Abstract
Advanced-stage cutaneous T-cell lymphoma (CTCL) is usually a fatal malignancy despite optimal use of currently available treatments. In this preclinical study of novel CTCL therapy, we performed in vitro and ex vivo experiments to determine the efficacy of combination treatment with a panel of BET bromodomain inhibitors (BETi) (JQ1, OTX015, CPI-0610, I-BET762) and HDAC inhibitors (HDACi) (SAHA/Vorinostat, Romidepsin). BETi/HDACi combinations were synergistic (combination index <1) against cell viability and induced G0/G1 cell cycle arrest. Apoptosis was uniformly enhanced. From a mechanistic standpoint, proliferative drivers c-Myc, Cyclin D1, NFkB, and IL-15Rα were reduced. Inhibitory CDKN1A was increased. CDKN1B, IL-7R, IL-17Rα, STAT3, and STAT5 alterations varied. There were significant increases in extrinsic apoptotic pathway death receptors and ligands (FasL, DR4, DR5, TRAIL, and TNFR1). At clinically tolerable levels of single agents, Romidepsin (1 nM) + OTX015 (125 nM) induced the greatest apoptosis (60%_80%) at 96 hours. Ex vivo studies of leukemic CTCL cells obtained from patients with Sezary syndrome also showed higher levels of apoptosis (about 60%-90%) in response to combination treatments relative to single agents. In contrast, combination treatment of normal CD4+ T cells induced only minimal apoptosis (<10%). Our findings show that the mechanism of action of BETi/HDACi therapy in CTCL involves induction of both cell cycle arrest and apoptosis with reduced proliferative drivers and enhanced expression of apoptotic extrinsic pathway death receptors and ligands. Relative to single agents, the superior anti-CTCL effects of BETi/HDACi combinations in vitro and ex vivo provide a rationale for clinical trials exploring their efficacy as therapy for CTCL.
Introduction
Cutaneous T-cell lymphoma (CTCL) is the most common form of primary cutaneous lymphoma [1], [2], [3]. It is a CD4+ T-cell neoplasm that includes mycosis fungoides and its leukemic variant, the Sezary syndrome. Although some patients never progress beyond early-stage indolent disease, others develop advanced stages that are often fatal. Multiple systemic therapies exist for advanced CTCL. With the recent exception of brentuximab vedotin which has a 65% objective response rate for CD30+ CTCL [4], most systemic CTCL therapies have at best a 30%-40% objective response rate. They are rarely curative, and each has its own unique set of clinically significant side effects. Various combinations of these agents have been used with added benefit for some patients; however, there remains a distinct need for more effective novel therapies.
Histone deacetylase inhibitors (HDACi) are group of small molecules that block the action of histone deacetylases (HDAC) [5], [6]. HDACi result in the accumulation of acetylated lysine residues on histones important for maintaining an open chromatin structure and promoting gene expression. Two HDACi that are FDA-approved specifically for CTCL include SAHA/vorinostat and romidepsin. They are relatively selective for class I and II HDAC and tend to promote expression of genes that enhance cell differentiation and apoptosis.
Featuring a tandem repeat of bromodomains at their N-terminus, BET (bromodomain and extra terminal) proteins are “super-reader” transcriptional regulators that play an important role during normal cell growth and development [7], [8]. They include BRD2, -3, -4, and -T. BET inhibitors (BETi) are typified by their prototype, JQ1 [9]. BETi bind to acetylated lysine recognition motifs on BET proteins, thereby blocking their interaction with acetylated histones. This leads to disruption of chromatin remodeling and inhibits expression of genes, some of which promote neoplastic cell proliferation. No BETi are currently FDA-approved, although several are in early-phase clinical trials for various solid and hematologic malignancies and metabolic disorders. These next-generation BETi include OTX015, CPI-0610, and I-BET762 [10], [11], [12].
We have studied the modulation of gene expression in malignancies including CTCL using HDACi [13], novel DNA methylation inhibitors like methotrexate [14], [15], and transcriptional knockdown of c-CBL, an E3 ubiquitin ligase that impedes T-cell receptor signaling and downstream activation-induced cell death [16]. Because of their respective effects on cell proliferation and differentiation, we were interested in exploring the possible benefits of epigenetic modulation of CTCL cells using a combination of BETi and HDACi. Our in vitro and ex vivo findings show that this combination is very effective against CTCL cells by inhibiting proliferation and promoting apoptosis.
Matetials and Methods
Reagents
SAHA/Vorinostat, Romidepsin, I-BET762 (all from Sigma-Aldrich, St. Louis, MO), CPI-0610, OTX015, and JQ1 (all from Cayman Chemicals, Ann Arbor, MI) were dissolved in DMSO to stock concentration of 10-50 mM, stored at −20°C. Serial dilutions were freshly made in DMSO for all cellular assays. The final concentration of DMSO in the medium was less than 0.1%, which did not show any effect on cell growth.
CTCL Cell Lines and Sezary Syndrome Blood Samples
All clinical samples were obtained with institutional review board approval. All patients gave written informed consent, and all protocols adhered to the Declaration of Helsinki principles.
PBMCs were isolated from Sézary syndrome blood samples via Ficoll (GE, Pittsburgh, PA) gradient centrifugation. All four cases were stage IV and had the following blood tumor burden based on CD3+4+7−26− phenotype and in some cases also Sezary cell cytology preparations: SS-1 (60,000 lymphocytes; 99% tumor cells), SS-2 (3000 lymphocytes; 92% tumor cells), SS-3 (40,000 lymphocytes; 72% tumor cells), and SS-4 (10,000 lymphocytes; 82% tumor cells). None had received systemic CTCL therapy for at least 1 month prior to blood sample collection. Normal CD4+ T cells were purchased from Precision for Medicine (Bethesda, MD) from two different donors. Cell were cultured in RPMI 1640 medium supplemented with 10% heat-inactivated fetal bovine serum (FBS, HyClone Laboratories, Logan, UT), 2 mM HEPES, 1% penicillin-streptomycin, 1 μg/ml phytohemagglutinin (PHA, Sigma-Aldrich, St. Louis, MO), and 20 ng/ml human IL-2 (San Diego, CA) with 5% CO2 at 37°C.
Human CTCL lines derived from patients with MF (MyLa, HH) or SS (SZ4, Hut-78, SeAx) were studied. All have been described previously [14], [16]. Cells were cultured in RPMI 1640 medium (Sigma Chemical, St. Louis, MO) supplemented with 10% FBS, 2 mM HEPES, and 1% penicillin-streptomycin with 5% CO2 at 37°C.
