Abstract
Background
Associations of adult height with cardiometabolic and pulmonary traits have been studied in majority European ancestry populations using Mendelian randomization and polygenic risk score (PRS) analysis. The standard PRS approach entails creating a PRS for height using variants identified in prior genome-wide association studies (GWAS). It is unclear how well the standard PRS approach performs in non-European populations and whether height–trait associations observed in Europeans are also observed in other populations.
Methods
In the Hispanic Community Health Study/Study of Latinos (HCHS/SOL), we used: (i) the standard approach to create a PRS for height (PRS1) and (ii) a novel approach to optimize the selection of variants from previously established height association loci to better explain height in HCHS/SOL (PRS2). We also estimated the extent to which PRS–trait associations were independent or mediated by the PRS effect on height.
Results
In 7539 women and 5245 men, PRS1 and PRS2 explained 9 and 29% of the variance in measured height, respectively. Both PRS1 and PRS2 were associated with forced expiratory volume in 1 second (FEV1), forced vital capacity (FVC), FEV1/ FVC ratio, total cholesterol and 2-hour oral glucose-tolerance test insulin levels. Additionally, PRS2 was associated with estimated glomerular filtration rate and ankle brachial index. Both PRS1 and PRS2 had pleiotropic associations with FEV1/ FVC ratio in mediation analyses.
Conclusions
Associations of polygenic scores of height with measures of lung function and cholesterol were consistent with those observed in prior studies of majority European ancestry populations. Mediation analysis may augment standard PRS approaches to disentangle pleiotropic and mediated effects.
Keywords: genome-wide association study, cardiovascular system, pulmonary elimination, genetic pleiotropy, causality, body height
Key Messages
Associations of adult height with cardiometabolic and pulmonary traits have been studied in majority European ancestry populations using Mendelian randomization (MR) and polygenic risk score (PRS) analyses.
It is unclear how well PRS analysis performs in non-European populations.
In the Hispanic Community Health Study/Study of Latinos, we used the standard PRS approach and a novel approach designed to better explain height in the study.
Associations of height with measures of lung function and cholesterol were consistent with those observed in prior studies of majority European ancestry populations.
Novel approaches to calculate stronger PRSs and mediation analysis may supplement standard approaches in non-European populations.
Introduction
There is increasing evidence that adult height predicts cardiovascular disease risk. For example, in a recent meta-analysis of 130 cohort studies, shorter stature was associated with increased risk of cardiovascular death, and this association was robust to adjustment for a wide range of demographic, behavioural and clinical factors such as blood pressure, lipids and diabetes.1 Similar associations of shorter stature with increased cardiovascular morbidity and mortality have also been reported in other studies.2–7
Despite the consistency of associations observed in the abovementioned studies, there remains a concern that associations of height with cardiovascular disease are explained by biases that can occur in conventional epidemiologic study designs (e.g. reverse causation).8,9 To address this possibility, several groups recently used either the MR or polygenic risk score (PRS) approach to reconsider associations of adult height with cardiovascular disease in majority European ancestry populations.10–12 Both approaches construct a PRS—a weighted or unweighted sum of trait-associated genotype alleles. The MR approach considers a causal analysis framework, in which genotypes that constitute the PRS are assumed to be causally associated only with height, but not with other traits of interest (genetic instrumental variable, IV), and used to estimate and/or test the causal effect of height itself. However, if the MR assumption on causal structure is violated,13,14 then a test of the genetic IV effect on cardiovascular disease does not test a height–cardiovascular disease relationship. In contrast, the PRS approach does not require testing causal assumptions and is now used to study a wide variety of biologic traits in the context of diverse study designs.
In this investigation, we used PRS analysis to study associations of genetic determinants of height with cardiometabolic and pulmonary traits in a diverse US Hispanic/Latino population from the Hispanic Community Health Study/Study of Latinos (HCHS/SOL). Cardiovascular disease events are not yet available in large numbers in HCHS/SOL, but two of the prior studies that used a PRS approach to consider the height–cardiovascular disease relationship in majority European ancestry populations also considered the effect of a PRS for height on cardiometabolic and pulmonary traits.10,11 To further characterize the shared genetic basis of height and cardiometabolic and pulmonary traits and to consider whether associations observed in Europeans are also seen in Hispanics/Latinos: (i) we used two different methods in an attempt to create a PRS that strongly represents the genetic components of height in HCHS/SOL and (ii) we conducted formal mediation analyses to calculate indirect (mediated through height) and direct (not mediated through height, reflecting pleiotropy) effects of genetic determinants of height on cardiometabolic and pulmonary traits.
Materials and methods
Study population
The HCHS/SOL is a study of 16 415 Hispanic/Latino adults recruited from four US communities (Bronx, Miami, Chicago and San Diego) during 2008–11.15,16 The HCHS/SOL study population was recruited using population-based survey methodology and is representative of the Hispanic/Latino populations residing in the four US sampling areas. Participants completed physical examinations and in-person interviews, and provided blood samples. The HCHS/SOL protocol was approved by each local institutional review board, and all participants provided informed consent. Genetic studies in HCHS/SOL are restricted to the 12 803 participants who provided informed consent for genetic testing and passed follow-up quality-control testing of genetic material.
