Abstract
While genetic variants are known to be associated with overall gene abundance in stimulated immune cells, less is known about their effects on alternative isoform usage. By analyzing RNA-seq profiles of monocyte-derived dendritic cells from 243 individuals, we uncovered thousands of unannotated isoforms synthesized in response to influenza infection and type 1 interferon stimulation. We identified more than a thousand quantitative trait loci (QTLs) associated with alternate isoform usage (isoQTLs), many of which are independent of expression QTLs (eQTLs) for the same gene. Compared with eQTLs, isoQTLs are enriched for splice sites and untranslated regions, but depleted of sequences upstream of annotated transcription start sites. Both eQTLs and isoQTLs explain a significant proportion of the disease heritability attributed to common genetic variants. At the ERAP2 locus, we shed light on the function of the gene and how two frequent, highly differentiated haplotypes with intermediate frequencies could be maintained by balancing selection. At baseline and following type 1 interferon stimulation, the major haplotype is associated with low ERAP2 expression caused by nonsense-mediated decay, while the minor haplotype, known to increase Crohn's disease risk, is associated with high ERAP2 expression. In response to influenza infection, we found two uncharacterized isoforms expressed from the major haplotype, likely the result of multiple perfectly linked variants affecting the transcription and splicing at the locus. Thus, genetic variants at a single locus could modulate independent gene regulatory processes in innate immune responses and, in the case of ERAP2, may confer a historical fitness advantage in response to virus.
An important aspect of eukaryotic gene regulation is the usage of alternative gene isoforms. This is achieved through several mechanisms at the transcript level, including alternative promoters for transcription initiation, alternative splicing of pre-messenger RNA, alternative polyadenylation, and selective degradation of isoforms. These processes regulate the relative abundances of multiple coding and noncoding RNAs from the same underlying DNA sequence, often resulting in altered function of the gene products in response to developmental or environmental changes (Black 2003; Maquat 2004; Matlin et al. 2005; Wang et al. 2008).
A case in point is the critical role of alternative isoform usage across many immune processes, such as the balance between IgM and IgD immunoglobulin isoforms in B cells (Enders et al. 2014), naïve and memory T-cell proportions controlled by PTPRC isoforms (Berard and Tough 2002), and innate immune responses to pathogens regulated by different isoforms of MYD88 (Martinez and Lynch 2013). Genetic variants that affect isoform usage have been associated with immune disorders (Xiong et al. 2015), including single-nucleotide polymorphisms (SNPs) that alter the relative splicing of two IRF5 isoforms (Graham et al. 2007) associated with systemic lupus erythematosus (SLE).
Previous studies have identified shared and divergent transcriptional programs in the antibacterial and antiviral responses of innate immune cells (Amit et al. 2009; Lee et al. 2014), with genetic variants imparting stimulation-specific effects on the total transcript abundances of thousands of genes (Barreiro et al. 2012; Fairfax and Knight 2014; Lee et al. 2014; Quach et al. 2016). While maps of genetic determinants of alternative isoform usage are beginning to emerge, most notably in lymphoblastoid cell lines (Lappalainen et al. 2013; Li et al. 2016), across post-mortem human tissues (The GTEx Consortium 2015; Rivas et al. 2015), and in macrophages stimulated with bacteria (Nedelec et al. 2016), differential isoform usage in the human antiviral response, its natural variability, and its genetic basis have not been studied.
Here, we integrate bulk RNA-sequencing with dense genotyping to systematically investigate the genetic control of isoform usage in monocyte-derived dendritic cells (MoDCs) at rest, and in response to influenza infection or type 1 interferon stimulation. Because the type 1 interferon pathway is known to be engaged by a broad array of microbial products, our study design is unique in allowing the separation of the universal and influenza-specific interferon-induced responses. Since the human transcriptome has never been annotated under these conditions, we first used de novo assembly to catalog and expectation maximization to quantify all synthesized isoforms in resting and stimulated MoDCs. Then, by harnessing the natural transcriptomic and genetic variation in the ImmVar cohort (Lee et al. 2014; Raj et al. 2014a; Ye et al. 2014), we mapped quantitative trait loci (QTLs) associated with alternate isoform usage (isoQTLs). Systematic characterization of isoQTLs, especially in comparison to expression quantitative trait loci (eQTLs), provides mechanistic insights into the genetic control of different aspects of gene regulation and enables the functional interpretation of loci associated with immune-related diseases and under natural selection.
Results
Influenza infection and type I interferon stimulation induce widespread alternate isoform usage
We used paired-end RNA-seq to profile the transcriptomes of primary MoDCs from healthy donors at rest (N = 99), and following stimulation with either recombinant interferon beta (IFNB1), a type 1 interferon that stimulates anti-viral effectors (N = 227), or influenza ΔNS1 (a strain engineered to maximize the type 1 interferon-induced response to infection by the deletion of a key virulence factor; N = 250) (Shapira et al. 2009). A total of 552 pass-filter samples (out of 576), 84 from all three conditions, 127 from both stimulation conditions, and 46 from only one condition, were analyzed (Supplemental Table S1). To define the corpus of transcribed isoforms in human MoDCs at rest and in response to stimulation, we assembled the transcriptome de novo in each sample (individual-condition pair) from RNA-seq alignments, retained only expressed isoforms (more than five transcripts per million [TPM] in any sample), and then combined isoforms across all samples to enable direct comparisons between conditions. We noted that unannotated isoforms that did not match current GENCODE, UCSC, or RefSeq annotations (Harrow et al. 2006; O'Leary et al. 2016; Casper et al. 2018) were enriched in genes expressed at less than 25 TPMs across all three conditions (Supplemental Fig. S1), and thus removed these genes and their corresponding transcripts from downstream analyses.
Our final assembled transcriptome contained 15,754 transcripts corresponding to 8194 genes (Supplemental Table S2), 64.5% of which had transcriptional structures that exactly matched an annotated transcript; 5204 transcripts (33.0%) contained a novel splice site, and 389 transcripts (2.5%) did not harbor novel splice sites but contained novel splice junctions. These novel splice events were well supported by the presence of spliced reads (more than 10 mapped reads) spanning exon junctions of our assembled transcriptome. Of the 25,099 novel splice events, 11,093 (44%) were within 5′ UTRs and 7251 (28.9%) were within 3′ UTRs, echoing a previous deep sequencing analysis that reported that the majority of novel isoforms are due to alternative splicing of UTRs (Deveson et al. 2018). An additional 2663 transcripts (16.9%) were annotated with a novel transcription start site (TSS). To assess the accuracy of these new TSSs, we aligned CAGE sequencing reads from resting MoDCs (Noguchi et al. 2017) to the set of unique TSSs (TSS + 500 bp downstream) from assembled transcripts in the baseline condition. Compared with all TSSs, 69% (vs. 84%) of the new TSSs were supported by the presence of at least one mapped CAGE read, and 40% (vs. 61%) were supported by more than five mapped reads. By visual inspection, we found the most common misannotations to be isoform reconstructions that began within the gene body, downstream from the annotated TSS. While some of these fragments may truly exist, we were unable to verify them with short read sequencing and thus conservatively estimated the false annotation rate to be ∼20% of new TSSs (difference between new and all TSSs) or 3.3% of our entire transcriptome.
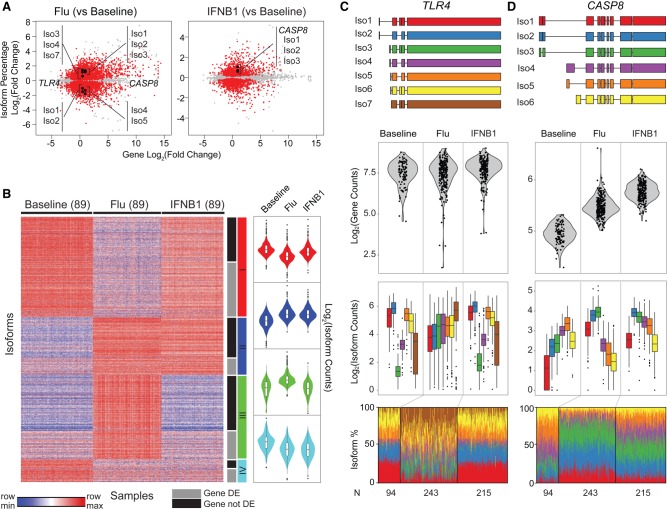
We next compared changes in isoform usage—estimated as the ratio of isoform abundance over the total gene abundance—in response to each stimulus. Relative to baseline, the usages of approximately twice as many isoforms (5326 vs. 2509) were altered in flu-infected compared with IFNB1-stimulated cells (beta regression, FDR < 0.05) (Supplemental Table S3; Supplemental Fig. S1A). This was corroborated by directly comparing flu-infected and IFNB1-stimulated cells where the usages of 5072 isoforms were altered (beta regression, FDR < 0.05) (Supplemental Table S3; Supplemental Fig. S2). In response to either stimulus, more than a third of the isoforms with differential usage were previously unannotated, highlighting the inadequacy of current transcriptome annotations in describing the full repertoire of gene isoforms in the human antiviral response. Of the differentially expressed genes (DESeq; FDR < 0.05, gene abs(log2[fold change]) > 1) (Supplemental Table S4) with more than one isoform, 54% (2233/4120) in flu-infected cells and 29% (1122/3898) in IFNB1-stimulated cells had at least one isoform that differed in usage (Fig. 1A).
Figure 1.
