Abstract
Background
Metabolite networks are suggested to reflect biological pathways in health and disease. However, it is unknown whether such metabolite networks are reproducible across different populations. Therefore, the current study aimed to investigate similarity of metabolite networks in four German population-based studies.
Methods
One hundred serum metabolites were quantified in European Prospective Investigation into Cancer and Nutrition (EPIC)-Potsdam (n = 2458), EPIC-Heidelberg (n = 812), KORA (Cooperative Health Research in the Augsburg Region) (n = 3029) and CARLA (Cardiovascular Disease, Living and Ageing in Halle) (n = 1427) with targeted metabolomics. In a cross-sectional analysis, Gaussian graphical models were used to construct similar networks of 100 edges each, based on partial correlations of these metabolites. The four metabolite networks of the top 100 edges were compared based on (i) common features, i.e. number of common edges, Pearson correlation (r) and hamming distance (h); and (ii) meta-analysis of the four networks.
Results
Among the four networks, 57 common edges and 66 common nodes (metabolites) were identified. Pairwise network comparisons showed moderate to high similarity (r = 63–0.96, h = 7–72), among the networks. Meta-analysis of the networks showed that, among the 100 edges and 89 nodes of the meta-analytic network, 57 edges and 66 metabolites were present in all the four networks, 58–76 edges and 75–89 nodes were present in at least three networks, and 63–84 edges and 76–87 edges were present in at least two networks. The meta-analytic network showed clear grouping of 10 sphingolipids, 8 lyso-phosphatidylcholines, 31 acyl-alkyl-phosphatidylcholines, 30 diacyl-phosphatidylcholines, 8 amino acids and 2 acylcarnitines.
Conclusions
We found structural similarity in metabolite networks from four large studies. Using a meta-analytic network, as a new approach for combining metabolite data from different studies, closely related metabolites could be identified, for some of which the biological relationships in metabolic pathways have been previously described. They are candidates for further investigation to explore their potential role in biological processes.
Keywords: metabolomics, Gaussian graphical models, network analysis, reproducibility, meta-analysis, biological pathways
Key Messages
Metabolite networks constructed with Gaussian graphical models showed similar structures across four population-based studies.
We suggest meta-analysis of metabolite networks as a novel approach to identifying biological pathways.
The identified associations between metabolites in the meta-analytic network, particularly for phospholipids and amino acids, are candidates for further investigation to explore their role in health and disease.
Introduction
Metabolomic profiling is increasingly used to discover biomarkers that reflect early perturbations linked to disease risk or to objectively measure food intake and other environmental exposures.1–3 Thereby, many novel biomarkers have been identified that may improve assessment of various exposures or predict disease risk.2,4–6 One important step in the process of biomarker discovery is usually the replication of results in different study populations to reduce the chance of type one error.
High-throughput metabolomics is often analysed using correlation-based networks to infer biological relationships in the data.7,8 This approach has been successfully applied in several single studies to identify novel metabolic pathways.9–11 However, little is known about whether these metabolite networks can be replicated across different populations. So, the question arises as to whether the correlation structure of the identified metabolites is similar across different studies that include study participants with different characteristics (e.g. age and lifestyle). This should be a prerequisite to replicating metabolomic results in different populations.
Moreover, metabolic profiles from different studies are frequently assessed, but meta-analysis of metabolite networks has not been conducted in the metabolomics field. Partial-correlation-based network comparisons and meta-analysis of such networks can help to identify consistent relationships between metabolites, which may be further investigated for their potential role in biological processes.
Probabilistic graphical models such as Gaussian graphical models (GGMs) are interesting methods proposed for analysis of metabolomics data.12 A GGM is an undirected graph that identifies independence between two variables conditional on all others and has been suggested as an effective tool to recover metabolic pathways from metabolite concentrations.12 This approach can be further used to combine metabolomics data of different studies by meta-analysing network edges (partial correlations between two metabolites adjusted for the other metabolites) and constructing a meta-analytic metabolite network to represent the association between metabolites and their underlying metabolic pathways. This meta-analytic network may identify metabolites that are linked in certain metabolic pathways.
Against this background, the present study aimed to compare and meta-analyse the metabolite correlation networks to assess their stability and identify closely related metabolites in four large German population studies, including the European Prospective Investigation into Cancer and Nutrition (EPIC)-Potsdam, EPIC-Heidelberg, KORA (Cooperative Health Research in the Augsburg Region) and CARLA (Cardiovascular Disease, Living and Ageing in Halle).
Methods
This study was based on metabolomic measurements of participants from four German population-based studies (EPIC-Potsdam, EPIC-Heidelberg, KORA and CARLA). Ethical approval for all four studies was obtained from relevant ethical-approval committees. Written informed consent was obtained from all participants in the included studies.
