422775 | MeGRXC3 | CDMC | AIWV | 105 |
| cassava4.1_027058m | Manes.01G215100.1 | Chr01:30426232..30427641 | MeGRXC4 | CCMS | GIWI | 103 |
| cassava4.1_018918m | Manes.03G049400.1 | Chr03:4265637..4266041 | MeGRXC5 | CCMC | ALWA | 135 |
| cassava4.1_027873m | Manes.03G192100.1 | Chr03: 27589330..27589919 | MeGRXC6 | CAMC | ALWV | 102 |
| cassava4.1_025892m | Manes.05G066700.1 | Chr05:5107836..5108531 | MeGRXC7 | CDMC | AIWV | 106 |
| cassava4.1_021286m | Manes.05G066900.1 | Chr05:5120724..5122777 | MeGRXC8 | CCMC | AIWL | 103 |
| cassava4.1_019954m | Manes.05G067000.1 | Chr05:5123236..5123953 | MeGRXC9 | CCMC | – | 102 |
| cassava4.1_024608m | Manes.11G083800.1 | Chr11:11464621..11464992 | MeGRXC10 | CCMC | AIWF | 124 |
| cassava4.1_032936m | Manes.12G007000.1 | Chr12:652588..653004 | MeGRXC11 | CCMC | ALWL | 139 |
| cassava4.1_032796m | Manes.13G007200.1 | Chr13:771539..771955 | MeGRXC12 | CCMC | ALWL | 139 |
| cassava4.1_018177m | Manes.13G141400.1 | Chr13:26980309..26981228 | MeGRXC13 | CCMS | ALWL | 157 |
| cassava4.1_026496m | Manes.15G015500.1 | Chr15:1268904..1269746 | MeGRXC14 | CCMC | ALWL | 123 |
| cassava4.1_024232m | Manes.15G015600.1 | Chr15:1265181..1265486 | MeGRXC15 | CCMC | ALWV | 102 |
| cassava4.1_028408m | Manes.15G124000.1 | Chr15:9399566..9399949 | MeGRXC16 | CCMC | ALWL | 128 |
| cassava4.1_024091m | Manes.16G081400.1 | Chr16:23848065..23848499 | MeGRXC17 | CCMC | ALWV | 145 |
| cassava4.1_018360m | Manes.17G050200.1 | Chr17:18791502..18792218 | MeGRXC18 | CCMC | ALWL | 151 |
Fig. 2.
Protein sequences alignment of cassava CC-type GRXs. Black boxes indicate conserved identify positions. The letters above the sequence indicate motif name
Identification of drought-inducible CC-type GRX genes in cassava cultivar Arg7 and SC124
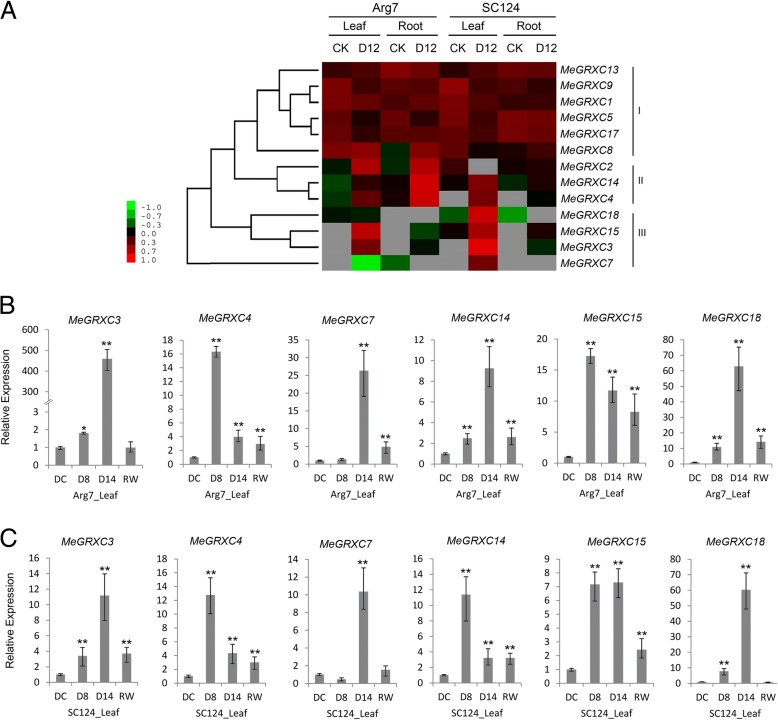
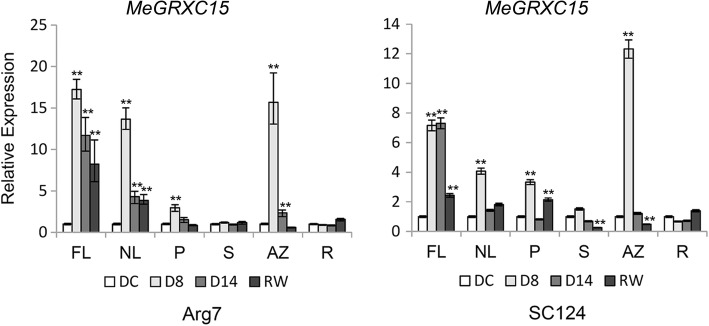
Previous studies used RNA-seq datasets to examine genes responsive to drought resistance in cassava [26–30]. RNA-seq datasets are available at NCBI and the accession numbers are listed in Additional file 2: Table S2. To investigate the expression of CC-type GRXs in response to drought in cassava, we used our previously reported transcriptomic data from cassava cultivar Arg7 and SC124 (Additional file 2: Table S3). Hierarchical expression clustering (FPKM) result shows that the CC-type GRX expression patterns in response to drought in cassava cultivars Arg7 and SC124 grouped in three clusters (Fig. 3a). Cluster II include CC-type GRXs induced by drought in both leaf and root. Cluster III include CC-type GRXs induced by drought only in leaves. To detail the expression of CC-type GRXs in the drought response in cassava leaves, we performed a qPCR analysis to investigate the expression changes of genes in cluster II and III under drought and re-water treatments. For this analysis, we selected six drought-inducible CC-type GRXs (MeGRXC3, C4, C7, C14, C15, and C18) from cluster II and III. We collected leaves from plants of two cassava cultivars under drought stress for eight (D8) or 14 days (D14), and rewatered D14 plants 24 h after rehydration (RW). We used leaves from well-watered cassava plants as control (DC). The qPCR results show that drought stress up-regulated the expression of all six CC-type GRXs in both Arg7 and SC124 leaves (Fig. 3b, c). The expression of MeGRXC3, C7 and C18 was the highest in D14 plants of both two cultivars. In contrast, the expression of MeGRXC4 and C15 was the highest in D8 plants of both two cultivars (Fig. 3b, c). Additionally, the expression of MeGRXC14 was the highest in D14 plants of Arg7 (Fig. 3b), while it was the highest in D8 plants of SC124 (Fig. 3c).
Fig. 3.
Expression analysis of drought-responsive CC-type GRX genes in cassava cultivar Arg7 and SC124. a Heat map represent expression of CC-type GRXs in drought stressed cassava Arg7 and SC124. b and (c) qPCR analysis of MeGRXC3, C4, C7, C14, C15, and C18 in drought stressed leaves of cassava Arg7and SC124. DC: control; D8: eight days after water withholding; D14: 14 days after water withholding; RW: rewatered D14 plants 24 h after rehydration. Expression levels of the six CC-type GRXs were normalized against DC. Biological triplicates were averaged and significance of differences between treatments and control were analyzed using the Student’s t-test (**, p ≤ 0.01). Bars represent the mean ± standard error
Cassava drought-responsive CC-type GRXs are induced by ABA in leaves
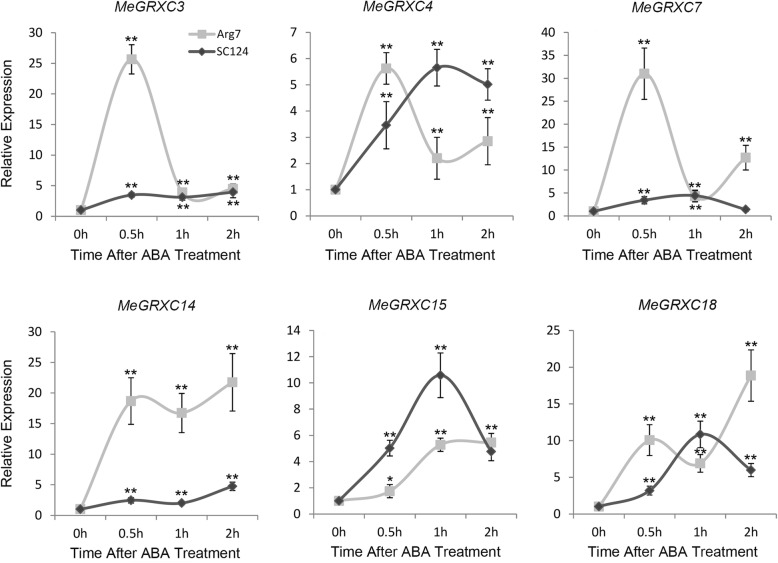
Numerous drought-responsive genes were described as ABA-inducible [31]. We performed qPCR analysis to investigate whether our drought-responsive CC-type GRXs are regulated by ABA in cassava leaves. We found that ABA application up-regulated the expression of six drought-responsive CC-type GRXs in leaves of Arg7 and SC124 (Fig. 4). The MeGRXC3, C14, and C15 showed similar ABA-induced expression patterns between Arg7 and SC124 (Fig. 4). This indicates that CC-type GRXs involving in ABA-dependent pathway during cassava response to drought.
