Abstract
The enterically transmitted hepatitis A (HAV) and hepatitis E viruses (HEV) are the leading causes of acute viral hepatitis in humans. Despite the discovery of HAV and HEV 40–50 years ago, their evolutionary origins remain unclear. Recent discoveries of numerous nonprimate hepatoviruses and hepeviruses allow revisiting the evolutionary history of these viruses. In this review, we provide detailed phylogenomic analyses of primate and nonprimate hepatoviruses and hepeviruses. We identify conserved and divergent genomic properties and corroborate historical interspecies transmissions by phylogenetic comparisons and recombination analyses. We discuss the likely non-recent origins of human HAV and HEV precursors carried by mammals other than primates, and detail current zoonotic HEV infections. The novel nonprimate hepatoviruses and hepeviruses offer exciting new possibilities for future research focusing on host range and the unique biological properties of HAV and HEV.
Hepatitis A virus (HAV) and hepatitis E virus (HEV) are the most common causes of acute viral hepatitis in humans, infecting millions and causing about 11,000 and 44,000 deaths worldwide, respectively (WHO 2017). HAV and HEV were identified in the early days of hepatitis virus discovery about 40–50 years ago (Feinstone et al. 1973; Balayan et al. 1983). During the route to discovery of HAV, an epidemiological link to animals was suspected (Robertson 2001). However, zoonotic transmission is not known for HAV. In contrast, infections with certain HEV genotypes can be zoonotic in temperate climates (Dalton et al. 2008). However, the majority of HAV and HEV infections in the world are acquired through contaminated water and food (Sattar et al. 2000; Guthmann et al. 2006; Renou et al. 2014).
HAV is classified within the genus Hepatovirus of the family Picornaviridae (Cristina and Costa-Mattioli 2007), whereas HEV is within the genus Orthohepevirus of the family Hepeviridae (Smith et al. 2014). Although these viruses are classified within different virus families, they share unique biological properties. Recent studies reveal that HAV and HEV occur as typical nonenveloped viruses in feces. In contrast, in blood they can exist as lipid-layered particles, challenging the concept that viruses are either enveloped or nonenveloped, and implying special cellular entry mechanisms (Takahashi et al. 2010; Feng et al. 2013).
Nonprimate HEV-related viruses have been identified in even-toed ungulates, rodents, bats, and fish over recent decades (Meng 2010; Drexler et al. 2012b; Smith et al. 2014). In contrast, the existence of nonprimate hepatoviruses was only discovered recently (Drexler et al. 2015). The recent expansion of the spectrum of nonprimate hepatoviruses and hepeviruses enables new insights into the evolutionary history of HAV and HEV.
EXPANSION OF THE KNOWN GENETIC DIVERSITY OF HEPATOVIRUSES AND HEPEVIRUSES
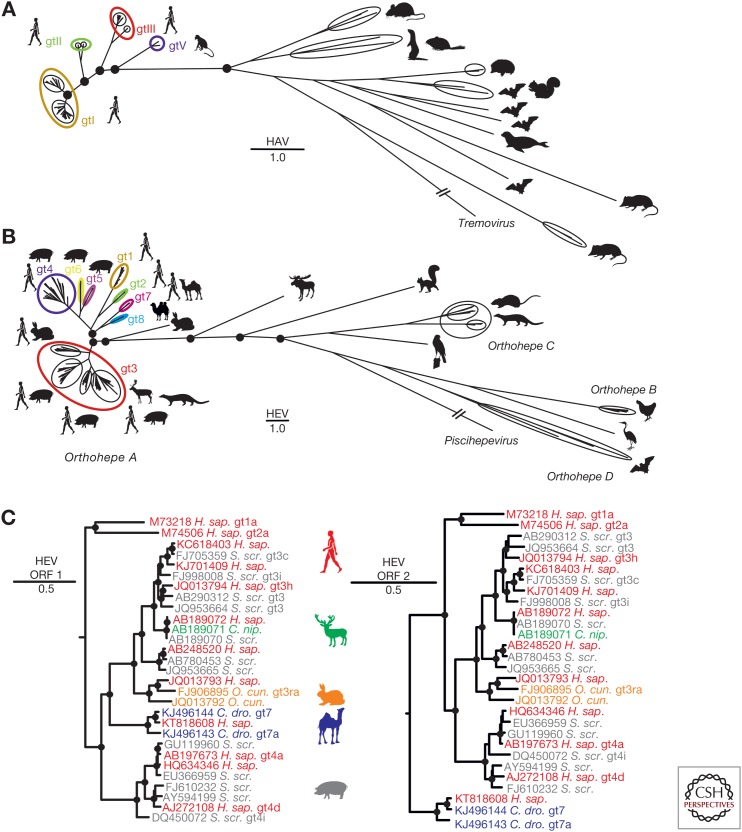
Until recently, the genus Hepatovirus contained only the species HAV comprising six genotypes. Genotypes (gt)I–III infect humans, and gtIV–gtVI were found sporadically in different species of Old World monkeys (Cristina and Costa-Mattioli 2007). As shown in Figure 1A, primate HAVs now represent only a small fraction of the hepatovirus genetic diversity. In 2015, 13 new putative Hepatovirus species were reported from various small mammals, representing >20 different mammalian species and three orders (Drexler et al. 2015). Another two novel hepatoviruses were described from seals in the United States (Anthony et al. 2015) and from Chinese woodchucks (Yu et al. 2016). Finally, another new hepatovirus was identified in a tree shrew during transcriptome sequencing (National Library of Medicine (US) NCBI, 1988b, Accession No. KT_877158.1). The expanded genus Hepatovirus now comprises nine species named Hepatovirus A (HAV) through Hepatovirus I according to the International Committee on the Taxonomy of Viruses (ICTV) (Zell et al. 2017). Another seven putative Hepatovirus species can be predicted using partial genomic data as a surrogate criterion for species delineation (Drexler et al. 2015). These 16 putative virus species were recovered from six different orders of mammalian hosts, including humans and monkeys (order Primates), rodents (Rodentia), a tree shrew (Scandentia), bats (Chiroptera), hedgehogs and shrews (Eulipotyphla), and seals (Carnivora). Despite the hugely increased diversity of the genus, classification of all novel hepatoviruses within a single genus is warranted by considerably larger sequence distances to the next closely related Picornaviridae genus Tremovirus (Drexler et al. 2015) and criteria for picornavirus classification from the ICTV (Zell et al. 2017).
Figure 1.
Genetic diversity of hepatoviruses and hepeviruses and evidence for zoonotic transmission of hepeviruses to humans. (A) Bayesian phylogeny of the full genome of hepatoviruses. A GenBank search with the term “Hepatovirus” was performed on August 3, 2017, and all sequences longer than 6000 nucleotides were selected. Duplicates, cell culture-adapted strains, or viruses isolated from experimentally infected animals were excluded from the dataset, resulting in 124 final hepatitis A virus (HAV) sequences. Complete polyprotein coding sequences were translation aligned, then nonhomologous regions (px/3A) were deleted. HAV genotype (gt)IV and gtVI are defined only by partial sequence information and were, thus, not included in A (Nainan et al. 1991; Robertson et al. 1992). (B) Bayesian phylogeny of the full genome of hepeviruses. A GenBank search using the term “Hepeviridae” was performed on August 4, 2017, and all sequences exceeding 6000 nucleotides were selected. Duplicates, cell culture-adapted strains, or viruses isolated from experimentally infected animals were excluded from the dataset unless they were described as reference sequences according to Smith et al. (2016), resulting in final hepatitis E virus (HEV) 317 sequences. Complete open reading frame (ORF)1 and 2 sequences were concatenated and translationally aligned, and the nonhomologous hypervariable region within ORF1 was deleted. Unique hepeviruses from fox and mink were not included in B because they are only partially sequenced (Bodewes et al. 2013; Krog et al. 2013). Orthohepe, Orthohepevirus. (C) Bayesian phylogenies of the complete ORF1 and 2 of representative human and nonhuman hepeviruses showing zoonotic origins of human HEV. The moose HEV was used as an outgroup (KF951328). All Bayesian phylogenies were generated at the nucleotide level from translation alignments excluding all ambiguous data or gaps using MrBayes V3.1 (Ronquist and Huelsenbeck 2003). A general time-reversible (GTR) model with a γ distribution (G) across sites and a proportion of invariant sites (I) (GTR + G + I) was used as the substitution model. Trees were run for two million generations, sampled every 100 steps. After an exclusion of 5000 of the total 20,000 trees as burn-in, final trees were annotated with TreeAnnotator from the BEAST package (Drummond and Rambaut 2007) and visualized with FigTree. Bayesian posterior probability support above 0.9 at nodes is highlighted by filled circles. The scale bar indicates genetic distance. HEV genotypes (gt) are indicated by arabic numerals and subtypes by letters; HAV genotypes are indicated by roman numerals. H. sap., Homo sapiens; S. scr., Sus scrofa; C. nip., Cervus nippon; O. cun., Oryctulagus cuniculus; C. dro., Camelus dromedarius.
