Abstract
In eukaryotic organisms, replication initiation follows a temporal program. Among the parameters that regulate this program in Saccharomyces cerevisiae, chromatin structure has been at the center of attention without considering the contribution of transcription. Here, we revisit the replication initiation program in the light of widespread genomic noncoding transcription. We find that noncoding RNA transcription termination in the vicinity of autonomously replicating sequences (ARSs) shields replication initiation from transcriptional readthrough. Consistently, high natural nascent transcription correlates with low ARS efficiency and late replication timing. High readthrough transcription is also linked to increased nucleosome occupancy and high levels of H3K36me3. Moreover, forcing ARS readthrough transcription promotes these chromatin features. Finally, replication initiation defects induced by increased transcriptional readthrough are partially rescued in the absence of H3K36 methylation. Altogether, these observations indicate that natural noncoding transcription into ARSs influences replication initiation through chromatin regulation.
DNA replication is a fundamental process occurring in all living organisms and ensuring accurate duplication of the genome. Eukaryotic replication initiation takes place at several dispersed locations termed replication origins. Origins are defined by a specific chromatin structure consisting of a nucleosome-depleted region (NDR) and the binding of specific replication initiation factors. In Saccharomyces cerevisiae, replication origins or ARSs (autonomously replicating sequences) are specified by an 11bp T-rich ARS consensus sequence (ACS) (Stinchcomb et al. 1979; Nieduszynski et al. 2006). ARSs also contain more degenerate A-rich B elements proposed to contribute to origin function by excluding nucleosomes (Bell 1995; Segal and Widom 2009; Eaton et al. 2010). Despite the occurrence of thousands of ACSs in the genome, only 200–300 are efficient for the recruitment of the AAA+ ATPase origin recognition complex (ORC) (Raghuraman et al. 2001; Hawkins et al. 2013; McGuffee et al. 2013). During the G1-phase, the ORC in conjunction with Cdt1 and Cdc6 promotes the binding of the MCM2-7 double hexamer helicase complex giving rise to the prereplication complex (pre-RC) (Deegan and Diffley 2016). The resulting ORC/MCM2-7-bound ARSs are said to be licensed for replication initiation and have the ability to initiate replication during the subsequent S-phase (Aparicio 2013).
Replication follows a temporal program of activation during S-phase. ARSs are defined by an activation timing based on the observation that some ARSs replicate earlier than others (Raghuraman et al. 2001; Hawkins et al. 2013). Moreover, using DNA combing, it appears that the fraction of cells in a population initiating replication at a given ARS is variable, defining a firing efficiency probability for each ARS (Czajkowsky et al. 2008; Hawkins et al. 2013; McGuffee et al. 2013). Timing and efficiency are linked, as inefficient origins tend to fire late during S-phase or to be replicated passively through the use of a neighboring origin (Yang et al. 2010). These two interdependent measurements of timing and efficiency are usually used to describe the replication initiation properties of ARSs.
In S. cerevisiae, many parameters affect ARS activity including limiting trans-acting factors, different chromosomal location, and/or subnuclear localization (Yoshida et al. 2013). ARS activity also depends on the chromatin context and histone modifications. First, early origins have a wider NDR than late ARSs and adjacent nucleosomes are more precisely positioned (Soriano et al. 2014; Rodriguez et al. 2017). Moreover, the strength of ORC recruitment correlates with ARS activity and is itself important for NDR establishment (Eaton et al. 2010; Belsky et al. 2015). Second, early ARS activation during S-phase depends on histone acetylation (Vogelauer et al. 2002; Unnikrishnan et al. 2010). Indeed, the Class I Histone Deacetylase (HDAC) Rpd3 delays initiation of a huge number of replication origins (Vogelauer et al. 2002; Aparicio et al. 2004; Knott et al. 2009) and narrows their nucleosome-depleted regions (Soriano et al. 2014).
Numerous studies attempting to consider transcription as another parameter to define replication initiation led to conflicting results. On one hand, transcription revealed some positive links with replication initiation, as highly transcribed genes were proposed to replicate earlier than lowly expressed genes (Fraser 2013). Furthermore, stalled RNA polymerase II (RNA Pol II) was involved in the recruitment of the ORC at the rDNA locus, and the activity of many replication origins depends on the presence of specific transcription factor binding sites (Knott et al. 2012; Mayan 2013). On the other hand, active ARSs are excluded from annotated ORFs and tend to localize after 3′-transcription terminators, suggesting that transcription and replication initiation do not coexist (Nieduszynski et al. 2006). Furthermore, natural or artificial induction of transcription through origins leads to replication defects via dissociation or sliding of the pre-RC and MCMs, respectively (Snyder et al. 1988; Nieduszynski et al. 2005; Mori and Shirahige 2007; Blitzblau et al. 2012; Gros et al. 2015). In this study, we aimed at clarifying the role of transcription in replication initiation considering the widespread genomic noncoding transcription.
The analysis of appropriate mutants and the development of new tools to examine nascent transcription have revealed that RNA Pol II occurs pervasively and that the transcriptional landscape in eukaryotic genomes extends far beyond mRNAs and stable noncoding RNAs (Churchman and Weissman 2011; Schaughency et al. 2014). One source of noncoding transcription stems from initiation at NDRs, an event controlled through early termination by the Nrd1-Nab3-Sen1 (NNS) complex recruited at the 5′ end of all RNA Pol II transcription units via interaction with the RNA Pol II C-terminal domain (CTD) (Steinmetz et al. 2001; Arigo et al. 2006). Recognition by Nrd1/Nab3 of specific motifs on the nascent RNA induces RNA Pol II termination usually within the first kilobase of transcription in a process coupled to degradation by the nuclear exosome component Rrp6 (Tudek et al. 2014). These cryptic unstable transcripts (CUTs) are revealed in the absence of Rrp6 (Wyers et al. 2005; Neil et al. 2009; Xu et al. 2009; van Dijk et al. 2011; Jensen et al. 2013). Depletion of Nrd1 results in transcriptional readthrough and accumulation of Nrd1 unterminated transcripts (NUTs), most of which correspond to extended CUTs (Schulz et al. 2013; Schaughency et al. 2014). In the presence of inefficient early termination signals, loss of Rrp6 can also favor readthrough transcription and elongation of CUTs (Castelnuovo et al. 2013). Another source of noncoding transcription is linked to mRNA 3′ end formation. The cleavage and polyadenylation (CPF) and cleavage factor (CF) complexes cleave the nascent mRNA just upstream of RNA Pol II. RNA Pol II release depends on a CPF-induced allosteric modification of the elongation complex as well as on the digestion of the generated 3′ fragment by the 5′-3′ exonuclease Rat1 (for a recent review, see Porrua et al. 2016). Thus, before being caught up by the so-called “torpedo,” RNA Pol II continues transcription, leading to an average 160-bp termination window after the polyadenylation site. Inefficient cleavage and polyadenylation can increase the level of this natural source of pervasive transcription (Kim et al. 2004; Luo et al. 2006; Baejen et al. 2017).
Using nascent transcription and replication analyses in strains depleted for early termination activities, we delineate how noncoding transcription negatively influences replication initiation by shaping the chromatin structure of ARS. Our study clearly defines genome-wide noncoding transcription as a new parameter regulating replication initiation.
Results
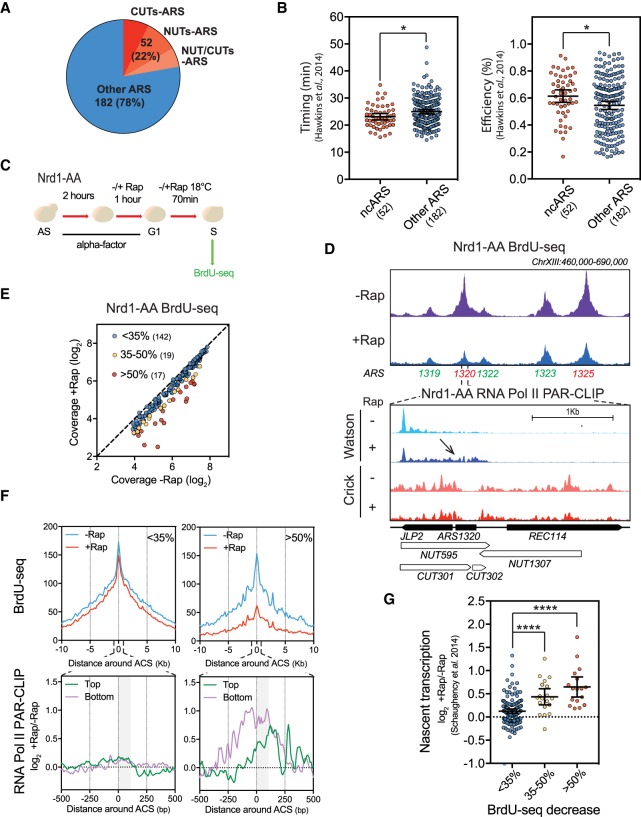
CUTs and NUTs are enriched at early and efficient ARSs
To determine the overlap of NUTs and CUTs with replication origins, we defined a list of 234 ARSs (Supplemental Table S1) with previously annotated ACSs (Nieduszynski et al. 2006; Soriano et al. 2014) and for which replication timing and efficiency had been established (Hawkins et al. 2013). Among the 234 well-defined ARSs used in this analysis, 52 (22%) overlap with a CUT (as already observed in Looke et al. 2010), a NUT, or both (Fig. 1A; Supplemental Fig. S1A; Xu et al. 2009; Schulz et al. 2013). CUTs and NUTs are defined in a mutant context when early termination is compromised. The presence of CUTs and NUTs over 52 replication origins indicates that termination of noncoding transcription through the NNS pathway is robust around these ARSs. Notably, the 52 ARSs overlapping with NUTs and/or CUTs (ncARSs) tend to be replicated earlier and more efficiently, on average, than the remaining 182 ARSs (Other ARSs) (Fig. 1B). These observations suggest that noncoding transcription termination may be a determinant of ARS replication timing and efficiency.