Fig. 4.
Effects of ABA on expression of the six drought-responsive CC-type GRXs in cassava cultivar Arg7 and SC124 leaves. Expression levels of the six CC-type GRXs were normalized against control. Biological triplicates were averaged and significance of differences between treatments and control were analyzed using the Student’s t-test (**, p ≤ 0.01; *, 0.01 < p ≤ 0.05). Bars represent the mean ± standard error
Cassava drought-responsive CC-type GRXs are localized in the nucleus and cytoplasm
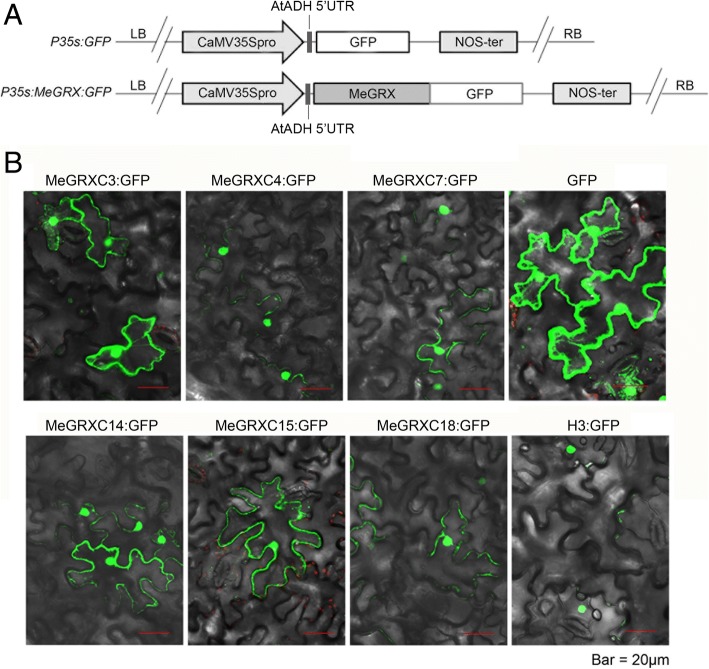
Most Arabidopsis CC-type GRX proteins localize in the cytosol or in the nucleus [10, 25, 32]. We respectively tagged the cDNA of MeGRXC3, C4, C7, C14, C15, and C18 with GFP at the C-terminus to analyze their cellular localization (Fig. 5a). We imaged the MeGRX:GFP fusion proteins in transiently transformed N.benthamiana leaf epidermis, detecting fluorescence in both the cytosol and the nucleus (Fig. 5b). The nuclear localization of MeGRX:GFP fusion proteins indicates that these drought-responsive CC-type GRXs may play roles in the nucleus during drought responses.
Fig. 5.
Protein localization analysis of the six drought-responsive CC-type GRXs. a Schematic diagram represent the design of 35S:MeGRX:GFP constructs. b Subcellular localization of MeGRX:GFP fusion proteins transiently expressed in N. benthamiana leaves. GFP was used as expression control, H3:GFP (H3, histone 3) was used as nuclear expression control
MeGRXC15 is tissue specifically induced by drought in cassava cultivars
For the MeGRXC15 shows similar expression pattern in leaves of both cassava cultivars Arg7 and SC124 under drought and ABA treatments (Figs. 3 and 4), this gene was selected for further investigation. Regulation of temporal and spatial expression patterns of specific stress-responsive is an important part of plant drought responses [33]. We performed qPCR analysis to examine the expression pattern of MeGRXC15 in different tissues of drought stressed cassava. We found that expression of MeGRXC15 was dramatically up-regulated by drought in functional leaf (FL), new leaf (NL), petiole (P), and abscission zone (AZ) of both cassava cultivars Arg7 and SC124 (Fig. 6). Abscission zone initiation in drought stressed cassava is based on reactive oxygen species (ROS) and ethylene (ET) signal transduction [3]. The MeGRXC15 shown higher induced expression level in AZ at D8 suggesting its potential roles in ROS or ET signal transduction pathway.
Fig. 6.
Expression analyses of MeGRXC15 in different tissues from drought stressed cassava cultivar Arg7 and SC124. FL, functional leaf; NL, new leaf; P, petiole; S, stem; AZ, abscission zone; R, root; DC: control; D8: eight days after water withholding; D14: 14 days after water withholding; RW: rewatered D14 plants 24 h after rehydration. Expression levels of the MeGRXC15 were normalized against DC. Biological triplicates were averaged and significance of differences between treatments and control were analyzed using the Student’s t-test (**, p ≤ 0.01). Bars represent the mean ± standard error
MeGRXC15 confers seedling development insensitive to ABA and drought hypersensitivity in soil-grown plants
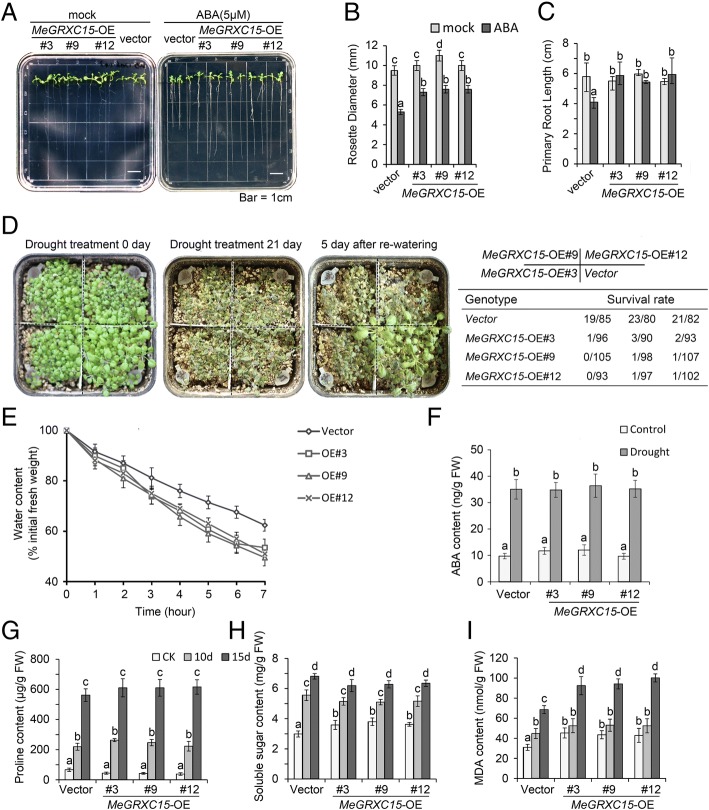
To investigate the function of MeGRXC15 in plant, we overexpressed this gene in Arabidopsis. Three independent lines of MeGRXC15-OE transgenic Arabidopsis were used in ABA and drought tolerance analyses and the transgenic Arabidopsis lines contained the empty vector (Fig. 5a) were used as controls. We found that ABA did not affect the seed germination of transgenic plants. We infer that overexpression of MeGRXC15 may cause ABA insensitivity in Arabidopsis. Next, 5-day-old seedlings of MeGRXC15-OE transgenic Arabidopsis were grown on MS medium supplement with 0 μM (mock) or 5 μM ABA, respectively. After 10 days grown on MS medium, no visible phenotypic differences between MeGRXC15-OE and control plants were observed (Fig. 7a). On ABA-supplement medium, the growth of control plants was significantly inhibited, while the growth of MeGRXC15-OE plants was less inhibited (Fig. 7a). The rosette diameter of MeGRXC15-OE plants was ~ 28% higher than that of control plants (Fig. 7b). Also, the primary root of MeGRXC15-OE plants was ~ 30% longer than that of control plants (Fig. 7c). Our data support the possibility that overexpression of MeGRXC15 caused ABA insensitivity in Arabidopsis.
Fig. 7.
