SUMMARY
RNA molecules are folded into structures and complexes to perform a wide variety of functions. Determination of RNA structures and their interactions is a fundamental problem in RNA biology. Most RNA molecules in living cells are large and dynamic, posing unique challenges to structure analysis. Here we review progress in RNA structure analysis, focusing on methods that use the “cross-link, proximally ligate, and sequence” principle for high-throughput detection of base-pairing interactions in living cells. Beginning with a comparison of commonly used methods in structure determination and a brief historical account of psoralen cross-linking studies, we highlight the important features of cross-linking methods and new biological insights into RNA structures and interactions from recent studies. Further improvement of these cross-linking methods and application to previously intractable problems will shed new light on the mechanisms of the “modern RNA world.”
1. THE EVERLASTING RNA STRUCTURE PROBLEM AND EVER-EVOLVING SOLUTIONS
Ever since the discovery of the DNA structure, specific base-pairing has been seen as a perfect mechanism of heredity, as Watson and Crick postulated “a possible copying mechanism for the genetic material” (Watson and Crick 1953). Although DNA structures are relatively limited in variety, RNA structures are more diverse, and their essential roles have been repeatedly discovered and shown in many fundamental RNA-centric processes that make up life on Earth. As the driving force in the formation of helices, RNA base-pairing underlies both intramolecular structures and intermolecular interactions. In the early 1950s and 1960s, it was shown that both transfer of information from DNA to RNA (transcription) and from RNA to protein (translation) use the same sequence complementarity mechanism. In the past three decades, structural analysis showed that it is the complex and dynamic structures in the spliceosome and the ribosome, rather than the protein components, that catalyze chemical reactions (Cech and Steitz 2014). RNA–RNA interactions also form the molecular basis of small RNA (sRNA)-mediated gene regulation in eukaryotes (small interfering RNAs [siRNAs] and microRNAs [miRNAs]) and bacteria (sRNAs). These classical studies have largely dispelled the long-held notion of RNA as passive carriers of genetic information and put RNA-based regulation at center stage of molecular biology.
A large variety of methods that use fundamentally different principles have been developed to study RNA structures (Table 1). X-ray crystallography provided the first atomic resolution picture of an RNA molecule, when the first transfer RNA (tRNA) crystal structure was solved in the 1970s (Kim et al. 1973; Robertus et al. 1974). Since then, X-ray crystallography has been the dominating tool in studying both protein and RNA structures (Shi 2014). Nuclear magnetic resonance (NMR) probes the conformation of molecules in solution, providing an alternative means to analyze RNA, especially flexible ones (Bothe et al. 2011). Cryogenic electron microscopy (cryo-EM), on the other hand, examines single frozen-hydrated particles in their native state, and therefore can be applied to larger and more dynamic molecules and assemblies. With new technological innovations in electron detection and image processing, the resolution of cryo-EM has dramatically improved in the past few years and in many cases approaches that of X-ray crystallography (Bai et al. 2015). Some of the most spectacular advances using cryo-EM include the structures of spliceosomes corresponding to several intermediary steps in splicing, each containing multiple small nuclear RNAs (snRNAs) and proteins (Kastner et al. 2019; Plaschka et al. 2019; Yan et al. 2019). Messenger RNAs (mRNAs) and long noncoding (lnc)RNAs, which make up the majority of distinct RNA species, are generally much more flexible, with well-folded regions interspersed with amorphous linker regions, and folded together with a large collection of nonstoichiometric RNA partners, RNA-binding proteins (RBPs), as well as indirect binders. Although it is likely that further technological innovations will greatly enhance the power of X-ray crystallography, NMR, and cryo-EM, currently the vast majority of RNA molecules are beyond the reach of these in vitro methods.
Table 1.
Major categories of methods for the analysis of RNA structures and interactions
| Categories | Variety of methods | Features | General reference(s) |
|---|---|---|---|
| X-ray crystallography | In vitro, atomic resolution, smaller, and nearly static molecules | Shi 2014 | |
| NMR | In vitro, atomic resolution, smaller molecules, can be either static or flexible | Bothe et al. 2011 | |
| Cryo-EM | In vitro, near atomic resolution, large, and heterogeneous complexes | Bai et al. 2015 | |
| Thermodynamic methods | Nussinov algorithm, Zuker algorithm, etc. RNA structure, Sfold, Mfold, UNAfold, ViennaRNA Package, etc. | Guarantees theoretically optimal structures, slow when permitting pseudoknotted structures | Eddy 2004; Mathews et al. 2010 |
| Comparative sequence analysis | Fold and Align, Fold then Align, Align then Fold, etc. | Implies functional relevance, limited by alignments and efficiency (local structures) | Mathews et al. 2010 |
| Chemical probing | DMS-seq, Structure-seq, SHAPE, icSHAPE, Mutate and Map, etc. | In vitro and in vivo, one-dimension averaged reactivity/flexibility/accessibility profile, can be used to probe secondary, tertiary structures and RNA–protein interactions | Kladwang et al. 2011; Ding et al. 2014; Rouskin et al. 2014; Spitale et al. 2015 |
| Enzymatic probing | PARS, Frag-seq, etc. | In vitro, one-dimension averaged reactivity/flexibility/accessibility profile | Kertesz et al. 2010; Underwood et al. 2010 |
| Cross-linking (based on physical proximity) | PARIS, LIGR-seq, SPLASH, CLASH, hiCLIP, MARIO | In vitro and in vivo, direct physical base-pairing contacts (except MARIO), captures alternative conformations | Helwak et al. 2013; Sugimoto et al. 2015; Aw et al. 2016; Lu and Chang 2016; Nguyen et al. 2016; Sharma et al. 2016 |
NMR, Nuclear magnetic resonance, Cryo-EM, cryogenic electron microscopy; DMS, dimethyl sulfate; SHAPE, selective 2′-hydroxyl acylation by primer extension; icSHAPE, in vivo click SHAPE; PARS, parallel analysis of RNA structures; Frag-seq, fragmentation sequencing; PARIS, psoralen analysis of RNA interactions and structures; LIGR-seq, ligation of interacting RNA and high-throughput sequencing; SPLASH, sequencing of psoralen cross-linked, ligated, and selected hybrids; CLASH, cross-linking, ligation, and sequencing of hybrids; hiCLIP, RNA hybrid and individual nucleotide resolution cross-linking and immunoprecipitation; MARIO, mapping RNA interactome in vivo.
In parallel with the in vitro experimental methods, two classes of computational algorithms were developed to address the RNA-folding problem, using thermodynamics and comparative sequence analysis, respectively (Table 1) (Fallmann et al. 2017). As RNA molecules are assumed to most likely fold into lowest free energy states, stable structures can be predicted based on free energy terms for each unit of stacked base pairs and loops, also called the Turner energy rules (Mathews et al. 2010). Given the time and space complexity of these algorithms, restrictions are commonly applied, such as prohibiting the formation of pseudoknots and long-range structures. In addition, our understanding of the basic energy rules in RNA structure formation, as well as the contributions from the cellular environment, is limited. Together, these issues have resulted in structure models that are “elegant and too often wrong” (Eddy 2004; Mathews et al. 2010).
RNA structures that have biological functions are under evolutionary pressure to preserve specific secondary or tertiary interactions, often in the form of invariant or covariant base pairs (Rivas and Eddy 2001). Therefore, another approach to determine RNA structures is by identifying these signatures of selection in multiple sequence alignments (Table 1). Comparative sequence analysis is not restricted by our limited understanding of the thermodynamic rules and, therefore, can reveal both standard base pairs and tertiary interactions. By the very nature of this approach, the structures derived from sequence comparison are probably functional. However, comparative sequence analysis is only applicable to high-quality alignments with certain sequence variation levels, and often restricted to small sequence windows in the interest of efficiency.
RNA folding brings nucleotides into specific chemical environments that change their reactivity properties (or solvent accessibility, flexibility). Therefore, reading out the reactivity of individual nucleotides reveals information about the specific folding. Based on this principle, a large number of methods have been developed in the past several decades, using various chemicals and enzymes to probe nucleotide reactivity (Table 1) (Ehresmann et al. 1987; Lu and Chang 2016). The chemicals and enzymes differentially react with the base, sugar, or the phosphodiester backbone, either in vitro or in vivo, resulting in modifications that can be read out as reverse transcription stops in gel electrophoresis or high-throughput sequencing. Reactivity profiles obtained from such experiments correlate with base-pairing status and can be used as constraints in secondary structure modeling, often as additional pseudoenergy terms. Probing data have been shown to improve the accuracy of structure modeling to some extent yet are still limited to simple structures. These experiments provide averaged signals for each RNA species, which does not reflect the dynamic nature of RNA in living cells (Lu and Chang 2016). Many of the inherent problems in RNA structure prediction persist even with the addition of chemical probing data. Statistically correlated modification patterns can be obtained from reverse transcriptase-induced mutations but are very low in efficiency and limited in length (Siegfried et al. 2014). A mutate-and-map strategy was recently developed to derive information about base pairs but is only applicable to one RNA at a time in vitro (Kladwang et al. 2011).