Figure 1.
NUTs- and CUTs-containing ARSs are down-regulated when early termination of noncoding RNAs is abrogated. (A) Numbers and proportions of ARSs overlapping with CUTs only (red), NUTs only (orange), or both CUTs and NUTs (pink). ARS annotations used in this study are listed in Supplemental Table S1. CUTs and NUTs were considered as overlapping when showing 50 bp of overlap in the ACS to +100 bp area according to the ACS T-rich sequence. (B) Scatter dot-plot indicating the timing and efficiency of the noncoding RNA-containing ARSs (ncARSs) compared to replication origins devoid of overlapping CUTs and NUTs (Other ARSs). Timing and efficiency data were retrieved from Hawkins et al. (2013). The mean and the 95% confidence interval are indicated. (C) Nrd1-AA cells were synchronized in G1-phase with alpha-factor for 3 h at 30°C. During the last hour, rapamycin (Rap) was added or not in the medium. Cells were then washed and released into the cell cycle at 18°C in the presence of BrdU and −/+Rap. After 70 min, cells were collected for DNA extraction and BrdU-seq. (D) Snapshot depicting a part of Chromosome XIII for the BrdU-seq. Affected ARSs are indicated in red and nonaffected in green. Bottom panel shows a zoom around ARS1320 of the RNA Pol II PAR-CLIP in the Nrd1-AA strain (Schaughency et al. 2014). Transcriptional readthrough is indicated by an arrow. (E) Plot depicting the mean coverage of BrdU nascent DNA in a 5-kb window around ACS in −Rap versus +Rap. The 17 red dots and 19 yellow dots represent the ARSs showing at least 50% and 35%–50% decrease in BrdU incorporation in +Rap, respectively. Blue dots represent the ARSs defined as nonaffected in BrdU incorporation. (F) Top: Metagene analysis of the BrdU-seq for the 142 nonaffected ARSs (<35%) and the 17 most affected ARSs (>50%). Profiles represent the mean coverage smoothed by a 200-bp moving window. ARSs were oriented according to their ACS T-rich sequence. Bottom: Metagene profiles of the ratio log2 +Rap/−Rap of the RNA Pol II PAR-CLIP signal 500 bp around the oriented ACS of the least and most affected ARS (Schaughency et al. 2014). Plots were smoothed by a 10-bp moving window. Since ARSs are oriented, nascent transcription going toward replication origins is defined as Top and Bottom. The gray box represents the window in which transcriptional readthrough was analyzed in G. (G) Scatter dot-plots representing the ratio log2 +Rap/−Rap of the RNA Pol II PAR-CLIP signal over the three classes of ARSs defined in E. Total nascent transcription in +Rap and −Rap was defined on oriented ARSs by adding the RNA Pol II PAR-CLIP mean densities between the ACS to +100 bp on the top strand to the signal over the same region on the bottom strand in each condition using the data from Schaughency et al. (2014). Each 100-bp segment was considered as 1 bin (see Methods). (*) P-value < 0.05; (****) P-value < 0.0001.
Noncoding transcription readthrough affects replication initiation
One hypothesis is that NNS termination in the vicinity of a subset of ARSs may shield them from pervasive transcription potentially deleterious for replication initiation. To investigate the effect of noncoding transcription readthrough on replication, early termination of noncoding RNAs was abrogated by rapid nuclear depletion of Nrd1 through anchor away (AA) (Haruki et al. 2008). Nrd1 depletion, induced by addition of rapamycin (Rap) to the engineered Nrd1-AA strain, is accompanied by transcription elongation and accumulation of NUTs (Schulz et al. 2013). To examine the effect on replication, the Nrd1-AA strain was treated with alpha factor to synchronize the cells in G1-phase and incubated an additional hour −/+ Rap to induce noncoding transcription. Cells were then released from G1 arrest in the presence of BrdU (Fig. 1C). FACS analyses indicate a slight cell cycle delay at 80 min in cells depleted for Nrd1, with an increased number of cells in G1 in +Rap compared to –Rap (Supplemental Fig. S1B). Samples were harvested for BrdU-seq at 70 min after G1-phase release. Visualization of the data revealed a number of well-defined peaks centered on specific ARSs (Fig. 1D; Supplemental Fig. S1C,D). Global analysis of the BrdU-seq showed that, out of the 178 selected early ARSs, 36 (20.2%) present a reproducible, more than 35% decrease in BrdU incorporation in +Rap versus −Rap (17 show >50% decrease and 19 between 35%–50% decrease) (Fig. 1E). Consistently, metagene analysis from −10 to +10 Kb around the ACS of the >50% affected ARSs revealed a substantial reduction in the BrdU-seq profile in +Rap, while the curves including the <35% affected ARSs presented only a slight change in +Rap versus −Rap (Fig. 1F). Affected ARSs showed a nice overlap with the NUTs-containing ARSs defined in Figure 1A (Supplemental Fig. S1E). To define whether the decreased BrdU-seq signal of affected ARSs upon Nrd1 depletion was linked to transcription, RNA Pol II PAR-CLIP data from the Corden lab (Schaughency et al. 2014) were used to examine the level of nascent transcription over ARS when depleting Nrd1. The ARSs with the strongest decrease in BrdU incorporation in +Rap versus −Rap also showed the highest increase in nascent RNA Pol II transcription into the ACSs to +100 bp ORC-footprinting area (Fig. 1E–G; Belsky et al. 2015). Thus, the replication defect observed following Nrd1 depletion is not due to the slightly slower cell cycle progression in +Rap but is directly linked to increased nascent transcription through the affected ARS.
RNA and BrdU analyses were also performed with the Rrp6-AA strain. Anchor away of Rrp6 has already been described by our lab to result in CUT elongation (Castelnuovo et al. 2014). Depletion of Rrp6 resulted in similar effects on ncRNA accumulation and replication initiation (Supplemental Fig. S2).
Overall, these data suggest that replication initiation may be hindered by noncoding readthrough transcription.
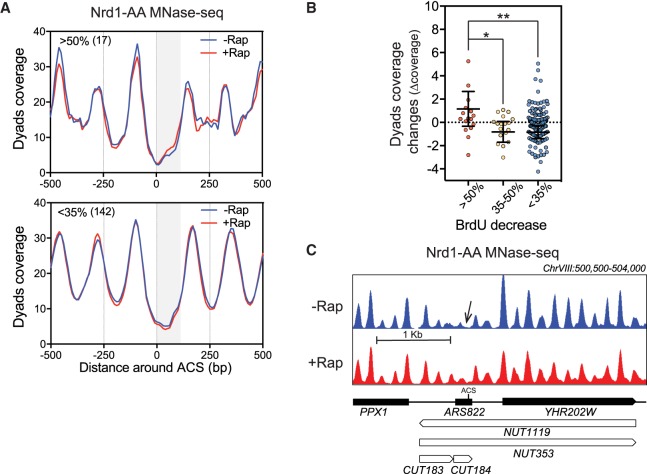
High noncoding readthrough transcription leads to ARS chromatin regulation
Given the links between replication initiation and chromatin structure, we then analyzed the effects of the Nrd1 depletion-induced transcription readthrough into replication origins on nucleosome positioning. Chromatin was extracted from Nrd1-AA cells either untreated or treated for 1 h with rapamycin and digested with micrococcal nuclease (MNase). First, sequencing of the 120- to 200-bp fragments protected by nucleosomes revealed the typical NDR around the ACS as previously described (Fig. 2A; Eaton et al. 2010). Second, analysis of nucleosome positioning at the classes of ARS defined in Figure 1E revealed a statistically significant increased density of nucleosome dyads in the NDRs of the ARSs that are the most affected for replication initiation and present a higher increase in readthrough transcription (Figs. 1G, 2A–C). Additional analyses reveal that the >50% class is significantly different from the <35% group not only when taking the mean of two replicates but also when considering each individual experiment (Fig. 2B; Supplemental Fig. S3). These results suggest that high levels of noncoding transcription into replication origins leads to chromatin closing, which may in turn perturb replication initiation. However, additional parameters are likely to influence replication since the mildly affected ARSs (35%–50%) do not show significant nucleosome shifting although they are affected in replication initiation.
