Abstract
Scope:
We have previously shown that loss of miR-140 has a pro-fibrotic effect in the mammary gland.
This study aims to investigate whether miR-140 loss and obesity act synergistically to promote non-alcoholic fatty liver disease (NAFLD), and to identify the underlying mechanisms.
Methods and Results:
Liver tissues were isolated from lean-fat-diet and high-fat-diet fed wild-type and miR-140 knockout mice. Using molecular staining and immunohistochemistry techniques, increased development of NAFLD and fibrotic indicators in miR-140 knockout mice were identified. Utilizing an in vitro model system, miR-140 was demonstrated to target TLR-4, and miR-140 overexpression was shown to be sufficient to inhibit palmitic acid signaling through the TLR-4/NFκB pathway.
Conclusion:
These findings demonstrate that loss of miR-140 results in increased expression of TLR-4, sensitizing cells to palmitic acid signaling and in increased inflammatory activity through the TLR4/NFκB pathway. This signaling axis promotes NAFLD development in a high-fat diet context and indicates the potential utility of miR-140 rescue as a therapeutic strategy in NAFLD.
Keywords: miR-140, non-alcoholic fatty liver disease (NAFLD), obesity, palmitic acid, TLR-4
1. Introduction
One of the most common causes of liver disease is obesity, with over 80% of obese individuals presenting with non-alcoholic fatty liver disease (NAFLD).[1] The prevalence of NAFLD has increased dramatically in the past 50 years, matching the obesity epidemic, and is believed to affect between 75 and 100 million people in the United States.[2]
Hepatic lipid buildup results in steatosis and NAFLD. NAFLD is divided into two categories: nonalcoholic fatty liver, and nonalcoholic steatohepatitis. While nonalcoholic fatty liver (NAFL) is believed to be generally benign, with patients exhibiting mild inflammation and steatosis, nonalcoholic steatohepatitis (NASH) is characterized by significant inflammation and liver damage resulting from the build-up of fat within the liver. However, NASH occurs in only approximately 20% of patients with NAFLD. It is likely that between 3 and 12% of U.S. adults have NASH,[3] and while NAFL is thought to progress to NASH, the potential mechanisms of progression remain under investigation.[4] Similarly, NASH may progress to hepatic fibrosis and cirrhosis, which results in 30 000 deaths every year in the United States. Additionally, cirrhosis is a dominant risk factor in the development of liver cancer, resulting in an additional 10 000 deaths per year.[5]
The predominant factors leading to NAFLD are a diet high in fat, obesity, the metabolic syndrome, and type II.[6,7] Eighty percent of obese individuals have NAFLD,[1] and the livers of patients with type II diabetes have been demonstrated to have up to 80% higher fat contents compared to non-diabetic patients.[6] The predominance of NAFLD in obese individuals has led to the “multiple-hit” hypothesis, describing NAFLD development from the accumulation of multiple factors, including obesity, insulin resistance, genetic and epigenetic factors, hormones, nutritional factors, and the microbiome.[8]
One of the mechanisms by which obesity affects cell signaling is through the actual constituents of the obesity-causing high-fat diet. A high-fat diet results in increased intake of free-fatty acids and obesity has also been shown to cause an increase in free fatty acid flux, meaning high levels of endogenous free fatty acids are released from adipose tissue.[9] In addition to their metabolic and energy storage functions, these free fatty acids directly impact cell signaling and cell phenotypes. One of the most common free fatty acids in the high-fat diet is palmitic acid (PA).[10,11] PA is a highly prevalent lipid in the human body, and makes up 20–30% of human depot fat.[12] It is the second most abundant circulating free fatty acid and is increased in obese adults. Treatment of obesity through metabolic surgery significantly decreases circulating PA, indicating that increases in PA are directly related to patient obesity.[13,14] While PA acts as a mode of energy storage, it is also an active component of signaling pathways, and is a ligand for toll-like receptor 4 (TLR-4).[15] PA has been shown to promote NAFLD through several mechanisms, including upregulating inflammatory signaling, inhibiting insulin signaling, and activating apoptotic pathways in hepatocytes.[16,17]
TLR-4 is a member of the pattern recognition receptor family, which recognize infectious agents and induce inflammatory signaling to activate the innate immune system. The most common ligand for TLR-4 is LPS, a bacterial lipopolysaccharide. Following ligand binding, TLR-4 activation results in phosphorylation of NFκB, as well as activation of the MAPK cascade, resulting in the production of cytokines including TNFα, IL-1β, IL-18, IL-6, IL-10, and TGFβ.[15,18–20] TLR-4 is expressed in a variety of cell types within the liver, including Kupffer cells, hepatocytes, hepatic stellate cells, and sinusoidal endothelial cells.[21]
In hepatocytes, treatment with PA induce hepatocyte steatosis,[22] as well as increase hepatocyte production of the proinflammatory cytokine IL-8.[23] Free saturated fatty acids including PA are incorporated into the membrane and can lead to ER stress. Moreover, PA results in JNK-dependent hepatocyte apoptosis, which is a common characteristic of NAFLD and leads to increased inflammation as macrophages are recruited to clear the apoptotic cells.[16,24] PA-treated hepatocytes have also been demonstrated to produce higher levels of exosomes compared to control hepatocytes, and exosomes secreted by PA-treated hepatocytes contain increased levels of hepatic fibrosis promoting miRNAs including miR-192, which increases expression of SMA, TGFβ, and collagen. Treatment of hepatic stellate cells with PA-hepatocyte exosomes was demonstrated to promote hepatic stellate cell activation to a greater extent than non-treated exosomes.[25] Moreover, hepatocyte TLR4 deficiency effectively prevents obesity-induced inflammation and insulin resistance in vivo, indicating the crucial role that the PA/TLR4 hepatocyte pathway plays in NAFLD.[26]
miR-140 was first defined as a regulator of cartilage differentiation.[27] Through utilization of a diet-induced obesity mouse model, we found that obesity downregulates miR-140 expression in the stromal mammary gland through TGFβ induced SMAD3 signaling. Moreover, we demonstrated that loss of miR-140 results in a feedback loop that drives mammary myofibroblast differentiation, promoting the development of a fibrotic mammary microenvironment.[28] As we demonstrated that high-fat diet resulted in miR-140 downregulation, we wanted to determine whether miR-140 loss is sufficient to serve as one of the necessary “hits” in the development of NAFLD. To this end, we performed a diet-induced obesity mouse model study using both wild-type and miR-140 knockout mice.[27] In this article, we demonstrate that miR-140 knockout sensitizes the mouse liver to NAFLD, and NASH, indicating that miR-140 loss may be a key “hit” in the development of NAFLD. Furthermore, we identify a potential mechanism for how miR-140 loss sensitizes the liver to NAFLD. We demonstrate that miR-140 targets and degrades TLR4, preventing PA-induced NFκB signaling. When miR-140 is lost, increased expression of TLR4 and cellular response to the increased levels of PA seen in obesity result in an increased risk of NAFLD development.