Contrary to the conventional chemical and enzymatic probing experiments, cross-linking-based methods, which are the topic of the current review, aim to directly identify the base-pairing regions and limit the “guesswork” of structure modeling to very short sequences that typically fold into a single possible duplex (Table 1). Originally developed in the 1970s, photoactivated psoralen cross-linking has provided mechanistic insights into many important and challenging problems in RNA biology. Recently, new technical improvements have led to a revival of this class of methods, bringing superior sensitivity, resolution, and throughput. Starting from a historical perspective, in this review we will discuss the experimental and analytical aspects of several psoralen cross-linking techniques that use the same principle of “cross-link, proximally ligate, and sequence.” Current applications of psoralen cross-linking have resulted in the surprising discoveries of long range, alternative, and dynamic structures, new RNA–RNA interactions, and principles in the high-level organization and networks of the transcriptome. As no tool is perfect for any task, it is essential to integrate cross-linking methods with other types of structure determination techniques. We will discuss the limitations of current implementations and how to push the technologies to the next level.
2. HISTORICAL USE OF PSORALEN TO ANALYZE RNA STRUCTURES AND INTERACTIONS
Psoralen is a group of planar tricyclic compounds that can intercalate in double-stranded nucleic acids (Fig. 1A) (Hearst 1981). The extensively conjugated system undergoes cycloaddition with pyrimidines upon long-wavelength ultraviolet (UV) irradiation, forming monoadducts and then diadducts, or interstrand cross-links (Fig. 1B). Short-wavelength UV reverses the cross-link, permitting analysis of cross-linked RNA using various methods. Cross-linking presents definitive and specific evidence for base-pairing and has provided the first mechanistic insights into a variety of noncoding (nc)RNAs including the ribosomal RNAs (rRNAs), spliceosomal snRNAs, and small nucleolar and small Cajal body RNAs (sno/scaRNAs). Hearst and colleagues have extensively discussed the chemical properties and early applications of psoralen on nucleic acid structures (Hearst 1981; Cimino et al. 1985). Here we briefly review some of the classical studies that paved the way for recent breakthroughs in throughput, sensitivity, and resolution of cross-linking-based methods.
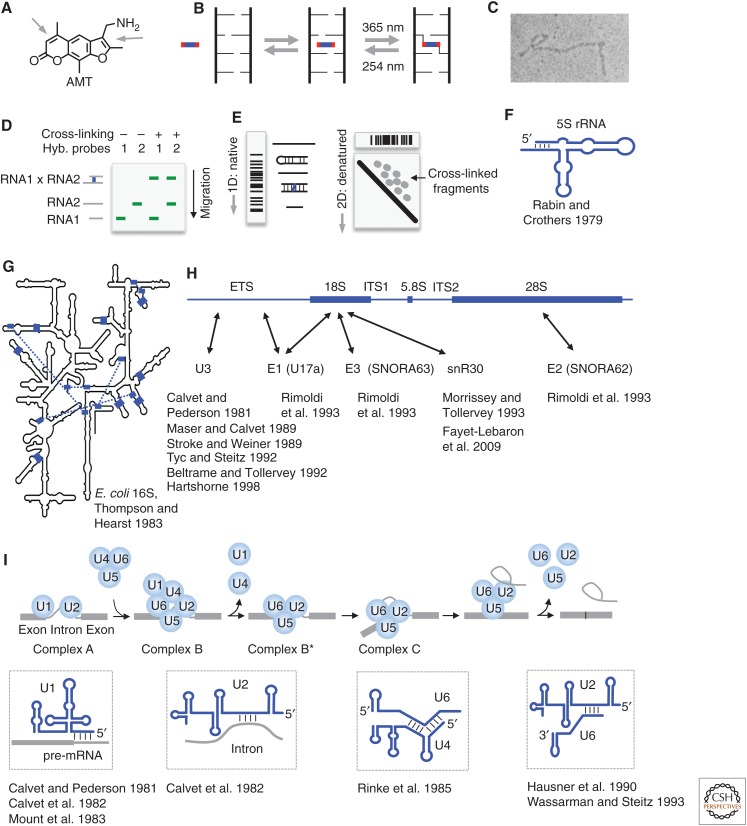
Figure 1.
Psoralen chemistry and historical applications. (A) Structure of the commonly used psoralen AMT (4′-aminomethyltrioxsalen). Arrows point to the double bonds that undergo cycloaddition to pyrimidines. (B) Model of psoralen intercalation into nucleic acid helices and reversible cross-linking. (C) Detection of psoralen cross-linked RNA structures by electron microscopy. (Image courtesy of Paul L. Wollenzien et al.; reprinted, with permission, from Wollenzien et al. 1978, © National Academy of Sciences.) (D) Detection of RNA–RNA interactions using gel electrophoresis and northern blotting. Cross-linked RNA pairs can be identified by the simultaneous hybridization of probes to both RNAs. (E) The “native-denatured” 2D gel method for selecting psoralen cross-linked RNA. The first-dimension gel is native. In the second dimension, the first-dimension gel slices are embedded in a urea-denatured gel and run at high temperature, in which the cross-linked fragments assume an “X” shape and migrate much more slowly, above the diagonal, forming scattered dots for simple RNA species or a smear for complex RNA mixtures, such as total RNA. (Adapted from Lu et al. 2016, with permission, from Elsevier.) (F) Secondary structure of the Escherichia coli 5S ribosomal RNA (rRNA), based on psoralen cross-linking and enzymatic sequencing (Rabin and Crothers 1979). (G) Psoralen cross-linked duplexes (blue blocks and the blue dashed lines) superimposed on the Escherichia coli 16S rRNA model derived from crystal structure (Petrov et al. 2014). (Redrawn, with permission, from Thompson and Hearst 1983.) (H) Summary of small nucleolar RNA (snoRNA)–rRNA interactions based on psoralen cross-linking. ETS/ITS: external/internal transcribed sequence. Arrows point to cross-linking sites. (I) Summary of several RNA–RNA interactions in precursor messenger RNA (pre-mRNA) splicing based on psoralen cross-linking. (The splicing model was redrawn, with permission, from Will and Luhrmann 2011.)
Initially, electron microscopy (EM) was used to detect psoralen cross-linking sites in denatured and psoralen cross-linked nucleic acids (Fig. 1C) (Cech and Pardue 1976; Wollenzien et al. 1978). EM is limited in resolution to dozens of base pairs and only shows the overall contour of the secondary structure. To analyze RNA–RNA interactions, denatured gel electrophoresis was used to resolve psoralen cross-linked samples, in which cross-linked RNAs were identified as slower bands by northern blotting (Fig. 1D) (Calvet and Pederson 1981; Calvet et al. 1982). Reversal of cross-linking further confirms the interaction. This setup works well for interactions involving sRNAs in which the partners can be exhaustively tested. To analyze complex interactions and structures, Brimacombe and colleagues developed the 2D gel method (Fig. 1E) (Zwieb and Brimacombe 1980). The 2D gel uses the differences in migration of the cross-linked RNA fragments in denatured gel versus native gel. After the first-dimension native gel, cross-linked fragments assume an “X” shape in the denatured gel and migrate slower than non-cross-linked fragments. The retarded fragments can be isolated from above the diagonal for sequencing. Together, these three methods were extensively used in the early studies of RNA structures and interactions.
The ribosome is a large RNA–protein complex that makes proteins from genetic information stored in mRNA. Several groups used psoralen cross-linking to study the folding of the rRNAs before crystallization was possible. Rabin and Crothers applied psoralen cross-linking to the 5S rRNA folded in solution and showed the existence of a terminal stem (Fig. 1F) (Rabin and Crothers 1979). Thompson and Hearst combined the in-solution psoralen cross-linking, 2D gel purification, photoreversal, and enzymatic sequencing to discover 13 base-paired regions in the Escherichia coli 16S rRNA (Fig. 1G) (Thompson and Hearst 1983). Although most of the duplexes from cross-linking are consistent with the evolutionary model built by Noller and Woese and the crystal structure model (Noller and Woese 1981; Petrov et al. 2014), a few of them could be artifacts of in vitro folding (for review, see Sergiev et al. 2001 and Whirl-Carrillo et al. 2002). Nevertheless, these heroic efforts showed the possibility of using psoralen to study complex RNAs.
The rRNAs are transcribed as a polycistronic precursor, then extensively processed and modified into the mature subunits. Psoralen cross-linking elucidated several critical steps in rRNA biogenesis and revealed functions of an abundant class of sRNAs, snoRNAs (Fig. 1H). Calvet and Pederson showed that the U3 snoRNA could be cross-linked to the large rRNAs in vivo, providing the first direct evidence of snoRNAs involvement in rRNA processing (Calvet and Pederson 1981). This interaction is required for the cleavage of the rRNAs to release the mature 18S rRNA (Kass et al. 1990). Later studies have continued to refine the cross-linking and mapping methods to identify the precise binding sites on rRNAs for U3, E1 (U17a), E2 (SNORA62), E3 (SNORA63), snR30, etc. (Calvet and Pederson 1981; Maser and Calvet 1989; Stroke and Weiner 1989; Beltrame and Tollervey 1992; Tyc and Steitz 1992; Morrissey and Tollervey 1993; Rimoldi et al. 1993; Hartshorne 1998; Fayet-Lebaron et al. 2009). Together with genetic experiments that specifically edit the identified binding sites, these studies showed the essential roles of snoRNAs in rRNA processing.
