Abstract
Proper control of immune responses by Foxp3+ regulatory T cells at inflamed sites is crucial for the prevention of immunopathology. TGF‐β‐induced Foxp3+ regulatory T (Treg) cells are generated in inflammatory environments as well as in steady‐state conditions. Inflammatory cytokines such as IFN‐γ and IL‐4 have an antagonistic effect on Treg cell conversion. However, it is not known how naive CD4+ T cells overcome the inhibitory environment in inflamed sites to differentiate into Treg cells. Here, we show that CCAAT/enhancer‐binding protein (C/EBP) functions as a safeguard that enhances Treg cell generation by dampening the inhibitory effect of IFN‐γ and IL‐4 on Foxp3 expression. We find that C/EBPβ is induced by retinoic acid and binds to the methyl‐CRE sequence in the Foxp3 TSDR to sustain its expression. C/EBPβ‐transduced iT reg cells show more potent suppressive activity in mouse disease models. We also reveal that C/EBPβ‐transduced human iT reg cells exhibit more enhanced suppressor function. These results establish C/EBP as a new molecular target for enhancing the formation and stability of Treg cells in inflammatory environments.
Keywords: C/EBP, Foxp3, inflammatory cytokines, iTreg
Subject Categories: Chromatin, Epigenetics, Genomics & Functional Genomics; Immunology; Transcription
Introduction
Control of immune responses between inflammatory versus regulatory responses is essential for ensuring immune defense without pathologic consequences. Regulatory T cells (Treg) play an indispensable role in maintaining the balance between immunity and regulation in inflamed tissues 1, 2. Foxp3 is a lineage‐specific factor required for Treg differentiation and function 3, 4, 5.
Foxp3+ regulatory T cells are primarily generated in the thymus (tTreg) or extrathymically at peripheral tissues (pTreg). They may also be induced in vitro (iTreg) in the presence of transforming growth factor β (TGF‐β). Treg cells can be generated in inflammatory conditions as well as in the steady states 6. Treg cells generated at inflammatory sites play an essential role in immune homeostasis and in controlling the extents of immune‐mediated pathology 7. Excessive contraction of the number and/or function of Treg cells can lead to immunopathology 8. Peripheral differentiation of Treg cells is profoundly antagonized by inflammatory cytokines, such as IFN‐γ and IL‐4 9, 10, 11. However, how naive CD4+ T cells overcome the inhibitory environment and differentiate into Treg cells in inflamed sites remains unclear.
Peripheral differentiation of Treg cells occurs mainly in the gut‐associated lymphoid tissues (GALT). Enhanced conversion of Treg in the GALT is dependent on the ability of GALT dendritic cells to produce TGF‐β and retinoic acid 12, 13. Retinoic acid (RA) serves as a potent cofactor for the peripheral induction of Treg cells 14, 15. In addition to increasing the frequency and stability of Treg, RA allows peripheral generation of Treg to occur in conditions that repress it, e.g., in the presence of inhibitory cytokines such as IFN‐γ and IL‐4 16, 17, 18. However, it is not known yet how RA functions in these contexts.
CCAAT/enhancer‐binding protein (C/EBP) comprises a family of basic region–leucine zipper (bZIP) transcription factors with six members: C/EBPα, β, γ, δ, ε, and ζ. Among the members that have transcriptional activation domain are those including C/EBPα, β, δ, and ε. C/EBPγ and ζ isoforms lack transactivation domain and function as dominant‐negative inhibitors. It is well established that there exists functional redundancy among C/EBPα, β, δ, and ε in a variety of gene transcriptions. In this study, we identified new roles of C/EBP in Treg cells for enhancing their stability and generation in inflammatory environments.
Results
Upregulation of the C/EBPs expression by RA
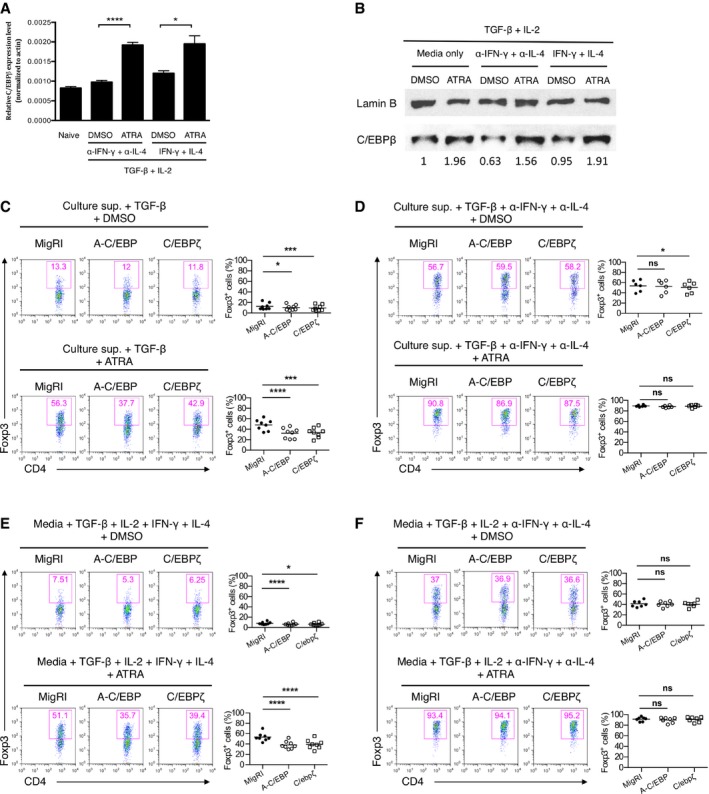
We first investigated whether retinoic acid affects the expression level of C/EBPβ during TGFβ‐induced Foxp3+ Treg differentiation. Given the fact that RA can sustain peripheral generation of Treg cells by overcoming the negative effects of inhibitory cytokines such as IFN‐γ and IL‐4 16, 17, 18, we decided to observe the effect of RA on C/EBPβ expression in the conditions blocking or containing IFN‐γ and IL‐4. Purified CD4+CD25−CD44− naïve T cells from C57BL/6 mice were cultured in iTreg‐polarizing conditions in vitro for 24 h in the presence or absence of all‐trans retinoic acid (ATRA) with or without inhibitory cytokines, IFN‐γ and IL‐4. Treatment of TGF‐β plus ATRA enhanced the expression of C/EBPβ compared with stimulation with TGF‐β alone at both mRNA and protein levels (Fig 1A and B). However, the addition of exogenous IFN‐γ and IL‐4 made little difference in the expression of C/EBPβ compared to the addition of anti‐IFN‐γ and anti‐IL‐4 antibodies. In addition, we found that RA also increases the expression levels of C/EBPα and C/EBPδ (Fig EV1).
Figure 1. C/EBP functions in the presence of inhibitory cytokines.
-
AReal‐time qRT–PCR analysis of C/EBPβ mRNA in CD4+ naïve T cells inactivated (naive) or cultured for 24 h under conditions as indicated with DMSO or ATRA (all‐trans retinoic acid). Data are representative of two independent experiments with consistent results and normalized with β‐actin (mean and s.e.m. of quadruplicates).
-
BImmunoblot analysis of C/EBPβ in the nuclear fraction of naïve CD4+ T cells cultured for 2 days under conditions as indicated. The immunoblot was quantified using ImageJ.
-
C–FFlow cytometry of intracellular Foxp3 staining in CD4+ naïve T cells transduced with control retrovirus (MigRI) or retrovirus encoding A‐C/EBP or C/EBPζ and cultured for 2 days under conditions as indicated. Representative experiments are shown in the left panel and pooled data with mean values from 8 (C, E), 6 (D), or 7 (F) independent experiments are shown on the right. Dot plots are gated for CD4+GFP+ and numbers indicate percent Foxp3+ cells in the gate.
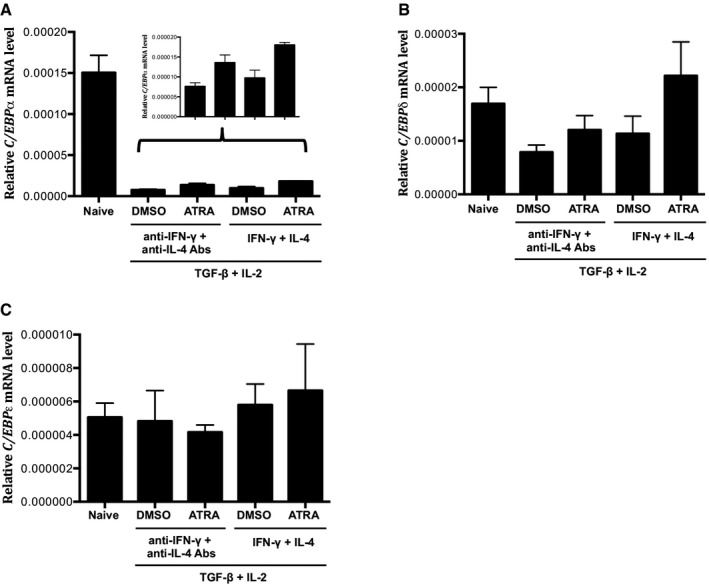
Figure EV1. C/EBPα and C/EBPδ are also upregulated by RA .
