Abstract
More than 40 different neurological diseases are caused by microsatellite repeat expansions that locate within translated or untranslated gene regions, including 5′ and 3′ untranslated regions (UTRs), introns, and protein-coding regions. Expansion mutations are transcribed bidirectionally and have been shown to give rise to proteins, which are synthesized from three reading frames in the absence of an AUG initiation codon through a novel process called repeat-associated non-ATG (RAN) translation. RAN proteins, which were first described in spinocerebellar ataxia type 8 (SCA8) and myotonic dystrophy type 1 (DM1), have now been reported in a growing list of microsatellite expansion diseases. This article reviews what is currently known about RAN proteins in microsatellite expansion diseases and experiments that provide clues on how RAN translation is regulated.
Microsatellite repeats, or short repetitive stretches of DNA containing 2–10 nucleotides are common in the human genome. A subset of these sequences has been shown to be unstable, and when expanded too many times, can cause disease. Microsatellite expansions are known to cause >40 different neurodegenerative diseases, including Huntington disease (HD), fragile X tremor ataxia syndrome (FXTAS), C9ORF72 amyotrophic lateral sclerosis (ALS)/frontotemporal dementia (FTD), and spinocerebellar ataxia types 1, 2, 3, 6, 7, 8, 10, 12, and 17 (Ranum and Cooper 2006; Orr and Zoghbi 2007; Lopez Castel et al. 2010).
A framework for understanding the pathogenic mechanisms of these diseases has been built on inferring the mechanisms of these diseases based on the position of the mutations within their corresponding genes. For example, for diseases such as HD, in which the mutation is translated as a glutamine stretch that is part of a long open reading frame (ORF), most research has been focused on how the mutant protein, which contains a long polyGln repeat tract, disturbs various cellular functions that result in cellular toxicity and disease. In contrast, the deleterious effects of mutant expansion RNAs or RNA gain-of-function effects have been the primary mechanistic focus for diseases characterized by expansion RNAs located outside canonical ORFs. Mutant RNAs produced from these noncoding expansion mutations have been shown to accumulate as nuclear RNA foci that sequester RNA-binding proteins (RBPs) and lead to a loss of their normal function (Ranum and Cooper 2006; Scotti and Swanson 2016). The myotonic dystrophies are the classic example of this type of disorder. For example, the myotonic dystrophy type 1 (DM1) and type 2 (DM2) mutations, which express CUG or CCUG expansion RNAs, respectively, sequester the muscleblind-like (MBNL) family of RNA-binding proteins within the nucleus as RNA foci. This sequestration prevents MBNL proteins from binding to their normal RNA-processing targets, and the resulting MBNL loss-of-function leads to RNA-processing abnormalities including dysregulation of alternative splicing and changes in polyadenylation (Ranum and Cooper 2006; Scotti and Swanson 2016).
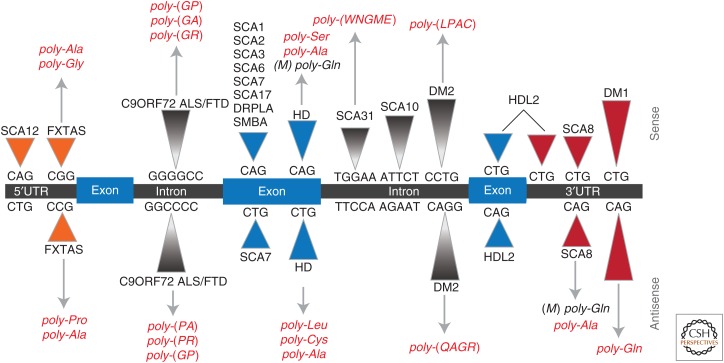
The discovery of repeat-associated non-ATG (RAN) translation challenged these models because it showed that repeat expansion mutations can express unexpected repetitive proteins in all three reading frames without an AUG initiation codon (Zu et al. 2011). RAN translation, which was initially discovered in spinocerebellar ataxia type 8 (SCA8) and DM1, has now been reported in seven different repeat expansion diseases, including C9orf72 ALS/FTD, FXTAS, HD, SCA31, and DM2 (Zu et al. 2011, 2013, 2017; Ash et al. 2013; Mori et al. 2013b; Todd et al. 2013; Banez-Coronel et al. 2015; Ishiguro et al. 2017). In addition to RAN translation, the bidirectional transcription of microsatellite expansion mutations (Cho et al. 2005; Moseley et al. 2006; Ladd et al. 2007; Rudnicki et al. 2007; Batra et al. 2010; Ranum et al. 2010) adds further mechanistic complexity to these disorders, and raises the possibility that one or both expansion transcripts and up to six RAN proteins contribute to many of these diseases (Fig. 1). For the translation field, the discovery that expansion mutations express proteins without a canonical AUG-initiation codon in multiple reading frames raises mechanistic questions about how RAN proteins are initiated and whether these mutant proteins can be therapeutically targeted (Cleary and Ranum 2017).
Figure 1.
Bidirectional transcription and repeat-associated non-ATG (RAN) translation in repeat expansion disorders. Antisense transcripts and RAN proteins have been observed in a growing number of microsatellite expansions, including spinocerebellar ataxia type 8 (SCA8), myotonic dystrophy type 1 (DM1), myotonic dystrophy type 2 (DM2), fragile X tremor ataxia syndrome (FXTAS), C9ORF72 amyotrophic lateral sclerosis (ALS)/frontotemporal dementia (FTD), Huntington disease (HD), and spinocerebellar ataxia type 31 (SCA31). RAN proteins that have been identified in various expansion diseases and confirmed in patient tissues are shown in red.
RAN TRANSLATION
The Discovery of RAN Translation in SCA8 and DM1
SCA8 is an autosomal dominant, neurodegenerative disease caused by a CTG•CAG repeat expansion. In 2006, it was recognized that the SCA8 expansion mutation is bidirectionally transcribed and produces both CUG (ATXN8OS) and CAG (ATXN8) expansion RNAs, which are expressed in opposite directions across the expansion mutation (Moseley et al. 2006). The CUG expansion transcripts form RNA foci, which have been shown to sequester muscleblind proteins and are thought to cause RNA gain-of-function problems (Daughters et al. 2009). In addition, the expanded CAG ATXN8 transcript expressed in the opposite direction produces a nearly pure polyGln protein from an unusual short ORF that contains an AUG-initiation codon directly upstream of the CAG repeat (Moseley et al. 2006).
