Abstract
MRAS is the closest relative to the classical RAS oncoproteins and shares most regulatory and effector interactions. However, it also has unique functions, including its ability to function as a phosphatase regulatory subunit when in complex with SHOC2 and protein phosphatase 1 (PP1). This phosphatase complex regulates a crucial step in the activation cycle of RAF kinases and provides a key coordinate input required for efficient ERK pathway activation and transformation by RAS. MRAS mutations rarely occur in cancer but deregulated expression may play a role in tumorigenesis in some settings. Activating mutations in MRAS (as well as SHOC2 and PP1) do occur in the RASopathy Noonan syndrome, underscoring a key role for MRAS within the RAS-ERK pathway. MRAS also has unique roles in cell migration and differentiation and has properties consistent with a key role in the regulation of cell polarity. Further investigations should shed light on what remains a relatively understudied RAS family member.
The RRAS subgroup (RRAS, TC21/RRAS2, and MRAS/RRAS3) of the RAS family GTPases (RFGs) are the closest relatives to the classical RAS oncogenes (H/N/KRAS, hereafter referred to collectively as RAS). These GTPases share many regulatory and effector proteins with RAS as well as transforming abilities (Chan et al. 1994; Saez et al. 1994; Kimmelman et al. 1997; Rodriguez-Viciana et al. 2006). Members of this group have distinct functions, and MRAS has been shown to play a number of roles in cellular processes such as differentiation, cytoskeletal remodeling, and cell migration. Uniquely among RFGs, MRAS is part of a phosphatase complex that positively regulates RAF kinase activation and is required to cooperate with RAS proteins for efficient ERK pathway activation.
SEQUENCE FEATURES OF MRAS
The RRAS subgroup lies within a distinct branch of the tree of all small GTPases with classical RAS and ERAS (Fig. 1A). MRAS is highly conserved between vertebrates and has considerable similarity to the ras-2 gene in Caenorhabditis elegans (Fig. 1B). Interestingly, although classical RAS orthologs exist in fly, fish, and nematode, they are absent in ascidian, which does have orthologs to MRAS and RRAS (Fig. 1B). Therefore, MRAS evolved independently of RRAS in metazoans and has been suggested to compensate for the lack of classical RAS in ascidian (Keduka et al. 2009).
Figure 1.
MRAS phylogeny. (A) Phylogenetic relationship of human RAS family proteins. (B) Phylogenetic analysis of RAS family proteins in human (Hs), zebrafish (Dr), fruitfly (Dm), nematode (Ce), and ascidian Ciona intestinalis (Ci). Sequence alignment was performed using MUSCLE (see ebi.ac.uk/Tools/msa/muscle) and tree generation with iTOL (see itol.embl.de).
The G domain of MRAS is similar in sequence to the classical RAS proteins (it shares 53% amino acid identity with KRAS), and like RRAS and TC21, it has a short amino-terminal extension. The terminal residue of MRAS is lysine, which differs from the terminal residues of RAS (Fig. 2A), and consequently rather than being farnesylated, MRAS is predicted to be posttranslationally modified instead by geranylgeranylation (Zhang and Casey 1996). The hypervariable region (HVR) of MRAS has similarities to KRAS4B in that carboxy-terminal cysteines capable of palmytoylation are absent and instead a polybasic region is found (Fig. 2A). Like KRAS4B, MRAS is found in disordered membrane domains rather than organized lipid rafts (Ehrhardt et al. 2002), which suggests these proteins may signal in similar pathways and/or be similarly regulated.
Figure 2.
Comparison of MRAS amino acid sequence with other RAS family GTPases (RFGs). (A) Alignment of RAS protein hypervariable regions with features shaded according to property. (B) Sequence alignment (of Switch I and II containing regions) of MRAS with H/N/KRAS, RRAS, and TC21.
MRAS REGULATION AND CONTROL OF DOWNSTREAM PATHWAYS
MRAS shares many regulatory proteins with other RFGs; it can be activated by SOS1, RASGRF1, RASGRP1, and RASGRP3 guanine nucleotide exchange factors (GEFs) and inactivated by p120 RASGAP, neurofibromin, and GAP1m GTPase-activating proteins (GAPs) (Mitin et al. 2005). Although some of these proteins also regulate RRAS/TC21, full analysis of this regulation links MRAS closely to classical RAS in terms of GEF/GAP specificity (Ohba et al. 2000). This implies that most physiological signals that activate RAS will also activate MRAS simultaneously and is consistent with MRAS functioning together with RAS to provide coordinate inputs for efficient RAF activation (see below).