HCHS/SOL examination methods
Participants were asked to fast and to refrain from smoking for 12 hours prior to the HCHS/SOL examination and to avoid vigorous physical activity on the morning of the visit. Height was measured to the nearest centimetre and body weight to the nearest 0.1 kg. Body mass index (BMI) was computed as weight divided by height squared (kg/m2). Following a 5‐minute rest period, three seated blood pressure (BP) measurements were obtained using an automatic sphygmomanometer; the second and third readings were averaged. Standard digitized spirometric measurements of timed pulmonary function [forced expiratory volume in 1 second (FEV1); forced vital capacity (FVC)] were performed based on Epidemiology Standardization Project17 and American Thoracic Society18 recommendations by using a SensorMedics model 1022 dry-rolling seal volume spirometer (CareFusion, Yorba Linda, CA). Blood samples were collected and shipped to a central laboratory for analysis according to standardized protocols. Quality-control procedures for the study, including blind replicate measurements on 5% of samples, have been described previously16 and all laboratory protocols are published online.19 Low-density lipoprotein (LDL) cholesterol was calculated using the Friedewald equation20 and the estimated glomerular filtration rate (eGFR) was calculated using the combined creatinine–cystatin C equation.21 After the initial venipuncture, all participants except those with self-reported diabetes or measured (glucose meter) fasting plasma glucose above 150 mg/dL (8.4 mmol/L) underwent a standard 75-g 2-hour oral glucose-tolerance test (OGTT), from which a 2-hour post-oral glucose-tolerance test of plasma glucose was obtained. Protocols to determine the ankle brachial index (ABI) in HCHS/SOL have also been published previously.22 ABI ≤0.9 is diagnostic of peripheral artery disease (PAD) and is a predictor of cardiovascular events and death, but risk of mortality is also elevated with ‘borderline’ ABI ≤1.0 and with ABI ≥1.4, which may reflect stiff (calcified) ankle arteries that may mask underlying PAD.23 Participants younger than 45 years were not examined by the ABI and those with ABI ≥1.4 were excluded from ABI analyses.
Genotyping, quality control and imputation
DNA extracted from blood was genotyped on the HCHS Custom 15041502 array (Illumina Omni2.5M + custom content). Quality-control procedures in HCHS/SOL, including methods used to construct principal components (PCs) of genetic ancestry and a kinship matrix reflecting genetic relatedness between study participants (some of whom were sampled from the same household), have been described previously.24 Genotyping and downstream quality-control procedures yielded 2 232 944 genetic variants for genotyped HCHS/SOL participants.
Genotype imputation was performed with the 1000 Genomes Project phase 3 reference panel25 as previously described.24 Variants with at least two copies of the minor allele and present in any of the four 1000 Genomes continental panels were imputed (about 50 million imputed variants prior to quality filtering). For each variant, we calculated the quality scores ‘info’ (outputted by IMPUTE226) and ‘oevar’, defined as the ratio between the observed variance of the allelic dosage to the expected variance based on the binomial distribution. Oevar was set to 1 for genotyped variants. We also defined the effN measure by effN = 2N × MAF × (1 – MAF) × oevar, which approximates the effect count of the minor alleles for rare variants, adjusted for imputation quality.
Statistical methods
PRSs based on previously reported height loci
We generated two PRSs for adult height in HCHS/SOL. For both PRSs, we used findings of the Genetic Investigation of ANthropometric Traits (GIANT) consortium. Specifically, we included 697 single-nucleotide polymorphisms (SNPs) that had P < 5 × 10−8 in a GWAS meta-analysis of adult height from 79 studies consisting of 253 288 individuals of European ancestry.27 Also included in the PRSs were an additional 71 and 36 SNPs that had P < 5 × 10–8 in GWAS meta-analyses of adult height in 93 926 individuals from East Asia28 and 20 427 individuals of African ancestry.29 Of these 804 identified SNPs, 801 were included in the first PRS (PRS1); 2 SNPs were unavailable in the HCHS/SOL-imputed genetic dataset and 1 SNP was of poor quality (info <0.7).
To construct PRS1, we used these 801 previously reported SNPs. The value of PRS1 for an individual HCHS/SOL participant was obtained by summing the counts of alleles associated with increased height of all 801 SNPs (SNPs within the same genomic regions were allowed). The second PRS (PRS2) was created to potentially serve as a stronger surrogate genetic basis for height in the HCHS/SOL study population (see Supplementary Methods, available as Supplementary data at IJE online). We first defined a genomic region of 5 × 105 base pairs centred at each of the 804 previously reported height-associated SNPs. These regions sometimes overlapped, e.g. when multiple reported height variants were tightly clustered. Second, we ranked all common (effN ≥ 250) variants (138–1368 independent SNPs and indels per region) in each region according to their p-values in a stratified GWAS of height (performed stratified by the Hispanic/Latino background group, followed by meta-analysis) and took the variant with the highest rank from each region (the ‘lead SNP’ for this region). Since genomic regions sometimes overlapped, this led to the selection of 604 unique lead SNPs, as some lead SNPs were the same for multiple regions. Finally, for each participant, the value of PRS2 was the sum of the height-increasing alleles of these 604 lead SNPs. Supplementary Table 4, available as Supplementary data at IJE online, shows the variants used to create PRS1 (801 SNPs) and PRS2 (604 SNPs and indels).
We report the variance in height explained by the PRSs. The variance in height explained by a PRS was calculated as %, where denotes the total variance in the model adjusted for all covariates but without the PRS and denotes the total variance in the model after further adjustment for the PRS.
Association testing of height and PRSs with cardiometabolic and pulmonary traits
We calculated associations of measured adult height, PRS1 and PRS2 with cardiometabolic and pulmonary traits using mixed-effects regression models. All associations were adjusted for age, sex and five PCs of ancestry to prevent biases due to population stratification. To account for the HCHS/SOL sampling design that included genetically related individuals and individuals living in the same household and community block unit, we used linear or logistic mixed models (for quantitative and binary traits, respectively), with correlations modelled via kinship, household sharing and community block unit sharing matrices. Other participant characteristics (e.g. income, educational attainment, markers of acculturation, diet, physical activity, medications) that might confound or modify associations observed in conventional epidemiologic study designs were not included in the regression models. For each of the tested variables (height, PRS1 and PRS2), we report the p-value (P), with P < 0.05/21 being the Bonferroni-corrected p-value based on the number of traits. In secondary analyses, we additionally stratified by sex.