To understand the relative contributions of the effects of the ATXN8 encoded polyGln protein (Moseley et al. 2006), Ranum and colleagues attempted to block its expression by mutating the upstream ATG initiation codon. Unexpectedly, this mutation did not block expression of the polyGln protein in transfected cells (Zu et al. 2011). This finding raised the question of whether proteins could be made in other reading frames. To answer this question, CAG expansion expression vectors with two stop codons in each reading frame upstream of the CAG tract were generated to ensure AUG initiation could not occur in any of the reading frames. Additionally, epitope tags in each of the three reading frames downstream from the repeat were used to tag proteins expressed from any of the three frames. Surprisingly, homopolymeric polyGln, polyAla, and polySer proteins were expressed in all three reading frames even when the transcripts lacked AUG or near-cognate AUG codons in any reading frame (Zu et al. 2011). Subsequent analyses of CAG expansion transcripts isolated from polyribosomes showed no evidence that editing of the repeat RNAs had introduced an AUG start codon (Zu et al. 2011).
Additional cell-culture experiments showed that hairpin-forming CAG and CUG expansions express RAN proteins without an AUG initiation codon (Zu et al. 2011). Steady-state levels of RAN proteins were differentially affected by repeat length in transfected cells with no RAN protein accumulation detectable with 20 CAG repeats or less, polyGln accumulation evident at ≥42 repeats, polySer at ≥58, and polyAla at ≥73 repeats (Zu et al. 2011). Because repeat length is often correlated with anticipation or worsening of disease, it is possible that the accumulation of RAN proteins in multiple reading frames with longer repeats exacerbates disease. Finally, Zu et al. developed antibodies to detect the carboxy-terminal regions of these proteins in vivo and showed that RAN proteins are expressed in vivo in both mouse and human disease tissue (Zu et al. 2011). In SCA8, a novel polyAla RAN protein was shown to accumulate in Purkinje cells and in DM1 a novel polyGln RAN protein expressed from the antisense CAG expansion transcript was detected in heart, skeletal muscle, and blood.
In summary, CAG and CUG microsatellite expansion RNAs undergo a novel form of translation that leads to the production of homopolymeric expansion proteins from all three reading frames without an AUG-initiation codon. The RAN proteins predicted by cell-culture experiments accumulate in vivo in both SCA8 and DM1 disease tissue.
RAN Proteins in C9ORF72 ALS/FTD
In 2011, a GGGGCC (G4C2) hexanucleotide repeat expansion located in intron 1 of the chromosome 9 open reading frame 72 (C9ORF72) gene was reported by two research groups as the most frequent known genetic cause of both familial ALS and FTD (C9ORF72 ALS/FTD) (DeJesus-Hernandez et al. 2011; Renton et al. 2011). This discovery was the first to link ALS and FTD to a larger group of microsatellite expansion mutation disorders. Although, initially, haploinsufficiency of the C9ORF72 protein and gain-of-function mechanisms of the GGGGCC expansion RNA were proposed in 2013, several investigators reported that, similar to SCA8 and DM1, the C9ORF72 expansion mutation also undergoes RAN translation (Ash et al. 2013; Mori et al. 2013b; Zu et al. 2013). In this case, the hexanucleotide G4C2 expansion mutations form RNA foci and also produce dipeptide repeat proteins (DPRs) with repetitive glycine-proline (GP), glycine-alanine (GA), and glycine-arginine (GR) repeat motifs in the sense direction. Also, similar to SCA8, DM1 and a number of other expansion diseases, this mutation was found to be expressed in the antisense direction and these antisense transcripts also produce three additional RAN proteins: proline-alanine (PA), proline-arginine (PR), and GP (Gendron et al. 2013; Mori et al. 2013a; Zu et al. 2013). Immunological evidence using antibodies directed at the predicted carboxy-terminal regions shows that proteins made in five of the six reading frames include unique carboxy-terminal ends that extend beyond the repeat tract (Zu et al. 2013). Although the contribution of the C9-RAN proteins in disease is not yet fully understood, there is substantial evidence that all six C9-RAN proteins are toxic and accumulate in C9ORF72 ALS/FTD autopsy tissue as cytoplasmic aggregates in multiple regions of the brain that are affected by the disease. These regions include the frontal cortex, hippocampus, and spinal cord (Ash et al. 2013; Gendron et al. 2013; Mackenzie et al. 2013; Mori et al. 2013a,b; Zu et al. 2013; Mackenzie 2016).
RAN Translation across CGG•CCG Expansion in FXTAS
Expansion of a CGG repeat tract in the 5′ untranslated region (UTR) of the FMR1 gene causes two distinct diseases, depending on the length of the repeat. Larger expansions (>200 repeats) cause transcriptional silencing of the FMR1 gene resulting in fragile X syndrome. In contrast, shorter alleles within the premutation range (55–200 repeats) cause FXTAS, a late-onset neurodegenerative disorder (Nelson et al. 2013).
Todd et al. (2013) reported RAN proteins are expressed in FXTAS. This work was initiated because these authors noticed puzzling green fluorescence protein (GFP)-positive aggregates in a fly model expressing 90 CGG repeats but not GFP alone. The CGG repeats are located in what was assumed to be a noncoding 5′UTR upstream of the GFP reporter. The formation of these aggregates was later explained after a series of experiments showed that FXTAS CGG expansion mutations containing 90 repeats undergo RAN translation and that the resulting poly(Gly)90-GFP fusion protein forms GFP-positive aggregates. Using mammalian cell culture, Todd et al. went on to show that CGG expansions lead to the production of polyGly-GFP and polyAla-GFP fusion proteins in the absence of an ATG initiation codon. RAN translation in the polyAla reading frame is length-dependent, occurring with constructs containing 88 but not 30 CGG repeats. In contrast, expression of the polyGly protein occurred with 88, 50, and 30 CGGs. Additional experiments showed initiation of translation begins upstream of the CGG repeat in the polyGly reading frame and, in contrast to SCA8, DM1 and C9ORF72 ALS/FTD repeat expansions, requires near-cognate initiation codons (Kearse et al. 2016). The requirement for near-cognate codons upstream of the repeat in the polyGly frame may explain the expression of polyGly protein, even at relatively short expansion sizes. Finally, the authors showed that polyGly aggregates accumulate in both model systems and human FXTAS brains using several custom carboxy-terminal antibodies (Todd et al. 2013). More recently, Krans et al. (2016) showed that homopolymeric proteins are also expressed in the antisense direction, generating polyPro, polyArg, and polyAla in cell culture, and provided evidence that at least two of the antisense proteins (polyPro and polyAla) accumulate in human brains.
Huntington Disease
HD is a progressive neurodegenerative disorder characterized by severe movement, cognitive, and behavioral changes. The disease is caused by a CAG•CTG expansion located within a long ORF of the huntingtin (HTT) gene. Although previous work on HD showed both sense CAG and antisense CUG expansion RNAs are expressed in HD (Rudnicki et al. 2007; Ross and Tabrizi 2011), most research has focused on understanding the deleterious effects of the mutant huntingtin protein, which contains a long polyGln expansion repeat tract.