Given its sequence similarity and identical effector domain (Fig. 2B), it is unsurprising that MRAS can bind many of the same effectors as RAS such as A-, B-, and CRAF, AFDN/AF6, RASSF5, RalGEFs, and PI3K (Quilliam et al. 2001; Ortiz-Vega et al. 2002; Rodriguez-Viciana et al. 2004). Through binding RGL2/RLF, MRAS activates RAL and ELK1 in MCF-7 cells in an ERK-independent manner (Ehrhardt et al. 2001; Castro et al. 2012). MRAS also controls activation of RAP activity through binding MR-GEF/RAPGEF5 (Rebhun et al. 2000) and RA-GEF2/RAPGEF6 (Gao et al. 2001), the latter being specifically linked to control of cell adhesion through tumor necrosis factor (TNF)-α-triggered integrin activation in haematopoietic cells (Yoshikawa et al. 2007).
MRAS is a weak activator of the ERK pathway compared with RAS (Kimmelman et al. 1997; Rodriguez-Viciana et al. 2004), which, at least in part, could be attributed to the lower affinity of MRAS for RAF compared with RAS. One reason for this may be the differences in sequence of MRAS, particularly in the Switch I region (Fig. 2B). Both the GppNHp- and guanosine diphosphate (GDP)-bound forms of MRAS have an open Switch I conformation, and mutation of these residues to those of RAS result in a higher proportion of MRAS in the closed state. In addition, these mutations increase the affinity of MRAS for the RAS-binding domain (RBD) of RAF (Ye et al. 2005), which implies that there are regions unique to MRAS (with others yet to be identified) that govern its affinity and specificity for various effectors.
MRAS REGULATION OF RAF ACTIVITY THROUGH SHOC2/PROTEIN PHOSPHATASE 1 (PP1)
In addition to sharing interactions with many of RAS effectors, MRAS also has its own unique effector interactions with RAPGEFs (Gao et al. 2001) as well as SHOC2 and PP1 (Rodriguez-Viciana et al. 2006). SHOC2 is a ubiquitously expressed protein comprised almost exclusively of leucine rich repeats that was originally identified in C. elegans as a positive modulator of the ERK pathway (Selfors et al. 1998; Sieburth et al. 1998). Active MRAS forms a ternary complex with SHOC2 and PP1 to form a phosphatase holoenzyme that specifically dephosphorylates a conserved inhibitory site in RAF kinases (S259 in CRAF, S365 in BRAF, and S214 in ARAF, hereby referred to as S259) that functions as a 14-3-3 binding site and plays a key role in the RAF-activation cycle (Rodriguez-Viciana et al. 2006).
The consensus model of RAF activation stipulates that RAF is maintained in an autoinhibited inactive state in the cytosol by an intramolecular interaction between the amino-terminal region and the catalytic domain, which is in part mediated by a 14-3-3 dimer bound to two phosphorylated residues (S259 and S621 in CRAF) (Fig. 3A) (Tzivion et al. 1998; Matallanas et al. 2011; Lavoie and Therrien 2015). RAS-guanosine triphosphate (GTP) binding to the RBD of RAF results in RAF translocation to the plasma membrane in which other activating steps then take place. Chief among these is the dephoshphorylation of the “S259” site, which leads to 14-3-3 displacement from this site, destabilizes the closed conformation of RAF, allows the cysteine-rich domain (CRD) to further anchor RAF to the membrane, and facilitates RAF dimerization (Fig. 3B) (Lavoie and Therrien 2015). The MRAS–SHOC2–PP1 complex functions as a key phosphatase promoting this dephosphorylation step, which occurs preferentially on the RAF that has been recruited to the plasma membrane by RAS proteins (Fig. 3B) (Rodriguez-Viciana et al. 2006).
Figure 3.
Coordinate inputs from RAS and MRAS–SHOC2–protein phosphatase 1 (PP1) are required for effective RAF activation. (A) Under basal conditions, in which RAS and MRAS are guanosine diphosphate (GDP)-bound and inactive, SHOC2 interacts with PP1 via SCRIB. BRAF and CRAF exist as autoinhibited monomers, which are phosphorylated at S259. (B) After pathway stimulation by a mitogen such as epidermal growth factor (EGF), guanosine triphosphate (GTP)-bound RAS recruits BRAF/CRAF to the membrane, and SHOC2 forms a complex with MRAS-GTP and PP1. Here, the complex dephosphorylates the S259/S365 site on RAF and the 14-3-3 dimer can now mediate RAF dimerization (shown here as a heterodimer of BRAF and CRAF). RBD, RAS-binding domain; CRD, cysteine-rich domain; BRS, BRAF-specific region.
PP1 is known to interact with hundreds of regulatory proteins that confer substrate specificity and unique properties to each resulting holoenzyme. By analogy with other PP1 holoenzymes as well as the heterotrimeric PP2A complex (Cho and Xu 2007; Shi 2009; Peti et al. 2013), MRAS and SHOC2 are predicted to “remodel” the substrate recognition surface within the complex, altering the physiochemical landscape to create a surface specific for recognition of P-S259 RAF but not other phosphorylation sites, even on the same target (Rodriguez-Viciana et al. 2006).