Transcriptome changes in response to stimulation. (A) Scatter plot of log2 fold change of gene abundance (x-axis) versus log2 fold change of isoform usage percentage (y-axis) in flu-infected (left) and IFNB1-simulated (right) cells compared with baseline. Each dot represents one isoform. Isoforms that significantly differed in their usage (beta regression, FDR < 0.05) are highlighted in red. (B) Clustering of isoform usage ratios in baseline, flu-infected, and IFNB1-stimulated cells. Heatmap colors are row scaled (red indicates row maximum, blue indicates row minimum; left). Violin plots (right) summarize the usages of all isoforms within a cluster separated by condition. Only isoforms (one per gene) that most significantly changed (beta regression, FDR < 0.05) in usage are shown. (C,D) De novo constructed isoforms (top), gene abundance (second row), and isoform abundance (third row) and usage percentage (bottom) for TLR4 (C) and CASP8 (D).
Isoforms that differed in usage partitioned into four prominent clusters (Fig. 1B, k-means clustering of the most significant isoforms, one per gene; Supplemental Table S5). Isoforms with increased usage in response to both stimuli (Cluster II) were highly enriched for innate system processes (GO:0002376, q < 6.41 × 10−6), defense response to virus (GO:0051607, q < 9.56 × 10−6), and type 1 interferon signaling pathway (GO:0060337, q < 1.25 × 10−3). Isoforms with increased usage in response to flu but not IFNB1 (Cluster III) were enriched for regulators of gene expression (GO:0010468, q < 6.23 × 10−3), including genes involved in the regulation of MAP kinase cascade (GO:0043408, q < 1.13 × 10−2) and inflammatory response (GO:0006954, q < 1.88 × 10−2). Isoforms with decreased usage in response to flu (Cluster I) were enriched for oxidoreductase activity (GO:0016616, q < 2.47 × 10−2); isoforms with decreased usage in response to both conditions (Cluster IV) were not significantly enriched for known Gene Ontology entries. TLR4, the toll-like receptor classically associated with sensing bacterial ligands but also shown to sense viral products (Doyle et al. 2002), was among the genes that had flu-specific isoform usage despite little change in total gene abundance (Fig. 1C; Supplemental Fig. S3). In flu-infected cells, the usages of longer isoforms with an upstream alternative start site (TLR4/Iso1 and TLR4/Iso2) were decreased, while the usages of TLR4/Iso3, TLR4/Iso4, and TLR4/Iso7 were increased. TLR4/Iso4 encodes the annotated 839-amino-acid product, while isoforms TLR4/Iso3 and TLR4/Iso7 encode shorter, 799-amino-acid products each with a truncated extracellular domain missing a predicted signal peptide. We also found decreased usage of short CASP8 isoforms (CASP8/Iso4, CASP8/Iso5, CASP8/Iso6) (Fig. 1D; Supplemental Fig. S3) only in flu-infected cells. CASP8 is best known to induce apoptosis via the Fas-associated protein with death domain (FADD) in response to extrinsic cytokine signals. CASP8/Iso4 has a unique N-terminal extension of 59 amino acids, which has been reported to allow for selective recruitment to the endoplasmic reticulum (Breckenridge et al. 2002). These results demonstrate that changes in isoform usage independent of overall gene abundance are pervasive and affect prominent innate immune sensors and regulators in viral versus interferon response.
Genetic variants associated with isoform usage are enriched for distinct gene regulatory elements
While it is known that common genetic variants modulate gene expression in both resting and stimulated MoDCs (Lee et al. 2014), we assessed if they could also affect isoform usage under these conditions. We associated over 10 million imputed variants with two transcriptional traits, isoform usage ratio and log of total gene abundance, to identify isoQTLs and eQTLs, respectively. After adjusting for unwanted variation that likely tracked with technical and biological confounders (Supplemental Figs. S4, S5), we identified 2763 isoforms corresponding to 1425 genes (linear regression, permutation FDR < 0.05) (Supplemental Table S6) with local isoQTLs (±500 kb of TSS) and 6694 genes (linear regression, permutation FDR < 0.05) (Supplemental Table S7) with local eQTLs in at least one condition. A substantial proportion of leading isoQTL SNPs (63% baseline, 40% flu, 41% IFNB1) were not significant eQTLs, suggesting that the genetic control of isoform usage and overall gene abundance are largely independent (Methods; Supplemental Figs. S6, S7).
Genetic variants could modulate isoform usage through several mechanisms, including perturbing the usage of alternate promoters, splice sites, or regulatory elements in the UTRs. We compared the genomic properties of isoQTLs and eQTLs to identify the mechanisms by which each class of variants acts. When normalized by exon and intron lengths, leading SNPs for local isoQTLs (one per isoform) were enriched across the entire gene body (Fig. 2A), in distinct contrast to leading SNPs for local eQTLs (one per gene), which were enriched near TSSs and transcription end sites (TESs). Further, compared with a set of eQTLs matched for allele frequency and distance to TSS, leading SNPs for local isoQTLs were most enriched for splice sites (baseline: 3.7×, flu: 3.0×, IFNB1: 4.0×), synonymous (baseline: 1.9×, IFNB1: 2.1×) and missense variants (baseline: 1.9×, flu: 1.8×), and 5′ UTRs (baseline: 1.3×, IFNB1: 1.2×) (Fig. 2B). Compared with eQTLs, isoQTLs were not enriched for binding sites of key transcription factors involved in myeloid cell response (Supplemental Fig. S8). These results suggest that genetic variants associated with isoform usage likely do so via cis regulatory sequences that modulate alternative splicing and transcript stability.
Figure 2.
Properties of local eQTLs and isoQTLs. (A) Frequency (y-axis) of the location of leading SNPs for local eQTLs and isoQTLs (permutation FDR < 0.05) with respect to meta gene structure (x-axis). Genes are normalized to five exonic regions (E1–E4 indicate exons 1 through 4; E* indicates exon 5 to the last exon) and four intronic regions (introns 1, 2, 3, and from intron 4 to the last intron). Upstream and downstream sequences are divided into 100-kb windows. (B) Log2 fold enrichment (x-axis) of leading SNPs for local eQTLs and isoQTLs (permutation FDR < 0.05) for genomic annotations. eQTL enrichments are calculated using a background set of SNPs matched for distance to TSS and allele frequency. IsoQTL enrichments are calculated with respect to a background set of eQTLs matched for distance to TSS and allele frequency. Multiple testing significance is indicated. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001.
Genetic control of alternative isoform usage in responses to virus infection and IFNB1 stimulation
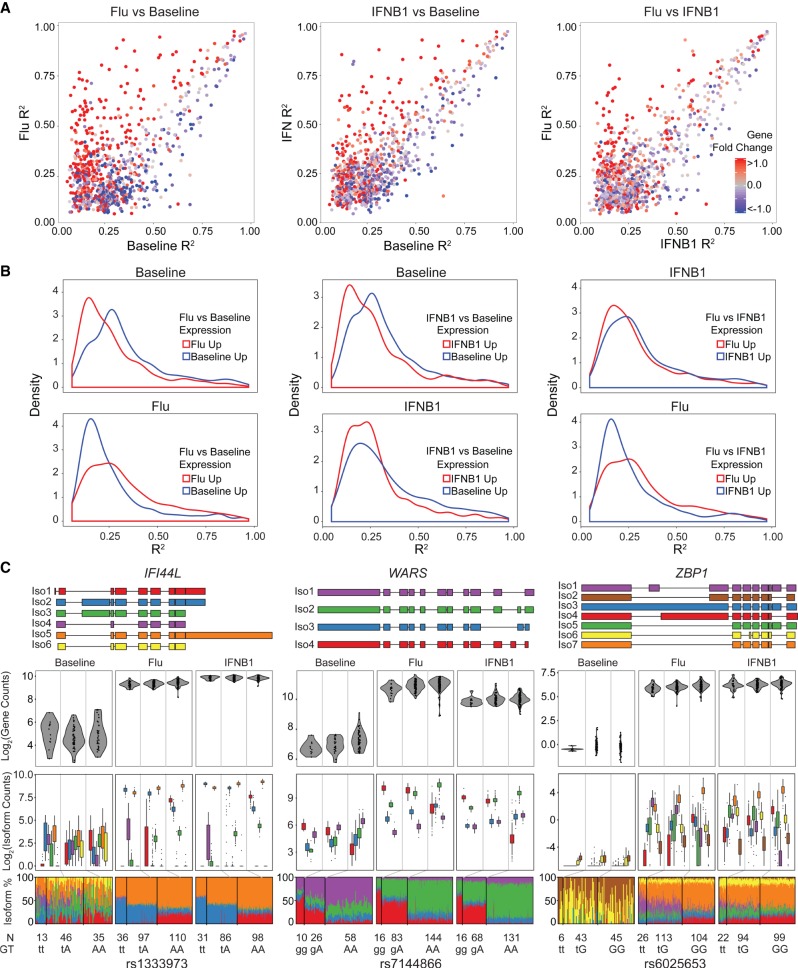
To assess how the genetic control of isoform usage differs in response to stimuli, we analyzed 84 donors whose cells were assayed in all three conditions to enable equally powered comparisons. At these sample sizes, we detected more eQTLs in cells stimulated with IFNB1 than in cells at rest or infected with flu (baseline: 1715, flu: 1755, IFNB1: 2108; permutation FDR < 0.05) (Supplemental Table S6) but similar numbers of isoQTLs across conditions (baseline: 717, flu: 644, IFNB1: 692; permutation FDR < 0.05) (Supplemental Table S7). For the 1164 isoforms with isoQTLs in at least one condition, we compared the effect sizes (Riso2) of associations across conditions (Fig. 3A). The correlation of Riso2s was lowest between flu-infected and resting (baseline) cells (Pearson's rflu.baseline = 0.61 compared with rIFNB1.baseline = 0.76 and rIFNB1.flu = 0.74). The corresponding genes of isoforms with higher Riso2 in stimulated cells were up-regulated in response to stimuli, suggesting that the genetic control of isoform usage is sensitive to activation of gene regulatory programs that control overall gene abundance (Fig. 3A,B).