Recently, Banez-Coronel et al. (2015) showed that both sense and antisense expansion transcripts undergo RAN translation and that additional homopolymeric RAN proteins (polyAla, polySer, polyLeu, and polyCys) accumulate in HD patient brains. These HD-RAN proteins accumulate in brain regions most affected in HD, including the striatum, and the severely affected cerebellar tissue from juvenile-onset cases. Brain regions with robust RAN protein accumulation show pathologic features of HD, including apoptosis, neuroinflammation, and white matter abnormalities. Cell-culture studies show that HD-RAN proteins decrease viability and increase death of neural cells. Additionally, these authors showed that repeat length differentially affects RAN protein aggregation and accumulation with increased polyAla levels and aggregation of polySer occurring in cells expressing >52 repeats, which are typically associated with severe juvenile HD. Additionally, it is interesting to note that polySer accumulation is first detectable in cells expressing ≥35 repeats, which is close to the repeat length at which HD can manifest (Ross and Tabrizi 2011).
In summary, four novel RAN proteins accumulate in HD and they can be expressed across repeats in multiple reading frames even when the repeat is located within a canonical ORF.
RAN Translation in Tetra- and Pentanucleotide Expansion Disorders
More recently, RAN proteins have been reported in tetra- and pentanucleotide repeat diseases. Ishiguro et al. (2017) reported the accumulation of a UGGAA expansion-encoded Trp-Asn-Gly-Met-Glu pentapeptide repeat protein (PPR) in a Drosophila SCA31 model and SCA31 human autopsy brains. Additionally, overexpression of several RNA-binding proteins (TDP-43, FUS, and hnRNPA2B1) reduce PPR protein levels in Drosophila, suggesting that these RBPs may play a role in RNA quality control and regulation of repeat-associated translation.
In DM2, Zu et al. (2017) showed that the intronic CCTG•CAGG expansion mutation located in the cellular nucleic acid binding protein (CNBP) gene produces tetrapeptide expansion proteins in both the sense (leucine-proline-alanine-cysteine [LPAC]) and antisense (glutamine-alanine-glycine-arginine [QAGR]) directions. Codon-replacement studies show that these proteins are toxic to neural cells independent of RNA gain-of-function effects. Additionally, the authors provide evidence that RNA gain-of-function and RAN mechanisms are linked by showing that nuclear sequestration of CCUG expansion transcripts into RNA foci decreases steady-state levels of LPAC RAN protein by preventing export of CCUG expansion RNAs to the cytoplasm.
RAN Protein Toxicity
RAN proteins with homopolymeric, dipeptide, tetrapeptide, and pentapeptide repeat motifs have now been reported in seven different microsatellite expansion diseases. Although the role of individual RAN proteins in each of these disorders is not yet clear, there is a growing body of evidence that RAN proteins expressed in multiple contexts are toxic to cells independent of RNA gain-of-function effects and, when overexpressed, can induce neurodegeneration in model systems. For example, codon-replacement strategies have shown that RAN proteins are toxic independent of RNA gain-of-function effects in mammalian cell systems for SCA8 (Zu et al. 2011), HD (Banez-Coronel et al. 2015), and DM2 (Zu et al. 2017) repeat expansions. Using mouse models with or without a near-cognate ACG codon in the 5′UTR, Sellier et al. (2017) showed that mice expressing both the polyGly RAN protein and CGG expansion RNAs develop behavioral deficits not found in FXTAS animals expressing CGG expansion RNAs alone. The mice that express both the CGG RNA and the polyGly protein contain a near-cognate ACG codon, which allows polyGly expression; in contrast, mice in which the ACG has been deleted express only the CGG RNA.
The most extensive data on the biophysical properties and toxicity of RAN proteins come from a large number of studies of C9ORF72 ALS/FTD. In several different in vitro and in vivo overexpression experiments, GA, GR, and PR DPRs have been reported to be toxic by multiple investigators. GA has the biophysical properties to aggregate and form amyloid-like fibrils that stain positive with Congo red and thioflavin-T (May et al. 2014; Chang et al. 2016). Additionally, mass spectrometry has shown that p62, Unc119, ubiquilin-1,2, and some proteosomal subunits are found in GA complexes (May et al. 2014; Schludi et al. 2017). Neuronal cell culture studies show GA overexpression causes impaired dendritic branching, endoplasmic reticulum (ER) stress, and apoptosis (May et al. 2014; Zhang et al. 2016). AAV-mediated overexpression of GA DPR causes cognitive and motor deficits in mice (Chew et al. 2015; Zhang et al. 2016). In contrast, several other studies have reported minimal toxicity of GA in primary neuronal cell culture and Drosophila models (Mizielinska et al. 2014; Wen et al. 2014). In fact, several independent studies performed using a non-hairpin-forming alternative codon strategy to distinguish RNA gain-of-function effects from RAN protein toxicity, showed that PR and GR are the most toxic in the Drosophila eye (Mizielinska et al. 2014; Wen et al. 2014; Freibaum et al. 2015; Lee et al. 2016). Toxicity of GR and PR has been attributed to (1) nucleolar localization in overexpression systems (Kwon et al. 2014; Tao et al. 2015); (2) association with low-complexity domain (LCD) proteins that cause phase separation into membraneless organelles (Lee et al. 2016; Lin et al. 2016); and (3) plugging the nuclear pore and impairing nucleocytoplasmic transport (Freibaum et al. 2015; Jovicic et al. 2015; Zhang et al. 2015; Shi et al. 2017).
Although a number of independent investigators provide strong support that specific DPRs can be toxic when overexpressed, additional studies that recapitulate the temporal and spatial expression of these proteins in mammalian systems will be needed to understand their impact on the disease. Recently, four research groups developed bacterial artificial chromosome (BAC) mouse models to better understand C9ORF72 ALS/FTD (O’Rourke et al. 2015; Peters et al. 2015; Jiang et al. 2016; Liu et al. 2016). Further studies in which RAN translation can be blocked or modulated without affecting the levels of the RNAs in these models will be useful for sorting out the role of RAN proteins in disease.
MECHANISM OF RAN TRANSLATION
RAN translation is now known to occur across a variety of different repeat motifs including CAG, CUG, CGG, CCG GGGGCC, CCCCGG, and, most recently, CCUG and CAGG for DM2 and UGGAA for SCA31 (Fig. 1) (Cleary and Ranum 2017; Ishiguro et al. 2017; Zu et al. 2017). Microsatellite expansion mutations in these diseases are present in ORFs, 3′UTRs, 5′UTRs, and intronic regions. Although the mechanisms whereby RAN proteins are produced are still poorly understood, several common themes are emerging, including the importance of repeat length, RNA structure, RNA-binding proteins, and translation initiation by canonical and noncanonical mechanisms.