A ROLE FOR MRAS IN POLARITY
Active MRAS can also associate with the polarity protein SCRIB, although this interaction is indirect and mediated by SHOC2 (Young et al. 2013). SCRIB is also a PP1-interacting protein and MRAS has the potential to “rearrange” the PP1 molecules within the SCRIB–SHOC2 complex. In the absence of MRAS, SHOC2 has very low affinity for PP1, and the PP1 in the complex is bound to SCRIB (Fig. 3A). In the presence of active MRAS, SHOC2 and PP1 form a ternary complex with high affinity and the PP1 interaction is now independent of SCRIB (Fig. 3B) (Young et al. 2013).
Through its interaction with SHOC2, MRAS is expected to recruit SCRIB and its associated interactome to sites of activation (Richier et al. 2010; Anastas et al. 2012; Young et al. 2013). SCRIB antagonizes SHOC2-mediated RAF “S259” dephosphorylation, at least partly, through a mechanism involving competition for PP1 molecules within the same macromolecular complex. SCRIB recruitment, through its interaction with the PIX–GIT complex, SGEF, and VANGL proteins, would also allow for the regulation of RAC/CDC42, ARF, RHOG, and the planar cell polarity pathways, respectively (Ellerbroek et al. 2004; Frank and Hansen 2008; Tada and Kai 2012). Furthermore, MRAS impairs the association of NOS1AP and the exchange factor βPIX with the SHOC2–SCRIB complex and thus has the ability to regulate the SCRIB interactome. MRAS is also expected to recruit to the same signaling platforms as other effectors, such as RAPGEFs, RALGEFs, and PI3K, which are also known to be involved in polarity adhesion and migration (Kimmelman et al. 2000; Ehrhardt et al. 2002; Rodriguez-Viciana et al. 2004). MRAS is therefore an excellent candidate to behave as a master regulator of polarity (Fig. 4).
Figure 4.
MRAS regulates cell polarity and migration through coordination of multiple signaling pathways (see text for details).
MRAS IN HUMAN DISEASE
Despite being the most closely related to RAS by sequence similarity, and given its shared interaction with many of the same effector proteins (as well as GAPs and GEFs), it is somewhat surprising that activating mutations in MRAS are rarely found in cancer, in clear contrast to RAS genes (Catalog of Somatic Mutations in Cancer [COSMIC]). Considering the key role of the RAF-ERK pathway in mediating RAS oncogenic properties, MRAS's lower affinity for RAF and its considerably weaker activation of the ERK pathway may at least partly account for this observation. Additionally, it is also possible that MRAS’s unique role in polarity may make constitutive activation disfavored in some contexts (Young et al. 2013). However, MRAS up-regulation may be linked to cancer in other ways.
MRAS expression is up-regulated in estrogen receptor (ER)-negative breast carcinomas compared with ER-positive in three independent studies (van de Vijver et al. 2002; Chin et al. 2006; Hess et al. 2006), and overexpression of constitutively active MRAS enables MCF-7 breast carcinoma cells to proliferate in the absence of estrogen (Castro et al. 2012). MRAS is part of the epithelial-to-mesenchymal (EMT) signature (Huang et al. 2012) and expression of active mutants causes EMT and oncogenic transformation in mouse scp-2 cells. These cells can grow in the absence of serum, lose contact inhibition, gain the ability to grow in an anchorage-independent manner, and form tumors in mice that have elevated levels of P-ERK and P-AKT (Ward et al. 2004). Many of these MRAS-induced characteristics are dependent on an Hepatocyte growth factor (HGF) autocrine mechanism (Zhang et al. 2004), which is interesting given that invasive growth and metastasis of mammary tumors correlates with HGF secretion (which itself occurs in the majority of tumors of that type) (Jeffers et al. 1996; Nagy et al. 1996).
The prominence of MRAS overexpression in cancer may also be context specific. For example, MRAS (along with MET) is overexpressed in multiple tumor types in a cytokine-driven/signal transducers and activators of transcription (STAT)3 activity-dependent manner (Yang et al. 2005). The Cancer Genome Atlas (TCGA) studies show that MRAS is amplified in 17% of lung squamous cell carcinomas but very rarely in lung adenocarcinoma (Fig. 5), which correlates inversely with the frequency of RAS mutations, which is high in adenocarcinomas but low in squamous cell carcinomas (cBioportal) (Cerami et al. 2012; Gao et al. 2013). MRAS was also overexpressed and/or amplified in 11% of ovarian serous cystadenocarcinoma and in 10% of head and neck squamous cell carcinoma (cBioportal). MRAS mutation frequency may be linked to specific subsets of cancers, for example, Borrmann type IV gastric cancer, which carries particularly poor prognoses and has a higher frequency of nonsynonymous MRAS mutations (17%) compared with overall gastric cancers (0.7% MRAS mutations) (Yasumoto et al. 2017). However, functional assays are still required to show that these mutations are activating.
Figure 5.
cBioportal oncoprints depicting genetic alterations in the classical RAS and RRAS subgroups of the RAS family of GTPases. TCGA, The Cancer Genome Atlas.