Immunoblotting
After drug treatment, cells were harvested by centrifugation and solubilized in RIPA buffer containing protease inhibitors (50 mM Tris, 150 mM NaCl, 1% Nonidet P-40, 0.1% sodium dodecylsulfate, and 10 μg/ml aprotinin). Protein concentrations of cell lysates were determined using a Bio-Rad DC Protein Assay kit. Approximately 15-30 μg of proteins from each sample was separated on 4%-20% Mini-PROTEAN TGX precast gels (Bio-Rad) and transferred to PVDF membrane. Proteins of interest were detected by immunoblotting using the following primary antibodies: anti–c-Myc (#9402), anti-NFkB1 (#12540), anti-NFkB2 (#4882), anti-RelB (#4922), anti-p65 (SC-372, Santa Cruz), anti-IkBα (#4814), anti–phospho-Stat3 (#9145), anti-Stat3 (#9139), anti–phospho-Stat5 (#9351), anti-Stat5 (#25656), anti-CyclinD1 (SC-450, Santa Cruz), anti-CDKN1A (SC-817, Santa Cruz), anti-CDKN1B (SC-1641, Santa Cruz), anti–clv-Caspase3 (#9664), anti–clv-Caspase8 (#9496), and anti–clv-Caspase9 (#9501). Unless otherwise specified, all are from Cell Signaling (Beverly, MA) with 1:1000 dilution. Mouse anti–β-actin (1:5000) from Sigma-Aldrich served as a loading control. After incubation with HRP-conjugated secondary antibodies (1:5000, goat anti-rabbit or mouse, Cell Signaling), specific protein bands on the blots were visualized by applying enhanced chemiluminescence reagents according to the manufacturer's instructions (Pierce, Rockford, IL) and then recorded with a LAS-4000 Mini imager (GE, Piscataway, NJ).
Cell Viability Assays and Caspase Activity Assays
CTCL cell lines were seeded onto 96-well plates at a density of 10,000 cells/well with SAHA or JQ1 at serial dilutions (0, 0.2, 0.5, 1, 5, 10 μM), respectively. After various times of incubation, cells were analyzed for cell viability by the addition of CellTiter Glo (Promega, Madison, WI) to detect ATP in the assay plates. After 30-minute incubation at 37°C, the luminescent signal from the viable cells was analyzed on a FlexStation 3 Benchtop Multi-Mode Microplate Reader (Molecular Devices, Sunnyvale, CA).
For Caspase activity assays, cells were seeded onto 96-well plates at a density of 5×104 cells/well with JQ1 (1 μM), SAHA (1 μM), or combination (1 μM each) treatment for 24 or 48 hours. Caspase-Glo 3/7, 8, and 9 assay kits (all from Promega, Madison, WI) were used to determine the relative activity of caspase according to manufacturer's instructions. After incubation at room temperature for 1 hour, plates were read in a FlexStation 3 Benchtop Multi-Mode Microplate Reader (Molecular Devices, Sunnyvale, CA).
Synergistic Assay and Combination Index (CI) Calculation
Cells were seeded in 96-well plates at 10,000 cells per well treated in duplicate with single agents and their fixed-ratio combination. A factorial dose matrix was used to sample all mixtures of two serially diluted single agents as previously described [17]. After 48 or 72 hours, cell viability was measured by CellTiter Glo (Promega, Madison, WI) as noted above.
CI scores were calculated as previously described [18] using Compusyn software version 1.0 (ComboSyn, Inc.). This process is a quantitative and theoretical method to determine the synergism or antagonism in the drug combination experiments. If the CI value is lower than 1, the drug combination is synergistic; if the CI value is higher than 1, the combination is antagonistic; if the CI value is equal to 1, the combination is additive.
Flow Cytometry
Cell apoptosis was measured using fluorescein isothiocyanate (FITC)–Annexin V and propidium iodide staining kit (# 556547, BD Biosciences) following the manufacturer's protocols. Briefly, cells (1×106/ml) with different treatments were collected, washed twice with cold PBS, and then resuspended in 1× binding buffer from the kit. FITC-Annexin V and propidium iodide were added and incubated for 15 mintes at room temperature in the dark before flow cytometry. Data were analyzed by FlowJo software (Treestar, Ashland, OR).
For cell cycle analyses, cells (1×106/ml) with different treatments were collected and washed with PBS, fixed with 70% ethanol overnight, and washed with PBS. RNA was degraded with RNase (QIAGEN, Valencia, CA), and DNA was stained with propidium iodide.
For analysis of cell surface molecules, cells (1×106/ml) were collected and washed twice with cold PBS. Cells were stained with FITC-conjugated anti-FAS/CD95 (#555673), biotin-conjugated anti-FAS ligand/CD178 (#556374), biotin-conjugated anti-TNF-R1/CD120a (#552536), PE-conjugated anti-DR4/TRAIL-R1/CD261 (#564180), and PE-conjugated anti-DR5/TRAIL-R2/CD262 (#565499, all from BD Biosciences) antibodies for 30 minutes on ice in the dark. Then, cells were washed with FACS buffer (2% FBS in PBS). For intracellular staining, cell were fixed in BD Cytofix buffer (#554655, BD Biosciences) for 20 minutes on ice, washed with FACS buffer containing 0.25% saponin (Sigma-Aldrich, St. Louis, MO), and then permeabilized with 0.25% saponin for 20 minutes. PE-conjugated anti-TRAIL/CD253 (#550516) and APC-conjugated anti-TNFα (#551384, BD Biosciences) antibodies were incubated with cells for 30 minutes on ice in the dark. If needed, secondary staining with FITC-conjugated avidin (BD Biosciences) was then performed, followed by further washing. Cells were then resuspended in FACS buffer and analyzed with a LSRII flow cytometer (BD Biosciences). Data analysis was performed using FlowJo software (Treestar, Ashland, OR).
Real-Time Quantitative PCR Assay
RNA was extracted from CTCL cells using Trizol (QIAGEN, Valencia, CA) following the manufacturer's instructions. Purified messenger RNA (mRNA) (2 μg) was used for the first-strand complementary DNA (cDNA) synthesis using iScript cDNA synthesis kit (Bio-Rad), and quantitative RT-PCR was performed using the CFX96 Real-Time PCR System. Each cDNA template was amplified in triplicate using SYBR Green PCR Master Mix (Bio-Rad).
Statistics
Statistically significant differences between treatment groups were determined by one-way ANOVA (GraphpadPrism 7.0, La Jolla, CA) using the Bonferroni multiple comparison post hoc test or a two-tailed t test for grouped comparison. */+ P < .05, **/++ P < .01, and ***/+++ P < .001 were used to show statistical significance throughout the article. Error bars in histograms show standard deviations of triplicate measurements.
Results
IC50 of BETi and HDACi in CTCL Lines
To establish drug dosing ranges relevant to CTCL lines, we used an ATP detection assay to determine the 50% maximal inhibitory concentration (IC50) of BETi and HDACi in five CTCL lines at 72 hours. As shown in Supplemental Figure 1, the approximate IC50s were 1 nM (Romidepsin), 1 μM (SAHA, JQ1), and 4-30 μM (other BETi). Values were highest in SeAx more often than in any other cell line. Notably, this CTCL line lacks the Fas death receptor (CD95) [19].