Description of the study populations
EPIC-Potsdam and EPIC-Heidelberg comprise 27 548 and 25 540 study participants, respectively. Study design and methods in EPIC-Potsdam and EPIC-Heidelberg were similar and have been described in detail elsewhere.13,14 For measurements of serum metabolites, a random subcohort was established in 2006 in EPIC-Potsdam (n = 2483) and 2009 in EPIC-Heidelberg (n = 843).15 The KORA study is conducted in Southern Germany16 and included 3044 participants, who took part in the survey (KORA F4) from 2006 to 2008. The CARLA study included 1779 participants with baseline examinations between 2002 and 2006. Serum metabolites were assessed for 1427 participants. Details of KORA and CARLA were described elsewhere.17,18 After exclusion of participants with missing data on any covariate (n = 23) or metabolites (n = 48), 2458 participants in EPIC-Potsdam, 812 in EPIC-Heidelberg, 3029 in KORA and 1427 in CARLA were available for analysis.
Blood-sample collection and assessment of covariates
Blood samples from all participants were collected at baseline or follow-up (KORA) using standard protocols as described elsewhere.4,18–20 Age, sex, weight and height were collected at baseline in all studies. Body mass index (BMI) was estimated as: (weight in kilogrammes)/(height in metres)2.
Metabolomic profiling
Metabolites were quantified in all four populations in serum blood samples using the AbsoluteIDQTM p150 and p150 Kits (Biocrates Life Scienes AG, Innsbruck Austria) together with FIA- and LC-ESI-MS/MS (flow injection analysis/liquid chromatography-electrospray ionization-tandem mass spectrometry) as described in detail by Römisch-Margl et al.21 and Zukunft et al.22 The AbsoluteIDQTM p150 Kit was applied for samples of EPIC-Potsdam23 and CARLA,24 the AbsoluteIDQTM p180 Kit for samples of the KORA F4 study25 and the MetaDisIDQ™ Kit for samples of EPIC-Heidelberg.13–15 Metabolite measurements of EPIC-Potsdam, KORA and CARLA samples were performed in the Genome Analysis Center at the Helmholtz Zentrum München and for EPIC-Heidelberg in Leipzig. To ensure comparability, only those metabolites were included in the analysis, which were quantified by all three metabolite kits. In addition, metabolites below the limit of detection and those with very high analytic variance in any of the four studies were excluded, leaving 100 metabolites for the present analysis. The final metabolite set contained hexose (sum of six-carbon monosaccharides without distinction of isomers), 2 acylcarnitines (Cx:y; x = number of carbon atoms, y = number of double bonds), 10 sphingolipids, 12 amino acids, 35 acyl-alkyl-, 32 diacyl- and 8 lyso-phosphatidylcholines (PC)s (Supplementary Table 1, available as Supplementary data at IJE online).
Statistical analysis
Metabolite concentrations were log-transformed to approximate normality. Distributions of the metabolite concentrations were visually assessed using QQ-Plot and Histogram, which showed approximately normal distribution. However, long tails and potential outliers were detected for some metabolites. QQ-Plots for the top 20 metabolites from the study sample are shown in Supplementary Figures 1–5, available as Supplementary data at IJE online. Therefore, a non-parametric approach was also used to confirm that major findings do not vary by choice of method. Means, standard deviations and coefficients of variation (CV) were calculated for each metabolite in all studies adjusted for age, sex and BMI. For comparison of metabolic profiles, each individual study metabolite network was estimated using the GGM approach. In the first step, a partial-correlation matrix of the 100 metabolites was estimated for each study sample. In the second step, the top 100 highly correlated metabolite pairs (edges) were selected to construct networks in the respective samples. The minimum partial correlation was 0.19 in the network of Heidelberg, 0.25 in the networks of Potsdam, 0.24 in the network of KORA and 0.26 in the network of CARLA. We selected the first 100 edges with the highest correlation, so that the identified networks are similar but interpretable, and the correlations are high enough to have biological relevance, since only highly correlated metabolites have been suggested to be biologically related.26 Identified networks were exported to Cytoscape for visualization.27 The same analyses were repeated using Spearman’s rank partial correlation. First, Spearman’s rank correlation for all the metabolites was estimated and then the top 100 highly correlated metabolite pairs (edges) were selected from each sample to construct respective networks.
To assess the similarity between network structures, correlations between each pair of networks were estimated using gcor function from R-package sna.28 For this purpose, the networks (edge lists) were converted into adjacency matrices, which in turn were used to estimate product–moment correlation. To estimate structural similarity between the four networks, hamming distance was determined using the same R-package. Hamming distance is the number of changes required to transform one network into another,29 e.g. if the hamming distance between two networks X and Y is 1, then one change (i.e. an addition or deletion of one edge) will result in an identical structure of the two (X and Y) networks. A lower hamming distance reflects a similarity in network structures. The hamming distance was estimated by transforming the networks into adjacency matrices. The adjacency matrices were then used to estimate the hamming distance with (code) hdist in sna R-package. For easier comparison, the numbers of common edges in each combination of the four metabolite networks were visualized using a Venn diagram, which was constructed using R-package VennDiagram.29 Commonality of the four networks was reflected by visualizing the common edges of the four networks estimated by both the Pearson and Spearman partial correlations. As metabolites in EPIC-Heidelberg were quantified in a different laboratory, though using a standardized approach, a second network of common edges was constructed for EPIC-Potsdam, CARLA and KORA only. For meta-analysis of the four networks, a random-effect meta-analysis of partial-correlation coefficients was conducted using all common edges. For meta-analysis, partial-correlation coefficients were transformed to fisher Z-scores and back-transformed after analysis. The correlation coefficients from meta-analysis were used to construct a meta-analytic metabolite network by selecting 100 highly correlated metabolite pairs, as was done for the individual networks. Due to high heterogeneity among studies, a combined metabolite network of all studies was constructed and common edges of all the metabolites observed in each study were visualized over the meta-analytic network in Cytoscape. Network analyses were adjusted for age, sex and BMI, which are related to metabolite differences.