ABA and drought tolerance analyses of MeGRXC15-OE transgenic Arabidopsis. a Post-germinated seedlings development of transgenic plants on MS medium supplemented with 0 (mock) and 5 μM ABA, respectively. The plants that contained empty vector (pG1300) were used as control. b and (c) Rosette diameter and primary root length of transgenic Arabidopsis under ABA treatment. d Drought responses of transgenic plants. Survival rates were calculated from three independent experiments. e Water loss rate analysis of transgenic Arabidopsis. f Endogenous ABA content in transgenic Arabidopsis under normal and drought conditions. Proline content (g), soluble sugar (h), and MDA content (i) in transgenic plants under drought treatment. Biological triplicates were averaged and significance of difference between treatments and control was analyzed using the Duncan’s multiple range tests. Different letters represent a significant difference at p < 0.05. Bars represent the mean ± standard error
To further investigate the roles of MeGRXC15 in drought tolerance, the control and three independent lines of MeGRXC15-OE plants were grown in soil in one pot under normal conditions. After planted in soil for 21 days, the plants were treated by withholding water (Fig. 7d). When exposed to water deficit for 21 days, all treated plants displayed severe wilting (Fig. 7d). The stressed plants were re-watered for five more days and then calculated the survival rate. The MeGRXC15-OE lines display a significantly lower survival rate than control plants (Fig. 7d). This indicates that overexpression of MeGRXC15 caused drought hypersensitivity in Arabidopsis under soil culture conditions. We monitored water loss rates of leaves from transgenic Arabidopsis. The leaves of MeGRXC15-OE plants lost ~ 10% more water than leaves of control plants did at seven hours after excised (Fig. 7e). However, the MeGRXC15 overexpression shows no effects on biosynthesis of endogenous ABA in Arabidopsis under normal and drought conditions (Fig. 7f).
Drought stress also leads to obviously physiological changes in plants. We monitored three stress responsive metabolites including proline, soluble sugar, and malondialdehyde (MDA). We found that overexpression of MeGRXC15 slightly affected proline and soluble sugar content, but dramatically increased MDA content in MeGRXC15-OE Arabidopsis, compared to that in the control plants under normal conditions (Fig. 7g, h, i). Prolonged (15 days) drought significantly induced the content of proline, soluble sugar, and MDA in both control and MeGRXC15-OE Arabidopsis (Fig. 7g, h, i). However, after drought treatment, only MDA content showed significant difference between MeGRXC15-OE and control plants. We found ~ 25% more MDA content in MeGRXC15-OE plants than that in control plants after drought treatment for 15 days (Fig. 7i). MDA is considered to be the final product of lipid peroxidation in the plant cell membrane and is an important indicator of membrane system injuries and cellular metabolism deterioration [34]. Our results indicate that overexpression of MeGRXC15 led to cell damage sensitivity to drought in Arabidopsis. It may partly explain the drought hypersensitivity in MeGRXC15-OE Arabidopsis.
MeGRXC15 regulates a group of genes involved in stress response and ABA, JA/ET signalling in Arabidopsis
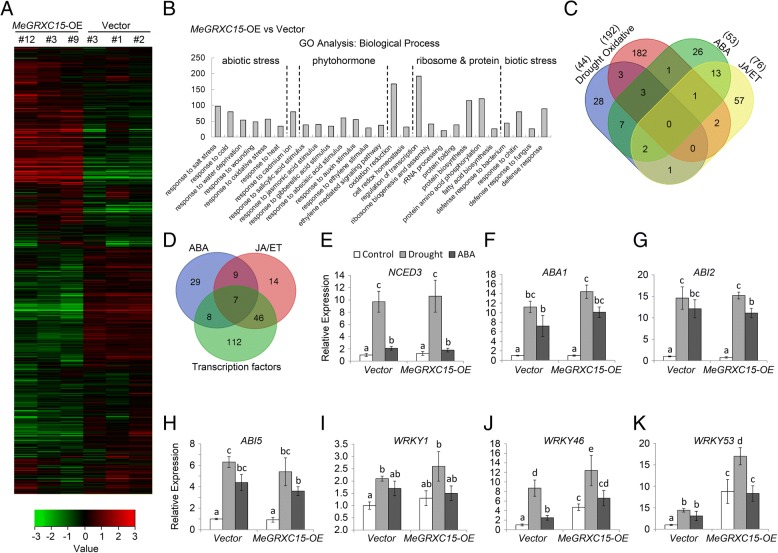
To understand the effects of the MeGRXC15 overexpression on gene expression in Arabidopsis, a microarray analysis was performed using the Affymetrix Arabidopsis ATH1 Genome Array. Three independent lines of MeGRXC15-OE and control Arabidopsis grown in soil under normal conditions were used. We found that transcription levels of 2674 genes were altered significantly (with more than a twofold change; P value < 0.05) in MeGRXC15-OE lines compared with control lines under normal conditions (Additional file 2: Table S2). 1264 genes were up-regulated, whereas 1410 genes were down-regulated. The relative expression levels of these genes were shown by the heat map (Fig. 8a). Gene ontology (GO) analysis shows that many stress-responsive genes are affected by MeGRXC15-OE Arabidopsis (Fig. 8b). 27 more abundant GO categories (q-value < 10− 5) including categories of response to abiotic, biotic stress, and phytohormone stimulus in MeGRXC15-OE Arabidopsis are exhibited here. Interestingly, nearly two hundred transcription factors were affected by MeGRXC15-OE Arabidopsis (Fig. 8b). We found that 192 oxidative stress-related, 44 drought-related, and 53 ABA-related genes were significantly altered in MeGRXC15-OE plants. Nevertheless, there are three members overlapping with the genes involved in response to drought, oxidative stress and ABA (Fig. 8c), indicating that a specific regulatory mechanism dependent on ABA-oxidative crosstalk conferred by MeGRXC15 is presented in response to drought. Moreover, three drought-related and three oxidative stress-related genes overlapping with genes that involved in JA/ET signal transduction respectively (Fig. 8c), suggesting the MeGRXC15 may play roles in drought response depending on regulation of JA/ET pathway. There are seven transcription factors overlapping with the genes involved in response to ABA or JA/ET (Fig. 8d). This suggests the regulatory roles of MeGRXC15 in ABA and JA/ET crosstalk.
Fig. 8.
Gene expression profiles in transgenic Arabidopsis. a Heat map represent gene expression differences between MeGRXC15-OE and control (Vector) plants. The data was processed and normalized as described in Materials and Methods. Hierarchical clustering of significantly expressed genes is displayed by average linkage. The figure was drawn by TreeView software. b GO analysis of MeGRXC15-OE induced genes in Arabidopsis. Comparison of GO terms identified from the differentially expression genes identified in SAM analysis. GO tags were selected according to the significance (p-value < 10− 5). Numbers on y-axis indicate gene numbers of the GO tag. c Venn diagram showing the overlap between MeGRXC15-OE regulated genes in response to different stress and signals. d Venn diagram showing the MeGRXC15-OE induced transcription factors which involve in ABA and JA/ET signalling. Expression analysis of NCED (e), ABI1 (f), ABI2 (g), ABI5 (h), WRKY1 (i), WRKY46 (j), and WRKY53 (k) in transgenic Arabidopsis under drought and ABA treatments. Expression levels of target genes were normalized against vector control. Biological triplicates were averaged and significance of difference between treatments and control was analyzed using the Duncan’s multiple range tests. Different letters represent a significant difference at p < 0.05. Bars represent the mean ± standard error
To clarify gene expression in MeGRXC15-OE Arabidopsis during drought and ABA treatments, we analyzed genes that are related to ABA- or drought-responses, including NCED3, ABI1, ABI2, ABI5, WRKY1, WRKY46, and WRKY53. The qPCR results show that expression of NCED3, ABI1, ABI2, and ABI5 was not affected by MeGRXC15-OE Arabidopsis under normal conditions (Fig. 8e-h). When exposed to drought or ABA, expression of these four genes was up-regulated in both control and MeGRXC15-OE plants (Fig. 8e-h). We found that expression of WRKY1, WRKY46, and WRKY53 was up-regulated by MeGRXC15-OE Arabidopsis under normal conditions (Fig. 8i-k). Furthermore, drought or ABA treatments both up-regulated WRKY1, WRKY46, and WRKY53 transcription in control plants (Fig. 8i-k). Likewise, drought significantly up-regulated these three WRKYs transcription in MeGRXC15-OE plants (Fig. 8i-k). However, ABA treatment did not affect transcription of WRKY1 and WRKY53 (Fig. 8i-k), it only slightly up-regulated WRKY46 transcription in MeGRXC15-OE plants (Fig. 8 i-k).