As shown in Figure 1B, the genetic diversity of hepeviruses exceeds the known diversity of hepatoviruses. This is consistent with their classification as a family (note that the scale of the hepevirus phylogeny is about twice that of the hepatovirus phylogeny). As for HAV, hepeviruses infecting humans represent only a small proportion of the known virus diversity. They are classified within a single species termed Orthohepevirus A, which currently comprises eight HEV genotypes. HEV gt1 and gt2 are found in humans only, whereas HEV gt3 and gt4 infect humans, even-toed ungulates (mainly pigs), rabbits, and small carnivores. HEV gt5 and gt6 are associated with pigs and wild boars (Smith et al. 2016), and HEV gt7 and gt8 are associated with camelids (Woo et al. 2014, 2016; Rasche et al. 2016a). As for HAV, a huge diversity of nonprimate viruses defines the genetic space of the family Hepeviridae, including at least three additional Orthohepevirus species and as-yet-unclassified hepeviruses from a moose, falcon, and tree shrew (National Library of Medicine (US) NCBI, 1988a, Accession No. KR_905549.1; Lin et al. 2014; Reuter et al. 2016). In sum, unique orthohepeviruses have been found in eight distinct mammalian host orders, including several orders in which hepatoviruses have also been found (Primates, Scandentia, Rodentia, Carnivora, Chiroptera), as well as even-toed ungulates including pigs, camels, deer, cattle, swine, moose (Artiodactyla), rabbits (Lagomorpha), and birds (Aves). A sister genus termed Piscihepevirus is defined by a trout hepevirus (Batts et al. 2011; Smith et al. 2014).
THE DIFFERENT RELEVANCE OF ANIMAL RESERVOIRS FOR HUMAN INFECTIONS
Despite the diversity of animal hepatoviruses, so far no zoonotic transmission of HAV has been reported. In contrast, the animal sources of human infections with HEV are diverse. As shown in Figure 1C, there is considerable evidence of zoonotic transmission for HEV gt3, gt4, and gt7, as indicated by viruses from humans grouping within viruses recovered from animals.
Major animal reservoirs of HEV gt3 and gt4 include domestic pigs and wild boars (Meng 2010). A number of human HEV infections could be linked to the consumption of pork meat and its products (Yazaki et al. 2003; Li et al. 2005; Colson et al. 2010; Kim et al. 2011; Renou et al. 2014; Riveiro-Barciela et al. 2015). Of note, for areas in which mixed farming of livestock species, including pigs, is a common practice (e.g., China), gt4 infection of other livestock species (such as sheep and cattle) were described. In such a scenario, virus shedding into milk was reported in cows infected with HEV gt4. These data may point to milk as a putative additional source of human food-borne HEV infections (Wu et al. 2015; Huang et al. 2016).
Zoonotic transmission of gt3 can also involve recently identified viruses from rabbits (Zhao et al. 2009; Cossaboom et al. 2012; Izopet et al. 2012). Cross-species transmission of HEV between deer and humans was reported in 2003 from Japan (Tei et al. 2003). Further studies suggested initial virus transmission between boar and deer (Takahashi et al. 2004).
Epidemiologic data are scarce for the recently identified HEV gt7 and gt8 recovered from camels. However, HEV infection in an immunocompromised patient was linked to the consumption of dromedary camel milk and meat in the United Arab Emirates (Lee et al. 2016). Because camels are a major livestock species in regions in which there is little or no rearing of swine (Bourn 2003; Kadim et al. 2013; Rasche et al. 2016a), they may be important regional sources of zoonotic infections. This hypothesis is consistent with the role of camels as sources of human infections with Middle East Respiratory Syndrome (MERS) coronavirus (Reusken et al. 2013; Memish et al. 2014) and the existence of close relatives of other human coronaviruses in camels (Corman et al. 2016).
GENOMIC VARIABILITY OF HEPATOVIRUSES AND HEPEVIRUSES
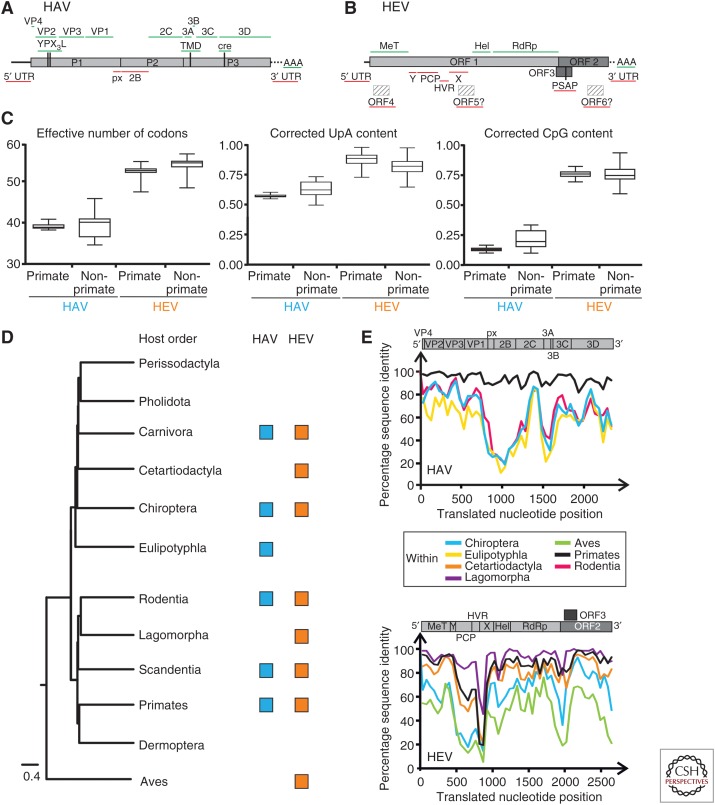
As shown in Figure 2A, nonprimate and primate HAV generally share the same genome organization. Conserved genomic properties (shown above the genome plot) include the absence of a leader (L) protein, YPX3L late domain motifs in VP2 likely involved in quasi-envelope acquisition (Feng et al. 2014), as well as the existence of a predicted transmembrane domain (TMD) in the 3A domain (Beneduce et al. 1997) and a cis-acting replication element (cre) in RNA–encoding the 3Dpol domain (Yang et al. 2008). Untranslated regions (UTRs) at the genome termini are generally variable in length but share characteristic features such as suggestive internal ribosome entry site (IRES) structures within the 5′UTR and a 3′ terminating poly(A) tail. However, the type of IRES may not be conserved among different hepatoviruses. Human HAVs have type III IRESs (Brown et al. 1991). As described previously, certain bat and rodent hepatoviruses contain sequence elements characteristic of type IV IRESs, presumably acquired by ancient recombination events involving different viral families (Drexler et al. 2015). Distinct from other picornaviruses, the VP1 capsid protein of HAV has a carboxy-terminal extension termed pX (often referred to as 2A), which is involved in capsid assembly and possibly quasi-envelopment (Cohen et al. 2002; Feng et al. 2014). Interestingly, this unique genomic feature could not be unambiguously identified in some of the recently found bat hepatoviruses (Drexler et al. 2015), raising new questions about the role and origin of pX in hepatoviruses.
Figure 2.
Genomic variability among hepatoviruses and hepeviruses. Genome organization showing conservation of putative functional domains within (A) hepatoviruses, and (B) hepeviruses. Conserved domains (green) are depicted above, and nonconserved domains (red) are depicted below graphs. (C) Relative CpG, UpA dinucleotide content and effective number of codons in hepatoviruses and hepeviruses calculated using SSE 1.3 software (Simmonds 2012). Median (bar) and quartiles (box and whiskers) are shown. (D) Phylogenetic relationships of boreoeutherian vertebrate orders, including an avian outgroup. (Phlyogeny adapted from Foley et al. 2016.) Squares indicate vertebrate orders in which hepatoviruses or orthohepeviruses were found. (E) Amino acid sequence identities within hepatoviruses (top) and hepeviruses (bottom) of different host orders. Generally, representative viruses from each host order were tagged and sequence identities within families were plotted using a fragment length of 400 and a step size of 200 amino acid residues. Alignment gaps were excluded from the analysis. A schematic representation of the hepatitis A virus (HAV)/hepatitis E virus (HEV) genome organization is depicted at the top for orientation. For HAV, complete coding sequences of the polyproteins were translationally aligned. Accession numbers of representative sequences were, within Primates: AB020564, AY644676, AB279732, D00924; Chiroptera: KT452742, KT452730, KT452729, KT452714; Rodentia: KT452735, KT452685, KT229611, KT452644, KT452637; Eulipotyphla: KT452691, KT452658. For HEV, the complete open reading frame (ORF)1 and ORF2 were concatenated and translationally aligned. ORF3 is only shown for indication of its position. Accession numbers of representative sequences were, within Lagomorpha: FJ906895, KJ013415; Primates: M73218, M74506, AP003430, AB197673; Cetartiodactyla: AF082843, AB189071, AB573435, AB602441, KF951328, KJ496143, KX387865; Chiroptera: JQ001749, KJ562187, KX513953; Aves: KX589065, KU670940, AY535004. Hel, Helicase; HVR, hypervariable region; MT, methyltransferase; PCP, papain-like cysteine protease; RdRp, RNA-dependent RNA polymerase; TMD, transmembrane domain; UTR, untranslated region; X, X domain/ADP-ribose-binding module; Y, Y-like domain.
As shown in Figure 2B, all nonhuman hepeviruses share the general genome organization of human HEV, and have at least three different open reading frames (ORFs). Additional ORFs that overlap with ORF1 have been hypothesized in nonhuman hepeviruses (Johne et al. 2010, 2012; Drexler et al. 2012b). However, the functional activity of these putative ORFs remains to be confirmed.