Repeat Length and Repeat Sequence
It has been shown by several investigators that RAN translation is repeat length-dependent with the accumulation of RAN proteins more likely to occur for expansions with longer repeat tracts. This length dependence applies across multiple diseases including SCA8, HD, DM1, DM2, and C9ORF72 ALS/FTD and for FXTAS in the polyAla reading frame (Zu et al. 2011, 2013, 2017; Todd et al. 2013; Banez-Coronel et al. 2015; Cleary and Ranum 2017). In general, longer repeats in these diseases are more likely to result in the accumulation of a cocktail of RAN proteins expressed from multiple reading frames and also more likely to result in increased disease severity.
A curious observation from studies of RAN translation across CAG•CTG expansions in HD and CCTG•CAGG expansion mutations in DM2 is that RAN translation and subsequent RAN protein accumulation are cell-type-dependent in some, but not all, reading frames, suggesting the possibility that RAN translation in these cells is modulated by sequence-specific effects such as secondary structures and near-cognate initiation codons. Additionally, cell-specific expression of specialized initiation factors that favor RAN translation could cause cell-type-specific expression of specific RAN proteins (Banez-Coronel et al. 2015; Zu et al. 2017).
Previous studies using CAG repeats with different 5′ flanking sequences cloned from the SCA3, HD, DM1, or HDL2 loci, showed that flanking sequences can affect RAN protein expression (Zu et al. 2011). Additionally, these investigators showed that insertion of a TAG stop immediately preceding the CAG repeat tract in the glutamine frame prevented translation of polyGln but not of polyAla or polySer. These data suggest translation initiation in the polyGln reading frame occurs upstream of the repeat in this construct. Similarly, Todd et al. (2013) and Scoles et al. (2015) have shown that 5′ and 3′ flanking sequences affect RAN translation in FXTAS and SCA2, respectively. These data suggested that RAN translation can be influenced by both the repeat expansion motif and flanking sequences.
Hairpin and G-Quadruplex RNA Structures
Single-stranded CAG, CUG, CGG, CCUG, and G4C2 repeats have all been reported to form stable secondary structures including hairpins and G-quadruplexes (Tian et al. 2000; Sobczak et al. 2003; Mirkin 2007; Fratta et al. 2012; Krzyzosiak et al. 2012; Reddy et al. 2013; Haeusler et al. 2014). When RAN translation was first discovered for hairpin-forming CAG repeats, Zu et al. (2011) hypothesized that hairpin formation is required for RAN translation. Consistent with this prediction, these authors showed hairpin-forming CAG, but not non-hairpin-forming CAA repeat constructs, undergo RAN translation.
In C9ORF72 ALS/FTD, it has also been shown that codon replacement RNAs, which encode various DPRs but do not form secondary RNA structures, do not undergo RAN translation but rather require an ATG initiation codon for expression (Mizielinska et al. 2014). Because G4C2 and CGG repeats can form both G-quadruplex and hairpin structures (Fratta et al. 2012; Reddy et al. 2013; Haeusler et al. 2014), it is possible that the transition and/or equilibrium between G-quadruplex and hairpin conformations may play a role in RAN translation across these expansions.
Additionally, SCA31 is distinct from other expansion diseases previously reported to undergo RAN translation in two important ways: (1) the repeat expansion mutation is not GC-rich and not predicted to form hairpin structures; (2) repeated iterations of the UGGAA repeat contain AUG initiation codons throughout the repeat tract (e.g., UGGAAUGGAAUG GAAUGGAA…) in all three reading frames (Ishiguro et al. 2017). It will be interesting to determine the frequency of RAN protein aggregates in SCA31. It will also be interesting to determine whether AUG and RAN mechanisms are both at play in this repeat expansion disorder and if one or multiple AUG initiation codons embedded within the repeat tract are used.
Cap-Dependent Initiation across CGG Repeats in FXTAS
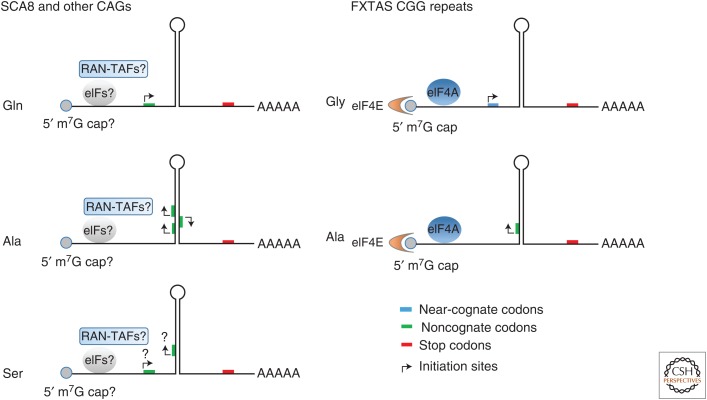
Kearse et al. (2016) recently showed that translation of the FMR polyGly initiates preferentially at close-cognate start codons upstream of the repeat, in a repeat-length-independent manner. In contrast, translation in the polyAla reading frame for FXTAS is length dependent and does not have a similar close-cognate initiation codon. Additionally, using cell-free in vitro translation protocols, these authors show that the polyGly and polyAla reading frames require an m7G cap, the eIF4A helicase, and 40S ribosomal scanning. These findings support a model involving canonical preinitiation complex loading and ribosome scanning and suggest ribosome stalling caused by the CGG RNA secondary structure facilitates translation in both the polyAla and polyGly reading frames (Fig. 2). Additionally, mass spectrometry performed by Sellier et al. (2017) on the FMR polyGly protein showed that initiation occurs at an upstream ACG close-cognate start codon and that the amino-terminal amino acid is a methionine in mammalian cells. These data suggest that for FXTAS, a canonical protein initiation mechanism is used in the polyGly reading frame but with the use of a close-cognate initiation codon. Additional studies in FXTAS will be required to determine the specific initiation site or sites within the polyAla reading frame.
Figure 2.
Repeat-associated non-ATG (RAN) translation of CAG and CGG repeats. Model of RAN translation showing repeat-length dependence and preference for hairpin or secondary RNA structures. RAN translation can occur in multiple reading frames and, depending on the disease locus, at start sites upstream of the repeats or at multiple sites within the repeat tract. Experiments on CAG repeats suggest that translation initiation in the polyGln frame occurs upstream of the repeat tract, although initiation in the polyAla reading frame occurs throughout the repeat. Translation of FXTAS polyGly and polyAla have been shown to be cap-dependent and require eIF4E and eIF4A. RAN-TAFs, putative RAN-translation associated factors.
For CAG•CTG expansions, several experiments showed that, although RAN translation is highly permissive across repeats in mammalian cell culture, RAN translation in a variety of cell-free translation systems, including rabbit reticulocyte lysates (RRLs), occurs less robustly and only in reading frames with close-cognate initiation codons (Zu et al. 2011). These data indicate that RRLs do not necessarily recapitulate the more highly permissive RAN translation, which occurs in mammalian cells. Because the sequence flanking the repeat tracts differs dramatically between disease loci and for individual reading frames, it will be important to determine what mechanistic features are shared between different repeat expansion diseases.