MeGRXC15 interacts with TGA5 or MeTGA074
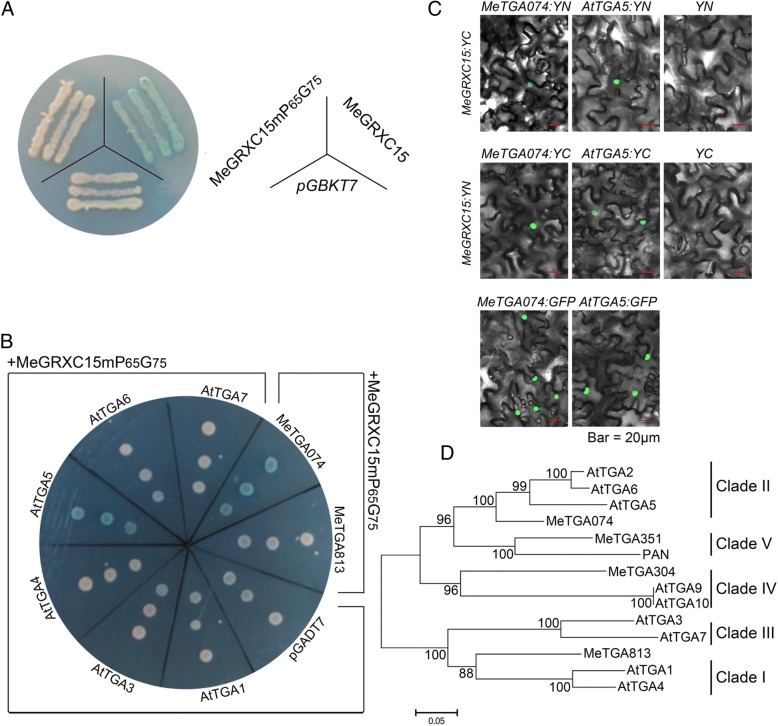
Several CC-type GRXs play roles in organ development or plant defense via interaction with TGA transcription factors [15, 17, 35, 36]. TGA transcription factors regulate genes that involved in both biotic and abiotic stress [37]. It is necessary to identify the interactors of MeGRXC15 in Arabidopsis and cassava. We fused MeGRXC15 with the GAL4 DNA-binding domain (BD) in pGBKT7 (Clontech) and then transformed the resulting construct into yeast strain Y187. The pGBKT7 vector was used as negative control. However, yeast cells harboring MeGRXC15:pGBKT7 activated X-α-gal on SD/−Trp / X-α-gal medium (Fig. 9a), suggesting that MeGRXC15 has transcriptional activation ability. CC-type GRXs need to interact with glutathione (GSH) to catalyze essential biosynthesis reactions by its redox regulation [25]. Therefore we created a MeGRXC15 mutant by replacing the GSH binding site. As is shown in Fig. 9a, the MeGRXC15 mutant MeGRXC15mP65G75 did not activate X-α-gal on the medium. This suggests that the GSH binding site is required for the transcriptional activation ability of MeGRXC15. A possible explanation is that MeGRXC15 may bind and modify the transcription factor depending on GSH in yeast.
Fig. 9.
Identification of protein interacts with MeGRXC15 in cassava and Arabidopsis. a Autonomous transactivation analysis of MeGRXC15 in yeast. MeGRXC15mP65G65 indicate mutant in MeGRXC15 GSH binding site. b Analysis of interaction between MeGRXC15 mP65G65 and TGA factors by yeast two-hybrid system. c BiFC analysis of the interactions between MeGRXC15 and TGAs identified by yeast two-hybrid system in transiently transformed N. benthamiana leaves. Green fluorescence in nucleus was detected for interactions of MeGRXC15 with MeTGA074 and AtTGA2, respectively. As a negative control, co-expression of MeGRXC15:YN with free YC, and MeGRXC15:YC with free YN failed to reconstitute a fluorescent YFP chromophore. Expression of MeTGA074:GFP and AtTGA2:GFP in transiently transformed N. benthamiana as positive controls. d Phylogenic analysis of TGA factors from Arabidopsis and cassava. A neighbor-joining tree was constructed with MEGA5.0 software based on sequences alignment with ClustalX
Subsequently, six TGA transcription factors including TGA1, 3, 4, 5, 6, 7 in Arabidopsis and two TGA transcription factors (MeTGA074 and MeTGA813) in cassava were respectively fused with GAL4 activation domain (AD) sequence in pGADT7 (Clontech). The resulting AD:TGA constructs and BD:MeGRXC15mP65G75 were pairwise co-transformed into yeast Y187, respectively. Yeast cells that harbored both AD:TGA and BD:MeGRXC15mP65G75 pair plasmids were grown on SD/ -Trp/ -Leu/ X-α-gal medium. The yeast cells containing pairwise plasmids AD:TGA5 / BD:MeGRXC15mP65G75 and AD:MeTGA074 / BD:MeGRXC15mP65G75 activated X-α-gal (Fig. 9b). This suggests that MeGRXC15 could respectively interact with TGA5 or MeTGA074.
To further investigate the interactions between MeGRXC15 and AtTGA5 / MeTGA074 in planta, we employed Bimolecular Fluorescence Complementation (BiFC) analysis. Nuclear fluorescence co-expression of MeGRXC15 and AtTGA5/MeTGA074 was detected in epidermal cells (Fig. 9c). The in planta nuclear interactions of MeGRXC15 with AtTGA5 / MeTGA074 suggest that this CC-type GRX might function in Arabidopsis and cassava by nuclear interaction with AtTGA5 / MeTGA074. We created a phylogenetic tree based on TGA protein sequences in Arabidopsis and cassava (Fig. 9d). We found that MeTGA074 is a member of clade II TGAs, closely related to AtTGA5. Together, our data suggest that MeGRXC15 may regulate drought response via interaction with AtTGA5 / MeTGA074.
Discussion
In Arabidopsis, only four of 21 CC-type GRXs (GRXC7/ROXY1, GRXC8/ROXY2, GRXC9/ GRX480/ROXY19, and GRXS13) were functionally characterized by genetic approaches [14–16, 25]. With such short knowledge identifying of the genomic and EST sequences in several plant species is a promising approach, which may allow expanding the knowledge of plant GRXs by the comparative and evolutionary analysis. Herein, we identified 38 putative GRX genes from cassava genome (Fig. 1), they are classified in five subgroups as in Arabidopsis and rice [6, 7]. CC-type GRXs are a land plant specific subgroup of the GRX family, derived from the CPYC subgroup and expanded from basal to higher land plant mainly through paleopolyploidy duplication and tandem duplication events [11]. Our result demonstrated that all cassava CC-type GRXs evolved from three cassava CPYC GRXs (Fig. 1). And we found MeGRXC1 and MeGRXC2, MeGRXC3 and MeGRXC4 are two pairs neighboring genes in cassava chromosome 1 (Table 1). Also MeGRXC8 and MeGRXC9 are neighboring genes in chromosome 5, MeGRXC14 and MeGRXC15 are neighboring genes in chromosome 15 (Table 1). These results indicate that tandem duplication events contributed to the expansion of CC-type GRXs in cassava.
To date, no CC-type GRX was characterized as a regulator of drought response in ABA dependent manner. Based on the RNA-seq and qPCR data, we found that six CC-type GRX genes were induced by drought stress in leaves of both Arg7 and SC124 (Fig. 3). Under drought stress, ABA concentrations increase and, in turn, induce gene expression [38]. Overexpression of the rice CC-type GRX OsGRX8 enhances tolerance to ABA and abiotic stresses in Arabidopsis, but the expression of this gene was induced by auxin, instead of ABA in rice [18]. However, exogenous ABA application induced the expression of the six drought-responsive CC-type GRX genes in leaves of both Arg7 and SC124 (Fig. 4), suggesting that CC-type GRXs regulated drought response probably in an ABA-dependent manner in cassava. This is further supported by our data showing that overexpression of MeGRXC15 in Arabidopsis resulted seedling development insensitivity to ABA (Fig. 7a) and induced overexpression of several genes which involved in the ABA signalling (Fig. 8).