Figure 3.
Comparison of local isoQTLs between conditions. (A) Correlation of effect sizes (R2) for significant local isoQTLs (permutation FDR < 0.05) between pairs of conditions. Transcripts are colored by differential expression (red indicates up-regulated in condition 2, y-axis; blue indicates up-regulated in condition 1, x-axis) for each pair of conditions. (B) Distributions of effect sizes (R2) for significant local isoQTLs (permutation FDR < 0.05) for each pair of conditions segregating genes based on expression in each condition. (C) De novo constructed transcript structure (top) and box-whisker plots (bottom three panels) between transcript quantitative traits (y-axis: log2(normalized gene abundance), log2(normalized isoform abundance), or isoform usage percentage) and genotype (x-axis) for three genes (IFI44L, WARS, and ZBP1) with risoQTLs.
To directly assess how stimulation modifies the effects of genetic variants on isoform usage, we mapped SNPs associated with the difference in isoform usage between conditions, herein referred to as local response-isoQTLs (risoQTLs). We identified, compared to resting cells, 53 (flu) and 30 (IFNB1) isoforms, corresponding to 31 and 14 genes, with at least one local risoQTLs (permutation FDR < 0.05) (Supplemental Table S8). Among the seven genes that shared local risoQTLs in both stimulated conditions were IFI44L and WARS (Fig. 3C). IFI44L is a type 1 interferon-stimulated gene known to moderately inhibit human hepatitis virus replication in vitro (Schoggins et al. 2011) and whose splicing has been shown to be influenced by rs1333973 (Lalonde et al. 2011). Rs1333973 is the most significant risoQTL associated with the usages of two isoforms differentiated by exon 2 in flu-infected and IFNB1-stimulated cells (flu vs. baseline: IFI44L/Iso1: +8.9%, P < 1.37 × 10−9; IFI44L/Iso2: −13.9%, P < 1.53 × 10−9; IFNB1 vs. baseline: IFI44L/Iso1: +9.6%, P < 1.21 × 10−10; IFI44L/Iso2: −15.7%, P < 2.14 × 10−10) (Supplemental Fig. S9). WARS is a tryptophanyl-tRNA synthetase primarily involved in protein synthesis. While WARS isoforms are known to encode for catalytic null enzymes (Lo et al. 2014) and have anti-angiogenic activity inducible by interferon gamma (Wakasugi et al. 2002), there have been no previous reports of the genetic control of these isoforms. The most significant risoQTL rs7144866 was associated with the usages of two isoforms differentiated by exon 2: While rs7144866A increases the usage of WARS/Iso1 in cells at rest, it increases the usage of WARS/Iso2 in flu-infected and IFNB1-stimulated cells resulting in the risoQTL (flu vs. baseline: −19.1%, 2.50 × 10−29; IFNB1 vs. baseline: −17.7%, 7.03 × 10−26) (Supplemental Fig. S10). There were 40 isoforms, corresponding to 30 genes, that have at least one risoQTL in flu-infected cells compared with interferon-stimulated cells (permutation FDR < 0.05) (Supplemental Table S8). Among these was ZBP1, a sensor of influenza infection that triggers cell death and inflammation and contributes to virus-induced lethality (Kuriakose et al. 2016). We found rs6025653t increases the usage of ZBP1/Iso1 by 9.67% (P < 4.16 × 10−16) (Fig. 3C; Supplemental Fig. S11) in flu-infected compared with interferon-stimulated cells. ZBP1/Iso1 is differentiated from all other ZBP1 isoforms by the retention of exon 9. These results suggest that while influenza-infected and interferon-stimulated cells expectedly share some genetic control of isoform usage (as interferons are induced by viral infection), influenza infection also confers specific genetic control of isoform usage of previously unknown genes, likely reflecting antiviral mechanisms independent of downstream effector (type 1 interferon) signaling.
Association of eQTLs and isoQTLs with immune-related diseases
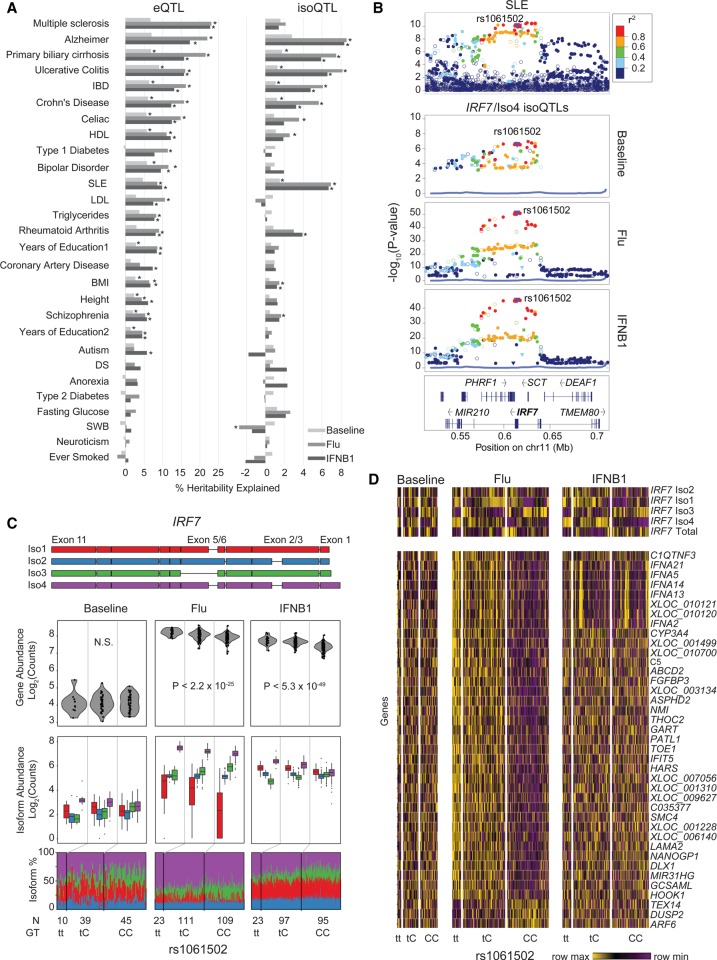
Previous analyses of the overlap between expression QTLs and genome-wide association studies (GWAS) have aided the localization and functional interpretation of causal variants at GWAS loci. Because disease-causing variants that affect isoform usage could have more profound effects on gene regulation by altering protein structure, we jointly analyzed disease-associated variants and local isoQTLs in addition to local eQTLs using two approaches. First, compared with SNPs from the latest GWAS catalog (MacArthur et al. 2017), local eQTLs in stimulated cells were enriched in multiple diseases, including inflammatory bowel disease (flu: P < 5.46 × 10−7, IFNB1: P < 5.21 × 10−5), rheumatoid arthritis (flu: P < 2.49 × 10−5, IFNB1: P < 0.03), and Parkinson's disease (flu: P < 4.06 × 10−7, IFNB1: P < 7.44 × 10−5) (Supplemental Fig. S12), while local isoQTLs were enriched in late onset Alzheimer's disease (flu: P < 1.15 × 10−6, IFNB1: P < 1.75 × 10−4), vitiligo (flu: P < 4.94 × 10−5, IFNB1: P < 0.42), and SLE (flu: P < 0.1, IFNB1: P < 3.62 × 10−3) (Supplemental Fig. S12). Notably, the overlap of isoQTLs with Alzheimer's loci included genes with known variants that affect splicing: CD33 (Hernandez-Caselles et al. 2006; Raj et al. 2014b) and CD46 (Russell et al. 1992). Corroborating this, we performed partitioned heritability analysis using linkage disequilibrium (LD) score regression (Finucane et al. 2015). For 28 traits with available summary statistics, both local eQTLs and local isoQTLs explained a statistically significant percentage of the SNP heritability of autoimmune diseases (e.g., ulcerative colitis, SLE) and some neurodegenerative diseases (e.g., Alzheimer's) and did not explain much of the SNP heritability of diseases with no known relationship to the innate immune system (e.g., type 2 diabetes) (Fig. 4A; Supplemental Table S9). These results suggest a role for both variants that affects isoform usage and gene expression in mediating autoimmune and neurodegenerative disease risk.
Figure 4.