Results
EPIC-Potsdam and EPIC-Heidelberg study populations were similar with respect to age, sex and BMI, whereas the study populations in KORA and CARLA were older and had a lower percentage of women and a higher BMI compared with the two EPIC studies (Table 1).
Table 1.
Sample characteristics of the included studiesa
| Characteristicsb | EPIC-Potsdam | EPIC-Heidelberg | KORA | CARLA |
|---|---|---|---|---|
| (n = 2458) | (n = 812) | (n = 3029) | (n = 1427) | |
| Age (years) | 50.3 (9.0) | 50.7 (7.9) | 56 (13.3) | 63.3 (9.7) |
| Sex (women %) | 61.2 | 54.9 | 51.5 | 44.9 |
| BMI (kg/m²) | 26.1 (4.3) | 25.6 (4.2) | 27.6 (4.8) | 28.1 (4.5) |
Shown are mean values and standard deviations.
Blood samples from EPIC-Potsdam, KORA and CARLA were analysed in the same laboratory. Samples from KORA were analysed using a different kit.
Considerable differences were found for metabolite concentrations (mean and CV) between the four studies (Supplementary Table 1, available as Supplementary data at IJE online). Overall, 29 metabolites in Potsdam, 50 metabolites in Heidelberg, 20 metabolites in KORA and 59 metabolites in CARLA showed high variation (≥ 30% CV) in concentration.
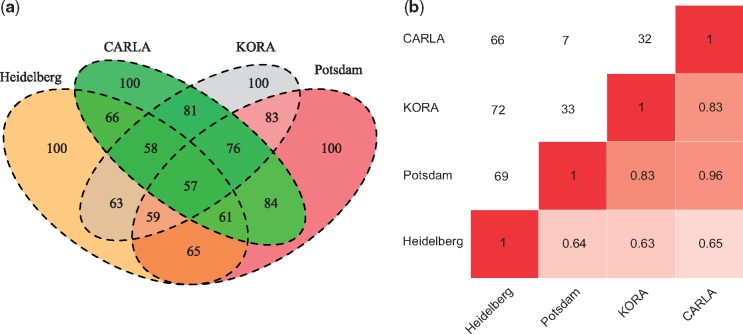
The metabolite networks of the four studies are shown in Supplementary Figures 5–8, available as Supplementary data at IJE online. All networks identified clusters of sphingolipids, lyso-PCs, diacyl-PCs and acyl-alkyl-PCs, albeit with large variation in network topologies, i.e. connection between metabolites. Amino acids showed the highest variation in network connectivity, although with consistent clustering of tryptophan, tyrosine and phenylalanine in all networks. Hexoses (represented as a single metabolite) were connected with amino acids valine and tryptophan only in CARLA. Two acylcarnitines were connected as a pair in all the studies except in EPIC-Potsdam. The highest variation in metabolites topology was observed in the network of EPIC-Heidelberg as compared with other networks (Figure 1a and Table 2). Pairwise comparison of the networks showed the greatest similarity represented by the lowest hamming distance and the highest correlation between EPIC-Potsdam and KORA. EPIC-Heidelberg’s metabolite network was the most dissimilar from all other networks, as it showed a high hamming distance and lower correlation (Figure 1b).
Figure 1.
(a) Edges overlap among four studies included in the study. Shown are the numbers of edges. (b) Pearson’s correlation and hamming distance between metabolite networks of the studies included in the study. The upper triangle shows the hamming distance and the lower triangle shows correlation among the networks. The lower values of the hamming distance show greater similarity whereas the lower value of correlation shows less similarity between the networks.
Overlap of the common edges among the different combinations of the four studies is shown in Figure 1b. The highest overlap of the edges was observed between EPIC-Potsdam and CARLA. The metabolite network of EPIC-Heidelberg showed the smallest overlap of edges with the other networks. Overall, 66 edges were consistently detected in all four networks, interlinking 80 metabolites (Figure 2). The other 20 out of 100 metabolites were unconnected and are not shown. Lyso-PCs, diacyl-PCs and sphingolipids consistently grouped together across all studies (Supplementary Figures 1–4, available as Supplementary data at IJE online). Among the four networks, in EPIC-Potsdam (nodes = 91), CARLA (nodes = 95) and KORA (nodes = 96), a relatively large number of metabolites were integrated in the networks, whereas 20 metabolites (mainly amino acids and acyl-alkyl PCs) remained unconnected in EPIC-Heidelberg (Table 2).
Figure 2.