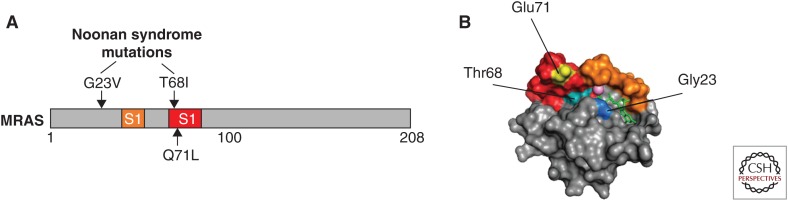
Two mutations in MRAS (p.Gly23Val and p.Thr68Ile) (Fig. 6) have been identified in patients with Noonan syndrome ([NS] MIM 163950), a developmental disorder that is part of the RASopathies family of related syndromes, which are driven by mutations in components of the RAS-ERK pathway. NS typically features facial dysmorphisms, slow growth rates, skeletal anomalies, mental retardation, predisposition to malignancies, and, often, cardiac defects. Of the two MRAS mutation-positive patients so far described, both had developmental delays, facial dysmorphisms, and cardiac hypertrophy. Similar clinical phenotypes are observed in patients with NS with loose anagen hair (NS-LAH) driven by mutations in MRAS-binding partners—SHOC2 and PP1 (Cordeddu et al. 2009; Gripp et al. 2016). MRAS p.Gly23Val corresponds to Gly13 of H/N/KRAS—a known oncogenic mutation. G23V-MRAS is primarily GTP-bound and activates ERK in cells (Higgins et al. 2017), and although not functionally tested as yet, mutation of position T68 is predicted to be activating given that it is within the Switch II region and lies within the GTP/GDP binding pocket (Fig. 6). Based on the observed phenotypes and what is known about their role in ERK pathway regulation, MRAS-driven NS is likely to be functioning through SHOC2–PP1 complex activity.
Figure 6.
Activating mutations in MRAS. (A) Position of Noonan syndrome mutations (upper) and activating mutation equivalent to codon 61 in RAS (lower). (B) Structure of MRAS bound to guanosine diphosphate (GDP) (green sticks) (PDB 1X1R). Switch I (orange), Switch II (red), and Mg2+ (pink sphere) are shown, as well as positions of key residues Gly23 (blue), Thr68 (turquoise), and Glu71 (yellow).
Gain-of-function CRAF mutations are also found in NS and cluster around S259 to disrupt 14-3-3 binding (Pandit et al. 2007; Razzaque et al. 2007; Kobayashi et al. 2010; Molzan et al. 2010), Of note, S259F as well as others in this region (S257L, 261S, and V263A), have been identified in cancer as well as NS. These results underscore the important role of MRAS–SHOC2–PP1 and RAF S259 dephosphorylation on RAS-ERK pathway activation.
Large-scale genome-wide association studies have identified the MRAS locus as a risk factor in cardiovascular disease (Erdmann et al. 2009; Schunkert et al. 2011; Liu et al. 2013). Intriguingly, although MRAS is widely expressed, it has particularly high expression in the heart (see biogps.org). Although a role for MAP kinase pathways in the heart is known (Rose et al. 2010), the precise role of MRAS remains to be elucidated.
OTHER FUNCTIONS OF MRAS
MRAS signaling is linked to differentiation and development in a variety of mammalian cell types. It is activated by nerve growth factor (NGF) and is required for ERK-dependent neuronal differentiation of rat pheochromocytoma PC12 cells (Kimmelman et al. 2002; Sun et al. 2006) and, in developing mouse bone, BMP2 treatment not only activates MRAS, but also increases its expression levels. Active MRAS stimulates transdifferentiation of C2C12 mouse myoblasts into osteoblasts, and causes osteoblast differentiation in a p38- and c-Jun amino-terminal kinase (JNK)-dependent manner (Watanabe-Takano et al. 2010). MRAS is up-regulated during dendrite development and contributes to dendrite growth via the ERK pathway. In these cells, semaphorin-4D (or Sema4D, a repulsive guidance molecule in the developing nervous system) binds to its receptor PLEXIN-B1, the cytoplasmic domain of which acts as a GAP on MRAS (and RRAS, but not TC21 or RAS). When MRAS activity is suppressed in this way, it results in growth cone collapse and remodeling of dendrite morphology (Saito et al. 2009). In the absence of Sema4D, active MRAS binds to an effector, LPD (lamellopodin), and recruits it to the membrane of growing dendrites where it participates in actin cytoskeleton remodeling via Ena/VASP proteins (Tasaka et al. 2012). However, other studies have suggested that sempahorins function as GAPs for RAPs not MRAS (Wang et al. 2012).