The psoralen cross-linking methods made critical contributions in elucidating the mechanisms of eukaryotic mRNA splicing (Fig. 1I). Steitz and colleagues hypothesized that the U-rich snRNAs are involved in the removal of introns from eukaryotic heterogeneous nuclear RNAs (hnRNAs; precursor messenger RNAs [pre-mRNAs]) based on sequence complementarity of U1 with the 5′ splice site (Lerner et al. 1980). Shortly after that, Calvet and Pederson showed that U1 and U2 small nuclear ribonucleoprotein complexes (snRNPs) can be reversibly cross-linked to pre-mRNAs, providing the first physical evidence for the direct interactions (Calvet and Pederson 1981; Calvet et al. 1982). Since then, a series of studies from several groups pinpointed the interaction sites between pre-mRNAs and snRNAs, revealing a complex and dynamic ribonucleoprotein complex (RNP) machine (Mount et al. 1983; Rinke et al. 1985; Hausner et al. 1990; Wassarman and Steitz 1993). The sequential formation and reorganization of distinct RNA–RNA interactions ensure efficient and precise removal of introns (Fig. 1I). The splicing mechanisms revealed by psoralen cross-linking have since been validated and refined by genetic analysis, X-ray crystallography, and cryo-EM (Will and Luhrmann 2011).
3. THE NEXT GENERATION: CROSS-LINKING WITH PROXIMITY LIGATION AND HIGH-THROUGHPUT SEQUENCING
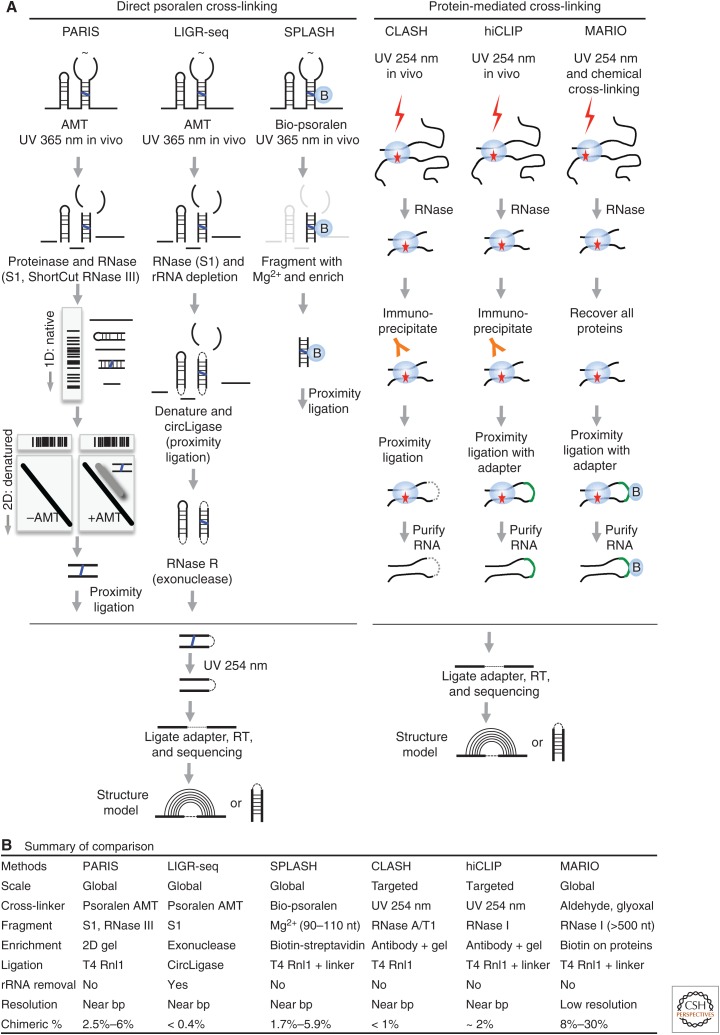
Traditional methods for the analysis of cross-linked RNA, such as EM, 1D and 2D gels, and enzymatic sequencing, are laborious and inefficient (Fig. 1C,D). As a result, applications were limited to highly abundant RNAs one at a time, like the rRNAs, snRNAs, and snoRNAs (Fig. 1). Several recent technological advancements have transformed classical psoralen cross-linking into a powerful transcriptome-scale discovery tool (Fig. 2A, left side). In the new generation of methods, including psoralen analysis of RNA interactions and structures (PARIS), sequencing of psoralen cross-linked, ligated, and selected hybrids (SPLASH), and ligation of interacting RNA and high-throughput sequencing (LIGR-seq), cells are treated with psoralens and UV irradiation; then cross-linked RNA fragments are enriched and proximally ligated so that each RNA duplex can be uniquely identified via high-throughput sequencing (Aw et al. 2016; Lu et al. 2016; Sharma et al. 2016; Lu et al. 2018). This strategy determines RNA structure and interactions with near base-pair resolution and single molecule accuracy at the transcriptome level. Three related methods that use a similar strategy, but without psoralen cross-linking, were also reported recently (Fig. 2A, right side) (Kudla et al. 2011; Helwak et al. 2013; Sugimoto et al. 2015; Nguyen et al. 2016). Here these methods are compared side by side to illustrate their similarities and differences.
Figure 2.
Recently developed cross-linking-based methods for the direct detection of RNA interactions and structures. Only the methods that use the “cross-link, proximally ligate, and sequence” principle are listed here. (A) Workflow of the methods. (B) Major differences among the methods. (Part of panel A is adapted from Lu et al. 2016, with permission, from Elsevier.)
Several major differences exist among the psoralen cross-linking methods (Fig. 2B). First, biotinylated psoralen was used in SPLASH instead of the more commonly used 4′-aminomethyltrioxsalen (AMT), thus enabling convenient isolation of RNA fragments covalently bound to psoralen, either with monoadducts (cycloaddition on one side) or cross-links. The bigger size of the bio-psoralen molecule also reduces efficiency in the intercalation of RNA duplexes. The concentrations of psoralens used vary considerably. Conventional chemical probing usually selects chemical concentrations to aim for “single-hit kinetics” to avoid the induction of structural changes during probing (Spitale et al. 2015). However, high concentrations of psoralen may not be an issue because cross-linking fixes structures to prevent further changes.
Given the low efficiency of cross-linking, several different strategies were used to enrich cross-linked fragments from total RNA. The 2D gel technique used in PARIS, although laborious, enriches cross-linked duplexes up to 100% in purity. In LIGR-seq, an exoribonuclease, RNase R, was used to remove nonlinked single-stranded fragments. This approach is easier to perform but does not efficiently remove non-cross-linked fragments, especially highly structured ones. In SPLASH, purification of bio-psoralen-reacted fragments is simple with the streptavidin beads, but both monoadduct and cross-linked RNA fragments are extracted. The three methods select RNA fragments at different sizes, affecting both proximity ligation efficiency and structure model resolution. Longer fragments promote ligation but also result in ambiguous base-pairing models.
Short-wavelength UV and bifunctional chemical cross-linkers were used in cross-linking, ligation, and sequencing of hybrids (CLASH), RNA hybrid and individual nucleotide resolution cross-linking and immunoprecipitation (hiCLIP), and mapping RNA interactome in vivo (MARIO; Table 1, Fig. 2). In CLASH and hiCLIP, UV–cross-linked protein–RNA complexes were selected using antibodies; therefore, only the targets of a single RBP are identified. These methods achieve deeper coverage of specific interactions given the limited scope. MARIO fixes both specific (from UV cross-linking) and nonspecific (from chemical cross-linking) interactions in physical proximity, and therefore the identified RNA fragment pairs are not necessarily base-paired. The detection of all protein-associated interactions is possible with MARIO because all RNA–protein complexes are recovered from cell lysate. However, the large size of the RNA fragments in MARIO precludes accurate structure modeling (Fig. 2B).
A commonly used strategy in the analysis of chromatin conformations, proximity ligation joins two restriction-digested DNA fragments using DNA ligases to enable the identification of their physical proximity in 3D space in the nucleus (Dekker et al. 2002). Proximity ligation in RNA was first noticed in the analysis of RNA–protein cross-linking studies and then intentionally used to identify potential RNA–RNA interactions (Kudla et al. 2011). In contrast to DNA, in which longer fragments and the sticky ends ensure highly efficient ligation, the ends of shorter RNA duplexes are under steric constraint, resulting in much lower ligation efficiency (Fig. 2B). The ligation efficiency has not improved much despite extensive optimizations. The typical T4 RNA ligase 1 (Rnl1) is used in most applications, whereas in LIGR-seq, a thermostable T4 Rnl1 homolog CircLigase was used (Blondal et al. 2005). In hiCLIP and MARIO, a linker oligonucleotide was used to bridge the two ends, but with no obvious improvement, except in MARIO in which much longer fragments are ligated.
The various technical approaches used in these methods provide flexible choices that can be used in a “mix-and-match” manner depending on the nature of the specific problem to be solved. Users can choose psoralen derivatives, RNA fragmentation methods, enrichment techniques, ligases with or without linkers, and antibodies or antisense oligos that select specific subsets of RNAs. Together these methods have been applied to multiple species and cell lines, providing a rich resource for future studies (Gong et al. 2018).