2. Experimental Section
2.1. Cell Culture, Reagents, Transfection
L929 (ATCC) was grown in Eagle’s Minimum Essential Medium supplemented with 10% horse serum. Cells were incubated in 5% CO2 at 37 °C. Cells were infected with miR-140 expression vector or control vectors using lentiviral infections as previously described.[28,29]
2.2. Human Primary Hepatocytes
Human primary hepatocytes were isolated using a modified two-step perfusion protocol[30,31] from human liver specimens obtained from the University of Maryland Medical Center with prior approval by the Institutional Review Board at the University Maryland at Baltimore or obtained from BIOIVT (Baltimore, MD). Hepatocytes with viability over 85% were seeded at 0.75 × 106 cells per well in 12-well collagen-coated plates in DMEM supplemented with 5% FBS, 100 U mL−1 penicillin, 100 μg mL−1 streptomycin, 4 μg mL−1 insulin, and 1 μm dexamethasone.[30,31]
2.3. Mouse Primary Macrophages
Ninety-six hours prior to isolation of primary mouse macrophages, 2.5 mL of 3% thioglycolate was injected into the peritoneal cavity. Mice were sacrificed, and 8–10 mL of 0.9% sodium chloride irrigation (Baxter) was injected into the peritoneal cavity to gather cells. Saline/cell mixture was centrifuged at 1000 RPM for 10 min, and cell pellet was resuspended for 2–3 min in Ammonium-Chloride-Potassium lysis buffer to induce red blood cell lysis. Macrophages were immediately harvested for RNA and qRT-PCR analysis.
2.4. Animal Models
Wild-type C57BL/6 mice were obtained from University of Maryland Baltimore Veterinary Resources. miR-140 knock-out mice on a C57BL/6 background were used as previously described.[27] Four-week-old female C57BL/6 mice were randomized and fed a lean-fat diet (Research Diets: D12450J 10% kcal from fat) or high-fat diet (Research Diets: D12492, 60% kcal from fat) ad libitum for 16 weeks (n = 5). Livers were resected and formalin fixed followed by paraffin embedding for immunohistochemistry. Percent weight gained was calculated by the formula [(final weight − original weight)/original weight] × 100. All studies were done in accordance with federal guidelines and institutional policies of University of Maryland Animal Care and Use Committee, IACUC protocol number 0717015.
2.5. Immunohistochemistry
Paraffin-embedded tissue sections were stained as previously described.[28] Slides were visualized using Nikon Eclipse Ti-U microscope.
2.6. Hematoxylin and Eosin Staining
Paraffin-embedded tissue slides were stained as previously described.[28] Slides were mounted and coverslipped usingD. P. X. and visualized using Nikon Eclipse Ti-U microscope.
2.7. Dual Luciferase Assay
The TLR4 3′-UTR was amplified by PCR using the primers 5′-ACTGATGCTAGCAAGACGTGCTTCAAATATCCA–3′and 5′-AGCAGTGAAGAAGGGTTCCACTCGAG ATCAGT-3′and cloned into the Nhe1 and Xho1 sites of the pGL3 luciferase vector. HEK293T cells were seeded in six-well plates and transfected with the TLR4-pGL3 Luciferase vector using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). Cells were co-transfected with miR-140 overexpression plasmid. After 48 h, luciferase activity was determined using the Dual-Luciferase assay system (Promega, Madison, WI). Activity was normalized to Renilla luciferase activity.
2.8. Masson Trichrome Stain
Modified Masson’s stain was performed according to manufacturers’ instructions (ScyTek Laboratories, West Logan, Utah; category number TRM-1) as previously described.[28] Sections were visualized using a Nikon Eclipse Ti microscope (Nikon Instruments Inc.; Melville, NY).
2.9. Western Blotting
Total cell lysates were separated by SDS-PAGE and blotted onto polyvinylidene difluoride membrane. The membrane was incubated with specific primary antibody overnight at 4 °C, followed by the horseradish peroxidase (HRP)-conjugated secondary antibody, and visualized by the ECL western blotting detection system (Thermo Scientific; Rockford, IL).
2.10. qRT-PCR
qRT-PCR analysis of miRNA expression was performed as described previously with normalization to U6 small nuclear RNA.[29] The following primers were used for examination of PD-L1 expression: forward primer: 5′-GCCTGCAGGGCATTC CAGA-3′, reverse primer: 5′-GTCCTTGGGAACCGTGACAG-3′.
2.11. Statistical Analysis
Statistical analysis was performed by Student’s t test or Ordinary one-way ANOVA with Tukey’s multiple comparison test. p-values of <0.05 were considered significant. Data are presented as mean value. Data were analyzed using GraphPad Prism software (version 6.0). When ANOVA analysis was used, significance between pairs is indicated as such: *, comparison with WTLFD; ˆ, comparison with WTHFD; #, comparison with KOLFD; +, comparison with KOHFD. p-Value is indicated as: *, p ≤ 0.05. **, p ≤ 0.01. *** p ≤ 0.001. **** p ≤ 0.0001.
