Abstract
Motivation: There is a growing need in bioinformatics for easy-to-use software implementations of algorithms that are usable across platforms. At the same time, reproducibility of computational results is critical and often a challenge due to source code changes over time and dependencies.
Results: The approach introduced in this paper addresses both of these needs with AlgoRun, a dedicated packaging system for implemented algorithms, using Docker technology. Implemented algorithms, packaged with AlgoRun, can be executed through a user-friendly interface directly from a web browser or via a standardized RESTful web API to allow easy integration into more complex workflows. The packaged algorithm includes the entire software execution environment, thereby eliminating the common problem of software dependencies and the irreproducibility of computations over time. AlgoRun-packaged algorithms can be published on http://algorun.org, a centralized searchable directory to find existing AlgoRun-packaged algorithms.
Availability and implementation: AlgoRun is available at http://algorun.org and the source code under GPL license is available at https://github.com/algorun
Contact: laubenbacher@uchc.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 Introduction
Using software tools developed by the bioinformatics community presents several challenges, beginning with the installation of the software itself and its dependencies. Reproduction of computational results by others faces the challenge of frequent modifications in either the implementation of the algorithm itself or in one of its dependencies, or both (Boettiger, 2015; Peng, 2011). Although code sharing is possible through online code repositories like GitHub, it is hard to share the entire computational environment that a given software implementation of an algorithm depends on. Boettiger (2015) investigated the two main approaches to overcome these issues. The first approach is workflow software (Altintas et al., 2004; Hull et al., 2006). Although it provides technical solutions to the challenges of dissemination, it is marked by relatively low total adoption rates (Gil et al., 2007). The second approach is the use of virtual machines (VMs) running on cloud services (Dudley and Butte, 2010; Howe, 2012). While this approach addresses the problem by capturing the entire software environment, it is sometimes perceived as being a ‘black box’ not well suited to ensuring reproducibility. Also, researchers cannot easily build on virtual machines in a consistent and scalable way (Boettiger, 2015). A more recent approach to solve this problem is to use software containers, particularly Docker, an open source technology used to package applications and their dependencies into a standardized software container. Boettiger (2015) showed that using Docker will overcome the problems related to dependency, documentation inaccuracy and code changes. In this paper, we present an enhancement to the software container approach by introducing AlgoRun. AlgoRun is a Docker-based software container template designed to provide traditional command-line algorithms with standard web capabilities: a web user interface and a RESTful web API to allow easy integration of algorithms into complex workflows. In the following sections, we introduce AlgoRun and we show how it can be used to package Bowtie (Langmead et al., 2009), a well-known bioinformatics software. For better readability, the term ‘AlgoRun container’ will now be used to refer to a ‘Docker-based software container created with the AlgoRun template’.
2 Methods
AlgoRun is built on top of Docker, an open source technology used to package applications and their dependencies into a standardized and environment-agnostic software container. AlgoRun allows the efficient creation of software containers dedicated to making algorithms re-usable and making computational results reproducible by the scientific community. AlgoRun requires no or minimal source code changes, and is compatible with all programming languages. To aid the dissemination of implemented algorithms, AlgoRun containers can be published on http://algorun.org. Building an AlgoRun container requires installing Docker, providing information about the algorithm and its dependencies in the package template, and, finally, building the AlgoRun container. Supplementary File 1 provides detailed instructions.
AlgoRun Features
AlgoRun containers provide implemented algorithms with:
Remote execution ability: AlgoRun enables users to run implemented algorithms via the web API or via the web user interface (Fig. 1).
Fig. 1.
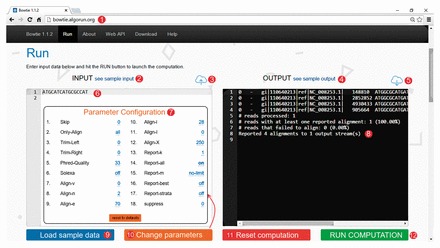
Example of web interface for the AlgoRun Bowtie software package: (1) Easy access through web browser instead of command line; (2) load a sample input; (3) upload dataset file; (4) click to see a sample of the output format; (5) save computation result to a file; (6) input box filled with data; (7) parameter configuration window; (8) algorithm output box filled with result; (9) input box with sample data; (10) parameter configuration window; (11) clear the input and output boxes; (12) run the algorithm on the given dataset
Ease-of-use: AlgoRun containers automatically generate a graphical web interface that requires minimal technical expertise.
Portability and compatibility: AlgoRun inherits Docker's main benefits by removing issues related to code changes, dependencies and backward compatibility over time.
Programmability and reusability: AlgoRun favors implemented algorithms’ reusability to create more complex workflows by providing a standard RESTful web API.
3 Application examples
As examples of how AlgoRun makes packaging, running and publishing algorithms a straightforward process, Supplementary File 2 shows the AlgoRun container creation process for the following implemented algorithms: Bowtie (Langmead et al., 2009), REACT (Vera-Licona, 2014) and KS (Kavvadias and Stavropoulos, 2005). To run these today, users need to download and install the necessary packages, their dependencies and run them from the command line. With AlgoRun however, users simply need to navigate to the dedicated AlgoRun web URL to start running computations, with no installation required. Instructions are provided for the option to run the container on the user’s local machine.
4 Conclusions
AlgoRun containers enable authors of algorithms to build a fully reusable software package for their work. The software package can be used immediately over the web or deployed in any computer environment, providing a friction-less access to computational capabilities. Taking the well-known Bowtie open-source software as an example, along with REACT and KS algorithms, we have demonstrated that the creation of such a software package requires minimal effort.
AlgoRun, with its robust web architecture, paves the way for building pipelines and networks of parallel and sequential computations. AlgoRun containers can be used in a distributed environment, which will greatly improve collaboration. Thus, AlgoRun has the potential to become the core architecture for computational software tools in the future.
Supplementary Material
Funding
R. Laubenbacher has been partially supported by Grant NSF DBI-1146819.
Conflict of Interest: none declared.
References
- Altintas I. et al. (2004). Kepler: an extensible system for design and execution of scientific workflows. In: Proceedings 16th int. conference on scientific and statistical database management. Washington, DC: IEEE Computer Society.
- Boettiger C. (2015) An introduction to Docker for reproducible research. ACM SIGOPS Oper. Syst. Rev., 49, 71–79. [Google Scholar]
- Dudley J.T., Butte A.J. (2010) In silico research in the era of cloud computing. Nat. Biotechnol., 28, 1181–1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil Y. et al. (2007) Examining the challenges of scientific workflows. Computer, 40, 24–32. [Google Scholar]
- Howe B. (2012) Virtual appliances, cloud computing, and reproducible research. Comput. Sci. Eng., 14, 36–41. [Google Scholar]
- Hull D. et al. (2006) Taverna: a tool for building and running workflows of services. Nucleic Acids Res., 34, W729–W732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavvadias D., Stavropoulos E. (2005) An efficient algorithm for the transversal hypergraph generation. J. Graph. Alg. Appl., 9, 239–264. [Google Scholar]
- Langmead B. et al. (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol., 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng R.D. (2011) Reproducible research in comp science. Science, 334, 1226–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vera-Licona P. (2014) An algebra-based method for inferring gene regulatory networks. BMC Syst. Biol., 8, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