Common edges of the serum metabolite network of the four studies: EPIC-Heidelberg, EPIC-Potsdam, CARLA and KORA. Nodes represent metabolites and edges are partial correlations between two metabolites adjusted for the other metabolites as well as age, sex and BMI. Continuous black lines represent positive and dashed lines represent inverse partial correlations. The thicknesses of the edges are proportional to the strength of the correlations. Nodes with different border colours represent different metabolite classes: black: amino acids; purple: lyso-phosphatidylcholines; sky-blue: sphingolipids; green: diacyl-phosphatidylcholines; red: acyl-alkyl-phosphatidylcholines.
Table 2.
Number of connected nodes (metabolites) in individual and combined metabolite networks in the four studies
| Name of study | Metabolitesa (number) |
|||||||
|---|---|---|---|---|---|---|---|---|
| Hexoses | AC | AA | LysoPC | DiA-PC | AA-PC | SL | Total | |
| (1) | (2) | (12) | (8) | (32) | (35) | (10) | (100) | |
| Heidelberg (H) | 00 | 02 | 06 | 08 | 30 | 29 | 10 | 85 |
| Potsdam (P) | 01 | 02 | 09 | 08 | 30 | 32 | 10 | 92 |
| CARLA (C) | 00 | 02 | 06 | 08 | 30 | 33 | 10 | 89 |
| KORA (K) | 00 | 00 | 08 | 08 | 29 | 31 | 10 | 86 |
| HP | 00 | 02 | 05 | 08 | 30 | 27 | 10 | 82 |
| HC | 00 | 02 | 03 | 08 | 30 | 27 | 10 | 80 |
| HK | 00 | 00 | 03 | 08 | 29 | 26 | 10 | 76 |
| PK | 00 | 00 | 07 | 08 | 29 | 30 | 10 | 84 |
| PC | 00 | 02 | 06 | 08 | 30 | 31 | 10 | 87 |
| CK | 00 | 00 | 05 | 08 | 29 | 31 | 10 | 83 |
| HPK | 00 | 00 | 03 | 08 | 29 | 25 | 10 | 75 |
| HPC | 00 | 02 | 03 | 08 | 30 | 26 | 10 | 79 |
| HCK | 00 | 00 | 02 | 08 | 29 | 26 | 10 | 75 |
| PCK | 00 | 00 | 05 | 08 | 29 | 30 | 10 | 82 |
| HPCK (Common network) | 00 | 00 | 02 | 08 | 29 | 17 | 10 | 66 |
| Meta-analytic network | 00 | 02 | 08 | 08 | 30 | 31 | 10 | 89 |
AC, acylcarnitines; AA, amino acids; LysoPC, lyso-phosphatidylcholines, DiA-PC, diacyl-phosphatidylcholines; AA-PC, acyl-alkyl- phosphatidylcholines; SL, sphingolipids.
A structural comparison of the four networks showed 57 common edges and 66 commonly connected nodes (Table 3), which are shown in a common network (Figure 2). The common network showed smaller clustering of similar classes of metabolites (Figure 2). Notably, sphingolipids, lyso-PCs and subgroups of acyl-alkyl-PCs and tryptophan, tyrosine and phenylalanine were clustered together. Due to differences between EPIC-Heidelberg and the other studies, we also constructed a common network of EPIC-Potsdam, CARLA and KORA, which showed higher similarity of the metabolite network structures among the three studies (Figure 3).
Table 3.
Number of connected nodes (metabolites) and edges in individual and common metabolite networks in the four studies
| Name of study | Metabolitesa (number) |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Hexoses | AC | AA | LysoPC | DiA-PC | AA-PC | SL | Total | No of Edges | |
| (1) | (2) | (12) | (8) | (32) | (35) | (10) | (100) | (100) | |
| Pearson r-based networks | |||||||||
| Heidelberg (H) | 00 | 02 | 06 | 08 | 30 | 29 | 10 | 85 | 100 |
| Potsdam (P) | 00 | 00 | 08 | 08 | 29 | 31 | 10 | 86 | 100 |
| CARLA (C) | 01 | 02 | 09 | 08 | 30 | 32 | 10 | 92 | 100 |
| KORA (K) | 00 | 02 | 06 | 08 | 30 | 33 | 10 | 89 | 100 |
| Common network | 00 | 00 | 02 | 08 | 29 | 17 | 10 | 66 | 57 |
| Spearman’s rank-based networks | |||||||||
| Heidelberg (H) | 00 | 02 | 07 | 08 | 30 | 29 | 10 | 86 | 100 |
| Potsdam (P) | 00 | 00 | 08 | 08 | 30 | 31 | 10 | 87 | 100 |
| CARLA (C) | 01 | 02 | 08 | 08 | 30 | 32 | 10 | 91 | 100 |
| KORA (K) | 00 | 02 | 05 | 08 | 30 | 32 | 10 | 87 | 100 |
| Common network | 00 | 00 | 00 | 07 | 26 | 20 | 10 | 65 | 56 |
AC, acylcarnitines; AA, amino acids; LysoPC, lyso-phosphatidylcholines, DiA-PC, diacyl-phosphatidylcholines; AA-PC, acyl-alkyl- phosphatidylcholines; SL, sphingolipids.
Figure 3.