3. Results
3.1. miR-140 Knockout Decreases Hepatic Steatosis
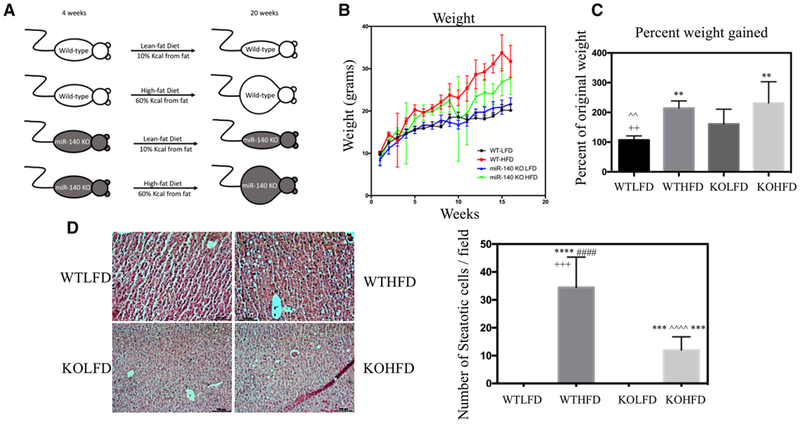
Following our previous study demonstrating the role of high-fat diet–induced miR-140 loss in fibrosis,[28] we next wanted to investigate the impact of HFD in combination with miR-140 loss on fibrosis development. We predicted that miR-140 loss alone may be sufficient to induce fibrosis in a lean-fat diet mouse model and performed an HFD-induced obesity mouse model to determine whether the combination of steatosis resulting from an HFD and miR-140 knockout was sufficient to fulfill the “multiple-hits” model of NAFLD development. Starting at 4 weeks of age, we fed female C57BL/6 (WT) and miR-140 knockout mice on a C57BL/6 background (KO) lean-fat (10% kcal from fat) or high-fat (60% kcal from fat) diet for 16 weeks ad libitum (Figure 1A). Mice were weighed weekly, and both WTHFD and KOHFD mice had a significant weight gain compared to LFD mice (Figure 1B). While the final weights of the KOHFD mice were approximately 5 g lower than WTHFD, we observed a non-significant increase in the percent weight gained of KOHFD mice compared to WTHFD. Interestingly, the final weight of KOLFD was higher than WTLFD, and KOLFD exhibited a significant increase in percent weight gained over WTLFD (Figure 1C). While the increased percent weight gained seen in KO mice is indicative of the small size of immature miR-140-KO mice, the increased final weight of KOLFD mice implies a metabolic effect of miR-140 knockout.
Figure 1.
miR-140 knockout decreases hepatic steatosis. A) Female wild-type C57BL/6 and miR-140 knockout on a C57BL/6 background mice were fed a lean-fat diet (10% kCal from fat, WTLFD/KOLFD) or high-fat diet (60% kCal from fat, WTHFD/KOHFD) ad libitum for 16 weeks. B) Average weight at each week. C) Percent weight gained after 16 weeks lean-fat or high-fat diet (p < 0.005). D) H&E staining of liver tissue sections and quantification of the number of steatotic cells (p < 0.0001). *, comparison with WTLFD. ˆ, comparison with WTHFD. #, comparison with KOLFD.+, comparison with KOHFD. p-Value is indicated as: *p ≤ 0.05. **p ≤ 0.01. ***p ≤ 0.001. ****p ≤ 0.0001.
To examine the development of hepatic steatosis, we performed Hematoxylin & Eosin staining. Both WTHFD and KOHFD liver slides exhibited increased steatotic cells compared to their LFD counterparts, and the WTHFD liver was significantly more steatotic than KOHFD (Figure 1D). These data are consistent with our findings that KOHFD mice had a decreased final weight. It is possible that this observation is related to our previous findings demonstrating that miR-140 expression is necessary for the adipogenic process, and that preadipocytes isolated from miR-140 knockout mice exhibited inhibited adipogenic capabilities;[32] however, further examination of the role of miR-140 in hepatic lipid metabolism is beyond the scope of this project.
3.2. miR-140 Knockout Promotes Collagen Deposition in the Hepatic Microenvironment
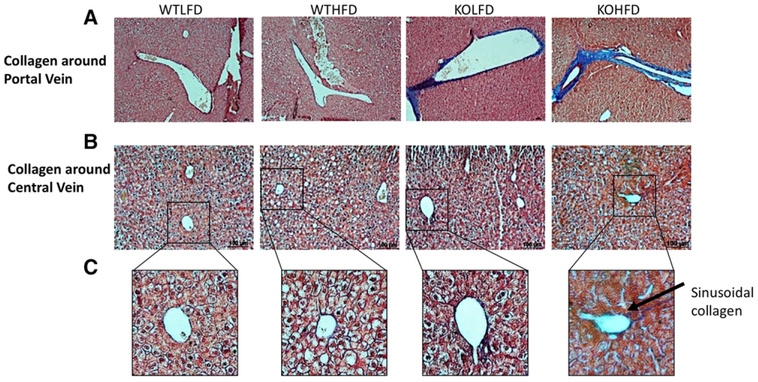
We next examined miR-140 KO and WT livers for signs of NAFLD and fibrosis. The fibrotic microenvironment contains high levels of extracellular matrix components such as collagen, and early stages of NASH often exhibit perisinusoidal/pericellular fibrosis, meaning that collagen deposition is limited to within/around sinusoids and hepatocytes.[33] We performed a modified Masson’s Trichrome stain to examine collagen deposition in the liver sections isolated from our diet-induced obesity study. We observed an almost complete absence of collagen deposition in both WTLFD and WTHFD mice. While no collagen was seen surrounding the portal veins of either mice, a small amount of collagen is seen around the central vein (Figure 2A,B). However, these data indicate a complete lack of NASH development in the wild-type mice. In contrast, both KOLFD and KOHFD liver exhibited significant collagen deposition. While we observed high levels of both portal/periportal and bridging fibrosis in mice under both diets, KOHFD mice demonstrated an increased amount of collagen over KOLFD mice (Figure 2A). Similarly, both KOLFD and KOHFD livers exhibited central vein collagen deposition, albeit at a decreased level compared to portal vein collagen, and small amounts of sinusoidal/pericellular collagen were found in KOHFD mice (Figure 2B,C). While portal fibrosis is less common than sinusoidal in cases of NASH, it typically occurs in cases where fatty deposition and other additional characteristics of liver disease are missing. Indeed, hepatic diseases in which portal fibrosis predominate can be difficult to detect, as biopsies often focus on lobular characteristics, making “silent NAFLD” difficult to treat. Moreover, it has been suggested that hepatic diseases that primarily exhibit portal fibrosis may be due to an alternative pathogenic mechanism from more common fatty liver disease.[33,34]
Figure 2.