Figure 2.
Noncoding transcription readthrough into replication origins alters nucleosome occupancy. (A) Metagene analysis of MNase-seq from Nrd1-AA cells treated or not with rapamycin for 1 h at the three classes of ARSs defined in Figure 1. Only paired-end reads from 120 to 200 bp length were considered to define nucleosome dyad coverage. The gray box represents the window in which transcriptional readthrough was analyzed. (B) Scatter dot-plot representing the difference of dyads coverage (Δcoverage = coverage [+Rap] − coverage [−Rap]) between the ACS to +100 of oriented ARSs when comparing +Rap and −Rap conditions. (C) Snapshot of the MNase-seq around ARS822 belonging to the class of most affected ARSs. (*) P-value < 0.05; (**) P-value < 0.01.
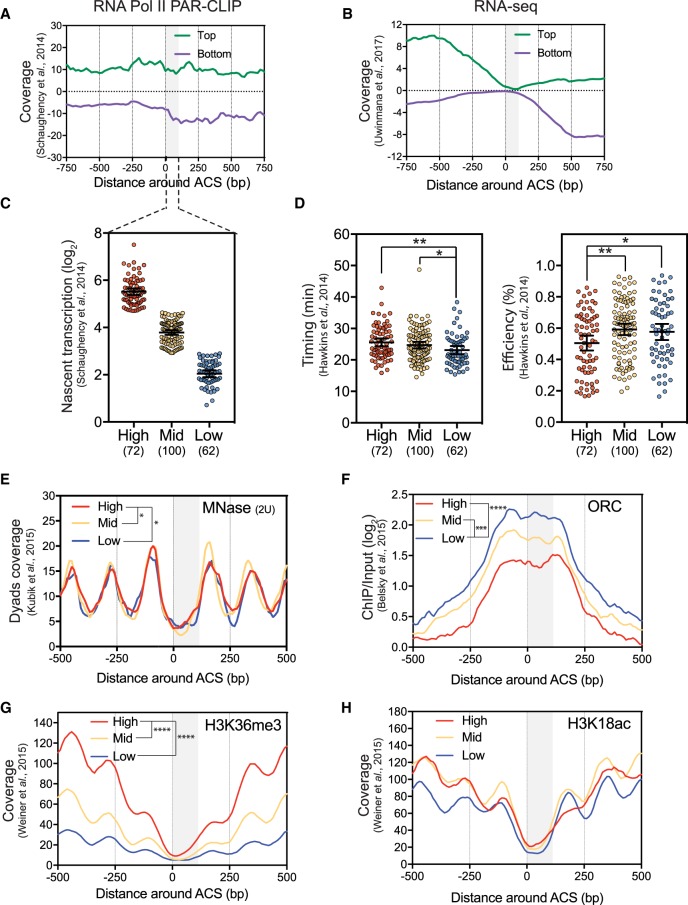
ARSs with a high basal level of readthrough transcription are late and inefficient
Recent data show that noncoding transcription occurs all over eukaryotic genomes (Jensen et al. 2013), and our results indicate that this transcription is detrimental for replication initiation. These observations led to the hypothesis that differences in nascent transcription between ARSs may influence both their activity and chromatin structure at steady-state. First, we confirmed that the production of stable transcripts, defined by RNA-seq, strongly drops in the vicinity of the 234 replication origins (including both early and late origins), while profiles of nascent transcription indicate that RNA Pol II density stays relatively constant through these ARSs (Fig. 3A,B). These observations establish that noncoding transcription through replication origins is a frequent event. The same set of ARS was then analysed for the level of natural basal readthrough transcription over a 100-bp segment between the ACS and the B elements using published RNA Pol II PAR-CLIP data (Schaughency et al. 2014). ARSs were subdivided into three groups according to their natural readthrough transcription levels into this region using a nonbiased k-means clustering approach (Fig. 3C). The highly transcribed ARSs were significantly enriched in ARS lying between two convergent genes (Supplemental Fig. S4A). Importantly, nascent transcription toward the ARSs measured upstream of and downstream from the oriented ACS was also significantly different for the three groups (Supplemental Fig. S4B), indicating that high ARS readthrough mainly stems from higher levels of nascent transcription from the adjacent convergent genes. Running these three groups of ARSs through replication timing and efficiency data (Hawkins et al. 2013) revealed that the 72 ARSs with high readthrough transcription have a significantly delayed replication timing and reduced replication efficiency compared to the ARSs with lower transcriptional readthrough (Fig. 3D), although the global correlation was low (Supplemental Fig. S4C). This suggests that once a certain threshold of pervasive transcription readthrough is reached, ARS activity is significantly reduced. Notably, the 52 NUTs/CUTs-containing ARSs present significantly lower natural readthrough transcription than the highly transcribed ARSs and appear more similar to the mildly transcribed ARSs, further supporting that NNS-mediated termination positively contributes to replication timing and efficiency by shielding ARSs from pervasive transcription (Supplemental Fig. S4D). Consistent with these results, earlier-defined ORC-bound ARSs (ORC-ARS) exhibit significantly less readthrough transcription compared to non-ORC-bound and nonreplicated ARSs (nr-ARSs) (Supplemental Fig. S4E; Eaton et al. 2010).
Figure 3.
Natural pervasive transcription correlates with timing/efficiency and specific chromatin features of replication origins. (A) Metagene analysis of Top and Bottom RNA Pol II PAR-CLIP (Schaughency et al. 2014) and (B) RNA-seq (Uwimana et al. 2017) data in the vicinity of 234 oriented replication origins. Data were smoothed by a 20-bp moving window. The gray box represents the window in which transcriptional readthrough was analyzed. (C) The 234 replication origins were divided into three classes (High, Mid, or Low) according to their level of total natural readthrough transcription using a nonbiased k-means clustering approach. This basal nascent transcription was calculated on oriented ARSs by adding the RNA Pol II PAR-CLIP mean densities between the ACS and +100 bp on the top strand to the signal over the same region on the bottom strand. Each 100-bp segment was considered as 1 bin. Natural nascent transcription data were taken from Schaughency et al. (2014). (D) Replication timing and efficiency of the three classes of ARSs defined as in C. (E–H) Nucleosome positioning, ORC, H3K36me3, and H3K18ac levels considering the three classes of replication origins. Data for nucleosome positioning, ORC recruitment, and histone marks were retrieved from Kubik et al. (2015), Belsky et al. (2015), and Weiner et al. (2015), respectively. For these plots, ARSs were oriented and aligned according to their ACS T-rich sequence. The significance of differences between the three classes was calculated over the ACS to +100 bp region considered as 1 bin and is indicated in the top left of each panel. No annotation is considered as nonsignificant. (*) P-value < 0.05; (**) P-value < 0.01; (***) P-value < 0.001; (****) P-value < 0.0001.
Thus, natural pervasive transcription into the ORC-binding area appears to be anti-correlated with ARS function as defined by its timing and efficiency as well as its ability to bind the ORC complex.
Steady-state highly transcribed ARSs present distinctive chromatin features
Using recently published nucleosome occupancy data (Kubik et al. 2015; Weiner et al. 2015), the 72 ARSs with high pervasive transcription levels appear to be associated with significantly increased nucleosome dyad density over the ORC-binding area compared to the 162 with lower transcription levels (Fig. 3E). This observation appears to be independent of the MNase sensitivity of the replication origins NDRs since less digested chromatin leads to a similar conclusion (Supplemental Fig. S5A; Kubik et al. 2015). Thus, high levels of nascent transcription into the ACS to +100 bp area correlate with higher nucleosome occupancy and are, as expected, also associated with lower ORC binding (Fig. 3F; Hoggard et al. 2013; Looke et al. 2013; Belsky et al. 2015; Das et al. 2015; Peace et al. 2016).
We also analyzed the correlations between natural pervasive transcription into ARSs and histone modifications (Weiner et al. 2015). We found that H3K36me3 levels over the ACS-100-bp area positively correlate with the levels of nascent transcription (Fig. 3G). We did not detect a significant correlation between nascent transcription and H3K18, H3K14, H4K12, and H4K5 acetylation over the ORC-binding region (Fig. 3H; Supplemental Fig. S5B–D). However, we detected a significant lack of these acetylation marks at the downstream nucleosome of the highly transcribed class (Supplemental Fig. S5F).
H3K36me3 is deposited by the Set2 histone methyltransferase and leads to the recruitment of the Rpd3 HDAC known to de-acetylate the lysines cited above and described as being involved in replication control (Rundlett et al. 1996; Knott et al. 2009). In contrast, the three groups of ARSs present no difference in H3K4me2, another mark promoting de-acetylation of the chromatin via binding of the Set3 HDAC (Supplemental Fig. S5E; Woo et al. 2017).
Together these observations support the view that noncoding transcription through an ARS promotes the formation of a closed chromatin structure reducing its ability to interact with an ORC, thereby decreasing its efficiency and/or delaying its replication timing.