BETi and HDACi Are Synergistic in CTCL Lines
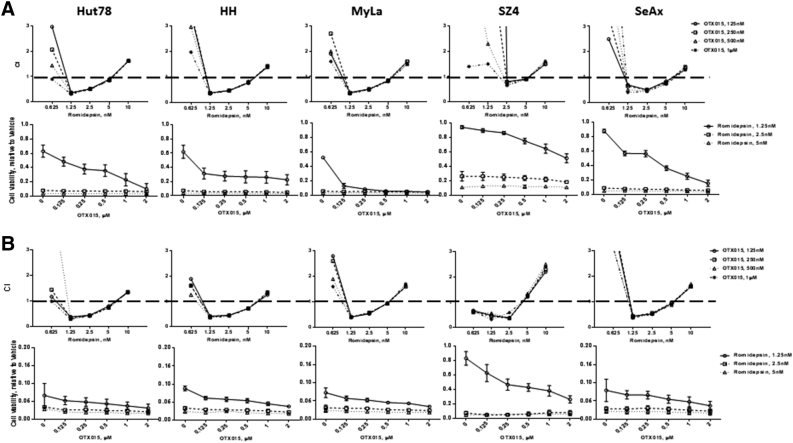
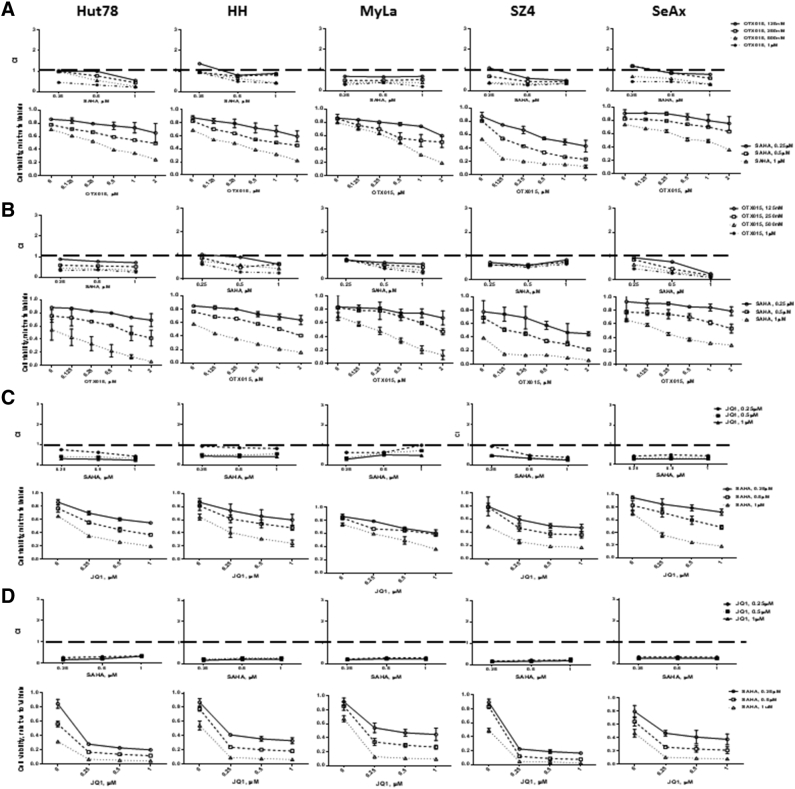
To determine whether there was synergy between BETi and HDACi in CTCL, cell viability was measured at various time points and drug dose combinations using an ATP detection assay. The CI was calculated with CompuSyn software (see Methods). If the CI value is <1, the drug combination is synergistic; if the CI value is >1, the combination is antagonistic. BETi and HDACi combinations showed a consistent synergistic effect (CI <1) in all five CTCL lines after 48 and 72 hours (see Figures 1 and 2 and Supplemental Figures 2 and 3). Among the next-generation BETi, OTX015 was the most effective agent overall. When combined with SAHA, it reduced viability to 15%-40%. When combined with Romidepsin, it reduced viability to <10% in 4 out of 5 CTCL lines.
Figure 1.
Synergistic assay of BETi and Romidepsin.
Cell viability was measured after 48 and 72 hours of drug treatment using a luminescent ATP detection assay. (A) Romidepsin and OTX015, 48 hours; (B) Romidepsin and OTX015, 72 hours; CI value <1 indicates the drug combination is synergistic; CI value >1 indicates the combination is antagonistic.
Figure 2.
Synergistic assay of BETi and SAHA.
Cell viability was measured after 48 and 72 hours of drug treatment using a luminescent ATP detection assay. (A) SAHA and OTX015, 48 hours; (B) SAHA and OTX015, 72 hours; (C) SAHA and JQ1, 48 hours; (D) SAHA and JQ1, 72 hours. CI value <1 indicates the drug combination is synergistic; CI value >1 indicates the combination is antagonistic.
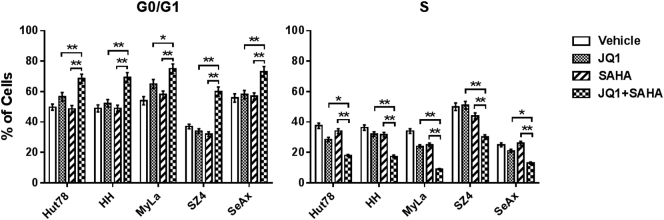
BETi and HDACi Induce G0/G1 Arrest in CTCL Lines
We performed cell cycle analysis using propidium iodide flow cytometry after treating CTCL lines with JQ1 and SAHA individually and in combination at 1 μM each. As shown in Figure 3 and Supplemental Figure 4, at 24 hours, these drugs induced statistically significant increases in G0/G1 and decreases in S phase, with the greatest effects observed with combination treatment in all five CTCL lines.
Figure 3.
Quantification of cell cycle analysis.
PI staining of total DNA content after 24 hours of JQ1 (1 μM), SAHA (1 μM), or combination (1 μM each) treatment. Combination of JQ1 with SAHA induces increased G0/G1 phase and reduced S phase in all CTCL cell lines. Data are presented as mean ± SD, n = 3, *P < .05, **P < .01 compared to either JQ1 or SAHA group.