In Arabidopsis, seven ROXY members under the control of ROXY1 promoter could complement the roxy1 mutant [35]. This indicates that the expression pattern is particularly important for the function of ROXY genes. When exposure to prolonged drought stress, the Arg7 plant display faster senescence in older leaves than the SC124 plants [2]. MeGRXC3, C7, and C15 shown fairly consistent difference in expression levels between Arg7 and SC124 in both drought and ABA treatments (Fig. 3, Figs 4 and 6), suggesting that the expression patterns of these genes are correlated with the different response of leaves in Arg7 and SC124 under drought stress. The GRXS13 is a CC-type GRX, which could be induced by oxidative stress in Arabidopsis, repression of this gene resulted higher accumulation of the superoxide anion O2− [16]. High ROS accumulation is required for abscission zone formation in cassava during drought stress [3]. Our data also shown that MeGRXC15 has highest expression levels in abscission zone (AZ) at D8 in both two cassava cultivars (Fig. 6), indicating the expression of MeGRXC15 maybe correlated to ROS accumulation. ABA induced ROS production is a required process in plant drought response [39, 40]. The gene expression profile in transgenic Arabidopsis shows that MeGRXC15 overexpression induced three genes overlapping with the genes involved in response to drought, oxidative stress and ABA (Fig. 8c), suggesting that MeGRXC15 regulates drought response likely through ABA and ROS signalling pathway. Overexpression of MeGRXC15 did not affect endogenous ABA synthesis (Fig. 7e) and NCED3 transcription (Fig. 8e), while it affected WRKY46 and WRKY53 transcription in Arabidopsis (Fig. 8j, k). In Arabidopsis, WRKY1, WRKY46, and WRKY53 negatively regulate drought tolerance by inhibition of ABA-induced stomatal closure [41–43]. Additionally, drought dramatically up-regulated WRKY46 and WRKY53 in MeGRXC15-OE plants (Fig. 8j, k), this may partly ascribe to the drought sensitivity of these plants. WRKY46 and WRKY53 are also involve in signalling transduction of other phytohormone, such as Brassinosteroid [44], Jasmonic acid and Salicylic acid [45]. ABA treatments did not affect the expression of WRKY46 and WRKY53 in transgenic Arabidopsis (Fig. 8j, k), indicating that MeGRXC15 may regulate WRKYs through an ABA-independent pathway during drought response. Furthermore, regulation on WRKYs expression under ABA treatment perhaps contributes to the ABA insensitivity in MeGRXC15-OE Arabidopsis.
The nuclear localization of ROXY1 is required for its function in petal development [35]. However, not all members of CC-type GRX subgroup have the same subcellular localization, unlike ROXY1, ROXY18 and ROXY20 are localized in cytosol [7]. All of our six drought-responsive CC-type GRXs are located in both nucleus and cytosol (Fig. 5b), suggesting the possibility that these genes could function in nucleus. The nuclear functions of ROXYs in Arabidopsis are partly dependent on its interaction with TGA transcription factors [15, 35, 36]. In Arabidopsis, TGA transcription factors have been classified to five subgroups, clade I, II, III, IV, and V. TGA2, 5, 6 are members of clade II TGAs, which are essential activators of jasmonic acid/ethylene-induce defense responses [15, 46–48] and act as key regulators in plant responses of abiotic stresses such as drought, cold, and oxidative stress [37]. Arabidopsis CC-type GRX GRX480/ROXY19 could interact with TGA2, 5, 6 [15]. TGA2 could interact with GRXS13, and act as repressors of GRXS13 expression in response to biotic stress [14]. Here, we found that MeGRXC15 could interact with Arabidopsis TGA5 and cassava MeTGA074 in the nucleus respectively (Fig. 9b, c). In Arabidopsis, GRX480 regulated the expression of ERF (Ethylene Response Factor) factors through interaction with TGA2/5/6 [15, 49]. We found 53 transcription factors including ERFs were induced by MeGRXC15 in Arabidopsis (Fig. 8d). A NES (Nuclear Export Signal) could be tagged to MeGRXC15 to eliminate its nuclear localization to investigate whether the MeGRXC15 regulates the transcription factors through nuclear interaction with TGA5 in Arabidopsis. It will be of interest to further study the mechanism by which MeGRXC15 respond to drought in ABA dependent manner via interaction with MeTGA074 in cassava.
In our study, when MeGRXC15 was fused to GAL4 binding domain (BD), the fusion protein exhibited strong autonomous transactivation activity in yeast (Fig. 9a), indicating that MeGRXC15 could recruit transcription factor in yeast nucleus and probably generate a complex protein like GAL4BD-MeGRXC15-TF (Activation Domain). Thus, the recombination protein was able to function as a transcription factor promoting the transcription of reporter gene in yeast strain Y187. Substitution mutants in GSH binding site of MeGRXC15 caused autonomous transactivation activity loss in yeast (Fig. 9a). The GRXs are generally reduced by GSH, and the GHS binding ability is required for the function of ROXY1 and ROXY2 in Arabidopsis [25]. The CC-type GRXs interact with TGA transcription factors dependent on its C-terminal L**LL and ALWL motifs [15, 36]. The mutation in GSH binding site probably will not affect the CC-type GRX interaction with TGA transcription factors. However, the ROXY1 negatively regulates the PAN activity, and positively regulates the other TGAs activity during the petal development in Arabidopsis [35]. Thus, modification of the GSH binding site of MeGRXC15 may affect its regulation on TGAs activity and therefore caused transcriptional activation losing of GAL4BD-MeGRXC15-TF complex protein.
Conclusion
Our study demonstrates that CC-type GRXs may function in ABA-mediated drought signalling in cassava. As a CC-type GRX, MeGRXC15 could interact with Arabidopsis TGA5 or cassava MeTGA074. Overexpression of MeGRXC15 results drought hypersensitivity in Arabidopsis. It will contribute to an enhanced understanding of the specific mechanisms that elucidate the roles of CC-type GRXs involved in drought response in cassava.
Methods
Bioinformatics analysis
The protein sequences of cassava GRXs were predicted using a TBLASTN search against the cassava genome database in Phytozome (https://phytozome.jgi.doe.gov/pz/portal.html, Manihot esculenta v6.1) with the protein sequence from Arabidopsis GRXs as a query. All Arabidopsis GRX protein sequences were downloaded from GenBank. Multiple sequence alignments were conducted using ClustalW [50] based on GRX protein sequences in Additional file 1: Table S1. An unrooted phylogenetic tree showing cassava GRXs and Arabidopsis GRX family was generated via the neighbor joining method using MEGA5.0 [51]. Editing of aligned sequences of cassava CC-type GRXs was performed using AlignX (Vector NTI suite 10.3, Invitrogen).
Transcriptome data analysis
For drought-responsive CC-type GRXs identification, we used our previously reported RNA-seq data [24]. We used data that included two tissues (leaf and root) under drought treatment and a control. The accession number of RNA-seq data is listed in Additional file 2: Table S2. Gene expression levels were normalized using FPKM. We selected the data of CC-type GRX genes (Additional file 3: Table S3), and generated a heat map and hierarchical clustering using Cluster 3.0.
Drought and ABA treatments on cassava
Two cassava cultivars, Arg7 and SC124, were used in this study. Stems of cassava Arg7 and SC124 were cultured in same pots (36 cm in diameter × 30 cm in height) containing well-mixed soil (nutrient soil: vermiculite: sand, 1:1:1) for 90 days in greenhouse at the Institute of Tropical Bioscience and Biotechnology (Haikou, China). For drought treatment, plants were treated by withholding water for eight or 14 days. Different tissues were collected from three Arg7 and SC124 plants at eight or 14 days after withholding water and 24 h after re-watering at the ending of treatment. Plants watered as normal were used as controls. Different tissues including Functional Leaf (FL), New Leaf (NL), Petiole (P), Stem (S), Abscission Zone (AZ) and Root ® from each plant were collected. For ABA treatments, mature leaves with petiole were excised from Arg7 and SC124 plants, treated by dipping the leaves in water (control) or in water with 20 μM ABA. The samples were collected after treated for 0, 0.5, 1, or 2 h.
Quantitative real-time PCR (qPCR) analysis
Total RNA was isolated from cassava or Arabidopsis leaves using RNAprep Pure Plant Kit (TIANGEN). The cDNA synthesis was performed with FastQuant RT Kit (TIANGEN). Expression analysis of CC-type GRXs in cassava after drought and exogenous ABA treatment was performed by qPCR with gene-specific primers (Additional file 2: Table. S4). All qPCR reactions were carried out in triplicates, with SYBR® Premix Ex Taq™ II Kit (Takara) on StepOne™ Real-Time PCR system (Applied Biosystems), and the comparative ΔΔCT method employed to evaluate amplified product quantities in the samples.
Protein subcellular localization
Full-length coding sequence without stop-codon of MeGRXC3, C4, C7, C14, C15, and C18 was isolated from cDNA of drought stressed leaves by RT-PCR respectively. Fragments were identified by sequencing and fused to GFP in front of the CaMV 35S promoter in the modified plant expression vector pG1300 (eGFP:pCAMBIA1300) to make 35S:MeGRXC3:GFP, 35S:MeGRXC4:GFP, 35S:MeGRXC7:GFP, 35S:MeGRXC14:GFP, 35S:MeGRXC15:GFP, and 35S:MeGRXC18:GFP. The 5’UTR of ATADH gene was inserted between 35S promoter and MeGRX coding sequence to enhance the expression of MeGRX:GFP. The resulting constructs and empty vector were transformed into Agrobacterium LBA4404. Leaves from four-week-old Nicotiana benthamiana plants were transformed by infiltration of Agrobacterium cells (OD600 = 1.2) harboring appropriate DNA construct using 5-mL syringe without needle. The empty vector (GFP) and 35S:MeHistone3:GFP (H3:GFP) were used as the positive controls. After three days, infiltrated N. benthamiana leaves were imaged for reconstitution of GFP fluorescence by confocal laser scanning microscope (Olympus FluoView FV1100).