Similar to HAV, several genomic properties are conserved among hepeviruses (shown above the genome plot). Within ORF1, the domains encoding the putative methyltransferase (MeT), helicase (Hel), and RNA-dependent RNA polymerase (RdRp) (Koonin et al. 1992; Rozanov et al. 1992) can be unambiguously identified in all hepeviruses. However, none of the other putative ORF1 domains described for HEV (see Kenney and Ming 2018) can be unambiguously identified in all of the nonhuman hepeviruses, including the X (ADP-ribose-1′-monophosphatase-like domain) and Y (unknown function) domains, and the predicted papain-like cysteine protease (PCP) domain (shown below the genome plot). Although an ORF3 can be predicted in all hepeviruses, this ORF varies considerably in size. For example, the ORF3 of the trout hepevirus defining the genus Piscihepevirus contains 226 translatable amino acid (aa) residues, whereas the predicted ORF3 of the tree shrew hepevirus may only contain 62 aa residues. Whether all predicted hepevirus ORF3 have similar functions, thus, remains to be confirmed. It was previously reported that the ORF3 of human and avian hepeviruses contains at least one PSAP “late domain” motif potentially involved in hepevirus envelopment and cell egress (Surjit et al. 2006; Nagashima et al. 2011; Feng et al. 2014; Kenney et al. 2015). Interestingly, a P(S/T)AP motif was not identifiable in the ORF3 of tree shrew, bat, rat, falcon, and trout hepeviruses. In some of these viruses, a PSAP motif can instead be found within the ORF1. However, the hepeviruses identified in a tree shrew, Rhinolophus bat, and a rat contain neither an identifiable P(S/T)AP nor other late-domain motifs such as YPX3L, PPXY, SDP, or GPPX3Y in any ORF (summarized in Ren and Hurley 2011). Whether other motifs involved in cell egress exist in these viruses and how divergent hepeviruses form quasi-enveloped particles facilitating cell egress in general is, thus, an open question.
CONSERVED CODON USAGE PATTERNS OF HEPATOVIRUSES AND HEPEVIRUSES
Codon usage varies among RNA viruses and is one of distinctive features of certain taxonomic groups. It is largely a product of pressure on the genomic nucleotide and dinucleotide content, rather than pressure to use abundant transfer RNAs (tRNAs) (Jenkins and Holmes 2003; Belalov and Lukashev 2013). The extent of codon usage bias can be approximated by a single number termed the effective number of codons (ENCs) (Wright 1990). The maximum possible value is 61 (all codons used equally), and the lowest possible is 20 (one codon used per each aa). HAV has been well known for very low ENC values (Pintó et al. 2007; Lukashev et al. 2012). This was a conserved genomic feature of the expanded genus Hepatovirus, because all nonprimate hepatoviruses also had relatively low ENC values, although the variation was greater than among primate HAVs (Fig. 2C).
Codon usage bias in HAV has been attributed to a specific replication strategy aimed at avoiding virus protein overexpression triggering an immune response (Pintó et al. 2007) or avoidance of specific dinucleotides, namely, CpG and UpA (Karlin et al. 1994; Rima and McFerran 1997; see Pintó et al. 2018). These dinucleotides are underrepresented in many mammalian RNA viruses, but to a variable extent (Jenkins and Holmes 2003; Belalov and Lukashev 2013). Depletion of CpG was attributed to pressure from a recently discovered zinc-finger antiviral protein (ZAP) (Takata et al. 2017), whereas UpA may be a target of antiviral ribonuclease (RNase) L (Player and Torrence 1998). HAV is known to have an extremely low relative CpG content (even after correction for genomic C and G content), typically between 0.1 and 0.16 of the statistically expected value (Fig. 2C). The avoidance of CpG is conserved within nonprimate hepatoviruses, although it is slightly less pronounced than in HAV (median 0.2, maximum 0.33). UpA dinucleotides are moderately underrepresented in the genomes of both HAV and nonprimate hepatoviruses (Fig. 2C). Therefore, codon usage bias and dinucleotide content bias, putatively related to interactions of the virus with host cell innate immune responses, are similar in all hepatoviruses.
In contrast, HEV and all other hepeviruses have a low codon usage bias, as is evident from their high ENC value, a minimal CpG bias with typical values around 0.75, and almost no UpA bias (Fig. 2C). Therefore, hepevirus genomes are apparently not subjected to the same pressure as hepatovirus genomes. The different mechanisms by which hepatoviruses and hepeviruses evade intracellular immunity, thus, require further investigation.
HOT SPOTS OF VARIABILITY WITHIN HEPATOVIRUS AND HEPEVIRUS GENOMES
Nonprimate HAV and HEV are now known to exist in several mammalian orders (Fig. 2D). Surprisingly, patterns of genetic variability in these newly discovered hepatitis viruses mirror those among HAV and HEV despite the genetic differences of their hosts. In hepatoviruses, the level of sequence identity within different host orders was generally lowest in the genomic region encompassing the pX-2B domains (Fig. 2E). The 2B domain of hepatoviruses infecting chiropteran, rodent, and eulipotyphlan hosts did not show any homology with other nonprimate hepatoviruses, and may represent a hot spot for acquisition of heterologous sequence from unknown sources, including host genetic material (Emerson et al. 1992; Beard et al. 2001; Drexler et al. 2015). Also, a distinct decrease in sequence identity occurred in the genomic region encompassing the 3A domain. Because 3A is a membrane protein (Beneduce et al. 1997), the decrease in sequence similarity may be tentatively linked to host-specific viral adaptation processes.
Similar to hepatoviruses, a genomic hot spot evident as a sharp decrease in sequence identity can be identified in hepeviruses. As shown in Figure 2E, hepevirus genera have little sequence identity (and no obvious homology) in the putative protease and hypervariable regions (HVRs) of ORF1. Again, this may hint at heterologous acquisitions of genetic material from unknown sources, because BLAST searches found this genome region to be neither homologous to any known sequence, nor to contain any identifiable functional domain. Indeed, the possibility of such external acquisitions was shown experimentally, when a fragment of 174 nucleotides of a human messenger RNA (mRNA) became inserted into the HEV HVR during adaptation of an HEV strain to replication in cell culture (Shukla et al. 2011). Interestingly, insertions within the HVR have also been tentatively associated with the failure of ribavirin treatment, the standard treatment for immunocompromised patients chronically infected with HEV (Debing et al. 2016; Todt et al. 2016). Nonhomologous proteins encoded by otherwise closely related viruses are often involved in suppression of innate antiviral mechanisms (Agol and Gmyl 2010). Indeed, the protease domain of HEV was suggested as an interferon antagonist (Nan et al. 2014). Whether these highly diverse genome regions have convergent functions is, thus, an intriguing question.
THE ROLE OF RECOMBINATION IN THE EVOLUTION OF HAV AND HEV
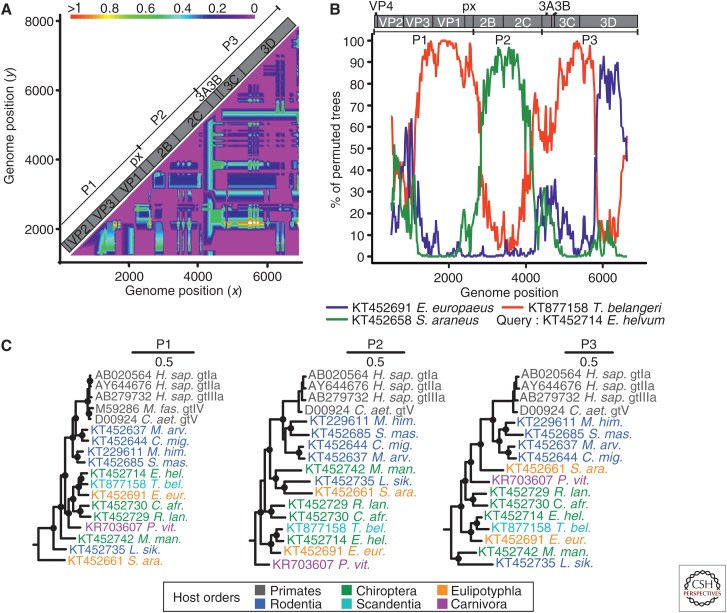
Recombination has been reported in HAV, both in cell culture (Lemon et al. 1991) and in vivo (Colina et al. 2004; Belalov et al. 2011). Recombination can also be detected in nonprimate hepatoviruses. Compatibility matrix analysis of distantly related hepatoviruses suggested several gene transfers (Fig. 3A). A bootscan analysis confirmed the mosaic nature of a nonprimate hepatovirus from Eidolon helvum bats and revealed typical picornavirus breakpoints mapping roughly to the borders of the P1, P2, and P3 domains, as well as a predicted breakpoint within the 3Dpol domain (Fig. 3B). These recombination events are also clearly evident from conflicts in phylogenetic trees of these three major hepatovirus polyprotein domains (Fig. 3C). Importantly, the primate HAVs do not appear to have been involved in recombination with other animal hepatoviruses. Multiple instances of recombination involving viruses that were found in distantly related hosts, on different continents, suggest common non-recent host shifts (or, at least, spillover infections) during hepatovirus evolution. Of note, the evidence for such ancestral recombination events (which require a host switch by one of the parental viruses) is consistent with the lack of high-order cosegregation of the viral and host phylogenies (Figs. 1A and 2D).
