Abstract
Glioblastomas (GBM) are aggressive brain tumors that inevitably recur despite surgical resection, chemotherapy, and radiation. The degree to which recurrent GBM retains its initial immunophenotype is incompletely understood. We generated tissue microarrays of paired initial and posttreatment GBM (3 pairs positive and 17 negative for IDH1 R132H ) from the same patients and made comparisons in the IDH1 R132H -negative group for immunohistochemical and gene expression differences between primary and recurrent tumors. In initial tumors, immunopositivity for Ki-67 in > 20% of tumor cells was associated with shorter progression-free and overall survival. Recurrent tumors showed decreased staining for CD34 suggesting lower vessel density. A subset of tumors showed increased staining for markers associated with the mesenchymal gene expression pattern, including CD44, phosphorylated STAT3, and YKL40. Recurrent tumors with the greatest increase in mesenchymal marker expression had rapid clinical progression, but no difference in overall survival after second surgery. Comparison of protein and gene expression data from the same samples revealed a poor correlation. A subset of tumors (15%) showed loss of neurofibromin protein in both initial and recurrent tumors. These data support the notion that GBM progression is associated with a shift toward a mesenchymal phenotype in a subset of tumors and this may portend a more aggressive behavior.
Keywords: Glioblastoma, Glioblastoma molecular subtype, Immunohistochemistry, Mesenchymal transition, Neurofibromin, Recurrent glioblastoma.
INTRODUCTION
Glioblastomas (GBM) are the most common primary malignant central nervous system (CNS) tumors in adults, accounting for 15.1% of all primary CNS neoplasms and 46.1% of malignant primary brain tumors overall ( 1 ). With an estimated incidence of 3.2 cases per 100 000, GBM account for over half of all primary CNS gliomas and 60%–75% of astrocytic tumors ( 1 , 2 ). Complete surgical resection of GBM is not attainable due to their diffusely infiltrative nature, and GBM invariably recur despite aggressive surgery, chemotherapy, and radiation therapy ( 3 ). Recent studies have revealed significant genetic heterogeneity in GBM and efforts to classify genetic subtypes of GBM are ongoing ( 4–7 ). Transcriptional profiling has identified a subgroup of GBM designated as “mesenchymal”; this group of tumors is characterized by absence of IDH mutations, lack of the CpG island methylator phenotype, and frequent mutation or loss of the NF1 tumor suppressor gene ( 5 ).
High expression of YLK40 and CD44 is associated with the mesenchymal subgroup, whereas oligodendrocyte transcription factor 2 (OLIG2) expression is typically low in mesenchymal subgroup tumors and high in proneural subgroup tumors ( 5 ). When they recur, nonmesenchymal GBM occasionally shift to the mesenchymal gene expression pattern, suggesting that transcriptional subtype may not be a stable tumor trait ( 4 , 8 ). Signaling through the nuclear factor (NF)-κB signaling pathway is implicated in the mesenchymal transition and promotes radiation resistance and may be mediated by factors from the tumor microenvironment ( 9 , 10 ). In addition, individual cells within a single GBM can exhibit a spectrum of gene expression profiles so that selection of a tumor cell subclone may occur upon treatment ( 11 ). The evolution of GBM at recurrence has important therapeutic implications. Several factors could impact the correlation of protein and gene expression, including posttranscriptional and posttranslational regulation and the sensitivity of immunohistochemical analyses. Thus, recurrent GBM frequently harbors different gene expression patterns compared with the initial tumor and how differences in gene expression translate into immunophenotypic marker stability in disease progression remain poorly understood.
This study aims for a better understanding how protein expression of select GBM markers changes in relation to recurrent posttreatment tumors. We generated tissue microarrays from 20 patients with paired initial and posttreatment samples and examined a panel of immunohistochemical markers. We used this platform to assess patterns of mesenchymal immunophenotypic marker expression in posttreatment recurrent tumors, focusing the analysis on tumors that are negative for IDH1 R132H . Gene expression analysis was performed on formalin-fixed paraffin-embedded samples using a Nanostring platform to correlate the immunohistochemical findings with gene expression levels. This study tests the hypothesis that protein expression changes occur in the course of GBM progression/recurrence, and that such changes predict recurrence rate and/or overall survival (OS).
MATERIALS AND METHODS
Case Selection and Tissue Microarray Generation
Candidate cases were identified by electronic search of the University of California San Francisco surgical neuropathology records for diagnostic lines or clinical histories containing the terms “residual” and/or “recurrent” and “glioblastoma.” We identified 24 patients who had material from multiple GBM resections. Hematoxylin and eosin (H&E)–stained slides were reviewed to select blocks with adequate viable tumor. Up to 3 1.5-mm cores were selected and placed in tissue microarray format, as previously described ( 12 ). Four of the patients were excluded from the analysis due to lack of adequate material, leaving a total of 20 cases with paired initial and first recurrence posttreatment specimens on the array. All samples were obtained in accordance with the Committee on Human Research at University of California, San Francisco (10-01318).