-
A–CReal‐time qRT–PCR analysis of (A) C/EBPα, (B) C/EBPδ, (C) C/EBPε mRNA in CD4+ naïve T cells inactivated (naive) or cultured for 24 h under conditions as indicated. Data are representative of two independent experiments with consistent results and normalized with β‐actin (mean and s.e.m. of quadruplicates).
C/EBP functions in the presence of inhibitory cytokines
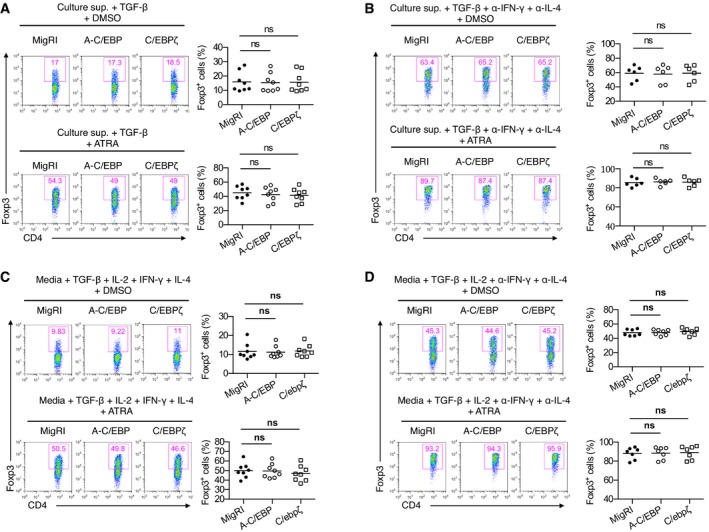
Next, we sought to determine which roles C/EBP plays in iTreg differentiation. Based on our observation that C/EBP expression is upregulated by RA, we hypothesized that C/EBP would regulate Foxp3 expression in the presence of RA. Since there is a high possibility of redundancy among C/EBP isoforms, we sought to disrupt functional redundancy among family members by expressing dominant‐negative A‐C/EBP, which inhibits the DNA binding of potentially all C/EBP family members 19 and C/EBPζ that is also a dominant‐negative inhibitor of C/EBP 20. We ectopically expressed dominant‐negative C/EBP (A‐C/EBP) and C/EBPζ in naïve CD4+ T cells by retroviral transduction and cultured them in iTreg‐polarizing conditions with or without ATRA in the presence of culture supernatant from CD4+CD25−CD44+ memory T cells stimulated with anti‐CD3 and anti‐CD28 (hereafter referred to as culture supernatant). Culture supernatant restrains Foxp3 induction as it contains inhibitory cytokines, such as IFN‐γ and IL‐4 16, 21. Enforced expression of A‐C/EBP or C/EBPζ slightly but significantly reduced iTreg differentiation in the presence of culture supernatant (Fig 1C). The addition of ATRA in the culture supernatant resulted in a more significant reduction in iTreg differentiation of A‐C/EBP or C/EBPζ‐transduced cells (Fig 1C). However, in the presence of neutralizing antibodies against IFN‐γ and IL‐4, the downregulation of Foxp3 expression by ectopic expression of A‐C/EBP or C/EBPζ in the presence of culture supernatant was not observed in both the presence and absence of ATRA (Fig 1D). This suggests that C/EBP functions mainly in the presence of inhibitory cytokines, IFN‐γ and IL‐4. The function of C/EBP in iTreg differentiation was also confirmed when culture supernatant was replaced with fresh media containing exogenous IFN‐γ, IL‐4, and IL‐2 (Fig 1E and F). No such effects were found on GFP‐negative cells, indicating a cell‐intrinsic regulation (Fig EV2).
Figure EV2. GFP‐negative cell fraction of Fig 1C–F.
C/EBPβ confers complete resistance to inhibitory cytokines during iTreg generation
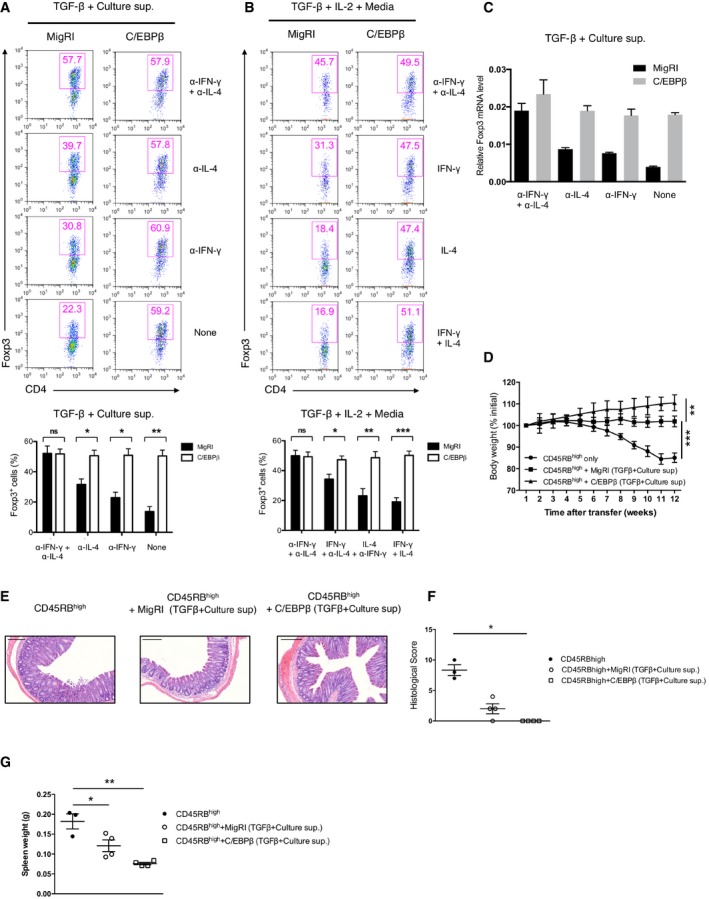
We investigated the effect of ectopic expression of C/EBP in the iTreg differentiation, reasoning that its enforced expression should attenuate the effect of inhibitory cytokines on iTreg generation. As expected, ectopic expression of C/EBPβ resulted in complete resistance to the suppression of Foxp3 expression by culture supernatant (Fig 2A). This effect was observed as long as IFN‐γ, IL‐4, or both of them were present in culture supernatant, which was also confirmed by addition of exogenous inhibitory cytokines (Fig 2B). The increased expression of Foxp3 in C/EBPβ‐transduced cells was again confirmed by quantitative reverse transcription–polymerase chain reaction (qRT–PCR; Fig 2C). We compared the ability of C/EBPα, β, δ, and ε to regulate Foxp3 expression (Fig EV3). Whereas C/EBPδ and C/EBPε caused only a modest resistance, C/EBPα also conferred resistance to inhibitory cytokines as C/EBPβ. With C/EBPα expression, Foxp3 expression was little downregulated in the presence of inhibitory cytokines compared to that in the presence of anti‐IFN‐γ and anti‐IL‐4 antibodies.
Figure 2. C/EBPβ confers complete resistance to inhibitory effect of IFN‐γ and IL‐4 on iT reg generation.
-
A, BFlow cytometry of intracellular Foxp3 staining in CD4+ naïve T cells transduced with control retrovirus (MigRI) or retrovirus encoding C/EBPβ and cultured for 2 days under conditions as indicated. Representative experiments are shown in the upper panel and pooled data from 4 (A), 3 (B) independent experiments are shown in the bottom. Dot plots are gated for CD4+GFP+ and numbers indicate percent Foxp3+ cells in the gate. Error bars represent mean ± s.e.m.
-
CReal‐time qRT–PCR analysis of FACS‐sorted GFP+ cells from CD4+ naïve T cells transduced with control retrovirus (MigRI) or retrovirus encoding C/EBPβ and cultured for 2 days in the conditions as indicated. Data are representative of two independent experiments with consistent results and normalized with β‐actin (mean ± s.e.m. of triplicates).
-
DBody weight of RAG‐2−/− mice after transfer with 5 × 105 CD4+CD45RBhigh cells alone (circles) or with 2 × 105 FACS‐sorted CD4+GFP+ cells transduced with control retrovirus (MigRI) (squares) or retrovirus encoding C/EBPβ (triangles) and cultured for 2 days in the presence of TGF‐β and culture supernatant. Each time point contains three (circles) or four (squares and triangles) mice in each group. Error bars represent mean ± s.d.
-
EHematoxylin and eosin staining of the distal colons of mice (scale bars, 200 μm)
-
F, GEach symbol represents an individual mouse. Small horizontal lines indicate the mean (error bars, s.e.m.). (F) Histological scores. (G) Comparison of the spleen weight.
Figure EV3. C/EPBα, δ, and ε also counteract the effect of inhibitory cytokines on Foxp3 induction.