Figure 3.
Evidence of recombination in hepatoviruses. (A) Phylogenetic compatibility scan of the full polyprotein genes of hepatoviruses isolated from humans, rodents, tree shrew, and bats. GenBank Accession numbers of sequences used were: AB279735; KT452644; KT452685; KT452729; KT877158; KT452742; KT452735. The graph was created using SSE 1.3 (Simmonds 2012), a sliding window of 500 nt, and a step size of 50 nt, with a bootstrap cutoff of 70%. (B) The bootscan graph shows percent of bootstrap replicates (y axis) that support grouping of the query sequence with each of three test sequences in a 1000 nt window sliding over the genome at a 20 nt step (x axis, window center position); the plot was done using Simplot 3.5 (Lole et al. 1999) and the Kimura substitution model; dotted line shows the 70% reliable bootstrap support cutoff. As a result of alignment shifts, the genome plots do not precisely correspond to raw genome positions. (C) Bayesian phylogenies of hepatovirus domains P1, P2 (only 2C), and P3 (only 3CD) showing reliable evidence of several recombination events. Viruses are colored according to their host order. Phylogenies were generated at the amino acid level from translation alignments excluding all ambiguous data or gaps using MrBayes V3.1 (Huelsenbeck and Ronquist 2001) and a WAG amino acid substitution model. Trees were generated as described above and rooted by the avian encephalomyelitis virus (genus Tremovirus). Bayesian posterior probabilities above 0.9 are marked by filled circles at nodes. The scale bar indicates genetic distance. M. fas., Macaca fascicularis; C. aet., Chlorocebus aethiops; M. arv., Microtus arvalis; C. mig., Cricetulus migratorius; M. him., Marmota himalayana; S. mas., Sigmodon mascotensis; E. hel., Eidolon helvum; T. bel., Tupaia belangeri chinensis; E. eur., Erinaceus europeaensis; C. afr., Coleura afra; R. lan., Rhinolophus landeri; P. vit., Phoca vitulina; M. man., Miniopterus cf. manavi; L. sik., Lophuromys sikapusi; S. ara., Sorex araneus.
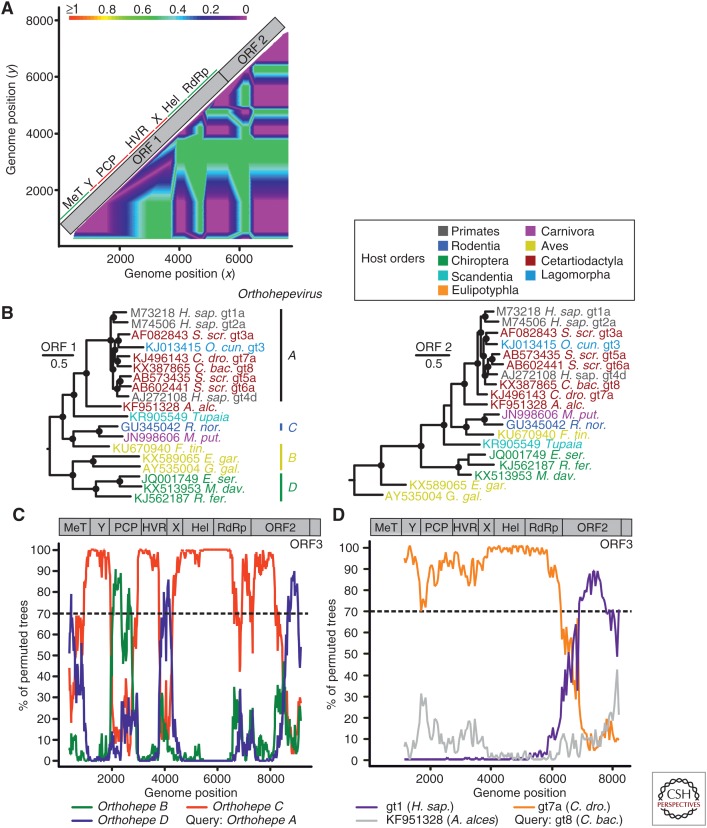
Recombination in HEV has been reported both within a genotype (van Cuyck et al. 2005) and between different genotypes (van Cuyck et al. 2005; Chen et al. 2012). Moreover, it has been suggested that the emergence of the family Hepeviridae involved recombination between Alphavirus-like viruses, which provided replication complex proteins, and astrovirus-related viruses, which provided structural proteins (Kelly et al. 2016). Compatibility matrix analysis suggests several recombination events within orthohepeviruses (Fig. 4A). The major phylogenetic grouping incongruences are observed in the papain-like cysteine protease domain and proline-rich genome region, and may result from nonhomologous recombination events with unknown partners. Phylogenies of the full ORF1 and 2 sequences are in general congruent, except for the camel hepevirus forming HEV gt7 (Fig. 4B). However, a bootscan analysis suggests several recombination events throughout the genome among orthohepeviruses A–D (Fig. 4C). The strong evidence for a recombinant origin of the camelid-associated HEV gt7 may point to a large diversity of parental viruses likely circulating in camels. Recombination data obtained for gt7 suggest that the ORF1 encoding nonstructural proteins may acquire diverse capsids from unknown recombination partners, similar to what has been observed in human parechoviruses (Drexler et al. 2011). This hypothesis will require investigations of more complete gt7 hepeviruses sampled from diverse geographic regions (Woo et al. 2014; Rasche et al. 2016a). Of note, phylogenetic signals in RNA viruses generally degrade over time because of high sequence variation and saturation of mutations. The fact that these recombination events between distinct hepevirus species remain detectable nonetheless implies that they involved highly divergent viruses. This is in contrast with a hypothetical emergence of HEV ancestors from a population of recombinant closely related viruses such as the camelid-associated HEV gt7 and gt8. From an ecological perspective, such recombination events indicate that the replication of orthohepeviruses may not be tightly restricted to their host species, as recombination requires coreplication of two highly divergent viruses in the same cell.
Figure 4.
Evidence of recombination in hepeviruses. (A) Phylogenetic compatibility scan of concatenated open reading frame (ORF)1 + ORF2 alignment of hepeviruses isolated from human (X98292), tree shrew (KR905549), Falco tinnunculus (KU670940), Rattus rattus (AB847306), chicken (KF511397), Eptesicus serotonius (JQ001749), and Rhinolophus ferrumequinum (KJ562187) was generated as described in the legend to Figure 3A. (B) Bayesian phylogenies of complete ORF1 and 2 of orthohepeviruses. Viruses are colored according to their host order. Phylogenies were generated at the nucleotide level from translation alignments excluding all ambiguous data or gaps using MrBayes V3.1. A general time-reversible (GTR) model with a γ distribution (G) across sites and a proportion of invariant sites (I) (GTR + G + I) was used as the substitution model. Trees were generated as above. Trees were rooted by the sister genus Piscihepevirus. Bayesian posterior probabilities above 0.9 are marked by filled circles at nodes. The scale bar indicates genetic distance. C. bac., Camelus bactrianus; A. alc., Alces alces; R. nor., Rattus norvegicus; M. put., Mustela putorius furo; F. tin., Falco tinnunculus; E. gar., Egretta garzetta; E. ser., Eptesicus serotinus; G. gal., Gallus gallus; M. dav., Myotis davidii; R. fer., Rhinolophus ferrumequinum. (C) Evidence of recombination in the species Orthohepevirus A relative to species B–D. The scan was performed similarly as described in the legend for Figure 3B. Window size was 800 nt, step size at 20 nt. Accession numbers of sequences used: JQ013793; JN998606; AY535004; KJ562187. As a result of alignment shifts, the genome plots do not precisely correspond to raw genome positions. Orthohepe, Orthohepevirus. (D) Evidence of recombination in species Orthohepevirus A genotype (gt) 8 (C. bactrianus) relative to gt1 (Homo sapiens), gt7a (Camelus dromedarius), and the moose (A. alces) hepevirus. Window size was 2500 nt, step size 50 nt. Accession numbers of sequences used: KX387865; KJ496143; X98292; KF951328. ORF1–ORF3 were concatenated for bootscan analyses, as indicated by genomic representations above panels C and D. Hel, Helicase; HVR, hypervariable genome region; PCP, papain-like cysteine protease.