INTERNAL RIBOSOME ENTRY SITES (IRES)
Previous studies showed that translational initiation is affected by secondary structures downstream from start codons (Kozak 1989). In addition, hairpin structures downstream from start codons at suboptimal sites can enhance translational initiation efficiency, possibly by slowing the 40S ribosomal subunit and enhancing the time for interaction between the Met-tRNAi anticodon and the suboptimal AUGs or close-cognate AUG-like start site (Kochetov et al. 2007).
Some viral mRNAs containing highly structured RNA elements at the 5′UTR have been shown to recruit initiation factors and ribosomal subunits to IRESs (Cullen 2009; Sonenberg and Hinnebusch 2009). Additionally, type IV IRESs in the cricket paralysis virus (CrPV) can facilitate translation initiation without eIFs or Met-tRNAiMet. In these cases, eukaryotic elongation factors 1 and 2 (eEF1 and eEF2) are involved in translation initiation. The PKI hairpin of the CrPV IRES functions as a tRNAiMet to initiate translation at non-AUG or noncognate AUG codons, including GCU or GCA that encode an alanine (Hellen and Sarnow 2001; Hellen 2009; Fernandez et al. 2014). Because RAN translation most often occurs across repeat expansions that form stable secondary structures, an IRES-like mechanism has also been proposed (Zu et al. 2011; Cleary and Ranum 2017). Mass spectrometry for CAG repeat expansions suggests RAN translation in the GCA/alanine frame initiates at multiple GCA codons in the repeats without incorporating an amino-terminal methionine (Fig. 2) (Zu et al. 2011). Interestingly, some repeat expansion-binding proteins, such as CUGBP1 and hnRNPs (Timchenko et al. 1996; Conlon et al. 2016), can serve as IRES translation associated factors (ITAFs) (Fox and Stover 2009). Taken together, these data suggest that RAN translation of specific repeats may involve IRES-like mechanisms.
RNA-Binding Proteins and RAN Translation
RNA foci and RAN proteins are found in a growing number of diseases including SCA8, C9ORF72 ALS/FTD, and now DM2. This raises the question of whether RNA gain-of-function and RAN mechanisms are linked.
DM2 is caused by an intronic CCTG expansion in the CNBP gene (Liquori et al. 2001). The clinical similarities between DM1 and DM2, the formation of CUG and CCUG repeat-containing nuclear RNA foci, and similar MBNL/CELF protein-related RNA-processing changes provide strong support that RNA gain-of-function and corresponding MBNL protein loss-of-function contribute to DM2 (Liquori et al. 2001; Cleary and Ranum 2013). In a recent study, Zu et al. (2017) showed that in DM2, antisense CAGG transcripts are expressed and elevated compared with antisense RNA levels found in control DM2 autopsy tissue. Additionally, poly(LPAC) and poly(QAGR) tetrapeptide RAN proteins produced from sense and antisense transcripts accumulate in human DM2 brains. Similar to RAN proteins for several other repeat expansion mutations, steady-state levels of the LPAC and QAGR proteins increased in a repeat length-dependent manner in transfected cells. RNA foci and LPAC RAN protein expression are inversely correlated, and steady-state levels of RAN proteins expressed from CCUG, CUG, and CAG expansion RNAs are reduced by Mbnl1 overexpression. These data suggest a nuclear sequestration model of disease. In this model, initially, RNA-binding proteins sequester expansion RNAs in the nucleus and play a protective role by preventing cytoplasmic RAN translation. During later stages of the disease, nuclear sequestration fails and cytoplasmic expansion RNAs produce RAN proteins that exacerbate disease (Zu et al. 2017). Because RNA foci and RAN proteins are found in a growing number of diseases (SCA8, DM1, DM2, FXTAS, and C9ORF72 ALS/FTD), nuclear sequestration of expansion RNAs by specific RNA-binding proteins into nuclear foci may be a general protective mechanism.
An additional puzzle is how transcripts of intronic expansion mutations like the DM2 and C9ORF72 expansions make it out into the cytoplasm to be translated in the first place. A recent study reported that increased binding of GGGGCC-expanded C9ORF72 repeats to the nuclear export adaptor SRSF1 may increase nuclear export through the nuclear RNA export factor 1 (NXF1) pathway (Hautbergue et al. 2017). Additionally, intron retention, which is increasingly recognized in a wide range of mammalian transcripts, and also in microsatellite expansion diseases (Gipson et al. 2013; Sathasivam et al. 2013; Braunschweig et al. 2014; Niblock et al. 2016), may provide a plausible alternative mechanism that would allow intronic expansion mutations to be retained and exported into the cytoplasm where they could undergo RAN translation. Finally, disruptions in the nuclear pore complex, which have been reported in both C9ORF72 ALS/FTD and HD (Freibaum et al. 2015; Jovicic et al. 2015; Zhang et al. 2015; Grima et al. 2017), may provide another route for the accumulation of expansion RNAs in the cytoplasm and RAN protein expression.
CONCLUSIONS AND PERSPECTIVES
RAN proteins have now been reported in seven out of more than 40 diseases known to be caused by microsatellite expansion mutations. Although there has been substantial progress in this field, there remain an even larger number of questions and future experiments. First, additional studies that rigorously define where these proteins begin and end are needed. These studies face two major challenges. Currently, mass spectrometry is challenging because of the repetitive nature of these proteins, and it is not clear that cell-free in vitro systems will accurately recapitulate the highly permissive initiation of RAN proteins in cells. Second, it is important to understand the molecular factors that are required for RAN translation and also to understand whether the mechanisms vary depending on the availability or absence of close-cognate codons and RNA secondary structures. The cell-type differences in the accumulation patterns of RAN proteins may reflect the requirements for various cellular factors that may differ depending on the sequence of both the repeat expansion and flanking sequence. Alternatively, the patchy accumulation of RAN protein aggregates seen in human autopsy tissue (Zu et al. 2013) may result from prion-like seeding and spreading mechanisms or localized differences in RAN protein production or turnover. It is also possible that stress pathways, known to play a critical role in canonical translational regulation, may also play a role in RAN protein diseases. Third, it is critical to understand the role that RAN proteins have in disease. Although many studies show that various RAN proteins are toxic when overexpressed, it is less clear whether and how these proteins contribute to disease when expressed in the spatial and temporal patterns and levels that are found in disease. Finding ways to target RAN proteins will be important for understanding their role in disease.
Understanding the mechanisms and impact of RAN proteins in neurological diseases is important and may lead to therapies that can turn off a pathogenic pathway common to a large number of neurodegenerative disorders. Moreover, because nearly half of the human genome consists of repetitive DNA elements, it is possible that RAN translation of repeats in normal mammalian genomes may lead to the production of an abundant category of novel proteins. Elucidating the mechanisms underlying RAN translation has the potential to shift the paradigm and our current views on the complexity of the proteome and how perturbations at this novel level impact fundamental aspects of biology.