miR-140 knockout promotes collagen deposition in the hepatic microenvironment. Modified Massons’ staining was performed to detect collagen deposition. A) Collagen deposition around the hepatic portal vein (10×). B) Collagen deposition around the central vein (20×). C) Digital zoom on 20× picture demonstrating sinusoidal collagen.
3.3. miR-140 Knockout Results in Phenotypic Characteristics of NAFLD
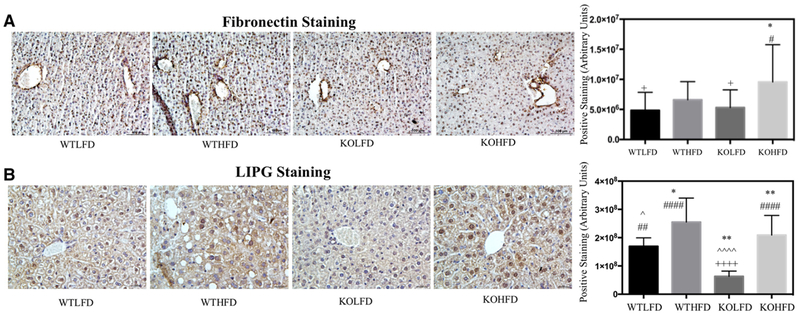
We next sought to characterize additional features of NAFLD. To further examine extracellular matrix deposition, we performed IHC staining for fibronectin, a primary component of the basement membrane. We recently identified fibronectin as a direct target of miR-140,[35] and found that chow-fed miR-140 knock-out mice had upregulated expression of fibronectin compared to WT.[28] As demonstrated in Figure 3A, there was a small, insignificant increase in central vein fibronectin expression in WTHFD mice compared to WTLFD. KOLFD mice exhibited similar expression of fibronectin to WTHFD, again with a small yet insignificant increase in expression. Despite this mild increase in expression in the KOLFD mice, KOHFD mice had significantly increased fibronectin expression (Figure 3A).
Figure 3.
miR-140 knockout results in phenotypic characteristics of non-alcoholic fatty liver disease. A) Immunohistochemistry staining of fibronectin. Quantification performed in ImageJ, average amount of positive staining per field, p < 0.001. B) Immunohistochemistry staining of LIPG. *, comparison with WTLFD. ˆ, comparison with WTHFD. #, comparison with KOLFD. +, comparison with KOHFD. p-Value is indicated as: *p ≤ 0.05. **p ≤ 0.01. ***p ≤ 0.001. ****p ≤ 0.0001.
Finally, we examined expression of the endothelial lipase LIPG. LIPG is an enzyme that aids in the cellular import and catabolism of lipoproteins. It is expressed in hepatic endothelial cells,[36] Kupffer cells,[37] and hepatocytes (human protein atlas, www.proteinatlas.org).[38] LIPG has been shown to promote inflammation through aiding in macrophage infiltration,[39] downregulating anti-inflammatory cytokines such as IL-10 and the upregulation of IL-12, IL-1β, IL-6, MCP-1, and TNF-α in macrophages.[37,40] Moreover, LIPG transcription has been shown to be activated through the NFκB pathway, potentially indicating a relationship to TLR4 signaling.[41] LIPG expression was previously demonstrated to be upregulated by an HFD,[36] potentially indicating a role in obesity. While LIPG has been relatively well characterized in endothelial cells and macrophages, few groups have studied its functions in hepatocytes, and the roles of LIPG’s pro-inflammatory signaling in NAFLD remain to be defined. To examine LIPG expression, we performed immunohistochemistry staining (Figure 3B). We found that, as previously described, LIPG expression was induced by HFD in both the wild-type miR-140 knockout livers. Interestingly, we found that miR-140 knockout resulted in a significant downregulation of LIPG expression in the KOLFD tissue, and a non-significant decrease in expression in the KOHFD liver; however, the mechanism of this downregulation remains to be defined. As LIPG plays a role in cellular lipid uptake, it is possible that LIPG downregulation found in miR-140 KO tissues plays a role in the decreased hepatocyte lipid uptake and resulting abrogation of steatosis observed in the KOHFD liver.