GWAS enrichment of local eQTLs and isoQTLs. (A) Partitioned heritability analysis: proportion of SNP heritability explained (x-axis) for 28 traits (y-axis) by eQTLs (left) and isoQTLs (right). (DS) Depressive symptoms; (SWB) subject well-being; (IBD) inflammatory bowel disease; (SLE) systemic lupus erythematosus; (BMI) body mass index; (HDL) high density lipoprotein; (LDL) low density lipoprotein. (B) LocusZoom plots of the IRF7 region for SLE GWAS associations (top) and IRF7/Iso4 isoQTLs for baseline, flu-infected, or IFNB1-stimulated cells (bottom three panels). (y-axis) −log10(P-value) of association; (x-axis) genomic location. Points are colored based on LD to rs1061502. (C) Transcript structure (top) and box-whisker plots (bottom three panels) between IRF7 transcript quantitative traits (y-axis: log2(normalized gene abundance), log2(normalized isoform abundance), or isoform usage percentage) and rs1061502 genotype (x-axis). (D) Heatmap of genes distally associated (permutation FDR < 0.05) with risoQTL rs1061502. Heatmap colors are row-scaled TPM values (yellow indicates row maximum; purple indicates row minimum).
The IRF7 locus harbors an isoQTL within an extended haplotype previously known to be associated with SLE (rs58688157 lead SNP, P < 2.97 × 10−11) (Fig. 4B; Morris et al. 2016). A linked SNP (rs1061502; LD R2 = 0.93, D′ = 0.97 to rs58688157) was the most significant association to overall IRF7 expression in IFNB1-stimulated (P < 2.87 × 10−49) and flu-infected cells (P < 2.21 × 10−25) (Fig. 4C; Supplemental Fig. S13). IsoQTL analysis further revealed that rs1061502T also increased the usage of IRF7/Iso4 (flu: beta = 5.3%, P < 9.42 × 10−52; IFNB1: beta = 5.8%, P < 4.81 × 10−46) (Fig. 4C, panel 3, purple; Supplemental Fig. S14) while it decreased the usage of IRF7/Iso3 (flu: beta = −3.4%, 1.15 × 10−15; IFNB1: beta = −7.0%, 2.04 × 10−29) (Fig. 4C, panel 3, green; Supplemental Fig. S14). Further, although overall IRF7 abundances were similar between the two stimulated conditions, IRF7/Iso4 (purple) was the dominant isoform in flu-infected cells but not in IFNB1-stimulated cells (10.7× fold, P < 10−306) (Fig. 4C, bottom panel; Supplemental Fig. S14). Rs1061502 was also a distal eQTL (permutation FDR < 0.2) for a cluster of genes, including NMI, IFNA2, IFIT5, and C5, only in flu-infected but not in IFNB1-stimulated cells (Fig. 4D). These results replicate and expand our previous findings that rs12805435 (LD R2 = 0.95, D′ = 0.98 to rs1061502) is associated in cis with IRF7 expression in IFNB1-stimulated and flu-infected cells and in trans with a cluster of IRF7-regulated genes only in flu-infected cells (Lee et al. 2014). The flu-specific trans associations could be due to the additive effects of flu-specific induction of IRF7/Iso4 independent of IFNB1 signaling and induction of overall IRF7 expression by rs1061502T. IRF7/Iso4 encodes a 516-amino-acid protein product and differs from other abundant isoforms in IFNB1-stimulated cells (IRF7/Iso1 and IRF7/Iso3) in the 5′ UTR and the coding sequence in the DNA-binding domain. Given the known link between type 1 interferons and SLE, our results suggest that SNPs affecting a specific IRF7 isoform could impact viral responses and autoimmune inflammation through similar mechanisms.
An ERAP2 risoQTL controls differential transcript usage during influenza infection
The ERAP2 locus is characterized by two frequent and highly differentiated (40 SNPs in perfect LD) haplotypes observed in every major human population (B: 53% and A: 47%) (Supplemental Fig. S15). The minor Haplotype A encodes a 965-amino-acid protein and is associated with Crohn's disease (Jostins et al. 2012) but not ulcerative colitis (Fig. 5A). The major allele (G) of rs2248374, a splice-site variant tagging Haplotype B, creates an alternate 3′ donor splice site inducing the alternative splicing of an extended exon 10 with two premature termination codons (Andres et al. 2010). As a result, transcripts from Haplotype B are degraded by nonsense-mediated decay, resulting in one of the most significant eQTLs and isoQTLs in most tissues and cell types (Lappalainen et al. 2013; Lee et al. 2014; Ye et al. 2014; The GTEx Consortium 2015). The ERAP2 locus has been maintained by long-term balancing selection (between 1.4 million [Andres et al. 2010] and 5.1 million yr [Cagliani et al. 2010]), raising the important question: In what environmental conditions does balancing selection act to maintain the seemingly loss-of-function (LOF) Haplotype B and the disease-causing Haplotype A in humans?
Figure 5.
Genetics of ERAP2 regulation. (A) LocusZoom plot of Crohn's disease and ulcerative colitis associations at the ERAP2 locus. (y-axis) −log10(P-value) of association; (x-axis) genomic location. (B) Structures of transcripts derived from each haplotype (top) and box-whisker plots (bottom three panels) between ERAP2 transcript quantitative traits (y-axis: log2(normalized gene abundance), log2(normalized isoform abundance), or isoform usage percentage)) and genotype (x-axis). (C) Correlation between ERAP2/Iso1 (orange), ERAP2/Iso3 (light blue), and ERAP2/Iso4 (dark blue) abundance (y-axis) and abundance of flu segment 4 (x-axis) segregated by rs2248374 genotype (squares indicate aa; triangles, aG; circles, GG). (D) Western blot of MoDCs before and after flu-infection from five Haplotype B homozygotes and two heterozygotes. A full-length ERAP2 protein isoform is expected at 120 kDa. At least one flu-specific ERAP2 protein isoform is expected at 49 kDa. (E) A schematic of the hypothesized regulation and function of two ERAP2 haplotypes. (N) N terminal domain; (AP) amino peptidase domain; (H) hinge domain; (C) C terminal domain; (PTC) premature termination codon.
Given the known role of ERAP2 in antigen presentation (Saveanu et al. 2005), we examined the genetic control of ERAP2 transcripts in the human antiviral response. In resting and IFNB1-stimulated cells, we confirmed the known genetic association of rs2248374G allele with lower ERAP2 expression (Fig. 5B). In flu-infected but not IFNB1-stimulated cells, two previously uncharacterized short isoforms (ERAP2/Iso3, ERAP2/Iso4) (Fig. 5B; Supplemental Fig. S16) were transcribed from Haplotype B, resulting in the partial rescue of ERAP2 expression. The short isoforms differed from the constitutive full-length isoform (ERAP2/Iso1 transcribed from Haplotype A) by the initiation of transcription at exon 9 and the alternative splicing of an extended exon 10, and they differed from each other by alternative splicing at a secondary splice site at exon 15. The initiation of transcription at exon 9 results in an alternate in-frame translation start site at exon 11, thus rendering the premature termination codon in exon 10 inactive. The influenza-dependent genetic control of ERAP2 isoform usage is supported by two additional lines of evidence. First, there was significant correlation between overall flu transcript abundance, a proxy for degree of infection, and ERAP2/Iso3 and ERAP2/Iso4 transcript abundances and usages in heterozygotes and Haplotype B homozygotes (Fig. 5C; Supplemental Fig. S17). Second, the transcription of an extended exon 10, a hallmark of flu-specific short isoforms, was observed in monocyte-derived macrophages infected by H3N2 over a time course in an independent RNA-seq data set (fluomics, GEO GSE97672) (Supplemental Fig. S18).
The predicted protein products of either ERAP2 short isoforms would be missing the N-terminal (N) and aminopeptase (AP) domains. From ERAP2/Iso3, only one protein product is expected to be translated, which maintains a partial hinge (H) domain and the full alpha helical C-terminal (C) domain. Although ERAP2/Iso4 is predicted to harbor a premature termination codon that could lead to NMD, the presence of the transcript suggests the alternate translation of an isoform starting at exon 16 that maintains a partial alpha helical C-terminal domain. This calls into question whether the short ERAP2 isoforms would function as an RNA or protein product. Western blotting with a full-length ERAP2 antibody detects at least one short protein isoform (∼50 kDa) in flu-infected cells from Haplotype B homozygotes and heterozygotes, suggesting the translation of the short influenza-specific ERAP2 isoforms (Fig. 5D; Supplemental Fig. S19).
We present a model of ERAP2 regulation and function consistent with our findings and previous results (Fig. 5E). The genetic signals at the ERAP2 locus suggest at least three perfectly linked variants on Haplotype B affecting ERAP2 transcription and splicing in response to viral infection. Rs2548538, an intronic variant that overlaps chromatin marks from lymphoblastoid cell lines (The ENCODE Project Consortium 2012), is a candidate SNP that causes alternate transcript initiation at exon 9. Rs2248374G, the known splice site mutation, creates an alternate preferred splice site, resulting in alternative splicing of an extended exon 10. Rs2549797G, a splice-site mutation that creates a competing alternate splice site, results in ∼40% of the transcripts with an extended exon 15. Previous work has shown that full-length ERAP2 is a prototypical aminopeptidase that homodimerizes and heterodimerizes with ERAP1 (Saveanu et al. 2005) to perform the final peptide trimming step prior to MHC class I loading in the ER. The translation of the flu-specific ERAP2 isoforms that lack the aminopeptidase domain suggests that it could act as a dominant negative to either ERAP2 or ERAP1 to disrupt normal antigen processing, creating a more immunogenic MHC peptide repertoire that could confer a fitness advantage in response to virus.