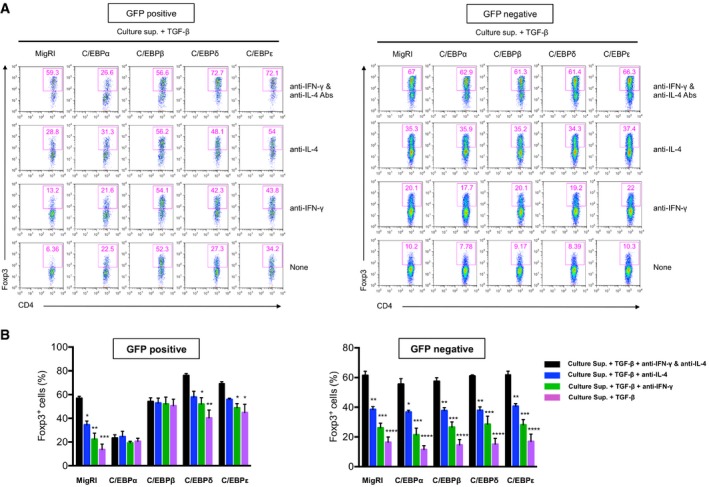
- Flow cytometry of intracellular staining for Foxp3 in CD4+ naïve T cells transduced with control retrovirus (MigRI) or retrovirus encoding C/EBPα, C/EBPβ, C/EBPδ, or C/EBPε and stimulated with anti‐CD3 and anti‐CD28 for 2 days in the presence of TGF‐β and culture supernatant with anti‐IFN‐γ and anti‐IL‐4 Abs, anti‐IL‐4 Ab, anti‐IFN‐γ Ab, or none. Histograms are gated for GFP+ (left panel) and GFP− (right panel) cells. Numbers indicate percent Foxp3+.
- Frequency of Foxp3+ cells from independent three experiments (mean and s.e.m.). Statistical analysis was performed using one‐way ANOVA (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001).
Next, to test whether the Foxp3+ populations in C/EBPβ‐transduced cells that had been resistant to the inhibitory cytokines are the real regulatory T cells with intact suppressive activity, we performed an adoptive transfer experiment using CD45RBhighCD4+ T cells that induce colitis in immune‐deficient mice 22. Mice that received CD45RBhighCD4+ T cells alone developed a wasting disease defined by progressive weight loss (Fig 2D), severe colitis (Fig 2E and F), and splenomegaly (Fig 2G). Mice co‐transferred with control vector‐transduced CD4+ T cells cultured in the presence of TGF‐β and culture supernatant were partially protected from the disease. In contrast, mice co‐transferred with C/EBPβ‐transduced CD4+ T cells cultured under the same conditions did show significant relief from the disease, suggesting considerable attenuation of the pathogenic activity of CD45RBhighCD4+ T cells.
We also tested whether C/EBPβ alone was sufficient to increase Foxp3 expression in the absence of TGF‐β. Less than 3% of C/EBPβ‐transduced CD4+ T cells were Foxp3+, suggesting that C/EBPβ alone is not sufficient for inducing Foxp3 expression (Appendix Fig S1).
Taken together, our findings suggest that C/EBPβ is not responsible for the de novo generation of Foxp3+ iTreg cells, but rather confers resistance to the inhibitory cytokines, IFN‐γ and IL‐4 and thereby ensures stable induction of Foxp3.
Regulation of Foxp3 expression by C/EBP in vivo
To demonstrate the effect of regulating C/EBP expression on peripheral generation of Treg cells in vivo, we investigated the effect of C/EBP on Foxp3 induction using OVA323‐339‐specific DO11.10 TCR‐transgenic T cells deficient in recombination‐activating gene 2 (Rag2). We transduced naive CD4+ T cells from Rag2−/− DO11.10 TCR‐transgenic mice with the retrovirus and adoptively transferred these cells into syngeneic wild‐type recipient mice followed by intravenous injection of the OVA323‐339 peptide. In this model, de novo induction of Foxp3 is significantly interfered by IFN‐γ and IL‐4 9. We observed that A‐C/EBP expression repressed Foxp3 expression in the transferred DO11.10 TCR‐transgenic CD4+ T cells, whereas C/EBPβ expression robustly upregulated the level of Foxp3 expression in the mesenteric lymph nodes (Fig 3A).
Figure 3. C/EBP is critical for the regulation of Foxp3 expression in vivo during iT reg generation.
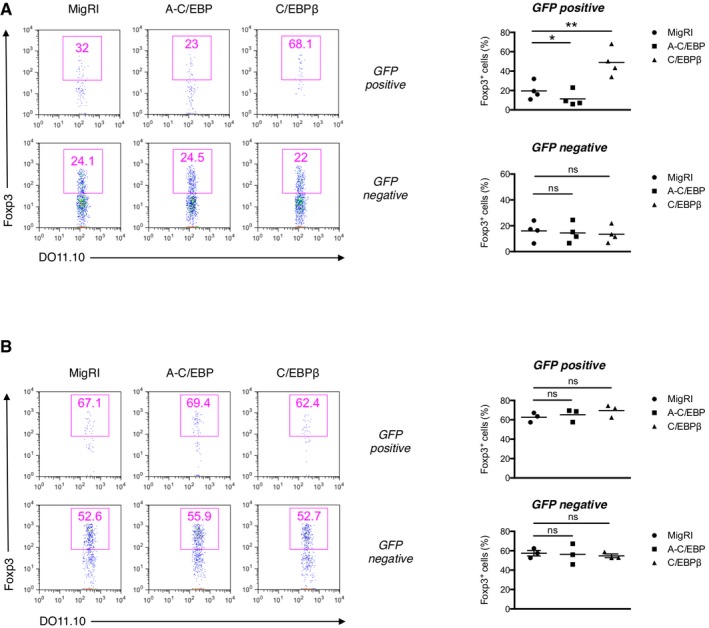
-
A, BFlow cytometry of intracellular Foxp3 staining of mesenteric lymph node cells of Balb/c mice that received RAG2−/−DO11.10 CD4+ naïve T cells transduced with control retrovirus (MigRI) or retrovirus encoding A–C/EBP or C/EBPβ. (A) The recipients were immunized via intravenous injection with OVA323–339 peptide (20 μg) 1 and 3 days after transfer of transduced cells and analyzed 5 days after the first immunization. (B) The recipients were fed 1% OVA solution in drinking water for 5 consecutive days. Representative experiments are shown in the left panel and pooled data from 4 (A) or 3 (B) independent experiments with mean values shown on the right. Dot plots are gated for CD4+KJ.1.26+GFP+ (top) or CD4+KJ.1.26+GFP− (bottom) and numbers indicate percent Foxp3+ cells in the gate. Statistical analysis was performed using one‐way ANOVA (*P < 0.05, **P < 0.01; ns, not significant).
Given the pronounced effect of C/EBP on iTreg differentiation in the presence of inhibitory cytokines in vitro, we evaluated the association between the function of C/EBP and inhibitory cytokines in vivo. Oral antigen administration impairs the development of effector/memory TH2 cells and creates more favorable conditions for Treg cell differentiation 23. To address the Foxp3 expression after oral administration in Rag2−/−DO11.10 adoptive transfer system, the recipient mice were fed OVA in drinking water. When comparing the Foxp3 expression in control vector‐transduced cells, OVA feeding led to 2–3 times higher Foxp3 expression than did intravenous administration (Fig 3A and B). In OVA‐fed mice, contrary to mice i.v injected with OVA, A‐C/EBP or C/EBPβ expression had little effect on Foxp3 expression (Fig 3B), which supports that C/EBP functions in the inhibitory environments in vivo.
C/EBPβ binds to the methyl‐CRE sequence in the Foxp3 TSDR and acts in a methylation‐dependent manner
Recently, it has been shown that the DNA methylation status of the Treg‐specific demethylated region (TSDR) in the Foxp3 locus is critical for maintenance of Foxp3 expression 24, 25, 26, 27, 28. Specifically, the TSDR in tTreg cells is widely demethylated and pTreg also bears gradually demethylated TSDR. In contrast, in vitro induced iTreg cells carried methylated TSDR, which renders them unstable upon restimulation in the absence of exogenous TGF‐β.