Generation of MeGRXC15-OE transgenic Arabidopsis
Wild type (Col-0) Arabidopsis plants for transformation were grown in 12 h light/12 h dark at 20–23 °C until the primary inflorescence was 5–15 cm tall and secondary inflorescence appeared at the rosette. Arabidopsis was transformed using the floral dip method [52] and A. tumefaciens strain LBA4404 carrying the DNA constructs 35S:MeGRXC15:GFP and the pG1300 empty vector control, respectively. More than three homozygous lines of each construct were selected for further phenotypic analyses. The MeGRXC15:GFP fusion protein subcellular localization in transgenic Arabidopsis epidermal cell was examined for reconstitution of green fluorescence by confocal laser scanning microscope (Olympus FluoView FV1100).
ABA tolerance assays of transgenic Arabidopsis
To study the response of MeGRXC15-OE transgenic plants to ABA, 5-d-old seedlings were transferred to MS medium containing with 0 μM (mock) and 5 μM ABA grown for 10 days. Rosette diameter, primary root length and lateral root number were measured. The transgenic plant that contained pG1300 vector was used as the empty vector control.
Drought stress tolerance assays of transgenic Arabidopsis
Post-germinated seedlings of MeGRXC15-OE and empty vector transgenic plants were grown in soil in one pot for 15 days under normal conditions. For drought stress, the plants were treated by water withholding for 21 days, then re-watering. Survival rates were calculated at five days after re-watering. Proline and soluble sugar, indicator of the drought response in plants, were measured. Lipid peroxidation in transgenic Arabidopsis leaf tissues was measured in terms of malondialdehyde (MDA) in the samples as described in reference [5] during drought stress. For water loss rate measurement, excised leaves from 28-d-old unstressed transgenic plants were kept on plastic dishes at room temperature. Their weight was measured after one hour, up to seven hour followed by calculation of water loss percentage.
Determination of endogenous ABA content
Endogenous ABA content was determined by extraction and detection using LC-ESI-MS/MS according to methods described previously [53]. 28-d-old leaves from five control or drought stressed plants of each line were mixed to constitute one biological replicate. 0.1 g mixed leaf sample was extracted with 1.5 mL methanol formic acid solution (Methanol: formic acid: water = 7.8: 0.2: 2). Results from three biological replicates were averaged.
Microarray analysis of transgenic Arabidopsis
Microarray experiments were conducted using Affymetrix Arabidopsis ATH1 Genome Array. Experiments were performed as three biological repeats using cDNAs prepared independently from three individual homozygous lines of MeGRXC15 overexpression Arabidopsis that were phenotypic analyzed in plant growth. The transgenic Arabidopsis plants that carried the pG1300 empty vector were used as controls. The experiments and data analysis were performed under the instruction of Affymetrix. Total microarray data were deposited in the NCBI GEO database with the accession number: GSE81136 (MeGRX232-OE). Gene ontology (GO) analyses for significant enrichments of various categories (Additional file 2: Table S5) were performed using MAS 3.0 (http://bioinfo.capitalbio.com/mas3/). The Venn diagrams were created by online tool (http://bioinformatics.psb.ugent.be/webtools/Venn/).
Identification and phylogenetic analysis of TGA transcription factors
The Arabidopsis TGA transcription factors protein sequences were download from GenBank database. The cassava TGA transcription factors were identified using TBLASTN against the Phytozome database website (https://phytozome.jgi.doe.gov, Manihot esculenta v6.1) with the protein sequences from Arabidopsis TGA transcription factors. Four TGA transcription factors were cloned with the accession number Manes.04G157200.1 (MeTGA074), Manes.04G004100.1 (MeTGA304), Manes.14G099100.1 (MeTGA351), and Manes.12G140100.1 (MeTGA813). An unrooted phylogenetic tree showing cassava and Arabidopsis TGA transcription factors was generated based on protein sequences (Additional file 3: Table S6) with a neighbor joining method using MEGA5.0 [51].
Transactivation analysis and yeast two hybrid assay
Before analyzing the interaction between MeGRXC15 and TGA transcription factors, an autonomous transactivation analysis was performed in yeast strain Y187. The MeGRXC15 was in frame fused to GAL4 BD (binding domain) in pGBKT7, and then transformed into yeast Y187. Because MeGRXC15 shows “autonomous transactivation” in yeast, a MeGRXC15 GSH binding site mutant MeGRXC15mP65G75 was produced by replacing P65AVFIGGILVG75 to A65AVFIGGILVA75. Next, for identification the interaction between MeGRXC15 and TGA transcription factors, a yeast two-hybrid assay has been performed in yeast strain Y187 based on the Matchmaker ™ GAL4 two-hybrid system 3 manual (Clontech). The MeGRXC15 GSH binding site mutant DNA construct MeGRXC15mP65G75:pGBKT7 was used as bait. The cDNA sequences of TGA transcription factors from Arabidopsis and cassava were introduced into the pGADT7, respectively in frame fused to GAL4 activation domain (AD). The MeGRXC15mP65G75:pGBKT7 and TGA:pGADT7 constructs were pairwise co-transformed into yeast strain Y187. The presence of transgenes was confirmed by growth on SD/ -Trp/−Leu plates. Interactions between two proteins were checked by examining β-galactosidase activity as the manual instructed.
Bimolecular fluorescence complementation analysis
To confirm the interactions between MeGRXC15 and TGA2 / MeTGA074 factors, a bimolecular fluorescence complementation assay was performed using the N.benthamiana transient system as previously report [54]. The full-length coding sequence without stop-codon of MeGRXC15 was in frame fused to N- or C-terminus to yellow fluorescent protein (YFP) fragments (YN/YC) respectively to produce 35S:MeGRXC15:YN:pBiFC and 35S:MeGRXC15:YC:pBiFC. The full-length coding sequence without stop-codon of TGA2 and MeTGA074 were in frame fused to YC or YN respectively to produce 35S:TGA2:YC:pBiFC, 35S:TGA2:YN:pBiFC, 35S:MeTGA074:YC:pBiFC, and 35S:MeTGA074:YN:pBiFC. The resulting constructs were then introduced into A. tumefaciens LBA4404 strains. Constructs were pair-wise transiently expressed in epidermal cells of tobacco leaves. Three days after agrobacterium co-transformation of leaves, reconstitution of YFP fluorescence was examined by confocal microscopy using GFP filter. Then the assays were performed as the method of proteins subcellular localization described. As positive controls, full-length green fluorescent protein (eGFP) was tagged to the C-terminus of TGA2 and MeTGA074 respectively, transiently expressed in tobacco leaves.
Additional files
Table S1. The protein sequences of GRX from cassava and Arabidopsis. (DOC 61 kb)
Table S2. The accession number of cassava drought related transcriptome data. Table S3. The RNA-seq data of GRXs in drought stressed cassava. Table S4. The list of primers. Table S5. GO results of MeGRXC15 regulated genes in transgenic Arabidopsis. (XLS 287 kb)
Table S6. Protein sequences of TGA transcription factors from cassava. and Arabidopsis. (DOC 35 kb)
Acknowledgements
We thank Prof. Wei-Cai Yang at the Institute of Genetics and Developmental Biology, Chinese Academy of Sciences for providing the plasmid containing eGFP gene. And we also thank Prof. Peng Zhang at the Institute of Plant Physiology and Ecology, Shanghai Institutes for Biological Science, Chinese Academy of Science for proof-reading.
Funding
This work was supported by the national key technology R&D program of China (grant no. 2015BAD15B01), and the Central Public-interest Scientific Institution Basal Research Fund for Chinese Academy of Tropical Agricultural Sciences (No.1630052016004).
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Abbreviations
- ABA
Abscisic acid
- AD
Activation domain
- Arg7
Argentina 7
- BD
Binding domain
- BLAST
Basic local alignment search tool
- ET
Ethylene
- GFP
Green fluorescent protein
- JA
Jasmonate acid
- NES
Nuclear export signal
- qPCR
Quantitative real-time polymerase chain reaction
- ROS
Reactive oxygen species
- SC124
South china 124
- YFP
Yellow fluorescent protein
Authors’ contributions
MBR carried out the experimental studies including qPCR analysis, microscopic studies, bimolecular fluorescence complementation analysis, microarray analysis, and drafted the manuscript. YLY carried out in transgenic Arabidopsis phenotype and yeast two-hybrid analysis. XG carried out cassava drought and ABA treatments. BW carried out bioinformatics and statistical analysis. XLY carried out molecular cloning and created the transgenic Arabidopsis. KML designed the research on MeGRXC15 and helped drafting the manuscript. MBR and MP planned the study. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The author declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Meng-Bin Ruan, Email: ruanmengbin@itbb.org.cn.