These ties to differentiation have also been studied in mouse embryonic stem cells (mESCs). Of all the Ras family members, Mras is the only gene whose expression is limited to undifferentiated mESCs. Before differentiation, expression is controlled by the cytokine LIF, and, as such, Mras is described as a marker of stemness (Mathieu et al. 2013). Persistent Mras down-regulation by removal of LIF affects the normal balance of expression of core pluripotency markers such as OCT4 and CAECAM1, and overexpression of MRAS leads to sustained expression of OCT4 and NANOG. In Xenopus, Mras is expressed throughout the embryo, maintaining pluripotency until the blastula stage and, as in mice, it is required for neuronal differentiation, which indicates that this protein has conserved functions across vertebrates (Mathieu et al. 2013).
At the organismal level, Mras null mice were initially described as phenotypically normal (Nunez Rodriguez et al. 2006), but further studies since then link Mras to the control of normal urinary function (Ehrhardt et al. 2015b) and normal olfactory and/or social processes in adult male mice because Mras−/− males are phenotypically more aggressive and show increased sexual behavior (Ehrhardt et al. 2015a).
CONCLUDING REMARKS
Although MRAS mutations are rarely found in cancer, it is likely that dysregulation of MRAS at the expression level may be a contributing factor to tumorigenesis in some contexts. Considering this and the studies of MRAS in cell-fate determination and stemness, understanding how MRAS expression is regulated in different cell types is of interest. The recent identification of activating mutations in MRAS in NS (as well as SHOC2 and PPP1CB) highlights a key role for the MRAS–SHOC2–PP1 complex in the regulation of the ERK pathway. MRAS has the ability to regulate multiple signaling pathways, including many that directly or indirectly regulate other GTPases of the RAS superfamily, and that suggests a role for MRAS as a master regulator of polarity. More comprehensive biochemical and structural studies will deepen our understanding of how MRAS-specific signaling contributes to cell behavior and will help to explain the phenotypes observed at the organismal level in which MRAS function is modified.
Footnotes
Editors: Linda VanAelst, Julian Downward, and Frank McCormick
Additional Perspectives on Ras and Cancer in the 21st Century available at www.perspectivesinmedicine.org
REFERENCES
- Anastas JN, Biechele TL, Robitaille M, Muster J, Allison KH, Angers S, Moon RT. 2012. A protein complex of SCRIB, NOS1AP and VANGL1 regulates cell polarity and migration, and is associated with breast cancer progression. Oncogene 31: 3696–3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro AF, Campos T, Babcock JT, Armijo ME, Martinez-Conde A, Pincheira R, Quilliam LA. 2012. M-Ras induces Ral and JNK activation to regulate MEK/ERK-independent gene expression in MCF-7 breast cancer cells. J Cell Biochem 113: 1253–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. 2012. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov 2: 401–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan AM, Miki T, Meyers KA, Aaronson SA. 1994. A human oncogene of the RAS superfamily unmasked by expression cDNA cloning. Proc Natl Acad Sci 91: 7558–7562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin K, DeVries S, Fridlyand J, Spellman PT, Roydasgupta R, Kuo WL, Lapuk A, Neve RM, Qian Z, Ryder T, et al. 2006. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell 10: 529–541. [DOI] [PubMed] [Google Scholar]
- Cho US, Xu W. 2007. Crystal structure of a protein phosphatase 2A heterotrimeric holoenzyme. Nature 445: 53–57. [DOI] [PubMed] [Google Scholar]
- Cordeddu V, Di Schiavi E, Pennacchio LA, Ma’ayan A, Sarkozy A, Fodale V, Cecchetti S, Cardinale A, Martin J, Schackwitz W, et al. 2009. Mutation of SHOC2 promotes aberrant protein N-myristoylation and causes Noonan-like syndrome with loose anagen hair. Nat Genet 41: 1022–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrhardt GR, Korherr C, Wieler JS, Knaus M, Schrader JW. 2001. A novel potential effector of M-Ras and p21 Ras negatively regulates p21 Ras-mediated gene induction and cell growth. Oncogene 20: 188–197. [DOI] [PubMed] [Google Scholar]
- Ehrhardt A, Ehrhardt GR, Guo X, Schrader JW. 2002. Ras and relatives—Job sharing and networking keep an old family together. Exp Hematol 30: 1089–1106. [DOI] [PubMed] [Google Scholar]