4. BASIC ANALYSIS OF RNA STRUCTURE CROSS-LINKING DATA
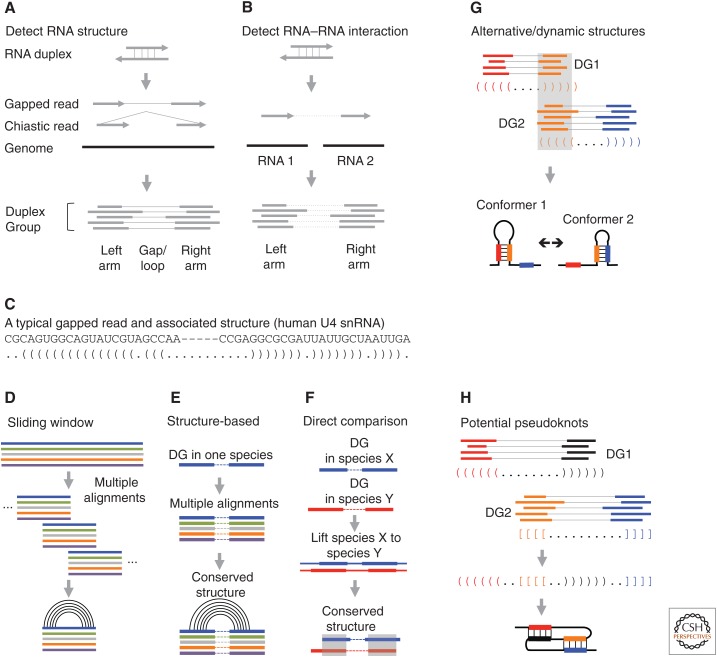
The cross-linking (PARIS-like) experiments produce a small percentage of noncontinuous reads as a result of proximity ligation of base-paired duplexes (Fig. 3A,B). The noncontinuous reads can be regularly gapped or chiastic, depending on which side proximity ligation occurs (Fig. 3A,B). Short-read mappers, like the spliced transcripts alignment to a reference (STAR) software, are capable of directly mapping both types of sequences with appropriate parameters (Dobin et al. 2013). Although the noncontinuous reads can be directly used, assembly of duplex groups (DGs) is a critical step that standardizes and simplifies subsequent analysis of both structures and interactions (Lu et al. 2016, 2018). A DG is a group of reads from RNA molecules in the same conformation, corresponding to one specific RNA duplex. Many structure prediction programs can be used to build the base-pairing model from the DGs (Table 1). Given the small arm size and the specific psoralen cross-linking of staggered uridines, a unique and unambiguous structure model can be established (Fig. 3C). The structure models can be validated using various orthogonal methods, including selective 2′-hydroxyl acylation by primer extension (SHAPE)-like chemical probing and conservation analysis (Lu et al. 2016). In addition, DGs enable facile analysis of significance, assembly of complex structures, including dynamic conformations, pseudoknots and high-level architectures, and analysis of RNA–protein interactions (Fig. 3C–H).
Figure 3.
RNA structure and interaction analysis based on cross-linking data. (A) Detecting RNA structures. Discontinuous sequencing reads, including “regularly” gapped and chiastic, are assembled into duplex groups (DGs). (B) Detecting RNA–RNA interaction. The two arms of discontinuous reads mapped to two RNA molecules are assembled into DGs. (C) An example of a discontinuous read from the U4 small nuclear RNA (snRNA). Pairs of parentheses indicate base pairs. (D) Conventional sliding window screen for conserved structures from whole genome alignments. Multiple genome alignments are split into windows with fixed width and step. Conservation and covariation are then scored in each window. (E,F) DGs guide comparative sequence analysis. (E) DGs from one species are used to extract two blocks of alignments separated by arbitrary distance for structure conservation analysis. (F) Structures can be directly compared with DGs from multiple species, using sequence alignments. (G) Alternative/dynamic RNA structures are identified based on the conflicts of the base-pairing models between two DGs. (H) Interlocked DGs suggest either alternative conformations or pseudoknot structures. (Panels E–G are adapted from Lu et al. 2016, with permission, from Elsevier.)
Whereas intramolecular structures are straightforward, intermolecular interactions are more difficult to determine, in part because of the complexity of the transcriptome, which contains homologous and repetitive sequences and frequent errors in the gene annotation. Genome-mapped reads need to be extensively filtered to remove artifacts. Alternatively, carefully curated transcriptome annotations can be used as reference to avoid spurious mapping but may miss ones not in the annotation. The significance of detected structures has been determined using different approaches. After DG assembly, a threshold was applied to the ratio of the number of reads in the DG divided by sequencing depth at the two arms (Lu et al. 2016). Alternatively, the Blencowe group used a probabilistic model to assess the significance of the detected interactions based on the observed and expected connections between the two arms (Sharma et al. 2016).
5. PREVALENT LONG-RANGE STRUCTURES IN THE TRANSCRIPTOME
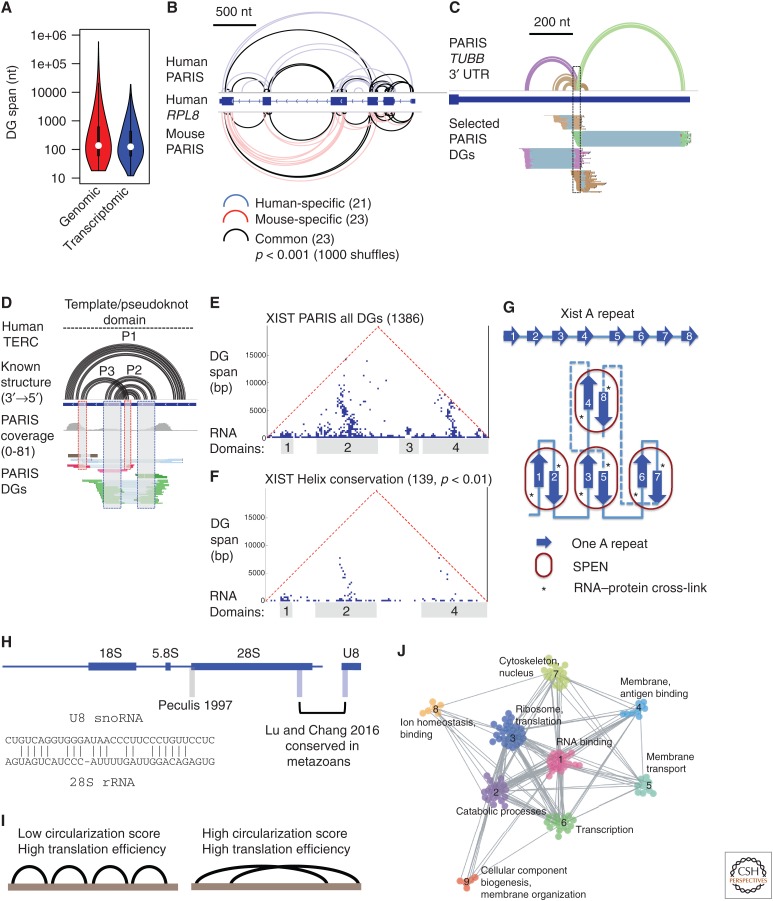
One surprising observation in the psoralen cross-linking and hiCLIP studies is the prevalence of long-range structures (Fig. 4A). (Here we use “structure” to denote all intramolecular base-pairing interactions, and “interaction” to refer to only intermolecular interactions [Sugimoto et al. 2015; Aw et al. 2016; Lu et al. 2016; Sharma et al. 2016].) Many structures span hundreds to thousands of nucleotides, and in mRNAs, often span multiple exons. Computational predictions, including ones that incorporate chemical probing data, have focused on local structures in the interest of efficiency. The resulting models should now be viewed with caution in light of evidence showing prevalent long-range structures.
Figure 4.
New insights into the RNA structurome and interactome. (A) Long-range structures are prevalent in the transcriptome. Duplex group (DG) spans were calculated from Henrietta Lacks cell line psoralen analysis of RNA interactions and structures (HeLa PARIS) data on genomic and transcriptomic coordinates. (B) Example of conserved structures in RPL8 messenger RNA (mRNA) based on direct comparison of human (human embryonic kidney cells 293 [HEK293]) and mouse (J1 embryonic stem cells) PARIS data. Significance of the overlap was tested using random shuffling of DGs in the exons. (C) Example of alternative/dynamic structures in the 3′ untranslated region (UTR) of tubulin β class 1 (TUBB) mRNA from HeLa PARIS data. The corresponding structure models (first track) and DGs are color coded. (D) PARIS detects pseudoknot structure (interlocked DGs) in the telomerase RNA component (TERC). (E) PARIS determines the architecture of X-inactive specific transcript (XIST). Each point in the heat map shows the connection between the two regions indicated by the feet of the triangle. Four major domains are obvious from the clustered DGs. (F) About 10% of PARIS-determined duplexes are conserved in eutherian mammals (p value < 0.01). (G) Model of SPEN-A-repeat complex in the XIST ribonucleoprotein (RNP). The base-pairing interactions among the 8.5 repeats are stochastic and only one specific family of conformations from PARIS data is shown here. SPEN binding involves both single- and double-stranded regions but is only cross-linked to the single-stranded region 3–5 nt upstream of the interrepeat duplex unit. (H) PARIS in human and mouse cells revealed the precise U8:28S interaction, which is conserved in metazoans. PARIS-determined interaction sites in blue, and the previously reported site in gray (Peculis 1997). The base-pairing model is shown in the inset. (I) Long-span structures as detected by sequencing of psoralen cross-linked, ligated, and selected hybrids (SPLASH) correlates with higher translation efficiency. (J) RNAs of related functions tend to interact more frequently than nonrelated ones. Global RNA–RNA interaction networks can be organized into modules and the network topology changes in response to stem cell differentiation. (Panels A–H are adapted from Lu et al. 2016; panels I–J are adapted from Aw et al. 2016, both, with permission, from Elsevier.)