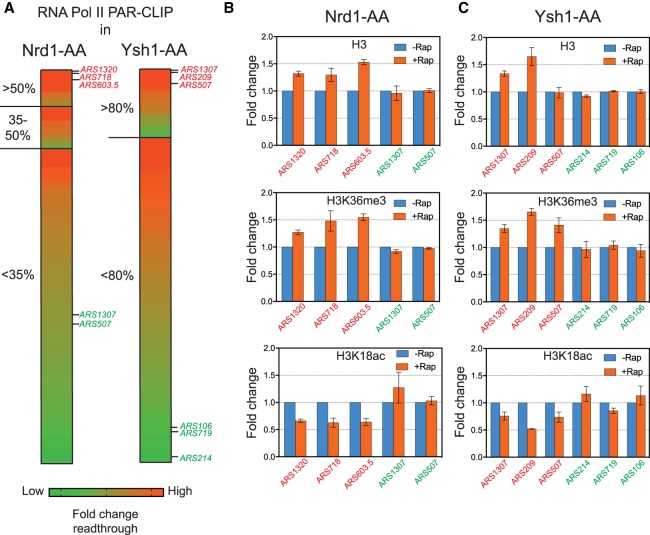
Noncoding and mRNA readthrough transcription over ARSs induce H3K36 methylation, histone deacetylation, and increased nucleosome occupancy
To strengthen the causal relationship between nascent transcription, changes in chromatin organization and ARS activity, we examined the effect of induced transcription on ARS chromatin changes. We first ranked the replication origins according to their increase in transcriptional readthrough levels in the Nrd1-AA strain and to their decrease in BrdU incorporation during early S-phase. We picked three ARSs belonging to the most affected ARSs in BrdU incorporation and presenting high levels of induced transcriptional readthrough (Fig. 4A). As a control, we took two replication origins belonging to the least affected ARSs in replication and showing no or weak induced readthrough. To relate the replication defect induced by noncoding readthrough transcription to changes in chromatin structure, chromatin immunoprecipitation was used to compare histone H3 occupancy, H3K36 methylation, and H3K18 acetylation in −/+ Rap. Primers for qPCR were designed to target the NDR of the ARSs. At the three affected ARSs, rapamycin treatment resulted in increased H3 occupancy and H3K36 methylation as well as a decrease in H3K18 acetylation, while no changes were observed at the nonaffected ARSs (Fig. 4B).
Figure 4.
Noncoding and mRNA transcription readthrough at replication origins leads to chromatin changes. (A) Heat map representing the fold change of transcriptional readthrough at replication origins in Nrd1-AA and Ysh1-AA mutants. The 178 ARSs were classified according to their BrdU incorporation defects and ranked by their total readthrough increase over the ACS to +100 bp region. The three ARSs indicated in red for each mutant present a high increase in total readthrough. The ARSs depicted in green show mid or low total readthrough and have been used as controls for the following experiments. (B,C) Chromatin immunoprecipitation (ChIP) of H3, H3K36me3, or H3K18ac at ARSs with high (red) and low (green) readthrough transcription. Asynchronous cells were treated 1 h or 30 min with rapamycin to induce Nrd1 and Ysh1 depletion from the nucleus, respectively. ChIP was performed as described in Methods. Immunoprecipitated ARS loci were normalized to immunoprecipitated SPT15 ORF after qPCR amplification. Fold enrichment was artificially set to 1 for the −Rap condition (n = 3). Error bars represent the standard error of the mean (SEM).
Since 86% of replication origins are located in the vicinity of a convergent coding gene, we decided to anchor away the essential mRNA 3′ cleavage and polyadenylation factor (CPF/CF) endonuclease Ysh1. As expected, nuclear depletion of Ysh1 has a major impact on replication progression and more specifically on replication initiation, as most of the 178 considered ARSs showed reduced BrdU incorporation in the presence of rapamycin, with 31 ARSs presenting more than an 80% decrease (Supplemental Fig. S6A–D). Combining BrdU-Seq in Ysh1-AA −/+ Rap with published RNA Pol II PAR-CLIP data of this strain (Schaughency et al. 2014) revealed that the 31 ARSs with the strongest replication defect also present the highest increase in readthrough transcription, suggesting a direct involvement of nascent transcription readthrough in this massive replication defect (Supplemental Fig. S6D,E). However, due to the general role of Ysh1 as an mRNA termination factor, we cannot fully rule out an indirect effect of its depletion on completion of replication. Replication origins were then ranked according to their induced levels of readthrough and defects in early replication as performed for the Nrd1-AA strain (Fig. 4A). Chromatin immunoprecipitation revealed increased H3 occupancy for the ARSs showing high levels of mRNA readthrough (with the exception of ARS507), while H3K36me3 levels increased and H3K18ac levels decreased (Fig. 4C).
Taken together, these experiments demonstrate that increased noncoding and mRNA readthrough transcription causes increased nucleosome occupancy and histone deacetylation at the downstream ARSs, two parameters described to interfere with the efficiency of ORC binding and ARS licensing (Vogelauer et al. 2002; Aparicio et al. 2004; Knott et al. 2009; Soriano et al. 2014).
Replication defects induced by noncoding readthrough transcription are partially rescued in the absence of H3K36 methylation
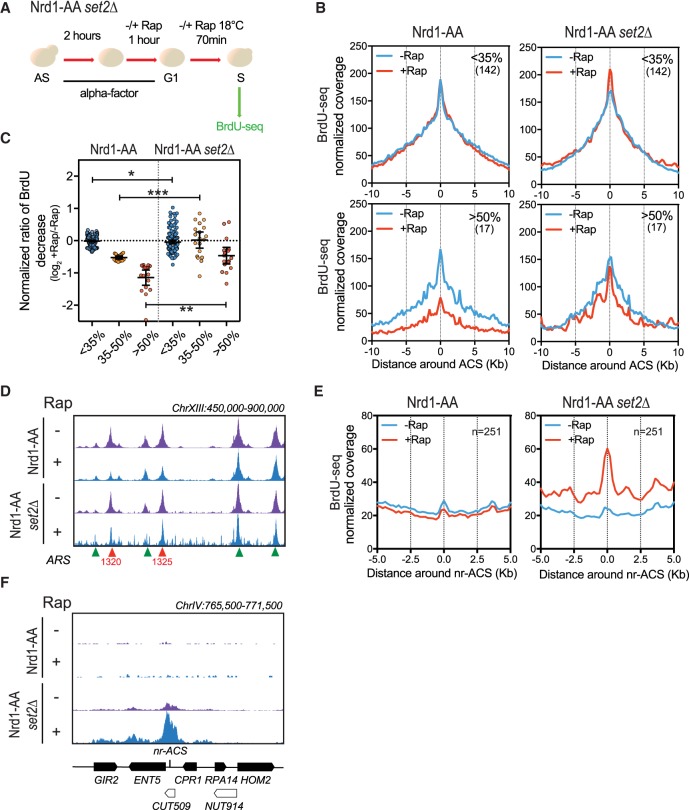
To decipher the molecular cascade of events, we analyzed the effects of noncoding transcription readthrough on replication initiation in a Nrd1-AA set2Δ strain (Fig. 5A). Global analysis of replication by flow cytometry indicates that replication is still delayed in the absence of H3K36 methylation in the Nrd1-AA background (Supplemental Fig. S7A). However, by taking the same classes of defective ARSs as defined in Figure 1, we observed a partial rescue of BrdU incorporation (Fig. 5B–D), although noncoding RNAs were still produced in the absence of Set2 (Supplemental Fig. S7B). These observations support the view that nascent transcription drives chromatin regulation of replication origins, which in turn defines, at least in part, ARS activity.
Figure 5.
Absence of Set2 H3K36 methyltransferase partially rescues replication defects due to noncoding transcription readthrough. (A) Experimental scheme as described in Figure 1C. (B) Metagene analysis of the BrdU-seq for the 142 nonaffected ARSs (<35%) and the 17 most affected ARSs (>50%) for the Nrd1-AA and Nrd1-AA set2Δ strains. Plots for the Nrd1-AA strain were already presented in Figure 1F, with the exception of the normalized coverage, for which calculation is detailed in Methods. Profiles represent the mean coverage smoothed by a 200-bp moving window. ARSs were oriented according to their ACS T-rich sequence. (C) Scatter dot-plot presenting the normalized BrdU ratio for the different classes of ARS affected in BrdU incorporation in an Nrd1-AA strain and for the same classes of ARSs in the Nrd1-AA set2Δ strain. (D) Snapshot depicting the BrdU-seq reads for a part of Chromosome XIII. ARSs that are rescued in BrdU incorporation in the Nrd1-AA set2Δ +Rap condition are depicted in red, while nonaffected ARSs are in green. (E) Metagene analysis of BrdU incorporation 5 kb around nonreplicating ACSs (nr-ACS) in the indicated strains grown in −/+Rap. The representation is smoothed over a 200-bp moving window. (F) Snapshot illustrating the activation of a dormant nr-ACS in the Nrd1-AA set2Δ +Rap condition. (*) P-value < 0.05; (**) P-value < 0.01; (***) P-value < 0.001.