Common edges of the serum metabolite network of the three studies: EPIC-Potsdam, CARLA and KORA. Nodes represent metabolites and edges are partial correlations between two metabolites adjusted for the other metabolites as well as age, sex and BMI. Continuous black lines represent positive and dashed lines represent inverse partial correlations. The thicknesses of the edges are proportional to the strength of the correlations. Nodes with different border colours represent different metabolite classes: black: amino acids; purple: lyso-phosphatidylcholines; sky-blue: sphingolipids; green: diacyl-phosphatidylcholines; red: acyl-alkyl-phosphatidylcholines.
The meta-analytic network of the partial-correlation coefficients represented by the 100 highly correlated metabolite pairs (edges) across the four studies is shown in Figure 4. Meta-analysis of the networks revealed that, among the 100 edges connecting 89 nodes of the meta-analytic network, 57 edges connecting 66 metabolites were present in all the four networks, 58–76 edges connecting 75–89 nodes were present in at least three networks and 63–84 edges connecting 76–87 nodes were present in at least two networks. The meta-analytic network showed clear clusters of the paired acylcarnitines, sphingolipids, lyso-PCs and three clusters of amino acids. Large but differently connected clusters of acyl-alkyl-PCs and diacyl-PCs formed the dominant structure of the networks. Comparison of this network with the common network of four studies showed dissimilarity in a number of edges (Figure 5). However, it was very similar to the combined network of Potsdam, KORA and CARLA (Supplementary Figure 9, available as Supplementary data at IJE online).
Figure 4.
Meta-analytic serum metabolite network of the four studies: EPIC-Heidelberg, EPIC-Potsdam, CARLA and KORA. Nodes represent metabolites and edges are partial correlations between two metabolites adjusted for the other metabolites as well as age, sex and BMI. Continuous black lines represent positive and dashed lines represent inverse partial correlations. The thicknesses of the edges are proportional to the strength of the correlations. Nodes with different border colours represent different metabolite classes: yellow: acylcarnitines; black: amino acids; purple: lyso-phosphatidylcholines; sky-blue: sphingolipids; green: diacyl-phosphatidylcholines; red: acyl-alkyl-phosphatidylcholines.
Figure 5.
Comparative network of the common network and the meta-analytic network of the four studies: EPIC-Heidelberg, EPIC-Potsdam, KORA and CARLA. Nodes represent metabolites and edges are partial correlations between two metabolites adjusted for the other metabolites as well as age, sex and BMI. Black edge colours represent common edges in the common network and the meta-analytic network, whereas the grey colour represents edges present only in the meta-analytic network. Similarly, the white colour of nodes represents common nodes in the compared networks, whereas the red colour represents nodes present only in the meta-analytic network. Nodes with different border colours represent different metabolite classes: yellow: acylcarnitines; black: amino acids; purple: lyso-phosphatidylcholines; sky-blue: sphingolipids; green: diacyl-phosphatidylcholines; red: acyl-alkyl-phosphatidylcholines.
The networks constructed using Spearman’s rank partial correlations are shown in Supplementary Figures 10–14, available as Supplementary data at IJE online. All the individual networks and common networks showed high similarity to the corresponding networks constructed using Pearson’s partial correlations (Table 3).
Discussion
In this study, we generated and compared the metabolite networks of four German population-based studies. Moreover, we applied a novel meta-analytic approach to combine metabolite networks to identify potentially stable correlation structures across all studies. Comparison of metabolite networks revealed overall considerable heterogeneity in network topologies. However, specific metabolite subgroups showed high consistency in the networks. Consistent network structures were detected for sphingolipids, lyso-PCs, acyl-alkyl-PCs and diacyl-PCs and among the amino acids tryptophan, tyrosine and phenylalanine. The meta-analytic network also showed clear grouping of the metabolite classes and was, in addition, sensitive for further plausible biological links. Consistent links between metabolites from the same group may reflect the same underlying metabolic pathways as the common determinants of the correlation structure across the study populations.
In the identified common as well as meta-analytic networks, we observed connections of sphingolipids with PCs, which could be related to the biosynthesis pathway of the sphingolipids. The synthesis of sphingolipids require enzymatic transfer of phosphocholines from PCs to ceramide, which in turn is converted to sphingolipids.30 The linkage between these two classes could also be due to limitation of the measurement kit owing to possible interference in the measurement of different metabolites.31 In addition, we observed a consistent connection between the aromatic amino acids phenylalanine, tryptophan and tyrosine. Phenylalanine is a substrate for tyrosine biosynthesis32 and, with tryptophan, the two are also precursors of catecholamines.33 In these networks, we also observed that the majority of stable edges connected metabolites that are known to be directly related by a single metabolic reaction step. This supports the idea that the reproducible correlation structure of metabolites likely reflects linkage in metabolic pathways.34 Our results are supported by an earlier KORA study which showed that the GGM has high sensitivity and specificity in identifying reactions that are one step apart.12 The same study, which is also included in this analysis, likewise reported that reactions that were two steps apart were reflected by negative correlations in the network. This was also observed in our study, e.g. SM.C16.1 and SM.C18.0 were negatively correlated in the common and meta-analysed networks.