Discussion
Although maps of genetic variants associated with overall gene abundances have been generated in many tissue types, the genetic control of alternate isoform usage has not been extensively studied. By using de novo transcript reconstruction, we found a large number of previously uncharacterized transcripts in human MoDCs, especially in response to influenza and interferon stimulation, indicating that the current reference human transcriptome is far from complete. However, de novo reconstruction using short reads has fundamental limitations especially for longer genes with multiple splice junctions (Steijger et al. 2013). A hybrid approach that uses long-read sequencing (Byrne et al. 2017; Tardaguila et al. 2018) to scaffold and short-read sequencing to assemble and quantify transcripts should result in higher-quality reference transcriptomes.
We found that genetic variants (isoQTLs) associated with alternate isoform usage are widespread, >40% of which are not associated with the overall abundance of the corresponding gene, indicative of independent genetic control of gene regulation at these loci. The enrichments of isoQTLs for known splice sites and autoimmune and neurodegenerative disease loci suggest a highly clinically relevant set of candidate variants that induce changes in protein sequence. To further assess the landscape of genetic control of isoform usage across immune cell types and stimuli, bulk sequencing in sorted cell populations or cost-effective single-cell sequencing (Kang et al. 2018) could be performed.
To conclusively establish the consequences of isoQTLs on protein function would require profiling of protein isoforms in large population cohorts, which remains a challenging task. Indeed, other studies have shown that much of the genetic effects on overall transcript abundance has little effect on overall protein abundance, likely due to buffering by post-transcriptional processes (Li et al. 2016). One independent approach to assess isoform function would be to compare transcript structure and abundances across species. Although several studies have begun to undertake this task across mammals for a number of tissues (Barbosa-Morais et al. 2012; Merkin et al. 2012), studies in immune cell types in response to specific stimuli are still lacking.
IsoQTLs, like eQTLs, can affect gene expression at distal loci in the genome, suggesting important downstream effects on gene regulation. The most striking example is at the IRF7 locus, where a splice-site SNP affects IRF7 splicing in response to influenza and interferon but only affects the expression of downstream genes in response to flu. This suggests that both genetic effects on isoform usage and stimulation-dependent regulation of IRF7 expression are necessary for the observed trans effects. Although C-terminal splice forms of IRF7 have been shown to differentially transactivate type 1 interferons and chemokines (Lin et al. 2000), IRF7/Iso4 is not known to have specific antiviral properties in vivo even though its ectopic expression is known to activate IFNAs in fibroblasts (Au et al. 1998). The association of the variant with SLE indicates a possible role for viral exposure to prime the immune system of individuals carrying the risk allele toward autoimmunity.
Genetic variants at a single locus could affect multiple facets of gene regulation in response to stimulation to establish variability in transcript structure and abundance. This was clearly demonstrated at the ERAP2 locus, where multiple variants result in differential expression and splicing of short isoforms in response to influenza but not interferon. The lack of expression in response to IFNB1 suggests the transcription of novel ERAP2 isoforms is likely initiated by viral sensing pathways upstream of type 1 interferon signaling. Furthermore, balancing selection at ERAP2 suggests that transcripts derived from both haplotypes could confer fitness advantages, likely in different environments. We provide evidence of the expression of short ERAP2 isoforms encoded by Haplotype B in response to influenza, suggesting viral infection as a possible selective agent. One can speculate that the long ERAP2 isoform encoded by Haplotype A could be selected under different environmental conditions that favor an overactive autoinflammatory response or a primed T-cell response. Altogether, the genetic analysis of isoforms under physiologically relevant conditions can help reveal new gene regulatory mechanisms by which alleles associated with disease and under natural selection function in response to environment.
Methods
Study subjects and sample preparation
Donors were recruited from the Boston community and gave written informed consent for the studies. Individuals were excluded if they had a history of inflammatory disease, autoimmune disease, chronic metabolic disorders, or chronic infectious disorders. Donors were between 18 and 56 yr of age (average 29.9). As previously described (Lee et al. 2014), 35–50 mL of peripheral blood from fasting subjects was collected between 7:30 and 8:30 am. The blood was drawn into sodium heparin tubes, and peripheral blood mononuclear cells (PBMCs) were isolated by Ficoll-Paque (GE Healthcare Life Sciences) centrifugation. PBMCs were frozen in liquid N2 in 90% FBS (Sigma-Aldrich) and 10% DMSO (Sigma-Aldrich). Monocytes were isolated from PBMCs by negative selection using the Dynabeads untouched human monocytes kit (Life Technologies) modified to increase throughput and optimize recovery and purity of CD14+CD16lo monocytes: The FcR blocking reagent was replaced with Miltenyi FcR blocking reagent (Miltenyi); per milliliter of antibody mix, an additional 333 µg biotinylated anti-CD16 (Biolegend, catalog no. 302004), 167 µg biotinylated anti-CD3 (Biolegend, catalog no. 344820), and 167 µg biotinylated anti-CD19 (Biolegend, catalog no. 302204) antibodies were added; the antibody labeling was modified to be performed in 96-well plates; and Miltenyi MS columns or multi-96 columns (Miltenyi) were used to separate magnetically labeled cells from unlabeled cells in an OctoMACS separator or MultiMACS M96 separator (Miltenyi), respectively. The number of PBMCs and monocytes was estimated using a CellTiter-Glo luminescent cell viability assay (Promega). A subset of the isolated monocytes was stained with PE-labeled anti-CD14 (M5E2; BD Biosciences) and FITC-labeled anti-CD16 (3G8; Biolegend) and was subjected to flow cytometry analysis using an Accuri C6 flow cytometer (BD Biosciences). A median of 94% CD14+ cells and 99% CD16lo cells was obtained.
Differentiation and stimulation of primary human MoDCs
Monocytes were cultured for 7 d in RPMI (Life Technologies) supplemented with 10% FBS, 100 ng/mL GM-CSF (R&D Systems), and 40 ng/mL IL-4 (R&D Systems) to differentiate the monocytes into MoDCs; 4 × 104 MoDCs were seeded in each well of a 96-well plate. Cells were left unstimulated or were stimulated either with Influenza A (PR8 ΔNS1, prepared as previously described) (Shapira et al. 2009) for 10 h or with 100 U/mL recombinant IFNB1 (PBL Assay Science) for 6.5 h. Cells were then lysed in RLT buffer (Qiagen) supplemented with 1% B1-mercaptoethanol (Sigma-Aldrich).
RNA isolation and sequencing
RNA from all samples was extracted using the RNeasy 96 kit (Qiagen, catalog no. 74182), according to the manufacturer's protocols. Five hundred seventy-six total samples were sequenced (99 baseline, 250 influenza infected, and 227 IFNB1 stimulated). Five hundred fifty-two pass-filter samples (94 baseline, 243 influenza, and 215 interferon) were sequenced to an average depth of 38 million 76-bp paired-end reads using the Illumina TruSeq kit. Samples were filtered if the genotypes estimated from the RNA-sequencing did not match the genotypes obtained from genotyping (19 samples) or if self-reported ethnicity did not match the ethnicity estimated from genotypes (five samples) (Supplemental Table S1). Reads were aligned to hg19 genome with TopHat v1.4.1 (--mate-inner-dist 300 --mate-std-dev 500) (Trapnell et al. 2009) with 86% mapping to transcriptome and 97% mapping to the genome (Supplemental Table S1). Realigning our data to hg38 (GRCh38) would not significantly affect our conclusions: hg38 (GRCh38) primarily improves from hg19 (GRCh37) in centromeric and other repetitive regions in the human genome that were not included in our analysis due to ambiguity from mapping short RNA-seq reads. It is estimated that hg38 increases the number of mappable mRNAs by 3% (Schneider et al. 2017) and thus minimally impacts our RNA-seq–based study. Further, as all our samples were mapped to hg19, any sequence errors that introduced artifacts into read mapping would have affected all the samples equally and would not have yielded significant differential expression or splicing results.
Transcriptome reconstruction
After aligning reads to the genome, transcriptomes were assembled for each sample individually using StringTie (Pertea et al. 2015) and default parameters. Abundances of annotated transcripts were quantified using Kallisto (Bray et al. 2016). For genes expressed at more than five TPM in any sample, isoforms expressed at less than five TPM across all samples were removed. In order to reduce the number of transfrags, transcriptomes across the same condition (e.g., baseline, flu, IFNB1) were first merged using cuffmerge (--overhang-tolerance 0) (Trapnell et al. 2012). Merged transcriptomes across all three conditions were then combined, and redundant isoforms were removed using cuffcompare (Trapnell et al. 2012).
Transcriptome comparison
For each reconstructed isoform, the position of each splice site, as well as the 5′ and 3′ position of each splice junction, was compared with annotated isoforms from GENCODE (v27) (Harrow et al. 2006), UCSC (hg19) (Casper et al. 2018), and RefSeq (hg19) (O'Leary et al. 2016), and comparison statistics for the most similar annotated isoform were reported. To quantify the coverage of spliced reads across splice junctions, each TopHat alignment was inputted into LeafCutter (Li et al. 2018), which uses the CIGAR strings in alignment BAM files to count the number of high-quality aligning reads at each splice junction. To detect novel TSSs, we checked the first 100 bp of our reconstructed isoforms for overlap with TSSs of annotated isoforms. Novel TSSs detected in our data set were further compared with CAGE reads from MoDCs from the FANTOM5 database (FF:11227-116C3, F:11308-117C3, FF:11384-118B7) (Noguchi et al. 2017) in the following way: First, a specialized transcriptome consisting of only the first 500 bp of each de novo assembled transcript (stranded, and after splicing) was created. Then, CAGE reads were aligned to this specialized transcriptome using Bowtie v0.12.7 with default parameters (Langmead 2010). Finally, the presence of each TSS was quantified by counting the number of mapped reads.