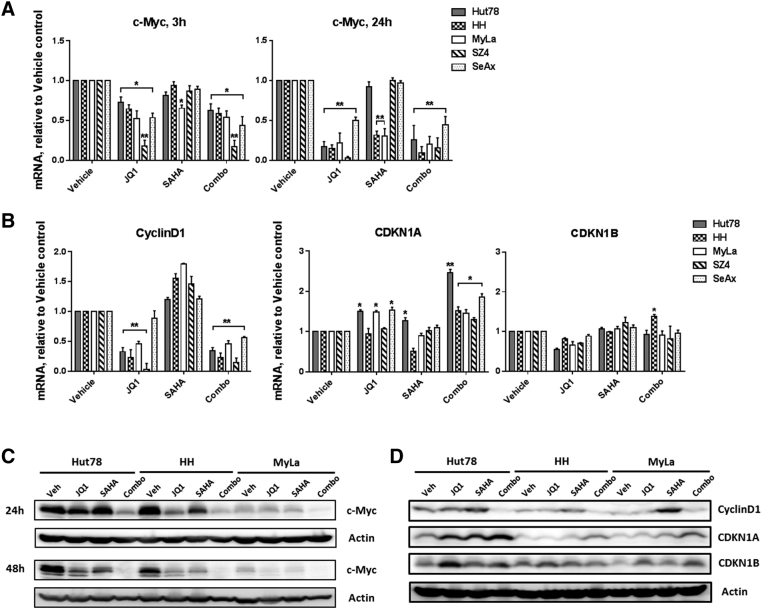
BETi and HDACi Reduce Some Proliferative Drivers and Variably Affect Others
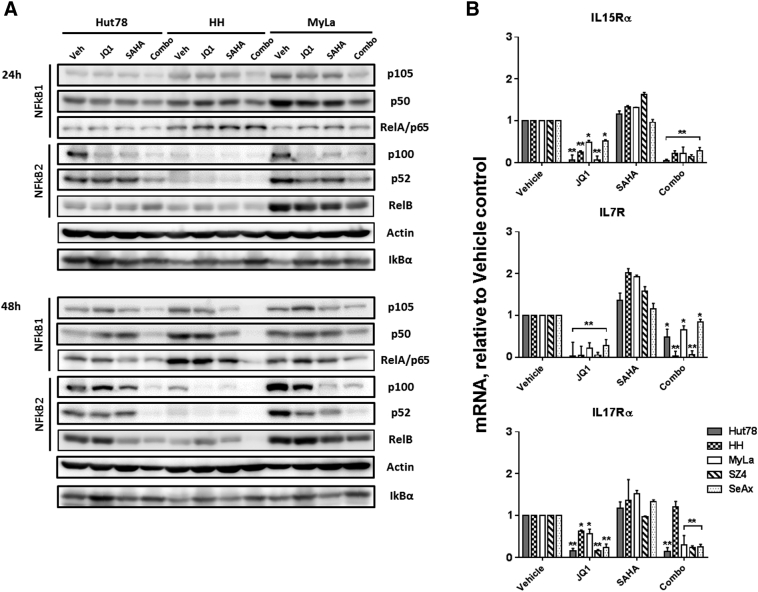
To explore the mechanisms underlying the reduced proliferation observed with BETi/HDACi combination treatment, we used CTCL lines to measure effects on the expression of CTCL proliferative drivers at the mRNA and protein levels. As shown in Figure 4, Figure 5 and Supplemental Figure 5, there were consistent reductions in the expression of c-Myc, Cyclin D1, NFkB, and IL-15Rα. NFkB family members NFkB1, RelA, NFkB2, and RelB were all decreased, and IkB was modestly increased. Some reductions were greater at 48 hours than at 24 hours.
Figure 4.
BETi and HDACi reduce c-Myc and downstream proliferative drivers.
(A and B) mRNA was prepared from CTCL cells under different conditions and used for qRT-PCR. Quantification: normalization to GAPDH mRNA. (C and D) Immunoblot analysis of c-Myc CyclinD1, CDKN1A, and CDKN1B protein in three CTCL cell lines; β-actin served as a loading control; cells were treated with JQ1 (1 μM), SAHA (1 μM), or combination (1 μM each). Mean ± SEM; n = 3, *P < .05, **P < .01 compared to vehicle control.
Figure 5.
BETi and HDACi reduce NFkB and cytokine receptors.
(A) Immunoblot analysis of NFkB1, NFkB2, and IkBα protein in three CTCL cell lines; β-actin served as a loading control. (B) mRNAs were prepared from CTCL cells under different conditions and used for qRT-PCR. Quantification: normalization to GAPDH mRNA; Cells were treated with JQ1 (1 μM), SAHA (1 μM), or combination (1 μM each). Mean ± SEM; n = 3, *P < .05, **P < .01 compared to vehicle control.
In contrast, other known proliferative drivers of CTCL were affected more variably among the CTCL lines studied. As shown in Figure 4, Figure 5 and Supplemental Figure 5, some IL-7R and IL-17Rα reductions were minimal, and IL-17Rα even showed a slight increase in one case. By 48 hours, STAT3, STAT5, and their phosphorylated forms showed minimal changes or distinct increases, depending on the particular CTCL line. As potent cyclin-dependent kinase inhibitors, CDKN1A but not CDKN1B exhibited consistent increased expression in CTCL lines, indicating that CDKN1A is the main target of BETi/HDACi combination treatment.
When comparing the relative contribution of BETi versus HDACi to the net effect of combination treatments on the expression of proliferative drivers, alterations appeared to be due mainly to BETi for most factors except the NFkB family proteins that showed mixed dominance of BETi and HDACi single agents.
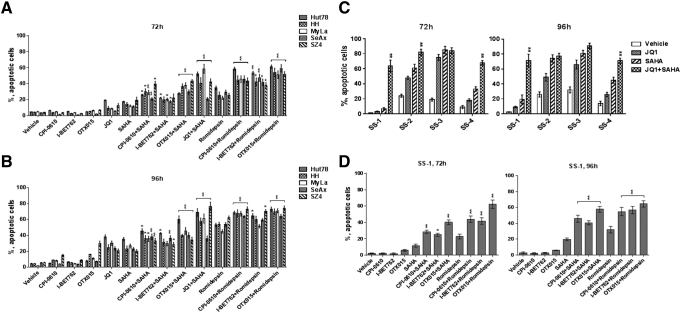
BETi and HDACi Induce Apoptosis in CTCL Lines and CTCL Leukemic Blood Samples
We determined the effects of single agent and combination drug treatment on apoptosis using Annexin V/propidium iodide flow cytometry. To confirm and extend our in vitro results using CTCL lines, we performed ex vivo studies of leukemic CTCL cells obtained from four patients with Sezary syndrome (SS). As shown in Figure 6 and Supplemental Figures 6-9, relative to vehicle controls, single agents and their combinations induced significant levels of early and late apoptosis in all CTCL lines and leukemic blood samples at 72 and 96 hours. Furthermore, all CTCL lines showed significantly greater apoptosis with combination treatment than with single agents. The full time course for induction of apoptosis by JQ1/SAHA after 24, 48, 72, and 96 hours is shown in Supplemental Figure 9. For each CTCL sample, combination treatment typically induced the highest levels of apoptosis. At levels of single agents known to be clinically tolerable, Romidepsin (1 nM) + OTX015 (125 nM) induced the greatest apoptosis (about 60%-80%) at 96 hours.
Figure 6.
BETi and HDACi induce marked apoptosis in CTCL cell lines and leukemic CTCL blood samples.