The compared networks also showed a clear separation of amino acids and acylcarnitines and separate clustering of sphingolipids and diacyl- and acyl-alkyl-PCs. These findings are in agreement with earlier results observed in KORA and EPIC-Potsdam.4 The modular structure of the metabolites may reflect metabolic pathways including biosynthesis, degradation and metabolism and interaction between the different classes of metabolites. Such biological interrelations were shown to be detectable in metabolomics data in observational studies, which could be reproduced across different populations.12 For example, PC.ae.C32: 1 and PC.ae.C32: 2 reflect Steaoryl-CoA desaturase/ Steaoryl-CoA desaturase 5 desaturation and a pair of PC.aa.C38: 5 and PC.aa.C40: 5 reflects various fatty acid elongations.35 Likewise, correlation between phenylalanine, tryptophan and tyrosine denotes amino-acid-associated pathways.36 Some of the consistent relationships between metabolites identified in the networks might hint towards so far unknown links. Such metabolites might be better candidates for further investigation to identify their role on the metabolic pathways.
In addition, for comparison of the four metabolite networks, we also constructed a meta-analytic metabolite network that shared higher similarity with the common networks from the three studies including EPIC-Potsdam, KORA and CARLA and less similarity with common networks including EPIC-Heidelberg. The heterogeneity among the identified networks may partly be attributed to the differences in health/disease status of the four populations, differences in diet, fasting status, medication/supplement use or lifestyle factors. Technical differences related to metabolite-concentration measurements in different laboratories or use of different kits could also have partly resulted in the observed differences. Indeed, it is already known that biochemical assay assessments, sample handling and other factors such as storage etc. are some of the reasons affecting reliability and are often addressed in metabolomic measurements.37–39 In addition, EPIC-Potsdam and EPIC-Heidelberg had similar study protocols, sample preparation, storage conditions and relatively similar population characteristics. However, large differences in the networks of the two populations were observed. Therefore, the difference between EPIC-Heidelberg and the other studies may partly be related to smaller sample size and technical issues such as metabolomic measurement in a different laboratory with different kits.
We also observed differences between the common network and the meta-analytic network. However, it must be noted that these differences are not unexpected, as the two were created using two different approaches, i.e. (i) by combining the common edges in all the networks in one network and (ii) meta-analysis of the four networks. It is important to underline that the meta-analytic network was created using an inverse variance approach, which gives higher weight to the studies with large sample sizes, i.e. KORA, EPIC-Potsdam and CARLA, respectively. This had a large influence on the effect size (partial correlations). Consequently, it resulted in a network that is more similar to the common network of the three larger studies. Nevertheless, the random meta-analytic approach is advantageous over the approach that was based on simple structural similarity, as the former takes both within- and between-studies variation into account. It should also be noted that many of the additional edges that were detected by the meta-analytic approach again corresponded to known metabolic reaction steps.
A major strength of this study was that, for the first time, metabolite networks between several large population-based studies were compared using an innovative meta-analytic networks approach. Thereby, the meta-analytic network was based on metabolomic measurements of almost 8000 participants, which represents a very large sample size for the application of these sophisticated metabolomic technologies. Metabolites were measured in different population samples, which might have slightly different environmental exposures despite living in the same country. However, the aim of this study was to see how similar these metabolite networks are in free-living populations with less restricted conditions. This approach was also chosen to better grasp the feasibility of replicating metabolomic results in different populations, which is often demanded when validating metabolomics data. In addition, relatively similar analytic methods were used to quantify the concentration of the metabolites, which makes the data more comparable than data from other platforms. Moreover, we reduced technical variation by including only those metabolites that were above the detection level and showed good reliability in any of the four studies.23
This study also had certain limitations. We found differences in mean metabolite concentrations between the cohorts, which are attributable to technical aspects (e.g. different laboratories and kits, sample processing and storage, etc.) as well as biological aspects (e.g. cohort differences such as age, sex, BMI, etc.). CVs of metabolite measurements were similar between the cohorts and comparable to other metabolomic studies that measured the same metabolites.23,40 However, a general limitation of metabolomic studies is that many metabolites are measured simultaneously and CVs of metabolites are usually higher than CVs of single biomarkers. Metabolite measurements were assessed in two different laboratories and using different kits, although the kits were from the same company. Further, metabolites were identified using a targeted approach, which has limited coverage. It might have resulted in missing many metabolites that are sharing similar metabolic pathways with the investigated metabolites. However, to conduct a similar study with untargeted metabolomic measurements may be challenging, as different metabolites may be detected in different populations and a number of metabolites will remain unidentified, which may complicate comparison across several studies. Another limitation of our study is that only one metabolite measurement per sample was available for the current study, so metabolite reliability could not be tested. However, two earlier EPIC studies showed moderate to good reliability of included metabolites over 4 months23 and over 2 years.40 In addition, participants with prevalent medical conditions were not excluded, which might have affected the metabolomics profiles. Similarly, we did not account for fasting state, as, due to logistic reasons in large studies, the majority were non-fasting samples, which may affect metabolite reliability.40
The existing methods of network construction employ either regression-based approaches or some thresholding criteria for edge inclusions in the respective networks. Nevertheless, using these approaches, the identified networks in different sample could be different, as the correlation between variables may vary due to a number of factors such as sample size, etc. Therefore, in order to construct networks of similar sizes for comparison in our study, we retained the 100 edges with the highest correlation in all four cohorts. As we do not perform any model selection, this method may result in inclusion of edges that may not reflect important biological relationships or exclusion of edges that could be important in some biological pathways. It is also important to note that GGM works under the assumption of the Gaussian distribution of the study variables. Therefore, we compared the QQ-plots against normal distribution to ensure log-normality of the metabolites concentrations. We observed some deviations in the tails of several metabolites. Nevertheless, the results were comparable with the non-parametric approach in identifying highly correlated metabolites.