Cardiometabolic and pulmonary traits in the following groups were considered in this investigation: anthropometry (BMI); blood pressure (systolic BP, diastolic BP and pulse pressure); lung function (FEV1, FVC and FEV1/FVC ratio); lipid measures [total cholesterol, high-density lipoprotein (HDL) cholesterol, LDL cholesterol and triglycerides]; glycemic traits (fasting glucose, 2h-OGTT glucose, fasting insulin, 2h-OGTT insulin, HOMA-IR, haemoglobin A1c); inflammation, kidney and PAD [hsCRP, eGFR and ABI (ABI > 1.0 vs ABI ≤ 1.0 and ABI > 1.0 vs ABI ≤ 0.9)].
Sensitivity analysis for PRSs
We performed two sensitivity analyses to address potential limitations of the PRSs—inclusion of highly correlated genetic variants in PRS1 due to the merging multiple external sources and potential over-fitting of PRS2 due to the high number of SNPs tested in each region. First, for both PRS1 and PRS2, we generated linkage disequilibrium (LD)-pruned counterparts, in which we computed the LD (r2) between all genetic variants composing them, and removed SNPs in a random order to generate a list of SNPs with pairwise r2 < 0.5. Second, for PRS2, we took the number of tests in each of the association regions into account, as follows. For each association region of size 1 Mbp, we took all genotyped SNPs (to reduce the number of tests, compared with genotyped + imputed) and calculated the effective number of SNPs (effSNP) in the region using the simpleM method.30 Then, we selected the (genotyped) SNP with the smallest p-value in the HCHS/SOL height GWAS, and used it in the PRS if its p-value waseffSNP. We calculated the explained variance of height by these PRSs and tested their associations with cardiometabolic and pulmonary traits.
Mediation analysis
Because HCHS/SOL recruited multiple members of households and mediation formulae were developed under the assumption of independent and identically distributed (IID) observations, we first generated a restricted data set of genetically unrelated (kinship coefficient <2–11 in the kinship matrix) individuals not living in the same household (8060 or 63% of the study population). Estimated direct (not mediated through height, reflecting pleiotropy) and indirect (mediated through height) effects of PRS1 and PRS2 on a given trait were then calculated using outputs from two regression models. In the first regression model, a trait was regressed on both measured height and the PRS of interest, adjusting for covariates (sampling weight, age, sex, centre, five PCs of ancestry) to obtain the effect of height () and the effect of the PRS () on the trait. In the second model, we regressed height on the PRS (adjusted for covariates) to obtain the effect of the PRS on height (). For continuous traits, the direct effect of the PRS on the cardiometabolic or pulmonary trait was and the indirect effect was For binary traits, the same equation was used after scaling the coefficients,31 although this approach has not been widely validated. We used a bootstrap with 5000 iterations to estimate confidence intervals for mediation analysis of continuous and binary traits.
Results
Study population and genetic risk scores
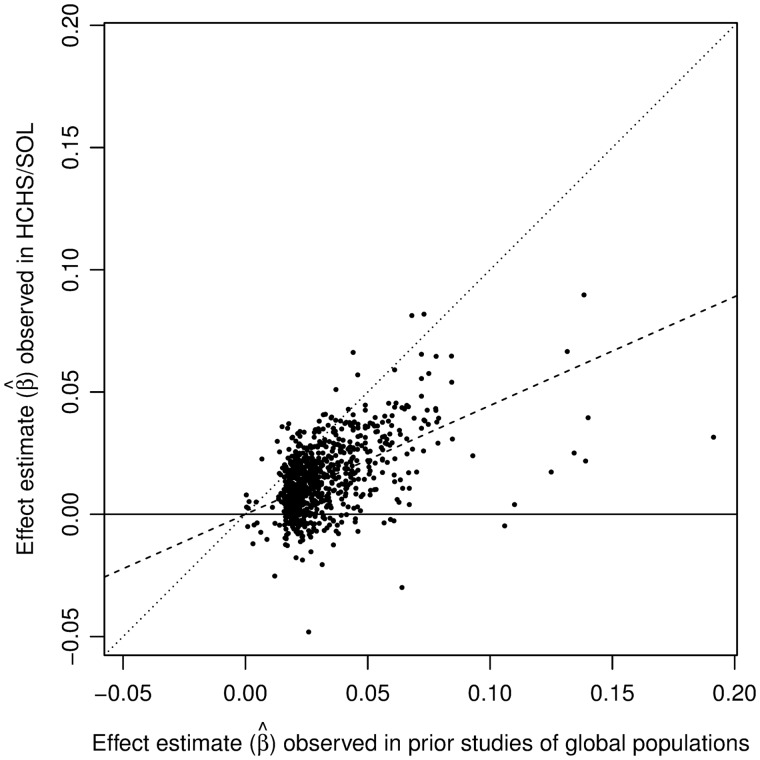
The study population included Hispanics/Latinos (7539 women and 5245 men) for whom there were data on measured height and genetic markers. Depending on trait-specific exclusion criteria, some analyses had lower sample sizes. The distribution of cardiometabolic and pulmonary traits in these participants is shown in Table 1. Associations of 801 SNPs that were reported as having P < 5 × 10−8 in prior GWAS studies of adult height in global populations of European, East Asian and African ancestry (see ‘Methods’ section) with height in HCHS/SOL are shown in Figure 1. The effect estimates (β) observed in prior GWAS studies for these 801 SNPs were correlated with those observed in HCHS/SOL with Pearson r = 0.51. PRS1, which was calculated with these 801 SNPs, was a strong predictor of height in HCHS/SOL in analyses adjusted for age, sex, five PCs of ancestry, kinship and HCHS/SOL recruitment site and sampling design [β = 1.86 per 1 standard deviation (SD) of PRS1 on height (cm); 95% confidence interval (CI) = 1.75–1.97; P = 1.4 × 10−264], explaining 9% of the variance in measured height. As expected by design, the association of PRS2 with measured height was stronger than that of PRS1 (β = 3.29 per 1 SD of PRS2 on height (cm); 95% CI = 3.20–3.38; P < 2.2 × 10−308) explaining 29% of the variance in measured height.