Quantification of flow cytometric analysis of apoptotic cell death (early and late apoptosis, right two quadrants of each Annexin V/PI scatter plot panel) after 3 and 4 days of CPI-0610 (125 nM), I-BET762 (125 nM), OTX015 (125 nM), JQ1 (1 μM), SAHA (1 μM), Romidepsin (1 nM), or combination treatment. (A and B) Five CTCL cell lines; (C and D) 4 leukemic CTCL blood samples. Data are presented as mean ± SEM, n = 3, “*” means combination group compared to HDACi alone group respectively. *P < .05, **P < .01.
To determine the effects on normal CD4+ T cells, samples from two healthy donors were subjected to similar treatment and analysis. As shown in Supplemental Figure 10, relative to no-treatment controls, apoptosis was only 2% greater in one sample and 9% greater in the other at 96 hours. In neither sample was the apoptosis induced by BETi/HDACi combination treatment greater than that observed for single agents.
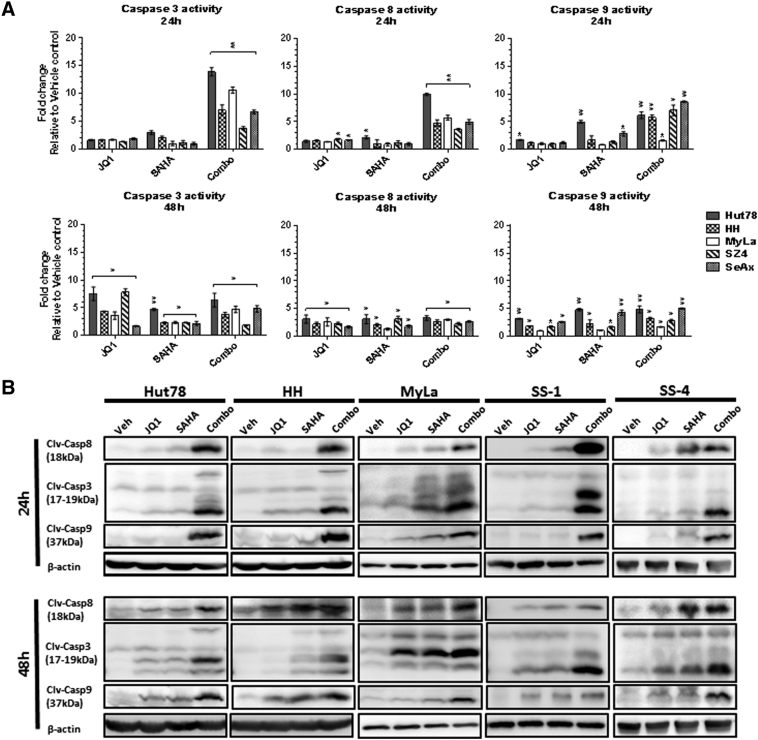
BETi and HDACi Induce Increased Caspase Cleavage in CTCL Lines
To dissect the mechanisms underlying the observed increase in apoptosis, we explored the relative contributions of the extrinsic apoptotic pathway (assessed by caspase 8 catalytic activity) and the intrinsic/mitochondrial apoptotic pathway (assessed by caspase 9 catalytic activity) as well as total apoptosis (assessed by caspase 3 catalytic activity). As shown in Figure 7A, JQ1/SAHA combination treatment showed the greatest induction of activation of all three caspases. At 24 hours, this was statistically significant not only relative to vehicle controls as shown in Figure 7 but also relative to JQ1 or SAHA alone. This occurred earlier (24 hours) than the maximal effects observed for the single agents (48 hours). Furthermore, the increase in caspase 8 activation was more consistent than the increase in caspase 9, suggesting that induction of the extrinsic apoptotic pathway is dominant in this setting. As shown in Figure 7B, immunoblot analysis of caspase 3, 8, and 9 cleavage in three CTCL lines and two leukemic CTCL blood samples confirmed that the JQ1/SAHA combination induced the greatest amount of caspase cleavage compared to single agent treatment.
Figure 7.
Combination of JQ1 and SAHA induce caspase cleavage in CTCL cell lines.
(A) Caspases 3, 8, and 9 activations were assessed by measurement of catalytic activity after treatment with JQ1 (1 μM), SAHA (1 μM), or combination (1 μM each) for 24 and 48 hours. Data are presented as mean ± SEM, n = 3, *P < .05, **P < .01 compared to vehicle control. (B) Immunoblot analysis of cleaved caspase 3, 8, and 9 in three CTCL cell lines and two Sézary syndrome blood samples. Cells were treated with JQ1 (1 μM), SAHA (1 μM), or combination (1 μM each) after 24 and 48 hours. β-Actin served as a loading control.
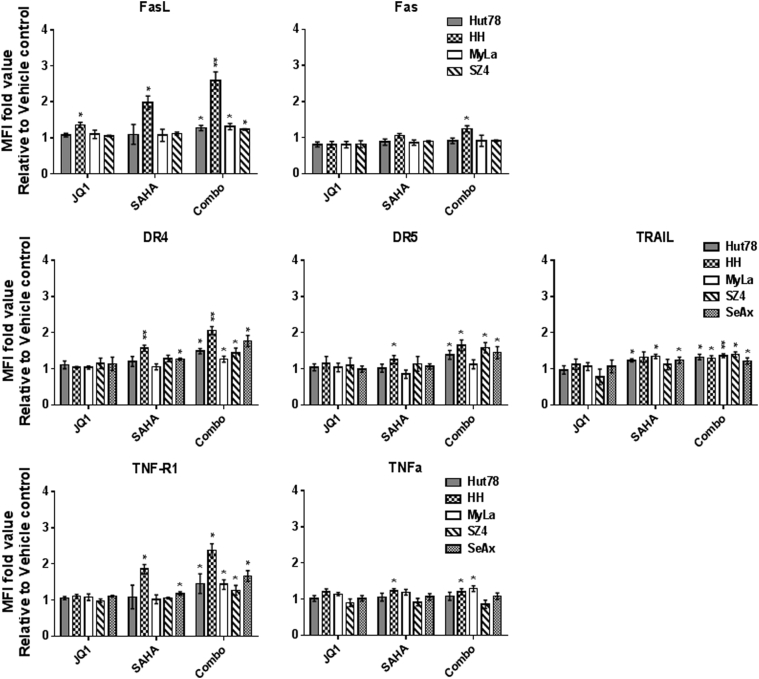
BETi and HDACi Induce Increased Death Receptors/Ligands in CTCL Lines
To determine the effects of BETi/HDACi treatment on mediators of extrinsic apoptosis, we used flow cytometry of immunostained cells to determine JQ1/SAHA effects the following death receptors and their ligands: Fas, FasL (Fas pathway); DR4, DR5, TRAIL (TRAIL pathway); and TNFR1, TNFα (TNF pathway). As shown in Figure 8, relative to vehicle controls, combination treatment for 72 hours showed significant increases in FasL, DR4, DR5, TRAIL, and TNFR1 that were more consistent across the CTCL lines than with the single agents. When combination treatment was compared to single agents, there was not much difference for SAHA, whereas JQ1 showed significantly less effect in many cases. This implied that most of the increases were mediated by SAHA with a minor contribution by JQ1.