In summary, we observed considerable similarities in metabolite sub-networks of sphingolipids, lyso-PCs, acyl-alkyl-PCs and diacyl-PCs and amino acids across the four populations, although large variations were observed in overall networks. Variation may partly be explained by technical issues, such as different laboratories and measurement kits. These technical difficulties should be investigated further and also be taken into account when replicating metabolomic results in different population-based studies. Stable links observed within groups of biochemically related metabolites may likely reflect close interdependency of the connected metabolites in metabolic pathways. Using the meta-analytic network as a new approach for combining metabolic data from different studies, closely related metabolites could be identified, for some of which the biological relationships in metabolic pathways had been previously described. The metabolites with observed relationships in the meta-analytic network may be candidates for further investigation to explore their potential role in biological processes.
Supplementary Material
Funding
EPIC-Potsdam was supported by a grant from the German Federal Ministry of Education and Research (BMBF) to the German Center for Diabetes Research (DZD) and the State of Brandenburg. The CARLA study was funded by a grant from the German Research Foundation (DFG) as part of the Collaborative Research Centre ‘Heart failure in the elderly—cellular mechanisms and therapy’, by three grants of the Wilhelm-Roux Programme of the Martin Luther University of Halle-Wittenberg (FKZ 14/41, 16/19 and 28/21), by the Federal Employment Office and by the Ministry of Education and Cultural Affairs of Saxony-Anhalt (MK-CARLA-MLU-2011).
Conflict of interest: Authors declare no conflict of interest.
References
- 1. Wittenbecher C, Muhlenbruch K, Kroger J. et al. Amino acids, lipid metabolites, and ferritin as potential mediators linking red meat consumption to type 2 diabetes. Am J Clin Nutr 2015;101:1241–50. [DOI] [PubMed] [Google Scholar]
- 2. Aleksandrova K, di Giuseppe R, Isermann B. et al. Circulating omentin as a novel biomarker for colorectal cancer risk: data from the EPIC-Potsdam cohort study. Cancer Res 2016;76:3862–71. [DOI] [PubMed] [Google Scholar]
- 3. Jenab M, Slimani N, Bictash M, Ferrari P, Bingham SA.. Biomarkers in nutritional epidemiology: applications, needs and new horizons. Hum Genet 2009;125:507–25. [DOI] [PubMed] [Google Scholar]
- 4. Floegel A, Stefan N, Yu Z. et al. Identification of serum metabolites associated with risk of type 2 diabetes using a targeted metabolomic approach. Diabetes 2013;62:639–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wang TJ, Larson MG, Vasan RS. et al. Metabolite profiles and the risk of developing diabetes. Nat Med 2011;17:448–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dietrich S, Floegel A, Weikert C. et al. Identification of serum metabolites associated with incident hypertension in the European prospective investigation into cancer and nutrition-potsdam study. Hypertension 2016;68:471–77. [DOI] [PubMed] [Google Scholar]
- 7. Krumsiek J, Bartel J, Theis FJ.. Computational approaches for systems metabolomics. Curr Opin Biotechnol 2016;39:198–206. [DOI] [PubMed] [Google Scholar]
- 8. Batushansky A, Toubiana D, Fait A.. Correlation-based network generation, visualization, and analysis as a powerful tool in biological studies: a case study in cancer cell metabolism. Biomed Res Int 2016;2016:1.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yazdani A, Yazdani A, Saniei A, Boerwinkle E.. A causal network analysis in an observational study identifies metabolomics pathways influencing plasma triglyceride levels. Metabolomics 2016;12:104.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fukushima A, Kusano M, Redestig H, Arita M, Saito K.. Metabolomic correlation-network modules in Arabidopsis based on a graph-clustering approach. BMC Syst Biol 2011;5:1.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Krumsiek J, Mittelstrass K, Do KT. et al. Gender-specific pathway differences in the human serum metabolome. Metabolomics 2015;11:1815–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Krumsiek J, Suhre K, Illig T, Adamski J, Theis FJ.. Gaussian graphical modeling reconstructs pathway reactions from high-throughput metabolomics data. BMC Syst Biol 2011;5:21.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Boeing H, Korfmann A, Bergmann MM.. Recruitment procedures of EPIC-Germany. Ann Nutr Metab 1999;43:205–15. [DOI] [PubMed] [Google Scholar]
- 14. Bergmann MM, Bussas U, Boeing H.. Follow-up procedures in EPIC-Germany—data quality aspects. Ann Nutr Metab 1999;43:225–34. [DOI] [PubMed] [Google Scholar]