3.4. A miR-140/TLR4/Palmitic Acid Molecular Mechanism Promotes NAFLD
We next explored the mechanisms by which miR-140 knockout promotes NAFLD-like phenotypes as well as to investigate mechanisms of HFD in the promotion of NAFLD. PA is a highly prevalent fatty acid in western high fat diets.[10] It is the second most abundant circulating free fatty acid and circulating levels of PA are increased in obese adults. Treatment of obesity through metabolic surgery significantly decreases circulating PA, indicating that increases in PA are directly related to patient obesity.[14,15,22] Moreover, PA has been demonstrated to be a highly active ligand for the toll-like receptor 4 signaling pathway. Toll-like receptor 4 (TLR4) activates pro-inflammatory signaling through multiple pathways, including the NFκB and AP-1 pathways. TLR4 signaling has been demonstrated to have a pro-fibrogenic effect in the liver, as it is a key component of macrophage recruitment and inflammatory pathways.[18] Moreover, TLR4 deficiency was recently shown to protect against hepatic fibrosis in pre-clinical mouse models of chronic liver disease.[42] TLR4 is expressed and active in most cell types of the liver, including both hepatocytes and non-parenchymal cells such as sinusoidal endothelial cells, Kupffer cells, and hepatic stellate cells.[21]
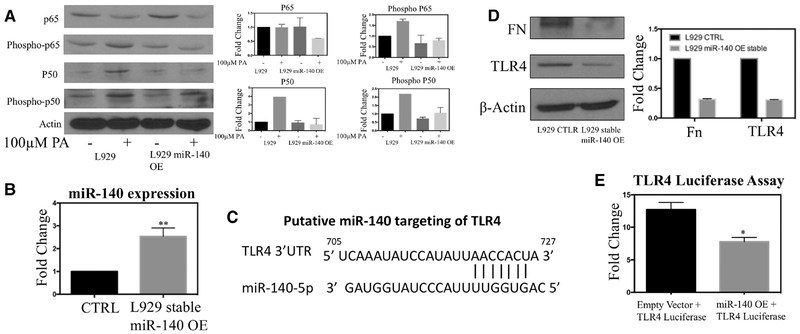
First, we examined whether miR-140 regulates PA-mediated signaling. Due to the difficulty of culturing and transfecting primary hepatocytes, we utilized the mouse fibroblast cell line L929. To examine the impact of miR-140, we created a stable miR-140 overexpressing L929 cell line. We treated wild-type and miR-140 overexpressing cells with 100 um PA for 24 h, followed by western blotting for the NFκB subunits p50 and p65 as well as their activated forms phospho-p50 and phospho-p65. As shown in Figure 4A, when wild-type cells were treated with 100 um PA, we observed no change in p65 but saw an increase in the expression of phospho-p65. Interestingly, we found that PA treatment increased the expression of p50 in general, while also increasing the expression of its phosphorylated form phospho-p50 (Figure 4A). In both cases, miR-140 overexpression inhibited palmitic acid–mediated activation of the NFκB pathway.
Figure 4.
A miR-140/TLR4/palmitic acid molecular mechanism promotes NAFLD in the absence of miR-140. A) Western blotting demonstrating that miR-140 overexpression inhibits palmitic acid mediated NFκB signaling activation. B) Expression of miR-140 after generation of a stable overexpression cell line using lentiviral vector. C) The putative binding site of miR-140 in the TLR4 3′UTR was identified using targetscan.org. D) Western blotting demonstrating miR-140 degradation of known miR-140 target fibronectin and TLR4. E) Dual luciferase assay verifying the targeting of TLR4 by miR-140.
Through in silico analysis (www.targetscan.org), we found that TLR4 was a putative target for miR-140 (Figure 4C). Moreover, miR-140 was recently demonstrated to target TLR4 in synovial fibroblasts.[43] Based on this, we hypothesized that miR-140 knockout results in increased expression of TLR4, resulting in an inflammatory microenvironment in both lean-fat and high-fat diet mice, and that increased palmitic acid in HFD mice results in the formation of the highly inflammatory microenvironment demonstrated in Figures 2 and 3. Utilizing our stable miR-140 overexpression mouse fibroblast cell line, we performed western blotting for both TLR4 and known miR-140 target protein fibronectin (Figure 4D). Both were highly downregulated, demonstrating that the miR-140 transfection was effective and that it likely targets TLR4 for degradation. Finally, we performed a luciferase assay to verify that miR-140 actively targets TLR4. We generated a TLR4-luciferase construct and co-transfected it and our miR-140 overexpression vector or an empty vector into HEK-2937 cells (in addition to phGR-TK Renilla luciferase vectors for normalization). We found that co-transfection with the miR-140 overexpression vector was sufficient to decrease luciferase expression by 38% (Figure 4E).
3.5. High-Fat Diet and miR-140 Loss Promote an Inflammatory Hepatic Microenvironment
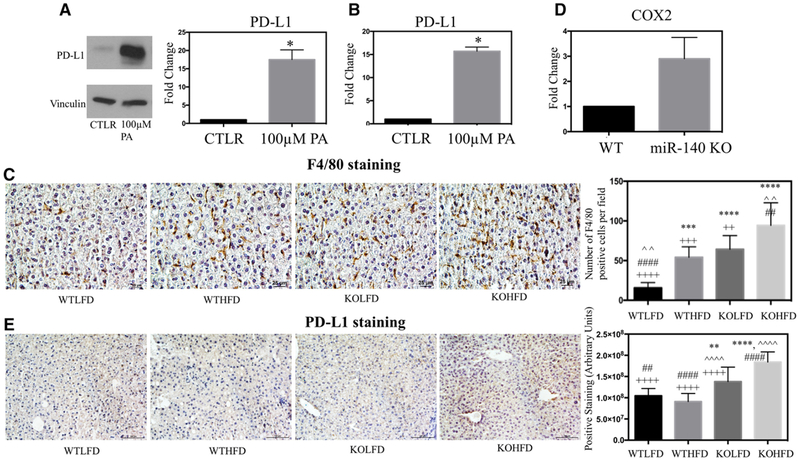
We next wanted to examine how HFD and loss of miR-140 may influence the inflammatory hepatic microenvironment. Recent research has demonstrated that obesity-induced lipid accumulation in the liver results in loss of CD4+ T cells, and that this loss may correspond to the increased risk of hepatocellular carcinoma in patients with NAFLD and NASH.[44] One mechanism for this effect may be the increased production of programmed death ligand 1 (PD-L1), which inhibits T cell proliferation and induces apoptosis. PD-L1 and other immunosuppressive proteins have been demonstrated to be upregulated in Kupffer cells, leukocytes, and hepatocytes isolated from chronically inflamed livers, and may serve as a mechanism to protect the liver in the event of high levels of inflammation.[45] To examine the effect of palmitic acid on the immunological and inflammatory liver microenvironment, we treated primary human hepatocytes with PA. We found that PA treatment significantly increased both RNA and protein PD-L1 expression (Figure 5A,B). It is possible that this is indicative of a novel pathway in which the increased concentration of PA in the obese liver promotes immunosuppressive pathways in parallel with increasing the inflammatory signaling found in the microenvironment and may be a key factor in the progression from NASH to hepatocarcinoma. To examine the impact of HFD on hepatic inflammation in vivo, and investigate the effect of miR-140 loss, we performed IHC staining for the pan-macrophage cell marker F4/80, a commonly used marker of Kupffer cells.