Figure 8.
Immunostaining and flow cytometric analysis of CTCL lines for death receptor/ligand partners.
Histograms of mean fluorescence intensities (MFIs) shows upregulation at 72 hours after JQ1 (1 μM), SAHA (1 μM), or combination (1 μM each) treatment. Data are presented as mean ± SD, n = 3, *P < .05, **P < .01, compared to vehicle control.
Discussion
In this study, the results of our in vitro experiments show that the combination of BETi and HDACi acts synergistically to decrease the viability of CTCL cells. This occurred at concentrations of agents suitable for clinical use and was accompanied by cell cycle arrest and induction of apoptosis. Mechanistic studies indicated that these effects are associated with a reduction in the expression of several factors known to be proliferative drivers in CTCL in conjunction with an upregulation of multiple apoptotic death receptors and ligands. Although both extrinsic and intrinsic apoptotic pathways were enhanced as evidenced by increases in activation of both caspases 8 and 9, respectively, alterations were more consistently observed for caspase 8. This is consistent with our observed upregulation of multiple death receptors and ligands found in the Fas, TRAIL, and TNF extrinsic apoptosis subpathways. Importantly, the superior apoptosis induced by combination treatment was also seen in ex vivo experiments using leukemic CTCL cells enriched from patients with Sezary syndrome. In contrast, normal CD4+ T cells (the subset from which CTCL is derived) were minimally affected by combined BETi/HDACi treatment under similar conditions. These last two observations provide strong support for the clinical relevance of our findings.
Although all five CTCL lines we studied showed significant increases in apoptosis, not surprisingly, SeAx generally showed the least effects. This is consistent with its lack of the Fas (CD95) death receptor gene involved in extrinsic apoptosis [19] and suggests that engagement of Fas by FasL may be a relatively important contributor to extrinsic pathway apoptosis in at least some CTCL cases. In fact, our prior research has shown that impairment of Fas/FasL-mediated “activation-induced cell death” occurs in CTCL and can be overcome by restoring signal transduction through the T-cell receptor complex in a manner that involves upregulation of FasL leading to apoptosis in CTCL cells that express its receptor, Fas [16]. In this context, another drug that might be suitable for use with BETi/HDACi combinations is methotrexate (MTX). We have shown that MTX can act as an epigenetic modulator by inhibiting DNA methylation by depleting cells of S-adenosylmethionine, the main methyl donor for DNA methyltranferases [14]. This can result in reduced methylation of the promoter regions of tumor suppressor genes like Fas, thereby leading to their de-repression and enhanced expression by CTCL cells. We have shown previously that combination treatments that increase both the Fas death receptor and its ligand can clearly enhance CTCL apoptosis [15], [16], and some have proven useful clinically as therapies for CTCL patients [20].
BETi and HDACi are both epigenetic modulators that can affect a wide range of gene expression. Based on our comparison of the effects of these agents on CTCL individually and in combination, it appears that BETi generally play a dominant role in the suppression of proliferative drivers, while HDACi are more crucial for upregulation of apoptotic factors. Nevertheless, their maximal impact on killing CTCL cells in vitro and ex vivo occurs when they are used in combination. Despite FDA approval for CTCL, SAHA's exact mechanisms of action in CTCL are not clear. It inhibits histone deacetylases, resulting in the accumulation of acetylated histones important for the expression of genes including those that induce cell apoptosis and differentiation [21], [22]. IL-15 has been long known to be expressed by CTCL and to support CTCL proliferation and survival [23]. ZEB-1 is a known repressor of IL-15; however, hypermethylation of the IL-15 promoter in CTCL prevents ZEB-1 binding and its repressive effects [24]. IL-15 leads to upregulation of HDACs and oncogenic miR21. HDACi interrupt IL-15 signaling and suppress CTCL progression [25]. Furthermore, our findings show that BETi/HDACi combination treatment enhances reduction of IL-15Rα expression. These observations provide further justification for treatment of CTCL not only with HDACi but also with BETi and DNA methylation inhibitors such as MTX.
In preliminary studies published in abstract form, BRD4 was reported to be highly expressed in CTCL, and IL-15 increased it, while miR29b decreased it [25], [26]. In various tumor systems, cytokines and receptors increased by BRD4 include IL-1α, IL-1β, IL-15Rα, IL-15Rβ, and IL-15Rγ [26], [27]. Known effects of BETi in neoplasia include reductions in c-Myc [28], [29], NFkB [30], [31], IL-7, and IL-15 pathways [26], [32], all factors that have been implicated in the growth of CTCL cells. Our findings show similar effects specifically for CTCL. Another study demonstrated that JQ1 can decrease CTCL cell proliferation as well as CD30 and CCR4 expression [33]. Based on these studies and our current data, it is likely that HDACi exert dominant effects on CTCL cell differentiation and apoptosis, while BETi and HDACi both block proliferative drivers of CTCL growth, thereby exerting synergistic beneficial effects when used in combination. From a mechanistic standpoint, our findings confirm their negative impact on CTCL proliferation and provide detailed information about their positive impact on tumor cell apoptosis. Future studies will be required to further delineate additional specific mechanisms of action for BETi/HDACi combinations in CTCL and the most effective drug combinations.
In any case, our preclinical findings provide proof-of-principle that epigenetic modulation with a combination of BETi and HDACi could be beneficial therapy for CTCL. It has shown efficacy against acute myelogenous leukemia and pancreatic carcinoma in preclinical studies [27], [34]. To our knowledge, the combination BETi/HDACi therapy has not been explored clinically in CTCL. However, since the completion of our studies, another group has published their own preclinical study of BETi/HDACi in CTCL [35]. Our current work provides significantly more detail about alterations in the expression of proliferative drivers and apoptotic receptors/ligands. In regard to overall impacts on cell viability and survival, their in vitro and ex vivo findings generally paralleled ours. However, their most effective BETi/HDACi combination potentially suitable for clinical use (ABBV075/romidepsin) required concentrations of 400 nM/1.7 nM to achieve roughly 2/3 apoptosis. In contrast, our most effective BETi/HDACi combination potentially suitable for clinical use (OTX015/romidepsin) required much lower concentrations of only 125 nM/1 nM to achieve roughly 2/3-3/4 apoptosis. This difference could prove to be clinically significant in regard to drug efficacy, toxicity, and tolerability. In this context, it is worth noting that one of our patients (SS-1) did not show a clinical response to SAHA, and her cells exhibited only 20% apoptosis after 96-hour exposure to SAHA alone. However, apoptosis rose to 64% when tumor cells were exposed to Romidepsin/OTX015 combination therapy ex vivo (Supplemental Figure 7). The combination clearly accounted for this effect since OTX015 or Romidepsin alone resulted in only 6% or 31% apoptosis, respectively. Because Romidepsin is generally more effective clinically in CTCL as a single agent compared to SAHA, it is probable that greater apoptosis will be achieved by using Romidepsin in future clinical studies of BETi/HDACi combinations. Patients SS-2 and SS-3 both achieved partial clinical responses to Romidepsin (at least 50% clinical improvement) in contrast to the lack of clinical response to SAHA in Patient SS-1; however, in our apoptosis assays, their tumor cells were also considerably more sensitive to SAHA than those of Patient SS-1.