Differential expression analysis
Isoform level differential expression testing was carried out with sleuth (Pimentel et al. 2017) using 100 bootstraps per sample. Gene-level quantification was estimated by summing isoform counts from Kallisto, and differential expression testing was carried out with DESeq2 (Anders and Huber 2010).
Differential isoform usage analysis
Differential isoform usage testing was carried out in R using the beta regression package betareg (Cribari-Neto and Zeileis 2010), and P-values were calculated using a likelihood ratio test and adjusted with a false-discovery rate adjustment.
Gene Ontology enrichment analysis
Gene Ontology (GO) enrichment analysis was carried out using GOrilla (Eden et al. 2009) and tested against a background of only the set of genes that were expressed in MoDCs and that we recovered during the transcriptome reconstruction (see above).
DNA extraction and genotyping
As previously described (Lee et al. 2014), genomic DNA was extracted from 5 mL whole blood (DNeasy blood & tissue kit; Qiagen) and quantified by NanoDrop. Each subject was genotyped using Illumina infinium human OmniExpress exome BeadChip, which includes genome-wide genotype data as well as genotypes for rare variants from 12,000 exomes as well as common coding variants from the whole genome. In total, 951,117 SNPs were genotyped, of which 704,808 SNPs are common variants (minor allele frequency [MAF] > 0.01) and 246,229 are part of the exomes. The genotype success rate was ≥97%. We applied rigorous subject and SNP quality control (QC) that includes (1) gender misidentification, (2) subject relatedness, (3) Hardy-Weinberg equilibrium testing, (4) use concordance to infer SNP quality, (5) genotype call rate, (6) heterozygosity outlier, and (7) subject mismatches. In the European population, we excluded 1987 SNPs with a call rate <95%, 459 SNPs with Hardy-Weinberg equilibrium P-value <10−6, 234 SNPs with a MisHap P-value <10−9, and 63,781 SNPs with MAF < 1% from a total of 66,461 SNPs excluded. In the African-American population, we excluded 2161 SNPs with a call rate <95%, 298 SNPs with Hardy-Weinberg equilibrium P-value <10−6, 50 SNPs with a MisHap P-value <10−9, and 17,927 SNPs with MAF <1% from a total of 20,436 SNPs excluded. In the East Asian population, we excluded 1831 SNPs with a call rate <95%, 213 SNPs with Hardy-Weinberg equilibrium P-value <10−6, 47 SNPs with a MisHap P-value <10−9, and 84,973 SNPs with MAF <1% from a total of 87,064 SNPs excluded. After QC, approximately 18,000–88,000 SNPs in each population were filtered out from our analysis. Underlying genetic stratification in the population was assessed by multidimensional scaling using data from The International HapMap Project (The International HapMap Consortium 2003) (CEU, YRI and CHB samples) combined with IBS cluster analysis using EIGENSTRAT 3.0 software (Price et al. 2006). The QC of the genotyping data was performed using PLINK (Purcell et al. 2007).
Genotype imputation
To accurately evaluate the evidence of association signal at variants that are not directly genotyped, we used BEAGLE (Browning and Browning 2016) software (v3.3.2) to impute the post-QC genotyped markers using reference haplotype panels from The 1000 Genomes Project (The 1000 Genomes Project Consortium Phase I Integrated Release Version 3) (Siva 2008), which contain a total of 37.9 million SNPs in 1092 individuals with ancestry from West Africa, East Asia, and Europe. For subjects of European and East Asian ancestry, we used haplotypes from Utah residents (CEPH) with Northern and Western European ancestry (CEU) and combined panels from Han Chinese in Beijing (CHB) and Japanese in Tokyo (JPT), respectively. For imputing African American subjects, we used a combined haplotype reference panel consisting of CEU and Yoruba in Ibadan, Nigeria (YRI). For the admixed African American population, using two reference panels substantially improves imputation performance. After genotype imputation, we filtered out poorly imputed (BEAGLE r2 < 0.1) and low MAF SNPs (MAF < 0.01), which resulted in 7.7 million, 6.6 million, and 12.7 million common variants in European, East Asian, and African American, respectively. This set of genotyped and imputed markers was used for all the subsequent association analysis.
Local eQTL and isoQTL mapping
QTL mapping was performed using the Matrix eQTL (Shabalin 2012) package using empirically determined number of principal components (PCs) as covariates for each analysis. The isoform usage ratio or log of normalized total gene abundance was regressed against all genetics variants with a MAF >5% in a 1-Mb (±500 kb) window, and the most significant association is kept. Zero to 44 PCs (local eQTLs) and zero to 12 PCs (local isoQTLs) in increments of two were tested, and the number of PCs was chosen to maximize the number of local e/isoQTLs detected (Supplemental Figs. S2, S3). Because of the smaller number of individuals in the baseline condition, the number of PCs adjusted was fewer (Supplemental Fig. S2). Because the isoform usage percentage implicitly adjusts for confounders that affect overall gene abundance and isoform abundance levels (i.e., other eQTLs), the number of adjusted PCs was also fewer (Supplemental Fig. S3). Experiment-wide empirical P-values were calculated by comparing the nominal P-values with null P-values determined by permuting each isoform/gene 1000 times (Churchill and Doerge 1994). The permutation P-values were not pooled to calculate the empirical P-values (i.e., the minimum possible P-value is 0.001). False-discovery rates were calculated using the qvalue package (https://github.com/StoreyLab/qvalue) as previously described (Storey and Tibshirani 2003).
Independence of eQTLs and isoQTLs
We examined the overlap between eQTLs and isoQTLs to better understand the underlying mechanisms by which differential isoform usage is achieved. There could be two genetic architectures that result in an isoQTL. Suppose we have two transcripts A1 and A2 for gene A. Consider the first architecture, SNP G1, affects the transcription of A1 but not A2: and . Compared with using log(transcript A1 abundance) as a trait, detecting an eQTL by fitting linear regression using log(total abundance) and detecting an isoQTL using isoform usage ratio would both have reduced power (Supplemental Figure S6). Consider the second architecture, SNP G2, affects the splicing of A1 versus A2: and . Compared with using log(transcript A1 abundance) as a trait, detecting an eQTL by fitting linear regression using log(total abundance) would have no power, and detecting an isoQTL using isoform usage ratio would have increased power (Supplemental Figure S7). We obtained similar results by performing negative binomial regression. The parameters used for the simulation were N = 100, MAF = 0.5, μ0 = 500. The size parameter for the negative binomial was chosen based on published recommendations (Frazee et al. 2015).
QTL annotation
QTLs were annotated using Variant Effect Predictor and Ensembl release 79 (McLaren et al. 2016). Exonic and intronic locations of QTLs were determined using UCSC's canonical transcripts (table knownCanonical) as a reference (Karolchik et al. 2004). Enrichments were calculated against background set of SNPs that were matched in allele frequency (binned by 4%) and distance to nearest TSS (binned by 10 kb).
Overlap with GWAS associations
The GREGOR suite (Schmidt et al. 2015) was used for calculating the enrichment of eQTLs and isoQTLs containing a GWAS loci across baseline, flu, and IFNB1 stimulations. GWAS associations for disease with FDR < 0.1 were reported.
Partitioned heritability analysis
We used ldsc with default parameters, which implements LD score regression (Finucane et al. 2015) to calculate the proportion of SNP heritability explained by eQTL/isoQTLs. We obtained summary statistics from 28 human traits/diseases from https://data.broadinstitute.org/alkesgroup/sumstats_formatted/.
Estimating flu transcript abundance
Flu transcript abundance was estimated by using RSEM (Li and Dewey 2011) to map RNA-seq data to a custom reference of the influenza PR8 genome (Supplemental Material).
ERAP2 western blot
Protein extracts were fractionated by SDS-PAGE (4%–12% Bis-Tris gel, Thermo Fisher Scientific, NP0335BOX) and transferred to PVDF membrane (Bio-Rad, catalog no. 162-0177). After blocking with 2% BSA in TBST (Tris buffered saline containing 0.1% Tween 20) for 1 h, membranes were incubated with primary antibody (either ERAP2, R&D Systems, catalog no. AF3830, 1:3000) or actin beta (Abcam, catalog no. ab6276, 1:15,000) overnight at 4°C. Membranes were then washed and incubated with a 1:5000 dilution of HRP conjugated secondary antibody (either donkey anti-goat from Santa Cruz Biotech, catalog no. sc2020, or with goat anti-mouse from Jackson Immune Research, catalog no. 115-035-146) for 1 h. Membranes were washed and developed with ECL system (VWR, catalog no. 89168-782) according to the manufacturer's protocol.
Data access
RNA-seq raw data from this study have been submitted to the NCBI database of Genotypes and Phenotypes (dbGaP; https://www.ncbi.nlm.nih.gov/gap) under accession number phs000815.v1.p1. RNA-seq processed data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE92904.
Competing interest statement
N.H. is a founder and SAB member of Neon Therapeutics and a SAB member of IFM Therapeutics. A.R. is a SAB member of Thermo Fisher Scientific, Syros Pharmaceuticals, and Driver group and a founder of Celsius Therapeutics.
Supplementary Material
Acknowledgments
We thank the ImmVar participants. We also thank the Ye, Regev, and Hacohen laboratories for discussions. R.E.G., M.S., and C.J.Y. are supported by National Institutes of Health (NIH) R01AR071522 and R01AI136972 to C.J.Y. N.H. is supported by NIH P50HG006193 and RC2GM093080. Special thanks to Genentech Inc. for additional funding support.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.240390.118.
