Abstract
Long non‐coding RNAs (lncRNAs) have emerged as important regulators of cell biology. The mechanisms by which lncRNAs function are likely numerous, and most are poorly understood. Currently, the mechanisms of functional lncRNAs include those that directly involve the lncRNA transcript, the process of their own transcription and splicing, and even underlying transcriptional regulatory elements within the genomic DNA that encodes the lncRNA. As our understanding of lncRNA biology evolves, so have the methods that are utilized to elucidate their functions. In this review, we survey a collection of different methods used to modulate lncRNA expression levels for the assessment of biological function. From RNA‐targeted strategies, genetic deletions, to engineered gene regulatory systems, the advantages and caveats of each method will be discussed. Ultimately, the selection of tools will be guided by which potential lncRNA mechanisms are being investigated, and no single method alone will likely be sufficient to reveal the function of any particular lncRNA.
Keywords: CRISPR, expression, lncRNA, methods
Subject Categories: Methods & Resources, RNA Biology
Glossary
- 2′‐MOE
2′‐O‐methoxyethylribose
- Airn
antisense Igf2r RNA non‐coding
- ASO
antisense oligonucleotide
- BAC
bacterial artificial chromosome
- Blustr
bivalent locus upregulated by the splicing and transcription of an RNA
- BRD4
bromodomain containing 4
- CAGE
cap analysis gene expression
- CCAT1
colon cancer‐associated transcript 1
- Cdkn1a
cyclin‐dependent kinase inhibitor 1A
- CMV
cytomegalovirus
- CRISPRa
clustered regularly interspaced short palindromic repeats activation
- CRISPR
clustered regularly interspaced short palindromic repeats
- CRISPRi
clustered regularly interspaced short palindromic repeats interference
- dCas9
nuclease‐dead Cas9
- Evf2
embryonic ventral forebrain 2
- FDA
United States food and drug administration
- Fendrr
FOXF1 adjacent non‐coding developmental regulatory RNA
- H3K9me3
histone‐3 lysine‐9 trimethylation
- hnRNP‐K
heterogeneous nuclear ribonucleoprotein K
- Igfr2
insulin‐like growth factor receptor 2
- Indel
insertions/deletions
- KRAB
Krüppel‐associated box
- LET
low expression in tumor
- LNA
locked nucleic acid
- lncRNA
long non‐coding RNA
- MALAT1
metastasis‐associated lung adenocarcinoma transcript 1
- MECP2
methyl‐CpG binding protein 2
- MYC
myelocytomatosis
- nt
nucleotides
- PAM
protospacer adjacent motif
- PARIS
psoralen analysis of RNA interactions and structures
- Plscr4
phospholipid scramblase 4
- Pnky
Pinky
- polyA
polyadenylation
- PVT1
plasmacytoma variant translocation 1
- RNAi
RNA interference
- sgRNA
single guide RNA
- SHAPE
selective 2′‐hydroxyl acylation and primer extension
- shRNA
short hairpin RNA
- siRNA
small interfering RNA
- SPEN
Spen family transcriptional repressor
- TALEN
transcription activator‐like effector nuclease
- TSS
transcription start site
- Ube3a‐ATS
ubiquitin protein ligase E3A antisense
- VP64
tetrameric viral protein 16 transcription activator domain
- XIST
X‐inactive‐specific transcript
Introduction
After completion of the human genome project—which revealed that only approximately 2% of our DNA code for proteins—the advent of next‐generation sequencing technologies enabled the surprising discovery that a substantial proportion of our non‐coding genome is transcribed into RNA. Bioinformatic studies have annotated tens of thousands of lncRNAs—transcripts > 200 nt in length that do not appear to code for proteins—and such lncRNAs can be mapped to every chromosome 1, 2, 3, 4, 5, 6, 7. While certain lncRNAs are now known to play key roles in critical cellular processes 1, 2, 3, 4—such as lncRNA XIST in X chromosome inactivation 6, 7, 8—the vast majority of lncRNAs have not been demonstrated to have significant biological functions. For this emerging field of research, an important next step is to identify which lncRNAs regulate important aspects of cell and molecular biology, and lncRNA loss‐of‐function and gain‐of‐function approaches are a mainstay for such discovery.
Broadly, the methods used to manipulate the levels of an RNA transcript involve either alterations at the level of the corresponding genomic DNA (e.g., gene modification or local recruitment of transcriptional regulators) or molecular strategies that directly involve the RNA transcript (e.g., RNA knockdown or transfection of RNA molecules). Importantly, not all functional lncRNA loci exert their biological effects through the transcribed RNA molecule itself. While some lncRNA loci do indeed function in trans, producing a lncRNA transcript that functions at locations genetically unlinked and spatially distant from their site of production (e.g., NORAD, HOTAIR, 5, 6) (Fig 1A), other lncRNA loci regulate gene expression in cis (Fig 1B), having transcriptional enhancer‐like function for genes on the same chromosome (e.g., Blustr, linc‐p21 7, 8, 9). Both the process of lncRNA transcription as well as transcript splicing can regulate the expression of a protein coding gene neighbor 7. While the level of lncRNA expression may predict biological function within a particular cell type 10, 11, lncRNA loci can even have enhancer‐like function in the absence of transcription 8. Furthermore, for lncRNA loci known to function in cis, its lncRNA transcripts can have additional biological functions in trans 12. Given the diversity of currently known lncRNA mechanisms (and also those still yet to be discovered), the tools used for functional studies should be carefully considered in the context of how the lncRNA may function.
Figure 1. Mechanisms of lncRNA function.
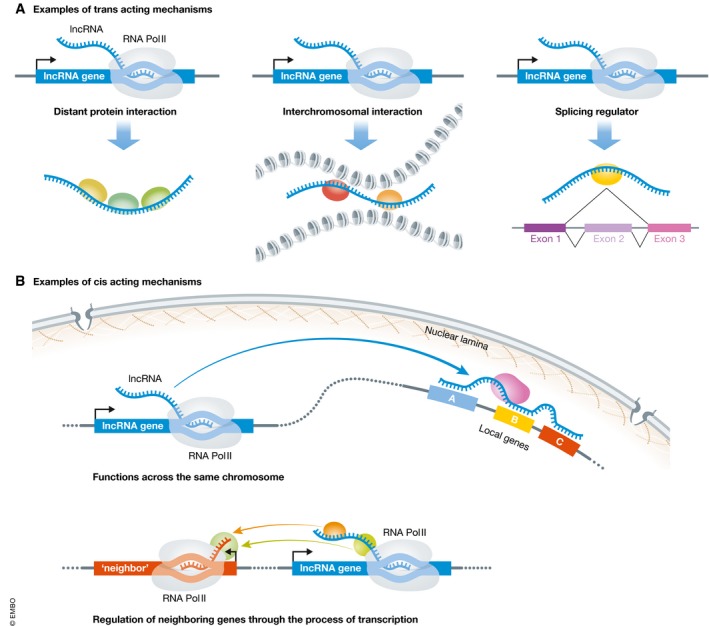
(A) trans‐acting mechanisms of lncRNA function include distal lncRNA–protein interactions (examples include NORAD, HOTAIR), interchromosomal interactions (e.g., FIRRE), and regulation of mRNA splicing (e.g., Pnky, MALAT1). (B) cis‐acting mechanisms of lncRNA function. The lncRNA transcript functions along the same chromosome from which it is transcribed (e.g., XIST). lncRNAs can also function in cis through the process of their own transcription (e.g., Blustr).