Rapamycin treatment of the Nrd1-AA set2Δ strain led to the appearance of small BrdU incorporation peaks all over the genome (Fig. 5D), suggesting that replication initiation loses its specificity when both noncoding transcription termination and H3K36 methylation are abrogated. Moreover, analysis of BrdU incorporation around non-ORC-bound and non-replicated ACSs (Eaton et al. 2010) revealed an increase of replication initiation at noncanonical sites when noncoding transcription readthrough is induced in the absence of Set2 (Fig. 5E,F). Thus, while noncoding transcription interferes with replication in the presence of Set2 by favoring a closed chromatin structure, transcription over dormant ARSs in the absence of Set2 activates replication, probably as a result of nucleosome instability.
Discussion
We have shown that ncRNA early termination by the Nrd1-dependent pathway in the vicinity of a subset of ARSs protects these origins from transcription and replication initiation defects. These observations suggest that inefficient noncoding transcription termination may influence replication origin activity. Consistently, our analyses reveal that natural readthrough transcription correlates with reduced ARS activity and a specific replication origin chromatin structure. Moreover, using mRNA or cryptic transcription termination mutants, we have established that nascent transcription is the causal link defining chromatin organization at a number of ARSs. Finally, we have presented evidence that transcription-induced chromatin modifications and not only nascent transcription per se control ARS activity. Thus, we propose widespread noncoding transcription as a novel primordial parameter defining replication initiation features (Fig. 6).
Figure 6.
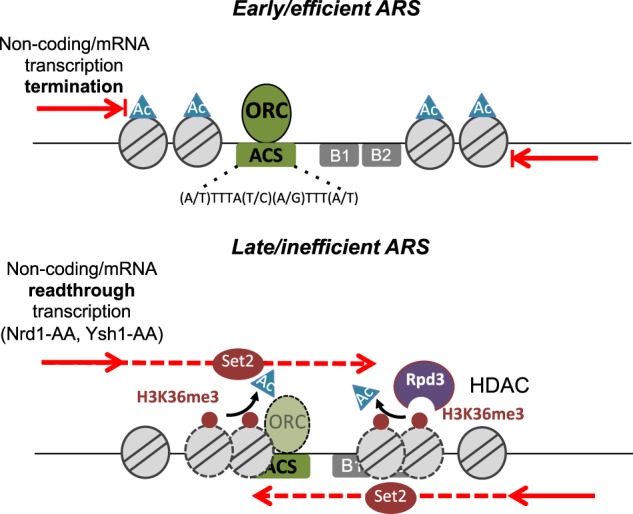
Noncoding transcription influences replication timing/efficiency by modulating ARS chromatin structure and ORC binding. The chromatin structure of replication origins is defined, at least in part, by the level of pervasive readthrough transcription. In the presence of efficient noncoding (Nrd1/Nab3/Sen1-dependent) or mRNA (CPF/CF-dependent) transcription termination, ARSs present low H3K36 trimethylation, high downstream nucleosome acetylation (Ac), a wide NDR, and more ORC binding at the ACS, favoring early and efficient replication. If transcription termination is deficient, H3K36me3 by Set2 increases and histone acetylation decreases, likely through the recruitment of the Rpd3 histone deacetylase; these modifications increase nucleosome stability and occupancy over the ARS, lowering the level of ORC recruitment and resulting in late and inefficient ARS replication.
Nrd1-dependent transcription termination protects a subset of early/efficient replication origins from noncoding transcription
Previous studies on the relationship between transcription and replication have led to conflicting observations. Our analyses of pervasive transcription lead to the conclusion that transcriptional readthrough at ARSs is detrimental for replication initiation as already proposed by some reports (Snyder et al. 1988; Mori and Shirahige 2007; Blitzblau et al. 2012; Gros et al. 2015). Importantly, we show that natural nascent transcription per se is a criterion defining ARS activity genome-wide. Thus, the strategy for a replication origin to increase its activity would be to limit pervasive transcription. Accordingly, a subset of early and efficient ARSs are protected from pervasive transcription thanks to surrounding noncoding transcription termination by the Nrd1-dependent pathway (Figs. 1, 3; Supplemental Fig. S4D). Thus, we propose that Nrd1-dependent termination in the vicinity of a replication origin is an efficient way to decrease transcriptional readthrough. Since Nrd1-dependent termination is regulated under stress conditions, it is tempting to speculate that this might also affect replication origins usage (Bresson et al. 2017; van Nues et al. 2017). This scenario would define a novel role for noncoding transcription in the regulation of genome maintenance.
No mechanism similar to Nrd1-dependent termination has been described in other eukaryotes yet (Wittmann et al. 2017). In S. cerevisiae, Nrd1-protected origins reach a median firing efficiency peaking at 58%, while firing efficiency is around 30% in Schizosaccharomyces pombe (Heichinger et al. 2006). An attractive view is that Nrd1-dependent transcription termination represents an evolutionary pathway maximizing replication initiation efficiency.
Our results indicate that there is no significant linear correlation between nascent transcription and ARS activity. This connection appears when ARSs are divided into subsets (Fig. 3). Similar data were described in a recent paper (Candelli et al. 2018). These observations indicate that nascent transcription influences ARS activity only beyond a certain threshold and that other parameters contribute to origin function. Accordingly, a variety of molecular events have been involved in regulating ARS activity, which include MCM levels bound to ARSs, cell cycle regulated binding, and affinity of the ORC for the ACS, or the presence of Fkh1/2 proteins (Hoggard et al. 2013; Looke et al. 2013; Belsky et al. 2015; Das et al. 2015; Peace et al. 2016).
Replication origin chromatin structure is influenced by noncoding readthrough transcription
Previous work has involved the chromatin structure at replication origins as a parameter defining replication initiation. Early ARSs tend to show an open chromatin, low H3K36me3, and high histone acetylation levels (Pryde et al. 2009; Soriano et al. 2014). Our results indicate that these features may be directly related to the level of natural nascent transcription. Indeed, when compared to highly transcribed ARSs, origins with a low level of readthrough transcription present lower nucleosome occupancy, lower H3K36me3, and higher histone acetylation levels (Fig. 3). Of note, H3K36me3 is deposited by Set2, a histone methyltransferase (HMT) recruited through interaction with the elongating RNA Pol II CTD. H3K36me3 serves as a platform for the binding of Rpd3S, a histone de-acetylase complex described to de-acetylate and stabilize reassembled nucleosomes in the wake of the transcription machinery, suppressing initiation from cryptic sites within ORFs (Carrozza et al. 2005; for a recent review, see Woo et al. 2017). This molecular mechanism may represent the connection between pervasive transcription and chromatin structure of replication origins. Indeed, increasing transcriptional readthrough into replication origins is accompanied by higher levels of H3K36me3, lower levels of H3K18ac, increased nucleosome occupancy, and replication defects (Figs. 1, 2, 4; Supplemental Fig. S6). Importantly, BrdU-seq experiments cannot discriminate between timing and efficiency defects. However, since nucleosome methylations are relatively stable modifications, it would be appealing to propose that even rare events of noncoding transcription may stably inactivate replication origins until the subsequent S-phase dilutes or a histone de-methylase erases these methylation marks. In such a model, pervasive transcription may shape replication origin chromatin for inefficient usage.
These observations shed new light on earlier published results. First, loss of Rpd3 leads to a global increase in acetylation around ARSs and early firing of many late origins (Vogelauer et al. 2002; Knott et al. 2009). It was proposed that the “accelerated” replication in rpd3Δ is mainly due to a reduced titration of replication initiation factors by the rDNA origins (Yoshida et al. 2014); however, loss of Rpd3 also has a global impact on increasing the size of replication origins NDR (Soriano et al. 2014). Of note, loss of Rpd3 abrogates the function of both Rpd3S and Rpd3L, another histone de-acetylase complex involved in gene repression following recruitment to promoters via a Set2/H3K36me3-independent pathway (Woo et al. 2017). While loss of Rpd3L subunits was shown to result in increased BrdU incorporation at a number of ARSs, especially those adjacent to up-regulated genes, loss of Rpd3S subunits or Set2 led to a weaker but more generalized increase in ARS replication initiation (Knott et al. 2009). This difference could reflect a primary specific effect of loss of Rpd3L on rDNA replication and increased availability of replication factors in the absence of this HDAC. Although the replication phenotype was less pronounced when deleting Rpd3S, the data support the view that pervasive transcription promotes Set2/H3K36me3-mediated histone de-acetylation by Rpd3S and globally contributes to negatively regulate replication origin activity.
Our results further indicate that high readthrough transcription correlates with decreased ORC binding (Fig. 3; Supplemental Fig. S4E). It was proposed that replication initiation timing depends more on the surrounding chromatin than on the ORC-ACS in vitro affinity by itself (Hoggard et al. 2013). It would be interesting to further dissect the molecular events at origins and to define whether nascent transcription per se evicts the ORC, leading to nucleosome deposition and histone modifications, or whether RNA Pol II readthrough and nucleosome incorporation outcompete ORC turnover (Fig. 6).