Yi-Ling Yang, Email: luckystar1221@163.com.
Kai-Mian Li, Email: likaimian@sohu.com.
Xin Guo, Email: guoxinyxh@gmail.com.
Bin Wang, Email: wangbinatt@126.com.
Xiao-Ling Yu, Email: lingdang01@126.com.
References
- 1.Alves AA, Setter TL. Response of cassava leaf area expansion to water deficit: cell proliferation, cell expansion and delayed development. Ann Bot. 2004;94(4):605–613. doi: 10.1093/aob/mch179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhao LP, Shao J, Li C, Wang B, Guo X, Yan B, Xia Y, Peng M. Analysis of different strategies adapted by two cassava cultivars in response to drought stress: ensuring survival or continuing growth. J Exp Bot. 2014;66(5):1477–1488. doi: 10.1093/jxb/eru507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Liao W, Wang G, Li Y, Wang B, Zhang P, Peng M. Reactive oxygen species regulate leaf pulvinus abscission zone cell separation in response to water-deficit stress in cassava. Sci Rep. 2016;6:21542. doi: 10.1038/srep21542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu J, Duan X, Yang J, Beeching JR, Zhang P. Coupled expression of cu/Zn-superoxide dismutase and catalase in cassava improves tolerance against cold and drought stresses. Plant Signal Behav. 2013;8(6):e24525. doi: 10.4161/psb.24525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu J, Duan X, Yang J, Beeching JR, Zhang P. Enhanced reactive oxygen species scavenging by overproduction of superoxide dismutase and catalase delays postharvest physiological deterioration of cassava storage roots. Plant Physiol. 2013;161(3):1517–1528. doi: 10.1104/pp.112.212803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rouhier N, Couturier J, Jacquot JP. Genome-wide analysis of plant glutaredoxin systems. J Exp Bot. 2006;57(8):1685–1696. doi: 10.1093/jxb/erl001. [DOI] [PubMed] [Google Scholar]
- 7.Meyer Y, Belin C, Delorme-Hinoux V, Reichheld J-P, Riondet C. Thioredoxin and Glutaredoxin Systems in Plants: molecular mechanisms, Crosstalks, and functional significance. Antioxid Redox Signal. 2012;17(8):1124–1160. doi: 10.1089/ars.2011.4327. [DOI] [PubMed] [Google Scholar]
- 8.Kanda M, Ihara Y, Murata H, Urata Y, Kono T, Yodoi J, Seto S, Yano K, Kondo T. Glutaredoxin modulates platelet-derived growth factor-dependent cell signaling by regulating the redox status of low molecular weight protein-tyrosine phosphatase. J Biol Chem. 2006;281(39):28518–28528. doi: 10.1074/jbc.M604359200. [DOI] [PubMed] [Google Scholar]
- 9.Carroll MC, Outten CE, Proescher JB, Rosenfeld L, Watson WH, Whitson LJ, Hart PJ, Jensen LT, Cizewski Culotta V. The effects of glutaredoxin and copper activation pathways on the disulfide and stability of cu,Zn superoxide dismutase. J Biol Chem. 2006;281(39):28648–28656. doi: 10.1074/jbc.M600138200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xing S, Rosso MG, Zachgo S. ROXY1, a member of the plant glutaredoxin family, is required for petal development in Arabidopsis thaliana. Development. 2005;132(7):1555–1565. doi: 10.1242/dev.01725. [DOI] [PubMed] [Google Scholar]
- 11.Ziemann M, Bhave M, Zachgo S. Origin and diversification of land plant CC-type glutaredoxins. Genome Biol Evol. 2009;1:265–277. doi: 10.1093/gbe/evp025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Couturier J, Didierjean C, Jacquot JP, Rouhier N. Engineered mutated glutaredoxins mimicking peculiar plant class III glutaredoxins bind iron-sulfur centers and possess reductase activity. Biochem Biophys Res Commun. 2010;403(3–4):435–441. doi: 10.1016/j.bbrc.2010.11.050. [DOI] [PubMed] [Google Scholar]
- 13.Wang Z, Xing S, Birkenbihl RP, Zachgo S. Conserved functions of Arabidopsis and rice CC-type glutaredoxins in flower development and pathogen response. Mol Plant. 2009;2(2):323–335. doi: 10.1093/mp/ssn078. [DOI] [PubMed] [Google Scholar]
- 14.La Camera S, L’Haridon F, Astier J, Zander M, Abou-Mansour E, Page G, Thurow C, Wendehenne D, Gatz C, Metraux JP, et al. The glutaredoxin ATGRXS13 is required to facilitate Botrytis cinerea infection of Arabidopsis thaliana plants. Plant J. 2011;68(3):507–519. doi: 10.1111/j.1365-313X.2011.04706.x. [DOI] [PubMed] [Google Scholar]
- 15.Zander M, Chen S, Imkampe J, Thurow C, Gatz C. Repression of the Arabidopsis thaliana jasmonic acid/ethylene-induced defense pathway by TGA-interacting glutaredoxins depends on their C-terminal ALWL motif. Mol Plant. 2012;5(4):831–840. doi: 10.1093/mp/ssr113. [DOI] [PubMed] [Google Scholar]
- 16.Laporte D, Olate E, Salinas P, Salazar M, Jordana X, Holuigue L. Glutaredoxin GRXS13 plays a key role in protection against photooxidative stress in Arabidopsis. J Exp Bot. 2012;63(1):503–515. doi: 10.1093/jxb/err301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hong L, Tang D, Zhu K, Wang K, Li M, Cheng Z. Somatic and reproductive cell development in rice anther is regulated by a putative glutaredoxin. Plant Cell. 2012;24(2):577–588. doi: 10.1105/tpc.111.093740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sharma R, Priya P, Jain M. Modified expression of an auxin-responsive rice CC-type glutaredoxin gene affects multiple abiotic stress responses. Planta. 2013;238:871–884. doi: 10.1007/s00425-013-1940-y. [DOI] [PubMed] [Google Scholar]
- 19.El-Kereamy A, Bi YM, Mahmood K, Ranathunge K, Yaish MW, Nambara E, Rothstein SJ. Overexpression of the CC-type glutaredoxin, OsGRX6 affects hormone and nitrogen status in rice plants. Front Plant Sci. 2015;6:934. doi: 10.3389/fpls.2015.00934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garg R, Jhanwar S, Tyagi AK, Jain M. Genome-wide survey and expression analysis suggest diverse roles of glutaredoxin gene family members during development and response to various stimuli in rice. DNA Res. 2010;17(6):353–367. doi: 10.1093/dnares/dsq023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gutsche N, Thurow C, Zachgo S, Gatz C. Plant-specific CC-type glutaredoxins: functions in developmental processes and stress responses. Biol Chem. 2015;396(5):495–509. doi: 10.1515/hsz-2014-0300. [DOI] [PubMed] [Google Scholar]
- 22.Wang W, Feng B, Xiao J, Xia Z, Zhou X, Li P, Zhang W, Wang Y, Moller BL, Zhang P, et al. Cassava genome from a wild ancestor to cultivated varieties. Nat Commun. 2014;5:5110. doi: 10.1038/ncomms6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bredeson Jessen V, Lyons Jessica B, Prochnik Simon E, Wu G Albert, Ha Cindy M, Edsinger-Gonzales Eric, Grimwood Jane, Schmutz Jeremy, Rabbi Ismail Y, Egesi Chiedozie, Nauluvula Poasa, Lebot Vincent, Ndunguru Joseph, Mkamilo Geoffrey, Bart Rebecca S, Setter Tim L, Gleadow Roslyn M, Kulakow Peter, Ferguson Morag E, Rounsley Steve, Rokhsar Daniel S. Sequencing wild and cultivated cassava and related species reveals extensive interspecific hybridization and genetic diversity. Nature Biotechnology. 2016;34(5):562–570. doi: 10.1038/nbt.3535. [DOI] [PubMed] [Google Scholar]
- 24.Hu W, Yang H, Yan Y, Wei Y, Tie W, Ding Z, Zuo J, Peng M, Li K. Genome-wide characterization and analysis of bZIP transcription factor gene family related to abiotic stress in cassava. Sci Rep. 2016;6:22783. doi: 10.1038/srep22783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xing S, Zachgo S. ROXY1 and ROXY2, two Arabidopsis glutaredoxin genes, are required for anther development. Plant J. 2008;53(5):790–801. doi: 10.1111/j.1365-313X.2007.03375.x. [DOI] [PubMed] [Google Scholar]