Although we used JQ1 as the prototype BETi in our preclinical studies, the toxicity profile of JQ1 limits its clinical use in humans. However, several next-generation JQ1 analogs have been developed for clinical applications. Among them, our current study includes three next-generation BETi (OTX015, CPI-0610, I-BET762) that have been shown to be tolerated in about a half dozen early phase clinical trials as therapies for hematologic malignancies including lymphomas, leukemias, and multiple myeloma, although none of these trials is specific for CTCL (see ClinicalTrials.gov). Durable objective responses (complete and partial) as well as evidence of clinical activity not meeting objective response criteria have been observed [36], [37]. Doses have included the 125- to 250-nM range that showed significant efficacy in our current in vitro and ex vivo studies.
These clinical trials have generally established that BETi can be administered as a single daily oral dose. The side effects noted for single agents might be diminished if the synergy between them that we have demonstrated allows for improved clinical efficacy of BETi/HDACi combinations used at reduced doses. Additional preclinical studies followed by clinical trials will be needed to determine the best combination of BETi/HDACi likely to benefit CTCL patients while minimizing side effects. It is likely that these systemic drugs would be targeted to patients with advanced-stage CTCL rather than those with more indolent early-stage disease. However, there have been efforts to administer HDACi topically for patients with early-stage CTCL. In an ongoing Phase II study, the novel topical HDACi Remetinostat showed an objective response rate of 40% (partial and complete responses) (see ClinicalTrials.gov, Identifier: NCT02213861). If BETi also prove suitable for topical application, perhaps they might have a therapeutic role in early-stage disease as part of a topical BETi/HDACi combination regimen.
Footnotes
Supported by NIH grants R21CA206104 and P30AR066524, Merit Review funding from the Department of Veterans Affairs, the Spatz Foundation, and the Swatek Family Fund (Dr. Wood).
Competing interests: The authors declare no potential conflicts of interest.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.neo.2018.11.006.
Appendix A. Supplementary data
Supplementary figures
References
- 1.Olsen E, Vonderheid E, Pimpinelli N, Willemze R, Kim Y, Knobler R. Revisions to the staging and classification of mycosis fungoides and Sezary syndrome: a proposal of the International Society for Cutaneous Lymphomas (ISCL) and the cutaneous lymphoma task force of the European Organization of Research and Treatment of Cancer (EORTC) Blood. 2007;110(6):1713–1722. doi: 10.1182/blood-2007-03-055749. [DOI] [PubMed] [Google Scholar]
- 2.Olsen EA, Rook AH, Zic J, Kim Y, Porcu P, Querfeld C. Sézary syndrome: immunopathogenesis, literature review of therapeutic options, and recommendations for therapy by the United States Cutaneous Lymphoma Consortium (USCLC) J Am Acad Dermatol. 2011;64(2):352–404. doi: 10.1016/j.jaad.2010.08.037. [DOI] [PubMed] [Google Scholar]
- 3.Olsen EA, Whittaker S, Kim YH, Duvic M, Prince HM, Lessin SR. Clinical end points and response criteria in mycosis fungoides and Sézary syndrome: a consensus statement of the International Society for Cutaneous Lymphomas, the United States Cutaneous Lymphoma Consortium, and the Cutaneous Lymphoma Task Force of the European Organisation for Research and Treatment of Cancer. J Clin Oncol. 2011;29(18):2598–2607. doi: 10.1200/JCO.2010.32.0630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prince HM, Kim YH, Horwitz SM, Dummer R, Scarisbrick J, Quaglino P. Brentuximab vedotin or physician's choice in CD30-positive cutaneous T-cell lymphoma (ALCANZA): an international, open-label, randomised, phase 3, multicentre trial. Lancet. 2017;390(10094):555–566. doi: 10.1016/S0140-6736(17)31266-7. [DOI] [PubMed] [Google Scholar]
- 5.Tan M, Luo H, Lee S, Jin F, Yang JS, Montellier E. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011;146(6):1016–1028. doi: 10.1016/j.cell.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fraga MF, Ballestar E, Villar-Garea A, Boix-Chornet M, Espada J, Schotta G. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet. 2005;37(4):391–400. doi: 10.1038/ng1531. [DOI] [PubMed] [Google Scholar]
- 7.Qi J. Bromodomain and extraterminal domain inhibitors (BETi) for cancer therapy: chemical modulation of chromatin structure. Cold Spring Harb Perspect Biol. 2014;6(12):a018663. doi: 10.1101/cshperspect.a018663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Belkina AC, Denis GV. BET domain co-regulators in obesity, inflammation and cancer. Nat Rev Cancer. 2012;12(7):465–477. doi: 10.1038/nrc3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O. Selective inhibition of BET bromodomains. Nature. 2010;468(7327):1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berthon C, Raffoux E, Thomas X, Vey N, Gomez-Roca C, Yee K. Bromodomain inhibitor OTX015 in patients with acute leukaemia: a dose-escalation, phase 1 study. Lancet Haematol. 2016;3(4):e186–e195. doi: 10.1016/S2352-3026(15)00247-1. [DOI] [PubMed] [Google Scholar]
- 11.Siu K, Ramachandran J, Yee A, Eda H, Santo L, Panaroni C. Preclinical activity of CPI-0610, a novel small-molecule bromodomain and extra-terminal protein inhibitor in the therapy of multiple myeloma. Leukemia. 2017;31(8):1760. doi: 10.1038/leu.2016.355. [DOI] [PubMed] [Google Scholar]
- 12.Mirguet O, Gosmini R, Toum Jrm, Clément CA, Ml Barnathan, Brusq J-M. Discovery of epigenetic regulator I-BET762: lead optimization to afford a clinical candidate inhibitor of the BET bromodomains. J Med Chem. 2013;56(19):7501–7515. doi: 10.1021/jm401088k. [DOI] [PubMed] [Google Scholar]
- 13.Nihal M, Ahmad N, Wood GS. SIRT1 is upregulated in cutaneous T-cell lymphoma, and its inhibition induces growth arrest and apoptosis. Cell Cycle. 2014;13(4):632–640. doi: 10.4161/cc.27523. [DOI] [PubMed] [Google Scholar]