In both mRNAs and lncRNAs, structures that connect the 5′ and 3′ ends have been detected. Wan and colleagues showed that there is a positive correlation between the circularization score (tendency to form longer range structures) with translation efficiency for mRNAs (Fig. 4I) (Aw et al. 2016). In the case of mRNAs, the end-to-end connections are reminiscent of the mRNA circularization mechanism essential for translation, in which translation initiation factors on the 5′ end interact with the poly(A)- binding protein on the 3′ end (Wells et al. 1998). mRNA circularization can also be mediated by base-pairing, as shown in the p53 mRNA, and this long-range base-pairing between the 5′ and 3′ untranslated regions (UTRs) promotes translation (Chen and Kastan 2010). End-to-end base-paired structures are frequently observed in viruses and play critical roles in virus replication and translation (Nicholson and White 2014). As RNA structures form during 5′ to 3′ transcription, and are constantly reorganized by splicing, translation, and other processes, it is important to determine how long-range structures emerge given that local contacts are more likely to form by chance. More interestingly, how do long-range structures contribute to the various RNA metabolism events? Comprehensive dissection of each structure by mutagenesis is needed to study their functions.
6. CONSERVATION ANALYSIS OF THE RNA STRUCTURES
RNA structures form spontaneously as a result of thermodynamics; presence of RNA structures does not imply function. Base-pair covariation, however, is an important indicator of evolutionary pressure on functional structures. Sifting through sequence alignments for conserved and covaried base pairs is a classical strategy for identifying functional structures, and this can be applied to either individual RNAs or whole genomes (Washietl et al. 2005; Pedersen et al. 2006; Smith et al. 2013). In practice, the search is restricted to small sliding windows across the genome because of extraordinary computational requirements (Fig. 3D). Significance can be calculated by shuffling in each window (Gesell and von Haeseler 2006). However, these methods only examine local structures and lack experimental validation. Given the stringency of these methods, conserved structures cannot be identified from sequences lacking sufficient variation.
The direct determination of helices in the cross-linking methods enables targeted analysis of structure conservation without distance limit. Using the PARIS-determined duplexes as guides, we developed two approaches to directly search for structure conservation (Lu et al. 2016). First, the DGs from one species were used to extract alignment blocks from multiple genomes, in which the two arms can come from distant regions (Fig. 3E). Second, when structure data are available from more than one species, direct comparison of the DGs is performed on sequence alignments (Fig. 3F). The second approach is especially useful when variation is too low or too high for statistical analysis of significance. We found that a substantial fraction of structures that span more than 200 nt are conserved. These analyses also showed that overall architectures are conserved for both coding and noncoding long RNAs (Fig. 4B). On the other hand, structure data may also be helpful in correcting sequence-based alignments that often do not consider structure information. This approach will be especially useful for distantly related sequences that cannot be aligned properly by sequence alone.
Given the ubiquitous presence of RNA structures, they can be quickly adapted for specific functions without showing obvious signs of conservation. As a result, such functional structures would have been missed by conventional methods that rely on covariation. Bartel and colleagues showed that random structures in the 3′ UTR are required for mRNA stability (Wu and Bartel 2017). This study highlights the limits of conventional comparative sequence analysis and the need for direct determination of structures.
7. DYNAMIC AND COMPLEX STRUCTURES: CATCHING RNA IN ACTION
Original work in the 1950s by Anfinsen showed that protein structures are mostly determined by primary sequence (Anfinsen 1973). Dynamics in protein structures occurs locally, usually without breaking secondary structures. However, RNA folding is more promiscuous given the simpler alphabet and base-pairing rules. In addition to local structure dynamics, global rearrangements of structures are frequent in RNA and essential for their activities (Dethoff et al. 2012). As described earlier in this review, dramatic rearrangement of structures and interactions occurs multiple times during pre-mRNA splicing (Fig. 1H). Conventional chemical probing, although applicable to structure determination in vivo, results in averaged reactivity profiles that are hard to deconvolve into the individual conformations. In contrast, psoralen cross-linking and proximity ligation directly capture each conformation in a single sequencing read, thus providing a direct readout of all the “cross-linkable” conformations, like taking snapshots of the RNA in action.
Dynamic and alternative structures take the form of conflicting DGs, in which two DGs overlap on one arm (Figs. 3G and 4C). Significantly overlapped base pairs cannot form at the same time, with the exception of triplexes, and usually are the result of two mutually exclusive conformations. Using this simple criterion, a large number of dynamic structures were uncovered, some of which are conserved between human and mouse (Lu et al. 2016). Whereas some of the conformations could be simply the kinetically or thermodynamically trapped folding intermediates, others may represent important functional states in a dynamic regulatory process. Wan and colleagues applied the SPLASH method to the human embryonic stem cell differentiation process and found that the transition in cellular states is accompanied by alterations of both RNA structures and RNA–RNA interactions, suggesting regulatory mechanisms by the dynamic structures and interactions (Aw et al. 2016). The constant rearrangement of intra- and intermolecular base-pairing interactions and the dynamic protein interactions create a rugged free energy landscape that integrates genetic and environmental signals. Further investigation of RNA dynamics will likely uncover new “switches” in gene regulation pathways.
Pseudoknots are composite structures that involve at least two duplexes that are interlocked. Pseudoknots stabilize structures and participate in several critical cellular processes, including telomere maintenance and ribosomal frameshifting (Gilley and Blackburn 1999; Giedroc and Cornish 2009). Computational methods can be used to identify pseudoknots but are very slow (Rivas and Eddy 1999). Psoralen cross-linking directly identifies interlocked duplexes that could be either alternative conformations or pseudoknots (Fig. 3H). For example, PARIS identified the pseudoknots in telomerase RNA component (TERC; Fig. 4D), RNA component of mitochondrial RNA processing endoribonuclease (RMRP), and ribonuclease P RNA component H1 (RPPH1) RNAs in both human and mouse (Lu et al. 2016). Careful analysis and validation of the interlocked duplexes may reveal additional pseudoknots in the transcriptome.
8. ARCHITECTURE OF THE XIST RNP: MODULAR STRUCTURES FOR MODULAR FUNCTIONS
The psoralen cross-linking methods identify physical contacts without a distance limit and therefore, reveal the overall architecture of RNA molecules. With few exceptions, whole transcript architectures have remained elusive for the vast majority of the transcriptome (Zappulla and Cech 2004). X-inactive specific transcript (XIST) is a lncRNA essential for X-chromosome inactivation (XCI) in placental mammals (eutheria). XIST serves as a scaffold to recruit a variety of protein complexes to orchestrate the complex XCI process. Using PARIS, we showed that the XIST RNA folds into compact and discrete domains, each spanning hundreds to thousands of nucleotides (Fig. 4E) (Lu et al. 2016, 2017). Despite the low sequence conservation, roughly 10% of the structures are conserved (Fig. 4F). The conserved duplexes support the demarcation of domains and suggest functional relevance of the high-level architecture. The XIST RNA coordinates multiple steps in XCI, including XIST localization to the inactive X, membrane attachment of the inactive X, histone and DNA modifications, and chromatin compaction. Targeted genetic analysis of each of the XIST domains will uncover the structural basis of the specific functions.
Stable and discrete domains may not be a common feature for all RNAs; for example, highly stable structures would not persist in mRNA coding regions because of the constant action of ribosomes. Even for lncRNAs, architectures differ greatly; for example, metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) and XIST have more long-range structures that are organized into compact domains, whereas nuclear-enriched abundant transcript 1 (NEAT1) has more local structures, without clearly separated modular domains (Lu et al. 2016). Comparison of the duplex spans in general showed that mRNAs have more local structures than lncRNAs (Aw et al. 2016). These studies have revealed a complex picture for the high-level organization of RNA structures. However, with the exception of a few RNAs (such as XIST and MALAT1), the extent of domain formation in other RNAs is still unknown because of the limited sequencing coverage and the inherent bias of psoralen cross-linking.
The integration of PARIS with in vivo click SHAPE (icSHAPE), conservation analysis, and protein-binding assays (in vitro electrophoretic mobile shift assay [EMSA] and cross-linking and immunoprecipitation [CLIP]) led to the first structure-interaction model of the essential A-repeat domain complex in XIST RNP (Fig. 4G) (Lu et al. 2016). In this model, repeat units in the A-repeat region randomly form duplexes with each other, resulting in near identical structure units. These structure units associate with the essential XCI factor SPEN in a cooperative manner. The structure model is consistent with nucleotide resolution reactivity of the repeats determined by icSHAPE. Stringent tests for covariation showed no significance for any pair of repeats, consistent with the stochastic nature of the duplex formation, which dilutes and distributes the evolutionary pressure (Rivas et al. 2017). This study establishes a “top-down” approach for studying long RNAs, both coding and noncoding, in which modular RNA structure domains are identified and isolated for further studies.
9. NOVEL RNA–RNA INTERACTIONS: THE MOLECULAR SOCIAL NETWORK
Traditionally, the analysis of RNA–RNA interactions has required knowledge of at least one partner in the RNA–RNA complex, or the proteins involved. For example, in the CLASH and hiCLIP methods, the identification of new interactions was based on the proteins that bind the RNA duplexes (Fig. 2). Application of psoralen cross-linking methods has resulted in important de novo discoveries of RNA–RNA interactions, such as snoRNA:rRNA, scaRNA:snRNA, mRNA:mRNA, snoRNA:mRNA, and snRNA:lncRNA. These discoveries have both solved some long-standing questions in the field and revealed potentially new principles that govern coordinated gene expression programs. Similar to the analysis of intramolecular RNA structures, psoralen cross-linking achieves near base-pair level resolution and single-molecule accuracy, which greatly facilitates further mechanistic studies.