ACKNOWLEDGMENTS
This work is supported by the National Institutes of Health (R01 NS098819, R37 NS040389, and P01 NS058901), Target ALS, CHDI, National Ataxia Foundation, ALS Association, Packard Foundation, and the Muscular Dystrophy Association.
Footnotes
Editors: Michael B. Mathews, Nahum Sonenberg, and John W.B. Hershey
Additional Perspectives on Translation Mechanisms and Control available at www.cshperspectives.org
REFERENCES
- Ash PE, Bieniek KF, Gendron TF, Caulfield T, Lin WL, Dejesus-Hernandez M, van Blitterswijk MM, Jansen-West K, Paul JW III, Rademakers R, et al. 2013. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron 77: 639–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banez-Coronel M, Ayhan F, Tarabochia AD, Zu T, Perez BA, Tusi SK, Pletnikova O, Borchelt DR, Ross CA, Margolis RL, et al. 2015. RAN translation in Huntington disease. Neuron 88: 667–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batra R, Charizanis K, Swanson MS. 2010. Partners in crime: Bidirectional transcription in unstable microsatellite disease. Hum Mol Genet 19: R77–R82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunschweig U, Barbosa-Morais NL, Pan Q, Nachman EN, Alipanahi B, Gonatopoulos-Pournatzis T, Frey B, Irimia M, Blencowe BJ. 2014. Widespread intron retention in mammals functionally tunes transcriptomes. Genome Res 24: 1774–1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YJ, Jeng US, Chiang YL, Hwang IS, Chen YR. 2016. The glycine-alanine dipeptide repeat from C9orf72 hexanucleotide expansions forms toxic amyloids possessing cell-to-cell transmission properties. J Biol Chem 291: 4903–4911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chew J, Gendron TF, Prudencio M, Sasaguri H, Zhang YJ, Castanedes-Casey M, Lee CW, Jansen-West K, Kurti A, Murray ME, et al. 2015. C9ORF72 repeat expansions in mice cause TDP-43 pathology, neuronal loss, and behavioral deficits. Science 348: 1151–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho DH, Thienes CP, Mahoney SE, Analau E, Filippova GN, Tapscott SJ. 2005. Antisense transcription and heterochromatin at the DM1 CTG repeats are constrained by CTCF. Mol Cell 20: 483–489. [DOI] [PubMed] [Google Scholar]
- Cleary JD, Ranum LP. 2013. Repeat-associated non-ATG (RAN) translation in neurological disease. Hum Mol Genet 22: R45–R51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleary JD, Ranum LP. 2017. New developments in RAN translation: Insights from multiple diseases. Curr Opin Genet Dev 44: 125–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlon EG, Lu L, Sharma A, Yamazaki T, Tang T, Shneider NA, Manley JL. 2016. The C9ORF72 GGGGCC expansion forms RNA G-quadruplex inclusions and sequesters hnRNP H to disrupt splicing in ALS brains. eLife 5: e17820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen BR. 2009. Viral RNAs: Lessons from the enemy. Cell 136: 592–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daughters RS, Tuttle DL, Gao W, Ikeda Y, Moseley ML, Ebner TJ, Swanson MS, Ranum LP. 2009. RNA gain-of-function in spinocerebellar ataxia type 8. PLoS Genet 5: e1000600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, et al. 2011. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72: 245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez IS, Bai XC, Murshudov G, Scheres SH, Ramakrishnan V. 2014. Initiation of translation by cricket paralysis virus IRES requires its translocation in the ribosome. Cell 157: 823–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox JT, Stover PJ. 2009. Mechanism of the internal ribosome entry site-mediated translation of serine hydroxymethyltransferase 1. J Biol Chem 284: 31085–31096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fratta P, Mizielinska S, Nicoll AJ, Zloh M, Fisher EM, Parkinson G, Isaacs AM. 2012. C9orf72 hexanucleotide repeat associated with amyotrophic lateral sclerosis and frontotemporal dementia forms RNA G-quadruplexes. Sci Rep 2: 1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freibaum BD, Lu Y, Lopez-Gonzalez R, Kim NC, Almeida S, Lee KH, Badders N, Valentine M, Miller BL, Wong PC, et al. 2015. GGGGCC repeat expansion in C9orf72 compromises nucleocytoplasmic transport. Nature 525: 129–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gendron TF, Bieniek KF, Zhang YJ, Jansen-West K, Ash PE, Caulfield T, Daughrity L, Dunmore JH, Castanedes-Casey M, Chew J, et al. 2013. Antisense transcripts of the expanded C9ORF72 hexanucleotide repeat form nuclear RNA foci and undergo repeat-associated non-ATG translation in c9FTD/ALS. Acta Neuropathol 126: 829–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gipson TA, Neueder A, Wexler NS, Bates GP, Housman D. 2013. Aberrantly spliced HTT, a new player in Huntington’s disease pathogenesis. RNA Biol 10: 1647–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grima JC, Daigle JG, Arbez N, Cunningham KC, Zhang K, Ochaba J, Geater C, Morozko E, Stocksdale J, Glatzer JC, et al. 2017. Mutant Huntingtin disrupts the nuclear pore complex. Neuron 94: 93–107.e106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeusler AR, Donnelly CJ, Periz G, Simko EA, Shaw PG, Kim MS, Maragakis NJ, Troncoso JC, Pandey A, Sattler R, et al. 2014. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature 507: 195–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hautbergue GM, Castelli LM, Ferraiuolo L, Sanchez-Martinez A, Cooper-Knock J, Higginbottom A, Lin YH, Bauer CS, Dodd JE, Myszczynska MA, et al. 2017. SRSF1-dependent nuclear export inhibition of C9ORF72 repeat transcripts prevents neurodegeneration and associated motor deficits. Nat Commun 8: 16063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellen CU. 2009. IRES-induced conformational changes in the ribosome and the mechanism of translation initiation by internal ribosomal entry. Biochim Biophys Acta 1789: 558–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellen CU, Sarnow P. 2001. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev 15: 1593–1612. [DOI] [PubMed] [Google Scholar]