Table 1.
Distribution of cardiometabolic traits in HCHS/SOL participants
| Women (n = 7539) | Men (n = 5245) | |
|---|---|---|
| Median (IQR) | Median (IQR) | |
| Age, years | 48 (38–57) | 47 (35–56) |
| Anthropometry | ||
| Height, cm | 157 (152–161) | 170 (165–175) |
| Weight, kg | 72 (63–83) | 82 (73–94) |
| BMI, kg/m2 | 29 (26–34) | 29 (26–32) |
| Blood pressure | ||
| Systolic blood pressure, mm Hg | 117 (106–131) | 123 (114–133) |
| Diastolic blood pressure, mm Hg | 71 (65–79) | 74 (67–81) |
| Pulse pressure, mm Hg | 45 (38–54) | 49 (43–55) |
| Lung function | ||
| FEV1, mL | 2466 (2094–2833) | 3442 (2933–3925) |
| FVC, mL | 3037 (2623–3450) | 4310 (3762–4864) |
| FEV1/FVC | 82 (78–85) | 80 (76–84) |
| Lipid measures | ||
| Total cholesterol, mg/dL | 197 (71–226) | 195 (167–224) |
| HDL cholesterol, mg/dL | 50 (43–59) | 43 (37–51) |
| LDL cholesterol, mg/dL | 119.4 (97–145) | 121 (97–145) |
| log(Triglycerides), mg/dL | 4.7 (4.3–5) | 4.8 (4.5–5.2) |
| Glycemic traits | ||
| Glucose, fasting, mg/dL | 94 (88–102) | 97 (92–106) |
| Glucose, 2h-OGTT, mg/dL | 119 (99–146) | 110 (89–136) |
| log(Insulin, fasting), mU/L | 2.4 (1.9–2.8) | 2.3 (1.9–2.8) |
| log(Insulin, 2h-OGTT), mU/L | 4.3 (3.8–4.8) | 4 (3.3–4.6) |
| log(HOMA-IR) | 0.9 (0.5–1.4) | 1 (0.4–1.5) |
| log(Hemoglobin A1c), mmol/mol | 3.6 (3.5–3.7) | 3.6 (3.5–3.7) |
| Inflammation, kidney and peripheral artery disease | ||
| hsCRP, mg/L | 1 (0.2–1.7) | 0.5 (–0.2–1.2) |
| eGFR | 104 (90–117) | 105 (91–116) |
| N (%) | N (%) | |
| ABI ≤0.9 | 276 (6) | 120 (4) |
| 0.9 < ABI ≤ 1.0 | 1064 (23) | 319 (11) |
| 1.0 < ABI ≤ 1.4 | 3286 (70) | 2443 (81) |
IQR, interquartile range; BMI, body mass index; FEV1, forced expiratory volume in 1 second; FVC, forced vital capacity; HDL, high-density lipoprotein; LDL, low-density lipoprotein; 2h-OGTT, 2-hour post-oral glucose-tolerance test; HOMA-IR, homeostatic model assessment of insulin resistance; hsCRP, high-sensitivity C-reactive protein; eGFR, estimated glomerular filtration rate.
Figure 1.
Associations of 801 single-nucleotide polymorphisms (SNPs) that had P < 5 × 10−8 in prior studies of adult height in global populations of European, East Asian and African ancestry (see the ‘Methods’ section) with measured height in the Hispanic Community Health Study/Study of Latinos (HCHS/SOL). The dashed line is the estimated regression line (no intercept) of effect estimates observed in prior studies of global populations and effect estimates observed in HCHS/SOL. The dotted line is the line of identity. SNP effects were estimated according to the height-increasing alleles in prior studies, on the Z-score of height (normalized rank).
Relationship of measured height with cardiometabolic and pulmonary traits
Measured height had associations below the Bonferroni-corrected level of P < 0.05/21 with most investigated traits: systolic BP, pulse pressure, FEV1, FVC, FEV1/FVC ratio, total cholesterol, HDL cholesterol, LDL cholesterol, 2h-OGTT glucose, fasting insulin, 2h-OGTT insulin, HOMA-IR, eGFR and both ABI comparisons (Figure 2 and Supplementary Table 3, available as Supplementary data at IJE online). Most of these associations also had P < 0.05/21 in both men and women in sex-stratified analyses, aside from those for total cholesterol, HDL cholesterol, LDL cholesterol and ABI > 1.0 vs ABI ≤ 0.9 (Supplementary Figures 1 and 2, available as Supplementary data at IJE online).