Moreover, demethylated TSDR stabilizes Foxp3 expression in the presence of inflammatory cytokines 29, 30. Bisulfite sequencing, however, showed that ectopic expression of C/EBPβ did not affect the methylation status of the TSDR (Fig EV4A and B). CREB has been reported to bind to CpG islands within the TSDR when this region is demethylated 31 and stabilize Foxp3 expression in a TSDR‐dependent manner. Interestingly, CpG methylation of the CRE sequence (TGAmCGTCA) creates C/EBP binding sites 32. Thus, we postulated that the novel function of C/EBPβ might be mediated by C/EBPβ binding to the methyl‐CRE sequence in the Foxp3 TSDR. We performed electrophoretic mobility shift assay (EMSA) to examine the DNA binding of C/EBPβ to the methyl‐CRE sequence of the TSDR (Fig 4A). Jurkat cells transfected with C/EBPβ displayed robust DNA binding activity to the methyl‐CRE sequence, which was completely shifted by anti‐C/EBPβ antibodies. Chromatin immunoprecipitation (ChIP) analysis of C/EBPβ binding coupled with real‐time quantitative PCR showed its occupancy of the TSDR in TGF‐β‐stimulated CD4+ T cells (Fig 4B). In addition, C/EBPβ binding to the TSDR was increased in TGF‐β plus ATRA‐stimulated CD4+ T cells (Fig 4B). Furthermore, when we demethylated the CpG dinucleotides in the genome using 5‐azacytidine (5‐aza), a cytosine analog that cannot be methylated, C/EBPβ binding to the TSDR diminished compared to TGF‐β plus ATRA‐stimulated CD4+ T cells (Fig 4B). The observation that CpG methylation in the CRE sequence facilitates preferential binding of C/EBPβ to the TSDR led us to test whether DNA methylation is required for the C/EBP‐directed resistance to inhibitory cytokines. The addition of 5‐aza in the culture supernatant containing ATRA during iTreg generation abrogated the loss of Foxp3 expression by A‐C/EBP or C/EBPζ, which indicates that the methyl‐CRE sequence in the TSDR is critical for C/EBP to regulate Foxp3 expression (Figs 4C and EV5A). The importance of the DNA methylation was also confirmed when culture supernatant was replaced with fresh media containing exogenous IFN‐γ, IL‐4, and IL‐2 (Figs 4D and EV5B). We also confirmed decreased DNA methylation in the TSDR by 5‐aza (Fig EV5C).
Figure EV4. C/EBPβ does not alter the methylation status of the TSDR .
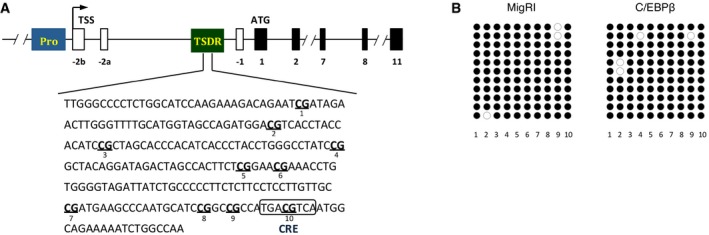
- The Foxp3 gene. Exon–intron structure and the TSDR are depicted (top). The position of 10 CpG motifs and CRE sequence within the TSDR is shown (bottom).
- Bisulfite sequencing of FACS‐sorted GFP+ cells from CD4+ naïve T cells transduced with control retrovirus (MigRI) or retrovirus encoding C/EBPβ and cultured for 2 days in the presence of TGF‐β and culture supernatant.
Figure 4. C/EBPβ binds to methyl‐CRE sequence in the Foxp3 TSDR and acts in a methylation‐dependent manner.
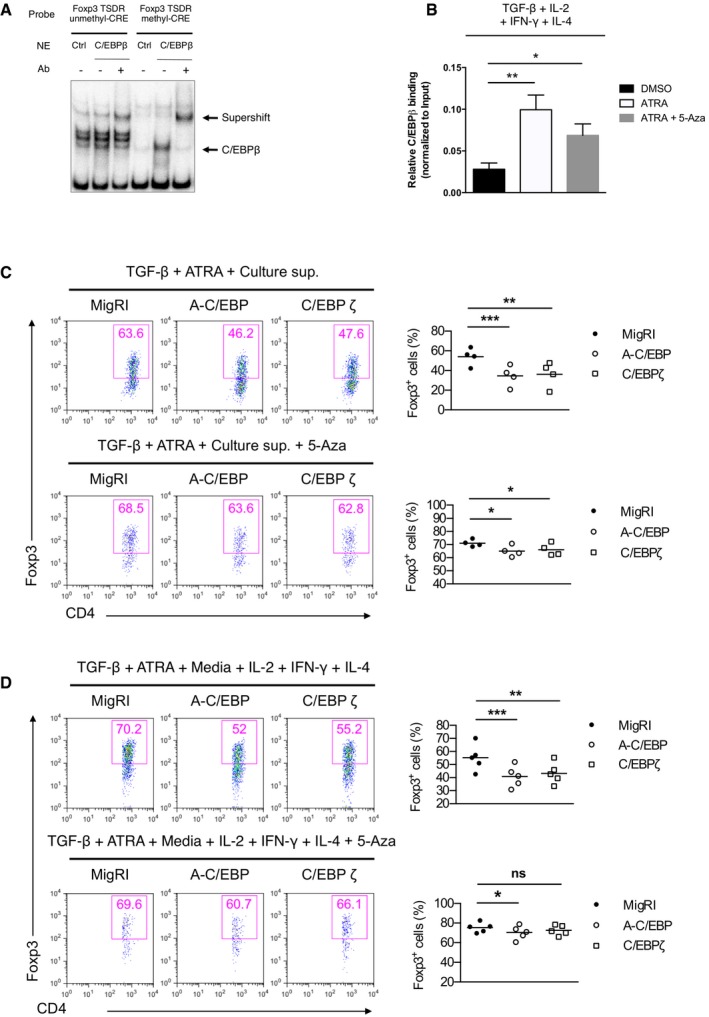
-
AEMSA of nuclear extracts of Jurkat cells transfected with indicated expression vectors and assessed with a probe covering the putative unmethyl‐ or methyl‐CRE sequence in the TSDR.
-
BChIP‐qPCR assay of C/EBPβ on the Foxp3 TSDR in naïve CD4+ T cells cultured for 2 days under conditions as indicated. Data are representative of two independent experiments with consistent results (mean ± s.e.m. of triplicates).
-
C, DFlow cytometry of intracellular Foxp3 staining in CD4+ naïve T cells transduced with control retrovirus (MigRI) or retrovirus encoding C/EBPβ and cultured for 2 days under conditions as indicated. Representative experiments are shown in the left panel and pooled data from 4 (C) or 5 (D) independent experiments with mean values are shown on the right. Dot plots are gated for CD4+GFP+ and numbers indicate percent Foxp3+ cells in the gate.
Figure EV5. GFP‐negative cell fraction of Fig 4C and D, and bisulfite sequencing after 5‐aza treatment.
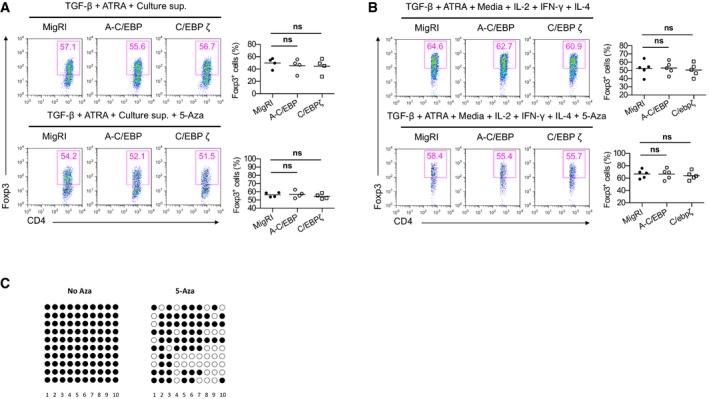
-
A, BGFP‐negative cell fraction of Fig 4C and D.
-
CBisulfite sequencing of TSDR of FACS‐sorted GFP+ cells from CD4+ naïve T cells transduced with control retrovirus (MigRI) and cultured for 2 days in the presence of TGF‐β, ATRA, 5‐aza, and culture supernatant.