We observe genome-wide appearance of BrdU peaks when SET2 is deleted in conjunction with the anchor away of Nrd1. These peaks are not detected in the set2Δ mutant alone, indicating that activation of dormant origins is not only related to the absence of the histone mark. Deletion of SET2 is known to drastically increase the level of intra-genic transcription initiation (Malabat et al. 2015). A fraction of this novel nascent transcription may be cleared by the Nrd1-dependent termination pathway. We propose that, in the absence of Nrd1, transcription may hit dormant origins, which, in the absence of H3K36 methylation, will promote replication initiation due to nucleosome instability and chromatin opening. Thus, when not associated with H3K36 methylation, transcription may have a positive effect on replication initiation.
Implications for a pervasive transcription-dependent replication initiation model in metazoans
Identification of replication origins in mouse embryonic stem cells showed that nearly half of them are contained in promoters (Sequeira-Mendes et al. 2009), and recent ORC binding data in human cells led to the same conclusion (Dellino et al. 2013; Miotto et al. 2016). In contrast, replication initiation in S. cerevisiae more frequently occurs next to gene terminators (Nieduszynski et al. 2006). Human promoters are bidirectional and lead to the production of highly unstable promoter upstream transcripts (PROMPTs), suggesting that pervasive transcription could also play a role in metazoan replication initiation (Preker et al. 2008; Mayer et al. 2015; Nojima et al. 2015). It has recently been shown that replication initiation in human cells occurs within broad, over-30-kb regions, flanked by ORC binding at one of the two ends (Petryk et al. 2016). Considering that MCM helicases can slide along the chromosome with the help of transcription (Gros et al. 2015), it is appealing to propose that promoter-associated noncoding transcription redistributes the MCM helicases from their ORC binding initial site of loading. Since we have established widespread noncoding transcription as a novel primordial parameter regulating replication initiation in S. cerevisiae, its importance for the metazoan replication program warrants future study.
Methods
Yeast strains
All strains were derived from W303 and Anchor-Away genetic backgrounds (see Supplemental Table S2; Haruki et al. 2008). Cells were cultivated as described in Supplemental Material.
G1-phase synchronization and BrdU labeling
Cells were grown as described in Soudet et al. (2014) with some modifications, indicated in Supplemental Material.
BrdU immunoprecipitation and sequencing
BrdU immunoprecipitation was mainly performed as described in Soudet et al. (2014) with some modifications, indicated in the Supplemental Material. Libraries of immunoprecipitated BrdU-containing DNA were constructed using the iDeal Library Preparation kit (Diagenode). Sequencing was performed on the HiSeq 4000 sequencer (Illumina). For specific loci analyses by quantitative PCR, BrdU-containing DNA was amplified using the SYBR Select Master Mix for CFX (Applied Biosciences) on a CFX96 real-time detection system (Bio-Rad).
MNase-seq
MNase treatment was performed as described in Weiner et al. (2015). Chromatin was extracted by breaking cells with bead beating in a magnalyser (Roche). Chromatin was then collected by centrifugation and resuspended in NP-buffer (0.5 mM spermidine, 1 mM β-ME, 0.075% NP-40, 50 mM NaCl, 10 mM Tris at pH 7.4, 5 mM MgCl2, 1 mM CaCl2). MNase (Thermo Fisher Scientific) treatment was performed at a previously optimized concentration to have comparable intensity of both mono- and di-nucleosomes within and between the samples. MNase treatment was followed by de-crosslinking and protease treatment, and DNA was extracted using NucleoSpin gel and PCR extraction columns (Macherey-Nagel). An iDeal Library Preparation kit (Diagenode) was then used for library construction. Sequencing was performed on the HiSeq 4000 sequencer (Illumina).
BrdU-seq and MNase-seq bioinformatic analyses
Fifty-base pair paired-end reads were aligned to sacCer3 genome assembly using HTSstation (David et al. 2014). PCR duplicates were removed from the analysis. For the MNase-seq, 120- to 200-bp fragments were filtered to detect molecules with nucleosome size using HTS Bioscript (David et al. 2014). BrdU-seq and MNase-seq density files (bigWig) were averaged for the two replicates of each condition. All subsequent analyses were performed using HTS Bioscript including metagene analyses. To assign one value of BrdU incorporation to each ARS, BrdU incorporation was measured 5 kb around ACSs considering this area as 1 bin. For the MNase-seq, nucleosome occupancy was quantified over the ACS to +100 bp area of oriented ARSs.
Since no spike-in was used in our experiments and since Nrd1-AA set2Δ +Rap substantially changes the density profile because of dormant origins firing, Figure 5 was normalized as follows: The <35% affected ARS class for BrdU incorporation was considered as equal in +Rap and −Rap in an average 5 kb around the ACS in both Nrd1-AA and Nrd1-AA set2Δ. This gave a normalization factor for each strain, which was then used to quantify the other classes.
Chromatin immunoprecipitation (ChIP)
ChIP experiments were performed as described previously with some modifications (Camblong et al. 2007), described in the Supplemental Material. ChIPs were repeated three times with different chromatin extracts from independent cultures. Immunoprecipitated DNA was then purified and quantified by qPCR. Immunoprecipitated ARS loci were normalized to immunoprecipitated SPT15 ORF after qPCR amplification.
List of noncoding RNAs and replication origins
The list of CUTs was obtained from Xu et al. (2009), while the list of NUTs was kindly provided by the Cramer lab (Schulz et al. 2013). Among the NUTs, only those showing at least a twofold increase in +Rap/−Rap were taken into account to unify the threshold of ncRNA definition between CUTs and NUTs. The list of ARS (Supplemental Table S1) consists of the 234 ACS taken from Soriano et al. (2014) that overlap with the replication origins described in Hawkins et al. (2013), for which replication timing and efficiency have been defined. Replication origins with an efficiency <15% were not taken into account.
Statistical analysis
All statistical analyses of this work were performed using Prism 7.0 (Graphpad). All tests are nonpaired tests (with the exception of Fig. 5C). t-tests or Mann–Whitney U tests were used according to the normality of the data analyzed, which was calculated using a d'Agostino-Pearson omnibus normality test.
Downloaded data sets
For RNA Pol II PAR-CLIP, RNA-seq, chromatin and ORC profiles, data were retrieved from Schaughency et al. (2014) (GEO: GSE56435), Uwimana et al. (2017) (GEO: GSE89601), Kubik et al. (2015) (GEO: GSE73337), Weiner et al. (2015) (GEO: GSE61888), and Belsky et al. (2015) (SRA: SRP041314).
Data access
All sequencing data from this study have been submitted to the NCBI Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) under accession number GSE111058.
Supplementary Material
Acknowledgments
We thank O. Aparicio, D. MacAlpine, and P. Cramer for sharing data and reagents and M. Strubin, F. Steiner, M. Shyian, G. Canal, T. Halazonetis, and all members from the Stutz lab for comments on the manuscript. We also thank D. Libri for communication of unpublished results. This work was funded by the Swiss National Science Foundation (31003A 153331), iGE3, and the Canton of Geneva, as well as a Polish Swiss Research Programme (PSRP NoCore 183/2010).
Author contributions: J.S. and F.S. conceived the study. J.S. performed most experiments and analyzed the results together with F.S.; J.K.G. performed the MNase-seq; J.S. and F.S. wrote the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.239582.118.
Freely available online through the Genome Research Open Access option.