EVOLUTION OF BAT HEPATOVIRUSES AND HEPEVIRUSES
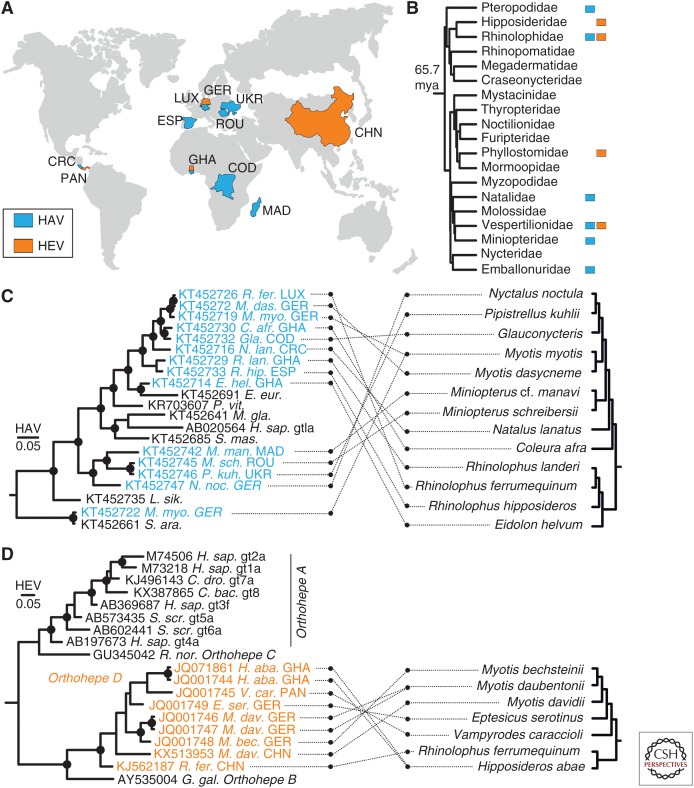
Recombination analysis and overall tree topology suggest frequent host shifts during both hepatovirus and hepevirus evolution. Bats represent an old mammalian order and are among the most speciose mammals with at least 1200 extant species (Teeling et al. 2005). These animals have been ascribed a prominent role for virus evolution, in general, and zoonotic viruses, in particular (Olival et al. 2017). Therefore, we analyzed bat-borne hepatoviruses and hepeviruses in the context of the evolutionary history of their hosts.
Strikingly, although bat hepatoviruses and hepeviruses were identified on different continents and in different host families (Fig. 5A,B), many of them are still monophyletic, suggesting a common viral ancestor (Fig. 5C,D). Given the diversity of hosts in other Orthohepevirus species and the evidence for frequent host shifts in hepatoviruses, this monophyletic clustering is surprising and may hint at a long-term association of specific clades (for hepatoviruses) or even virus species (for orthohepeviruses) with the order Chiroptera.
Figure 5.
Global distribution and phylogenetic relationships of bat hepatoviruses and hepeviruses. (A) Map showing the geographic origins of recently identified bat hepatoviruses (blue) and bat hepeviruses (orange). CHN, China; COD, Democratic Republic of the Congo; CRC, Costa Rica; ESP, Spain; GER, Germany; GHA, Ghana; LUX, Luxembourg; MAD, Madagascar; PAN, Panama; ROU, Romania; UKR, Ukraine. (B) Chiropteran phylogeny (phylogeny adapted from Foley et al. 2016) complemented manually by the family of Miniopteridae, which diverged around 43 million years ago (mya) from the Vespertilionidae (Miller-Butterworth et al. 2007). Bat families in which hepatitis A virus (HAV)- or hepatitis E virus (HEV)-related viruses have been found are tagged with blue or orange squares, respectively. (C) Bayesian phylogeny of a 863-nucleotide partial VP2/VP3 region of bat and representative nonchiropteran hepatoviruses. Analyzed region corresponds to positions 391–1253 in a prototype genotype (gt) Ia HAV strain (GenBank AB020564). M. gla., Myodes glareolus. See also the legend to Figure 3. (D) Bayesian phylogeny of a 324-nucleotide partial RNA-dependent RNA polymerase (RdRp) region of bat and representative nonchiropteran hepeviruses. The analyzed region corresponds to positions 4255–4577 in an HEV gt1 prototype strain (GenBank accession number M73218). Generally, Bayesian phylogenies were generated on translation alignments excluding all ambiguous data or gaps using MrBayes V3.1 (Huelsenbeck and Ronquist 2001) and a WAG amino acid substitution matrix. Trees were rooted by Tremovirus for HAV and Piscihepevirus for HEV, respectively. Bayesian posterior probabilities above 0.9 are marked by filled circles at nodes. The scale bar indicates genetic distance. Host trees were generated using complete cytochrome B coding sequences retrieved from GenBank and settings as described for virus phylogenies, including priors, to increase phylogenetic resolution above the level of host families.
There are several possible explanations for these findings. First, the monophyly of bat viruses could hint at cospeciation of ancestral viruses and bat hosts. However, this concept is currently subject to question even for taxa assumed to be prototypic examples of virus and host coevolution, such as the gammaherpesviruses (Escalera-Zamudio et al. 2016). In the particular case of bat hepato- and hepeviruses, virus–host coevolution is not consistent with conflicting phylogenetic clustering of host species and the corresponding viruses (Fig. 5C,D). Furthermore, the relatively short branches in viral phylogenies are difficult to reconcile with the formation of extant bat families, projected to 10–64 million years ago (Eick et al. 2005; Teeling et al. 2005) and their non-recent geographical dispersal.
Second, bat viruses may have been acquired from bridging hosts other than bats. However, for both virus families, the phylogenies do not suggest the existence of bridging hosts.
Third, bat populations may be connected and share viruses among each other, similarly to what has been hypothesized for European and Asian picorna- and coronaviruses (Lukashev et al. 2017). Connectivity over large distances may be facilitated by the ability of bats to fly and, indeed, some bat species are migratory. However, most bats only migrate across short distances rarely exceeding 100 km. Only very few species, for example, the African fruit bat E. helvum, some European bats (e.g., Pipistrellus leiseri, Nyctalus noctula), and some American bats (Leptonycteris species, Tadarida brasiliensis) can cover distances of up to 2000 km (Kunz and Fenton 2005). Most importantly, such migration normally does not include transoceanic flights. Additionally, there is no regular meeting site for bats from different continents, contrasting with birds meeting in certain wintering or breeding sites, where global exchange of viruses can take place (Global Consortium for H5N8 and Related Influenza Viruses 2016). As an alternative hypothesis, bats can be moved across continents accidentally through human activity (Constantine 2003), facilitating virus exchange with bats in new habitats. However, given the low rate of acute infections in bats (Drexler et al. 2012b, 2015), it is not likely that a translocated bat would shed virus and transmit it to other bats.
A similar, albeit less strict phylogenetic association between monophyletic clades of viruses infecting bats from different continents has been observed for Old World hepadnaviruses (Rasche et al. 2016b), suggesting that the observed association between bats and enteric hepatitis viruses may not be an isolated ecological phenomenon. In sum, the reasons for the striking association of bats with distinct hepatovirus and hepevirus clades remain obscure. Nonetheless, the observed phylogenies are consistent with non-recent host–virus associations in bat hepatovirus and hepeviruses.
CONCLUDING REMARKS
The recent discovery of previously unrecognized animal hepatitis viruses significantly expands the scope of prospective virological studies. Immediate applications for HAV include investigation of the role of nonprimate hepatovirus pX domains in capsid assembly, and the search for HAV and HEV receptors, because recent evidence suggests that HAVcr1/TIM-1 is not a canonical receptor for HAV (Das et al. 2017). For both HAV and HEV, it is intriguing to consider whether quasi-envelopment is an evolutionarily conserved feature. Recent evidence for nonenveloped hepatitis B–related viruses in fish reveal that envelopment can emerge or be lost during the adaptation of viruses to different vertebrate hosts (Lauber et al. 2017). The absence of identifiable late domains in several hepeviruses suggests that additional features facilitating quasi-envelopment may exist, or envelope acquisition may not be conserved among all hepeviruses.
Our data suggest that cross-species transmissions have occurred repeatedly during the evolution of ancestors of the enterically transmitted hepatitis viruses. Cell culture and infection experiments using viral isolates, once these become available, and recombinant viruses may provide fundamental insight concerning the barriers to cross-species transmission of hepatoviruses and hepeviruses. For the hepatoviruses, cleavage of mitochondrial antiviral signaling protein (MAVS) as a major marker of viral immune evasion and host specificity (Hirai-Yuki et al. 2016) merits immediate investigation. Finally, these viruses offer new possibilities for the development of animal and cell culture models, thereby allowing new insight into the immune control and pathogenesis of the enterically transmitted hepatitis viruses.
The expanded genetic diversity of enterically transmitted hepatitis viruses and their hosts suggests that the emergence of HAV and HEV in humans is a relatively recent evolutionary event. In both cases, a zoonotic origin seems likely, although the immediate ancestors of human viruses remain unknown. Bats have been implicated as particularly relevant animal reservoirs of zoonotic viruses (Luis et al. 2013; Olival et al. 2017). Whereas the available data suggest that bats can host relatives of enterically transmitted hepatitis viruses, bat viruses are not likely direct ancestors of human viruses, a situation that is strikingly different from other human viruses, including paramyxo- and coronaviruses (Drexler et al. 2012a, 2014). For HAV, ancestors of human viruses have been projected to rodents (Drexler et al. 2015). For HEV, the role of small mammals, including rodents, requires further investigation because the Orthohepevirus species—including hepeviruses from humans and rodents—are monophyletic, suggesting common ancestry.
We have hypothesized previously that human HEV gt3 and gt4 may have been introduced into livestock from humans, and human infections may be a retrograde event (Drexler et al. 2012b). This line of thought is compatible with the high genetic diversity of HEV gt1–gt4 in humans and the finding of a distantly related virus in moose (Lin et al. 2014), which may represent an authentic artiodactylan hepevirus lineage other than those viruses infecting swine and other ungulates. However, this hypothesis is at odds with the diversity of HEV genotypes in wild boars and camels, and the existence of diverse rabbit HEV lineages. Hypothetically, small mammals, such as rodents and lagomorphs, may have evolved hepevirus ancestors that were introduced into artiodactyl species reared as livestock, and that may now act as sources of human infections. This scenario is paralleled by the emergence of MERS-coronavirus in camels directly or through intermediate hosts from ancestors existing in bats (Corman et al. 2014, 2016) and, potentially, SARS-coronavirus through bats and civets (Drexler et al. 2014).