Elucidating lncRNA function is also complicated by the genomic arrangement of lncRNAs. Many lncRNAs overlap with coding genes (both in the sense and in the antisense directions) 13, making it often difficult to genetically disrupt the lncRNA without affecting local coding genes. As alluded to above, some lncRNA loci map to known enhancers, which similarly complicates experimental approaches and interpretations of results. Because there are relatively few lncRNAs known to have important functions—even fewer with described molecular mechanisms—and the relative lack of evolutionary conservation 14, 15, the function of lncRNA loci cannot yet be predicted from primary sequences. Furthermore, although the expression of lncRNAs is very cell type‐specific 10, 15, 16, the specificity of expression does not always predict critical biological function 17. For instance, despite being expressed in a diverse range of cell types, some lncRNAs can exhibit exquisitely cell type‐specific function 18. Therefore, loss‐ and gain‐of‐function experiments are paramount to understanding the function of lncRNAs.
To study lncRNA function by loss‐of‐function or gain‐of‐function methods, it is important to begin with a sound understanding of transcript properties (e.g., its primary sequence, potential isoforms, presence or lack of polyadenylation) as well as corresponding DNA loci (e.g., accurate mapping of the TSS, genomic relationship to protein coding genes and known enhancers). For the purposes of this review, we will assume that these basic aspects of lncRNA bioinformatics are known. Furthermore, we focus our discussion on the experimental tools used to modulate lncRNA expression for the purpose of demonstrating biological function. Experimental approaches for investigating the molecular mechanism of lncRNAs have been reviewed elsewhere 19, 20, 21. Since genome‐scale genetic screens have been extremely powerful for the discovery of protein coding gene networks as well as non‐coding DNA elements 22, 23, 24, and similar screens of lncRNA loci have recently proven useful 18, 25, 26, we also discuss the suitability of different methods for large‐scale screening.
Direct targeting of lncRNA transcripts
Building off its success in the knockdown of protein coding genes, RNAi has been frequently used to deplete lncRNAs 16, 27, 28, 29. Typically, after transfection of siRNAs or expression of shRNAs, RNAi triggers the degradation of target RNA molecules through direct complementarity, mediated by the RNA‐induced silencing complex (Fig 2A), and for mRNAs, protein translation can be inhibited as well 30, 31, 32. The efficiency of RNAi‐mediated knockdown is variable, depending in part on the subcellular localization of the target RNA 33, 34. Although mammalian RNAi is thought to predominantly occur in the cytoplasm, RNAi factors such as Argonaute and Dicer have been found in the nuclei of cells as well 35, which may explain how RNAi achieves knockdown of nuclear enriched lncRNAs such as MALAT1 36 and Pnky 16, 37. Knockdown efficiency also relates to the degree of secondary structure of the target RNA molecule, with the extent of knockdown anticorrelated with the amount of energy required to disrupt the local secondary structure 38, 39, 40. Therefore, consideration of the location of stem loops and helices, which can be facilitated by methods such as SHAPE and PARIS 41, 42, may facilitate the use of RNAi.
Figure 2. Methods of lncRNA loss of function.

(A) RNAi mediated by RISC cleavage of lncRNA. (B) ASO‐mediated recruitment of RNAse H to target lncRNA transcript for degradation. (C) Cas13‐based direct RNA cleavage of target lncRNA. (D) Insertion of polyA transcription termination signals into lncRNA gene locus. (E) Gene body deletion of lncRNA. (F) Genetic deletion of exon 1. (G) CRISPRi‐mediated knockdown of lncRNA expression.
Given the compact size of the RNA effectors, RNAi experiments have been successfully scaled up for high throughput and pooled genetic screens for lncRNA function 29, 43. However, RNAi‐based screens have a significant risk of false positives caused by off‐target effects 39, 40, 41. The specificity of both siRNAs and microRNAs depends primarily on a 7‐ to 8‐nt region called the “seed” sequence, which must be taken into account when designing RNAi experiments for lncRNAs, as mismatches of even one nucleotide in this region along with unintended complementarity to other genes can lead to inefficient knockdown of the lncRNA and/or extensive off‐target effects, respectively 44, 45, 46, 47, 48, 49. One way to counteract potential off‐target effects is to test multiple siRNA or shRNAs against the same lncRNA target and assess for concordance of phenotype. Whether or not the “pooling” of multiple siRNAs decreases off‐target effects is controversial 50. It should also be noted that ectopic expression of RNAi‐resistant lncRNA transcripts may not be a suitable rescue strategy, unless the lncRNA is thought to act in trans, since these rescue transcripts are not likely produced from their native loci and may not be produced at normal levels.
An alternative to RNAi for the degradation of lncRNAs are ASOs (Fig 2B). ASOs are 15–20‐nt single‐stranded DNA oligomers that are typically chemically modified to increase the efficacy of knockdown and decrease in vivo toxicity. In particular, the 2′‐MOE and LNA gapmer modifications have been shown to increase affinity toward target RNA transcripts and endow resistance to nucleases 51, 52, 53, allowing these modified ASOs to have half‐lives between days to several weeks in vivo 54, 55. ASOs hybridize with target RNA transcripts through complementarity and induce RNaseH‐mediated degradation of the target transcripts 56. Thus, in contrast to RNAi‐based methods, ASO‐mediated knockdown is very efficient in the nucleus, making this approach suitable for studying the function of both cis‐acting and trans‐acting lncRNAs 9, 57, 58. It remains unclear whether ASOs are suitable for identifying lncRNA genes that function through the act of transcription itself.
Through a process called gymnosis, ASOs can enter cells without the aid of transfection reagents, and there are now a number of ASOs used as pharmaceuticals to treat human disease 59, 60. While FDA‐approved ASOs currently target protein coding transcripts, ASOs that target specific lncRNAs also exhibit therapeutic promise. For instance, when injected into the mouse brain ventricle, ASOs can trigger knockdown of the Angelman's syndrome associated lncRNA Ube3a‐ATS, resulting in improvement of behavioral deficits associated with this genetic disorder 61. In a mouse model of breast cancer, intravenous injection of Malat1 ASOs decreases tumor metastases as compared to scrambled ASO controls 11. However, due to the structural modifications requiring direct chemical synthesis and their relatively high cost, ASOs are currently suboptimal for high throughput or pooled screening and have not been used for genome‐scale screens of non‐coding RNA function.
A more recently developed method that can directly degrade lncRNAs is one that utilizes the Cas13 family of CRISPR ribonucleases 62 (Fig 3C). When provided with sgRNAs complementary to the target RNA, CRISPR‐Cas13 can efficiently cleave the RNA target 52, 53, 54. Cas13 has been used to knockdown lncRNAs in mammalian cells 52, 53, 54, and because the sgRNAs can be stably expressed from viral vectors, Cas13‐based methods may be suitable for genome‐scale screening for lncRNA transcript function.