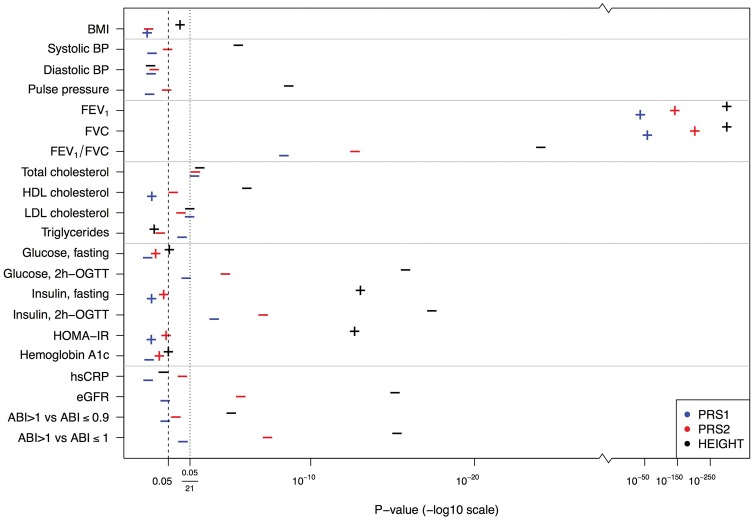
Figure 2.
P-values of estimated effect sizes of PRS1 (estimated from prior GWAS of European, African and Asian populations), PRS2 (created to explain adult height in HCHS/SOL) and measured height with cardiometabolic and pulmonary traits in HCHS/SOL men and women combined. Effects are for a one (1) standard deviation (SD) increase in PRS1, PRS2 or measured height. Plus (+) symbols indicate a positive association and minus (–) symbols indicate an inverse association. Two lines are shown—corresponding to P = 0.05 and for the Bonferroni-corrected P = 0.05/21, where 21 is the number of cardiometabolic and pulmonary traits.
Relationship of genetic risk scores with cardiometabolic and pulmonary traits
The strongest associations of both PRS1 (estimated from prior GWAS of European, African and Asian populations) and PRS2 (created to predict adult height in HCHS/SOL) were with measures of lung function. Specifically, PRS1 and PRS2 had trait-increasing associations (P < 0.001) with FEV1 and FVC in both sexes combined (Figure 2) and in women and men separately (Supplementary Figures 1 and 2, available as Supplementary data at IJE online). Furthermore, the direction of the PRS1 and PRS2 associations with FEV1 and FVC were the same as those for measured height (all positive associations). In contrast, PRS1 and PRS2 were inversely associated with the FEV1/FVC ratio [a marker in some patient populations of persistent airflow limitation and chronic obstructive pulmonary disease (COPD)32]—these associations had P < 0.05/21 in both sexes but were stronger in women as compared with men. Sample sizes, effect estimates, 95% CIs and p-values for combined and sex-stratified analyses of PRS1, PRS2 and measured height with cardiometabolic and pulmonary traits are shown in Supplementary Tables 1–3, available as Supplementary data at IJE online.
We also observed inverse associations of PRS1 and PRS2 with 2h-OGTT insulin and total cholesterol measures in both sexes combined (P < 0.05/21, Figure 2 and Supplementary Table 3, available as Supplementary data at IJE online). However, the 2h-OGTT insulin associations with PRS1 had P < 0.05/21 only in men (P < 0.001 in men vs P = 0.002 in women). Further, only PRS2 in women had total cholesterol association with P < 0.05/21 (P = 0.18 in men vs P = 0.001 in women; Supplementary Figures 1 and 2 and Supplementary Tables 1 and 2, available as Supplementary data at IJE online).
In addition, PRS2 (which explained a greater percentage of the variance in height than PRS1) was associated at the Bonferroni-corrected level with 2h-OGTT glucose, eGFR and ABI > 1.0 vs ABI ≤ 1.0 in the combined population of women and men. PRS2 was also associated with ABI > 1.0 vs ABI ≤ 1.0 in both women and men in sex-stratified analyses (Supplementary Figures 1 and 2 and Supplementary Tables 1 and 2, available as Supplementary data at IJE online) with P < 0.05/21. Associations of PRS1 and PRS2 with other cardiometabolic traits beyond those mentioned above were consistent with null associations.
Sensitivity analysis
Three alternate forms of PRS1 and PRS2 were considered: (i) an LD-pruned PRS1 with pairwise r2 < 0.5 (726 SNPs); (ii) an LD-pruned PRS2 with pairwise r2 < 0.5 (597 SNPs); and (iii) an LD-pruned PRS2 in which we also took the number of tests in each of the association regions into account (93 SNPs). Supplementary Table 6, available as Supplementary data at IJE online, provides for each of these alternate PRSs the explained variance in height. Supplementary Table 7 and Supplementary Figures 3–5, available as Supplementary data at IJE online, show associations of these alternate PRSs with cardiometabolic and pulmonary traits.
LD-pruned PRS1 had reduced variance in height explained as compared with PRS1 (8.83 vs 9%), but associations of LD-pruned PRS1 and PRS1 were similar with some qualitatively different findings (e.g. total cholesterol, ABI > 1.0—Supplementary Figure 3, available as Supplementary data at IJE online). LD-pruned PRS2 explained a similar amount of the variance in height as PRS2 (28.9 vs 29%) and had similar associations with cardiometabolic and pulmonary traits (Supplementary Figure 4, available as Supplementary data at IJE online). LD-pruned PRS2 additionally corrected for multiple testing explained 10.4% of the variance of height (Supplementary Table 6, available as Supplementary data at IJE online) and its associations with cardiometabolic and pulmonary traits were usually weaker than those of the primary PRS2. Nevertheless, LD-pruned PRS2 additionally corrected for multiple testing in association regions still reached P < 0.05/21 for pulmonary traits and 2h-OGTT insulin (Supplementary Figure 4, available as Supplementary data at IJE online).