In sum, recent insight into the genetic diversity that exists among the enterically transmitted hepatitis viruses reveals that the emergence of both HAV and HEV is likely to have been a relatively recent evolutionary event. The repertoire of nonprimate homologs of HAV and HEV offers unprecedented possibilities to unravel fundamental biological properties defining these unique pathogens in the coming years.
Footnotes
Editors: Stanley M. Lemon and Christopher Walker
Additional Perspectives on Enteric Hepatitis Viruses available at www.perspectivesinmedicine.org
REFERENCES
*Reference is also in this collection.
- Agol VI, Gmyl AP. 2010. Viral security proteins: Counteracting host defences. Nat Rev Microbiol 8: 867–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony SJ, St Leger JA, Liang E, Hicks AL, Sanchez-Leon MD, Jain K, Lefkowitch JH, Navarrete-Macias I, Knowles N, Goldstein T, et al. 2015. Discovery of a novel hepatovirus (Phopivirus of seals) related to human hepatitis A virus. MBio 6: e01180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balayan MS, Andzhaparidze AG, Savinskaya SS, Ketiladze ES, Braginsky DM, Savinov AP, Poleschuk VF. 1983. Evidence for a virus in non-A, non-B hepatitis transmitted via the fecal-oral route. Intervirology 20: 23–31. [DOI] [PubMed] [Google Scholar]
- Batts W, Yun S, Hedrick R, Winton J. 2011. A novel member of the family Hepeviridae from cutthroat trout (Oncorhynchus clarkii). Virus Res 158: 116–123. [DOI] [PubMed] [Google Scholar]
- Beard MR, Cohen L, Lemon SM, Martin A. 2001. Characterization of recombinant hepatitis A virus genomes containing exogenous sequences at the 2A/2B junction. J Virol 75: 1414–1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belalov IS, Lukashev AN. 2013. Causes and implications of codon usage bias in RNA viruses. PLoS ONE 8: e56642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belalov IS, Isaeva OV, Lukashev AN. 2011. Recombination in hepatitis A virus: Evidence for reproductive isolation of genotypes. J Gen Virol 92: 860–872. [DOI] [PubMed] [Google Scholar]
- Beneduce F, Ciervo A, Morace G. 1997. Site-directed mutagenesis of hepatitis A virus protein 3A: Effects on membrane interaction. Biochim Biophys Acta 1326: 157–165. [DOI] [PubMed] [Google Scholar]
- Bodewes R, van der Giessen J, Haagmans BL, Osterhaus AD, Smits SL. 2013. Identification of multiple novel viruses, including a parvovirus and a hepevirus, in feces of red foxes. J Virol 87: 7758–7764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourn D. 2003. Livestock dynamics in the Arabian peninsula: A regional review of national livestock resources and international livestock trade. Environmental Research Group Oxford Limited, Oxford, UK. [Google Scholar]
- Brown EA, Day SP, Jansen RW, Lemon SM. 1991. The 5′ nontranslated region of hepatitis A virus RNA: Secondary structure and elements required for translation in vitro. J Virol 65: 5828–5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Zhang Q, He C, Zhang L, Li J, Zhang W, Cao W, Lv YG, Liu Z, Zhang JX, et al. 2012. Recombination and natural selection in hepatitis E virus genotypes. J Med Virol 84: 1396–1407. [DOI] [PubMed] [Google Scholar]
- Cohen L, Benichou D, Martin A. 2002. Analysis of deletion mutants indicates that the 2A polypeptide of hepatitis A virus participates in virion morphogenesis. J Virol 76: 7495–7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colina R, Casane D, Vasquez S, Garcia-Aguirre L, Chunga A, Romero H, Khan B, Cristina J. 2004. Evidence of intratypic recombination in natural populations of hepatitis C virus. J Gen Virol 85: 31–37. [DOI] [PubMed] [Google Scholar]
- Colson P, Borentain P, Queyriaux B, Kaba M, Moal V, Gallian P, Heyries L, Raoult D, Gerolami R. 2010. Pig liver sausage as a source of hepatitis E virus transmission to humans. J Infect Dis 202: 825–834. [DOI] [PubMed] [Google Scholar]
- Constantine DG. 2003. Geographic translocation of bats: Known and potential problems. Emerg Infect Dis 9: 17–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman VM, Ithete NL, Richards LR, Schoeman MC, Preiser W, Drosten C, Drexler JF. 2014. Rooting the phylogenetic tree of Middle East respiratory syndrome coronavirus by characterization of a conspecific virus from an African bat. J Virol 88: 11297–11303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman VM, Eckerle I, Memish ZA, Liljander AM, Dijkman R, Jonsdottir H, Juma Ngeiywa KJ, Kamau E, Younan M, Al Masri M, et al. 2016. Link of a ubiquitous human coronavirus to dromedary camels. Proc Natl Acad Sci 113: 9864–9869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossaboom CM, Cordoba L, Cao D, Ni YY, Meng XJ. 2012. Complete genome sequence of hepatitis E virus from rabbits in the United States. J Virol 86: 13124–13125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristina J, Costa-Mattioli M. 2007. Genetic variability and molecular evolution of hepatitis A virus. Virus Res 127: 151–157. [DOI] [PubMed] [Google Scholar]
- Dalton HR, Bendall R, Ijaz S, Banks M. 2008. Hepatitis E: An emerging infection in developed countries. Lancet Infect Dis 8: 698–709. [DOI] [PubMed] [Google Scholar]
- Das A, Hirai-Yuki A, González-López O, Rhein B, Moller-Tank S, Brouillette R, Hensley L, Misumi I, Lovell W, Cullen JM, et al. 2017. TIM1 (HAVCR1) is not essential for cellular entry of either quasi-enveloped or naked hepatitis A virions. MBio 8: e00969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debing Y, Ramiere C, Dallmeier K, Piorkowski G, Trabaud MA, Lebosse F, Scholtes C, Roche M, Legras-Lachuer C, de Lamballerie X, et al. 2016. Hepatitis E virus mutations associated with ribavirin treatment failure result in altered viral fitness and ribavirin sensitivity. J Hepatol 65: 499–508. [DOI] [PubMed] [Google Scholar]
- Drexler JF, Grywna K, Lukashev A, Stocker A, Almeida PS, Wieseler J, Ribeiro TC, Petersen N, Ribeiro Hda C Jr, Belalov I, et al. 2011. Full genome sequence analysis of parechoviruses from Brazil reveals geographical patterns in the evolution of non-structural genes and intratypic recombination in the capsid region. J Gen Virol 92: 564–571. [DOI] [PubMed] [Google Scholar]
- Drexler JF, Corman VM, Muller MA, Maganga GD, Vallo P, Binger T, Gloza-Rausch F, Cottontail VM, Rasche A, Yordanov S, et al. 2012a. Bats host major mammalian paramyxoviruses. Nat Commun 3: 796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler JF, Seelen A, Corman VM, Fumie Tateno A, Cottontail V, Melim Zerbinati R, Gloza-Rausch F, Klose SM, Adu-Sarkodie Y, Oppong SK, et al. 2012b. Bats worldwide carry hepatitis E virus-related viruses that form a putative novel genus within the family Hepeviridae. J Virol 86: 9134–9147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler JF, Corman VM, Drosten C. 2014. Ecology, evolution and classification of bat coronaviruses in the aftermath of SARS. Antiviral Res 101: 45–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexler JF, Corman VM, Lukashev AN, van den Brand JM, Gmyl AP, Brunink S, Rasche A, Seggewibeta N, Feng H, Leijten LM, et al. 2015. Evolutionary origins of hepatitis A virus in small mammals. Proc Natl Acad Sci 112: 15190–15195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A. 2007. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol 7: 214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eick GN, Jacobs DS, Matthee CA. 2005. A nuclear DNA phylogenetic perspective on the evolution of echolocation and historical biogeography of extant bats (Chiroptera). Mol Biol Evol 22: 1869–1886. [DOI] [PubMed] [Google Scholar]
- Emerson SU, Huang YK, McRill C, Lewis M, Purcell RH. 1992. Mutations in both the 2B and 2C genes of hepatitis A virus are involved in adaptation to growth in cell culture. J Virol 66: 650–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escalera-Zamudio M, Rojas-Anaya E, Kolokotronis SO, Taboada B, Loza-Rubio E, Mendez-Ojeda ML, Arias CF, Osterrieder N, Greenwood AD. 2016. Bats, primates, and the evolutionary origins and diversification of mammalian Gammaherpesviruses. MBio 7: e01425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinstone SM, Kapikian AZ, Purceli RH. 1973. Hepatitis A: detection by immune electron microscopy of a viruslike antigen associated with acute illness. Science 182: 1026–1028. [DOI] [PubMed] [Google Scholar]