- Ishiguro T, Sato N, Ueyama M, Fujikake N, Sellier C, Kanegami A, Tokuda E, Zamiri B, Gall-Duncan T, Mirceta M, et al. 2017. Regulatory role of RNA chaperone TDP-43 for RNA misfolding and repeat-associated translation in SCA31. Neuron 94: 108–124.e107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Zhu Q, Gendron TF, Saberi S, McAlonis-Downes M, Seelman A, Stauffer JE, Jafar-Nejad P, Drenner K, Schulte D, et al. 2016. Gain of toxicity from ALS/FTD-linked repeat expansions in C9ORF72 is alleviated by antisense oligonucleotides targeting GGGGCC-containing RNAs. Neuron 90: 535–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovicic A, Mertens J, Boeynaems S, Bogaert E, Chai N, Yamada SB, Paul JW III, Sun S, Herdy JR, Bieri G, et al. 2015. Modifiers of C9orf72 dipeptide repeat toxicity connect nucleocytoplasmic transport defects to FTD/ALS. Nat Neurosci 18: 1226–1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse MG, Green KM, Krans A, Rodriguez CM, Linsalata AE, Goldstrohm AC, Todd PK. 2016. CGG repeat-associated non-AUG translation utilizes a cap-dependent scanning mechanism of initiation to produce toxic proteins. Mol Cell 62: 314–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochetov AV, Palyanov A, Titov II, Grigorovich D, Sarai A, Kolchanov NA. 2007. AUG_hairpin: Prediction of a downstream secondary structure influencing the recognition of a translation start site. BMC Bioinformatics 8: 318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. 1989. Context effects and inefficient initiation at non-AUG codons in eucaryotic cell-free translation systems. Mol Cell Biol 9: 5073–5080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krans A, Kearse MG, Todd PK. 2016. Repeat-associated non-AUG translation from antisense CCG repeats in fragile X tremor/ataxia syndrome. Ann Neurol 80: 871–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzyzosiak WJ, Sobczak K, Wojciechowska M, Fiszer A, Mykowska A, Kozlowski P. 2012. Triplet repeat RNA structure and its role as pathogenic agent and therapeutic target. Nucleic Acids Res 40: 11–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon I, Xiang S, Kato M, Wu L, Theodoropoulos P, Wang T, Kim J, Yun J, Xie Y, McKnight SL. 2014. Poly-dipeptides encoded by the C9orf72 repeats bind nucleoli, impede RNA biogenesis, and kill cells. Science 345: 1139–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladd PD, Smith LE, Rabaia NA, Moore JM, Georges SA, Hansen RS, Hagerman RJ, Tassone F, Tapscott SJ, Filippova GN. 2007. An antisense transcript spanning the CGG repeat region of FMR1 is upregulated in premutation carriers but silenced in full mutation individuals. Hum Mol Genet 16: 3174–3187. [DOI] [PubMed] [Google Scholar]
- Lee KH, Zhang P, Kim HJ, Mitrea DM, Sarkar M, Freibaum BD, Cika J, Coughlin M, Messing J, Molliex A, et al. 2016. C9orf72 dipeptide repeats impair the assembly, dynamics, and function of membrane-less organelles. Cell 167: 774–788.e717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Mori E, Kato M, Xiang S, Wu L, Kwon I, McKnight SL. 2016. Toxic PR poly-dipeptides encoded by the C9orf72 repeat expansion target LC domain polymers. Cell 167: 789–802.e712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liquori CL, Ricker K, Moseley ML, Jacobsen JF, Kress W, Naylor SL, Day JW, Ranum LP. 2001. Myotonic dystrophy type 2 caused by a CCTG expansion in intron 1 of ZNF9. Science 293: 864–867. [DOI] [PubMed] [Google Scholar]
- Liu Y, Pattamatta A, Zu T, Reid T, Bardhi O, Borchelt DR, Yachnis AT, Ranum LP. 2016. C9orf72 BAC mouse model with motor deficits and neurodegenerative features of ALS/FTD. Neuron 90: 521–534. [DOI] [PubMed] [Google Scholar]
- Lopez Castel A, Cleary JD, Pearson CE. 2010. Repeat instability as the basis for human diseases and as a potential target for therapy. Nat Rev Mol Cell Biol 11: 165–170. [DOI] [PubMed] [Google Scholar]
- Mackenzie IR. 2016. The role of dipeptide-repeat protein pathology in C9orf72 mutation cases. Neuropathol Appl Neurobiol 42: 217–219. [DOI] [PubMed] [Google Scholar]
- Mackenzie IR, Arzberger T, Kremmer E, Troost D, Lorenzl S, Mori K, Weng SM, Haass C, Kretzschmar HA, Edbauer D, et al. 2013. Dipeptide repeat protein pathology in C9ORF72 mutation cases: Clinico-pathological correlations. Acta Neuropathol 126: 859–879. [DOI] [PubMed] [Google Scholar]
- May S, Hornburg D, Schludi MH, Arzberger T, Rentzsch K, Schwenk BM, Grasser FA, Mori K, Kremmer E, Banzhaf-Strathmann J, et al. 2014. C9orf72 FTLD/ALS-associated Gly-Ala dipeptide repeat proteins cause neuronal toxicity and Unc119 sequestration. Acta Neuropathol 128: 485–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirkin SM. 2007. Expandable DNA repeats and human disease. Nature 447: 932–940. [DOI] [PubMed] [Google Scholar]
- Mizielinska S, Gronke S, Niccoli T, Ridler CE, Clayton EL, Devoy A, Moens T, Norona FE, Woollacott IO, Pietrzyk J, et al. 2014. C9orf72 repeat expansions cause neurodegeneration in Drosophila through arginine-rich proteins. Science 345: 1192–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K, Arzberger T, Grasser FA, Gijselinck I, May S, Rentzsch K, Weng SM, Schludi MH, van der Zee J, Cruts M, et al. 2013a. Bidirectional transcripts of the expanded C9orf72 hexanucleotide repeat are translated into aggregating dipeptide repeat proteins. Acta Neuropathol 126: 881–893. [DOI] [PubMed] [Google Scholar]
- Mori K, Weng SM, Arzberger T, May S, Rentzsch K, Kremmer E, Schmid B, Kretzschmar HA, Cruts M, Van Broeckhoven C, et al. 2013b. The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science 339: 1335–1338. [DOI] [PubMed] [Google Scholar]
- Moseley ML, Zu T, Ikeda Y, Gao W, Mosemiller AK, Daughters RS, Chen G, Weatherspoon MR, Clark HB, Ebner TJ, et al. 2006. Bidirectional expression of CUG and CAG expansion transcripts and intranuclear polyglutamine inclusions in spinocerebellar ataxia type 8. Nat Genet 38: 758–769. [DOI] [PubMed] [Google Scholar]
- Nelson DL, Orr HT, Warren ST. 2013. The unstable repeats—Three evolving faces of neurological disease. Neuron 77: 825–843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niblock M, Smith BN, Lee YB, Sardone V, Topp S, Troakes C, Al-Sarraj S, Leblond CS, Dion PA, Rouleau GA, et al. 2016. Retention of hexanucleotide repeat-containing intron in C9orf72 mRNA: Implications for the pathogenesis of ALS/FTD. Acta Neuropathol Commun 4: 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke JG, Bogdanik L, Muhammad AK, Gendron TF, Kim KJ, Austin A, Cady J, Liu EY, Zarrow J, Grant S, et al. 2015. C9orf72 BAC transgenic mice display typical pathologic features of ALS/FTD. Neuron 88: 892–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HT, Zoghbi HY. 2007. Trinucleotide repeat disorders. Annu Rev Neurosci 30: 575–621. [DOI] [PubMed] [Google Scholar]
- Peters OM, Cabrera GT, Tran H, Gendron TF, McKeon JE, Metterville J, Weiss A, Wightman N, Salameh J, Kim J, et al. 2015. Human C9ORF72 hexanucleotide expansion reproduces RNA foci and dipeptide repeat proteins but not neurodegeneration in BAC transgenic mice. Neuron 88: 902–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranum LP, Cooper TA. 2006. RNA-mediated neuromuscular disorders. Annu Rev Neurosci 29: 259–277. [DOI] [PubMed] [Google Scholar]
- Ranum LP, Daughters RS, Tuttle DL, Gao W, Ikeda Y, Moseley ML, Ebner T, Swanson MS. 2010. Double the trouble: Bidirectional expression of the SCA8 CAG/CTG expansion mutation - evidence for RNA and protein gain of function effects. Rinsho Shinkeigaku 50: 982–983. [DOI] [PubMed] [Google Scholar]
- Reddy K, Zamiri B, Stanley SY, Macgregor RB Jr, Pearson CE. 2013. The disease-associated r(GGGGCC)n repeat from the C9orf72 gene forms tract length-dependent uni- and multimolecular RNA G-quadruplex structures. J Biol Chem 288: 9860–9866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renton AE, Majounie E, Waite A, Simon-Sanchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, Myllykangas L, et al. 2011. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 72: 257–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross CA, Tabrizi SJ. 2011. Huntington’s disease: From molecular pathogenesis to clinical treatment. Lancet Neurol 10: 83–98. [DOI] [PubMed] [Google Scholar]
- Rudnicki DD, Holmes SE, Lin MW, Thornton CA, Ross CA, Margolis RL. 2007. Huntington’s disease–like 2 is associated with CUG repeat-containing RNA foci. Ann Neurol 61: 272–282. [DOI] [PubMed] [Google Scholar]
- Sathasivam K, Neueder A, Gipson TA, Landles C, Benjamin AC, Bondulich MK, Smith DL, Faull RL, Roos RA, Howland D, et al. 2013. Aberrant splicing of HTT generates the pathogenic exon 1 protein in Huntington disease. Proc Natl Acad Sci 110: 2366–2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schludi MH, Becker L, Garrett L, Gendron TF, Zhou Q, Schreiber F, Popper B, Dimou L, Strom TM, Winkelmann J, et al. 2017. Spinal poly-GA inclusions in a C9orf72 mouse model trigger motor deficits and inflammation without neuron loss. Acta Neuropathol 134: 241–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scoles DR, Ho MH, Dansithong W, Pflieger LT, Petersen LW, Thai KK, Pulst SM. 2015. Repeat associated non-AUG translation (RAN translation) dependent on sequence downstream of the ATXN2 CAG repeat. PLoS ONE 10: e0128769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotti MM, Swanson MS. 2016. RNA mis-splicing in disease. Nat Rev Genet 17: 19–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellier C, Buijsen RA, He F, Natla S, Jung L, Tropel P, Gaucherot A, Jacobs H, Meziane H, Vincent A, et al. 2017. Translation of expanded CGG repeats into FMRpolyG is pathogenic and may contribute to fragile X tremor ataxia syndrome. Neuron 93: 331–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi KY, Mori E, Nizami ZF, Lin Y, Kato M, Xiang S, Wu LC, Ding M, Yu Y, Gall JG, et al. 2017. Toxic PRn poly-dipeptides encoded by the C9orf72 repeat expansion block nuclear import and export. Proc Natl Acad Sci 114: E1111–E1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobczak K, de Mezer M, Michlewski G, Krol J, Krzyzosiak WJ. 2003. RNA structure of trinucleotide repeats associated with human neurological diseases. Nucleic Acids Res 31: 5469–5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG. 2009. Regulation of translation initiation in eukaryotes: Mechanisms and biological targets. Cell 136: 731–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Z, Wang H, Xia Q, Li K, Li K, Jiang X, Xu G, Wang G, Ying Z. 2015. Nucleolar stress and impaired stress granule formation contribute to C9orf72 RAN translation-induced cytotoxicity. Hum Mol Genet 24: 2426–2441. [DOI] [PubMed] [Google Scholar]
- Tian B, White RJ, Xia T, Welle S, Turner DH, Mathews MB, Thornton CA. 2000. Expanded CUG repeat RNAs form hairpins that activate the double-stranded RNA-dependent protein kinase PKR. RNA 6: 79–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timchenko LT, Miller JW, Timchenko NA, DeVore DR, Datar KV, Lin L, Roberts R, Caskey CT, Swanson MS. 1996. Identification of a (CUG)n triplet repeat RNA-binding protein and its expression in myotonic dystrophy. Nucleic Acids Res 24: 4407–4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd PK, Oh SY, Krans A, He F, Sellier C, Frazer M, Renoux AJ, Chen KC, Scaglione KM, Basrur V, et al. 2013. CGG repeat-associated translation mediates neurodegeneration in fragile X tremor ataxia syndrome. Neuron 78: 440–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen X, Tan W, Westergard T, Krishnamurthy K, Markandaiah SS, Shi Y, Lin S, Shneider NA, Monaghan J, Pandey UB, et al. 2014. Antisense proline-arginine RAN dipeptides linked to C9ORF72-ALS/FTD form toxic nuclear aggregates that initiate in vitro and in vivo neuronal death. Neuron 84: 1213–1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Donnelly CJ, Haeusler AR, Grima JC, Machamer JB, Steinwald P, Daley EL, Miller SJ, Cunningham KM, Vidensky S, et al. 2015. The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature 525: 56–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YJ, Gendron TF, Grima JC, Sasaguri H, Jansen-West K, Xu YF, Katzman RB, Gass J, Murray ME, Shinohara M, et al. 2016. C9ORF72 poly(GA) aggregates sequester and impair HR23 and nucleocytoplasmic transport proteins. Nat Neurosci 19: 668–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu T, Gibbens B, Doty NS, Gomes-Pereira M, Huguet A, Stone MD, Margolis J, Peterson M, Markowski TW, Ingram MA, et al. 2011. Non-ATG-initiated translation directed by microsatellite expansions. Proc Natl Acad Sci 108: 260–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu T, Liu Y, Banez-Coronel M, Reid T, Pletnikova O, Lewis J, Miller TM, Harms MB, Falchook AE, Subramony SH, et al. 2013. RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc Natl Acad Sci 110: E4968–E4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zu T, Cleary JD, Liu Y, Banez-Coronel M, Bubenik JL, Ayhan F, Ashizawa T, Xia G, Clark HB, Yachnis AT, et al. 2017. RAN translation regulated by muscleblind proteins in myotonic dystrophy type 2. Neuron 95: 1292–1305.e1295. [DOI] [PMC free article] [PubMed] [Google Scholar]