References
- Aparicio OM. 2013. Location, location, location: it's all in the timing for replication origins. Genes Dev 27: 117–128. 10.1101/gad.209999.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio JG, Viggiani CJ, Gibson DG, Aparicio OM. 2004. The Rpd3-Sin3 histone deacetylase regulates replication timing and enables intra-S origin control in Saccharomyces cerevisiae. Mol Cell Biol 24: 4769–4780. 10.1128/MCB.24.11.4769-4780.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arigo JT, Eyler DE, Carroll KL, Corden JL. 2006. Termination of cryptic unstable transcripts is directed by yeast RNA-binding proteins Nrd1 and Nab3. Mol Cell 23: 841–851. 10.1016/j.molcel.2006.07.024 [DOI] [PubMed] [Google Scholar]
- Baejen C, Andreani J, Torkler P, Battaglia S, Schwalb B, Lidschreiber M, Maier KC, Boltendahl A, Rus P, Esslinger S, et al. 2017. Genome-wide analysis of RNA polymerase II termination at protein-coding genes. Mol Cell 66: 38–49.e6. 10.1016/j.molcel.2017.02.009. [DOI] [PubMed] [Google Scholar]
- Bell SP. 1995. Eukaryotic replicators and associated protein complexes. Curr Opin Genet Dev 5: 162–167. 10.1016/0959-437X(95)80003-4 [DOI] [PubMed] [Google Scholar]
- Belsky JA, MacAlpine HK, Lubelsky Y, Hartemink AJ, MacAlpine DM. 2015. Genome-wide chromatin footprinting reveals changes in replication origin architecture induced by pre-RC assembly. Genes Dev 29: 212–224. 10.1101/gad.247924.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blitzblau HG, Chan CS, Hochwagen A, Bell SP. 2012. Separation of DNA replication from the assembly of break-competent meiotic chromosomes. PLoS Genet 8: e1002643 10.1371/journal.pgen.1002643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bresson S, Tuck A, Staneva D, Tollervey D. 2017. Nuclear RNA decay pathways aid rapid remodeling of gene expression in yeast. Mol Cell 65: 787–800.e5. 10.1016/j.molcel.2017.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camblong J, Iglesias N, Fickentscher C, Dieppois G, Stutz F. 2007. Antisense RNA stabilization induces transcriptional gene silencing via histone deacetylation in S. cerevisiae. Cell 131: 706–717. 10.1016/j.cell.2007.09.014 [DOI] [PubMed] [Google Scholar]
- Candelli T, Gros J, Libri D. 2018. Pervasive transcription fine-tunes replication origin activity. bioRxiv 10.1101/384859 [DOI] [PMC free article] [PubMed]
- Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, et al. 2005. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123: 581–592. 10.1016/j.cell.2005.10.023 [DOI] [PubMed] [Google Scholar]
- Castelnuovo M, Rahman S, Guffanti E, Infantino V, Stutz F, Zenklusen D. 2013. Bimodal expression of PHO84 is modulated by early termination of antisense transcription. Nat Struct Mol Biol 20: 851–858. 10.1038/nsmb.2598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castelnuovo M, Zaugg JB, Guffanti E, Maffioletti A, Camblong J, Xu Z, Clauder-Münster S, Steinmetz LM, Luscombe NM, Stutz F. 2014. Role of histone modifications and early termination in pervasive transcription and antisense-mediated gene silencing in yeast. Nucleic Acids Res 42: 4348–4362. 10.1093/nar/gku100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchman LS, Weissman JS. 2011. Nascent transcript sequencing visualizes transcription at nucleotide resolution. Nature 469: 368–373. 10.1038/nature09652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czajkowsky DM, Liu J, Hamlin JL, Shao Z. 2008. DNA combing reveals intrinsic temporal disorder in the replication of yeast chromosome VI. J Mol Biol 375: 12–19. 10.1016/j.jmb.2007.10.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das SP, Borrman T, Liu VW, Yang SC, Bechhoefer J, Rhind N. 2015. Replication timing is regulated by the number of MCMs loaded at origins. Genome Res 25: 1886–1892. 10.1101/gr.195305.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David FP, Delafontaine J, Carat S, Ross FJ, Lefebvre G, Jarosz Y, Sinclair L, Noordermeer D, Rougemont J, Leleu M. 2014. HTSstation: a web application and open-access libraries for high-throughput sequencing data analysis. PLoS One 9: e85879 10.1371/journal.pone.0085879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deegan TD, Diffley JF. 2016. MCM: one ring to rule them all. Curr Opin Struct Biol 37: 145–151. 10.1016/j.sbi.2016.01.014 [DOI] [PubMed] [Google Scholar]
- Dellino GI, Cittaro D, Piccioni R, Luzi L, Banfi S, Segalla S, Cesaroni M, Mendoza-Maldonado R, Giacca M, Pelicci PG. 2013. Genome-wide mapping of human DNA-replication origins: Levels of transcription at ORC1 sites regulate origin selection and replication timing. Genome Res 23: 1–11. 10.1101/gr.142331.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton ML, Galani K, Kang S, Bell SP, MacAlpine DM. 2010. Conserved nucleosome positioning defines replication origins. Genes Dev 24: 748–753. 10.1101/gad.1913210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser HB. 2013. Cell-cycle regulated transcription associates with DNA replication timing in yeast and human. Genome Biol 14: R111 10.1186/gb-2013-14-10-r111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gros J, Kumar C, Lynch G, Yadav T, Whitehouse I, Remus D. 2015. Post-licensing specification of eukaryotic replication origins by facilitated Mcm2-7 sliding along DNA. Mol Cell 60: 797–807. 10.1016/j.molcel.2015.10.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruki H, Nishikawa J, Laemmli UK. 2008. The anchor-away technique: rapid, conditional establishment of yeast mutant phenotypes. Mol Cell 31: 925–932. 10.1016/j.molcel.2008.07.020 [DOI] [PubMed] [Google Scholar]
- Hawkins M, Retkute R, Muller CA, Saner N, Tanaka TU, de Moura AP, Nieduszynski CA. 2013. High-resolution replication profiles define the stochastic nature of genome replication initiation and termination. Cell Rep 5: 1132–1141. 10.1016/j.celrep.2013.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heichinger C, Penkett CJ, Bähler J, Nurse P. 2006. Genome-wide characterization of fission yeast DNA replication origins. EMBO J 25: 5171–5179. 10.1038/sj.emboj.7601390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoggard T, Shor E, Muller CA, Nieduszynski CA, Fox CA. 2013. A link between ORC-origin binding mechanisms and origin activation time revealed in budding yeast. PLoS Genet 9: e1003798 10.1371/journal.pgen.1003798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen TH, Jacquier A, Libri D. 2013. Dealing with pervasive transcription. Mol Cell 52: 473–484. 10.1016/j.molcel.2013.10.032 [DOI] [PubMed] [Google Scholar]
- Kim M, Krogan NJ, Vasiljeva L, Rando OJ, Nedea E, Greenblatt JF, Buratowski S. 2004. The yeast Rat1 exonuclease promotes transcription termination by RNA polymerase II. Nature 432: 517–522. 10.1038/nature03041 [DOI] [PubMed] [Google Scholar]
- Knott SR, Viggiani CJ, Tavare S, Aparicio OM. 2009. Genome-wide replication profiles indicate an expansive role for Rpd3L in regulating replication initiation timing or efficiency, and reveal genomic loci of Rpd3 function in Saccharomyces cerevisiae. Genes Dev 23: 1077–1090. 10.1101/gad.1784309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knott SR, Peace JM, Ostrow AZ, Gan Y, Rex AE, Viggiani CJ, Tavaré S, Aparicio OM. 2012. Forkhead transcription factors establish origin timing and long-range clustering in S. cerevisiae. Cell 148: 99–111. 10.1016/j.cell.2011.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubik S, Bruzzone MJ, Jacquet P, Falcone JL, Rougemont J, Shore D. 2015. Nucleosome stability distinguishes two different promoter types at all protein-coding genes in yeast. Mol Cell 60: 422–434. 10.1016/j.molcel.2015.10.002 [DOI] [PubMed] [Google Scholar]
- Looke M, Reimand J, Sedman T, Sedman J, Jarvinen L, Varv S, Peil K, Kristjuhan K, Vilo J, Kristjuhan A. 2010. Relicensing of transcriptionally inactivated replication origins in budding yeast. J Biol Chem 285: 40004–40011. 10.1074/jbc.M110.148924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Looke M, Kristjuhan K, Varv S, Kristjuhan A. 2013. Chromatin-dependent and -independent regulation of DNA replication origin activation in budding yeast. EMBO Rep 14: 191–198. 10.1038/embor.2012.196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Johnson AW, Bentley DL. 2006. The role of Rat1 in coupling mRNA 3′-end processing to transcription termination: implications for a unified allosteric–torpedo model. Genes Dev 20: 954–965. 10.1101/gad.1409106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malabat C, Feuerbach F, Ma L, Saveanu C, Jacquier A. 2015. Quality control of transcription start site selection by nonsense-mediated-mRNA decay. eLife 4: e06722 10.7554/eLife.06722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayan MD. 2013. RNAP-II molecules participate in the anchoring of the ORC to rDNA replication origins. PLoS One 8: e53405 10.1371/journal.pone.0053405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer A, di Iulio J, Maleri S, Eser U, Vierstra J, Reynolds A, Sandstrom R, Stamatoyannopoulos JA, Churchman LS. 2015. Native elongating transcript sequencing reveals human transcriptional activity at nucleotide resolution. Cell 161: 541–554. 10.1016/j.cell.2015.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuffee SR, Smith DJ, Whitehouse I. 2013. Quantitative, genome-wide analysis of eukaryotic replication initiation and termination. Mol Cell 50: 123–135. 10.1016/j.molcel.2013.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miotto B, Ji Z, Struhl K. 2016. Selectivity of ORC binding sites and the relation to replication timing, fragile sites, and deletions in cancers. Proc Natl Acad Sci 113: E4810–E4819. 10.1073/pnas.1609060113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori S, Shirahige K. 2007. Perturbation of the activity of replication origin by meiosis-specific transcription. J Biol Chem 282: 4447–4452. 10.1074/jbc.M609671200 [DOI] [PubMed] [Google Scholar]