- Feng Z, Hensley L, McKnight KL, Hu F, Madden V, Ping L, Jeong SH, Walker C, Lanford RE, Lemon SM. 2013. A pathogenic picornavirus acquires an envelope by hijacking cellular membranes. Nature 496: 367–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Hirai-Yuki A, McKnight KL, Lemon SM. 2014. Naked viruses that aren’t always naked: Quasi-enveloped agents of acute hepatitis. Annu Rev Virol 1: 539–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley NM, Springer MS, Teeling EC. 2016. Mammal madness: Is the mammal tree of life not yet resolved? Philos Trans R Soc Lond B Biol Sci 371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Global Consortium for H5N8 and Related Influenza Viruses. 2016. Role for migratory wild birds in the global spread of avian influenza H5N8. Science 354: 213–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthmann JP, Klovstad H, Boccia D, Hamid N, Pinoges L, Nizou JY, Tatay M, Diaz F, Moren A, Grais RF, et al. 2006. A large outbreak of hepatitis E among a displaced population in Darfur, Sudan, 2004: The role of water treatment methods. Clin Infect Dis 42: 1685–1691. [DOI] [PubMed] [Google Scholar]
- Hirai-Yuki A, Hensley L, McGivern DR, Gonzalez-Lopez O, Das A, Feng H, Sun L, Wilson JE, Hu F, Feng Z, et al. 2016. MAVS-dependent host species range and pathogenicity of human hepatitis A virus. Science 353: 1541–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F, Li Y, Yu W, Jing S, Wang J, Long F, He Z, Yang C, Bi Y, Cao W, et al. 2016. Excretion of infectious hepatitis E virus into milk in cows imposes high risks of zoonosis. Hepatology 64: 350–359. [DOI] [PubMed] [Google Scholar]
- Huelsenbeck JP, Ronquist F. 2001. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17: 754–755. [DOI] [PubMed] [Google Scholar]
- Izopet J, Dubois M, Bertagnoli S, Lhomme S, Marchandeau S, Boucher S, Kamar N, Abravanel F, Guerin JL. 2012. Hepatitis E virus strains in rabbits and evidence of a closely related strain in humans, France. Emerg Infect Dis 18: 1274–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins GM, Holmes EC. 2003. The extent of codon usage bias in human RNA viruses and its evolutionary origin. Virus Res 92: 1–7. [DOI] [PubMed] [Google Scholar]
- Johne R, Heckel G, Plenge-Bonig A, Kindler E, Maresch C, Reetz J, Schielke A, Ulrich RG. 2010. Novel hepatitis E virus genotype in Norway rats, Germany. Emerg Infect Dis 16: 1452–1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johne R, Dremsek P, Kindler E, Schielke A, Plenge-Bonig A, Gregersen H, Wessels U, Schmidt K, Rietschel W, Groschup MH, et al. 2012. Rat hepatitis E virus: Geographical clustering within Germany and serological detection in wild Norway rats (Rattus norvegicus). Infect Genet Evol 12: 947–956. [DOI] [PubMed] [Google Scholar]
- Kadim IT, Mahgoub O, Faye B, Farouk MM. 2013. Camel meat and meat products. CAB International, Wallingford, UK. [Google Scholar]
- Karlin S, Doerfler W, Cardon LR. 1994. Why is CpG suppressed in the genomes of virtually all small eukaryotic viruses but not in those of large eukaryotic viruses? J Virol 68: 2889–2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly AG, Netzler NE, White PA. 2016. Ancient recombination events and the origins of hepatitis E virus. BMC Evol Biol 16: 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Kenney SP, Ming X-J. 2018. Hepatitis E virus genome structure and replication strategy. Cold Spring Harb Perspect Med 10.1101/cshperspect.a031724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenney SP, Wentworth JL, Heffron CL, Meng XJ. 2015. Replacement of the hepatitis E virus ORF3 protein PxxP motif with heterologous late domain motifs affects virus release via interaction with TSG101. Virology 486: 198–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YM, Jeong SH, Kim JY, Song JC, Lee JH, Kim JW, Yun H, Kim JS. 2011. The first case of genotype 4 hepatitis E related to wild boar in South Korea. J Clin Virol 50: 253–256. [DOI] [PubMed] [Google Scholar]
- Koonin EV, Gorbalenya AE, Purdy MA, Rozanov MN, Reyes GR, Bradley DW. 1992. Computer-assisted assignment of functional domains in the nonstructural polyprotein of hepatitis E virus: Delineation of an additional group of positive-strand RNA plant and animal viruses. Proc Natl Acad Sci 89: 8259–8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krog JS, Breum SO, Jensen TH, Larsen LE. 2013. Hepatitis E virus variant in farmed mink, Denmark. Emerg Infect Dis 19: 2028–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz TH, Fenton MB. 2005. Bat ecology. University of Chicago Press, Chicago. [Google Scholar]
- Lauber C, Seitz S, Mattei S, Suh A, Beck J, Herstein J, Borold J, Salzburger W, Kaderali L, Briggs JAG, et al. 2017. Deciphering the origin and evolution of hepatitis B viruses by means of a family of non-enveloped fish viruses. Cell Host Microbe 22: 387–399.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee GH, Tan BH, Teo EC, Lim SG, Dan YY, Wee A, Aw PP, Zhu Y, Hibberd ML, Tan CK, et al. 2016. Chronic infection with camelid hepatitis E virus in a liver transplant recipient who regularly consumes camel meat and milk. Gastroenterology 150: 355–357.e3. [DOI] [PubMed] [Google Scholar]
- Lemon SM, Murphy PC, Shields PA, Ping LH, Feinstone SM, Cromeans T, Jansen RW. 1991. Antigenic and genetic variation in cytopathic hepatitis A virus variants arising during persistent infection: Evidence for genetic recombination. J Virol 65: 2056–2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li TC, Chijiwa K, Sera N, Ishibashi T, Etoh Y, Shinohara Y, Kurata Y, Ishida M, Sakamoto S, Takeda N, et al. 2005. Hepatitis E virus transmission from wild boar meat. Emerg Infect Dis 11: 1958–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Norder H, Uhlhorn H, Belak S, Widen F. 2014. Novel hepatitis E like virus found in Swedish moose. J Gen Virol 95: 557–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lole KS, Bollinger RC, Paranjape RS, Gadkari D, Kulkarni SS, Novak NG, Ingersoll R, Sheppard HW, Ray SC. 1999. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol 73: 152–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luis AD, Hayman DT, O’Shea TJ, Cryan PM, Gilbert AT, Pulliam JR, Mills JN, Timonin ME, Willis CK, Cunningham AA, et al. 2013. A comparison of bats and rodents as reservoirs of zoonotic viruses: Are bats special? Proc Biol Sci 280: 20122753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukashev AN, Drexler JF, Belalov IS, Eschbach-Bludau M, Baumgarte S, Drosten C. 2012. Genetic variation and recombination in Aichi virus. J Gen Virol 93: 1226–1235. [DOI] [PubMed] [Google Scholar]
- Lukashev AN, Corman VM, Schacht D, Gloza-Rausch F, Seebens-Hoyer A, Gmyl AP, Drosten C, Drexler JF. 2017. Close genetic relatedness of picornaviruses from European and Asian bats. J Gen Virol 98: 955–961. [DOI] [PubMed] [Google Scholar]
- Memish ZA, Cotten M, Meyer B, Watson SJ, Alsahafi AJ, Al Rabeeah AA, Corman VM, Sieberg A, Makhdoom HQ, Assiri A, et al. 2014. Human infection with MERS coronavirus after exposure to infected camels, Saudi Arabia, 2013. Emerg Infect Dis 20: 1012–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng XJ. 2010. Hepatitis E virus: Animal reservoirs and zoonotic risk. Vet Microbiol 140: 256–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller-Butterworth CM, Murphy WJ, O’Brien SJ, Jacobs DS, Springer MS, Teeling EC. 2007. A family matter: Conclusive resolution of the taxonomic position of the long-fingered bats, Miniopterus. Mol Biol Evol 24: 1553–1561. [DOI] [PubMed] [Google Scholar]
- Nagashima S, Takahashi M, Jirintai, Tanaka T, Yamada K, Nishizawa T, Okamoto H. 2011. A PSAP motif in the ORF3 protein of hepatitis E virus is necessary for virion release from infected cells. J Gen Virol 92: 269–278. [DOI] [PubMed] [Google Scholar]
- Nainan OV, Margolis HS, Robertson BH, Balayan M, Brinton MA. 1991. Sequence analysis of a new hepatitis A virus naturally infecting cynomolgus macaques (Macaca fascicularis). J Gen Virol 72 Pt 7: 1685–1689. [DOI] [PubMed] [Google Scholar]
- Nan Y, Yu Y, Ma Z, Khattar SK, Fredericksen B, Zhang YJ. 2014. Hepatitis E virus inhibits type I interferon induction by ORF1 products. J Virol 88: 11924–11932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Library of Medicine (US) NCBI. 1988a. Hepatitis E virus isolate Yunnan-2013, complete genome, www.ncbi.nlm.nih.gov/nuccore/KR905549.1.
- National Library of Medicine (US) NCBI. 1988b. Tupaia hepatovirus A isolate TN1, complete genome, www.ncbi.nlm.nih.gov/nuccore/KT877158.