Freely available online through the Genome Research Open Access option.
References
- Amit I, Garber M, Chevrier N, Leite AP, Donner Y, Eisenhaure T, Guttman M, Grenier JK, Li W, Zuk O, et al. 2009. Unbiased reconstruction of a mammalian transcriptional network mediating pathogen responses. Science 326: 257–263. 10.1126/science.1179050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Genome Biol 11: R106 10.1186/gb-2010-11-10-r106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andres AM, Dennis MY, Kretzschmar WW, Cannons JL, Lee-Lin SQ, Hurle B, NISC Comparative Sequencing Program, Schwartzberg PL, Williamson SH, Bustamante CD, et al. 2010. Balancing selection maintains a form of ERAP2 that undergoes nonsense-mediated decay and affects antigen presentation. PLoS Genet 6: e1001157 10.1371/journal.pgen.1001157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Au WC, Moore PA, LaFleur DW, Tombal B, Pitha PM. 1998. Characterization of the interferon regulatory factor-7 and its potential role in the transcription activation of interferon A genes. J Biol Chem 273: 29210–29217. 10.1074/jbc.273.44.29210 [DOI] [PubMed] [Google Scholar]
- Barbosa-Morais NL, Irimia M, Pan Q, Xiong HY, Gueroussov S, Lee LJ, Slobodeniuc V, Kutter C, Watt S, Colak R, et al. 2012. The evolutionary landscape of alternative splicing in vertebrate species. Science 338: 1587–1593. 10.1126/science.1230612 [DOI] [PubMed] [Google Scholar]
- Barreiro LB, Tailleux L, Pai AA, Gicquel B, Marioni JC, Gilad Y. 2012. Deciphering the genetic architecture of variation in the immune response to Mycobacterium tuberculosis infection. Proc Natl Acad Sci 109: 1204–1209. 10.1073/pnas.1115761109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berard M, Tough DF. 2002. Qualitative differences between naïve and memory T cells. Immunology 106: 127–138. 10.1046/j.1365-2567.2002.01447.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black DL. 2003. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem 72: 291–336. 10.1146/annurev.biochem.72.121801.161720 [DOI] [PubMed] [Google Scholar]
- Bray NL, Pimentel H, Melsted P, Pachter L. 2016. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol 34: 525–527. 10.1038/nbt.3519 [DOI] [PubMed] [Google Scholar]
- Breckenridge DG, Nguyen M, Kuppig S, Reth M, Shore GC. 2002. The procaspase-8 isoform, procaspase-8L, recruited to the BAP31 complex at the endoplasmic reticulum. Proc Natl Acad Sci 99: 4331–4336. 10.1073/pnas.072088099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning BL, Browning SR. 2016. Genotype imputation with millions of reference samples. Am J Hum Genet 98: 116–126. 10.1016/j.ajhg.2015.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne A, Beaudin AE, Olsen HE, Jain M, Cole C, Palmer T, DuBois RM, Forsberg EC, Akeson M, Vollmers C. 2017. Nanopore long-read RNAseq reveals widespread transcriptional variation among the surface receptors of individual B cells. Nat Commun 8: 16027 10.1038/ncomms16027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cagliani R, Riva S, Biasin M, Fumagalli M, Pozzoli U, Lo Caputo S, Mazzotta F, Piacentini L, Bresolin N, Clerici M, et al. 2010. Genetic diversity at endoplasmic reticulum aminopeptidases is maintained by balancing selection and is associated with natural resistance to HIV-1 infection. Hum Mol Genet 19: 4705–4714. 10.1093/hmg/ddq401 [DOI] [PubMed] [Google Scholar]
- Casper J, Zweig AS, Villarreal C, Tyner C, Speir ML, Rosenbloom KR, Raney BJ, Lee CM, Lee BT, Karolchik D, et al. 2018. The UCSC Genome Browser database: 2018 update. Nucleic Acids Res 46: D762–D769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill GA, Doerge RW. 1994. Empirical threshold values for quantitative trait mapping. Genetics 138: 963–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cribari-Neto F, Zeileis A. 2010. Beta regression in R. J Stat Softw 34: 1–24. 10.18637/jss.v034.i02 [DOI] [Google Scholar]
- Deveson IW, Brunck ME, Blackburn J, Tseng E, Hon T, Clark TA, Clark MB, Crawford J, Dinger ME, Nielsen LK, et al. 2018. Universal alternative splicing of noncoding exons. Cell Syst 6: 245–255 e245. 10.1016/j.cels.2017.12.005 [DOI] [PubMed] [Google Scholar]
- Doyle S, Vaidya S, O'Connell R, Dadgostar H, Dempsey P, Wu T, Rao G, Sun R, Haberland M, Modlin R, et al. 2002. IRF3 mediates a TLR3/TLR4-specific antiviral gene program. Immunity 17: 251–263. 10.1016/S1074-7613(02)00390-4 [DOI] [PubMed] [Google Scholar]
- Eden E, Navon R, Steinfeld I, Lipson D, Yakhini Z. 2009. GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10: 48 10.1186/1471-2105-10-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74. 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enders A, Short A, Miosge LA, Bergmann H, Sontani Y, Bertram EM, Whittle B, Balakishnan B, Yoshida K, Sjollema G, et al. 2014. Zinc-finger protein ZFP318 is essential for expression of IgD, the alternatively spliced Igh product made by mature B lymphocytes. Proc Natl Acad Sci 111: 4513–4518. 10.1073/pnas.1402739111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fairfax BP, Knight JC. 2014. Genetics of gene expression in immunity to infection. Curr Opin Immunol 30: 63–71. 10.1016/j.coi.2014.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finucane HK, Bulik-Sullivan B, Gusev A, Trynka G, Reshef Y, Loh PR, Anttila V, Xu H, Zang C, Farh K, et al. 2015. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet 47: 1228–1235. 10.1038/ng.3404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazee AC, Jaffe AE, Langmead B, Leek JT. 2015. Polyester: simulating RNA-seq datasets with differential transcript expression. Bioinformatics 31: 2778–2784. 10.1093/bioinformatics/btv272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham RR, Kyogoku C, Sigurdsson S, Vlasova IA, Davies LR, Baechler EC, Plenge RM, Koeuth T, Ortmann WA, Hom G, et al. 2007. Three functional variants of IFN regulatory factor 5 (IRF5) define risk and protective haplotypes for human lupus. Proc Natl Acad Sci 104: 6758–6763. 10.1073/pnas.0701266104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The GTEx Consortium. 2015. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science 348: 648–660. 10.1126/science.1262110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow J, Denoeud F, Frankish A, Reymond A, Chen CK, Chrast J, Lagarde J, Gilbert JG, Storey R, Swarbreck D, et al. 2006. GENCODE: producing a reference annotation for ENCODE. Genome Biol 7(Suppl 1): S4.1–9. 10.1186/gb-2006-7-s1-s4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Caselles T, Martinez-Esparza M, Perez-Oliva AB, Quintanilla-Cecconi AM, Garcia-Alonso A, Alvarez-Lopez DM, Garcia-Penarrubia P. 2006. A study of CD33 (SIGLEC-3) antigen expression and function on activated human T and NK cells: Two isoforms of CD33 are generated by alternative splicing. J Leukoc Biol 79: 46–58. 10.1189/jlb.0205096 [DOI] [PubMed] [Google Scholar]
- The International HapMap Consortium. 2003. The International HapMap Project. Nature 426: 789–796. 10.1038/nature02168 [DOI] [PubMed] [Google Scholar]
- Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, et al. 2012. Host–microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 491: 119–124. 10.1038/nature11582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HM, Subramaniam M, Targ S, Nguyen M, Maliskova L, McCarthy E, Wan E, Wong S, Byrnes L, Lanata CM, et al. 2018. Multiplexed droplet single-cell RNA-sequencing using natural genetic variation. Nat Biotechnol 36: 89–94. 10.1038/nbt.4042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D, Hinrichs AS, Furey TS, Roskin KM, Sugnet CW, Haussler D, Kent WJ. 2004. The UCSC Table Browser data retrieval tool. Nucleic Acids Res 32: D493–D496. 10.1093/nar/gkh103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuriakose T, Man SM, Malireddi RK, Karki R, Kesavardhana S, Place DE, Neale G, Vogel P, Kanneganti TD. 2016. ZBP1/DAI is an innate sensor of influenza virus triggering the NLRP3 inflammasome and programmed cell death pathways. Sci Immunol 1: aag2045 10.1126/sciimmunol.aag2045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalonde E, Ha KC, Wang Z, Bemmo A, Kleinman CL, Kwan T, Pastinen T, Majewski J. 2011. RNA sequencing reveals the role of splicing polymorphisms in regulating human gene expression. Genome Res 21: 545–554. 10.1101/gr.111211.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B. 2010. Aligning short sequencing reads with Bowtie. Curr Protoc Bioinformatics Chapter 11: Unit 11 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappalainen T, Sammeth M, Friedlander MR, t Hoen PA, Monlong J, Rivas MA, Gonzalez-Porta M, Kurbatova N, Griebel T, Ferreira PG, et al. 2013. Transcriptome and genome sequencing uncovers functional variation in humans. Nature 501: 506–511. 10.1038/nature12531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MN, Ye C, Villani AC, Raj T, Li W, Eisenhaure TM, Imboywa SH, Chipendo PI, Ran FA, Slowikowski K, et al. 2014. Common genetic variants modulate pathogen-sensing responses in human dendritic cells. Science 343: 1246980 10.1126/science.1246980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12: 323 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YI, van de Geijn B, Raj A, Knowles DA, Petti AA, Golan D, Gilad Y, Pritchard JK. 2016. RNA splicing is a primary link between genetic variation and disease. Science 352: 600–604. 10.1126/science.aad9417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YI, Knowles DA, Humphrey J, Barbeira AN, Dickinson SP, Im HK, Pritchard JK. 2018. Annotation-free quantification of RNA splicing using LeafCutter. Nat Genet 50: 151–158. 10.1038/s41588-017-0004-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R, Mamane Y, Hiscott J. 2000. Multiple regulatory domains control IRF-7 activity in response to virus infection. J Biol Chem 275: 34320–34327. 10.1074/jbc.M002814200 [DOI] [PubMed] [Google Scholar]