Immunohistochemistry
Immunohistochemical stains were performed using antibodies to the following: anti-Ki67 antibody (MIB-1), CD44, OLIG2, epidermal growth factor receptor (EGFR), p53, ionized calcium-binding adapter molecule 1 (IBA1), chitinase-3-like protein 1 (YKL-40), phosphorylated S6 kinase (pS6K), phosphorylated STAT3 (pSTAT3), CD34, neurofibromin 1 (NF1), and IDH1 R132H . Antibody manufacturers, dilutions, and scoring criteria are listed in Table 1 . Because CD44 showed a more variable staining pattern than other stains, it was scored as 0 = negative, 1 = weak patchy staining, 2 = weak diffuse staining, 3 = moderate patchy staining, 4 = moderate diffuse staining, 5 = strong patchy staining, and 6 = strong diffuse staining ( Supplementary Data Fig. S1 ). We selected a cutoff of 10% strong nuclear staining for p53 because this cutoff has approximately 79% sensitivity and 97% specificity for a TP53 mutation ( 13 ). Neurofibromin immunostaining using antibody clone NFC was performed and scored according to previously published criteria ( 14 ). Arrays were independently reviewed by 2 of the investigators (M.D.W. and J.J.P.), and samples with a significant scoring discrepancy were reviewed together to achieve a final consensus. Increased mesenchymal marker protein expression was operationally defined as an increase of 1 or more points in the immunohistochemistry (IHC) grading scale averaged across recurrent tumor cores. This immunostaining panel could generate a maximum of 480 individual data points: 20 cases × 2 specimens per case × 12 immunostains. For 449 of 480 data points (93.5%), at least 1 core on the TMA was adequate for scoring, and 369 of these (82.2% of 449) had 2 or 3 cores present (overall 76.9% of data points with 2 or more cores). To obtain data in specimens where none of the cores survived TMA processing, IHC was performed on whole tissue sections from the same block and scored using identical criteria as for the TMA cores.
TABLE 1.
Antibody Dilutions, Manufacturers, and Scoring Criteria
| Antibody | Manufacturer, Catalog number | Dilution | Scoring Criteria |
|---|---|---|---|
| Ki-67 (clone MIB-1) | Ventana 790-4286 | Undiluted | % positive tumor nuclei |
| p53 | Dako M7001 | 1:25 |
|
| IDH1 R132H | Dianova H09 | 1:25 | Positive or negative in tumor cells |
| Ionized calcium-binding adapter molecule 1 (IBA1) | Wako 019-19741 | 1:200 |
|
| Oligodendrocyte transcription factor 2 (OLIG2) | Millipore AB9610 | 1:50 |
|
| YKL-40 | Quidel 4815 | 1:200 | As for OLIG2 |
| Phospho-S6K | Cell Signaling 2215 | 1:50 | As for OLIG2 |
| Phospho-STAT3 | Cell Signaling 9145 | 1:10 | As for OLIG2, nuclear staining only |
| CD34 | Millipore CBL496 | 1:800 |
|
| CD44 | BD 550392 | 1:1000 | See text and Supplementary Data |
| Epidermal growth factor receptor (EGFR) | Ventana 790-2988 | 1:1 |
|
| Neurofibromin 1 (NF1) | See text | See text | See text |
Ventana, Tucson, AZ; Dako, Carpinteria, CA; Dianova, Hamburg, Germany; Wako, Richmond, VA; Millipore, Billerica, MA; Quidel, San Diego, CA; Cell Signaling Technology, Danvers, MA.
Gene Expression Analysis
For NanoString analysis, total RNA was isolated from tumor cores (2- to 3 1-mm cores) from formalin-fixed paraffin-embedded blocks containing >75% tumor cells as determined by H&E staining, according to the manufacturer’s protocol (RNEasy FFPE kit; Qiagen, Valencia, CA). Concentrations were determined using NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Wilmington, DE), and RNA integrity was assessed using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). A custom code set was generated and probes for the analysis were synthesized by NanoString technologies. The data set included probes for 14 genes of interest, including 5 proneural genes ( DLL3 , NCAM1 , OLIG2 , SOX9 , and SOX2 ), 3 mesenchymal genes ( CHI3L1 , TIMP1 , and CD44 ), and 6 normalizing genes ( ACTB , B2M , GAPDH , POLR2A , SDHA , and TBP ). RNA (200 ng) was analyzed with the NanoString nCounter Analysis System at NanoString Technologies according to the manufacturer’s protocol (NanoString Technologies, Seattle, WA). This analysis included mRNA expression data for 9 paired tumors from the tissue microarray where CD44, OLIG2, and EGFR were also assessed at the protein level.