Mediation analysis
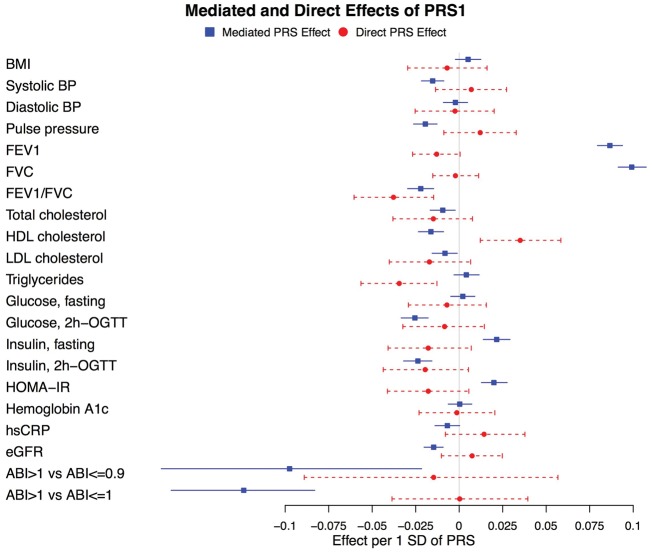
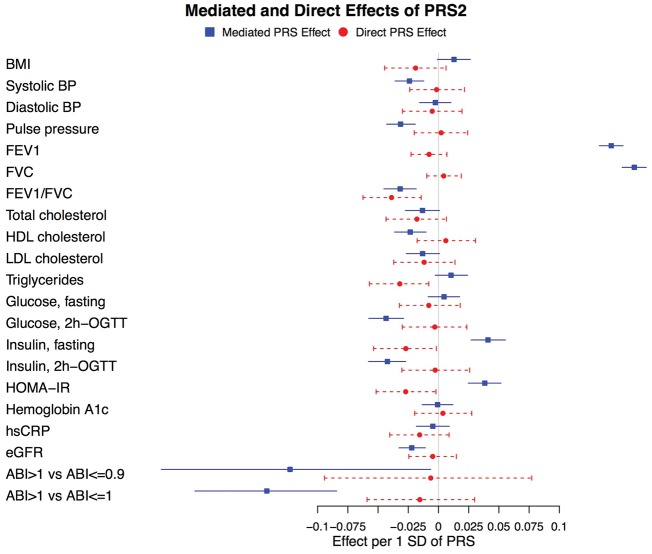
Indirect (mediated through height) and direct (not mediated through height, reflecting pleiotropy) effects of PRS1 and PRS2 along with their 95% CIs calculated by bootstrapping are shown in Figures 3 and 4. Whereas, for most traits, the direct effects of PRS1 and PRS2 were null, there were a few notable exceptions. Specifically, the direct effects of both PRS1 and PRS2 on the FEV1/FVC ratio and on triglycerides had P < 0.05/21. The direct effect of PRS1 on HDL cholesterol, as well as the direct effects of PRS2 on fasting insulin and HOMA-IR, had P < 0.5/21. As observed in Figures 3 and 4, associations of PRS1 with HDL cholesterol and PRS2 with fasting insulin and HOMA-IR had both indirect and direct effects in opposite directions with P < 0.05/21.
Figure 3.
Mediation analysis for associations of PRS1 with cardiometabolic and pulmonary traits in HCHS/SOL men and women combined. For quantitative traits, effects are for a one (1) standard deviation (SD) increase in PRS1 and a one (1) SD increase in the cardiometabolic trait. For the binary analyses of ankle brachial index (ABI), the outcome is a one (1) SD increase in the log odds ratio (OR). 95% confidence intervals (CIs) were calculated using a bootstrap (see the ‘Methods’ section).
Figure 4.
Mediation analysis for associations of PRS2 with cardiometabolic and pulmonary traits in HCHS/SOL men and women combined. For quantitative traits, effects are for a one (1) standard deviation (SD) increase in PRS2 and a one (1) SD increase in the cardiometabolic trait. For the binary analyses of ankle brachial index (ABI), the outcome is a one (1) SD increase in the log odds ratio (OR). 95% confidence intervals (CIs) were calculated using a bootstrap (see the ‘Methods’ section).
Discussion
In this study, we investigated the complex relationship of height and its genetic determinants with cardiometabolic and pulmonary traits in an admixed population of Hispanics/Latinos. We first calculated two polygenic risk scores (PRS1 and PRS2), both based on previously reported height loci, but PRS2 was specifically designed to more accurately reflect height in HCHS/SOL. We then used the PRSs, as has recently been used by other groups,10,11 to study the association of height with cardiometabolic and pulmonary traits. Finally, we also used mediation analysis to study pathways of associations between height, its PRSs, and cardiometabolic and pulmonary traits.
The strongest associations of the PRSs observed in our study were with FEV1, FVC (trait-increasing associations) and the FEV1/FVC ratio (trait-decreasing association). These observations are consistent with those of the prior height study of exclusively European ancestry participants that included measures of lung function.11 No direct effects (not mediated through height) of PRS1 or PRS2 with FEV1 and FVC were observed, suggesting that stature or related biological effects are directly related to these measures of lung function.
In contrast, a direct effect of both PRS1 and PRS2 with the FEV1/FVC ratio [a marker in some patient populations of persistent airflow limitation and chronic obstructive pulmonary disease (COPD)32] was also observed, suggesting pleiotropy (Figures 3 and 4). In fact, the mediation analyses suggested both direct and mediated (via height) inverse associations of the PRSs with FEV1/FVC ratio (Figures 3 and 4). Although it appears surprising that a direct PRS effect was observed for the composite FEV1/FVC ratio measure but not for FEV1 and FVC individually, this observation is consistent with the fact that a GWAS of lung function measures identified mostly different loci associated with FEV1/FVC ratio vs FEV1 (albeit with some overlap).33
Both PRS1 and PRS2 were associated with total cholesterol with P < 0.05/21 (Figure 2 and Supplementary Table 3, available as Supplementary data at IJE online). Associations of PRS1 and PRS2 with LDL cholesterol were in the same direction and had similar magnitudes (Supplementary Table 3, available as Supplementary data at IJE online) and no direct effects with total or LDL cholesterol were observed. These data, which are consistent with those of recent studies of majority European ancestry participants in which a height PRS had significant inverse associations with LDL cholesterol,10,11 support a potentially causal association of height [or an unmeasured mediator (X) caused by height] with total and/or LDL cholesterol. However, associations of PRSs with triglyceride levels observed in prior studies10,11 were not observed in HCHS/SOL and direct effects (not mediated through height) of PRS1 and PRS2 with triglyceride levels observed in mediation analyses suggest that caution is warranted in the interpretation of the height–triglyceride relationship. The interpretation of opposite direct and indirect associations for HDL cholesterol (PRS1) and fasting insulin and HOMA-IR (PRS2) is unclear, although it is conceivable that pleiotropic and height-mediated effects on these traits are qualitatively different.
Both PRS1 and PRS2 or PRS2 alone were also associated with 2h-OGTT insulin, 2h-OGTT glucose, eGFR and ABI > 1.0 vs ABI ≤ 1.0 with P < 0.05/21. No direct effects were observed in mediation analyses for these associations. To our knowledge, these associations have not been reported previously and require replication in other cohorts.
PRS1, based solely on SNPs discovered in other (majority European ancestry) study populations, explained 9% of the variance in height in HCHS/SOL, whereas PRS2 explained 29% of the variance. This improvement in the variance explained is expected given our methodology of selecting ‘lead SNPs’ in previously identified loci and it is likely that this methodology leads to over-fitting of genetic associations with height. In other words, PRS2 may have stronger associations with height in HCHS/SOL than it would in the general population of US Hispanics/Latinos, and may contain measurement error. However, we speculate that over-fitting, if it occurred, would not bias associations with cardiometabolic or pulmonary traits because measurement error in height would not be expected to affect cardiometabolic traits, although it could reduce power.
In sensitivity analysis, we considered this issue using an LD-pruned PRS2 in which we took the number of tests in each of the association regions into account. Associations of LD-pruned PRS2 adjusted for multiple testing (93 SNPs) were similar albeit sometimes attenuated as compared with those of primary PRS2 (604 SNPs—Supplementary Figure 4, available as Supplementary data at IJE online). This could be due to a loss of power or to less bias or both. A limitation of mediation analysis is that it assumes that there is no unmeasured confounding between measured height and the outcome (cardiometabolic and pulmonary traits). For most HCHS/SOL participants, this may be a reasonable assumption because adult height preceded the HCHS/SOL study examination. However, as noted above, unmeasured confounding (U) by factors such as childhood economic hardship that preceded attainment of adult height could bias the mediation analyses if they were also independently associated with cardiometabolic and pulmonary traits measured at the time of the HCHS/SOL clinic visit.
The concordance of our findings related to lung function and cholesterol with those of recent studies of majority European ancestry participants10,11 suggests that the PRS approach may be an important tool to investigate the underlying biology of cardiometabolic and pulmonary traits in global populations. When statistical power permits, our results suggest that it may also be advantageous to stratify by sex because PRS associations with cholesterol and 2h-OGTT insulin were not the same in women and men, despite the fact that our PRSs were not sex-specific. Finally, we suggest that mediation analysis may represent a helpful supplement to the PRS approach. Whereas, for most traits, the direct effects of the PRSs were null, for FEV1/FVC, triglycerides, fasting insulin and HOMA-IR, it is possible that genetic variants that predict height are associated with these traits through pathways unrelated to those that result in increased height.
Funding
The Hispanic Community Health Study/Study of Latinos was carried out as a collaborative study supported by contracts from the National Heart, Lung, and Blood Institute (NHLBI) to the University of North Carolina (N01-HC65233), University of Miami (N01-HC65234), Albert Einstein College of Medicine (N01-HC65235), Northwestern University (N01-HC65236) and San Diego State University (N01-HC65237). The following Institutes/Centers/Offices contribute to the HCHS/SOL through a transfer of funds to the NHLBI: National Center on Minority Health and Health Disparities, the National Institute of Deafness and Other Communications Disorders, the National Institute of Dental and Craniofacial Research, the National Institute of Diabetes and Digestive and Kidney Diseases, the National Institute of Neurological Disorders and Stroke, and the Office of Dietary Supplements. Support was also provided to the Genetic Analysis Center at the University of Washington via NHLBI and NIDCR contracts HHSN268201300005C AM03 and MOD03.
Supplementary Material
Acknowledgements
The authors thank the staff and participants of HCHS/SOL for their important contributions. Investigators website: http://www.cscc.unc.edu/hchs/.
Conflict of interest: None declared.
References
- 1. Emerging Risk Factors Collaboration. Adult height and the risk of cause-specific death and vascular morbidity in 1 million people: individual participant meta-analysis. Int J Epidemiol 2012;41:1419–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bjornsson E, Thorgeirsson G, Gudnason T.. Adult height associates with angiographic extent of coronary artery disease. Atherosclerosis 2016;254:237–41. [DOI] [PubMed] [Google Scholar]
- 3. Forsen T, Eriksson J, Qiao Q, Tervahauta M, Nissinen A, Tuomilehto J.. Short stature and coronary heart disease: a 35-year follow-up of the Finnish cohorts of The Seven Countries Study. J Intern Med 2000;248:326–32. [DOI] [PubMed] [Google Scholar]
- 4. Kelly RF, Mohanty J, Hashim AS, Parrillo JE.. Association between height and coronary artery disease in black men and women. Am J Cardiol 2000;85:1253–55. [DOI] [PubMed] [Google Scholar]
- 5. Paajanen TA, Oksala NK, Kuukasjarvi P, Karhunen PJ.. Short stature is associated with coronary heart disease: a systematic review of the literature and a meta-analysis. Eur Heart J 2010;31:1802–09. [DOI] [PubMed] [Google Scholar]
- 6. Silventoinen K, Zdravkovic S, Skytthe A. et al. Association between height and coronary heart disease mortality: a prospective study of 35, 000 twin pairs. Am J Epidemiol 2006;163:615–21. [DOI] [PubMed] [Google Scholar]
- 7. Lee CM, Barzi F, Woodward M. et al. Adult height and the risks of cardiovascular disease and major causes of death in the Asia-Pacific region: 21, 000 deaths in 510, 000 men and women. Int J Epidemiol 2009;38:1060–71. [DOI] [PubMed] [Google Scholar]
- 8. Batty GD, Shipley MJ, Gunnell D. et al. Height loss and future coronary heart disease in London: the Whitehall II study. J Epidemiol Community Health 2011;65:461–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Davey Smith G, Hart C, Upton M. et al. Height and risk of death among men and women: aetiological implications of associations with cardiorespiratory disease and cancer mortality. J Epidemiol Community Health 2000;54:97–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nelson CP, Hamby SE, Saleheen D. et al. Genetically determined height and coronary artery disease. N Engl J Med 2015;372:1608–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nuesch E, Dale C, Palmer TM. et al. Adult height, coronary heart disease and stroke: a multi-locus Mendelian randomization meta-analysis. Int J Epidemiol 2016;45:1927–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rosenberg MA, Kaplan RC, Siscovick DS. et al. Genetic variants related to height and risk of atrial fibrillation: the cardiovascular health study. Am J Epidemiol 2014;180:215–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Swerdlow DI, Kuchenbaecker KB, Shah S. et al. Selecting instruments for Mendelian randomization in the wake of genome-wide association studies. Int J Epidemiol 2016;45:1600–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Glymour MM, Tchetgen Tchetgen EJ, Robins JM.. Credible Mendelian randomization studies: approaches for evaluating the instrumental variable assumptions. Am J Epidemiol 2012;175:332–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lavange LM, Kalsbeek WD, Sorlie PD. et al. Sample design and cohort selection in the Hispanic Community Health Study/Study of Latinos. Ann Epidemiol 2010;20:642–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sorlie PD, Aviles-Santa LM, Wassertheil-Smoller S. et al. Design and implementation of the Hispanic Community Health Study/Study of Latinos. Ann Epidemiol 2010;20:629–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ferris BG Jr, Speizer FE, Bishop Y, Prang G, Weener J.. Spirometry for an epidemiologic study: deriving optimum summary statistics for each subject. Bull Eur Physiopathol Respir 1978;14:145–66. [PubMed] [Google Scholar]
- 18. American Thoracic Society (ATS) statement—Snowbird workshop on standardization of spirometry. Am Rev Respir Dis 1979;119:831–38. [DOI] [PubMed] [Google Scholar]
- 19. Manuals and Forms of the Hispanic Community Health Study/Study of Latinos. 2016. http://www2.cscc.unc.edu/hchs/manuals-forms (28 November 2016, date last accessed).
- 20. Friedewald WT, Levy RI, Fredrickson DS.. Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin Chem 1972;18:499–502. [PubMed] [Google Scholar]
- 21. Inker LA, Schmid CH, Tighiouart H. et al. Estimating glomerular filtration rate from serum creatinine and cystatin C. N Engl J Med 2012;367:20–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Allison MA, Gonzalez F 2nd, Raij L. et al. Cuban Americans have the highest rates of peripheral arterial disease in diverse Hispanic/Latino communities. J Vasc Surg 2015;62:665–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Criqui MH, Aboyans V.. Epidemiology of peripheral artery disease. Circ Res 2015;116:1509–26. [DOI] [PubMed] [Google Scholar]
- 24. Conomos MP, Laurie CA, Stilp AM. et al. Genetic diversity and association studies in US Hispanic/Latino populations: applications in the Hispanic Community Health Study/Study of Latinos. Am J Hum Genet 2016;98:165–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Abecasis GR, Auton A, Brooks LD. et al. An integrated map of genetic variation from 1,092 human genomes. Nature 2012;491:56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Howie BN, Donnelly P, Marchini J.. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet 2009;5:e1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wood AR, Esko T, Yang J. et al. Defining the role of common variation in the genomic and biological architecture of adult human height. Nat Genet 2014;46:1173–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. He M, Xu M, Zhang B. et al. Meta-analysis of genome-wide association studies of adult height in East Asians identifies 17 novel loci. Hum Mol Genet 2015;24:1791–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. N’Diaye A, Chen GK, Palmer CD. et al. Identification, replication, and fine-mapping of Loci associated with adult height in individuals of African ancestry. PLoS Genet 2011;7:e1002298.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Gao X, Becker LC, Becker DM, Starmer JD, Province MA.. Avoiding the high Bonferroni penalty in genome-wide association studies. Genet Epidemiol 2010;34:100–05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kenny DA. Mediation with Dichotomous Outcomes. 2013. http://davidakenny.net/doc/dichmed.pdf (22 December 2016, date last accessed).
- 32. Vestbo J, Hurd SS, Agusti AG. et al. Global strategy for the diagnosis, management, and prevention of chronic obstructive pulmonary disease: GOLD executive summary. Am J Respir Crit Care Med 2013;187:347–65. [DOI] [PubMed] [Google Scholar]
- 33. Soler Artigas M, Loth DW, Wain LV. et al. Genome-wide association and large-scale follow up identifies 16 new loci influencing lung function. Nat Genet 2011;43:1082–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.