As discussed in the beginning, early studies of snoRNAs and scaRNAs led to the discovery of snoRNA-guided rRNA processing and modifications. Yet many sno/scaRNAs are still “orphans,” with no identified targets. In certain cases, these RNAs may have more than one partner, but only one was known owing to the limited scope of conventional methods. An example of these poorly annotated snoRNAs is U8, which is highly expressed and conserved in metazoans (Fig. 4H). Previous studies suggested that the 5′ end of U8 base pairs with the 5′ end of the 28S rRNA to guide the cleavage of precursor ribosomal RNA (pre-rRNA; Fig. 4H, site labeled “Peculis 1997”). However, PARIS experiments in both human and mouse cells clearly identified an interaction site near the 3′ end of the 28S rRNA (Lu et al. 2016). The newly identified interaction is conserved in all metazoans and the duplex is more extensive than the previous site (Fig. 4H, inset). This result highlights the critical contribution of psoralen cross-linking in providing direct physical evidence for RNA–RNA interactions.
In addition to the intermolecular interactions that involve sRNAs (snRNAs, snoRNAs, scaRNAs, etc.), psoralen cross-linking also detected mRNA:mRNA interactions, although with much fewer reads given their low abundance. Interestingly, the mRNA:mRNA interactions form extended networks, in which functionally related mRNAs are clustered together, indicating co-regulation on the RNA level (Fig. 4J) (Aw et al. 2016). Even more surprisingly, differentiation of human embryonic stem (ES) cells alters the topology of the interaction networks. It remains unclear what factors drive the formation and the alteration of the interaction networks and what functional consequences such physical contacts bring. Given the limited coverage of current cross-linking data, it is likely that future work will uncover other new interactions and mechanisms. Quantitative information about the interactions is also needed to further understand their relevance. For example, are these interactions kiss-and-run encounters or long-term committed relationships?
10. LIMITATIONS OF THE PSORALEN CROSS-LINKING METHODS AND FUTURE DIRECTIONS
Psoralen cross-linking has provided novel insights into the RNA structurome and interactome in living cells. Despite more than 40 years of work, current cross-linking-based methods still suffer from several limitations. Here we discuss the major issues, which will hopefully facilitate data interpretation and point to directions for further improvement.
Psoralen cross-linking of nucleic acid duplexes is slow and inefficient, often taking 30 min or more to achieve effective cross-linking (Lu et al. 2016). Prolonged cross-linking can change the cellular physiology to alter RNA structures. For example, long exposure to low temperature (needed to avoid UV-induced heating) may induce stress response. Light-activated psoralens also modify other cellular components such as lipids and proteins (Cimino et al. 1985). These side effects should be considered when designing experiments that examine the effects of certain conditions on RNA structures. In addition, the slow reaction makes it difficult to obtain dynamic information about RNA structures in response to environmental signals.
Psoralen cross-linking has significant bias on both the sequence and structure levels. Psoralens are preferentially (>90%) cross-linked to staggered uridines (Cimino et al. 1985; Boyer et al. 1988). Compact RNP structures may block cross-linking, and this effect is even more difficult to estimate. Therefore, we can only make semiquantitative conclusions about the structures that we observe in these experiments. For example, it is not possible to directly calculate the relative abundance of alternative conformations. In addition, conclusions regarding the overall topology of intra- and inter-RNA interaction networks are likely to be inaccurate because of the bias.
Psoralens are the only class of photoreversible cross-linkers available now for the general analysis of nucleic acid structure and interactions. New reversible cross-linkers with improved reaction speed and reduced bias will be of great value for the RNA field. For example, reversible RNA-modifying groups can be added to other classes of compounds that intercalate the DNA/RNA duplex. In fact, many cancer chemotherapy drugs act by modifying DNA, and can be potentially adapted to reversibly cross-link RNA (Deans and West 2011). Chemicals that cross-link tertiary contacts will provide important new information currently missed by psoralen cross-linking in living cells. For example, nitrogen mustard has been used to successfully identify tertiary contacts in the ribosome and snRNAs (Datta and Weiner 1992; Sergiev et al. 2001).
Another major bottleneck in the cross-linking methods is the low efficiency of proximity ligation. In general, the highest efficiency achieved is only ∼5% for short fragments that permit near base-pair resolution structure modeling (Fig. 2B). Improving proximity ligation efficiency would greatly reduce sequencing cost and increase the amount of useful information obtained from high-throughput experiments. Potential solutions include new ligases that are less sensitive to the structural context, and ligation conditions that promote the “ligatable” conformations, such as higher temperature, chemical denaturation, and linkers that effectively increase the flexibility of the RNA ends.
Although sequencing cost will continue to drop, targeted analysis is a more cost-effective solution to analyzing RNAs of interest, especially low abundance RNAs from essential genes. In particular, the psoralen cross-linking-based methods could be combined with antibody enrichment of RBPs or antisense oligo enrichment of single or groups of RNAs.
11. INTEGRATING METHODS TO SOLVE COMPLEX RNA STRUCTURES AND INTERACTIONS
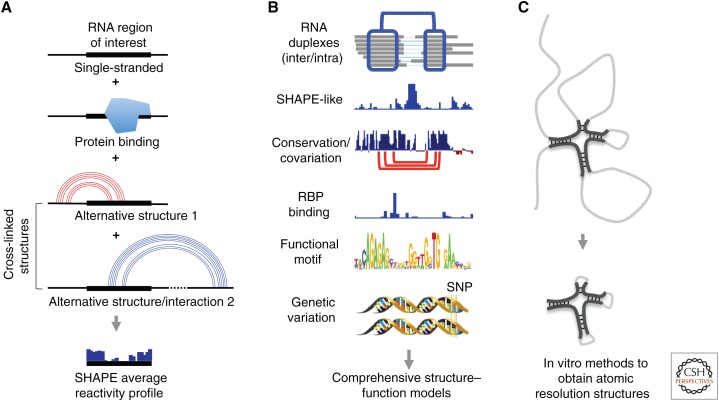
The cross-linking-based methods are particularly powerful in sorting through the complexities of RNA molecules in living cells, which are commonly depicted as a plate of “cooked spaghetti.” X-ray crystallography, NMR, cryo-EM, computational modeling, chemical/enzymatic probing, and cross-linking-based methods, as summarized in Table 1, each have their own merits and limitations; therefore, integration of diverse approaches will lead to the most comprehensive understanding of the basic properties of RNA (Fig. 5). In this context, the direct base-pairing information from cross-linking experiments provides an essential guide for the integrated analysis.
Figure 5.
Integrated RNA structure analysis using multiple methods. (A) Comparison of cross-linking-based methods and accessibility/flexibility measurement (here using selective 2′-hydroxyl acylation with primer extension [SHAPE] as an example). The measured average accessibility profile is the sum of all possible RNA structure and interaction conformations plus the effect of protein binding. In contrast, cross-linking methods directly identify the structure conformations. (B) Cross-linking data provide an essential guide to place various types of RNA data in a structural context, thereby facilitating integrated functional analysis of RNA molecules. (C) From the large and flexible RNA molecules (gray lines), cross-linking-based methods identify small, compact, and stable structure domains (black lines), which can be further studied using higher resolution methods like X-ray crystallography and nuclear magnetic resonance (NMR).
Many approaches can be envisioned to integrate structure data and other relevant information from the various methods described above. For example, incorporation of SHAPE-like data into cross-linking-based structure models achieves both high confidence and high resolution that is not possible by either method alone (Lu et al. 2016). As SHAPE-like methods provide a largely unbiased average reactivity profile of the structure ensemble and cross-linking methods nominate all cross-linkable conformations (see diagram in Fig. 5A), the integration of these two methods enables quantitative analysis of the alternative conformations. Analysis of RNA–protein interactions can now be performed in a structural context, which may explain the lack of apparent specificity on the sequence level (Fig. 5B). Also, as already shown (Fig. 3D,E), cross-linking-based structure models can guide comparative sequence analysis, and on the other hand, the conservation assigns functional significance to structures, prioritizing some of them for further studies (Fig. 5B). Over the years, extensive knowledge of RNA sequence motifs has been accumulated, such as ones that regulate splicing, translation, stability, and localization. Transcriptome-wide structures and interactions place such motifs in a larger context, which will help reveal previously unknown regulatory mechanisms and serve as examples for identifying novel functional motifs. A relevant example is the identification of long-range structures that bring RNA-binding Fox-1 Homolog 2 (Rbfox2) proteins to the vicinity of splicing motifs (Lovci et al. 2013). Genome-wide association studies have identified large numbers of genetic variations in the noncoding parts of the genome, and these are very likely to exert their effects through RNA structures (Halvorsen et al. 2010). Although conventional chemical probing has already been used in characterizing and interpreting genetic variations, cross-linking methods are more direct in revealing their structural contexts, especially for long-range, dynamic, and complex structures (Fig. 5B).