- Ehrhardt A, Wang B, Leung MJ, Schrader JW. 2015a. Absence of M-Ras modulates social behavior in mice. BMC Neurosci 16: 68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrhardt A, Wang B, Yung AC, Wang Y, Kozlowski P, van Breemen C, Schrader JW. 2015b. Urinary retention, incontinence, and dysregulation of muscarinic receptors in male mice lacking Mras. PLoS ONE 10: e0141493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellerbroek SM, Wennerberg K, Arthur WT, Dunty JM, Bowman DR, DeMali KA, Der C, Burridge K. 2004. SGEF, a RhoG guanine nucleotide exchange factor that stimulates macropinocytosis. Mol Biol Cell 15: 3309–3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann J, Grosshennig A, Braund PS, Konig IR, Hengstenberg C, Hall AS, Linsel-Nitschke P, Kathiresan S, Wright B, Tregouet DA, et al. 2009. New susceptibility locus for coronary artery disease on chromosome 3q22.3. Nat Genet 41: 280–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank SR, Hansen SH. 2008. The PIX-GIT complex: A G protein signaling cassette in control of cell shape. Semin Cell Dev Biol 19: 234–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Satoh T, Liao Y, Song C, Hu CD, Kariya Ki K, Kataoka T. 2001. Identification and characterization of RA-GEF-2, a Rap guanine nucleotide exchange factor that serves as a downstream target of M-Ras. J Biol Chem 276: 42219–42225. [DOI] [PubMed] [Google Scholar]
- Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. 2013. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 6: pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gripp KW, Aldinger KA, Bennett JT, Baker L, Tusi J, Powell-Hamilton N, Stabley D, Sol-Church K, Timms AE, Dobyns WB. 2016. A novel rasopathy caused by recurrent de novo missense mutations in PPP1CB closely resembles Noonan syndrome with loose anagen hair. Am J Med Genet A 170: 2237–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess KR, Anderson K, Symmans WF, Valero V, Ibrahim N, Mejia JA, Booser D, Theriault RL, Buzdar AU, Dempsey PJ, et al. 2006. Pharmacogenomic predictor of sensitivity to preoperative chemotherapy with paclitaxel and fluorouracil, doxorubicin, and cyclophosphamide in breast cancer. J Clin Oncol 24: 4236–4244. [DOI] [PubMed] [Google Scholar]
- Higgins EM, Bos JM, Mason-Suares H, Tester DJ, Ackerman JP, MacRae CA, Sol-Church K, Gripp KW, Urrutia R, Ackerman MJ. 2017. Elucidation of MRAS-mediated Noonan syndrome with cardiac hypertrophy. JCI Insight 2: e91225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Holzel M, Knijnenburg T, Schlicker A, Roepman P, McDermott U, Garnett M, Grernrum W, Sun C, Prahallad A, et al. 2012. MED12 controls the response to multiple cancer drugs through regulation of TGF-β receptor signaling. Cell 151: 937–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffers M, Rong S, Vande Woude GF. 1996. Enhanced tumorigenicity and invasion-metastasis by hepatocyte growth factor/scatter factor-met signalling in human cells concomitant with induction of the urokinase proteolysis network. Mol Cell Biol 16: 1115–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keduka E, Kaiho A, Hamada M, Watanabe-Takano H, Takano K, Ogasawara M, Satou Y, Satoh N, Endo T. 2009. M-Ras evolved independently of R-Ras and its neural function is conserved between mammalian and ascidian, which lacks classical Ras. Gene 429: 49–58. [DOI] [PubMed] [Google Scholar]
- Kimmelman A, Tolkacheva T, Lorenzi MV, Osada M, Chan AM. 1997. Identification and characterization of R-ras3: A novel member of the RAS gene family with a non-ubiquitous pattern of tissue distribution. Oncogene 15: 2675–2685. [DOI] [PubMed] [Google Scholar]
- Kimmelman AC, Osada M, Chan AM. 2000. R-Ras3, a brain-specific Ras-related protein, activates Akt and promotes cell survival in PC12 cells. Oncogene 19: 2014–2022. [DOI] [PubMed] [Google Scholar]
- Kimmelman AC, Nunez Rodriguez N, Chan AM. 2002. R-Ras3/M-Ras induces neuronal differentiation of PC12 cells through cell-type-specific activation of the mitogen-activated protein kinase cascade. Mol Cell Biol 22: 5946–5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Aoki Y, Niihori T, Cave H, Verloes A, Okamoto N, Kawame H, Fujiwara I, Takada F, Ohata T, et al. 2010. Molecular and clinical analysis of RAF1 in Noonan syndrome and related disorders: Dephosphorylation of serine 259 as the essential mechanism for mutant activation. Hum Mutat 31: 284–294. [DOI] [PubMed] [Google Scholar]
- Lavoie H, Therrien M. 2015. Regulation of RAF protein kinases in ERK signalling. Nat Rev Mol Cell Biol 16: 281–298. [DOI] [PubMed] [Google Scholar]