Significant attenuation of experimental autoimmune encephalitis and colitis by C/EBPβ‐transduced iTreg cells
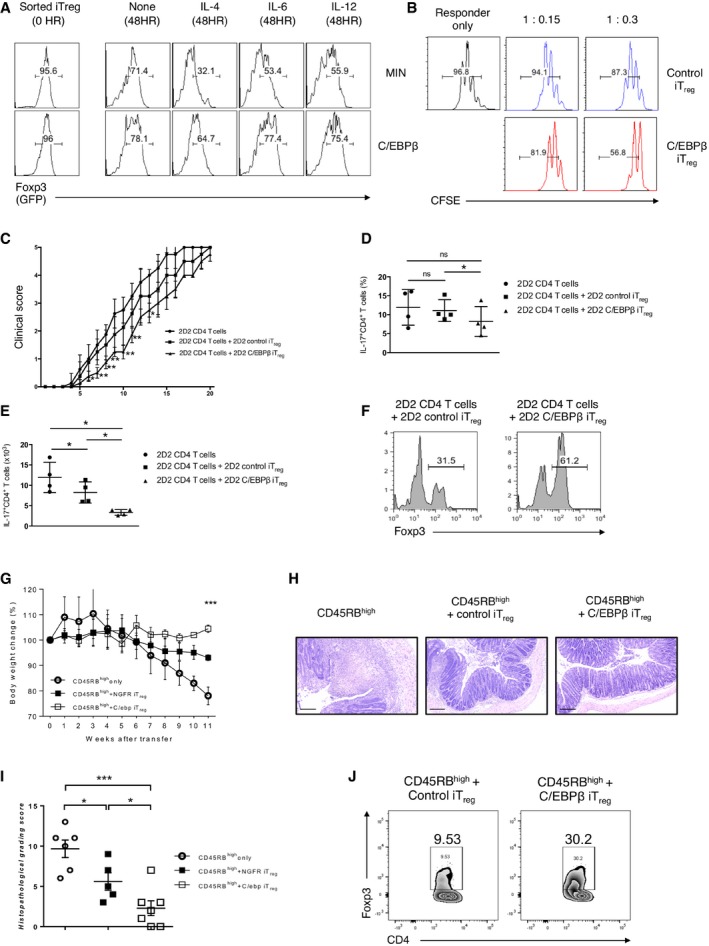
One obstacle to making therapeutic use of iTreg cells generated in vitro is that they have the propensity to be unstable and lose Foxp3 expression. In light of our observation that C/EBPβ confers complete resistance to inhibitory cytokines during the course of iTreg generation, we asked whether C/EBPβ has the capacity to protect Treg identity by preserving its stable Foxp3 expression. To address this, we ectopically expressed C/EBPβ in naïve CD4+ T cells from Foxp3GFP mice by retroviral transduction and cultured them in the presence of TGFβ, anti‐IFN‐γ, and anti‐IL‐4 Abs. We sorted Foxp3/GFP+ cells and cultured them without TGF‐β but in the presence of T‐cell‐polarizing cytokines for additional 2 days. The addition of IL‐4 greatly downregulated Foxp3 expression of control iTreg cells (Fig 5A). IL‐6 and IL‐12, albeit to a lesser degree, also destabilized Foxp3 expression. Of note, C/EBPβ‐transduced iTreg cells faithfully maintain Foxp3 expression.
Figure 5. Significant attenuation of experimental autoimmune encephalitis by C/EBPβ‐transduced iT reg cells.
-
ACD4+ naïve T cells from Foxp3EGFP mice were transduced with control retrovirus (MIN) or retrovirus encoding C/EBPβ (MIN‐C/EBPβ) and stimulated with anti‐CD3 and anti‐CD28 for 2 days in the presence of TGF‐β, anti‐IFN‐γ, and anti‐IL‐4 Abs. GFP+(Foxp3+) hNGFR+ T cells were sorted to high purity (0 h) and restimulated with anti‐CD3 and anti‐CD28 for 2 days in the presence of IL‐4, IL‐6, IL‐12, or none. Foxp3 expression was assessed based on GFP expression. Data are representative of two independent experiments with consistent results.
-
BFlow cytometry of CFSE‐labeled CD45.1+CD4+ T cells stimulated 3 days alone or together with CD45.2+GFP(Foxp3)+NGFR+‐sorted CD4+ naïve T cells from Foxp3EGFP mice transduced with control retrovirus (Min) or retrovirus encoding C/EBPβ and cultured for 3 days in the presence of TGF‐β, anti‐IFN‐γ, and anti‐IL‐4 Abs. Histograms are gated for CD45.1+. The ratios shown are responder to suppressor. Data are representative of two independent experiments with consistent results.
-
CEAE disease course of RAG2−/− mice that received CD45.1+2D2CD4+ naive T cells alone or together with CD4+GFP/Foxp3+ iTreg cells from CD45.1−2D2CD4+GFP(Foxp3)+ naïve T cells transduced with either control retrovirus (MIN) or retrovirus encoding C/EBPβ; cultured in the presence of TGF‐β, anti‐IFN‐γ, and anti‐IL‐4 Abs; and sorted based on GFP/Foxp3 expression. The recipients were immunized with myelin oligodendrocyte glycoprotein peptide (100 μg) 1 day after transfer. Data show mean ± s.e.m. of the EAE clinical score of 9 (day 1 to day 11), 5 (day 12 to day 16), or 4 (day 17 to day 20) mice of each group.
-
D, E(D) The percentage of IL‐17‐producing cells in CD45.1+Vβ11+ CD4+ T cells in the spinal cords 11 days after transfer. (E) The absolute number of IL‐17A+CD45.1+Vβ11+CD4+ T cells in the spinal cords 11 days after transfer. Each symbol represents an individual mouse. Small horizontal lines indicate the mean (error bars, s.d.).
-
FFlow cytometry of Foxp3 staining of splenic cells. Histograms are gated for CD45.1−CD4+Vβ11+. Data are representative of two independent experiments with consistent results.
-
GBody weight of RAG‐2−/− mice after transfer with 5 × 105 CD45.1+CD4+CD45RBhigh cells alone (circles) or with 2 × 105 FACS‐sorted CD45.1−CD4+GFP(Foxp3)+NGFR+ cells transduced with control retrovirus (Min)(filled rectangles) or retrovirus encoding C/EBPβ (empty rectangles) and cultured for 2 days in the presence of TGF‐β, anti‐IFN‐γ, and anti‐IL‐4 Abs. Each time point contains three mice in each group. Error bars represent mean ± s.e.m.
-
HHematoxylin and eosin staining of the distal colons of mice (scale bars, 200 μm).
-
IHistological scores. Each symbol represents an individual mouse. Small horizontal lines indicate the mean (error bars, s.e.m.).
-
JThe percentage of Foxp3+ cells in CD45.1−CD4+ T cells in colonic lamina propria 11 weeks after transfer.
To determine whether the increased stability of C/EBPβ‐transduced iTreg cells would benefit their suppressive function, we first investigated their capacity to suppress proliferation of responder CD4+ T cells in vitro. For this, CFSE‐labeled responder cells were cocultured with control or C/EBPβ‐transduced iTreg cells. Remarkably, C/EBPβ‐transduced iTreg cells exhibited a higher suppressive activity compared to control iTreg cells (Fig 5B).
Next, we assessed an enhanced suppressor capacity by C/EBPβ‐transduced iTreg cells in vivo during experimental autoimmune encephalitis, an animal model of multiple sclerosis. We transduced naïve CD4 T cells from CD45.2+ 2D2 Foxp3GFP mice with either an expression vector for C/EBPβ or a control vector and cultured these cells in the presence of TGF‐β, anti‐IFN‐γ, and anti‐IL‐4 Abs. T cells from 2D2 Foxp3GFP mice express a transgenic TCR with specificity for the myelin oligodendrocyte glycoprotein amino acids 35–55 (MOG35–55) as well as a Foxp3GFP reporter. We purified Foxp3/GFP+ populations and transferred those cells into RAG2−/− mice with naïve CD4 T cells from CD45.1+ 2D2 mice. After immunization with MOG35–55 peptide, mice that received CD45.1+ 2D2 CD4 T cells together with C/EBPβ‐transduced CD45.2+ 2D2 Foxp3/GFP+ iTreg cells exhibited an attenuated disease phenotype (Fig 5C), significantly lower IL‐17A production in CD4 T cells from spinal cords (Fig 5D) and considerable reduction in numbers of IL‐17A‐producing cells infiltrated in the spinal cords (Fig 5E) compared to mice that received CD45.1+ 2D2 CD4 T cells alone or with control vector‐transduced CD45.2+ 2D2 Foxp3/GFP+ iTreg cells. Strikingly, C/EBPβ‐transduced iTreg cells retained more of the Foxp3 expression (Fig 5F), suggesting that C/EBPβ‐mediated stabilization of Foxp3 expression acted as a major factor to attenuate the severity of the disease.
To further document the suppressive activity of C/EBPβ‐transduced iTreg cells, we induced colitis by transferring CD45RBhighCD4+ T cells to RAG2−/− mice and compared the suppressive activity between control and C/EBPβ‐transduced iTreg cells. As anticipated, we observed a substantially improved suppressive activity of C/EBPβ‐transduced iTreg cells, which was verified based on weight loss (Fig 5G) and histological scoring (Fig 5H and I). We also confirmed a higher level of Foxp3 expression in C/EBPβ‐transduced iTreg cells (Fig 5J).
C/EBPβ stabilizes FOXP3 expression in human CD4 T cells
To examine the possibility that C/EBPβ may confer resistance to inhibitory cytokines in human iTreg cells as well, we isolated naïve CD4+CD25−CD45RA+ T cells from human peripheral blood, ectopically expressed C/EBPβ by retroviral transduction and cultured them in iTreg‐polarizing conditions (Fig 6A). We found that, in the presence of TGF‐β, C/EBPβ‐transduced human CD4+ T cells were significantly resistant to the inhibitory effect of IFN‐γ and IL‐4 on the FOXP3 expression, whereas control vector‐transduced human CD4 T cells were more susceptible to the repressive effect of inhibitory cytokines on the FOXP3 expression.
Figure 6. Human C/EBPβ also confers resistance to inhibitory effect of IFN‐γ and IL‐4 on human iT reg generation.