- 14.Wu J, Wood GS. Reduction of Fas/CD95 promoter methylation, upregulation of Fas protein, and enhancement of sensitivity to apoptosis in cutaneous T-cell lymphoma. Arch Dermatol. 2011;147(4):443–449. doi: 10.1001/archdermatol.2010.376. [DOI] [PubMed] [Google Scholar]
- 15.Salva KA, Wood GS. Epigenetically enhanced photodynamic therapy (ePDT) is superior to conventional photodynamic therapy for inducing apoptosis in cutaneous T-cell lymphoma. Photochem Photobiol. 2015;91(6):1444–1451. doi: 10.1111/php.12521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu J, Salva KA, Wood GS. c-CBL E3 ubiquitin ligase is overexpressed in cutaneous T-cell lymphoma: its inhibition promotes activation-induced cell death. J Invest Dermatol. 2015;135(3):861–868. doi: 10.1038/jid.2014.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chou TC. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010;70(2):440–446. doi: 10.1158/0008-5472.CAN-09-1947. [DOI] [PubMed] [Google Scholar]
- 18.Chou TC. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol Rev. 2006;58(3):621–681. doi: 10.1124/pr.58.3.10. [DOI] [PubMed] [Google Scholar]
- 19.Wu J, Siddiqui J, Nihal M, Vonderheid EC, Wood GS. Structural alterations of the FAS gene in cutaneous T-cell lymphoma (CTCL) Arch Biochem Biophys. 2011;508(2):185–191. doi: 10.1016/j.abb.2010.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wood GS, Wu J. Methotrexate and pralatrexate. Dermatol Clin. 2015;33(4):747–755. doi: 10.1016/j.det.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.West AC, Johnstone RW. New and emerging HDAC inhibitors for cancer treatment. J Clin Invest. 2014;124(1):30–39. doi: 10.1172/JCI69738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rozati S, Cheng PF, Widmer DS, Fujii K, Levesque MP, Dummer R. Romidepsin and azacitidine synergize in their epigenetic modulatory effects to induce apoptosis in CTCL. Clin Cancer Res. 2016;22(8):2020–2031. doi: 10.1158/1078-0432.CCR-15-1435. [DOI] [PubMed] [Google Scholar]
- 23.Döbbeling U, Dummer R, Laine E, Potoczna N, Qin J-Z, Burg G. Interleukin-15 is an autocrine/paracrine viability factor for cutaneous T-cell lymphoma cells. Blood. 1998;92(1):252–258. [PubMed] [Google Scholar]
- 24.Mishra A, La Perle K, Kwiatkowski S, Sullivan LA, Sams GH, Johns J. Mechanism, consequences, and therapeutic targeting of abnormal IL15 signaling in cutaneous T-cell lymphoma. Cancer Discov. 2016;6(9):986–1005. doi: 10.1158/2159-8290.CD-15-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kohnken R, Wen J, Mundy-Bosse B, McConnell K, Keiter A, Grinshpun L. Diminished microRNA-29b level is associated with BRD4-mediated activation of oncogenes in cutaneous T-cell lymphoma. Blood. 2018;131(7):771–781. doi: 10.1182/blood-2017-09-805663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kohnken R, Wen J, Yano M, Hartlage A, Grinshpun L, Caligiuri MA. Targeting BRD4 disables IL-15 oncogenic signaling in cutaneous T-cell lymphoma via down regulation of IL-15 receptor complex. Blood. 2016;128:1097. [Google Scholar]
- 27.Mazur PK, Herner A, Mello SS, Wirth M, Hausmann S, Sánchez-Rivera FJ. Combined inhibition of BET family proteins and histone deacetylases as a potential epigenetics-based therapy for pancreatic ductal adenocarcinoma. Nat Med. 2015;21(10):1163–1171. doi: 10.1038/nm.3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146(6):904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mertz JA, Conery AR, Bryant BM, Sandy P, Balasubramanian S, Mele DA. Targeting MYC dependence in cancer by inhibiting BET bromodomains. Proc Natl Acad Sci. 2011;108(40):16669–16674. doi: 10.1073/pnas.1108190108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang G, Liu R, Zhong Y, Plotnikov AN, Zhang W, Zeng L. Down-regulation of NF-κB transcriptional activity in HIV-associated kidney disease by BRD4 inhibition. J Biol Chem. 2012;287(34):28840–28851. doi: 10.1074/jbc.M112.359505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ceribelli M, Kelly PN, Shaffer AL, Wright GW, Xiao W, Yang Y. Blockade of oncogenic IκB kinase activity in diffuse large B-cell lymphoma by bromodomain and extraterminal domain protein inhibitors. Proc Natl Acad Sci. 2014;111(31):11365–11370. doi: 10.1073/pnas.1411701111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ott CJ, Kopp N, Bird L, Paranal RM, Qi J, Bowman T. BET bromodomain inhibition targets both c-Myc and IL7R in high-risk acute lymphoblastic leukemia. Blood. 2012;120(14):2843–2852. doi: 10.1182/blood-2012-02-413021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kamijo H, Sugaya M, Takahashi N, Oka T, Miyagaki T, Asano Y. BET bromodomain inhibitor JQ1 decreases CD30 and CCR4 expression and proliferation of cutaneous T-cell lymphoma cell lines. Arch Dermatol Res. 2017:1–7. doi: 10.1007/s00403-017-1749-9. [DOI] [PubMed] [Google Scholar]
- 34.Fiskus W, Sharma S, Qi J, Valenta JA, Schaub LJ, Shah B. Highly active combination of BRD4 antagonist and histone deacetylase inhibitor against human acute myelogenous leukemia cells. Mol Cancer Ther. 2014;13(5):1142–1154. doi: 10.1158/1535-7163.MCT-13-0770. [DOI] [PubMed] [Google Scholar]
- 35.Kim SR, Lewis JM, Cyrenne BM, Monico PF, Mirza FN, Carlson KR. BET inhibition in advanced cutaneous T cell lymphoma is synergistically potentiated by BCL2 inhibition or HDAC inhibition. Oncotarget. 2018;9(49):29193–29207. doi: 10.18632/oncotarget.25670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Albrecht BK, Gehling VS, Hewitt MC, Vaswani RG, Cote A, Leblanc Y. Identification of a benzoisoxazoloazepine inhibitor (CPI-0610) of the bromodomain and extra-terminal (BET) family as a candidate for human clinical trials. J Med Chem. 2016;59(4):1330–1339. doi: 10.1021/acs.jmedchem.5b01882. [DOI] [PubMed] [Google Scholar]
- 37.Abramson JS, Blum KA, Flinn IW, Gutierrez M, Goy A, Maris M. BET inhibitor CPI-0610 is well tolerated and induces responses in diffuse large B-cell lymphoma and follicular lymphoma: preliminary analysis of an ongoing phase 1 study. Blood. 2015;126:1491. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures