Abstract
This review summarizes our current understanding of the major pathway for the initiation phase of protein synthesis in eukaryotic cells, with a focus on recent advances. We describe the major scanning or messenger RNA (mRNA) m7G cap-dependent mechanism, which is a highly coordinated and stepwise regulated process that requires the combined action of at least 12 distinct translation factors with initiator transfer RNA (tRNA), ribosomes, and mRNAs. We limit our review to studies involving either mammalian or budding yeast cells and factors, as these represent the two best-studied experimental systems, and only include a reference to other organisms where particular insight has been gained. We close with a brief description of what we feel are some of the major unknowns in eukaryotic initiation.
We present a summary of the current knowledge of the molecular mechanisms enabling the initiation of protein synthesis in eukaryotic cells. We focus on the major m7G cap-dependent pathway, and only briefly consider alternative initiation routes. We cannot cover all the work that has been performed and so we focus primarily on more recent studies. The evidence for the pathway derives from a wide variety of complementary biochemical, genetic, and structural biology approaches used to mainly study both mammalian translation initiation and that of the yeast Saccharomyces cerevisiae. The systems are generally highly similar, but there are some differences and increased complexity is found in mammalian cells. One advantage of the yeast system is its high amenability to genetics that, when combined with other complementary approaches, has provided deep mechanistic insight. We begin with a brief overview of the general initiation pathway, focusing on the roles of the initiation factors and then discuss each step in detail, highlighting key points of mechanistic understanding. The review concludes with a consideration of important questions that remain as yet unanswered.
AN OVERVIEW OF THE INITIATION PATHWAY
The initiation of protein synthesis is the process that results in bringing together an 80S ribosome with a messenger RNA (mRNA) and initiator methionyl-transfer RNA (Met-tRNAi). These three components combine such that Met-tRNAi makes codon–anticodon base-pair interactions with the correct initiation codon at the start of an open-reading frame (ORF) within the mRNA. AUG is the typical initiation codon, although near-cognate codons are used to initiate on some mRNAs (Starck and Shastri 2016; Kearse and Wilusz 2017). The codon–anticodon interaction is confined within the P site of an 80S ribosome. This topology then allows the elongation phase of protein synthesis to begin, by binding an elongator aminoacyl-tRNA to the vacant 80S A site to read the adjacent codon downstream. Although this may appear relatively simple, eukaryotes have evolved a complex translation initiation pathway. There are at least 12 dedicated proteins, termed eukaryotic initiation factors (eIFs), that each play critical roles in the process, and several of these factors are comprised of multisubunit proteins. Why so involved, when prokaryotes initiate translation using just three factors (Rodnina 2018)? Of course, this is a matter of conjecture, but it likely reflects the need to tightly control a process that consumes a large amount of cellular energy in the form of ATP and GTP and can cause serious consequences when out of balance, as evidenced by many disease states caused by aberrant translation (see Stern-Ginossar et al. 2018; Wek 2018). In addition to the discussion below, readers may find it informative to consult previous reviews covering the topics we discuss here (Hinnebusch and Lorsch 2012; Hinnebusch 2014, 2017; Dever et al. 2016).
The initiation pathway is shown schematically in Figure 1 and is envisaged as a series of steps (blue text) in which the eIFs guide the Met-tRNAi and ribosomal subunits to the mRNA AUG codon. Met-tRNAi is brought to the ribosome in a complex with eIF2•GTP. eIF2 is a GDP/GTP binding “G” protein and has high affinity for Met-tRNAi only when GTP-bound (Kapp and Lorsch 2004). The eIF2•GTP•Met-tRNAi complex is widely known as the ternary complex (TC). This is typically considered the first step in translation initiation. As eIF2•GDP is the stable form of eIF2 (Panniers et al. 1988; Erickson and Hannig 1996), it must first be activated to eIF2•GTP by the guanine nucleotide exchange factor (GEF) eIF2B before the TC can form (Rowlands et al. 1988; Pavitt et al. 1998). Next, the TC binds to the small ribosomal subunit (40S) with eIF5, eIF3, eIF1, and eIF1A to form a larger 43S preinitiation complex (PIC). There may be several routes to PIC formation. eIF1, eIF1A, and eIF3 can dissociate 80S complexes and bind to 40S subunits (Pisarev et al. 2007) and then recruit eIF5/TC. Alternatively, eIF3 and eIF1 can form an independent multifactor complex (MFC) with TC and eIF5 before binding the 40S subunit (Asano et al. 2000; Sokabe et al. 2012).
Figure 1.
Overview of the general eukaryotic translation initiation pathway. The pathway for recruiting initiator transfer RNA (tRNA) to the messenger RNA (mRNA) AUG codon in the context of an 80S ribosome (bottom right) is depicted as a series of major steps, labeled with blue text, linked with black arrows. Individual eukaryotic initiation factor (eIF) cartoons and complexes are labeled with black text and nucleotide hydrolysis/inorganic phosphate release reactions are shown by blue arrows. The broad green arrow indicates the direction of scanning toward the AUG codon. The regulatory reactions leading to eIF2 and eIF4E inhibition are shown with plum and red arrows. All steps are described in the main text, starting with eIF2 activation. The timing of release of some factors from initiating ribosomes/mRNA (eIF4F, eIF4B, or eIF3) is not yet clear, so this is not shown.
mRNAs are activated by binding eIF4F at the 5′ 7-methylguanosine cap (m7G cap) and the poly(A) binding protein (PABP) at the 3′ poly(A) tail (Jackson et al. 2010). eIF4F comprises a complex formed between eIF4E, eIF4G, and eIF4A. The large eIF4G subunit has binding sites for eIF4E, PABP, and RNA and can form a “closed-loop” circularized mRNA/RNA-binding protein (RPB) complex (Fig. 1). eIF4A is an RNA helicase and both eIF4F formation and the binding to eIF4A of an accessory factor eIF4B enhance eIF4A activity. eIF4A unwinds mRNA secondary structures to facilitate 43S PIC recruitment at, or close to, the m7G cap, forming an mRNA•43S complex (Fig. 1) (Kumar et al. 2016). eIFs 1 and 1A bound at the P and A sites of the 40S subunit, respectively, open a cleft in the 40S between its “head” and “body” that facilitates single-stranded mRNA binding and the subsequent scanning step. Further interactions between eIF3 in the 43S PIC and a region of eIF4G on the activated mRNA facilitate forming and stabilizing this intermediate complex (Villa et al. 2013). mRNAs have 5′ leaders, typically called 5′ untranslated regions (5′UTRs), that span the distance between the m7G cap and the initiation codon for the major ORF. 5′UTRs vary in length, sequence, and structure. Usually the AUG codon closest to the m7G cap is used to initiate protein synthesis, but there are exceptions.
Scanning describes the movement of the 43S PIC along the mRNA from the m7G cap, in a 3′ direction, searching for an AUG initiation codon in a suitable context to make a stable mRNA codon–tRNAi anticodon interaction. This requires energy in the form of ATP. In Figure 1, this is depicted with PIC/m7G cap complex interactions maintained. This “open” scanning form transitions to a “closed” complex on AUG recognition, in which base pairing is established between the P-site-bound Met-tRNAi anticodon and the mRNA AUG codon. Here scanning is halted and initiation complex reorganization is triggered. eIF2-bound GTP can be hydrolyzed to GDP + phosphate (Pi) during scanning in an eIF5-dependent reaction, but Pi is released from eIF2 only on AUG recognition (Algire et al. 2005; Majumdar and Maitra 2005). Following AUG recognition, eIF1 relocates, triggering an eIF2 conformational change that likely facilitates Pi release. This significantly lowers the affinity of eIF2 for Met-tRNAi (Algire et al. 2005). Together these changes facilitate release of eIF2•GDP and eIF5. eIF2 must be reactivated again to form the eIF2•GTP complex to participate in further rounds of initiation. Finally, eIF5B•GTP is recruited and facilitates the joining of the large ribosomal subunit (60S) and reorientation of the initiator tRNA (Yamamoto et al. 2014). Release of eIF5B•GDP and eIF1A allows formation of an 80S ribosome attached to an mRNA with Met-tRNAi bound to the AUG codon in the P site, poised to begin translation elongation.
43S PIC FORMATION
eIF2 Activation and TC Formation
eIF2 is the main Met-tRNAi carrier and is a key regulatory switch that modulates the global initiation pathway. Met-tRNAi has 20- to 50-fold higher affinity for eIF2•GTP than for eIF2•GDP (Kapp and Lorsch 2004; Jennings et al. 2017). However, eIF2•GTP is unstable, whereas eIF2•GDP is relatively stable (Panniers et al. 1988; Kapp and Lorsch 2004; Jennings et al. 2016), necessitating the action of a GEF (eIF2B) to promote GDP dissociation and thereby facilitate GTP and Met-tRNAi binding to form the TC. During periods of cell stress, eIF2 is subjected to tight control as several protein kinases can phosphorylate the α subunit of eIF2 (eIF2αP) on a conserved serine residue (serine 51 in humans). This converts eIF2 from being a substrate to an inhibitor of eIF2B GEF activity (Fig. 1) and provides a mechanism to impose a global brake on protein synthesis initiation at its outset (Rowlands et al. 1988; Pavitt et al. 1998). This is an important regulatory mechanism, because selective mRNAs are able to escape such global repression. These translational controls are described in greater depth elsewhere (Proud 2018; Wek 2018, but see also Hinnebusch et al. 2016; Young and Wek 2016).
Evidence from yeast suggests that there is little free eIF2•GDP in cells, as eIF5 associates with high affinity forming eIF2•GDP/eIF5 complexes that are produced on release of these factors at the end of each initiation cycle (Algire et al. 2005; Singh et al. 2006). eIF5 can stabilize GDP binding to eIF2•GDP and thereby ensure tight regulation of initiation by preventing any spontaneous release of GDP from eIF2 that might otherwise bypass the eIF2B-centered control mechanism. Thus, eIF5 is a GDP-dissociation inhibitor (GDI) (Jennings and Pavitt 2010). This activity requires the eIF5 carboxy-terminal domain (CTD) and evolutionarily conserved residues within the central linker region, but not the amino-terminal domain (NTD) that is necessary for eIF5’s role as a GTPase-activating protein (GAP). The CTD and linker interact with both eIF2γ and β subunits and may constrain eIF2, preventing eIF2B-independent GDP release (Alone and Dever 2006; Jennings and Pavitt 2010; Jennings et al. 2016). Thus, eIF2B displaces eIF5 from eIF2•GDP before performing nucleotide exchange. In yeast, eIF5 displacement requires the eIF2Bγ and ε subunits (Jennings et al. 2013). eIF5 displacement by eIF2B has not yet been studied with mammalian factors.
eIF2B is a large multifunctional protein. Encoded by five genes (subunits α–ε), its α, β, and δ subunits form an eIF2αP-sensing regulatory complex, whereas eIF2Bγ and ε perform eIF2 activation functions (Pavitt 2005; Jennings et al. 2013). eIF2B was recently shown to be a dimer of pentamers (Gordiyenko et al. 2014; Wortham et al. 2014), with the Schizosaccharomyces pombe structure revealing that the decamer contains a regulatory (αβδ)2 hexameric “core” with two separate εγ “arms,” so it can likely interact with two eIF2 molecules simultaneously. Consistent with prior genetic and biochemical evidence, in vitro cross-linking indicated that the eIF2α domain 1, including serine 51, makes direct contact with an interface composed of all three eIF2B regulatory subunits, whereas eIF2γ (to which GDP/GTP directly binds) interacts with both γ and ε (Kashiwagi et al. 2016). The eIF2Bε carboxy-terminal catalytic domain (Boesen et al. 2004; Mohammad-Qureshi et al. 2007) was not resolved in the eIF2B complex structure, so further studies will be required to elucidate both the mechanism of eIF2B GEF action and its major form of control by eIF2αP.
Following nucleotide exchange, eIF2•GTP binds Met-tRNAi to form the TC. eIF2 binds with 20-fold enhanced specificity to Met-tRNAi over the distinct elongator methionyl-tRNA (Met-tRNAm) that decodes internal AUG methionine codons (Kapp et al. 2006). Assessing the function of tRNAs where specific nucleotides have been swapped has identified Met-tRNAi elements that ensure it binds eIF2 and is excluded from elongation. In Met-tRNAi these include the A1:U72 base pair that is replaced by a G:C pair in Met-tRNAm. Substitutions here and at bases throughout the tRNAi contribute to eIF2 binding (Drabkin et al. 1993; Kapp et al. 2006), and make other contacts important for scanning and ensuring the fidelity of start codon recognition (Kolitz and Lorsch 2010; Dong et al. 2014). As eIF2γ is a structural mimic of tRNA-binding elongation factors SelB, EF1A (EF-Tu), and eEF1A, it was proposed that eIF2 would bind tRNAi in a similar manner to the elongation factors (Schmitt et al. 2010). However, cross-linking and structural studies of both yeast and archaeal eIF2 proteins reveal that the eIF2α and β subunits also contribute to a distinct Met-tRNAi binding mode where both eIF2γ and eIF2α make extensive contacts with the Met-tRNAi acceptor stem (Shin et al. 2011; Schmitt et al. 2012; Naveau et al. 2013). As described below, cryoelectron microscopy (cryo-EM) of partial 48S complexes has provided further insight into Met-tRNAi binding by eIF2 (Hussain et al. 2014; Llacer et al. 2015; Hinnebusch 2017; Jobe et al. 2018).
Although the TC is stable in isolation, it was recently shown that eIF2B can bind and disrupt the TC (Jennings et al. 2017). This unexpected finding suggests that eIF2B can both promote TC formation and destabilize the product of this GEF reaction. Hence, free TC may not be long-lived in cells and may not represent the final product of eIF2 activation. When eIF5 is bound to the TC, it impairs the ability of eIF2B to destabilize the TC, consistent with the idea that TC/eIF5 complexes represent a stable active form of eIF2 that can be recruited to the PIC (Jennings et al. 2017). Therefore, eIF2B can displace eIF5 from eIF2•GDP/eIF5 complexes to perform nucleotide exchange (Jennings et al. 2013), but cannot remove eIF5 from TC/eIF5. Hence, eIF5 binding to TC likely prevents eIF2B from antagonizing the next steps in protein synthesis initiation (Jennings et al. 2017).
eIFs-1, -1A, and -3 Promote TC Binding to 40S
Three additional factors, eIF1, eIF1A, and eIF3, are implicated in recruiting the TC to the 40S ribosomal subunit (Majumdar et al. 2003; Olsen et al. 2003; Kolupaeva et al. 2005; Cheung et al. 2007). Their cooperative binding induces conformational changes that rotate the head and open up a cleft between the head and body of the 40S to facilitate TC binding (Maag et al. 2005; Passmore et al. 2007; Weisser et al. 2013; Sokabe and Fraser 2014). eIF1 is a small protein that binds to the platform of the 40S subunit body close to the P site and mRNA channel (Weisser et al. 2013; Aylett et al. 2015). eIF1A, which is a homolog of bacterial IF1 (Rodnina 2018), sits adjacent to eIF1 in the 40S A site contacting both the head and body. Both eIF1 and eIF1A bind cooperatively to the 40S subunit (Maag et al. 2005; Sokabe and Fraser 2014) above rRNA helix 44, and eIF1A is located at the decoding center (Weisser et al. 2013). The decoding center monitors A-site tRNA-mRNA codon–anticodon pairing during elongation. Both factors play crucial roles in many steps in the initiation pathway and mutations in either factor affect TC recruitment, scanning, and the stringency of AUG recognition (Cheung et al. 2007; Fekete et al. 2007; Martin-Marcos et al. 2013; Hinnebusch 2014). eIF1A is also important for 60S subunit joining. These roles are described below.
Like eIF1 and 1A, eIF3 promotes TC binding to the 43S PIC (Valasek 2012; Sokabe and Fraser 2014; Aitken et al. 2016). The mammalian eIF3 complex is the largest initiation factor comprising 13 subunits (designated a→m) and ∼800 kDa. In contrast, yeast eIF3 is smaller, sharing only homologs of the a, b, c, g, i, and j subunits. It has been suggested that the yeast complex represents a minimal core eIF3 (Valasek 2012; Wagner et al. 2016; Valasek et al. 2017). In yeast, an eIF3ac dimer is connected to the eIF3bgij subcomplex via an eIF3a–CTD/3b interaction (Valasek 2012). This eIF3aCTD/bgij complex has been termed the “yeast-like core” of eIF3 (Fig. 2, left) (des Georges et al. 2015) or alternatively the “peripheral” subunits (Simonetti et al. 2016). Six of the additional mammalian subunits (eIF3d–f, h, and k–m) interact with the conserved eIF3ac module to form a large “octamer” structural core of the mammalian complex comprising subunits a, c, e, f, h, k, l, and m that form a five-lobed structure (des Georges et al. 2015) to which eIF3d binds (Fig. 2, right). It is likely that the enhanced complexity of mammalian eIF3 allows for more diverse regulatory inputs to control translation than in yeast.
Figure 2.
Structural models of eukaryotic initiation factor (eIF)3. A composite model of the structure of eIF3 showing the mammalian octamer (right) from PDB 5A5T (des Georges et al. 2015) and the associated eIF3d subunit (PDB 5K4D) (Lee et al. 2016) that is linked via eIF3a to the yeast-like core eIF3bgi complex (PDB 5A5U) (des Georges et al. 2015). The eIF3g is composed of an RNA-recognition motif (RRM) (PDB 2CQ0) and a β-propeller domain similar to eIF3b. eIF3j can interact with the eIF3b RRM. NTD, Amino-terminal domain.
Structural studies have shown, in progressively increasing detail, that the large eIF3 complex binds and wraps around the solvent exposed (back) surface of the body of the 40S subunit such that it can contact and monitor important events occurring at both the mRNA entry channel/A site as well as the exit channel/E site and factor binding to the intersubunit interface (Hashem et al. 2013b; Erzberger et al. 2014; Aylett et al. 2015; des Georges et al. 2015; Llacer et al. 2015; Simonetti et al. 2016). A cartoon model summarizing some of these factor–40S interactions is shown in Figure 3. The images are a composite derived from both yeast and mammalian studies and show positions for all eIF3 subunits. The structural studies are supported by many functional analyses of eIF3 interactions that indicate a complex network of interactions among the 43S PIC factors that promote its formation and also make critical contributions to later steps in initiation (Valasek 2012; Sokabe and Fraser 2014; Aitken et al. 2016). Specifically, the eIF3 octamer core of the mammalian complex binds around the 40S mRNA exit channel. In yeast, eIF3c binds eIF1 and eIF5 and these subunits make further links to the TC (Valasek et al. 2003, 2004; Yamamoto et al. 2005). In contrast, the eIF3aCTD-3bgij yeast-like core complex is located at the mRNA entry channel, where eIF3aCTD binds both eIF1 and eIF2, whereas 3a, 3b, 3g, and 3j contact 18S rRNA and ribosomal proteins (Fig. 3) (Valasek 2012). eIF3j appears more loosely associated with the other eIF3 subunits and has distinct roles suggesting that it should be viewed as an eIF3-associated factor (Valasek et al. 2017). eIF3j enhances the affinity of many interactions among the 43S components when part of eIF3 (Fraser et al. 2007; Sokabe and Fraser 2014). Recent analyses of yeast eIF3 mutants indicate that mutations in eIF3b, 3i, or 3g destabilize TC binding to the PIC and mutations within these subunits or in eIF3a also impair mRNA recruitment (Khoshnevis et al. 2014; Aitken et al. 2016).
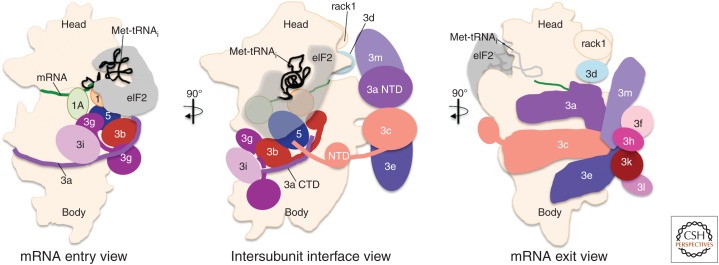
Figure 3.
Model for preinitiation complex (PIC) arrangement at AUG recognition. Three 90° rotated cartoon views of an idealized 48S PIC, modeled on recent structural and functional studies. This model was based on the partial yeast 48S closed complex structure (3JAP) (Llacer et al. 2015) and modified to include findings of others (Luna et al. 2012, 2013; Aylett et al. 2015; des Georges et al. 2015; Simonetti et al. 2016; Obayashi et al. 2017). The entry view (left) shows the eukaryotic initiation factor (eIF)3 yeast-like core subunits, whereas the exit view (right) shows only the mammalian eIF3 octamer complex. eIF2 is shown as semitransparent in the central intersubunit view (middle) to indicate the position of factors otherwise hidden below. For further details of eIF2 interactions, see Figure 4. The position of Rack1 on the ribosome head is shown for orientation purposes only and is not discussed in the text. Factor colors correspond to those used in other figures.
To sum up, eIF3 is critical for bringing the TC to the 40S subunit and for stabilizing mRNA interactions. It also plays a role bridging the 43S PIC to the mRNA m7G cap-binding complex as human eIF3c, 3d, and 3e subunits can be cross-linked to a region of eIF4G (Villa et al. 2013). Of these subunits, yeast eIF3 lacks 3d and 3e, suggesting 3c may fulfill this role, although the eIF5-CTD can provide further stabilizing interactions between the 43S PIC and eIF4G in place of the eIF4G/eIF3 contacts found for mammalian eIF3 (Yamamoto et al. 2005; Singh et al. 2012). As described below eIF3 also facilitates later steps in initiation and can play wider roles in translation including the recycling of posttermination ribosomes and in reinitiation (Pisarev et al. 2007; Beznoskova et al. 2015; Mohammad et al. 2017; Valasek et al. 2017; see also Hellen 2018; Wek 2018).
MFC Is an Alternative Route to the 43S PIC
Another route to form the 43S PIC has been proposed following the isolation of a complex containing the TC and eIFs 1, 3, and 5. Originally described in yeast and termed the MFC, it has now also been isolated from both plant and mammalian cells (Asano et al. 2000; Dennis et al. 2009; Sokabe et al. 2012). The MFC likely helps cooperative recruitment of these translation factors to 40S subunits as there are multiple interactions among components. The yeast eIF5-CTD makes independent contacts with eIF3c-NTD, eIF1, and eIF2β (Yamamoto et al. 2005; Luna et al. 2012). Thus, eIF5 bridges an interaction between the TC and eIF3c, in addition to a direct eIF2/eIF3a–CTD interaction (Valasek et al. 2002). In the human MFC, the eIF3/eIF5 interaction may be weaker than in yeast (Sokabe et al. 2012). There is evidence that in plants the affinities of these MFC components for each other is enhanced by phosphorylation (Dennis et al. 2009), but whether phosphorylation similarly affects human MFC interaction affinities has not yet been evaluated (Sokabe et al. 2012). Thus, as indicated in Figure 1, there are at least two routes to the formation of 43S complexes. The relative importance of these 43S PIC formation routes to individual mRNAs in different cells or in response to different stimuli remains to be resolved.
mRNA RECRUITMENT AND SCANNING
Having constructed the 43S PIC, the next step is mRNA recruitment. eIF4A, eIF4B, eIF4E, and eIF4G are all that is required in model in vitro systems. As these proteins have been purified and a range of mutants analyzed, some of the biochemical properties of these proteins are known as independent entities. However, there remains some uncertainty to their sequential utilization in the initiation pathway. eIF4E is the major m7G cap recognition factor (Gross et al. 2003) and is a site of translational control via a series of eIF4E-binding proteins (4E-BPs) that limit eIF4E access to eIF4G (Richter and Sonenberg 2005). Binding of 4E-BPs to eIF4E is regulated by phosphorylation that alters the structure of the binding protein so that it can no longer bind eIF4E (Bah et al. 2015). When not bound by a 4E-BP, eIF4E can interact with eIF4G (Gross et al. 2003; Gruner et al. 2016; Proud 2018). eIF4G enhances the affinity of eIF4E for the m7G cap, suggesting the interactions help reinforce each other to promote initiation (Gross et al. 2003; Yanagiya et al. 2009; O’Leary et al. 2013). eIF4E binds to a motif shared among the 4E-BPs and eIF4G: YX4Lφ (where X and φ denote any amino acid and any hydrophobic residue, respectively). There are structures available of eIF4E with short regions of eIF4G, including this motif, which reveal this interaction (Gross et al. 2003; Gruner et al. 2016). eIF4G is frequently referred to as a large protein scaffold because, as previously described, it contains multiple regions to which many translation factors and mRNA can bind (Fig. 1) (Yanagiya et al. 2009; Park et al. 2011), and can also bridge this eIF4F/mRNA/PABP complex to the 43S PIC via eIF3 (Villa et al. 2013).
At the heart of the mRNA activation process is eIF4A that is one of the best-characterized RNA helicases and one of the original founding members of the DEAD-box RNA helicases (Grifo et al. 1984; Linder et al. 1989). It is the only “traditional” initiation factor that is known to bind ATP and it functions during both mRNA activation and mRNA scanning. Characteristic activities for eIF4A include: ATP-dependent retention of RNA on nitrocellulose filters, RNA-dependent ATP hydrolysis, and RNA duplex unwinding (Merrick 2015). All of these activities are also found in the eIF4F complex where the level of activity is always greater (Abramson et al. 1987; Rozen et al. 1990; Feoktistova et al. 2013; Sokabe and Fraser 2017). Activity enhancements are a reflection of improved binding constants for mRNA as well as enhanced efficiency (as rate). The activities of both eIF4A and eIF4F can be further improved by the presence of eIF4B (Harms et al. 2014). What is unclear currently is whether the ATP-dependent action of eIF4A in the initiation pathway is exclusively through eIF4F or whether there are individual steps that use just eIF4A (Pause et al. 1994).
What is thought to be the most important function of eIF4A is its helicase activity that can be used to reduce or eliminate secondary structure in an mRNA or, by analogy to the DEAD-box RNA helicase NPH-II, to separate protein from RNA (Jankowsky et al. 2001). Both of these elements are important for loading a single strand of RNA into the mRNA channel on the 43S PIC. This conversion of an mRNA (as the mRNA initially exists having exited the nucleus) to an activated mRNA is catalyzed by the sequential recognition by eIF4F of the m7G cap and the generation of a single strand of RNA (eliminating secondary structure, protein, or both from the 5′ end of the mRNA) that can be placed on the 43S PIC. The first step is the recognition of the m7G cap by the eIF4E subunit of eIF4F. This recognition ensures that the bound RNA is an mRNA (has an m7G cap) and that eIF4E is bound to the 5′ end of the mRNA in an ATP-independent manner. The subsequent removal of RNA secondary structure and possible protein is dependent on ATP and is greatly enhanced by eIF4B (Abramson et al. 1987; Rozen et al. 1990). Based on the relatively slow turnover numbers for ATP hydrolysis (Lorsch and Herschlag 1998; Merrick 2015), presumed to be the result of the slow chemical step in the hydrolysis of ATP, the complex of eIF4F and eIF4B remains associated with the mRNA long enough to effect an interaction with eIF3 on the 43S PIC that leads to the binding of the mRNA to the 40S subunit via a protein–protein interaction (eIF4F and eIF3) although it is likely that the 40S subunit also contributes to the mRNA loading process.
A major player in mRNA recruitment is eIF3, both as an essential component in the formation of the 43S PIC as well as in stabilization of the bound mRNA at both the entry and exit channels (Aitken et al. 2016). Of particular importance are the NT and CT domains of eIF3a and the NTD of eIF3c, and this is consistent with the mapping of these portions of eIF3 to various cryo-EM structures of 48S complexes (Aylett et al. 2015; des Georges et al. 2015; Llacer et al. 2015; Jobe et al. 2018). Recently m7G cap-interacting domains were identified within both eIF3d (Lee et al. 2016) and eIF3l (Kumar et al. 2016) that may play roles in transitioning an eIF4E-bound mRNA m7G cap to one where the m7G cap is anchored by eIF3. Such a role for eIF3 would be consistent with the apparent loss of eIF4E when 48S complexes are formed (Kumar et al. 2016). Release of eIF4E would then permit the known regulation by 4E-BP under appropriate circumstances (Merrick 2015; Proud 2018). It should be noted that yeast eIF3 lacks homologs of the d and l subunits, and so further studies will be needed to resolve the relative importance of these m7G cap interactions for general protein synthesis initiation. eIF3j binds to the 40S decoding center and makes contact with eIF1A (Fig. 3) (Fraser et al. 2007; Aylett et al. 2015). eIF3j binding was shown to impair mRNA recruitment to 40S, except when TC was present, suggesting eIF3j may impair mRNA binding to 40S complexes lacking TC (Fraser et al. 2007; Mitchell et al. 2010). In summary, numerous studies suggest that a network of multiple interactions is established among initiation factors, tRNA, and the 40S ribosome, which act together to facilitate mRNA recruitment to the 48S PIC.
Perhaps the least understood step in the initiation process is scanning. This is in part because of the difficulty in obtaining a 48S PIC with the mRNA in its initial position when first bound to the 40S subunit, as the requirements for mRNA binding and scanning appear to be similar (requiring eIF4A, eIF4B, eIF4F, and ATP). The early work of Kozak established the requirement for ATP in scanning (Kozak 1980). eIF4A uses ATP and, in vitro, is sufficient as either eIF4A alone or within eIF4F to form 48S complexes positioned at the start codon (Pestova et al. 1998; Dmitriev et al. 2003; Aylett et al. 2015; Llacer et al. 2015). The question is how is the “RNA helicase” activity used to drive the scanning process? Some hints at this have been obtained from RNA helicase studies. For yeast eIF4F, there is about a 22-fold preference for a 5′ overhang on a RNA duplex substrate relative to a 3′ one (Rajagopal et al. 2012). This could conceivably orient the mRNA for movement in a 5′ to 3′ direction. However, in contrast, there is no equivalent preference shown by mammalian eIF4F (Rogers et al. 2001). A recent study using a single molecule, optical trapping assay for RNA duplex unwinding was able to show processivity with complexes of human eIF4A, eIF4B, and eIF4G (García-García et al. 2015), although this result has not been reproduced in standard assays of duplex unwinding with these proteins (Rogers et al. 2001; Gao et al. 2016). What is not yet clear in the scanning process is how the TC is able to inspect the mRNA for a start codon while the PIC is traversing the 5′UTR.
Translation of the mRNA is assumed to temporarily remove extensive secondary structure and RNA-associated proteins from both the 5′ leader and ORF. Thus, at termination the release of the 40S and 60S subunits frees them to initiate again (Hellen 2018). Kinetically, reuse of ribosomes to repeatedly translate the same mRNA is likely aided by what is termed the “closed-loop model” wherein PABP and eIF4G interact with each other to circularize the mRNA (Fig. 1) (Wells et al. 1998; Park et al. 2011; Archer et al. 2015; Costello et al. 2015, 2017). Thus, at termination, the ribosomal subunits are likely released in close proximity to the 5′ end of the mRNA and this facilitates subsequent initiation events in a more efficient manner. This may be especially important in cells in log phase growth where the availability of free 40S subunits is limiting (Chu et al. 2014), and is likely more important on some mRNAs than others as, at least in yeast, it is clear that the propensity of some mRNAs to form a closed loop is greater than others (Archer et al. 2015; Costello et al. 2015). Recent computational analyses support the idea that the closed-loop can enhance translation rates, particularly on shorter mRNAs (Rogers et al. 2017).
Some mRNAs have extensive secondary structure or length, or both, in their 5′UTRs and therefore require additional “helicase power,” which may come in the form of increased concentrations of eIF4A or eIF4B or of a variety of other RNA helicases, including mammalian Dhx29 and yeast Ded1 (Svitkin et al. 2001; Pisareva et al. 2008; Parsyan et al. 2011; Sen et al. 2015, 2016). At present, it is not clear whether these other helicases function entirely independently or as a partner/subunit of one of the initiation factors, as yeast Ded1 may function as an eIF4F subunit (Gao et al. 2016). The current interpretation is that such helicases regularly participate in many initiation events but are critically important for optimal initiation of mRNAs with highly structured 5′UTRs.
AUG RECOGNITION
AUG Recognition Causes Conformational Changes in the PIC
Typically, the first encountered AUG in an mRNA is used, providing it has a good sequence context. In mammals, the sequence GCCPuCCAUGG, generally called the Kozak consensus sequence, enhances AUG recognition (Jackson et al. 2010). Within this sequence, the purine (Pu = A or G) nucleotide at −3 and the G at +4 position are most important (both relative to the A of the AUG, designated +1). In contrast, AUG contexts differing significantly from this context can be bypassed, a process called leaky scanning. Leaky scanning is used by specific mRNAs to regulate expression levels and is used as an autoregulatory loop as described for the translation factor eIF1 and eIF5 mRNAs (Ivanov et al. 2010; Martin-Marcos et al. 2011; Loughran et al. 2012; Hinnebusch et al. 2016). Alternatively, leaky scanning may include or exclude an NTD signal sequence that can alter the destination of the final protein. For example, a single mRNA encodes an enzyme with both cytoplasmically and mitochondrially targeted forms where different start codons are used to generate the isoforms (Slusher et al. 1991). Similarly, leaky scanning of an upstream ORF can regulate the flow of ribosomes to the downstream ORF (see Wek 2018). It is also notable that mRNAs with long leaders can be efficiently translated, as can those with very short leaders (Berthelot et al. 2004). How close an AUG codon can be to the m7G cap and still be translated has been addressed recently in vitro. It was found that AUG codons immediately adjacent to the m7G cap can be used efficiently, providing that eIF1 was excluded from the reactions (Kumar et al. 2016). Hence, it is possible that eIF1 may prevent initiation very close to the m7G cap. A potential exception to this is the translation initiator of short 5′UTR (TISU) sequence SAASAUGGCGGC (S = G or C) in mRNAs that appears to be much more permissive for initiation events close to the m7G cap (Elfakess et al. 2011; Sinvani et al. 2015; Kwan and Thompson 2018).
To recognize an AUG initiation codon, the Met-tRNAi anticodon within the scanning PIC must base-pair with the mRNA and signal this interaction to the associated factors in the PIC. These signaling events drive conformational changes in the PIC that switch it from an “open” to a “closed” conformation wherein altered interactions between the components cause release of eIF1, eIF2•GDP, and eIF5 from the PIC (Unbehaun et al. 2004; Cheung et al. 2007). Many of the precise changes that occur are now becoming understood. These ultimately result in a PIC arrangement competent to recruit the 60S subunit. Both biochemistry and yeast genetics implicate eIF1, eIF1A, and eIF5 along with eIF2 in stringent AUG recognition. Highly informative to the mechanism of AUG recognition has been the study of yeast Sui− mutants that enhance inappropriate recognition of a mutated UUG start codon. Compensating Ssu− “suppressor of sui” mutations in these initiation factors have also been described that enhance the stringency of AUG selection, as recently reviewed (Dever et al. 2016).
In the scanning PIC, eIF1 is located close to the P site (Hashem et al. 2013a; Weisser et al. 2013; Hussain et al. 2014). Its location prevents complete Met-tRNAi anticodon–AUG pairing. Recent structural insights revealed that in the open scanning conformation, the eIF2β subunit extends between eIF1 and eIF1A connecting these factors with Met-tRNAi that is bound to the 40S head. It was proposed that contacts observed between eIF1, eIF2β, and the Met-tRNAi anticodon stem help to stabilize the scanning conformation (Fig. 4, “open”) (Llacer et al. 2015). Upon AUG recognition, there are a number of rearrangements that occur following the transition to the intermediate closed conformation that occurs following scanning arrest (Fig. 4, “closed”). These include movements of the 40S that define these conformations (Llacer et al. 2015), as well as changes in the relative positions of the bound factors that are highlighted in Figure 4. Met-tRNAi is repositioned and establishes codon–anticodon pairing with the mRNA. The eIF1A amino-terminal tail (NTT) appears to monitor or stabilize the closed codon–anticodon duplex in agreement with prior biochemical observations (Fekete et al. 2007). Likewise eIF2α contacts the conserved mRNA nucleotide at the –3 position using Arg54, which is close to the regulatory Ser51 residue, in agreement with prior cross-linking experiments (Pisarev et al. 2006). In contrast, eIF2β retracts from both the tRNA acceptor stem and eIF1A. In the closed conformation structure, eIF1 is partially displaced to enable codon–anticodon pairing between Met-tRNAi and the AUG codon of the mRNA.
Figure 4.
Altered conformation of initiation factors on AUG recognition. Cartoons derived from cryoelectron microscopy (cryo-EM) analyses of partial preinitiation complexes in a scanning mode (open, left panel) and AUG recognition mode (closed, right panel) (Llacer et al. 2015). Note, in particular, the movement of eukaryotic initiation factor (eIF)1, eIF1A (tail), and eIF2β. eIF3 and part of the 40S head structures were removed for clarity. Images were created with University of California San Francisco (UCSF) Chimera software from PDB files 3JAP and 3JAQ. See text for details.
Although not resolved in these PIC structures, eIF5 plays critical roles in AUG recognition and is thought to move during the PIC rearrangements mediating the open–closed transitions (Aylett et al. 2015; Llacer et al. 2015; Aitken et al. 2016; Obayashi et al. 2017). eIF1A and eIF5 interactions help retain eIF1 in the open scanning PIC (Maag et al. 2005; Luna et al. 2012, 2013) and eIF5–eIF3c interactions contribute to coordinating the rearrangements (Obayashi et al. 2017). It has also been shown that the 40S ribosomal proteins make important contacts with mRNA, Met-tRNAi, and translation factors that assist in determining accuracy of AUG codon recognition. Rps5/uS7 is located at the 40S mRNA exit channel and contacts TC and AUG –3/–4 context nucleotides (Pisarev et al. 2008), and mutations in uS7 were shown to impair AUG recognition analogous to mutations in translation initiation factors (Visweswaraiah et al. 2015) or in Met-tRNAi itself (Dong et al. 2014).
eIF2•GTP Hydrolysis and Factor Release
Although the eIF5-CTD makes a series of critical contacts during scanning, it is the Arg15 residue within the eIF5-NTD that stimulates hydrolysis of eIF2-bound GTP (Das and Maitra 2001; Paulin et al. 2001). It is thought that hydrolysis can occur during scanning, but that release of Pi to form eIF2•GDP occurs only at AUG recognition (Algire et al. 2005; Majumdar and Maitra 2005). (Note that in the structures shown in Fig. 4, a nonhydrolyzable variant of GTP [GDPCP] was used to stabilize the intermediate complexes.) It is proposed that the eIF1A carboxy-terminal tail (CTT) moves to contact eIF5-NTD on AUG recognition, movement that is coupled to both the dissociation of eIF1 from the PIC and to Pi release from eIF2 (Nanda et al. 2013; Saini et al. 2014). It appears likely that Met-tRNAi–AUG base-pairing and multiple factor interactions stimulate this series of events (Aitken et al. 2016; Obayashi et al. 2017). Although not shown in Figure 4, structural analysis of a late-stage 48S complex reveals that large movements of the eIF3 yeast-like core subunits occur before eIF2•GDP release (Simonetti et al. 2016).
Following GTP hydrolysis, eIF2•GDP has a low affinity for Met-tRNAi (Kapp and Lorsch 2004; Jennings et al. 2017), enabling eIF2•GDP to be released from the 48S complex. It is proposed that eIF5 leaves with eIF2 (Unbehaun et al. 2004) where it prevents premature release of GDP, as described above (Jennings and Pavitt 2010), before subsequent reactivation for further rounds of translation. In contrast, eIF1A remains ribosome-bound to stimulate 60S joining. The precise timing of when eIF3 and the eIF4 factors dissociate is not yet clear and so the release of these factors is not shown in Figure 1.
60S SUBUNIT JOINING
The ribosomal 40S and 60S subunits naturally bind to each other, especially in the presence of elevated concentrations of Mg2+. Thus, it was not too surprising that one of the early activities ascribed to eIF5B was its subunit-dependent GTPase activity, an activity not dependent on any other initiation factor or Met-tRNAi (Merrick et al. 1975). Early studies established that the use of GDPNP (or GDPCP) blocked initiation complex formation at the level of a 48S complex (with either artificial or natural mRNAs). Thus, release of eIF2 from the 40S subunit was an established early requirement for 60S joining. The rationale for this can be readily inferred from the recent 48S structures that provide extensive detail into the positioning of the various initiation factors, mRNA, and Met-tRNAi (Figs. 3 and 4). Factor release opens up the 40S surface for the binding of eIF5B•GTP, assisted by retaining 40S bound eIF1A (Schreier et al. 1977; Choi et al. 2000; Marintchev et al. 2003; Olsen et al. 2003). The interactions between eIF5B and eIF1A have been visualized both by crystallography (Zheng et al. 2014), and dynamically by nuclear magnetic resonance (NMR) (Nag et al. 2016). In these studies, the eIF1A-CTT is observed to interact with domain IV of eIF5B. Together these events are thought to stabilize Met-tRNAi–AUG interactions at the P-site and recruit the 60S subunit.
Uncertain in these events is whether eIF5B drives or facilitates the release of eIF2•GDP/eIF5 or the release of eIF3. Cross-linking studies with an mRNA containing 4-thioU at the –3 position indicate that there is a conformational change (and change in cross-linking to eIF2α) that occurs in the presence of eIF5 and eIF5B (Pisareva and Pisarev 2014). Yet in studies that have examined the formation of 80S complexes on model mRNAs, eIF3 was routinely associated with the 40S subunits, but little or none was found in the 80S complexes (Peterson et al. 1979; Pisareva and Pisarev 2014). This would suggest that there may be other mechanisms to promote the release of eIF3.
When trapped as a GDPNP (or GDPCP) complex, eIF5B has been located on the 80S ribosome (Fernandez et al. 2013; Yamamoto et al. 2014). It is positioned across the interface of the 40S subunit such that contact between eIF5B domain IV places the Met-tRNAi in the P/I conformation, which with hydrolysis of the GTP would lead the Met-tRNAi into the P position. There is a large change in the conformation of 80S-bound eIF5B, where domains III and IV are rotated 60°–65° relative to free eIF5B (Fernandez et al. 2013; Yamamoto et al. 2014). A recent “remodeling” performed by Kuhle and Ficner (2014) places the Met-tRNAi 3′ CCA end and the methionine in domain IV of eIF5B in a position equivalent to that seen with either IF2 or EF1A (EF-Tu) and their aminoacyl-tRNAs. It appears that the hydrolysis of GTP by eIF5B is sufficient to lead to its removal from the 80S ribosome (as is also true for bacterial IF2 (Rodnina 2018). eIF5B•GDP release is coupled to the removal of eIF1A (Fringer et al. 2007). The 80S ribosome is then ready to participate in the elongation phase of protein synthesis, with Met-tRNAi in the P site and a vacant A site.
ALTERNATIVE INITIATION EVENTS
For cells that are in log phase growth, almost all the translation is thought to proceed as described earlier. However, that does leave a small percentage of mRNAs translated by a different route. Characteristic of the expression of these mRNAs is that they are either cell-type-specific or they are expressed under conditions of stress. As such, these mechanisms usually have limited impact on overall expression because of their lack of competitiveness for the translational machinery and the consequence is that these systems have been difficult to recapitulate in vitro. Two of the best-studied alternative schemes are the internal ribosome entry site (IRES) promoted initiation and regulated-reinitiation on GCN4/ATF4 and related mRNAs with multiple upstream ORFs (Pelletier and Sonenberg 1988; Dever et al. 1992). IRESs provide an alternative 40S recruitment strategy, whereas GCN4/ATF4 and other mRNAs use the scanning mechanism described above (see Kwan and Thompson 2018 and Wek 2018, respectively, for coverage of these examples in greater detail). Further mechanisms include the previously mentioned TISU, an alternate 5′ sequence that promotes efficient initiation at AUG codons close to the 5′ end in the absence of scanning (Sinvani et al. 2015; Haimov et al. 2017; Kwan and Thompson 2018), and m6 methylation of 5′UTR adenine residues, which can promote cap-independent translation (Meyer et al. 2015; Peer et al. 2018).
The more curious events are those where eIF2 does not seem to be the protein responsible for directing the binding of the initiator Met-tRNAi. The most commonly cited player is eIF2A, which is a single polypeptide of 65 kDa (Merrick and Anderson 1975). eIF2A appears to play a major role in initiating translation for major histocompatibility complex (MHC) class I peptides (Starck and Shastri 2016), in the integrated stress response (Starck et al. 2016), tumor progression (Sendoel et al. 2017), and viral replication (Ventoso et al. 2006; Kim et al. 2011). Other proteins such as eIF2D (also known as ligatin), eIF5B, or MCT-1/DENR have been implicated in binding initiator tRNA to ribosomes (Pestova et al. 2008; Dmitriev et al. 2010; Skabkin et al. 2010; Weisser et al. 2017). Similarly, initiator Met-tRNAi may not always be used to initiate. Some initiation at CUG leucine codons has been found independent of eIF2 (Schwab et al. 2004). Biochemical analysis revealed that cells use an elongator leucine-bound transfer RNA (Leu-tRNA) to initiate translation at cryptic CUG start codons in an eIF2A-dependent manner (Starck et al. 2012). The ability to use these alternate proteins to initiate protein synthesis is generally assisted by the activation of one of the eIF2 kinases and the subsequent phosphorylation of serine 51 of the α subunit of eIF2, yielding a reduced concentration of ternary complexes (Fig. 1). The definition of the exact pathway of initiation for alternate initiating aminoacyl-tRNA-binding proteins remains to be elucidated.
The strangest and least understood to date is repeat associated non-ATG (RAN) translation, which occurs in microsatellite repeat diseases (Zu et al. 2013) where initiation occurs in all three reading frames. The ability to initiate in all three reading frames would be consistent with the properties of either eIF2D or eEF1A, but currently no mechanism is proposed. RAN translation is described elsewhere (Zu et al. 2018). It is likely that the study of various diseases in the future may yield yet additional mechanisms, although it is anticipated that these, like those above, would be dominated by the most common binder of the initiator tRNA, eIF2.
REGULATION OF THE 80S INITIATION PATHWAY
It is of interest to know the molar concentration of each initiation factor as this is also important for regulation where the alteration of the concentration of active initiation factors can influence both the amount of total protein made and mRNA selectivity. Shown in Table 1 is a subset of the data obtained by Kulak et al. (2014), using a label-free mass spectrometry protocol (numbers rounded off) for exponentially growing yeast and HeLa cell cultures, thereby enabling comparison across species from a single study. These values share consistencies with some earlier evaluations (Duncan and Hershey 1983; von der Haar and McCarthy 2002), although there are differences that may reflect specific difficulties with the estimation of one or more of the individual factors by various methods. If the numbers in Table 1 are roughly correct, then the following conclusions can be drawn:
Most initiation factors are less abundant than the number of ribosomes, except for eIF4A, which is roughly equimolar.
eIF2B levels are lower than eIF2 and eIF5, consistent with nucleotide exchange being a regulated step in initiation.
In yeast, eIF4G is limiting for the formation of eIF4F, whereas in HeLa cells eIF4E is limiting.
The concentration of “extra” RNA helicases that might assist in the initiation process is about one-fifth the concentration of eIF4A, consistent with their proposed more specialized role.
The level of the 4E-BP regulators is sufficient to completely shut down eIF4F-dependent protein synthesis (Korets et al. 2011), but the level of the eIF4A regulator Pdcd4 in HeLa cells is not (Lankat-Buttgereit and Goke 2009).
Table 1.
Number of protein molecules per cell
| Protein | Yeast (×103) | HeLa (×104) |
|---|---|---|
| Cytoplasmic ribosomes | 310 | 1064 |
| eIF1 | 41 | 198 |
| eIF1A | 60 | 101 |
| eIF2 | 31 | 210 |
| eIF2B | 6 | 75 |
| eIF2A | 11 | 72 |
| eIF3 | 29 | 164 |
| eIF4A | 196 | 801 |
| eIF4B | 18 | 131 |
| eIF4E | 36 | 41 |
| eIF4G (1 + 2) | 11 | 74 |
| eIF4H | - | 103 |
| eIF5 | 48 | 91 |
| eIF5B | 13 | 76 |
| PABP | 96 | 252 |
| DED1/DDX3 | 32 | 138 |
| DHX29 | - | 12 |
| Caf20 | 10 | - |
| Eap1 | 1.6 | - |
| 4E-BP1 | - | 33 |
| 4E-BP2 | - | 6 |
| 4E-T | - | 0.4 |
| Pdcd4 | - | 84 |
These data have been reorganized from mass spectrometry data reported previously (Kulak et al. 2014). For multisubunit factors and ribosomes, the value given corresponds to the most abundant subunit value reported.
Posttranslational modification of specific initiation factors renders it possible that any given factor might become more or less active. At present, the major global regulatory steps in initiation are understood as the phosphorylation of eIF2 (serine 51 of the α subunit) and the dephosphorylation of 4E-BPs, both of which are inhibitory (Fig. 1). These and other mechanisms are discussed elsewhere in the literature (Proud 2018; Wek 2018).
PERSPECTIVE: WHAT WE MIGHT LIKE TO KNOW
The size and complexity of the initiation factors and the ribosomal subunits have made detailed studies challenging, but methods continue to improve and offer hopes for a more complete understanding of protein synthesis in the near future. One advance would be to detail the precise timing and sequence of the binding, movement, and release events depicted in Figure 1, the 80S initiation pathway. With this, one would hope to define the rate-limiting step(s). From the observation of 43S and 48S complexes by sucrose gradient analysis, it would seem likely that there are at least two slow steps, the binding of the mRNA (hence 43S complexes) and either the scanning of the mRNA or subunit joining (hence the 48S complexes). Kinetic analyses of translation should address this point and recent progress is described elsewhere (Sokabe and Fraser 2018).
For a single complete initiation event, is only one copy of each initiation factor used or do some factors recycle multiple times or does initiation require multiple copies of some factors to be engaged? For the initiation factors eIF1, eIF1A, eIF2, eIF2B, eIF3, eIF5, and eIF5B it would seem likely that only a single copy of each factor is required. However, for eIF4A (and perhaps eIF4F), because multiple rounds of ATP hydrolysis are required, and as eIF4A is significantly more abundant than other initiation factors (Table 1), it is plausible that multiple copies are used, with the number likely dependent on the complexity of the mRNA 5′UTR (e.g., amount of secondary structure and length) (Svitkin et al. 2001). As has been noted for a few particular mRNAs, either in vitro or in vivo, if increased concentrations or other RNA helicases are required, multiple rounds of use for each of these may also be required (Pause et al. 1994).
During scanning on longer 5′ leaders, is contact maintained between eIF4F and the m7G cap until subunit joining or through multiple rounds of initiation? Energetically, it makes sense for eIF4F to remain bound to the m7G cap and assist with multiple rounds of initiation before detaching; however, it was recently proposed that eIF4E may detach from the mRNA during scanning (Kumar et al. 2016). Similarly, is contact between eIF4F and the PIC maintained during scanning? This tethered scenario is depicted in Figure 1, with the mRNA threaded though the channel between the head and body of the 40S subunit. However, other models of scanning propose that the PIC can detach from eIF4F (Archer et al. 2016).
There are several well-studied examples of the regulated expression of proteins via reinitiation, including GCN4, ATF4, and the glutamine amidotransferase subunit of Arg-specific carbamoyl phosphate synthetase (Elbarghati et al. 2008; Spevak et al. 2010; Dever et al. 2016). For the regulated expression of these mRNAs, what are the important translation factors or other trans-acting elements and what are the important cis-acting elements (such as upstream ORF [uORF] length or sequence, mRNA sequence, or structure contexts, etc.)? Current understanding of GCN4 translation, the most well-studied reinitiation system, implicates several sequences around its uORFs and eIF3 (Szamecz et al. 2008; Munzarova et al. 2011; Gunisova and Valasek 2014; Mohammad et al. 2017). Given the findings that a high percentage of mRNAs contain either bona fide uORFs or uORFs with non-AUG start codons (Calvo et al. 2009), it is highly probable that both nonstandard and reinitiation mechanisms will turn out to be more common than currently thought (Hinnebusch et al. 2016).
Finally, it is clear that there are rare (tissue- or disease-specific), but important initiation events for which we have little idea as to how they are accomplished: MHC class I peptide synthesis, tumor progression, and RAN translation being examples studied (Malys and McCarthy 2011; Starck et al. 2012; Zu et al. 2013; Sendoel et al. 2017). The ability to better understand each of these processes could enable the development of treatments to defeat these rare events with fewer side effects than drugs that are currently available. There is still a lot to do.
ACKNOWLEDGMENTS
Research in the Pavitt laboratory is funded by United Kingdom Biotechnology and Biological Sciences Research Council Grants BB/L000652/1, BB/L020157/1, and BB/M006565/1. W.C.M. is supported by the School of Medicine at Case Western Reserve University. We declare no competing interests.
Footnotes
Editors: Michael B. Mathews, Nahum Sonenberg, and John W.B. Hershey
Additional Perspectives on Translation Mechanisms and Control available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Abramson RD, Dever TE, Lawson TG, Ray BK, Thach RE, Merrick WC. 1987. The ATP-dependent interaction of eukaryotic initiation factors with mRNA. J Biol Chem 262: 3826–3832. [PubMed] [Google Scholar]
- Aitken CE, Beznoskova P, Vlckova V, Chiu WL, Zhou F, Valasek LS, Hinnebusch AG, Lorsch JR. 2016. Eukaryotic translation initiation factor 3 plays distinct roles at the mRNA entry and exit channels of the ribosomal preinitiation complex. eLife 5: e20934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Algire MA, Maag D, Lorsch JR. 2005. Pi release from eIF2, not GTP hydrolysis, is the step controlled by start-site selection during eukaryotic translation initiation. Mol Cell 20: 251–262. [DOI] [PubMed] [Google Scholar]
- Alone PV, Dever TE. 2006. Direct binding of translation initiation factor eIF2γ-G domain to its GTPase-activating and GDP-GTP exchange factors eIF5 and eIF2Bɛ. J Biol Chem 281: 12636–12644. [DOI] [PubMed] [Google Scholar]
- Archer SK, Shirokikh NE, Hallwirth CV, Beilharz TH, Preiss T. 2015. Probing the closed-loop model of mRNA translation in living cells. RNA Biol 12: 248–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archer SK, Shirokikh NE, Beilharz TH, Preiss T. 2016. Dynamics of ribosome scanning and recycling revealed by translation complex profiling. Nature 535: 570–574. [DOI] [PubMed] [Google Scholar]
- Asano K, Clayton J, Shalev A, Hinnebusch AG. 2000. A multifactor complex of eukaryotic initiation factors, eIF1, eIF2, eIF3, eIF5, and initiator tRNA(Met) is an important translation initiation intermediate in vivo. Genes Dev 14: 2534–2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aylett CH, Boehringer D, Erzberger JP, Schaefer T, Ban N. 2015. Structure of a yeast 40S-eIF1-eIF1A-eIF3-eIF3j initiation complex. Nat Struct Mol Biol 22: 269–271. [DOI] [PubMed] [Google Scholar]
- Bah A, Vernon RM, Siddiqui Z, Krzeminski M, Muhandiram R, Zhao C, Sonenberg N, Kay LE, Forman-Kay JD. 2015. Folding of an intrinsically disordered protein by phosphorylation as a regulatory switch. Nature 519: 106–109. [DOI] [PubMed] [Google Scholar]
- Berthelot K, Muldoon M, Rajkowitsch L, Hughes J, McCarthy JE. 2004. Dynamics and processivity of 40S ribosome scanning on mRNA in yeast. Mol Microbiol 51: 987–1001. [DOI] [PubMed] [Google Scholar]
- Beznoskova P, Wagner S, Jansen ME, von der Haar T, Valasek LS. 2015. Translation initiation factor eIF3 promotes programmed stop codon readthrough. Nucleic Acids Res 43: 5099–5111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boesen T, Mohammad SS, Pavitt GD, Andersen GR. 2004. Structure of the catalytic fragment of translation initiation factor 2B and identification of a critically important catalytic residue. J Biol Chem 279: 10584–10592. [DOI] [PubMed] [Google Scholar]
- Calvo SE, Pagliarini DJ, Mootha VK. 2009. Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc Natl Acad Sci 106: 7507–7512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung YN, Maag D, Mitchell SF, Fekete CA, Algire MA, Takacs JE, Shirokikh N, Pestova T, Lorsch JR, Hinnebusch AG. 2007. Dissociation of eIF1 from the 40S ribosomal subunit is a key step in start codon selection in vivo. Genes Dev 21: 1217–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi SK, Olsen DS, Roll-Mecak A, Martung A, Remo KL, Burley SK, Hinnebusch AG, Dever TE. 2000. Physical and functional interaction between the eukaryotic orthologs of prokaryotic translation initiation factors IF1 and IF2. Mol Cell Biol 20: 7183–7191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu D, Kazana E, Bellanger N, Singh T, Tuite MF, von der Haar T. 2014. Translation elongation can control translation initiation on eukaryotic mRNAs. EMBO J 33: 21–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello J, Castelli LM, Rowe W, Kershaw CJ, Talavera D, Mohammad-Qureshi SS, Sims PF, Grant CM, Pavitt GD, Hubbard SJ, et al. 2015. Global mRNA selection mechanisms for translation initiation. Genome Biol 16: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello JL, Kershaw CJ, Castelli LM, Talavera D, Rowe W, Sims PFG, Ashe MP, Grant CM, Hubbard SJ, Pavitt GD. 2017. Dynamic changes in eIF4F-mRNA interactions revealed by global analyses of environmental stress responses. Genome Biol 18: 201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Maitra U. 2001. Functional significance and mechanism of eIF5-promoted GTP hydrolysis in eukaryotic translation initiation. Prog Nucleic Acid Res Mol Biol 70: 207–231. [DOI] [PubMed] [Google Scholar]
- Dennis MD, Person MD, Browning KS. 2009. Phosphorylation of plant translation initiation factors by CK2 enhances the in vitro interaction of multifactor complex components. J Biol Chem 284: 20615–20628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- des Georges A, Dhote V, Kuhn L, Hellen CU, Pestova TV, Frank J, Hashem Y. 2015. Structure of mammalian eIF3 in the context of the 43S preinitiation complex. Nature 525: 491–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dever TE, Feng L, Wek RC, Cigan AM, Donahue TF, Hinnebusch AG. 1992. Phosphorylation of initiation factor 2 α by protein kinase GCN2 mediates gene-specific translational control of GCN4 in yeast. Cell 68: 585–596. [DOI] [PubMed] [Google Scholar]
- Dever TE, Kinzy TG, Pavitt GD. 2016. Mechanism and regulation of protein synthesis in Saccharomyces cerevisiae. Genetics 203: 65–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dmitriev SE, Pisarev AV, Rubtsova MP, Dunaevsky YE, Shatsky IN. 2003. Conversion of 48S translation preinitiation complexes into 80S initiation complexes as revealed by toeprinting. FEBS Lett 533: 99–104. [DOI] [PubMed] [Google Scholar]
- Dmitriev SE, Terenin IM, Andreev DE, Ivanov PA, Dunaevsky JE, Merrick WC, Shatsky IN. 2010. GTP-independent tRNA delivery to the ribosomal P-site by a novel eukaryotic translation factor. J Biol Chem 285: 26779–26787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Munoz A, Kolitz SE, Saini AK, Chiu WL, Rahman H, Lorsch JR, Hinnebusch AG. 2014. Conserved residues in yeast initiator tRNA calibrate initiation accuracy by regulating preinitiation complex stability at the start codon. Genes Dev 28: 502–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drabkin HJ, Helk B, RajBhandary UL. 1993. The role of nucleotides conserved in eukaryotic initiator methionine tRNAs in initiation of protein synthesis. J Biol Chem 268: 25221–25228. [PubMed] [Google Scholar]
- Duncan R, Hershey JW. 1983. Identification and quantitation of levels of protein synthesis initiation factors in crude HeLa cell lysates by two-dimensional polyacrylamide gel electrophoresis. J Biol Chem 258: 7228–7235. [PubMed] [Google Scholar]
- Elbarghati L, Murdoch C, Lewis CE. 2008. Effects of hypoxia on transcription factor expression in human monocytes and macrophages. Immunobiology 213: 899–908. [DOI] [PubMed] [Google Scholar]
- Elfakess R, Sinvani H, Haimov O, Svitkin Y, Sonenberg N, Dikstein R. 2011. Unique translation initiation of mRNAs-containing TISU element. Nucleic Acids Res 39: 7598–7609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson FL, Hannig EM. 1996. Ligand interactions with eukaryotic translation initiation factor 2: Role of the γ-subunit. EMBO J 15: 6311–6320. [PMC free article] [PubMed] [Google Scholar]
- Erzberger JP, Stengel F, Pellarin R, Zhang S, Schaefer T, Aylett CH, Cimermancic P, Boehringer D, Sali A, Aebersold R, et al. 2014. Molecular architecture of the 40S•eIF1•eIF3 translation initiation complex. Cell 158: 1123–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fekete CA, Mitchell SF, Cherkasova VA, Applefield D, Algire MA, Maag D, Saini AK, Lorsch JR, Hinnebusch AG. 2007. N- and C-terminal residues of eIF1A have opposing effects on the fidelity of start codon selection. EMBO J 26: 1602–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feoktistova K, Tuvshintogs E, Do A, Fraser CS. 2013. Human eIF4E promotes mRNA restructuring by stimulating eIF4A helicase activity. Proc Natl Acad Sci 110: 13339–13344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez IS, Bai XC, Hussain T, Kelley AC, Lorsch JR, Ramakrishnan V, Scheres SH. 2013. Molecular architecture of a eukaryotic translational initiation complex. Science 342: 1240585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser CS, Berry KE, Hershey JW, Doudna JA. 2007. eIF3j is located in the decoding center of the human 40S ribosomal subunit. Mol Cell 26: 811–819. [DOI] [PubMed] [Google Scholar]
- Fringer JM, Acker MG, Fekete CA, Lorsch JR, Dever TE. 2007. Coupled release of eukaryotic translation initiation factors 5B and 1A from 80S ribosomes following subunit joining. Mol Cell Biol 27: 2384–2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Putnam AA, Bowers HA, Guenther UP, Ye X, Kindsfather A, Hilliker AK, Jankowsky E. 2016. Coupling between the DEAD-box RNA helicases Ded1p and eIF4A. eLife 5: e16408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-García C, Frieda KL, Feoktistova K, Fraser CS, Block SM. 2015. Factor-dependent processivity in human eIF4A DEAD-box helicase. Science 348: 1486–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordiyenko Y, Schmidt C, Jennings MD, Matak-Vinkovic D, Pavitt GD, Robinson CV. 2014. eIF2B is a decameric guanine nucleotide exchange factor with a γ2ɛ2 tetrameric core. Nat Commun 5: 3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grifo JA, Abramson RD, Satler CA, Merrick WC. 1984. RNA-stimulated ATPase activity of eukaryotic initiation factors. J Biol Chem 259: 8648–8654. [PubMed] [Google Scholar]
- Gross JD, Moerke NJ, von der Haar T, Lugovskoy AA, Sachs AB, McCarthy JE, Wagner G. 2003. Ribosome loading onto the mRNA cap is driven by conformational coupling between eIF4G and eIF4E. Cell 115: 739–750. [DOI] [PubMed] [Google Scholar]
- Gruner S, Peter D, Weber R, Wohlbold L, Chung MY, Weichenrieder O, Valkov E, Igreja C, Izaurralde E. 2016. The structures of eIF4E–eIF4G complexes reveal an extended interface to regulate translation initiation. Mol Cell 64: 467–479. [DOI] [PubMed] [Google Scholar]
- Gunisova S, Valasek LS. 2014. Fail-safe mechanism of GCN4 translational control—uORF2 promotes reinitiation by analogous mechanism to uORF1 and thus secures its key role in GCN4 expression. Nucleic Acids Res 42: 5880–5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haimov O, Sinvani H, Martin F, Ulitsky I, Emmanuel R, Tamarkin-Ben-Harush A, Vardy A, Dikstein R. 2017. Efficient and accurate translation initiation directed by TISU involves RPS3 and RPS10e binding and differential eukaryotic initiation factor 1A regulation. Mol Cell Biol 37: e00150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms U, Andreou AZ, Gubaev A, Klostermeier D. 2014. eIF4B, eIF4G and RNA regulate eIF4A activity in translation initiation by modulating the eIF4A conformational cycle. Nucleic Acids Res 42: 7911–7922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashem Y, des Georges A, Dhote V, Langlois R, Liao HY, Grassucci RA, Hellen CU, Pestova TV, Frank J. 2013a. Structure of the mammalian ribosomal 43S preinitiation complex bound to the scanning factor DHX29. Cell 153: 1108–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashem Y, des Georges A, Dhote V, Langlois R, Liao HY, Grassucci RA, Pestova TV, Hellen CU, Frank J. 2013b. Hepatitis-C-virus-like internal ribosome entry sites displace eIF3 to gain access to the 40S subunit. Nature 503: 539–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Hellen CUT. 2018. Translation termination and ribosome recycling in eukaryotes. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG. 2014. The scanning mechanism of eukaryotic translation initiation. Annu Rev Biochem 83: 779–812. [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG. 2017. Structural insights into the mechanism of scanning and start codon recognition in eukaryotic translation initiation. Trends Biochem Sci 42: 589–611. [DOI] [PubMed] [Google Scholar]
- Hinnebusch AG, Lorsch JR. 2012. The mechanism of eukaryotic translation initiation: New insights and challenges. Cold Spring Harb Perspect Biol 4: a011544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch AG, Ivanov IP, Sonenberg N. 2016. Translational control by 5′-untranslated regions of eukaryotic mRNAs. Science 352: 1413–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain T, Llacer JL, Fernandez IS, Munoz A, Martin-Marcos P, Savva CG, Lorsch JR, Hinnebusch AG, Ramakrishnan V. 2014. Structural changes enable start codon recognition by the eukaryotic translation initiation complex. Cell 159: 597–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov IP, Loughran G, Sachs MS, Atkins JF. 2010. Initiation context modulates autoregulation of eukaryotic translation initiation factor 1 (eIF1). Proc Natl Acad Sci 107: 18056–18060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RJ, Hellen CU, Pestova TV. 2010. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol 11: 113–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowsky E, Gross CH, Shuman S, Pyle AM. 2001. Active disruption of an RNA-protein interaction by a DExH/D RNA helicase. Science 291: 121–125. [DOI] [PubMed] [Google Scholar]
- Jennings MD, Pavitt GD. 2010. eIF5 has GDI activity necessary for translational control by eIF2 phosphorylation. Nature 465: 378–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings MD, Zhou Y, Mohammad-Qureshi SS, Bennett D, Pavitt GD. 2013. eIF2B promotes eIF5 dissociation from eIF2*GDP to facilitate guanine nucleotide exchange for translation initiation. Genes Dev 27: 2696–2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings MD, Kershaw CJ, White C, Hoyle D, Richardson JP, Costello JL, Donaldson IJ, Zhou Y, Pavitt GD. 2016. eIF2β is critical for eIF5-mediated GDP-dissociation inhibitor activity and translational control. Nucleic Acids Res 44: 9698–9709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jennings MD, Kershaw CJ, Adomavicius T, Pavitt GD. 2017. Fail-safe control of translation initiation by dissociation of eIF2α phosphorylated ternary complexes. eLife 6: e24542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Jobe A, Liu Z, Gutierrez-Vargas C, Frank J. 2018. New insights into ribosome structure and function. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapp LD, Lorsch JR. 2004. GTP-dependent recognition of the methionine moiety on initiator tRNA by translation factor eIF2. J Mol Biol 335: 923–936. [DOI] [PubMed] [Google Scholar]
- Kapp LD, Kolitz SE, Lorsch JR. 2006. Yeast initiator tRNA identity elements cooperate to influence multiple steps of translation initiation. RNA 12: 751–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashiwagi K, Takahashi M, Nishimoto M, Hiyama TB, Higo T, Umehara T, Sakamoto K, Ito T, Yokoyama S. 2016. Crystal structure of eukaryotic translation initiation factor 2B. Nature 531: 122–125. [DOI] [PubMed] [Google Scholar]
- Kearse MG, Wilusz JE. 2017. Non-AUG translation: A new start for protein synthesis in eukaryotes. Genes Dev 31: 1717–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoshnevis S, Gunisova S, Vlckova V, Kouba T, Neumann P, Beznoskova P, Ficner R, Valasek LS. 2014. Structural integrity of the PCI domain of eIF3a/TIF32 is required for mRNA recruitment to the 43S pre-initiation complexes. Nucleic Acids Res 42: 4123–4139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Park SM, Park JH, Keum SJ, Jang SK. 2011. eIF2A mediates translation of hepatitis C viral mRNA under stress conditions. EMBO J 30: 2454–2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolitz SE, Lorsch JR. 2010. Eukaryotic initiator tRNA: Finely tuned and ready for action. FEBS Lett 584: 396–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolupaeva VG, Unbehaun A, Lomakin IB, Hellen CU, Pestova TV. 2005. Binding of eukaryotic initiation factor 3 to ribosomal 40S subunits and its role in ribosomal dissociation and anti-association. RNA 11: 470–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korets SB, Czok S, Blank SV, Curtin JP, Schneider RJ. 2011. Targeting the mTOR/4E-BP pathway in endometrial cancer. Clin Cancer Res 17: 7518–7528. [DOI] [PubMed] [Google Scholar]
- Kozak M. 1980. Role of ATP in binding and migration of 40S ribosomal subunits. Cell 22: 459–467. [DOI] [PubMed] [Google Scholar]
- Kuhle B, Ficner R. 2014. eIF5B employs a novel domain release mechanism to catalyze ribosomal subunit joining. EMBO J 33: 1177–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulak NA, Pichler G, Paron I, Nagaraj N, Mann M. 2014. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat Methods 11: 319–324. [DOI] [PubMed] [Google Scholar]
- Kumar P, Hellen CU, Pestova TV. 2016. Toward the mechanism of eIF4F-mediated ribosomal attachment to mammalian capped mRNAs. Genes Dev 30: 1573–1588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Kwan T, Thompson SR. 2018. Noncanonical translation initiation in eukaryotes. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lankat-Buttgereit B, Goke R. 2009. The tumour suppressor Pdcd4: Recent advances in the elucidation of function and regulation. Biol Cell 101: 309–317. [DOI] [PubMed] [Google Scholar]
- Lee AS, Kranzusch PJ, Doudna JA, Cate JH. 2016. eIF3d is an mRNA cap-binding protein that is required for specialized translation initiation. Nature 536: 96–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder P, Lasko PF, Ashburner M, Leroy P, Nielsen PJ, Nishi K, Schnier J, Slonimski PP. 1989. Birth of the D-E-A-D box. Nature 337: 121–122. [DOI] [PubMed] [Google Scholar]
- Llacer JL, Hussain T, Marler L, Aitken CE, Thakur A, Lorsch JR, Hinnebusch AG, Ramakrishnan V. 2015. Conformational differences between open and closed states of the eukaryotic translation initiation complex. Mol Cell 59: 399–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorsch JR, Herschlag D. 1998. The DEAD-box protein eIF4A. 1: A minimal kinetic and thermodynamic framework reveals coupled binding of RNA and nucleotide. Biochemistry 37: 2180–2193. [DOI] [PubMed] [Google Scholar]
- Loughran G, Sachs MS, Atkins JF, Ivanov IP. 2012. Stringency of start codon selection modulates autoregulation of translation initiation factor eIF5. Nucleic Acids Res 40: 2898–2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna RE, Arthanari H, Hiraishi H, Nanda J, Martin-Marcos P, Markus MA, Akabayov B, Milbradt AG, Luna LE, Seo HC, et al. 2012. The C-terminal domain of eukaryotic initiation factor 5 promotes start codon recognition by its dynamic interplay with eIF1 and eIF2β. Cell Rep 1: 689–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna RE, Arthanari H, Hiraishi H, Akabayov B, Tang L, Cox C, Markus MA, Luna LE, Ikeda Y, Watanabe R, et al. 2013. The interaction between eukaryotic initiation factor 1A and eIF5 retains eIF1 within scanning preinitiation complexes. Biochemistry 52: 9510–9518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maag D, Fekete CA, Gryczynski Z, Lorsch JR. 2005. A conformational change in the eukaryotic translation preinitiation complex and release of eIF1 signal recognition of the start codon. Mol Cell 17: 265–275. [DOI] [PubMed] [Google Scholar]
- Majumdar R, Maitra U. 2005. Regulation of GTP hydrolysis prior to ribosomal AUG selection during eukaryotic translation initiation. EMBO J 24: 3737–3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumdar R, Bandyopadhyay A, Maitra U. 2003. Mammalian translation initiation factor eIF1 functions with eIF1A and eIF3 in the formation of a stable 40S preinitiation complex. J Biol Chem 278: 6580–6587. [DOI] [PubMed] [Google Scholar]
- Malys N, McCarthy JE. 2011. Translation initiation: Variations in the mechanism can be anticipated. Cell Mol Life Sci 68: 991–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marintchev A, Kolupaeva VG, Pestova TV, Wagner G. 2003. Mapping the binding interface between human eukaryotic initiation factors 1A and 5B: A new interaction between old partners. Proc Natl Acad Sci 100: 1535–1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Marcos P, Cheung YN, Hinnebusch AG. 2011. Functional elements in initiation factors 1, 1A, and 2β discriminate against poor AUG context and non-AUG start codons. Mol Cell Biol 31: 4814–4831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Marcos P, Nanda J, Luna RE, Wagner G, Lorsch JR, Hinnebusch AG. 2013. β-Hairpin loop of eukaryotic initiation factor 1 (eIF1) mediates 40S ribosome binding to regulate initiator tRNA(Met) recruitment and accuracy of AUG selection in vivo. J Biol Chem 288: 27546–27562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrick WC. 2015. eIF4F: A retrospective. J Biol Chem 290: 24091–24099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrick WC, Anderson WF. 1975. Purification and characterization of homogeneous protein synthesis initiation factor M1 from rabbit reticulocytes. J Biol Chem 250: 1197–1206. [PubMed] [Google Scholar]
- Merrick WC, Kemper WM, Anderson WF. 1975. Purification and characterization of homogeneous initiation factor M2A from rabbit reticulocytes. J Biol Chem 250: 5556–5562. [PubMed] [Google Scholar]
- Meyer KD, Patil DP, Zhou J, Zinoviev A, Skabkin MA, Elemento O, Pestova TV, Qian SB, Jaffrey SR. 2015. 5′UTR m6A promotes cap-independent translation. Cell 163: 999–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell SF, Walker SE, Algire MA, Park EH, Hinnebusch AG, Lorsch JR. 2010. The 5′-7-methylguanosine cap on eukaryotic mRNAs serves both to stimulate canonical translation initiation and to block an alternative pathway. Mol Cell 39: 950–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad MP, Munzarova Pondelickova V, Zeman J, Gunisova S, Valasek LS. 2017. In vivo evidence that eIF3 stays bound to ribosomes elongating and terminating on short upstream ORFs to promote reinitiation. Nucleic Acids Res 45: 2658–2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad-Qureshi SS, Haddad R, Hemingway EJ, Richardson JP, Pavitt GD. 2007. Critical contacts between the eukaryotic initiation factor 2B (eIF2B) catalytic domain and both eIF2β and -2γ mediate guanine nucleotide exchange. Mol Cell Biol 27: 5225–5234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munzarova V, Panek J, Gunisova S, Danyi I, Szamecz B, Valasek LS. 2011. Translation reinitiation relies on the interaction between eIF3a/TIF32 and progressively folded cis-acting mRNA elements preceding short uORFs. PLoS Genet 7: e1002137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag N, Lin KY, Edmonds KA, Yu J, Nadkarni D, Marintcheva B, Marintchev A. 2016. eIF1A/eIF5B interaction network and its functions in translation initiation complex assembly and remodeling. Nucleic Acids Res 44: 7441–7456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanda JS, Saini AK, Munoz AM, Hinnebusch AG, Lorsch JR. 2013. Coordinated movements of eukaryotic translation initiation factors eIF1, eIF1A, and eIF5 trigger phosphate release from eIF2 in response to start codon recognition by the ribosomal preinitiation complex. J Biol Chem 288: 5316–5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naveau M, Lazennec-Schurdevin C, Panvert M, Dubiez E, Mechulam Y, Schmitt E. 2013. Roles of yeast eIF2α and eIF2β subunits in the binding of the initiator methionyl-tRNA. Nucleic Acids Res 41: 1047–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obayashi E, Luna RE, Nagata T, Martin-Marcos P, Hiraishi H, Singh CR, Erzberger JP, Zhang F, Arthanari H, Morris J, et al. 2017. Molecular landscape of the ribosome pre-initiation complex during mRNA scanning: Structural role for eIF3c and its control by eIF5. Cell Rep 18: 2651–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Leary SE, Petrov A, Chen J, Puglisi JD. 2013. Dynamic recognition of the mRNA cap by Saccharomyces cerevisiae eIF4E. Structure 21: 2197–2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen DS, Savner EM, Mathew A, Zhang F, Krishnamoorthy T, Phan L, Hinnebusch AG. 2003. Domains of eIF1A that mediate binding to eIF2, eIF3, and eIF5B and promote ternary complex recruitment in vivo. EMBO J 22: 193–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panniers R, Rowlands AG, Henshaw EC. 1988. The effect of Mg2+ and guanine nucleotide exchange factor on the binding of guanine nucleotides to eukaryotic initiation factor 2. J Biol Chem 263: 5519–5525. [PubMed] [Google Scholar]
- Park EH, Walker SE, Lee JM, Rothenburg S, Lorsch JR, Hinnebusch AG. 2011. Multiple elements in the eIF4G1 N-terminus promote assembly of eIF4G1*PABP mRNPs in vivo. EMBO J 30: 302–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsyan A, Svitkin Y, Shahbazian D, Gkogkas C, Lasko P, Merrick WC, Sonenberg N. 2011. mRNA helicases: The tacticians of translational control. Nat Rev Mol Cell Biol 12: 235–245. [DOI] [PubMed] [Google Scholar]
- Passmore LA, Schmeing TM, Maag D, Applefield DJ, Acker MG, Algire MA, Lorsch JR, Ramakrishnan V. 2007. The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Mol Cell 26: 41–50. [DOI] [PubMed] [Google Scholar]
- Paulin FE, Campbell LE, O'Brien K, Loughlin J, Proud CG. 2001. Eukaryotic translation initiation factor 5 (eIF5) acts as a classical GTPase-activator protein. Curr Biol 11: 55–59. [DOI] [PubMed] [Google Scholar]
- Pause A, Methot N, Svitkin Y, Merrick WC, Sonenberg N. 1994. Dominant negative mutants of mammalian translation initiation factor eIF-4A define a critical role for eIF-4F in cap-dependent and cap-independent initiation of translation. EMBO J 13: 1205–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavitt GD. 2005. eIF2B, a mediator of general and gene-specific translational control. Biochem Soc Trans 33: 1487–1492. [DOI] [PubMed] [Google Scholar]
- Pavitt GD, Ramaiah KV, Kimball SR, Hinnebusch AG. 1998. eIF2 independently binds two distinct eIF2B subcomplexes that catalyze and regulate guanine-nucleotide exchange. Genes Dev 12: 514–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Peer E, Moshitch-Moshkovitz S, Rechavi G, Dominissini D. 2018. The epitranscriptome in translation regulation. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier J, Sonenberg N. 1988. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature 334: 320–325. [DOI] [PubMed] [Google Scholar]
- Pestova TV, Borukhov SI, Hellen CU. 1998. Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature 394: 854–859. [DOI] [PubMed] [Google Scholar]
- Pestova TV, de Breyne S, Pisarev AV, Abaeva IS, Hellen CU. 2008. eIF2-dependent and eIF2-independent modes of initiation on the CSFV IRES: A common role of domain II. EMBO J 27: 1060–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson DT, Merrick WC, Safer B. 1979. Binding and release of radiolabeled eukaryotic initiation factors 2 and 3 during 80S initiation complex formation. J Biol Chem 254: 2509–2516. [PubMed] [Google Scholar]
- Pisarev AV, Kolupaeva VG, Pisareva VP, Merrick WC, Hellen CU, Pestova TV. 2006. Specific functional interactions of nucleotides at key −3 and +4 positions flanking the initiation codon with components of the mammalian 48S translation initiation complex. Genes Dev 20: 624–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev AV, Hellen CU, Pestova TV. 2007. Recycling of eukaryotic posttermination ribosomal complexes. Cell 131: 286–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev AV, Kolupaeva VG, Yusupov MM, Hellen CU, Pestova TV. 2008. Ribosomal position and contacts of mRNA in eukaryotic translation initiation complexes. EMBO J 27: 1609–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisareva VP, Pisarev AV. 2014. eIF5 and eIF5B together stimulate 48S initiation complex formation during ribosomal scanning. Nucleic Acids Res 42: 12052–12069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisareva VP, Pisarev AV, Komar AA, Hellen CU, Pestova TV. 2008. Translation initiation on mammalian mRNAs with structured 5′UTRs requires DExH-box protein DHX29. Cell 135: 1237–1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Proud CG. 2018. Regulation of translation by cap-binding proteins. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a033050. [DOI] [Google Scholar]
- Rajagopal V, Park EH, Hinnebusch AG, Lorsch JR. 2012. Specific domains in yeast translation initiation factor eIF4G strongly bias RNA unwinding activity of the eIF4F complex toward duplexes with 5′-overhangs. J Biol Chem 287: 20301–20312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter JD, Sonenberg N. 2005. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature 433: 477–480. [DOI] [PubMed] [Google Scholar]
- *.Rodnina MV. 2018. Translation in prokaryotes. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers GW Jr, Richter NJ, Lima WF, Merrick WC. 2001. Modulation of the helicase activity of eIF4A by eIF4B, eIF4H, and eIF4F. J Biol Chem 276: 30914–30922. [DOI] [PubMed] [Google Scholar]
- Rogers DW, Bottcher MA, Traulsen A, Greig D. 2017. Ribosome reinitiation can explain length-dependent translation of messenger RNA. PLoS Comput Biol 13: e1005592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowlands AG, Panniers R, Henshaw EC. 1988. The catalytic mechanism of guanine nucleotide exchange factor action and competitive inhibition by phosphorylated eukaryotic initiation factor 2. J Biol Chem 263: 5526–5533. [PubMed] [Google Scholar]
- Rozen F, Edery I, Meerovitch K, Dever TE, Merrick WC, Sonenberg N. 1990. Bidirectional RNA helicase activity of eucaryotic translation initiation factors 4A and 4F. Mol Cell Biol 10: 1134–1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini AK, Nanda JS, Martin-Marcos P, Dong J, Zhang F, Bhardwaj M, Lorsch JR, Hinnebusch AG. 2014. Eukaryotic translation initiation factor eIF5 promotes the accuracy of start codon recognition by regulating Pi release and conformational transitions of the preinitiation complex. Nucleic Acids Res 42: 9623–9640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt E, Naveau M, Mechulam Y. 2010. Eukaryotic and archaeal translation initiation factor 2: A heterotrimeric tRNA carrier. FEBS Lett 584: 405–412. [DOI] [PubMed] [Google Scholar]
- Schmitt E, Panvert M, Lazennec-Schurdevin C, Coureux PD, Perez J, Thompson A, Mechulam Y. 2012. Structure of the ternary initiation complex aIF2-GDPNP-methionylated initiator tRNA. Nat Struct Mol Biol 19: 450–454. [DOI] [PubMed] [Google Scholar]
- Schreier MH, Erni B, Staehelin T. 1977. Initiation of mammalian protein synthesis. I. Purification and characterization of seven initiation factors. J Mol Biol 116: 727–753. [DOI] [PubMed] [Google Scholar]
- Schwab SR, Shugart JA, Horng T, Malarkannan S, Shastri N. 2004. Unanticipated antigens: Translation initiation at CUG with leucine. PLoS Biol 2: e366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen ND, Zhou F, Ingolia NT, Hinnebusch AG. 2015. Genome-wide analysis of translational efficiency reveals distinct but overlapping functions of yeast DEAD-box RNA helicases Ded1 and eIF4A. Genome Res 25: 1196–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen ND, Zhou F, Harris MS, Ingolia NT, Hinnebusch AG. 2016. eIF4B stimulates translation of long mRNAs with structured 5′UTRs and low closed-loop potential but weak dependence on eIF4G. Proc Natl Acad Sci 113: 10464–10472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sendoel A, Dunn JG, Rodriguez EH, Naik S, Gomez NC, Hurwitz B, Levorse J, Dill BD, Schramek D, Molina H, et al. 2017. Translation from unconventional 5′ start sites drives tumour initiation. Nature 541: 494–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin BS, Kim JR, Walker SE, Dong J, Lorsch JR, Dever TE. 2011. Initiation factor eIF2γ promotes eIF2-GTP-Met-tRNAi(Met) ternary complex binding to the 40S ribosome. Nat Struct Mol Biol 18: 1227–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonetti A, Brito Querido J, Myasnikov AG, Mancera-Martinez E, Renaud A, Kuhn L, Hashem Y. 2016. eIF3 peripheral subunits rearrangement after mRNA binding and start-codon recognition. Mol Cell 63: 206–217. [DOI] [PubMed] [Google Scholar]
- Singh CR, Lee B, Udagawa T, Mohammad-Qureshi SS, Yamamoto Y, Pavitt GD, Asano K. 2006. An eIF5/eIF2 complex antagonizes guanine nucleotide exchange by eIF2B during translation initiation. EMBO J 25: 4537–4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh CR, Watanabe R, Chowdhury W, Hiraishi H, Murai MJ, Yamamoto Y, Miles D, Ikeda Y, Asano M, Asano K. 2012. Sequential eukaryotic translation initiation factor 5 (eIF5) binding to the charged disordered segments of eIF4G and eIF2β stabilizes the 48S preinitiation complex and promotes its shift to the initiation mode. Mol Cell Biol 32: 3978–3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinvani H, Haimov O, Svitkin Y, Sonenberg N, Tamarkin-Ben-Harush A, Viollet B, Dikstein R. 2015. Translational tolerance of mitochondrial genes to metabolic energy stress involves TISU and eIF1-eIF4GI cooperation in start codon selection. Cell Metab 21: 479–492. [DOI] [PubMed] [Google Scholar]
- Skabkin MA, Skabkina OV, Dhote V, Komar AA, Hellen CU, Pestova TV. 2010. Activities of Ligatin and MCT-1/DENR in eukaryotic translation initiation and ribosomal recycling. Genes Dev 24: 1787–1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slusher LB, Gillman EC, Martin NC, Hopper AK. 1991. mRNA leader length and initiation codon context determine alternative AUG selection for the yeast gene MOD5. Proc Natl Acad Sci 88: 9789–9793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokabe M, Fraser CS. 2014. Human eukaryotic initiation factor 2 (eIF2)-GTP-Met-tRNAi ternary complex and eIF3 stabilize the 43S preinitiation complex. J Biol Chem 289: 31827–31836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokabe M. Fraser CS. 2017. A helicase-independent activity of eIF4A in promoting mRNA recruitment to the human ribosome. Proc Natl Acad Sci 114: 6304–6309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Sokabe M, Fraser CS. 2018. Toward a kinetic understanding of eukaryotic translation. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokabe M, Fraser CS, Hershey JW. 2012. The human translation initiation multi-factor complex promotes methionyl-tRNAi binding to the 40S ribosomal subunit. Nucleic Acids Res 40: 905–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spevak CC, Ivanov IP, Sachs MS. 2010. Sequence requirements for ribosome stalling by the arginine attenuator peptide. J Biol Chem 285: 40933–40942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starck SR, Shastri N. 2016. Nowhere to hide: Unconventional translation yields cryptic peptides for immune surveillance. Immunol Rev 272: 8–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starck SR, Jiang V, Pavon-Eternod M, Prasad S, McCarthy B, Pan T, Shastri N. 2012. Leucine-tRNA initiates at CUG start codons for protein synthesis and presentation by MHC class I. Science 336: 1719–1723. [DOI] [PubMed] [Google Scholar]
- Starck SR, Tsai JC, Chen K, Shodiya M, Wang L, Yahiro K, Martins-Green M, Shastri N, Walter P. 2016. Translation from the 5′ untranslated region shapes the integrated stress response. Science 351: aad3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Stern-Ginossar N, Thompson SR, Mathews MB, Mohr I. 2018. Translational control in virus-infected cells. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a033001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkin YV, Pause A, Haghighat A, Pyronnet S, Witherell G, Belsham GJ, Sonenberg N. 2001. The requirement for eukaryotic initiation factor 4A (elF4A) in translation is in direct proportion to the degree of mRNA 5′ secondary structure. RNA 7: 382–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szamecz B, Rutkai E, Cuchalova L, Munzarova V, Herrmannova A, Nielsen KH, Burela L, Hinnebusch AG, Valasek L. 2008. eIF3a cooperates with sequences 5′ of uORF1 to promote resumption of scanning by post-termination ribosomes for reinitiation on GCN4 mRNA. Genes Dev 22: 2414–2425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unbehaun A, Borukhov SI, Hellen CU, Pestova TV. 2004. Release of initiation factors from 48S complexes during ribosomal subunit joining and the link between establishment of codon-anticodon base-pairing and hydrolysis of eIF2-bound GTP. Genes Dev 18: 3078–3093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valasek LS. 2012. “Ribozoomin”—Translation initiation from the perspective of the ribosome-bound eukaryotic initiation factors (eIFs). Curr Protein Pept Sci 13: 305–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valasek L, Nielsen KH, Hinnebusch AG. 2002. Direct eIF2-eIF3 contact in the multifactor complex is important for translation initiation in vivo. EMBO J 21: 5886–5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valasek L, Mathew AA, Shin BS, Nielsen KH, Szamecz B, Hinnebusch AG. 2003. The yeast eIF3 subunits TIF32/a, NIP1/c, and eIF5 make critical connections with the 40S ribosome in vivo. Genes Dev 17: 786–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valasek L, Nielsen KH, Zhang F, Fekete CA, Hinnebusch AG. 2004. Interactions of eukaryotic translation initiation factor 3 (eIF3) subunit NIP1/c with eIF1 and eIF5 promote preinitiation complex assembly and regulate start codon selection. Mol Cell Biol 24: 9437–9455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valasek LS, Zeman J, Wagner S, Beznoskova P, Pavlikova Z, Mohammad MP, Hronova V, Herrmannova A, Hashem Y, Gunisova S. 2017. Embraced by eIF3: Structural and functional insights into the roles of eIF3 across the translation cycle. Nucleic Acids Res 45: 10948–10968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventoso I, Sanz MA, Molina S, Berlanga JJ, Carrasco L, Esteban M. 2006. Translational resistance of late αvirus mRNA to eIF2α phosphorylation: A strategy to overcome the antiviral effect of protein kinase PKR. Genes Dev 20: 87–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villa N, Do A, Hershey JW, Fraser CS. 2013. Human eukaryotic initiation factor 4G (eIF4G) protein binds to eIF3c, -d, and -e to promote mRNA recruitment to the ribosome. J Biol Chem 288: 32932–32940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visweswaraiah J, Pittman Y, Dever TE, Hinnebusch AG. 2015. The β-hairpin of 40S exit channel protein Rps5/uS7 promotes efficient and accurate translation initiation in vivo. eLife 4: e07939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von der Haar T, McCarthy JE. 2002. Intracellular translation initiation factor levels in Saccharomyces cerevisiae and their role in cap-complex function. Mol Microbiol 46: 531–544. [DOI] [PubMed] [Google Scholar]
- Wagner S, Herrmannova A, Sikrova D, Valasek LS. 2016. Human eIF3b and eIF3a serve as the nucleation core for the assembly of eIF3 into two interconnected modules: The yeast-like core and the octamer. Nucleic Acids Res 44: 10772–10788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisser M, Voigts-Hoffmann F, Rabl J, Leibundgut M, Ban N. 2013. The crystal structure of the eukaryotic 40S ribosomal subunit in complex with eIF1 and eIF1A. Nat Struct Mol Biol 20: 1015–1017. [DOI] [PubMed] [Google Scholar]
- Weisser M, Schafer T, Leibundgut M, Bohringer D, Aylett CHS, Ban N. 2017. Structural and functional insights into human re-initiation complexes. Mol Cell 67: 447–456.e447. [DOI] [PubMed] [Google Scholar]
- *.Wek RC. 2018. Role of eIF2α kinases in translational control and adaptation to cellular stresses. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a032870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells SE, Hillner PE, Vale RD, Sachs AB. 1998. Circularization of mRNA by eukaryotic translation initiation factors. Mol Cell 2: 135–140. [DOI] [PubMed] [Google Scholar]
- Wortham NC, Martinez M, Gordiyenko Y, Robinson CV, Proud CG. 2014. Analysis of the subunit organization of the eIF2B complex reveals new insights into its structure and regulation. FASEB J 28: 2225–2237. [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Singh CR, Marintchev A, Hall NS, Hannig EM, Wagner G, Asano K. 2005. The eukaryotic initiation factor (eIF) 5 HEAT domain mediates multifactor assembly and scanning with distinct interfaces to eIF1, eIF2, eIF3, and eIF4G. Proc Natl Acad Sci 102: 16164–16169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H, Unbehaun A, Loerke J, Behrmann E, Collier M, Burger J, Mielke T, Spahn CM. 2014. Structure of the mammalian 80S initiation complex with initiation factor 5B on HCV-IRES RNA. Nat Struct Mol Biol 21: 721–727. [DOI] [PubMed] [Google Scholar]
- Yanagiya A, Svitkin YV, Shibata S, Mikami S, Imataka H, Sonenberg N. 2009. Requirement of RNA binding of mammalian eukaryotic translation initiation factor 4GI (eIF4GI) for efficient interaction of eIF4E with the mRNA cap. Mol Cell Biol 29: 1661–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young SK, Wek RC. 2016. Upstream open reading frames differentially regulate gene-specific translation in the integrated stress response. J Biol Chem 291: 16927–16935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng A, Yu J, Yamamoto R, Ose T, Tanaka I, Yao M. 2014. X-ray structures of eIF5B and the eIF5B-eIF1A complex: The conformational flexibility of eIF5B is restricted on the ribosome by interaction with eIF1A. Acta Crystallogr D Biol Crystallogr 70: 3090–3098. [DOI] [PubMed] [Google Scholar]
- Zu T, Liu Y, Banez-Coronel M, Reid T, Pletnikova O, Lewis J, Miller TM, Harms MB, Falchook AE, Subramony SH, et al. 2013. RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc Natl Acad Sci 110: E4968–E4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Zu T, Pattamatta A, Ranum LPW. 2018. RNA translation in neurological diseases. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a033019. [DOI] [PMC free article] [PubMed] [Google Scholar]