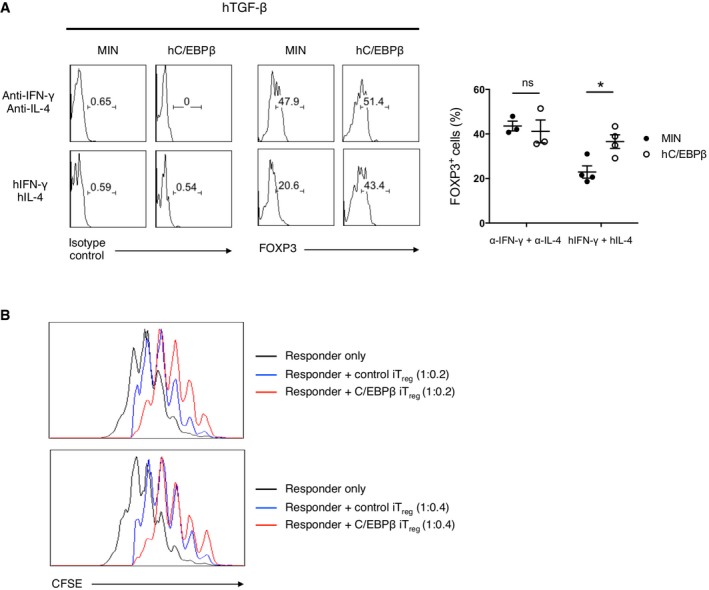
- Flow cytometry of intracellular FOXP3 staining (right) or isotype control (left) in human CD4+CD25−CD45RA+ naïve T cells transduced with control retrovirus (Min) or retrovirus encoding hC/EBPβ and cultured for 2 days under conditions as indicated. Representative experiments are shown in the left panel and pooled data with mean ± s.e.m. are shown on the right. Histograms are gated for CD4+NGFR+. Statistical analysis was performed using unpaired two‐tailed t‐test (*P < 0.05; ns, not significant).
- Flow cytometry of CFSE‐labeled human CD4+CD25− T cells (responder) stimulated 4 days alone or together with NGFR+‐sorted human CD4+CD25−CD45RA+ naïve T cells (suppressor) transduced with control retrovirus (Min) or retrovirus encoding hC/EBPβ and cultured for 5 days in the presence of hTGF‐β, anti‐hIFN‐γ, and anti‐hIL‐4 Abs. Histograms are gated for CFSE+. The ratio of responder to suppressor is 1:0.2 (top) or 1:0.4(bottom). Data are representative of two independent experiments with consistent results.
Next, we cocultured C/EBPβ‐transduced human iTreg cells with CFSE‐labeled responder naïve CD4 T cells to compare their suppressive activity to that of control vector‐transduced iTreg cells. We found that C/EBPβ‐transduced human iTreg cells exerted far more potent suppressor function than did control vector‐transduced iTreg cells (Fig 6B).
Discussion
The presence of inhibitory cytokines such as IFN‐γ and IL‐4 poses a significant obstacle to peripheral differentiation of Treg cells during inflammation. Evidence indicates that differentiating Treg cells are equipped with a self‐protective program that prevents the production of inhibitory cytokines by themselves and allows cells to commit to the Treg cell lineage 33. However, given that IFN‐γ and IL‐4 are secreted in multiple lineages of cells in inflamed sites, the mechanism that blocks the production of inhibitory cytokines by differentiating T cells does not ensure their commitment to the Treg cell lineage. Here, we have demonstrated that C/EBPβ ensures stable induction of Treg cells by overcoming the presence of inhibitory cytokines in both mouse and human systems.
Stabilization of Foxp3 expression is attributed to the epigenetic regulation of the TSDR in Foxp3 locus. The TSDR is substantially demethylated in tTreg cells, but is fully methylated in vitro derived iTreg cells. Although in vivo derived pTreg cells exhibit a demethylated TSDR, it appears to be fully demethylated in several weeks. Importantly, demethylated TSDR is responsible for protecting Foxp3 expression in inflammatory cytokine conditions that restrain Treg generation 30. A number of transcription factors such as Ets‐1, CREB/ATF, and Foxp3 itself bind to the TSDR only when the region is demethylated 27, 31, 34. Notably, all of these transcription factors fail to bind to the TSDR when the region is methylated.
Identification of the molecular mechanism underlying the C/EBPβ‐mediated resistance to the repression of Foxp3 expression by inhibitory cytokines is particularly important, considering the recent active debate on Treg lineage stability and Treg cell therapy. Although we cannot rule out the possibility of other indirect mechanisms by which C/EBPβ mediates the resistance to inhibitory cytokines, the specific association of C/EBPβ with the Foxp3 TSDR suggests that Foxp3 is a direct target gene of C/EBPβ. Interestingly, we found that the ability of C/EBPβ to counteract the repression of Foxp3 expression by inhibitory cytokines is dependent on the binding of C/EBPβ to the methylated Foxp3 TSDR. The TSDR of pTreg seems to be gradually demethylated during the course of differentiation. The complete development of pTreg equipped ultimately with demethylated TSDR might need to be preceded by the unique methylation‐dependent C/EBP activity to protect Foxp3 expression from inhibitory cytokines at the early stages of its development. Identification of the mechanisms that lead to the demethylation of the TSDR may be crucial for the generation of stable iTreg populations aimed at the treatment of a variety of diseases. Our findings, however, suggest a novel way of increasing the stability of iTreg cells without modulating the methylation status of the TSDR, but rather by regulating the level of C/EBP expression.
It has been reported that RA controls TGF‐β‐induced Treg differentiation by multiple mechanisms 14, 16, 17: that is, not only does RA enhance Foxp3 expression independently of inhibitory cytokines, but also attenuates the negative effect of inhibitory cytokines on Foxp3 induction. However, no mechanistic explanation on whether the dual roles of RA in Foxp3 expression originate from a common route or disparate pathways was provided. Regarding this complex problem, we have also uncovered the existence of a novel molecular mechanism by which C/EBPβ elevated by retinoic acid counteracts the repression of Foxp3 expression by inhibitory cytokines.
Our data presented in this study demonstrate that C/EBPα, δ, and ε are also able to confer resistance to inflammatory cytokines, IFN‐γ and IL‐4, albeit C/EBPβ is the most potent. This result implies the functional redundancy among them. In line with this result, we observed no differences in Foxp3 expression in C/EBPβ‐deficient CD4 T cells even in the presence of inflammatory cytokines and RA (Appendix Fig S2). Considering the functional redundancy among C/EBP family members, single‐gene knockout may not be sufficient to uncover the physiological roles of C/EBP family members. Thus, relatively normal differentiation of iTreg cells in C/EBPβ‐deficient CD4 T cells appears to be mainly due to the availability of C/EBPα, δ, and ε. The upregulation of C/EBPα and δ expression by RA makes them likely alternatives to C/EBPβ. However, the functional redundancy among C/EBP members remains to be investigated further in detail. Nevertheless, our findings suggest that C/EBP targeting can be applied for novel applications treating inflammatory diseases.
Materials and Methods
Mice
Balb/c and C57BL/6 mice were purchased from The Jackson Laboratory. Foxp3EGFP mice and 2D2 TCR‐transgenic mice were kindly provided by T. A. Chatila (the University of California at Los Angeles) and Dong‐Sup Lee (Seoul National University), respectively. All mice were bred and maintained in specific pathogen‐free barrier facilities at Seoul National University and were used according to protocols approved by the Institutional Animal Care and Use Committees (IACUC) of Seoul National University.
Antibodies and reagents
The Abs against CD3ε (2C11), CD28 (37.51), APC‐ or PE‐conjugated CD‐25 (PC61), PE‐ or biotin‐conjugated human CD271, PE‐conjugated DO11.10 (KJ1.26), PE‐conjugated Vβ11 (RR3‐15), and biotin‐conjugated Vα3.2 [b,c] (RR3‐16) from BD Pharmingen; PE‐Cy7‐conjugated CD4 (GK1.5), APC‐conjugated Foxp3 (FJK‐16s), biotin‐conjugated CD44 (IM7), FITC‐conjugated CD44 (IM7), IL‐4 (11B11), and IFN‐γ (XMG1.2) from eBiosciences; TGF‐β1,2,3 (1D11); C/EBPβ (C‐19), from Santa Cruz. RhTGF‐β1 from eBiosciences, mIL‐6, mIFN‐γ, mIL‐4 from Peprotech, and all‐trans retinoic acid, 5‐azacytidine from Sigma.
Cell sorting
Naïve CD4+ T cells (CD4+CD25−CD44−) and memory CD4+ T cells (CD4+CD25−CD44+) from spleen and lymph node cells of 8‐ to 12‐week‐old C57BL/6 mice 35 were purified using FACS Aria II (BD) or SH800 Cell Sorter (Sony). The purity of naïve and memory CD4+ T cells was consistently >98 and 95%, respectively.