[46] We found that levels of Kupffer cell infiltration were highly consistent with our previously examined markers of fibrosis. Kupffer cell infiltration was significantly increased in WTHFD tissues compared to WTLFD, demonstrating the formation of an inflammatory liver microenvironment as a result of a high-fat diet, as is commonly seen in NAFLD. Interestingly, we observed that Kupffer cell infiltration in KOLFD tissue was very close to that seen in WTHFD tissues, demonstrating a similar level of inflammation from miR-140 knockout alone as is caused by HFD. Finally, similarly to what we observed in ECM staining, the KOHFD tissue exhibited highly increased Kupffer cell infiltration compared to all other conditions (Figure 5C). This indicates the highly synergistic effects of an HFD and miR-140 knockout in promoting an inflammatory microenvironment, which in turn promotes the increased collagen and fibronectin deposition previously observed (Figures 2 and 3A). To further examine the impact of miR-140 on macrophages, we isolated primary mouse macrophages from the peritoneal cavity of wild-type and miR-140 knockout mice. After isolation, we performed qRT-PCR for several macrophage markers of both M1 and M2 polarization states. While we saw no difference in the polarization of unstimulated macrophages (data not shown), we observed a large increase in the expression of COX2 in miR-140 knockout macrophages (Figure 5D). The role of COX2 in macrophage polarization is contentious, with some studies identifying it as an M1 gene product and others showing it to be necessary for M2 polarization.[47,48] However, despite these conflicting data, COX2 is a well-known protein in the key prostaglandin inflammatory pathways.[49] Interestingly, COX2 is activated by the NFκB pathway, and therefore its upregulation in miR-140 knockout macrophages may be further evidence of the impact of miR-140 loss on activation the NFκB pathway and pro-inflammatory signaling.[50] Finally, we utilized IHC to examine the expression of PD-L1 in liver tissue. We found that both KOLFD and KOHFD demonstrated significant increases in PD-L1 expression. These data further demonstrate the importance of miR-140 in promoting an inflammatory hepatic microenvironment. While we were surprised to observe no increase in PD-L1 expression in the WTHFD liver compared to WTLFD liver, this may indicate that the palmitic acid content of the HFD was insufficient to induce similar PD-L1 expression to what we observed using in vitro palmitic acid treatment. Further studies utilizing a palmitic acid supplemented diet are necessary to interrogate this potential pathway.
Figure 5.
High-fat diet and miR-140 loss promote an inflammatory hepatic microenvironment. A) Western blotting demonstrating protein expression of PD-L1 in primary human hepatocytes following treatment with 100 um palmitic acid for 24 h. B) qRT-PCR of PD-L1 expression in primary hepatocytes following treatment with 100 um palmitic acid for 24 h. C) Immunohistochemistry staining for pan-macrophage marker F4/80. Quantification performed in ImageJ, average number of macrophage Kupffer cells per field. p < 0.0001. D) qPCR detection of COX-2 expression in primary peritoneal macrophages isolated from wild-type and miR-140 knockout mice. E) Immunohistochemistry staining for PD-L1. Quantification performed in ImageJ. p < 0.0001. *, comparison with WTLFD. ˆ, comparison with WTHFD. #, comparison with KOLFD.+, comparison with KOHFD. p-Value is indicated as: *p < 0.05. **p < 0.01. ***p < 0.001. ****p < 0.0001.
4. Discussion
In this article, we have identified an important role of miR-140 in the development of an inflammatory microenvironment and response to high-fat diet.[51–53] We previously demonstrated that an HFD-mediated TGFβ signaling resulted in downregulation of miR-140 in the mammary gland, and that loss of miR-140 expression resulted in increased myofibroblast differentiation.[28] This led us to next examine the impact of a high-fat diet in miR-140 knockout mice, and to question whether absence of miR-140 resulted in an exaggerated pathologic response to HFD compared to wild-type mice. In order to examine the impact on a tissue that would be in close contact with the free-fatty acid components of an HFD, we chose to focus on the development and progression of NAFLD. More than 80% of obese people present with NAFLD.[1]
While NAFLD in itself is a relatively minor disease, these individuals have a high likelihood of progressing to non-alcoholic steatohepatitis, liver fibrosis (cirrhosis), and liver cancer.[3,4] There are over 30 000 deaths in the United States resulting from cirrhosis-mediated loss of liver function, and an additional 10 000 deaths occur due to liver cancer, for which cirrhosis is a dominant risk factor.[5] It is believed that NAFLD and NASH develop due to a “multiple-hits”, a combination of lipid accumulation and inflammation as well as other factors including hormonal signaling, nutritional factors, and the microbiome.[8]
In the data presented above, we demonstrate that miR-140 loss is sufficient to the development of a highly inflammatory hepatic microenvironment, resulting in increased deposition of ECM molecules such as collagen and fibronectin and the potential development of fibrosis. This occurred without significant steatosis, and in fact was seen in the LFD liver as well as HFD liver samples.
Interestingly, we observed that while both miR-140 knockout LFD and HFD livers exhibited increases in ECM deposition and inflammation, further signaling pathway activation induced by a high-fat diet results in additional increases. This is consistent with the fact that microRNA regulation is predominantly through degradation of mRNA transcripts. This implies that miR-140 knockout lifts miR-140 mediated inhibition of specific pathways, resulting both in increased activation of pathways that increase ECM deposition and inflammation and in the sensitization of these pathways to high-fat diet–mediated signaling.
To interrogate the mechanism behind miR-140-induced sensitization to the HFD, we performed in silico analysis of miR-140 targets affiliated with HFD. We demonstrate that TLR4, which promotes inflammatory signaling and is expressed by most cells in the liver, is a target of miR-140. It is likely that this is a key mechanism for the inflammatory microenvironment observed in both KOLFD and KOHFD livers. Moreover, the highly prevalent free fatty acid PA is a ligand for TLR4. Loss of miR-140 inhibition of TLR4 may sensitize the liver to circulating palmitic acid and is a likely method by which a high-fat diet induces significantly increased inflammation and ECM deposition in the KOHFD liver. Finally, we found that treatment of hepatocytes with palmitic acid, one of the most highly prevalent free fatty acids in obese individuals, resulted in significantly upregulated expression of PD-L1, a key immunosuppressive factor in the development of hepatocellular carcinoma.