- Liu H, Huang XQ, Yang M, Ji XM, Du X, Zheng J. 2013. MRAS genetic variation is associated with atherothrombotic stroke in the Han Chinese population. J Clin Neurol 9: 223–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matallanas D, Birtwistle M, Romano D, Zebisch A, Rauch J, von Kriegsheim A, Kolch W. 2011. Raf family kinases: Old dogs have learned new tricks. Genes Cancer 2: 232–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieu ME, Faucheux C, Saucourt C, Soulet F, Gauthereau X, Fedou S, Trouillas M, Theze N, Thiebaud P, Boeuf H. 2013. MRAS GTPase is a novel stemness marker that impacts mouse embryonic stem cell plasticity and Xenopus embryonic cell fate. Development 140: 3311–3322. [DOI] [PubMed] [Google Scholar]
- Mitin N, Rossman KL, Der CJ. 2005. Signaling interplay in Ras superfamily function. Curr Biol 15: R563–R574. [DOI] [PubMed] [Google Scholar]
- Molzan M, Schumacher B, Ottmann C, Baljuls A, Polzien L, Weyand M, Thiel P, Rose R, Rose M, Kuhenne P, et al. 2010. Impaired binding of 14-3-3 to C-RAF in Noonan syndrome suggests new approaches in diseases with increased Ras signaling. Mol Cell Biol 30: 4698–4711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy J, Curry GW, Hillan KJ, McKay IC, Mallon E, Purushotham AD, George WD. 1996. Hepatocyte growth factor/scatter factor expression and c-met in primary breast cancer. Surg Oncol 5: 15–21. [DOI] [PubMed] [Google Scholar]
- Nunez Rodriguez N, Lee IN, Banno A, Qiao HF, Qiao RF, Yao Z, Hoang T, Kimmelman AC, Chan AM. 2006. Characterization of R-Ras3/M-Ras null mice reveals a potential role in trophic factor signaling. Mol Cell Biol 26: 7145–7154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohba Y, Mochizuki N, Yamashita S, Chan AM, Schrader JW, Hattori S, Nagashima K, Matsuda M. 2000. Regulatory proteins of R-Ras, TC21/R-Ras2, and M-Ras/R-Ras3. J Biol Chem 275: 20020–20026. [DOI] [PubMed] [Google Scholar]
- Ortiz-Vega S, Khokhlatchev A, Nedwidek M, Zhang XF, Dammann R, Pfeifer GP, Avruch J. 2002. The putative tumor suppressor RASSF1A homodimerizes and heterodimerizes with the Ras-GTP binding protein Nore1. Oncogene 21: 1381–1390. [DOI] [PubMed] [Google Scholar]
- Pandit B, Sarkozy A, Pennacchio LA, Carta C, Oishi K, Martinelli S, Pogna EA, Schackwitz W, Ustaszewska A, Landstrom A, et al. 2007. Gain-of-function RAF1 mutations cause Noonan and LEOPARD syndromes with hypertrophic cardiomyopathy. Nat Genet 39: 1007–1012. [DOI] [PubMed] [Google Scholar]
- Peti W, Nairn AC, Page R. 2013. Structural basis for protein phosphatase 1 regulation and specificity. FEBS J 280: 596–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quilliam LA, Rebhun JF, Zong H, Castro AF. 2001. Analyses of M-Ras/R-Ras3 signaling and biology. Methods Enzymol 333: 187–202. [DOI] [PubMed] [Google Scholar]
- Razzaque MA, Nishizawa T, Komoike Y, Yagi H, Furutani M, Amo R, Kamisago M, Momma K, Katayama H, Nakagawa M, et al. 2007. Germline gain-of-function mutations in RAF1 cause Noonan syndrome. Nat Genet 39: 1013–1017. [DOI] [PubMed] [Google Scholar]
- Rebhun JF, Castro AF, Quilliam LA. 2000. Identification of guanine nucleotide exchange factors (GEFs) for the Rap1 GTPase. Regulation of MR-GEF by M-Ras-GTP interaction. J Biol Chem 275: 34901–34908. [DOI] [PubMed] [Google Scholar]
- Richier L, Williton K, Clattenburg L, Colwill K, O’Brien M, Tsang C, Kolar A, Zinck N, Metalnikov P, Trimble WS, et al. 2010. NOS1AP associates with Scribble and regulates dendritic spine development. J Neurosci 30: 4796–4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Viciana P, Sabatier C, McCormick F. 2004. Signaling specificity by Ras family GTPases is determined by the full spectrum of effectors they regulate. Mol Cell Biol 24: 4943–4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Viciana P, Oses-Prieto J, Burlingame A, Fried M, McCormick F. 2006. A phosphatase holoenzyme comprised of Shoc2/Sur8 and the catalytic subunit of PP1 functions as an M-Ras effector to modulate Raf activity. Mol Cell 22: 217–230. [DOI] [PubMed] [Google Scholar]
- Rose BA, Force T, Wang Y. 2010. Mitogen-activated protein kinase signaling in the heart: Angels versus demons in a heart-breaking tale. Physiol Rev 90: 1507–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saez R, Chan AM, Miki T, Aaronson SA. 1994. Oncogenic activation of human R-Ras by point mutations analogous to those of prototype H-Ras oncogenes. Oncogene 9: 2977–2982. [PubMed] [Google Scholar]