T‐cell differentiation in vitro
All cultures for T cells used RPMI‐1640 medium supplemented with 10% FBS, 2 mM glutamine, 100 U/ml penicillin, 100 μg/ml streptomycin, and 55 μM 2‐mercaptoethanol 36. CD4+ T cells were activated with plate‐bound anti‐CD3 (3 μg/ml) and soluble anti‐CD28 (0.5 μg/ml) antibodies. Concentrations of cytokines and antibodies are as follows: 5 ng/ml TGF‐β, 10 U/ml IL‐2, 10 ng/ml IFN‐γ, 1 ng/ml IL‐4, 10 ng/ml IL‐6, 10 ng/ml IL‐12, 5 μg/ml anti‐IFN‐γ, 5 μg/ml anti‐IL‐4, 10 μg/ml anti‐TGF‐β, 1 μM 5‐aza, 100 nM ATRA. Culture supernatant was harvested after 48 h from cultures of memory CD4+ T cells activated with plate‐bound anti‐CD3 (3 μg/ml) and anti‐CD28 (3 μg/ml) antibodies.
Retroviral transduction
Naïve CD4+ T cells were stimulated for 24 h with plate‐bound anti‐CD3 (3 μg/ml) and soluble anti‐CD28 (1 μg/ml) together with soluble anti‐IFN‐γ (5 μg/ml), anti‐IL‐4 (5 μg/ml), and anti‐TGF‐β (10 μg/ml), and spin‐infected for 1 h with retroviral supernatant containing 6 μg/ml polybrene from Phoenix packaging cells transfected with retroviral expression plasmids.
Intracellular staining
CD4+ T cells were fixed with cytofix/permeabilization solution (BD), stained with anti‐Foxp3‐APC, and analyzed on FACS Canto II (BD). In human cells, Foxp3 staining kit (eBioscience) was used.
Quantitative real‐time PCR
Quantitative real‐time PCR was performed on ABI StepOnePlus real‐time PCR system using SYBR green PCR master mix (Applied Biosystems).
The PCR primers used are as follows: C/EBPα forward, 5′‐CGCAAGAGCCGAGATAAAGC‐3′; C/EBPα reverse, 5′‐CACGGCTCAGCTGTTCCA‐3′; C/EBPβ forward, 5′‐AGCGGCTGCAGAAGAAGGT‐3′; C/EBPβ reverse, 5′‐GGCAGCTGCTTGAACAAGTTC‐3′; C/EBPδ forward, 5′‐CATCGACTTCAGCGCCTACA‐3′; C/EBPδ reverse, 5′‐GCTTTGTGGTTGCTGTTGAAGA‐3′; C/EBPε forward, 5′‐CCTGCAGTCCCCTTCTCAAG‐3′; C/EBPε reverse, 5′‐TCACGTCGCAGTCGGTACTC‐3′; Foxp3 forward, 5′‐GGACAGACCACACTTCATGCA‐3′; Foxp3 reverse, 5′‐GCTGATCATGGCTGGGTTGT‐3′; β‐actin forward, 5′‐CAACGAGCGGTTCCGATG‐3′; β‐actin reverse, 5′‐GCCACAGGATTCCATACCCA‐3′.
Experimental colitis model
RAG‐2−/− mice were injected intravenously with CD4+CD45RBhigh cells (5 × 105) alone or together with FACS‐sorted CD4+GFP+ cells (2 × 105) infected with control retrovirus (MigRI) or C/EBPβ‐encoding retrovirus and cultured for 2 days in the presence of TGF‐β and culture supernatant. Mice were observed daily and weighed weekly. 12 weeks after cell transfer, mice were sacrificed for histological analysis. Tissue samples from distal portion of the large intestine were prepared and fixed in 10% formalin. Fixed tissues were embedded in paraffin and were cut into 6‐μm sections and stained with hematoxylin and eosin. Histopathology was scored by a pathologist in a blinded fashion as a combination of intestinal inflammation (score 0–5), neutrophil infiltration (score 0–4), and gland epithelial hyperplasia (score 0–4).
Oral or intravenous administration of OVA
Naive CD4+DO11.10+CD44−CD25− cells isolated from Rag2−/−DO11.10 mice were transduced with the retrovirus and were adoptively transferred into Balb/c recipient mice by i.v injection 1 day before antigen administration. For oral antigen administration, the recipient mice were fed 1% OVA (GradeV; Sigma‐Aldrich) solution dissolved in drinking water for 5 consecutive days for oral antigen administration, or immunized by i.v. injection of 20 μg of OVA323–339 peptide in PBS saline 1 and 3 days after adoptive transfer for intravenous administration. Six days after adoptive transfer, mesenteric lymph nodes were harvested from recipient mice and analyzed.
Experimental autoimmune encephalomyelitis
Experimental autoimmune encephalomyelitis (EAE) was induced by adoptive transfer of naive CD4+Vβ11+Vα3.2+CD44−CD25− cells isolated from 2D2 mice into Rag2−/− mice by i.v injection. One day later, the recipient mice were immunized by subcutaneous injection of 200 μg myelin oligodendrocyte glycoprotein, amino acids 35–55 in complete Freund's adjuvant.
Chromatin immunoprecipitation assays
ChIP assay was carried out according to the manufacturer's guide (Upstate/Millipore). The PCR primer set used is as follows:
Foxp3 TSDR forward, 5′‐ACAGAATCGATAGAACTTGGGTTTT‐3′; Foxp3 TSDR reverse, 5′‐CGAGAAGTGGCTAGTCTATCCTGTA‐3′.
Bisulfite sequencing
Methylation analysis was performed using Methyl Detector kit (Active Motif) according to manufacturer's protocols.
PCR products were performed using PfuTurbo Cx hotstart DNA polymerase (Stratagene) with the primer set (Foxp3 CNS2 forward, 5′‐TTTTGGGTTTTTTTGGTATTTAAGA‐3′; Foxp3 CNS2 reverse, 5′‐TTAACCAAATTTTTCTACCATTAAC‐3′). PCR products were T/A cloned into the pGEM‐T easy vector (Promega). Recombinant plasmids from the individual bacterial colonies were purified and sequenced.
Electrophoretic mobility shift assay
The DNA binding reaction was performed in a total volume of 20 μl containing nuclear extracts (3 μg), end‐labeled probe, and 2 μg of poly(dI‐dC) in binding buffer containing 10 mM HEPES (pH 7.9), 80 mM KCl, 0.05 mM EDTA, 1 mM MgCl2, 1 mM dithiothreitol, and 6% glycerol. The DNA–protein complex was resolved in a nondenaturing 5% polyacrylamide gel in 0.5× TBE buffer.
The following oligonucleotides were used as probes: Foxp3 CNS2 methyl‐CRE forward, 5′‐CCGGCCGCCATGA m CGTCAATGGCAGAAA‐3′; Foxp3 CNS2 methyl‐CRE reverse, 5′‐TTTCTGCCATTGA m CGTCATGGCGGCCGG‐3′; Foxp3 CNS2 unmethyl‐CRE forward, 5′‐CCGGCCGCCATGACGTCAATGGCAGAAA‐3′; Foxp3 CNS2 unmethyl‐CRE reverse, 5′‐TTTCTGCCATTGACGTCATGGCGGCCGG‐3′
Human T‐cell isolation and in vitro stimulation
Human adult blood samples were anonymously provided by the Blood Center of the Korean Red Cross, Seoul, under the approval of the Institutional Review Board with consent for research use. Experiments involving human blood were approved by the Institutional Review Board at Seoul National University (SNUIRB No. 1502/001‐013). PBMCs were prepared from venous blood from healthy adult donors by Ficoll‐Hypaque density gradient centrifugation. Human‐purified CD4 T cells were stimulated with Dynabeads Human T‐Activator CD3/CD28 in a bead‐to‐cell ratio of 1:1.
Statistics
Independent two‐tailed t‐test was used for analysis of two groups. When comparing more than two groups, one‐way ANOVA was used. In the case where the data are non‐parametric, Kruskal–Wallis test was performed.
Author contributions
SL performed most of the experiments with help from KP, JK and HM. KP and JK performed IBD colitis experiments. SL and RHS designed the research, analyzed results and wrote the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Review Process File
Acknowledgements
We thank T. A. Chatila for providing Foxp3EGFP mice, W. S. Pear for providing retroviral vectors, C. Vinson for providing DN‐C/EBP (AC/EBP) cDNA, and Dong‐Sup Lee for providing 2D2 TCR‐transgenic mice. This research was supported by grants from (i) the Korean Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (No. HI14C0311); (ii) the National Research Foundation of Korea (NRF) grant, funded by the Ministry of Science, ICT and Future Planning (MSIP) (No. 2016R1A2B3013865); and (iii) Korea Mouse Phenotyping Project (NRF‐2014M3A9D5A01073789) of the Ministry of Science, ICT and Future Planning through the National Research Foundation.