With the ability to define overall architectures of RNA by cross-linking, large RNA molecules refractory to atomic resolution structure analysis in vitro can be studied based on the “divide-and-conquer” principle (Fig. 5C). Stable and compact structure domains can be trimmed to remove flexible regions based on cross-linking data, purified and then subject to X-ray crystallography, NMR, and cryo-EM analysis. The identification of stable duplex units in the XIST A-repeat domain is one such example, in which the physiologically relevant structure units can be studied in great detail in vitro (see an example of the XIST A-repeat domain in Fig. 4G). De novo and data-assisted 3D structure modeling represent an alternative solution to directly solving the structure by in vitro atomic resolution methods. The modeling efforts will also benefit from the cross-linking methods that provide physical contact information.
The biogenesis and function of RNA molecules in cells are on a long journey, in which constant remodeling of structures impacts every aspect of their life. The dissection of these dynamic structures will certainly reveal new principles of gene regulation. Identification of RNA structure motifs that control gene expression may lead to new therapeutic opportunities by revealing potential drug targets. For example, RNA viruses represent a major health concern, and the mechanistic studies of viral RNA structures would provide important information for designing antiviral drugs. With this new generation of methods for direct identification of base-pairing interactions in nucleic acids, opportunities abound for many topics in the RNA field for both basic and translational research.
ACKNOWLEDGMENTS
This work was supported by National Institutes of Health (NIH) R01-HG004361, P50-HG007735 (H.Y.C.) and Stanford Jump Start Award (Z.L.). Z.L. is a Layton Family Fellow of the Damon Runyon-Sohn Foundation Pediatric Cancer Fellowship Award (DRSG-14–15). We thank Fan Liu for comments on the manuscript.
Footnotes
Editors: Thomas R. Cech, Joan A. Steitz, and John F. Atkins
Additional Perspectives on RNA Worlds available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Anfinsen CB. 1973. Principles that govern the folding of protein chains. Science 181: 223–230. [DOI] [PubMed] [Google Scholar]
- Aw JG, Shen Y, Wilm A, Sun M, Lim XN, Boon KL, Tapsin S, Chan YS, Tan CP, Sim AY, et al. 2016. In vivo mapping of eukaryotic RNA interactomes reveals principles of higher-order organization and regulation. Mol Cell 62: 603–617. [DOI] [PubMed] [Google Scholar]
- Bai XC, McMullan G, Scheres SH. 2015. How cryo-EM is revolutionizing structural biology. Trends Biochem Sci 40: 49–57. [DOI] [PubMed] [Google Scholar]
- Beltrame M, Tollervey D. 1992. Identification and functional analysis of two U3 binding sites on yeast pre-ribosomal RNA. EMBO J 11: 1531–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondal T, Thorisdottir A, Unnsteinsdottir U, Hjorleifsdottir S, Aevarsson A, Ernstsson S, Fridjonsson OH, Skirnisdottir S, Wheat JO, Hermannsdottir AG, et al. 2005. Isolation and characterization of a thermostable RNA ligase 1 from a Thermus scotoductus bacteriophage TS2126 with good single-stranded DNA ligation properties. Nucleic Acids Res 33: 135–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bothe JR, Nikolova EN, Eichhorn CD, Chugh J, Hansen AL, Al-Hashimi HM. 2011. Characterizing RNA dynamics at atomic resolution using solution-state NMR spectroscopy. Nat Methods 8: 919–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer V, Moustacchi E, Sage E. 1988. Sequence specificity in photoreaction of various psoralen derivatives with DNA: Role in biological activity. Biochemistry 27: 3011–3018. [DOI] [PubMed] [Google Scholar]
- Calvet JP, Pederson T. 1981. Base-pairing interactions between small nuclear RNAs and nuclear RNA precursors as revealed by psoralen cross-linking in vivo. Cell 26: 363–370. [DOI] [PubMed] [Google Scholar]
- Calvet JP, Meyer LM, Pederson T. 1982. Small nuclear RNA U2 is base-paired to heterogeneous nuclear RNA. Science 217: 456–458. [DOI] [PubMed] [Google Scholar]
- Cech TR, Pardue ML. 1976. Electron microscopy of DNA crosslinked with trimethylpsoralen: Test of the secondary structure of eukaryotic inverted repeat sequences. Proc Nal Acad Sci 73: 2644–2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech TR, Steitz JA. 2014. The noncoding RNA revolution—Trashing old rules to forge new ones. Cell 157: 77–94. [DOI] [PubMed] [Google Scholar]
- Chen J, Kastan MB. 2010. 5′-3′-UTR interactions regulate p53 mRNA translation and provide a target for modulating p53 induction after DNA damage. Genes Dev 24: 2146–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimino GD, Gamper HB, Isaacs ST, Hearst JE. 1985. Psoralens as photoactive probes of nucleic acid structure and function: Organic chemistry, photochemistry, and biochemistry. Ann Rev Biochem 54: 1151–1193. [DOI] [PubMed] [Google Scholar]
- Datta B, Weiner AM. 1992. Cross-linking of U1 snRNA using nitrogen mustard. Evidence for higher order structure. J Biol Chem 267: 4503–4507. [PubMed] [Google Scholar]
- Deans AJ, West SC. 2011. DNA interstrand crosslink repair and cancer. Nat Rev Cancer 11: 467–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dekker J, Rippe K, Dekker M, Kleckner N. 2002. Capturing chromosome conformation. Science 295: 1306–1311. [DOI] [PubMed] [Google Scholar]
- Dethoff EA, Chugh J, Mustoe AM, Al-Hashimi HM. 2012. Functional complexity and regulation through RNA dynamics. Nature 482: 322–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Y, Tang Y, Kwok CK, Zhang Y, Bevilacqua PC, Assmann SM. 2014. In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature 505: 696–700. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SR. 2004. How do RNA folding algorithms work? Nat Biotechnol 22: 1457–1458. [DOI] [PubMed] [Google Scholar]
- Ehresmann C, Baudin F, Mougel M, Romby P, Ebel JP, Ehresmann B. 1987. Probing the structure of RNAs in solution. Nucleic Acids Res 15: 9109–9128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallmann J, Will S, Engelhardt J, Gruning B, Backofen R, Stadler PF. 2017. Recent advances in RNA folding. J Biotechnol 261: 97–104. [DOI] [PubMed] [Google Scholar]
- Fayet-Lebaron E, Atzorn V, Henry Y, Kiss T. 2009. 18S rRNA processing requires base pairings of snR30 H/ACA snoRNA to eukaryote-specific 18S sequences. EMBO J 28: 1260–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesell T, von Haeseler A. 2006. In silico sequence evolution with site-specific interactions along phylogenetic trees. Bioinformatics 22: 716–722. [DOI] [PubMed] [Google Scholar]
- Giedroc DP, Cornish PV. 2009. Frameshifting RNA pseudoknots: Structure and mechanism. Virus Res 139: 193–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilley D, Blackburn EH. 1999. The telomerase RNA pseudoknot is critical for the stable assembly of a catalytically active ribonucleoprotein. Proc Natl Acad Sci 96: 6621–6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong J, Shao D, Xu K, Lu Z, Lu ZJ, Yang YT, Zhang QC. 2018. RISE: A database of RNA interactome from sequencing experiments. Nucleic Acids Res 46: D194–D201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halvorsen M, Martin JS, Broadaway S, Laederach A. 2010. Disease-associated mutations that alter the RNA structural ensemble. PLoS Genet 6: e1001074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartshorne T. 1998. Distinct regions of U3 snoRNA interact at two sites within the 5′ external transcribed spacer of pre-rRNAs in Trypanosoma brucei cells. Nucleic Acids Res 26: 2541–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hausner TP, Giglio LM, Weiner AM. 1990. Evidence for base-pairing between mammalian U2 and U6 small nuclear ribonucleoprotein particles. Genes Dev 4: 2146–2156. [DOI] [PubMed] [Google Scholar]
- Hearst JE. 1981. Psoralen photochemistry. Ann Rev Biophys Bioeng 10: 69–86. [DOI] [PubMed] [Google Scholar]
- Helwak A, Kudla G, Dudnakova T, Tollervey D. 2013. Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell 153: 654–665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass S, Tyc K, Steitz JA, Sollner-Webb B. 1990. The U3 small nucleolar ribonucleoprotein functions in the first step of preribosomal RNA processing. Cell 60: 897–908. [DOI] [PubMed] [Google Scholar]
- *.Kastner B, Will CL, Stark H, Lührmann R. 2019. Structural insights into nuclear pre-mRNA splicing in higher eukaryotes. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kertesz M, Wan Y, Mazor E, Rinn JL, Nutter RC, Chang HY, Segal E. 2010. Genome-wide measurement of RNA secondary structure in yeast. Nature 467: 103–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Quigley GJ, Suddath FL, McPherson A, Sneden D, Kim JJ, Weinzierl J, Rich A. 1973. Three-dimensional structure of yeast phenylalanine transfer RNA: Folding of the polynucleotide chain. Science 179: 285–288. [DOI] [PubMed] [Google Scholar]
- Kladwang W, VanLang CC, Cordero P, Das R. 2011. A two-dimensional mutate-and-map strategy for non-coding RNA structure. Nat Chem 3: 954–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla G, Granneman S, Hahn D, Beggs JD, Tollervey D. 2011. Cross-linking, ligation, and sequencing of hybrids reveals RNA–RNA interactions in yeast. Proc Natl Acad Sci 108: 10010–10015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner MR, Boyle JA, Mount SM, Wolin SL, Steitz JA. 1980. Are snRNPs involved in splicing? Nature 283: 220–224. [DOI] [PubMed] [Google Scholar]