Figure 3. Methods of lncRNA gain of function.

(A) Expression of lncRNA from a BAC transgene. (B) Direct transfection or injection of mature lncRNA transcripts. (C) Genetic knock‐in of strong promoter element upstream of lncRNA loci to activate lncRNA expression. (D) CRISPRa‐mediated upregulation of lncRNA expression. (E) CRISPR display mediating ectopic localization of lncRNAs through a dCas9/lncrna‐sgRNA chimera.
Genome modification‐based strategies
Genetic deletion of endogenous genes through homologous recombination is an established loss‐of‐function technique 63 (Fig 2E), and one that has been greatly accelerated by CRISPR/Cas9‐mediated gene editing 64, 65, 66. Two of the earliest studied mammalian lncRNAs, XIST and H19, have been deleted through replacement of the entire lncRNA locus with a drug selection cassette, revealing their roles in dosage compensation and imprinting, respectively 67, 68. More recently, 18 different lncRNAs were knocked out in mice through replacement of the lncRNA gene body with a lacZ reporter cassette 69. Furthermore, conditional knockout strategies have enabled lineage‐specific deletion of lncRNAs, which revealed a surprising role of XIST in hematopoietic malignancies 70. Caveats of genetic deletion approaches include the potential introduction of strong promoters, which can affect nearby transcription 71; residual loxP sites, which can trigger embryonic methylation of targeted genes 72; and the unintentional removal or disruption of local DNA regulatory elements such as transcription factor binding sites, enhancers, and CpG islands 73. Of note, these issues with genetic deletion are not unique to the study of lncRNAs (i.e., many genes that encode proteins also contain DNA regulatory elements 74, 75). Limiting the size of the genetic deletion can mitigate such concerns and provide additional insights into lncRNA mechanism. For instance, deletion of the A‐repeat region of XIST demonstrated that these lncRNA sequences mediate X chromosome inactivation by interacting with the transcriptional repressor SPEN, in addition to Lamin B receptor, leading to the recruitment of the inactive X to the nuclear lamina 76, 77, 78, 79, 80. Single exon deletions have also been performed on lncRNAs 7 (Fig 2F). However, the implementation of such size‐limited genetic deletions assumes some a priori knowledge of how the lncRNA functions. Another point of consideration in deletion experiments in vivo is the genetic background in which the deletion is performed, which can contribute to the penetration of subtle lncRNA phenotypes 76, 77, 78.
Premature termination of lncRNA transcription through knock‐in of polyA sites is also an effective loss‐of‐function strategy that may reduce the risk of disrupting known or potential DNA regulatory elements 79, 80 (Fig 2D). For instance, insertion of a triple polyA transcription stop site into exon 1 of the neural lncRNA Evf2 results in a truncated transcript while sparing known local cis regulatory regions 81. Similarly, the mesoderm‐specific lncRNA Fendrr was targeted by insertion of a polyA termination cassette into exon 1 of the lncRNA gene, resulting in embryonic lethality 82. While usually effective, the insertion of polyA sites does not always result in complete lncRNA knockdown. For instance, for lncRNA Dlx1as, despite the presence of multiple polyA sites after exon 1, levels of Dlx1as remain at ~40%, perhaps due to transcriptional read‐through 83.
One important advantage of polyA site insertion is that because it inhibits transcriptional elongation, mechanisms that involve the process of transcription itself can sometimes be distinguished from those that depend upon the intact, full‐length lncRNA transcript. Taking advantage of the ability to make this mechanistic distinction, by using progressively downstream polyA insertions and promoter repositioning, Latos and colleagues were able to show that the lncRNA Airn represses its neighboring gene Igfr2 through transcription over the Igf2r promoter 84. Similarly, the lncRNA Upperhand was shown to regulate its divergently transcribed coding neighbor gene Hand2 by the process of its transcription through a cardiac lineage super‐enhancer. While polyA cassette insertion into exon 2 of Upperhand resulted in diminished Hand2 expression, insertion of the tdTomato coding sequence into the same exon 2—thereby disrupting the primary sequence of Upperhand, did not affect Hand2 expression 85. By comparing polyA insertion mutants with exon 1 deletion of lncRNAs, Engreitz and colleagues were also able to demonstrate that certain lncRNAs regulate the expression of neighboring genes through processes that involve transcriptional initiation and elongation as well as RNA splicing 7. Interestingly, the transcription of certain coding genes was also found to positively regulate the levels of coding gene neighbors. However, the relative challenge of introducing polyA cassettes through current gene targeting methods may limit its scalability in genome‐scale screens.
Engineered CRISPR methods of decreasing lncRNA expression
The development of the CRISPR/Cas9 system for mammalian genome editing has greatly facilitated the interrogation of gene function, especially at large scale 22, 86, 87. However, Cas9‐mediated mutagenesis through the generation of double‐strand breaks and non‐homologous end joining is often not suitable for the study of lncRNAs, since by definition they do not produce proteins and therefore may have biological functions that are less likely to be perturbed by small indels that produce frameshift mutations. Double Cas9 excision of DNA sequences that flank lncRNAs are more likely to inactivate lncRNA gene function and has been used to delete up to hundreds of human lncRNAs, revealing the function of previously uncharacterized lncRNA loci 26, 88, 89.
Engineered Cas9 variants, in particular dCas9 fused to transcriptional activators or repressors, are highly effective for modulating the expression of lncRNAs without alterations to the underlying genomic DNA sequence 90, 91, 92, 93, 94, 95, 96. The CRISPRi system (Fig 2G), in which dCas9 is fused with the KRAB repressor domain (dCas9‐KRAB), silences transcription through steric blockade of RNA polymerase elongation and local deposition of H3K9me3, which is a characteristic heterochromatin mark 90, 91, 97. With a precise and relatively narrow window of activity between −50 and +300 bp relative to the TSS of the target gene, CRISPRi can site‐specifically knock down lncRNA expression while minimizing disruption to the activity of cis regulatory regions or neighboring genes 92, making it broadly useful for lncRNAs 10, 18, whose gene structures are often antisense, overlapping, or divergent to other nearby genes 13. Empirical determination of sgRNA mismatch tolerance for CRISPRi and CRISPR/Cas9 has demonstrated exquisite sensitivity in the 12 nt most proximal to the PAM (i.e., seed region), with one mismatch in this region decreasing activity of CRISPRi by up to 100% 92. Despite evidence of widespread binding of sgRNA/Cas9 complexes throughout the genome, it is thought that activity, whether nuclease or transcriptional modulation, requires more than transient interaction to have measurable effects on gene expression 91, 98, 99. Iterations of the engineered CRISPR/Cas9 system, such as addition of the repressor domain of the transcriptional regulator MECP2 to dCas9‐KRAB 96, may be useful for lncRNA knockdown. Furthermore, more accurate mapping of lncRNA TSSs from CAGE analysis 100 and optimizations to the design of sgRNA sequences 101 have augmented the CRISPRi toolbox for the study of lncRNAs, making large‐scale screening more readily accessible. For instance, in a pooled screening approach to discover lncRNA function in seven different human cell lines, of over 16,000 lncRNA loci targeted, 499 exhibit cell growth‐modifying phenotypes, and most of these lncRNA loci were previously unknown to have function 18. Furthermore, with the relatively large scale of this screen that was facilitated by the robustness of CRISPRi, machine learning could be applied to the data, revealing genomic features that predict essential lncRNA function 18.
With CRISPRi, because site‐specific gene repressors are recruited to the genomic DNA, it is possible that cis regulatory elements such as enhancers—embedded within or near the TSS of lncRNA genes—are also affected. CRISPRi is indeed capable of silencing transcriptional enhancers, enabling the identification and characterization of cis regulatory regions 23, 97. However, the narrow window of activity of CRISPRi complexes, their minimal off‐target effects, and restriction of sgRNAs to the TSS of lncRNAs all reduce the potential of unintentional perturbation of enhancers 92, 101. Furthermore, in the study of protein coding genes, the concordance of results from CRISPRi‐ and Cas9 nuclease‐mediated screens suggests that CRISPRi‐mediated results do not generally arise from the unintentional modulation of cis regulatory regions 101, 102. In any case, as best practice, multiple sgRNAs targeting the same lncRNA should be tested for validation studies, and orthogonal loss‐of‐function experiments (e.g., ASO‐mediated transcript knockdown or insertion of polyA termination sites) can also be performed to help decipher the function(s) of the lncRNA locus and its transcriptional product.
lncRNA gain‐of‐function strategies
The function of lncRNAs can also be discovered by their overexpression. For instance, viral vectors that produce specific lncRNAs have been used to overexpress lncRNA‐LET and Plscr4, demonstrating that increased levels of these lncRNAs can affect hypoxia signaling and cardiac hypertrophy, respectively 103, 104. Of note, many “standard” expression vectors contain sequence elements that target the RNA transcript to ribosomes and enhance translational efficiency, and the inclusion of these sequences may confer lncRNAs with functions that they do not have under physiological conditions. Furthermore, ectopic expression may not target the lncRNA products to their physiological subcellular locations, such as nuclear paraspeckles, chromatin, nuclear lamina, or cytoplasm. Direct injection or transfection of in vitro‐transcribed lncRNAs has also been performed to demonstrate lncRNA function (Fig 3B), and these methods may also be useful for the study of lncRNAs that are presumed to function in trans 105. However, neither the expression of lncRNAs from vectors/plasmids nor the introduction of in vitro‐transcribed lncRNAs preserves information encoded within and surrounding the lncRNA locus and therefore cannot be used to investigate potential cis‐acting mechanisms.
lncRNAs can also be expressed from the local genomic context. One method employs BAC transgenes that contain the lncRNA locus, and these experiments are especially useful for establishing trans‐acting lncRNA mechanisms in vivo (Fig 3A). For instance, a BAC transgene containing the intact lncRNA Fendrr gene can rescue certain phenotypes of Fendrr‐null mice 82. Knock‐in strategies can also activate transcription of lncRNAs at their endogenous loci (Fig 3C). For instance, overexpression of the colorectal cancer‐associated lncRNA CCAT1 has been achieved through TALEN‐mediated knock‐in of a CMV promoter upstream of CCAT1, resulting in 15‐ to 30‐fold increase in CCAT1 expression and, as a consequence, increased MYC expression and tumorigenesis in a colorectal cancer cell line 106.
Programmable transcriptional activation using engineered CRISPR/Cas9 systems has also enabled gain‐of‐function studies for lncRNAs (Fig 3D). By localizing gene activation domains such as VP64 just upstream of the TSS of lncRNAs (or any other gene transcribed by RNA polymerase II) 91, 93, 95, lncRNA genes can be specifically overexpressed 94. These CRISPRa approaches have been used for genome‐wide screens to identify lncRNAs that play roles in drug resistance in cancer cells 107, 108.
Another compelling gain‐of‐function strategy has been termed CRISPR display (Fig 3E). CRISPR display employs an engineered dCas9 that interacts with modified sgRNA‐lncRNA chimeras, allowing site‐specific delivery of lncRNA transcripts of up to several kilobases (or smaller lncRNA domains) to ectopic regions of the genome 109. In addition to testing a subset of different trans lncRNA mechanisms, CRISPR display can distinguish the function of the transcript from the act of transcription for potential cis mechanisms.
Box 1: In need of answers.
In the human genome, there are many thousands of loci that produce long non‐coding RNAs (lncRNAs), and understanding their function has been challenging. The loci that produce lncRNAs can function locally in cis—sometimes even independently of their RNA product—but other lncRNAs can function in trans. Therefore, the selection of tools to modulate lncRNA expression levels, and the interpretation of experimental results, must be carefully considered in the context of how lncRNAs carry out their function. The clearest examples of lncRNA function have utilized a combination of experimental methods, and continued development of these approaches will further aid in our understanding of these enigmatic members of the non‐coding genome.
Reconciling lessons learned from perturbation of lncRNA expression levels
Given the diversity of currently known lncRNA functions and fundamental differences in the methods used to produce lncRNA gain or loss of function, it is perhaps not surprising that our understanding of lncRNA biology has evolved over time. For instance, lincRNA‐p21 was initially described as a lncRNA that regulates the p53 transcriptional response through a global trans‐acting mechanism by interacting with hnRNP‐K to localize at p53 target genes 27. However, those phenotypes relied on RNAi‐mediated knockdown of lincRNA‐p21, which may have led to the overestimation of trans‐acting potential of the lncRNA. Subsequent experiments using allele‐specific deletion and ASO‐mediated degradation of lincRNA‐p21 demonstrated that this lncRNA positively regulates its neighboring gene, Cdkn1a (p21), with the lncRNA transcript being required for this cis regulatory function 9. Later, genetic deletion experiments demonstrated that the lincRNA‐p21 locus contains enhancers that regulate the expression of the neighboring gene Cdkn1a—even in tissues that do not express lincRNA‐p21—and these findings were further validated in luciferase enhancer reporter assays 8.
Another notable example of the mechanistic dichotomies that can exist within a lncRNA locus is the cancer‐related PVT1, which is ~55 kb away from MYC. PVT1 was previously shown to be a trans‐acting lncRNA that stabilizes the MYC protein, promoting oncogenesis 110. Consistent with its known tumor‐promoting properties, CRISPRa‐mediated activation of PVT1 conferred drug resistance to leukemia cells, as shown through pooled genetic screens 107, 108. Unexpectedly, CRISPRi‐mediated repression of PVT1 also enhanced the proliferation of leukemia and breast cancer cells 18. Cho and colleagues subsequently showed that the promoter of PVT1 acts as a DNA boundary element that competitively binds with distal enhancers of MYC 111. When the PVT1 promoter region is repressed by CRISPRi, these distal MYC enhancers interact with the MYC promoter, activating MYC transcription through a BRD4‐dependent manner. While PVT1 lncRNA transcript levels are repressed after CRISPRi, the pro‐proliferation phenotype is dominated by the cis regulatory mechanism.