- Lo WS, Gardiner E, Xu Z, Lau CF, Wang F, Zhou JJ, Mendlein JD, Nangle LA, Chiang KP, Yang XL, et al. 2014. Human tRNA synthetase catalytic nulls with diverse functions. Science 345: 328–332. 10.1126/science.1252943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur J, Bowler E, Cerezo M, Gil L, Hall P, Hastings E, Junkins H, McMahon A, Milano A, Morales J, et al. 2017. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res 45: D896–D901. 10.1093/nar/gkw1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maquat LE. 2004. Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat Rev Mol Cell Biol 5: 89–99. 10.1038/nrm1310 [DOI] [PubMed] [Google Scholar]
- Martinez NM, Lynch KW. 2013. Control of alternative splicing in immune responses: many regulators, many predictions, much still to learn. Immunol Rev 253: 216–236. 10.1111/imr.12047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matlin AJ, Clark F, Smith CW. 2005. Understanding alternative splicing: towards a cellular code. Nat Rev Mol Cell Biol 6: 386–398. 10.1038/nrm1645 [DOI] [PubMed] [Google Scholar]
- McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GR, Thormann A, Flicek P, Cunningham F. 2016. The Ensembl Variant Effect Predictor. Genome Biol 17: 122 10.1186/s13059-016-0974-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkin J, Russell C, Chen P, Burge CB. 2012. Evolutionary dynamics of gene and isoform regulation in mammalian tissues. Science 338: 1593–1599. 10.1126/science.1228186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris DL, Sheng Y, Zhang Y, Wang YF, Zhu Z, Tombleson P, Chen L, Cunninghame Graham DS, Bentham J, Roberts AL, et al. 2016. Genome-wide association meta-analysis in Chinese and European individuals identifies ten new loci associated with systemic lupus erythematosus. Nat Genet 48: 940–946. 10.1038/ng.3603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nedelec Y, Sanz J, Baharian G, Szpiech ZA, Pacis A, Dumaine A, Grenier JC, Freiman A, Sams AJ, Hebert S, et al. 2016. Genetic ancestry and natural selection drive population differences in immune responses to pathogens. Cell 167: 657–669 e621. 10.1016/j.cell.2016.09.025 [DOI] [PubMed] [Google Scholar]
- Noguchi S, Arakawa T, Fukuda S, Furuno M, Hasegawa A, Hori F, Ishikawa-Kato S, Kaida K, Kaiho A, Kanamori-Katayama M, et al. 2017. FANTOM5 CAGE profiles of human and mouse samples. Sci Data 4: 170112 10.1038/sdata.2017.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, Rajput B, Robbertse B, Smith-White B, Ako-Adjei D, et al. 2016. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res 44: D733–D745. 10.1093/nar/gkv1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. 2015. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 33: 290–295. 10.1038/nbt.3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimentel H, Bray NL, Puente S, Melsted P, Pachter L. 2017. Differential analysis of RNA-seq incorporating quantification uncertainty. Nat Methods 14: 687–690. 10.1038/nmeth.4324 [DOI] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. 2006. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38: 904–909. 10.1038/ng1847 [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81: 559–575. 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quach H, Rotival M, Pothlichet J, Loh YE, Dannemann M, Zidane N, Laval G, Patin E, Harmant C, Lopez M, et al. 2016. Genetic adaptation and Neandertal admixture shaped the immune system of human populations. Cell 167: 643–656.e617. 10.1016/j.cell.2016.09.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj T, Rothamel K, Mostafavi S, Ye C, Lee MN, Replogle JM, Feng T, Lee M, Asinovski N, Frohlich I, et al. 2014a. Polarization of the effects of autoimmune and neurodegenerative risk alleles in leukocytes. Science 344: 519–523. 10.1126/science.1249547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj T, Ryan KJ, Replogle JM, Chibnik LB, Rosenkrantz L, Tang A, Rothamel K, Stranger BE, Bennett DA, Evans DA, et al. 2014b. CD33: increased inclusion of exon 2 implicates the Ig V-set domain in Alzheimer's disease susceptibility. Hum Mol Genet 23: 2729–2736. 10.1093/hmg/ddt666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas MA, Pirinen M, Conrad DF, Lek M, Tsang EK, Karczewski KJ, Maller JB, Kukurba KR, DeLuca DS, Fromer M, et al. 2015. Human genomics. Effect of predicted protein-truncating genetic variants on the human transcriptome. Science 348: 666–669. 10.1126/science.1261877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell SM, Sparrow RL, McKenzie IF, Purcell DF. 1992. Tissue-specific and allelic expression of the complement regulator CD46 is controlled by alternative splicing. Eur J Immunol 22: 1513–1518. 10.1002/eji.1830220625 [DOI] [PubMed] [Google Scholar]
- Saveanu L, Carroll O, Lindo V, Del Val M, Lopez D, Lepelletier Y, Greer F, Schomburg L, Fruci D, Niedermann G, et al. 2005. Concerted peptide trimming by human ERAP1 and ERAP2 aminopeptidase complexes in the endoplasmic reticulum. Nat Immunol 6: 689–697. 10.1038/ni1208 [DOI] [PubMed] [Google Scholar]
- Schmidt EM, Zhang J, Zhou W, Chen J, Mohlke KL, Chen YE, Willer CJ. 2015. GREGOR: evaluating global enrichment of trait-associated variants in epigenomic features using a systematic, data-driven approach. Bioinformatics 31: 2601–2606. 10.1093/bioinformatics/btv201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider VA, Graves-Lindsay T, Howe K, Bouk N, Chen HC, Kitts PA, Murphy TD, Pruitt KD, Thibaud-Nissen F, Albracht D, et al. 2017. Evaluation of GRCh38 and de novo haploid genome assemblies demonstrates the enduring quality of the reference assembly. Genome Res 27: 849–864. 10.1101/gr.213611.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM. 2011. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472: 481–485. 10.1038/nature09907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabalin AA. 2012. Matrix eQTL: ultra fast eQTL analysis via large matrix operations. Bioinformatics 28: 1353–1358. 10.1093/bioinformatics/bts163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapira SD, Gat-Viks I, Shum BO, Dricot A, de Grace MM, Wu L, Gupta PB, Hao T, Silver SJ, Root DE, et al. 2009. A physical and regulatory map of host-influenza interactions reveals pathways in H1N1 infection. Cell 139: 1255–1267. 10.1016/j.cell.2009.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siva N. 2008. 1000 Genomes project. Nat Biotechnol 26: 256 10.1038/nbt0308-256b [DOI] [PubMed] [Google Scholar]
- Steijger T, Abril JF, Engstrom PG, Kokocinski F, RGASP Consortium, Hubbard TJ, Guigo R, Harrow J, Bertone P. 2013. Assessment of transcript reconstruction methods for RNA-seq. Nat Methods 10: 1177–1184. 10.1038/nmeth.2714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. 2003. Statistical significance for genomewide studies. Proc Natl Acad Sci 100: 9440–9445. 10.1073/pnas.1530509100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tardaguila M, de la Fuente L, Marti C, Pereira C, Pardo-Palacios FJ, Del Risco H, Ferrell M, Mellado M, Macchietto M, Verheggen K, et al. 2018. SQANTI: extensive characterization of long-read transcript sequences for quality control in full-length transcriptome identification and quantification. Genome Res 28: 396–411. 10.1101/gr.222976.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111. 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7: 562–578. 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakasugi K, Slike BM, Hood J, Otani A, Ewalt KL, Friedlander M, Cheresh DA, Schimmel P. 2002. A human aminoacyl-tRNA synthetase as a regulator of angiogenesis. Proc Natl Acad Sci 99: 173–177. 10.1073/pnas.012602099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. 2008. Alternative isoform regulation in human tissue transcriptomes. Nature 456: 470–476. 10.1038/nature07509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong HY, Alipanahi B, Lee LJ, Bretschneider H, Merico D, Yuen RK, Hua Y, Gueroussov S, Najafabadi HS, Hughes TR, et al. 2015. RNA splicing. The human splicing code reveals new insights into the genetic determinants of disease. Science 347: 1254806 10.1126/science.1254806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye CJ, Feng T, Kwon HK, Raj T, Wilson MT, Asinovski N, McCabe C, Lee MH, Frohlich I, Paik HI, et al. 2014. Intersection of population variation and autoimmunity genetics in human T cell activation. Science 345: 1254665 10.1126/science.1254665 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.