EMBO Reports (2018) 19: e45995
References
- 1. Josefowicz SZ, Lu LF, Rudensky AY (2012) Regulatory T cells: mechanisms of differentiation and function. Annu Rev Immunol 30: 531–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Belkaid Y, Tarbell K (2009) Regulatory T cells in the control of host‐microorganism interactions (*). Annu Rev Immunol 27: 551–589 [DOI] [PubMed] [Google Scholar]
- 3. Fontenot JD, Gavin MA, Rudensky AY (2003) Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat Immunol 4: 330–336 [DOI] [PubMed] [Google Scholar]
- 4. Hori S, Nomura T, Sakaguchi S (2003) Control of regulatory T cell development by the transcription factor Foxp3. Science 299: 1057–1061 [DOI] [PubMed] [Google Scholar]
- 5. Gavin MA, Rasmussen JP, Fontenot JD, Vasta V, Manganiello VC, Beavo JA, Rudensky AY (2007) Foxp3‐dependent programme of regulatory T‐cell differentiation. Nature 445: 771–775 [DOI] [PubMed] [Google Scholar]
- 6. Bilate AM, Lafaille JJ (2012) Induced CD4+Foxp3+ regulatory T cells in immune tolerance. Annu Rev Immunol 30: 733–758 [DOI] [PubMed] [Google Scholar]
- 7. Curotto de Lafaille MA, Kutchukhidze N, Shen S, Ding Y, Yee H, Lafaille JJ (2008) Adaptive Foxp3+ regulatory T cell‐dependent and ‐independent control of allergic inflammation. Immunity 29: 114–126 [DOI] [PubMed] [Google Scholar]
- 8. Oldenhove G, Bouladoux N, Wohlfert EA, Hall JA, Chou D, Dos Santos L, O'Brien S, Blank R, Lamb E, Natarajan S et al (2009) Decrease of Foxp3+ Treg cell number and acquisition of effector cell phenotype during lethal infection. Immunity 31: 772–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wei J, Duramad O, Perng OA, Reiner SL, Liu YJ, Qin FX (2007) Antagonistic nature of T helper 1/2 developmental programs in opposing peripheral induction of Foxp3+ regulatory T cells. Proc Natl Acad Sci U S A 104: 18169–18174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dardalhon V, Awasthi A, Kwon H, Galileos G, Gao W, Sobel RA, Mitsdoerffer M, Strom TB, Elyaman W, Ho IC et al (2008) IL‐4 inhibits TGF‐beta‐induced Foxp3+ T cells and together with TGF‐beta, generates IL‐9+ IL‐10+ Foxp3(‐) effector T cells. Nat Immunol 9: 1347–1355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mantel PY, Kuipers H, Boyman O, Rhyner C, Ouaked N, Ruckert B, Karagiannidis C, Lambrecht BN, Hendriks RW, Crameri R et al (2007) GATA3‐driven Th2 responses inhibit TGF‐beta1‐induced FOXP3 expression and the formation of regulatory T cells. PLoS Biol 5: e329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sun CM, Hall JA, Blank RB, Bouladoux N, Oukka M, Mora JR, Belkaid Y (2007) Small intestine lamina propria dendritic cells promote de novo generation of Foxp3 T reg cells via retinoic acid. J Exp Med 204: 1775–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Coombes JL, Siddiqui KR, Arancibia‐Carcamo CV, Hall J, Sun CM, Belkaid Y, Powrie F (2007) A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF‐beta and retinoic acid‐dependent mechanism. J Exp Med 204: 1757–1764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mucida D, Park Y, Kim G, Turovskaya O, Scott I, Kronenberg M, Cheroutre H (2007) Reciprocal TH17 and regulatory T cell differentiation mediated by retinoic acid. Science 317: 256–260 [DOI] [PubMed] [Google Scholar]
- 15. Hall JA, Grainger JR, Spencer SP, Belkaid Y (2011) The role of retinoic acid in tolerance and immunity. Immunity 35: 13–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nolting J, Daniel C, Reuter S, Stuelten C, Li P, Sucov H, Kim BG, Letterio JJ, Kretschmer K, Kim HJ et al (2009) Retinoic acid can enhance conversion of naive into regulatory T cells independently of secreted cytokines. J Exp Med 206: 2131–2139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Takaki H, Ichiyama K, Koga K, Chinen T, Takaesu G, Sugiyama Y, Kato S, Yoshimura A, Kobayashi T (2008) STAT6 Inhibits TGF‐beta1‐mediated Foxp3 induction through direct binding to the Foxp3 promoter, which is reverted by retinoic acid receptor. J Biol Chem 283: 14955–14962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Benson MJ, Pino‐Lagos K, Rosemblatt M, Noelle RJ (2007) All‐trans retinoic acid mediates enhanced T reg cell growth, differentiation, and gut homing in the face of high levels of co‐stimulation. J Exp Med 204: 1765–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chatterjee R, Bhattacharya P, Gavrilova O, Glass K, Moitra J, Myakishev M, Pack S, Jou W, Feigenbaum L, Eckhaus M et al (2011) Suppression of the C/EBP family of transcription factors in adipose tissue causes lipodystrophy. J Mol Endocrinol 46: 175–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ron D, Habener JF (1992) CHOP, a novel developmentally regulated nuclear protein that dimerizes with transcription factors C/EBP and LAP and functions as a dominant‐negative inhibitor of gene transcription. Genes Dev 6: 439–453 [DOI] [PubMed] [Google Scholar]
- 21. Hill JA, Hall JA, Sun CM, Cai Q, Ghyselinck N, Chambon P, Belkaid Y, Mathis D, Benoist C (2008) Retinoic acid enhances Foxp3 induction indirectly by relieving inhibition from CD4+CD44hi Cells. Immunity 29: 758–770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Izcue A, Coombes JL, Powrie F (2006) Regulatory T cells suppress systemic and mucosal immune activation to control intestinal inflammation. Immunol Rev 212: 256–271 [DOI] [PubMed] [Google Scholar]
- 23. Mucida D, Kutchukhidze N, Erazo A, Russo M, Lafaille JJ, Curotto de Lafaille MA (2005) Oral tolerance in the absence of naturally occurring Tregs. J Clin Invest 115: 1923–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Huehn J, Polansky JK, Hamann A (2009) Epigenetic control of FOXP3 expression: the key to a stable regulatory T‐cell lineage? Nat Rev Immunol 9: 83–89 [DOI] [PubMed] [Google Scholar]
- 25. Floess S, Freyer J, Siewert C, Baron U, Olek S, Polansky J, Schlawe K, Chang HD, Bopp T, Schmitt E et al (2007) Epigenetic control of the foxp3 locus in regulatory T cells. PLoS Biol 5: e38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Polansky JK, Kretschmer K, Freyer J, Floess S, Garbe A, Baron U, Olek S, Hamann A, von Boehmer H, Huehn J (2008) DNA methylation controls Foxp3 gene expression. Eur J Immunol 38: 1654–1663 [DOI] [PubMed] [Google Scholar]
- 27. Zheng Y, Josefowicz S, Chaudhry A, Peng XP, Forbush K, Rudensky AY (2010) Role of conserved non‐coding DNA elements in the Foxp3 gene in regulatory T‐cell fate. Nature 463: 808–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ohkura N, Hamaguchi M, Morikawa H, Sugimura K, Tanaka A, Ito Y, Osaki M, Tanaka Y, Yamashita R, Nakano N et al (2012) T cell receptor stimulation‐induced epigenetic changes and Foxp3 expression are independent and complementary events required for Treg cell development. Immunity 37: 785–799 [DOI] [PubMed] [Google Scholar]
- 29. Feng Y, Arvey A, Chinen T, van der Veeken J, Gasteiger G, Rudensky AY (2014) Control of the inheritance of regulatory T cell identity by a cis element in the Foxp3 locus. Cell 158: 749–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li X, Liang Y, LeBlanc M, Benner C, Zheng Y (2014) Function of a Foxp3 cis‐element in protecting regulatory T cell identity. Cell 158: 734–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kim HP, Leonard WJ (2007) CREB/ATF‐dependent T cell receptor‐induced FoxP3 gene expression: a role for DNA methylation. J Exp Med 204: 1543–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Rishi V, Bhattacharya P, Chatterjee R, Rozenberg J, Zhao J, Glass K, Fitzgerald P, Vinson C (2010) CpG methylation of half‐CRE sequences creates C/EBPalpha binding sites that activate some tissue‐specific genes. Proc Natl Acad Sci U S A 107: 20311–20316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Beal AM, Ramos‐Hernandez N, Riling CR, Nowelsky EA, Oliver PM (2011) TGF‐beta induces the expression of the adaptor Ndfip1 to silence IL‐4 production during iTreg cell differentiation. Nat Immunol 13: 77–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Polansky JK, Schreiber L, Thelemann C, Ludwig L, Kruger M, Baumgrass R, Cording S, Floess S, Hamann A, Huehn J (2010) Methylation matters: binding of Ets‐1 to the demethylated Foxp3 gene contributes to the stabilization of Foxp3 expression in regulatory T cells. J Mol Med (Berl) 88: 1029–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Jeong JH, Jo A, Park P, Lee H, Lee HO (2015) Brca2 deficiency leads to T cell loss and immune dysfunction. Mol Cells 38: 251–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nair VS, Song MH, Ko M, Oh KI (2016) DNA Demethylation of the Foxp3 Enhancer Is Maintained through Modulation of Ten‐Eleven‐Translocation and DNA Methyltransferases. Mol Cells 39: 888–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Review Process File