The tale of PVT1 is not to discount the utility of site‐specific gene regulatory methods for screens (both small‐ and genome‐scale) to discover lncRNA function. Rather, these methods allow the triaging of functional lncRNA loci, which then motivates further genetic dissection using orthologous techniques discussed here and elsewhere to elucidate their molecular functions 21, 73. As illustrated by these and other well‐characterized lncRNAs, the dissection of lncRNA mechanism(s) will likely involve multiple methods to modulate its expression level, and careful interpretation of results in the context of how each experimental approach functions at the molecular‐genetic level.
Conclusion
The tens of thousands of long non‐coding RNAs in the human genome represent a heterogenous class of genes that can function in a range of different ways. While they all share the property of not encoding proteins, they differ in how they regulate the genome and interact with the cellular machinery. It is now evident that lncRNAs can have cis or trans‐acting function, and in some cases, they can have both. Furthermore, enhancers and other DNA regulatory elements embedded within lncRNA loci can function independently of the lncRNA transcript. Our continued understanding of these genes will require a toolbox of different methods to modulate their expression and test their functions. To more fully understand lncRNA function, it will also be necessary to integrate additional experimental methods that build from our growing understanding of each lncRNA mechanism, such as lncRNA–protein binding assays, RNA structure interrogation, and higher order chromatin organization mapping. Nonetheless, the methods described in this review can also be applied to the study of lncRNA mechanisms, for instance through loss‐of‐function screening of putative lncRNA interacting proteins. The selection of tools will be guided by the unique characteristics and underlying mechanisms of each specific lncRNA. The development and use of these tools can also facilitate the study of protein coding genes and other non‐coding elements of the genome.
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgements
This study was supported by NIH grants 1R01NS091544, R21NS101395A, the Childhood Brain Tumor Foundation, the Broad Foundation, and the Hana Jabsheh Initiative to D.A.L.
EMBO Reports (2018) 19: e46955
See the Glossary for abbreviations used in this article.
Contributor Information
S John Liu, Email: liujohn@gmail.com.
Daniel A Lim, Email: daniel.lim@ucsf.edu.
References
- 1. Rinn JL, Chang HY (2012) Genome regulation by long noncoding RNAs. Annu Rev Biochem 81: 145–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Quinn JJ, Chang HY (2016) Unique features of long non‐coding RNA biogenesis and function. Nat Rev Genet 17: 47–62 [DOI] [PubMed] [Google Scholar]
- 3. Ransohoff JD, Wei Y, Khavari PA (2018) The functions and unique features of long intergenic non‐coding RNA. Nat Rev Mol Cell Biol 19: 143–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lee JT (2012) Epigenetic regulation by long noncoding RNAs. Science 338: 1435–1439 [DOI] [PubMed] [Google Scholar]
- 5. Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, Tsai M‐C, Hung T, Argani P, Rinn JL et al (2011) Long non‐coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature 464: 1071–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lee S, Kopp F, Chang T‐C, Sataluri A, Chen B, Sivakumar S, Yu H, Xie Y, Mendell JT (2016) Noncoding RNA NORAD regulates genomic stability by sequestering PUMILIO proteins. Cell 164: 69–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Engreitz JM, Haines JE, Perez EM, Munson G, Chen J, Kane M, McDonel PE, Guttman M, Lander ES (2016) Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature 539: 452–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Groff AF, Sanchez‐Gomez DB, Soruco MML, Gerhardinger C, Barutcu AR, Li E, Elcavage L, Plana O, Sanchez LV, Lee JC et al (2016) In vivo characterization of Linc‐p21 reveals functional cis‐regulatory DNA elements. Cell Rep 16: 2178–2186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dimitrova N, Zamudio JR, Jong RM, Soukup D, Resnick R, Sarma K, Ward AJ, Raj A, Lee JT, Sharp PA et al (2014) LincRNA‐p21 activates p21 in cis to promote Polycomb target gene expression and to enforce the G1/S checkpoint. Mol Cell 54: 777–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liu SJ, Nowakowski TJ, Pollen AA, Lui JH, Horlbeck MA, Attenello FJ, He D, Weissman JS, Kriegstein AR, Diaz AA et al (2016) Single‐cell analysis of long non‐coding RNAs in the developing human neocortex. Genome Biol 17: 67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Arun G, Diermeier S, Akerman M, Chang K‐C, Wilkinson JE, Hearn S, Kim Y, MacLeod AR, Krainer AR, Norton L et al (2016) Differentiation of mammary tumors and reduction in metastasis upon Malat1 lncRNA loss. Genes Dev 30: 34–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Paralkar VR, Taborda CC, Huang P, Yao Y, Kossenkov AV, Prasad R, Luan J, Davies JOJ, Hughes JR, Hardison RC et al (2016) Unlinking an lncRNA from its associated cis element. Mol Cell 62: 104–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, Guernec G, Martin D, Merkel A, Knowles DG et al (2012) The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res 22: 1775–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Marques AC, Ponting CP (2009) Catalogues of mammalian long noncoding RNAs: modest conservation and incompleteness. Genome Biol 10: R124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cabili MN, Trapnell C, Goff L, Koziol M, Tazon‐Vega B, Regev A, Rinn JL (2011) Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 25: 1915–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ramos AD, Diaz A, Nellore A, Delgado RN, Park K‐Y, Gonzales‐Roybal G, Oldham MC, Song JS, Lim DA (2013) Integration of genome‐wide approaches identifies lncRNAs of adult neural stem cells and their progeny in vivo. Cell Stem Cell 12: 616–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Oliver PL, Chodroff RA, Gosal A, Edwards B, Cheung AFP, Gómez‐Rodríguez J, Elliot G, Garrett LJ, Lickiss T, Szele F et al (2015) Disruption of Visc‐2, a brain‐expressed conserved long noncoding RNA, does not elicit an overt anatomical or behavioral phenotype. Cereb Cortex 25: 3572–3585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu SJ, Horlbeck MA, Cho SW, Birk HS, Malatesta M, He D, Attenello FJ, Villalta JE, Cho MY, Chen Y et al (2017) CRISPRi‐based genome‐scale identification of functional long noncoding RNA loci in human cells. Science 355: eaah7111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. McFadden EJ, Hargrove AE (2016) Biochemical methods to investigate lncRNA and the influence of lncRNA: protein complexes on chromatin. Biochemistry 55: 1615–1630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chu C, Spitale RC, Chang HY (2015) Technologies to probe functions and mechanisms of long noncoding RNAs. Nat Struct Mol Biol 22: 29–35 [DOI] [PubMed] [Google Scholar]