- Olival KJ, Hosseini PR, Zambrana-Torrelio C, Ross N, Bogich TL, Daszak P. 2017. Host and viral traits predict zoonotic spillover from mammals. Nature 546: 646–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pintó RM, Aragones L, Costafreda MI, Ribes E, Bosch A. 2007. Codon usage and replicative strategies of hepatitis A virus. Virus Res 127: 158–163. [DOI] [PubMed] [Google Scholar]
- *.Pintó RM, Pérez-Rodríguez F-J, D’Andrea L, de Castellarnau M, Guix S, Bosch A. 2018. Codon usage of HAV: Its implications on translation kinetics and capsid folding. Cold Spring Harb Perspect Med doi: 10.1101/cshperspect.a031781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Player MR, Torrence PF. 1998. The 2-5A system: Modulation of viral and cellular processes through acceleration of RNA degradation. Pharmacol Ther 78: 55–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasche A, Saqib M, Liljander AM, Bornstein S, Zohaib A, Renneker S, Steinhagen K, Wernery R, Younan M, Gluecks I, et al. 2016a. Hepatitis E virus infection in dromedaries, North and East Africa, United Arab Emirates, and Pakistan, 1983–2015. Emerg Infect Dis 22: 1249–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasche A, Souza B, Drexler JF. 2016b. Bat hepadnaviruses and the origins of primate hepatitis B viruses. Curr Opin Virol 16: 86–94. [DOI] [PubMed] [Google Scholar]
- Ren X, Hurley JH. 2011. Proline-rich regions and motifs in trafficking: From ESCRT interaction to viral exploitation. Traffic 12: 1282–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renou C, Roque-Afonso AM, Pavio N. 2014. Foodborne transmission of hepatitis E virus from raw pork liver sausage, France. Emerg Infect Dis 20: 1945–1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reusken CB, Haagmans BL, Muller MA, Gutierrez C, Godeke GJ, Meyer B, Muth D, Raj VS, Smits-De Vries L, Corman VM, et al. 2013. Middle East respiratory syndrome coronavirus neutralising serum antibodies in dromedary camels: A comparative serological study. Lancet Infect Dis 13: 859–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter G, Boros A, Matics R, Kapusinszky B, Delwart E, Pankovics P. 2016. Divergent hepatitis E virus in birds of prey, common kestrel (Falco tinnunculus) and red-footed falcon (F. vespertinus), Hungary. Infect Genet Evol 43: 343–346. [DOI] [PubMed] [Google Scholar]
- Rima BK, McFerran NV. 1997. Dinucleotide and stop codon frequencies in single-stranded RNA viruses. J Gen Virol 78: 2859–2870. [DOI] [PubMed] [Google Scholar]
- Riveiro-Barciela M, Minguez B, Girones R, Rodriguez-Frias F, Quer J, Buti M. 2015. Phylogenetic demonstration of hepatitis E infection transmitted by pork meat ingestion. J Clin Gastroenterol 49: 165–168. [DOI] [PubMed] [Google Scholar]
- Robertson BH. 2001. Viral hepatitis and primates: Historical and molecular analysis of human and nonhuman primate hepatitis A, B, and the GB-related viruses. J Viral Hepat 8: 233–242. [DOI] [PubMed] [Google Scholar]
- Robertson BH, Jansen RW, Khanna B, Totsuka A, Nainan OV, Siegl G, Widell A, Margolis HS, Isomura S, Ito K, et al. 1992. Genetic relatedness of hepatitis A virus strains recovered from different geographical regions. J Gen Virol 73 (Pt 6): 1365–1377. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- Rozanov MN, Koonin EV, Gorbalenya AE. 1992. Conservation of the putative methyltransferase domain: A hallmark of the “Sindbis-like” supergroup of positive-strand RNA viruses. J Gen Virol 73: 2129–2134. [DOI] [PubMed] [Google Scholar]
- Sattar SA, Jason T, Bidawid S, Farber J. 2000. Foodborne spread of hepatitis A: Recent studies on virus survival, transfer and inactivation. Can J Infect Dis 11: 159–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla P, Nguyen HT, Torian U, Engle RE, Faulk K, Dalton HR, Bendall RP, Keane FE, Purcell RH, Emerson SU. 2011. Cross-species infections of cultured cells by hepatitis E virus and discovery of an infectious virus–host recombinant. Proc Natl Acad Sci 108: 2438–2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmonds P. 2012. SSE: A nucleotide and amino acid sequence analysis platform. BMC Res Notes 5: 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DB, Simmonds P; International Committee on Taxonomy of Viruses Hepeviridae Study Group, Jameel S, Emerson SU, Harrison TJ, Meng XJ, Okamoto H, Van der Poel WH, Purdy MA. 2014. Consensus proposals for classification of the family Hepeviridae. J Gen Virol 95: 2223–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DB, Simmonds P, Izopet J, Oliveira-Filho EF, Ulrich RG, Johne R, Koenig M, Jameel S, Harrison TJ, Meng XJ, et al. 2016. Proposed reference sequences for hepatitis E virus subtypes. J Gen Virol 97: 537–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surjit M, Oberoi R, Kumar R, Lal SK. 2006. Enhanced α1 microglobulin secretion from hepatitis E virus ORF3-expressing human hepatoma cells is mediated by the tumor susceptibility gene 101. J Biol Chem 281: 8135–8142. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Kitajima N, Abe N, Mishiro S. 2004. Complete or near-complete nucleotide sequences of hepatitis E virus genome recovered from a wild boar, a deer, and four patients who ate the deer. Virology 330: 501–505. [DOI] [PubMed] [Google Scholar]
- Takahashi M, Tanaka T, Takahashi H, Hoshino Y, Nagashima S, Jirintai, Mizuo H, Yazaki Y, Takagi T, Azuma M, et al. 2010. Hepatitis E Virus (HEV) strains in serum samples can replicate efficiently in cultured cells despite the coexistence of HEV antibodies: Characterization of HEV virions in blood circulation. J Clin Microbiol 48: 1112–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takata MA, Goncalves-Carneiro D, Zang TM, Soll SJ, York A, Blanco-Melo D, Bieniasz PD. 2017. CG dinucleotide suppression enables antiviral defence targeting non-self RNA. Nature 550: 124–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeling EC, Springer MS, Madsen O, Bates P, O’Brien SJ, Murphy WJ. 2005. A molecular phylogeny for bats illuminates biogeography and the fossil record. Science 307: 580–584. [DOI] [PubMed] [Google Scholar]
- Tei S, Kitajima N, Takahashi K, Mishiro S. 2003. Zoonotic transmission of hepatitis E virus from deer to human beings. Lancet 362: 371–373. [DOI] [PubMed] [Google Scholar]
- Todt D, Gisa A, Radonic A, Nitsche A, Behrendt P, Suneetha PV, Pischke S, Bremer B, Brown RJ, Manns MP, et al. 2016. In vivo evidence for ribavirin-induced mutagenesis of the hepatitis E virus genome. Gut 65: 1733–1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Cuyck H, Fan J, Robertson DL, Roques P. 2005. Evidence of recombination between divergent hepatitis E viruses. J Virol 79: 9306–9314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. 2017. Global Hepatitis Report 2017. Geneva, Switzerland. [Google Scholar]
- Woo PC, Lau SK, Teng JL, Tsang AK, Joseph M, Wong EY, Tang Y, Sivakumar S, Xie J, Bai R, et al. 2014. New hepatitis E virus genotype in camels, the Middle East. Emerg Infect Dis 20: 1044–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo PC, Lau SK, Teng JL, Cao KY, Wernery U, Schountz T, Chiu TH, Tsang AK, Wong PC, Wong EY, et al. 2016. New hepatitis E virus genotype in Bactrian camels, Xinjiang, China, 2013. Emerg Infect Dis 22: 2219–2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright F. 1990. The “effective number of codons” used in a gene. Gene 87: 23–29. [DOI] [PubMed] [Google Scholar]
- Wu J, Si F, Jiang C, Li T, Jin M. 2015. Molecular detection of hepatitis E virus in sheep from southern Xinjiang, China. Virus Genes 50: 410–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Yi M, Evans DJ, Simmonds P, Lemon SM. 2008. Identification of a conserved RNA replication element (cre) within the 3Dpol-coding sequence of hepatoviruses. J Virol 82: 10118–10128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yazaki Y, Mizuo H, Takahashi M, Nishizawa T, Sasaki N, Gotanda Y, Okamoto H. 2003. Sporadic acute or fulminant hepatitis E in Hokkaido, Japan, may be food-borne, as suggested by the presence of hepatitis E virus in pig liver as food. J Gen Virol 84: 2351–2357. [DOI] [PubMed] [Google Scholar]
- Yu JM, Li LL, Zhang CY, Lu S, Ao YY, Gao HC, Xie ZP, Xie GC, Sun XM, Pang LL, et al. 2016. A novel hepatovirus identified in wild woodchuck Marmota himalayana. Sci Rep 6: 22361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zell R, Delwart E, Gorbalenya AE, Hovi T, King AMQ, Knowles NJ, Lindberg AM, Pallansch MA, Palmenberg AC, Reuter G, et al. 2017. ICTV virus taxonomy profile: Picornaviridae. J Gen Virol 98: 2421–2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Ma Z, Harrison TJ, Feng R, Zhang C, Qiao Z, Fan J, Ma H, Li M, Song A, et al. 2009. A novel genotype of hepatitis E virus prevalent among farmed rabbits in China. J Med Virol 81: 1371–1379. [DOI] [PubMed] [Google Scholar]