- 15. Kühn T, Floegel A, Sookthai D. et al. Higher plasma levels of lysophosphatidylcholine 18: 0 are related to a lower risk of common cancers in a prospective metabolomics study. BMC Med 2016;14:01–09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Rathmann W, Kowall B, Heier M. et al. Prediction models for incident type 2 diabetes mellitusin the older population: KORA S4/F4 cohort study. Diabet Med 2010;27:1116–23. [DOI] [PubMed] [Google Scholar]
- 17. Rathmann W, Strassburger K, Heier M. et al. Incidence of Type 2 diabetes in the elderly German population and the effect of clinical and lifestyle risk factors: KORA S4/F4 cohort study. Diabet Med 2009;26:1212–19. [DOI] [PubMed] [Google Scholar]
- 18. Greiser KH, Kluttig A, Schumann B. et al. Cardiovascular disease, risk factors and heart rate variability in the elderly general population: design and objectives of the CARdiovascular disease, Living and Ageing in Halle (CARLA) Study. BMC Cardiovasc Disord 2005;5:33.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Holle R, Happich M, Lowel H, Wichmann HE.. KORA—a research platform for population based health research. Gesundheitswesen 2005;67:19–25. [DOI] [PubMed] [Google Scholar]
- 20. Kühn T, Floegel A, Sookthai D. et al. Higher plasma levels of lysophosphatidylcholine 18: 0 are related to a lower risk of common cancers in a prospective metabolomics study. BMC Med 2016;14:13.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Römisch-Margl W, Prehn C, Bogumil R, Röhring C, Suhre K, Adamski J.. Procedure for tissue sample preparation and metabolite extraction for high-throughput targeted metabolomics. Metabolomics 2012;8:133–42. [Google Scholar]
- 22. Zukunft S, Sorgenfrei M, Prehn C, Möller G, Adamski J.. Targeted metabolomics of dried blood spot extracts. Chromatographia 2013;76:1295–305. [Google Scholar]
- 23. Floegel A, Drogan D, Wang-Sattler R. et al. Reliability of serum metabolite concentrations over a 4-month period using a targeted metabolomic approach. PLoS One 2011;6:e21103.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lacruz ME, Kluttig A, Tiller D. et al. Cardiovascular risk factors associated with blood metabolite concentrations and their alterations during a 4-year period in a population-based cohort. Circ Cardiovasc Genet 2016;9:487–94. [DOI] [PubMed] [Google Scholar]
- 25. Jourdan C, Petersen AK, Gieger C. et al. Body fat free mass is associated with the serum metabolite profile in a population-based study. PLoS One 2012;7:e40009.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Camacho D, de la Fuente A, Mendes P.. The origin of correlations in metabolomics data. Metabolomics 2005;1:53–63. [Google Scholar]
- 27. Shannon P, Markiel A, Ozier O. et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13:2498–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Butts CT. Social network analysis with SNA. J Stat Soft 2008;24:1–51. [Google Scholar]
- 29. Chen H, Boutros PC.. VennDiagram: a package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinformatics 2011;12:35.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gault CR, Obeid LM, Hannun YA.. An overview of sphingolipid metabolism: from synthesis to breakdown. Adv Exp Med Biol 2010;688:1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.AbsoluteIDQ(TM) Kit. Analytical Specifications p150 Innsbruck: BIOCRATES Life Sciences, 2006.
- 32. Matthews DE. An overview of phenylalanine and tyrosine kinetics in humans. J Nutr 2007;137:1549S–75S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Cansev M, Wurtman R.. 4 Aromatic Amino Acids in the Brain: Handbook of Neurochemistry and Molecular Neurobiology. Berlin, Heidelberg: Springer, 2007, pp. 59–97. [Google Scholar]
- 34. Floegel A, Wientzek A, Bachlechner U. et al. Linking diet, physical activity, cardiorespiratory fitness and obesity to serum metabolite networks: findings from a population-based study. Int J Obes 2014;38:1388–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Agbaga M-P, Mandal MNA, Anderson RE.. Retinal very long-chain PUFAs: new insights from studies on ELOVL4 protein. J Lipid Res 2010;51:1624–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kanehisa M, Goto S.. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 2000;28:27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Strnadova KA, Holub M, Muhl A. et al. Long-term stability of amino acids and acylcarnitines in dried blood spots. Clin Chem 2007;53:717–22. [DOI] [PubMed] [Google Scholar]
- 38. Zivkovic AM, Wiest MM, Nguyen UT, Davis R, Watkins SM, German JB.. Effects of sample handling and storage on quantitative lipid analysis in human serum. Metabolomics 2009;5:507–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zelena E, Dunn WB, Broadhurst D. et al. Development of a robust and repeatable UPLC-MS method for the long-term metabolomic study of human serum. Anal Chem 2009;81:1357–64. [DOI] [PubMed] [Google Scholar]
- 40. Carayol M, Licaj I, Achaintre D. et al. Reliability of serum metabolites over a two-year period: a targeted metabolomic approach in fasting and non-fasting samples from EPIC. PLoS One 2015;10:e0135437.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.