In summary, we demonstrate that miR-140 loss is a potential “hit” in the development of NAFLD/NASH, and that miR-140 knockout induces an inflammatory microenvironment with increased ECM in the absence of an HFD. Moreover, HFD and miR-140 knockout were synergistic in their promotion of liver inflammation and ECM deposition, an effect likely mediated in part by the sensitization of miR-140 knockout cells to palmitic acid mediated signaling pathways. Therefore, miR-140 may be an effective therapeutic target for the prevention of NAFLD/NASH, and miR-140 loss may be useful as a biomarker for increased risk of the development of liver disease.
Acknowledgments
B.W. collected data and wrote the manuscript, P.-K.L. contributed data, Y.Y. contributed data, L.L. contributed data, H.W. contributed data and reviewed the manuscript, Q.Z. designed the scope of the manuscript and reviewed the manuscript. This work was supported by grants from the National Cancer Institute (RO1) (CA163820A1 and CA157779A1) (Q.Z.) and the American Cancer Society (RSG-12-006-01-CNE) (Q.Z.).
Footnotes
Conflicts of Interest
The authors declare no conflict of interest.
Contributor Information
Benjamin Wolfson, Department of Biochemistry and Molecular Biology, Greenebaum Cancer Center, University of Maryland School of Medicine, Baltimore, Maryland 21201, USA.
Pang-Kuo Lo, Department of Biochemistry and Molecular Biology, Greenebaum Cancer Center, University of Maryland School of Medicine, Baltimore, Maryland 21201, USA.
Yuan Yao, Department of Biochemistry and Molecular Biology, Greenebaum Cancer Center, University of Maryland School of Medicine, Baltimore, Maryland 21201, USA.
Linhao Li, Department of Pharmaceutical Sciences, University of Maryland School of Pharmacy, Baltimore, MD, 21202, USA.
Hongbing Wang, Department of Pharmaceutical Sciences, University of Maryland School of Pharmacy, Baltimore, MD, 21202, USA.
Qun Zhou, Department of Biochemistry and Molecular Biology, Greenebaum Cancer Center, University of Maryland School of Medicine, Baltimore, Maryland 21201, USA.
References
- [1].Sanyal AJ, Gastroenterology 2002, 123, 1705. [DOI] [PubMed] [Google Scholar]
- [2].Rinella ME, JAMA 2015, 313, 2263. [DOI] [PubMed] [Google Scholar]
- [3].Spengler EK, Loomba R, Mayo Clin. Proc 2015, 90, 1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Sayiner M, Koenig A, Henry L, Younossi ZM, Clin. Liver Dis 2016, 20, 205. [DOI] [PubMed] [Google Scholar]
- [5].El-Serag HB, Rudolph KL, Gastroenterology 2007, 132, 2557. [DOI] [PubMed] [Google Scholar]
- [6].Lonardo A, Bellentani S, Argo CK, Ballestri S, Byrne CD, Caldwell SH, Cortez-Pinto H, Grieco A, Machado MV, Miele L, Targher G, Dig. Liver Dis 2015, 47, 997. [DOI] [PubMed] [Google Scholar]
- [7].Satapathy S, Sanyal A, Semin. Liver Dis 2015, 35, 221. [DOI] [PubMed] [Google Scholar]
- [8].Buzzetti E, Pinzani M, Tsochatzis EA, Metabolism 2016, 65, 1038. [DOI] [PubMed] [Google Scholar]
- [9].Mittendorfer B, Curr. Opin. Clin. Nutr. Metab. Care 2011, 14, 535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Ricchi M, Odoardi MR, Carulli L, Anzivino C, Ballestri S, Pinetti A,Fantoni LI, Marra F, Bertolotti M, Banni S, Lonardo A, Carulli N,Loria P, J. Gastroenterol. Hepatol 2009, 24, 830. [DOI] [PubMed] [Google Scholar]
- [11].Mancini A, Imperlini E, Nigro E, Montagnese C, Daniele A, Orru S, Buono P, Molecules 2015, 20, 17339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Kingsbury KJ, Paul S, Crossley A, Morgan DM, Biochem. J 1961, 78, 541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Ni Y, Zhao L, Yu H, Ma X, Bao Y, Rajani C, Loo LW, Shvetsov YB,Yu H, Chen T, Zhang Y, Wang C, Hu C, Su M, Xie G, Zhao A, Jia W, Jia W, EBioMedicine 2015, 2, 1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Ubhayasekera SJKA, Staaf J, Forslund A, Bergsten P, Bergquist J, Anal. Bioanal. Chem 2013, 405, 1929. [DOI] [PubMed] [Google Scholar]
- [15].Nicholas DA, Zhang K, Hung C, Glasgow S, Aruni AW, Unternaehrer J, Payne KJ, Langridge WHR, De Leon M, PLoS One 2017, 12, e0176793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Leamy AK, Egnatchik RA, Young JD, Prog. Lipid Res 2013, 52, 165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Ferramosca A, Wjg 2014, 20, 1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Yang L, Seki E, Front. Physiol 2012, 3, 138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Chow JC, Young DW, Golenbock DT, Christ WJ, Gusovsky F,J. Biol. Chem 1999, 274, 10689. [DOI] [PubMed] [Google Scholar]
- [20].Gallego C, Golenbock D, Gomez MA, Saravia NG, Infect. Immun 2011, 79, 2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Guo J, Friedman SL, Fibrogenesis Tissue Repair 2010, 3, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Moravcová A, Červinková Z, Kučera O, Mezera V, Rychtrmoc D,Lotkova H, Physiol. Res 2015, 64(Suppl 5), S627. [DOI] [PubMed] [Google Scholar]