- Neil H, Malabat C, d'Aubenton-Carafa Y, Xu Z, Steinmetz LM, Jacquier A. 2009. Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature 457: 1038–1042. 10.1038/nature07747 [DOI] [PubMed] [Google Scholar]
- Nieduszynski CA, Blow JJ, Donaldson AD. 2005. The requirement of yeast replication origins for pre-replication complex proteins is modulated by transcription. Nucleic Acids Res 33: 2410–2420. 10.1093/nar/gki539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieduszynski CA, Knox Y, Donaldson AD. 2006. Genome-wide identification of replication origins in yeast by comparative genomics. Genes Dev 20: 1874–1879. 10.1101/gad.385306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nojima T, Gomes T, Grosso AR, Kimura H, Dye MJ, Dhir S, Carmo-Fonseca M, Proudfoot NJ. 2015. Mammalian NET-seq reveals genome-wide nascent transcription coupled to RNA processing. Cell 161: 526–540. 10.1016/j.cell.2015.03.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peace JM, Villwock SK, Zeytounian JL, Gan Y, Aparicio OM. 2016. Quantitative BrdU immunoprecipitation method demonstrates that Fkh1 and Fkh2 are rate-limiting activators of replication origins that reprogram replication timing in G1 phase. Genome Res 26: 365–375. 10.1101/gr.196857.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petryk N, Kahli M, d'Aubenton-Carafa Y, Jaszczyszyn Y, Shen Y, Silvain M, Thermes C, Chen CL, Hyrien O. 2016. Replication landscape of the human genome. Nat Commun 7: 10208 10.1038/ncomms10208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porrua O, Boudvillain M, Libri D. 2016. Transcription termination: variations on common themes. Trends Genet 32: 508–522. 10.1016/j.tig.2016.05.007 [DOI] [PubMed] [Google Scholar]
- Preker P, Nielsen J, Kammler S, Lykke-Andersen S, Christensen MS, Mapendano CK, Schierup MH, Jensen TH. 2008. RNA exosome depletion reveals transcription upstream of active human promoters. Science 322: 1851–1854. 10.1126/science.1164096 [DOI] [PubMed] [Google Scholar]
- Pryde F, Jain D, Kerr A, Curley R, Mariotti FR, Vogelauer M. 2009. H3 K36 methylation helps determine the timing of Cdc45 association with replication origins. PLoS One 4: e5882 10.1371/journal.pone.0005882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghuraman MK, Winzeler EA, Collingwood D, Hunt S, Wodicka L, Conway A, Lockhart DJ, Davis RW, Brewer BJ, Fangman WL. 2001. Replication dynamics of the yeast genome. Science 294: 115–121. 10.1126/science.294.5540.115 [DOI] [PubMed] [Google Scholar]
- Rodriguez J, Lee L, Lynch B, Tsukiyama T. 2017. Nucleosome occupancy as a novel chromatin parameter for replication origin functions. Genome Res 27: 269–277. 10.1101/gr.209940.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rundlett SE, Carmen AA, Kobayashi R, Bavykin S, Turner BM, Grunstein M. 1996. HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc Natl Acad Sci 93: 14503–14508. 10.1073/pnas.93.25.14503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaughency P, Merran J, Corden JL. 2014. Genome-wide mapping of yeast RNA polymerase II termination. PLoS Genet 10: e1004632 10.1371/journal.pgen.1004632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz D, Schwalb B, Kiesel A, Baejen C, Torkler P, Gagneur J, Soeding J, Cramer P. 2013. Transcriptome surveillance by selective termination of noncoding RNA synthesis. Cell 155: 1075–1087. 10.1016/j.cell.2013.10.024 [DOI] [PubMed] [Google Scholar]
- Segal E, Widom J. 2009. Poly(dA:dT) tracts: major determinants of nucleosome organization. Curr Opin Struct Biol 19: 65–71. 10.1016/j.sbi.2009.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sequeira-Mendes J, Diaz-Uriarte R, Apedaile A, Huntley D, Brockdorff N, Gomez M. 2009. Transcription initiation activity sets replication origin efficiency in mammalian cells. PLoS Genet 5: e1000446 10.1371/journal.pgen.1000446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder M, Sapolsky RJ, Davis RW. 1988. Transcription interferes with elements important for chromosome maintenance in Saccharomyces cerevisiae. Mol Cell Biol 8: 2184–2194. 10.1128/MCB.8.5.2184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano I, Morafraile EC, Vázquez E, Antequera F, Segurado M. 2014. Different nucleosomal architectures at early and late replicating origins in Saccharomyces cerevisiae. BMC Genomics 15: 791 10.1186/1471-2164-15-791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soudet J, Jolivet P, Teixeira MT. 2014. Elucidation of the DNA end-replication problem in Saccharomyces cerevisiae. Mol Cell 53: 954–964. 10.1016/j.molcel.2014.02.030 [DOI] [PubMed] [Google Scholar]
- Steinmetz EJ, Conrad NK, Brow DA, Corden JL. 2001. RNA-binding protein Nrd1 directs poly(A)-independent 3′-end formation of RNA polymerase II transcripts. Nature 413: 327–331. 10.1038/35095090 [DOI] [PubMed] [Google Scholar]
- Stinchcomb DT, Struhl K, Davis RW. 1979. Isolation and characterisation of a yeast chromosomal replicator. Nature 282: 39–43. 10.1038/282039a0 [DOI] [PubMed] [Google Scholar]
- Tudek A, Porrua O, Kabzinski T, Lidschreiber M, Kubicek K, Fortova A, Lacroute F, Vanacova S, Cramer P, Stefl R, et al. 2014. Molecular basis for coordinating transcription termination with noncoding RNA degradation. Mol Cell 55: 467–481. 10.1016/j.molcel.2014.05.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unnikrishnan A, Gafken PR, Tsukiyama T. 2010. Dynamic changes in histone acetylation regulate origins of DNA replication. Nat Struct Mol Biol 17: 430–437. 10.1038/nsmb.1780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uwimana N, Collin P, Jeronimo C, Haibe-Kains B, Robert F. 2017. Bidirectional terminators in Saccharomyces cerevisiae prevent cryptic transcription from invading neighboring genes. Nucleic Acids Res 45: 6417–6426. 10.1093/nar/gkx242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk EL, Chen CL, d'Aubenton-Carafa Y, Gourvennec S, Kwapisz M, Roche V, Bertrand C, Silvain M, Legoix-Ne P, Loeillet S, et al. 2011. XUTs are a class of Xrn1-sensitive antisense regulatory non-coding RNA in yeast. Nature 475: 114–117. 10.1038/nature10118 [DOI] [PubMed] [Google Scholar]
- van Nues R, Schweikert G, de Leau E, Selega A, Langford A, Franklin R, Iosub I, Wadsworth P, Sanguinetti G, Granneman S. 2017. Kinetic CRAC uncovers a role for Nab3 in determining gene expression profiles during stress. Nat Commun 8: 12 10.1038/s41467-017-00025-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelauer M, Rubbi L, Lucas I, Brewer BJ, Grunstein M. 2002. Histone acetylation regulates the time of replication origin firing. Mol Cell 10: 1223–1233. 10.1016/S1097-2765(02)00702-5 [DOI] [PubMed] [Google Scholar]
- Weiner A, Hsieh TH, Appleboim A, Chen HV, Rahat A, Amit I, Rando OJ, Friedman N. 2015. High-resolution chromatin dynamics during a yeast stress response. Mol Cell 58: 371–386. 10.1016/j.molcel.2015.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann S, Renner M, Watts BR, Adams O, Huseyin M, Baejen C, El Omari K, Kilchert C, Heo DH, Kecman T, et al. 2017. The conserved protein Seb1 drives transcription termination by binding RNA polymerase II and nascent RNA. Nat Commun 8: 14861 10.1038/ncomms14861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo H, Dam Ha S, Lee SB, Buratowski S, Kim T. 2017. Modulation of gene expression dynamics by co-transcriptional histone methylations. Exp Mol Med 49: e326 10.1038/emm.2017.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyers F, Rougemaille M, Badis G, Rousselle JC, Dufour ME, Boulay J, Regnault B, Devaux F, Namane A, Seraphin B, et al. 2005. Cryptic Pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell 121: 725–737. 10.1016/j.cell.2005.04.030 [DOI] [PubMed] [Google Scholar]
- Xu Z, Wei W, Gagneur J, Perocchi F, Clauder-Münster S, Camblong J, Guffanti E, Stutz F, Huber W, Steinmetz LM. 2009. Bidirectional promoters generate pervasive transcription in yeast. Nature 457: 1033–1037. 10.1038/nature07728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SC, Rhind N, Bechhoefer J. 2010. Modeling genome-wide replication kinetics reveals a mechanism for regulation of replication timing. Mol Syst Biol 6: 404 10.1038/msb.2010.61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Poveda A, Pasero P. 2013. Time to be versatile: regulation of the replication timing program in budding yeast. J Mol Biol 425: 4696–4705. 10.1016/j.jmb.2013.09.020 [DOI] [PubMed] [Google Scholar]
- Yoshida K, Bacal J, Desmarais D, Padioleau I, Tsaponina O, Chabes A, Pantesco V, Dubois E, Parrinello H, Skrzypczak M, et al. 2014. The histone deacetylases Sir2 and Rpd3 act on ribosomal DNA to control the replication program in budding yeast. Mol Cell 54: 691–697. 10.1016/j.molcel.2014.04.032 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.