- Saito Y, Oinuma I, Fujimoto S, Negishi M. 2009. Plexin-B1 is a GTPase activating protein for M-Ras, remodelling dendrite morphology. EMBO Rep 10: 614–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schunkert H, Konig IR, Kathiresan S, Reilly MP, Assimes TL, Holm H, Preuss M, Stewart AF, Barbalic M, Gieger C, et al. 2011. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet 43: 333–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selfors LM, Schutzman JL, Borland CZ, Stern MJ. 1998. soc-2 encodes a leucine-rich repeat protein implicated in fibroblast growth factor receptor signaling. Proc Natl Acad Sci 95: 6903–6908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y. 2009. Serine/threonine phosphatases: Mechanism through structure. Cell 139: 468–484. [DOI] [PubMed] [Google Scholar]
- Sieburth DS, Sun Q, Han M. 1998. SUR-8, a conserved Ras-binding protein with leucine-rich repeats, positively regulates Ras-mediated signaling in C. elegans. Cell 94: 119–130. [DOI] [PubMed] [Google Scholar]
- Sun P, Watanabe H, Takano K, Yokoyama T, Fujisawa J, Endo T. 2006. Sustained activation of M-Ras induced by nerve growth factor is essential for neuronal differentiation of PC12 cells. Genes Cells 11: 1097–1113. [DOI] [PubMed] [Google Scholar]
- Tada M, Kai M. 2012. Planar cell polarity in coordinated and directed movements. Curr Top Dev Biol 101: 77–110. [DOI] [PubMed] [Google Scholar]
- Tasaka G, Negishi M, Oinuma I. 2012. Semaphorin 4D/plexin-B1-mediated M-Ras GAP activity regulates actin-based dendrite remodeling through lamellipodin. J Neurosci 32: 8293–8305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzivion G, Luo Z, Avruch J. 1998. A dimeric 14-3-3 protein is an essential cofactor for Raf kinase activity. Nature 394: 88–92. [DOI] [PubMed] [Google Scholar]
- van de Vijver MJ, He YD, van’t Veer LJ, Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, et al. 2002. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med 347: 1999–2009. [DOI] [PubMed] [Google Scholar]
- Wang Y, He H, Srivastava N, Vikarunnessa S, Chen YB, Jiang J, Cowan CW, Zhang X. 2012. Plexins are GTPase-activating proteins for Rap and are activated by induced dimerization. Sci Signal 5: ra6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward KR, Zhang KX, Somasiri AM, Roskelley CD, Schrader JW. 2004. Expression of activated M-Ras in a murine mammary epithelial cell line induces epithelial–mesenchymal transition and tumorigenesis. Oncogene 23: 1187–1196. [DOI] [PubMed] [Google Scholar]
- Watanabe-Takano H, Takano K, Keduka E, Endo T. 2010. M-Ras is activated by bone morphogenetic protein-2 and participates in osteoblastic determination, differentiation, and transdifferentiation. Exp Cell Res 316: 477–490. [DOI] [PubMed] [Google Scholar]
- Yang J, Chatterjee-Kishore M, Staugaitis SM, Nguyen H, Schlessinger K, Levy DE, Stark GR. 2005. Novel roles of unphosphorylated STAT3 in oncogenesis and transcriptional regulation. Cancer Res 65: 939–947. [PubMed] [Google Scholar]
- Yasumoto M, Sakamoto E, Ogasawara S, Isobe T, Kizaki J, Sumi A, Kusano H, Akiba J, Torimura T, Akagi Y, et al. 2017. Muscle RAS oncogene homolog (MRAS) recurrent mutation in Borrmann type IV gastric cancer. Cancer Med 6: 235–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye M, Shima F, Muraoka S, Liao J, Okamoto H, Yamamoto M, Tamura A, Yagi N, Ueki T, Kataoka T. 2005. Crystal structure of M-Ras reveals a GTP-bound “off” state conformation of Ras family small GTPases. J Biol Chem 280: 31267–31275. [DOI] [PubMed] [Google Scholar]
- Yoshikawa Y, Satoh T, Tamura T, Wei P, Bilasy SE, Edamatsu H, Aiba A, Katagiri K, Kinashi T, Nakao K, et al. 2007. The M-Ras-RA-GEF-2-Rap1 pathway mediates tumor necrosis factor-α dependent regulation of integrin activation in splenocytes. Mol Biol Cell 18: 2949–2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young LC, Hartig N, Munoz-Alegre M, Oses-Prieto JA, Durdu S, Bender S, Vijayakumar V, Vietri Rudan M, Gewinner C, Henderson S, et al. 2013. An MRAS, SHOC2, and SCRIB complex coordinates ERK pathway activation with polarity and tumorigenic growth. Mol Cell 52: 679–692. [DOI] [PubMed] [Google Scholar]
- Zhang FL, Casey PJ. 1996. Protein prenylation: Molecular mechanisms and functional consequences. Annu Rev Biochem 65: 241–269. [DOI] [PubMed] [Google Scholar]
- Zhang KX, Ward KR, Schrader JW. 2004. Multiple aspects of the phenotype of mammary epithelial cells transformed by expression of activated M-Ras depend on an autocrine mechanism mediated by hepatocyte growth factor/scatter factor. Mol Cancer Res 2: 242–255. [PubMed] [Google Scholar]