- [23].Joshi-Barve S, Barve SS, Amancherla K, Gobejishvili L, Hhill D,Cave M, Hote P, McClain CJ, Hepatology 2007, 46, 823. [DOI] [PubMed] [Google Scholar]
- [24].Malhi H, Bronk SF, Werneburg NW, Gores GJ, J. Biol. Chem 2006, 281, 12093. [DOI] [PubMed] [Google Scholar]
- [25].Lee Y-S, Kim SY, Ko E, Lee J-H, Yi H-S, Yoo Y-J, Je J, Suh S-J,Jung Y-K, Seo Y-S, Yim H-J, Jeong W-I, Yeon J-E, Um S-H, Byun K-S, Sci. Rep 2017, 7, 3710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Jia L, Vianna CR, Fukuda M, Berglund ED, Liu C, Tao C, Sun K,Liu T, Harper MJ, Lee CE, Lee S, Scherer PE, Elmquist JK, Nat. Commun 2014, 5, 3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Miyaki S, Sato T, Inoue A, Otsuki S, Ito Y, Yokoyama S, Kato Y, Takemoto F, Nakasa T, Yamashita S, Takada S, Lotz MK, Ueno-Kudo H, Asahara H, Genes & Development 2010, 24, 1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Wolfson B, Zhang Y, Gernapudi R, Duru N, Yao Y, Lo PK, Zhou Q, Mol. Cell. Biol 2017, 37, e00461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Eades G, Yao Y, Yang M, Zhang Y, Chumsri S, Zhou Q, J. Biol. Chem 2011, 286, 25992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Li L, Li D, Heyward S, Wang H, PLoS One 2016, 11, e0150587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Lecluyse EL, Alexandre E, Methods Mol. Biol 2010, 640, 57. [DOI] [PubMed] [Google Scholar]
- [32].Gernapudi R, Wolfson B, Zhang Y, Yao Y, Yang P, Asahara H, Zhou Q, Mol. Cell. Biol 2015, 36, MCB.00702–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Takahashi Y, Fukusato T, Wjg 2014, 20, 15539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Hübscher SG, Histopathology 2006, 49, 450. [DOI] [PubMed] [Google Scholar]
- [35].Duru N, Zhang Y, Gernapudi R, Wolfson B, Lo PK, Yao Y, Zhou Q, Sci. Rep 2016, 6, 39572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Yu KC-W, David C, Kadambi S, Stahl A, Hirata K, Ishida T,Quertermous T, Cooper AD, Choi SY, J. Lipid Res 2004, 45, 1614. [DOI] [PubMed] [Google Scholar]
- [37].Wang X, Jin W, Rader DJ, Circ. Res 2007, 100, 1008. [DOI] [PubMed] [Google Scholar]
- [38].Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P,Mardinoglu A, Sivertsson A, Kampf C, Sjostedt E, Asplund A, Olsson I, Edlund K, Lundberg E, Navani S, Szigyarto CA, Odeberg J, Djureinovic D, Takanen JO, Hober S, Alm T, Edqvist PH,Berling H, Tegel H, Mulder J, Rockberg J, Nilsson P, Schwenk JM,Hamsten M, von Feilitzken K, Forsberg M, Persson L, Johann-son F, Zwahlen M, von Heijne G, Nielsen J, Ponten F, Science 2015, 347, 1260419. [DOI] [PubMed] [Google Scholar]
- [39].Kojma Y, Hirata K-I, Ishida T, Shimokawa Y, Inoue N, Kawashima S, Quertermous T, Yokoyama M, J. Biol. Chem 2004, 279, 54032. [DOI] [PubMed] [Google Scholar]
- [40].Yasuda T, Hirata K-I, Ishida T, Kojima Y, Tanaka H, Okada T, Quertermous T, Yokoyama M, Jat 2007, 14, 192. [DOI] [PubMed] [Google Scholar]
- [41].Kivelä AM, Mäkinen PI, Jyrkkänen H-K, Mella-Aho E, Xia Y, Kansanen E, Leinonen H, Verma IM, Yla-Herttuala S, Levonen AL, Atherosclerosis 2010, 213, 122. [DOI] [PubMed] [Google Scholar]
- [42].Weber SN, Bohner A, Dapito DH, Schwabe RF, Lammert F, PLoS One 2016, 11, e0158819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Li H, Guan S-B, Lu Y, Wang F, Biomed. Pharmacother 2017, 96, 208. [DOI] [PubMed] [Google Scholar]
- [44].Ma C, Kesarwala AH, Eggert T, Medina-Echeverz J, Kleiner DE,Jin P, Stroncek DF, Terabe M, Kapoor V, ElGindi M, Han M, Thornton AM, Zhang H, Egger M, Luo J, Felsher DW, McVicar DW, Weber A, Heikenwalder M, Greten TF, Nature Publishing Group 2016, 531, 253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Kassel R, Cruise MW, Iezzoni JC, Taylor NA, Pruett TL, Hahn YS, Hepatology 2009, 50, 1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Nguyen-Lefebvre AT, Horuzsko A, J Enzymol Metab 2015, 1, 101. [PMC free article] [PubMed] [Google Scholar]
- [47].Na Y-R, Yoon Y-N, Son D-I, Seok S-H, PLoS One 2013, 8, e63451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Li H, Yang B, Huang J, Lin Y, Xiang T, Wan J, Li H, Chouaib S, Ren G, Oncotarget 2015, 6, 29637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Ricciotti E, FitzGerald GA, Arterioscler. Thromb. Vasc. Biol 2011, 31, 986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Wang N, Liang H, Zen K, Front. Immunol 2014, 5, 614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Hwang S, Park S-K, Lee HY, Kim SW, Lee JS, Choi EK, You D,Kim CS, Suh N, FEBS Lett 2014, 588, 2957. [DOI] [PubMed] [Google Scholar]
- [52].Li Q, Yao Y, Eades G, Liu Z, Li Q, Zhou Q, Oncogene 2014, 33, 2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Yang J, Qin S, Yi C, Ma G, Zhu H, Zhou W, Xiong Y, Zhu X, Wang Y, He L, Guo X, FEBS Lett 2011, 585, 2992. [DOI] [PubMed] [Google Scholar]