- Lovci MT, Ghanem D, Marr H, Arnold J, Gee S, Parra M, Liang TY, Stark TJ, Gehman LT, Hoon S, et al. 2013. Rbfox proteins regulate alternative mRNA splicing through evolutionarily conserved RNA bridges. Nat Struct Mol Biol 20: 1434–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Chang HY. 2016. Decoding the RNA structurome. Curr Opin Struct Biol 36: 142–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Zhang QC, Lee B, Flynn RA, Smith MA, Robinson JT, Davidovich C, Gooding AR, Goodrich KJ, Mattick JS, et al. 2016. RNA duplex map in living cells reveals higher-order transcriptome structure. Cell 165: 1267–1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Carter AC, Chang HY. 2017. Mechanistic insights in X chromosome inactivation. Philos Trans R Soc B 372: 20160356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Gong J, Zhang QC. 2018. PARIS: Psoralen analysis of RNA interactions and structures with high throughput and resolution. Methods Mol Biol 1649: 59–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maser RL, Calvet JP. 1989. U3 small nuclear RNA can be psoralen-cross-linked in vivo to the 5′ external transcribed spacer of pre-ribosomal-RNA. Proc Natl Acad Sci 86: 6523–6527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathews DH, Moss WN, Turner DH. 2010. Folding and finding RNA secondary structure. Cold Spring Harb Perspect Biol 2: a003665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissey JP, Tollervey D. 1993. Yeast snR30 is a small nucleolar RNA required for 18S rRNA synthesis. Mol Cell Biol 13: 2469–2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mount SM, Pettersson I, Hinterberger M, Karmas A, Steitz JA. 1983. The U1 small nuclear RNA-protein complex selectively binds a 5′ splice site in vitro. Cell 33: 509–518. [DOI] [PubMed] [Google Scholar]
- Nguyen TC, Cao X, Yu P, Xiao S, Lu J, Biase FH, Sridhar B, Huang N, Zhang K, Zhong S. 2016. Mapping RNA–RNA interactome and RNA structure in vivo by MARIO. Nat Commun 7: 12023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson BL, White KA. 2014. Functional long-range RNA–RNA interactions in positive-strand RNA viruses. Nat Rev Microbiol 12: 493–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noller HF, Woese CR. 1981. Secondary structure of 16S ribosomal RNA. Science 212: 403–411. [DOI] [PubMed] [Google Scholar]
- Peculis BA. 1997. The sequence of the 5′ end of the U8 small nucleolar RNA is critical for 5.8S and 28S rRNA maturation. Mol Cell Biol 17: 3702–3713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen JS, Bejerano G, Siepel A, Rosenbloom K, Lindblad-Toh K, Lander ES, Kent J, Miller W, Haussler D. 2006. Identification and classification of conserved RNA secondary structures in the human genome. PLoS Comput Biol 2: e33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrov AS, Bernier CR, Gulen B, Waterbury CC, Hershkovits E, Hsiao C, Harvey SC, Hud NV, Fox GE, Wartell RM, et al. 2014. Secondary structures of rRNAs from all three domains of life. PloS One 9: e88222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Plaschka C, Newman AJ, Nagai K. 2019. Structural basis of nuclear pre-mRNA splicing: Lessons from yeast. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabin D, Crothers DM. 1979. Analysis of RNA secondary structure by photochemical reversal of psoralen crosslinks. Nucleic Acids Res 7: 689–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rimoldi OJ, Raghu B, Nag MK, Eliceiri GL. 1993. Three new small nucleolar RNAs that are psoralen cross-linked in vivo to unique regions of pre-rRNA. Mol Cell Biol 13: 4382–4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinke J, Appel B, Digweed M, Luhrmann R. 1985. Localization of a base-paired interaction between small nuclear RNAs U4 and U6 in intact U4/U6 ribonucleoprotein particles by psoralen cross-linking. J Mol Biol 185: 721–731. [DOI] [PubMed] [Google Scholar]
- Rivas E, Eddy SR. 1999. A dynamic programming algorithm for RNA structure prediction including pseudoknots. J Mol Biol 285: 2053–2068. [DOI] [PubMed] [Google Scholar]
- Rivas E, Eddy SR. 2001. Noncoding RNA gene detection using comparative sequence analysis. BMC Bioinformatics 2: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas E, Clements J, Eddy SR. 2017. Lack of evidence for conserved secondary structure in long noncoding RNAs. Nat Methods 14: 45–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertus JD, Ladner JE, Finch JT, Rhodes D, Brown RS, Clark BF, Klug A. 1974. Structure of yeast phenylalanine tRNA at 3 Å resolution. Nature 250: 546–551. [DOI] [PubMed] [Google Scholar]
- Rouskin S, Zubradt M, Washietl S, Kellis M, Weissman JS. 2014. Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature 505: 701–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergiev PV, Dontsova OA, Bogdanov AA. 2001. Chemical methods for the structural study of the ribosome: Judgment day. Mol Biol 35: 472–496. [PubMed] [Google Scholar]
- Sharma E, Sterne-Weiler T, O’Hanlon D, Blencowe BJ. 2016. Global mapping of human RNA–RNA interactions. Mol Cell 62: 618–626. [DOI] [PubMed] [Google Scholar]
- Shi Y. 2014. A glimpse of structural biology through X-ray crystallography. Cell 159: 995–1014. [DOI] [PubMed] [Google Scholar]
- Siegfried NA, Busan S, Rice GM, Nelson JA, Weeks KM. 2014. RNA motif discovery by SHAPE and mutational profiling (SHAPE-MaP). Nat Methods 11: 959–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MA, Gesell T, Stadler PF, Mattick JS. 2013. Widespread purifying selection on RNA structure in mammals. Nucleic Acids Res 41: 8220–8236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitale RC, Flynn RA, Zhang QC, Crisalli P, Lee B, Jung JW, Kuchelmeister HY, Batista PJ, Torre EA, Kool ET, et al. 2015. Structural imprints in vivo decode RNA regulatory mechanisms. Nature 519: 486–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroke IL, Weiner AM. 1989. The 5′ end of U3 snRNA can be crosslinked in vivo to the external transcribed spacer of rat ribosomal RNA precursors. J Mol Biol 210: 497–512. [DOI] [PubMed] [Google Scholar]
- Sugimoto Y, Vigilante A, Darbo E, Zirra A, Militti C, D’Ambrogio A, Luscombe NM, Ule J. 2015. hiCLIP reveals the in vivo atlas of mRNA secondary structures recognized by Staufen 1. Nature 519: 491–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JF, Hearst JE. 1983. Structure of E. coli 16S RNA elucidated by psoralen crosslinking. Cell 32: 1355–1365. [DOI] [PubMed] [Google Scholar]
- Tyc K, Steitz JA. 1992. A new interaction between the mouse 5′ external transcribed spacer of pre-rRNA and U3 snRNA detected by psoralen crosslinking. Nucleic Acids Res 20: 5375–5382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underwood JG, Uzilov AV, Katzman S, Onodera CS, Mainzer JE, Mathews DH, Lowe TM, Salama SR, Haussler D. 2010. FragSeq: Transcriptome-wide RNA structure probing using high-throughput sequencing. Nat Methods 7: 995–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washietl S, Hofacker IL, Lukasser M, Huttenhofer A, Stadler PF. 2005. Mapping of conserved RNA secondary structures predicts thousands of functional noncoding RNAs in the human genome. Nat Biotechnol 23: 1383–1390. [DOI] [PubMed] [Google Scholar]
- Wassarman DA, Steitz JA. 1993. A base-pairing interaction between U2 and U6 small nuclear RNAs occurs in >150S complexes in HeLa cell extracts: Implications for the spliceosome assembly pathway. Proc Natl Acad Sci 90: 7139–7143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson JD, Crick FH. 1953. Molecular structure of nucleic acids: A structure for deoxyribose nucleic acid. Nature 171: 737–738. [DOI] [PubMed] [Google Scholar]
- Wells SE, Hillner PE, Vale RD, Sachs AB. 1998. Circularization of mRNA by eukaryotic translation initiation factors. Mol Cell 2: 135–140. [DOI] [PubMed] [Google Scholar]
- Whirl-Carrillo M, Gabashvili IS, Bada M, Banatao DR, Altman RB. 2002. Mining biochemical information: Lessons taught by the ribosome. RNA 8: 279–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will CL, Luhrmann R. 2011. Spliceosome structure and function. Cold Spring Harb Perspect Biol 3: a003707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollenzien PL, Youvan DC, Hearst JE. 1978. Structure of psoralen-crosslinked ribosomal RNA from Drosophila melanogaster. Proc Natl Acad Sci 75: 1642–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Bartel DP. 2017. Widespread influence of 3′-end structures on mammalian mRNA processing and stability. Cell 169: 905–917.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Yan C, Wan R, Shi Y. 2019. Molecular mechanisms of pre-mRNA splicing through structural biology of the spliceosome. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zappulla DC, Cech TR. 2004. Yeast telomerase RNA: A flexible scaffold for protein subunits. Proc Natl Acad Sci 101: 10024–10029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwieb C, Brimacombe R. 1980. Localisation of a series of intra-RNA cross-links in 16S RNA, induced by ultraviolet irradiation of Escherichia coli 30S ribosomal subunits. Nucleic Acids Res 8: 2397–2411. [DOI] [PMC free article] [PubMed] [Google Scholar]