- 21. Kopp F, Mendell JT (2018) Functional classification and experimental dissection of long noncoding RNAs. Cell 172: 393–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wang T, Birsoy K, Hughes NW, Krupczak KM, Post Y, Wei JJ, Lander ES, Sabatini DM (2015) Identification and characterization of essential genes in the human genome. Science 350: 1096–1101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fulco CP, Munschauer M, Anyoha R, Munson G, Grossman SR, Perez EM, Kane M, Cleary B, Lander ES, Engreitz JM (2016) Systematic mapping of functional enhancer‐promoter connections with CRISPR interference. Science 354: 769–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Horlbeck MA, Xu A, Wang M, Bennett NK, Park CY, Bogdanoff D, Adamson B, Chow ED, Kampmann M, Peterson TR et al (2018) Mapping the genetic landscape of human cells. Cell 174: 953–967.e22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Joung J, Engreitz JM, Konermann S, Abudayyeh OO, Verdine VK, Aguet F, Gootenberg JS, Sanjana NE, Wright JB, Fulco CP et al (2017) Genome‐scale activation screen identifies a lncRNA locus regulating a gene neighbourhood. Nature 548: 343–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhu S, Li W, Liu J, Chen C‐H, Liao Q, Xu P, Xu H, Xiao T, Cao Z, Peng J et al (2016) Genome‐scale deletion screening of human long non‐coding RNAs using a paired‐guide RNA CRISPR‐Cas9 library. Nat Biotechnol 34: 1279–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann‐Broz D, Khalil AM, Zuk O, Amit I, Rabani M et al (2010) A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell 142: 409–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A et al (2009) Many human large intergenic noncoding RNAs associate with chromatin‐modifying complexes and affect gene expression. Proc Natl Acad Sci USA 106: 11667–11672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, Young G, Lucas AB, Ach R, Bruhn L et al (2011) lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 477: 295–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Doench JG, Petersen CP, Sharp PA (2003) siRNAs can function as miRNAs. Genes Dev 17: 438–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zeng Y, Yi R, Cullen BR (2003) MicroRNAs and small interfering RNAs can inhibit mRNA expression by similar mechanisms. Proc Natl Acad Sci USA 100: 9779–9784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297 [DOI] [PubMed] [Google Scholar]
- 33. Zeng Y, Cullen BR (2002) RNA interference in human cells is restricted to the cytoplasm. RNA 8: 855–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lennox KA, Behlke MA (2016) Cellular localization of long non‐coding RNAs affects silencing by RNAi more than by antisense oligonucleotides. Nucleic Acids Res 44: 863–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gagnon KT, Li L, Chu Y, Janowski BA, Corey DR (2014) RNAi factors are present and active in human cell nuclei. Cell Rep 6: 211–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schmidt LH, Spieker T, Koschmieder S, Schäffers S, Humberg J, Jungen D, Bulk E, Hascher A, Wittmer D, Marra A et al (2011) The long noncoding MALAT‐1 RNA indicates a poor prognosis in non‐small cell lung cancer and induces migration and tumor growth. J Thorac Oncol 6: 1984–1992 [DOI] [PubMed] [Google Scholar]
- 37. Ramos AD, Andersen RE, Liu SJ, Nowakowski TJ, Hong SJ, Gertz CC, Salinas RD, Zarabi H, Kriegstein AR, Lim DA (2015) The long noncoding RNA Pnky regulates neuronal differentiation of embryonic and postnatal neural stem cells. Cell Stem Cell 16: 439–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shao Y, Chan CY, Maliyekkel A, Lawrence CE, Roninson IB, Ding Y (2007) Effect of target secondary structure on RNAi efficiency. RNA 13: 1631–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Heale BSE, Soifer HS, Bowers C, Rossi JJ (2005) siRNA target site secondary structure predictions using local stable substructures. Nucleic Acids Res 33: e30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yoshinari K, Miyagishi M, Taira K (2004) Effects on RNAi of the tight structure, sequence and position of the targeted region. Nucleic Acids Res 32: 691–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM (2005) RNA structure analysis at single nucleotide resolution by selective 2’‐hydroxyl acylation and primer extension (SHAPE). J Am Chem Soc 127: 4223–4231 [DOI] [PubMed] [Google Scholar]
- 42. Lu Z, Zhang QC, Lee B, Flynn RA, Smith MA, Robinson JT, Davidovich C, Gooding AR, Goodrich KJ, Mattick JS et al (2016) RNA duplex map in living cells reveals higher‐order transcriptome structure. Cell 165: 1267–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lin N, Chang K‐Y, Li Z, Gates K, Rana ZA, Dang J, Zhang D, Han T, Yang C‐S, Cunningham TJ et al (2014) An evolutionarily conserved long noncoding RNA TUNA controls pluripotency and neural lineage commitment. Mol Cell 53: 1005–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Farh KK‐H, Grimson A, Jan C, Lewis BP, Johnston WK, Lim LP, Burge CB, Bartel DP (2005) The widespread impact of mammalian MicroRNAs on mRNA repression and evolution. Science 310: 1817–1821 [DOI] [PubMed] [Google Scholar]
- 45. Birmingham A, Anderson EM, Reynolds A, Ilsley‐Tyree D, Leake D, Fedorov Y, Baskerville S, Maksimova E, Robinson K, Karpilow J et al (2006) 3′ UTR seed matches, but not overall identity, are associated with RNAi off‐targets. Nat Meth 3: 199–204 [DOI] [PubMed] [Google Scholar]
- 46. Jackson AL, Linsley PS (2010) Recognizing and avoiding siRNA off‐target effects for target identification and therapeutic application. Nat Rev Drug Discov 9: 57–67 [DOI] [PubMed] [Google Scholar]
- 47. Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136: 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Amarzguioui M, Holen T, Babaie E, Prydz H (2003) Tolerance for mutations and chemical modifications in a siRNA. Nucleic Acids Res 31: 589–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lin X, Ruan X, Anderson MG, McDowell JA, Kroeger PE, Fesik SW, Shen Y (2005) siRNA‐mediated off‐target gene silencing triggered by a 7 nt complementation. Nucleic Acids Res 33: 4527–4535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Parsons BD, Schindler A, Evans DH, Foley E (2009) A direct phenotypic comparison of siRNA pools and multiple individual duplexes in a functional assay. PLoS ONE 4: e8471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. McKay RA, Miraglia LJ, Cummins LL, Owens SR, Sasmor H, Dean NM (1999) Characterization of a potent and specific class of antisense oligonucleotide inhibitor of human protein kinase C‐alpha expression. J Biol Chem 274: 1715–1722 [DOI] [PubMed] [Google Scholar]
- 52. Kurreck J, Wyszko E, Gillen C, Erdmann VA (2002) Design of antisense oligonucleotides stabilized by locked nucleic acids. Nucleic Acids Res 30: 1911–1918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Grünweller A, Wyszko E, Bieber B, Jahnel R, Erdmann VA, Kurreck J (2003) Comparison of different antisense strategies in mammalian cells using locked nucleic acids, 2’‐O‐methyl RNA, phosphorothioates and small interfering RNA. Nucleic Acids Res 31: 3185–3193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Laxton C, Brady K, Moschos S, Turnpenny P, Rawal J, Pryde DC, Sidders B, Corbau R, Pickford C, Murray EJ (2011) Selection, optimization, and pharmacokinetic properties of a novel, potent antiviral locked nucleic acid‐based antisense oligomer targeting hepatitis C virus internal ribosome entry site. Antimicrob Agents Chemother 55: 3105–3114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Geary RS, Norris D, Yu R, Bennett CF (2015) Pharmacokinetics, biodistribution and cell uptake of antisense oligonucleotides. Adv Drug Deliv Rev 87: 46–51 [DOI] [PubMed] [Google Scholar]
- 56. Wu H, Lima WF, Zhang H, Fan A, Sun H, Crooke ST (2004) Determination of the role of the human RNase H1 in the pharmacology of DNA‐like antisense drugs. J Biol Chem 279: 17181–17189 [DOI] [PubMed] [Google Scholar]
- 57. Werner MS, Sullivan MA, Shah RN, Nadadur RD, Grzybowski AT, Galat V, Moskowitz IP, Ruthenburg AJ (2017) Chromatin‐enriched lncRNAs can act as cell‐type specific activators of proximal gene transcription. Nat Struct Mol Biol 24: 596–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sarma K, Levasseur P, Aristarkhov A, Lee JT (2010) Locked nucleic acids (LNAs) reveal sequence requirements and kinetics of Xist RNA localization to the X chromosome. Proc Natl Acad Sci USA 107: 22196–22201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Monteleone G, Neurath MF, Ardizzone S, Di Sabatino A, Fantini MC, Castiglione F, Scribano ML, Armuzzi A, Caprioli F, Sturniolo GC et al (2015) Mongersen, an oral SMAD7 antisense oligonucleotide, and Crohn's disease. N Engl J Med 372: 1104–1113 [DOI] [PubMed] [Google Scholar]
- 60. Chiriboga CA, Swoboda KJ, Darras BT, Iannaccone ST, Montes J, De Vivo DC, Norris DA, Bennett CF, Bishop KM (2016) Results from a phase 1 study of nusinersen (ISIS‐SMN(Rx)) in children with spinal muscular atrophy. Neurology 86: 890–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Meng L, Ward AJ, Chun S, Bennett CF, Beaudet AL, Rigo F (2015) Towards a therapy for Angelman syndrome by targeting a long non‐coding RNA. Nature 518: 409–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Shmakov S, Abudayyeh OO, Makarova KS, Wolf YI, Gootenberg JS, Semenova E, Minakhin L, Joung J, Konermann S, Severinov K et al (2015) Discovery and functional characterization of diverse class 2 CRISPR‐Cas systems. Mol Cell 60: 385–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Doyle A, McGarry MP, Lee NA, Lee JJ (2011) The construction of transgenic and gene knockout/knockin mouse models of human disease. Transgenic Res 21: 327–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Conibear J, Chia B, Ngai Y, Bates AT, Counsell N, Patel R, Eaton D, Faivre‐Finn C, Fenwick J, Forster M et al (2018) Study protocol for the SARON trial: a multicentre, randomised controlled phase III trial comparing the addition of stereotactic ablative radiotherapy and radical radiotherapy with standard chemotherapy alone for oligometastatic non‐small cell lung cancer. BMJ Open 8: e020690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J (2013) RNA‐programmed genome editing in human cells. Elife 2: e00471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM (2013) RNA‐guided human genome engineering via Cas9. Science 339: 823–826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Marahrens Y, Panning B, Dausman J, Strauss W, Jaenisch R (1997) Xist‐deficient mice are defective in dosage compensation but not spermatogenesis. Genes Dev 11: 156–166 [DOI] [PubMed] [Google Scholar]
- 68. Ripoche MA, Kress C, Poirier F, Dandolo L (1997) Deletion of the H19 transcription unit reveals the existence of a putative imprinting control element. Genes Dev 11: 1596–1604 [DOI] [PubMed] [Google Scholar]
- 69. Sauvageau M, Goff LA, Lodato S, Bonev B, Groff AF, Gerhardinger C, Sanchez‐Gomez DB, Hacisuleyman E, Li E, Spence M et al (2013) Multiple knockout mouse models reveal lincRNAs are required for life and brain development. Elife 2: e01749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Yildirim E, Kirby JE, Brown DE, Mercier FE, Sadreyev RI, Scadden DT, Lee JT (2013) Xist RNA is a potent suppressor of hematologic cancer in mice. Cell 152: 727–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Steshina EY, Carr MS, Glick EA, Yevtodiyenko A, Appelbe OK, Schmidt JV (2006) Loss of imprinting at the Dlk1‐Gtl2 locus caused by insertional mutagenesis in the Gtl2 5′ region. BMC Genet 7: 44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Rassoulzadegan M, Magliano M, Cuzin F (2002) Transvection effects involving DNA methylation during meiosis in the mouse. EMBO J 21: 440–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Bassett AR, Akhtar A, Barlow DP, Bird AP, Brockdorff N, Duboule D, Ephrussi A, Ferguson‐Smith AC, Gingeras TR, Haerty W et al (2014) Considerations when investigating lncRNA function in vivo. Elife 3: e03058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kowalczyk MS, Hughes JR, Garrick D, Lynch MD, Sharpe JA, Sloane‐Stanley JA, McGowan SJ, De Gobbi M, Hosseini M, Vernimmen D et al (2012) Intragenic enhancers act as alternative promoters. Mol Cell 45: 447–458 [DOI] [PubMed] [Google Scholar]
- 75. Kim T‐K, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara‐Haley K, Kuersten S et al (2010) Widespread transcription at neuronal activity‐regulated enhancers. Nature 465: 182–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Li L, Liu B, Wapinski OL, Tsai M‐C, Qu K, Zhang J, Carlson JC, Lin M, Fang F, Gupta RA et al (2013) Targeted disruption of Hotair leads to homeotic transformation and gene derepression. Cell Rep 5: 3–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Amândio AR, Necsulea A, Joye E, Mascrez B, Duboule D (2016) Hotair is dispensible for mouse development. PLoS Genet 12: e1006232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Li L, Helms JA, Chang HY (2016) Comment on “Hotair is dispensable for mouse development”. PLoS Genet 12: e1006406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Maxwell IH, Harrison GS, Wood WM, Maxwell F (1989) A DNA cassette containing a trimerized SV40 polyadenylation signal which efficiently blocks spurious plasmid‐initiated transcription. Biotechniques 7: 276–280 [PubMed] [Google Scholar]
- 80. Friedrich G, Soriano P (1991) Promoter traps in embryonic stem cells: a genetic screen to identify and mutate developmental genes in mice. Genes Dev 5: 1513–1523 [DOI] [PubMed] [Google Scholar]
- 81. Bond AM, VanGompel MJW, Sametsky EA, Clark MF, Savage JC, Disterhoft JF, Kohtz JD (2009) Balanced gene regulation by an embryonic brain ncRNA is critical for adult hippocampal GABA circuitry. Nat Neurosci 12: 1020–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Grote P, Wittler L, Hendrix D, Koch F, Währisch S, Beisaw A, Macura K, Bläss G, Kellis M, Werber M et al (2013) The tissue‐specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell 24: 206–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Kraus P, Sivakamasundari V, Lim SL, Xing X, Lipovich L, Lufkin T (2013) Making sense of Dlx1 antisense RNA. Dev Biol 376: 224–235 [DOI] [PubMed] [Google Scholar]
- 84. Latos PA, Pauler FM, Koerner MV, Şenergin HB, Hudson QJ, Stocsits RR, Allhoff W, Stricker SH, Klement RM, Warczok KE et al (2012) Airn transcriptional overlap, but not its lncRNA products, induces imprinted Igf2r silencing. Science 338: 1469–1472 [DOI] [PubMed] [Google Scholar]
- 85. Anderson KM, Anderson DM, McAnally JR, Shelton JM, Bassel‐Duby R, Olson EN (2016) Transcription of the non‐coding RNA upperhand controls Hand2 expression and heart development. Nature 539: 433–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Wang T, Wei JJ, Sabatini DM, Lander ES (2014) Genetic screens in human cells using the CRISPR‐Cas9 system. Science 343: 80–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, Heckl D, Ebert BL, Root DE, Doench JG et al (2014) Genome‐scale CRISPR‐Cas9 knockout screening in human cells. Science 343: 84–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Aparicio‐Prat E, Arnan C, Sala I, Bosch N, Guigó R, Johnson R (2015) DECKO: single‐oligo, dual‐CRISPR deletion of genomic elements including long non‐coding RNAs. BMC Genom 16: 846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Covarrubias S, Robinson EK, Shapleigh B, Vollmers A, Katzman S, Hanley N, Fong N, McManus MT, Carpenter S (2017) CRISPR/Cas‐based screening of long non‐coding RNAs (lncRNAs) in macrophages with an NF‐κB reporter. J Biol Chem 292: 20911–20920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, Lim WA (2013) Repurposing CRISPR as an RNA‐guided platform for sequence‐specific control of gene expression. Cell 152: 1173–1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern‐Ginossar N, Brandman O, Whitehead EH, Doudna JA et al (2013) CRISPR‐mediated modular RNA‐guided regulation of transcription in eukaryotes. Cell 154: 442–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, Guimaraes C, Panning B, Ploegh HL, Bassik MC et al (2014) Genome‐scale CRISPR‐mediated control of gene repression and activation. Cell 159: 647–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Tanenbaum ME, Gilbert LA, Qi LS, Weissman JS, Vale RD (2014) A protein‐tagging system for signal amplification in gene expression and fluorescence imaging. Cell 159: 635–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, Hsu PD, Habib N, Gootenberg JS, Nishimasu H et al (2015) Genome‐scale transcriptional activation by an engineered CRISPR‐Cas9 complex. Nature 517: 583–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Chavez A, Scheiman J, Vora S, Pruitt BW, Tuttle M, P R Iyer E, Lin S, Kiani S, Guzman CD, Wiegand DJ et al (2015) Highly efficient Cas9‐mediated transcriptional programming. Nat Meth 12: 326–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Yeo NC, Chavez A, Lance‐Byrne A, Chan Y, Menn D, Milanova D, Kuo C‐C, Guo X, Sharma S, Tung A et al (2018) An enhanced CRISPR repressor for targeted mammalian gene regulation. Nat Meth 31: 230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Thakore PI, D'Ippolito AM, Song L, Safi A, Shivakumar NK, Kabadi AM, Reddy TE, Crawford GE, Gersbach CA (2015) Highly specific epigenome editing by CRISPR‐Cas9 repressors for silencing of distal regulatory elements. Nat Meth 12: 1143–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. O'Geen H, Henry IM, Bhakta MS, Meckler JF, Segal DJ (2015) A genome‐wide analysis of Cas9 binding specificity using ChIP‐seq and targeted sequence capture. Nucleic Acids Res 43: 3389–3404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Wu X, Scott DA, Kriz AJ, Chiu AC, Hsu PD, Dadon DB, Cheng AW, Trevino AE, Konermann S, Chen S et al (2014) Genome‐wide binding of the CRISPR endonuclease Cas9 in mammalian cells. Nat Biotechnol 32: 670–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Hon C‐C, Ramilowski JA, Harshbarger J, Bertin N, Rackham OJL, Gough J, Denisenko E, Schmeier S, Poulsen TM, Severin J et al (2017) An atlas of human long non‐coding RNAs with accurate 5′ ends. Nature 543: 199–204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Horlbeck MA, Gilbert LA, Villalta JE, Adamson B, Pak RA, Chen Y, Fields AP, Park CY, Corn JE, Kampmann M et al (2016) Compact and highly active next‐generation libraries for CRISPR‐mediated gene repression and activation. Elife 5: 914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Evers B, Jastrzebski K, Heijmans JPM, Grernrum W, Beijersbergen RL, Bernards R (2016) CRISPR knockout screening outperforms shRNA and CRISPRi in identifying essential genes. Nat Biotechnol 34: 631–633 [DOI] [PubMed] [Google Scholar]
- 103. Yang F, Huo X‐S, Yuan S‐X, Zhang L, Zhou W‐P, Wang F, Sun S‐H (2013) Repression of the long noncoding RNA‐LET by histone deacetylase 3 contributes to hypoxia‐mediated metastasis. Mol Cell 49: 1083–1096 [DOI] [PubMed] [Google Scholar]
- 104. Lv L, Li T, Li X, Xu C, Liu Q, Jiang H, Li Y, Liu Y, Yan H, Huang Q et al (2018) The lncRNA Plscr4 controls cardiac hypertrophy by regulating miR‐214. Mol Ther Nucleic Acids 10: 387–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP (2011) Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell 147: 1537–1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Xiang J‐F, Yin Q‐F, Chen T, Zhang Y, Zhang X‐O, Wu Z, Zhang S, Wang H‐B, Ge J, Lu X et al (2014) Human colorectal cancer‐specific CCAT1‐L lncRNA regulates long‐range chromatin interactions at the MYC locus. Cell Res 24: 513–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Bester AC, Lee JD, Chavez A, Lee Y‐R, Nachmani D, Vora S, Victor J, Sauvageau M, Monteleone E, Rinn JL et al (2018) An integrated genome‐wide CRISPRa approach to functionalize lncRNAs in drug resistance. Cell 173: 649–664.e20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Boettcher M, Tian R, Blau JA, Markegard E, Wagner RT, Wu D, Mo X, Biton A, Zaitlen N, Fu H et al (2018) Dual gene activation and knockout screen reveals directional dependencies in genetic networks. Nat Biotechnol 36: 170–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Shechner DM, Hacisuleyman E, Younger ST, Rinn JL (2015) Multiplexable, locus‐specific targeting of long RNAs with CRISPR‐display. Nat Meth 12: 664–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Tseng Y‐Y, Moriarity BS, Gong W, Akiyama R, Tiwari A, Kawakami H, Ronning P, Reuland B, Guenther K, Beadnell TC et al (2014) PVT1 dependence in cancer with MYC copy‐number increase. Nature 512: 82–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Cho SW, Xu J, Sun R, Mumbach MR, Carter AC, Chen YG, Yost KE, Kim J, He J, Nevins SA et al (2018) Promoter of lncRNA gene PVT1 is a tumor‐suppressor DNA boundary element. Cell 173: 1398–1412.e22 [DOI] [PMC free article] [PubMed] [Google Scholar]