- 26.Xia J, Zeng C, Chen Z, Zhang K, Chen X, Zhou Y, Song S, Lu C, Yang R, Yang Z, et al. Endogenous small-noncoding RNAs and their roles in chilling response and stress acclimation in cassava. BMC Genomics. 2014;15:634. doi: 10.1186/1471-2164-15-634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zeng C, Chen Z, Xia J, Zhang K, Chen X, Zhou Y, Bo W, Song S, Deng D, Guo X, et al. Chilling acclimation provides immunity to stress by altering regulatory networks and inducing genes with protective functions in cassava. BMC Plant Biol. 2014;14:207. doi: 10.1186/s12870-014-0207-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu W, Xia Z, Yan Y, Ding Z, Tie W, Wang L, Zou M, Wei Y, Lu C, Hou X, et al. Genome-wide gene phylogeny of CIPK family in cassava and expression analysis of partial drought-induced genes. Front Plant Sci. 2015;6:914. doi: 10.3389/fpls.2015.00914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hu W, Wei Y, Xia Z, Yan Y, Hou X, Zou M, Lu C, Wang W, Peng M. Genome-wide identification and expression analysis of the NAC transcription factor family in cassava. PLoS One. 2015;10(8):e0136993. doi: 10.1371/journal.pone.0136993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu W, Hou X, Xia Z, Yan Y, Wei Y, Wang L, Zou M, Lu C, Wang W, Peng M. Genome-wide survey and expression analysis of the calcium-dependent protein kinase gene family in cassava. Mol Gen Genomics. 2016;291(1):241–53. [DOI] [PubMed]
- 31.Nakashima K, Yamaguchi-Shinozaki K. ABA signaling in stress-response and seed development. Plant Cell Rep. 2013;32(7):959–970. doi: 10.1007/s00299-013-1418-1. [DOI] [PubMed] [Google Scholar]
- 32.Rouhier N, Unno H, Bandyopadhyay S, Masip L, Kim SK, Hirasawa M, Gualberto JM, Lattard V, Kusunoki M, Knaff DB, et al. Functional, structural, and spectroscopic characterization of a glutathione-ligated [2Fe-2S] cluster in poplar glutaredoxin C1. Proc Natl Acad Sci U S A. 2007;104(18):7379–7384. doi: 10.1073/pnas.0702268104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shinozaki K, Yamaguchi-Shinozaki K. Gene networks involved in drought stress response and tolerance. J Exp Bot. 2006;58(2):221–227. doi: 10.1093/jxb/erl164. [DOI] [PubMed] [Google Scholar]
- 34.Dhindsa RS, Matowe W. Drought tolerance in two mosses: correlated with enzymatic Defence against lipid peroxidation. J Exp Bot. 1981;32(1):79–91. doi: 10.1093/jxb/32.1.79. [DOI] [Google Scholar]
- 35.Li S, Lauri A, Ziemann M, Busch A, Bhave M, Zachgo S. Nuclear activity of ROXY1, a glutaredoxin interacting with TGA factors, is required for petal development in Arabidopsis thaliana. Plant Cell. 2009;21(2):429–441. doi: 10.1105/tpc.108.064477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li S, Gutsche N, Zachgo S. The ROXY1 C-terminal L**LL motif is essential for the interaction with TGA transcription factors. Plant Physiol. 2011;157(4):2056–2068. doi: 10.1104/pp.111.185199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sham A, Al-Azzawi A, Al-Ameri S, Al-Mahmoud B, Awwad F, Al-Rawashdeh A, Iratni R, AbuQamar S. Transcriptome analysis reveals genes commonly induced by Botrytis cinerea infection, cold, drought and oxidative stresses in Arabidopsis. PLoS One. 2014;9(11):e113718. doi: 10.1371/journal.pone.0113718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seki M, Umezawa T, Urano K, Shinozaki K. Regulatory metabolic networks in drought stress responses. Curr Opin Plant Biol. 2007;10(3):296–302. doi: 10.1016/j.pbi.2007.04.014. [DOI] [PubMed] [Google Scholar]
- 39.Xiong L, Schumaker KS, Zhu JK. Cell signaling during cold, drought, and salt stress. Plant Cell. 2002;14(Suppl):S165–S183. doi: 10.1105/tpc.000596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Apel K, Hirt H. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu Rev Plant Biol. 2004;55:373–399. doi: 10.1146/annurev.arplant.55.031903.141701. [DOI] [PubMed] [Google Scholar]
- 41.Ding ZJ, Yan JY, Xu XY, Yu DQ, Li GX, Zhang SQ, Zheng SJ. Transcription factor WRKY46 regulates osmotic stress responses and stomatal movement independently in Arabidopsis. Plant J. 2014;79(1):13–27. doi: 10.1111/tpj.12538. [DOI] [PubMed] [Google Scholar]
- 42.Sun Y, Yu D. Activated expression of AtWRKY53 negatively regulates drought tolerance by mediating stomatal movement. Plant Cell Rep. 2015;34(8):1295–1306. doi: 10.1007/s00299-015-1787-8. [DOI] [PubMed] [Google Scholar]
- 43.Qiao Z, Li CL, Zhang W. WRKY1 regulates stomatal movement in drought-stressed Arabidopsis thaliana. Plant Mol Biol. 2016;91(1–2):53–65. doi: 10.1007/s11103-016-0441-3. [DOI] [PubMed] [Google Scholar]
- 44.Chen J, Nolan TM, Ye H, Zhang M, Tong H, Xin P, Chu J, Chu C, Li Z, Yin Y. Arabidopsis WRKY46, WRKY54, and WRKY70 Transcription Factors Are Involved in Brassinosteroid-Regulated Plant Growth and Drought Responses. Plant Cell. 2017;29(6):1425–1439. doi: 10.1105/tpc.17.00364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miao Y, Zentgraf U. The antagonist function of Arabidopsis WRKY53 and ESR/ESP in leaf senescence is modulated by the jasmonic and salicylic acid equilibrium. Plant Cell. 2007;19(3):819–830. doi: 10.1105/tpc.106.042705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kesarwani M, Yoo J, Dong X. Genetic interactions of TGA transcription factors in the regulation of pathogenesis-related genes and disease resistance in Arabidopsis. Plant Physiol. 2007;144(1):336–346. doi: 10.1104/pp.106.095299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zander M, La Camera S, Lamotte O, Metraux JP, Gatz C. Arabidopsis thaliana class-II TGA transcription factors are essential activators of jasmonic acid/ethylene-induced defense responses. Plant J. 2010;61(2):200–210. doi: 10.1111/j.1365-313X.2009.04044.x. [DOI] [PubMed] [Google Scholar]
- 48.Stotz HU, Mueller S, Zoeller M, Mueller MJ, Berger S. TGA transcription factors and jasmonate-independent COI1 signalling regulate specific plant responses to reactive oxylipins. J Exp Bot. 2013;64(4):963–975. doi: 10.1093/jxb/ers389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ndamukong I, Abdallat AA, Thurow C, Fode B, Zander M, Weigel R, Gatz C. SA-inducible Arabidopsis glutaredoxin interacts with TGA factors and suppresses JA-responsive PDF1.2 transcription. Plant J. 2007;50(1):128–139. doi: 10.1111/j.1365-313X.2007.03039.x. [DOI] [PubMed] [Google Scholar]
- 50.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22(22):4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Clough SJ, Bent AF. Floral dip: a simplified method for agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16(6):735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 53.Ross AR, Ambrose SJ, Cutler AJ, Feurtado JA, Kermode AR, Nelson K, Zhou R, Abrams SR. Determination of endogenous and supplied deuterated abscisic acid in plant tissues by high-performance liquid chromatography-electrospray ionization tandem mass spectrometry with multiple reaction monitoring. Anal Biochem. 2004;329(2):324–333. doi: 10.1016/j.ab.2004.02.026. [DOI] [PubMed] [Google Scholar]
- 54.Pazmino DM, Rodriguez-Serrano M, Romero-Puertas MC, Archilla-Ruiz A, Del Rio LA, Sandalio LM. Differential response of young and adult leaves to herbicide 2,4-dichlorophenoxyacetic acid in pea plants: role of reactive oxygen species. Plant Cell Environ. 2011;34(11):1874–1889. doi: 10.1111/j.1365-3040.2011.02383.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. The protein sequences of GRX from cassava and Arabidopsis. (DOC 61 kb)
Table S2. The accession number of cassava drought related transcriptome data. Table S3. The RNA-seq data of GRXs in drought stressed cassava. Table S4. The list of primers. Table S5. GO results of MeGRXC15 regulated genes in transgenic Arabidopsis. (XLS 287 kb)
Table S6. Protein sequences of TGA transcription factors from cassava. and Arabidopsis. (DOC 35 kb)
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files.