Statistical Analysis
The clinical outcomes were progression-free survival (PFS; date of first surgery to date of clinical or radiographic progression); OS (date of first surgery to date of death [n = 19] or referral to hospice care [n = 1]); and PFS after second surgery (date of second surgery to date of death [n = 19] or referral to hospice [n = 1]). Survival analysis was performed in PRISM software (La Jolla, CA) using a log-rank (Mantel-Cox) test. Immunohistochemical scores and gene expression values were compared using a paired 2-tailed t -test using PRISM software or Microsoft Excel, respectively. Statistical significance is reported as p values, designated in the figures as *p < 0.05, **p < 0.01, and ***p < 0.001.
Gene expression data were analyzed by the method of Bhat et al ( 9 ). Briefly, normalized gene expression values were converted to z scores and summed to produce metagene scores for proneural and mesenchymal genes. The overall mesenchymal score was obtained by subtracting the proneural metagene score from the mesenchymal metagene score. Samples were ranked by the mesenchymal gene metascore. A mesenchymal transition was operationally defined as tumors that increased their ranking by >2 points upon recurrence. Conversely, a proneural transition was defined as a decrease of >2 points in the mesenchymal rank list upon recurrence.
RESULTS
The tissue microarrays included 20 paired initial and posttreatment GBM specimens that were representative of the demographics of GBM, with a slight male predominance (60% male), age of onset between the 4th and 8th decade, and tumor location throughout the cerebral hemispheres with most tumors located in the temporal or frontal lobes ( Table 2 ). Surgical resection status was not available for 4 cases. The remaining patients underwent gross total (n = 10; 63%) or near total/subtotal (n = 6; 37%) tumor resection. Resection was followed by temozolomide and radiation therapy in all cases with available treatment histories; treatment data were not available for 2 patients. Approximately half of the patients had additional experimental treatments as part of clinical trials. Approximately half of the patients had residual tumor on postoperative imaging. At recurrence, tumor debulking to the maximum possible extent was documented for 14 of 17 patients with available clinical data, whereas the remaining 3 patients underwent subtotal resection.
TABLE 2.
Patient Clinical Characteristics
| Sex | N | % |
|---|---|---|
| Male | 12 | 60 |
| Female | 8 | 40 |
| Age at initial resection (years) | ||
| Mean | 52.3 | |
| Range | 31–73 | |
| Tumor location | ||
| Frontal lobe | 7 | 35 |
| Parietal lobe | 2 | 10 |
| Occipital lobe | 3 | 15 |
| Temporal lobe | 8 | 40 |
| IDH1 status | ||
| IDH1 R132H | 3 | 15% |
| Survival, days a (N) | Mean | Range |
| OS ( 17 ) | 838 | 346–2541 |
| PFS ( 14 ) | 519 | 87–1382 |
| PFS-SS ( 13 ) | 175 | 62–503 |
a Survival values are for non-IDH1 R132H tumors. Censored data points are not included. Overall survival (OS) is the time from initial resection to death (16 patients) or referral to palliative care (1 patient). Progression free survival (PFS) is the time from the initial resection to clinical or radiographic progression. Progression free survival after second surgery (PFS-SS) is the time from the second tumor resection to next clinical or radiographic progression.
Immunohistochemical analysis of tissue microarrays identified 3 initial tumors strongly positive for IDH1 R132H mutant protein and this was stable posttreatment at recurrence. Because the IDH1 R132H -negative tumors generally occurred in older patients where other IDH mutations would be uncommon, IHC-negative cases were not tested further for other IDH1 mutations, or for mutations in IDH2 . Based on a recently published model incorporating patient age, the presence or absence of a lower-grade precursor, World Health Organization grade IV histology, and IDH1 R132H IHC results, the probability of an alternate IDH mutation is <5% in the majority of our IDH1 R132H -negative cases (11 of 17; 65%) ( 15 ). No tumors initially negative with the IDH1 R132H antibody-acquired positivity at recurrence. IDH mutant GBM has a distinct biology and prognosis from that of IDH wild-type tumors ( 16 ). In accordance with the known features of IDH mutant GBM, in this cohort the IDH1 R132H -positive cases had a protracted clinical course, with 2 patients alive at 2638 and 3010 days after their initial resection and 1 patient lost to follow-up 213 days after initial resection. Thus, these 3 specimens were excluded from further immunohistochemical and gene expression analysis, with the exception of neurofibromin IHC.
Detailed clinical characteristics of the final cohort of 17 paired non-IDH1 R132H tumors are provided in Table 3 . In this relatively small cohort, patient age (≥60 years vs <60 years), gender, radiographically residual tumor (presence or absence), extent of tumor resection (gross total vs nongross total), and initial tumor size (above or below median value) had no effect on OS or PFS ( Supplementary Data Fig. S2 ). The initial Karnovsky performance status was not available for enough patients to warrant a meaningful analysis.
TABLE 3.
Clinical Characteristics of Non-IDH1 R132H Tumor Pairs
| Pair | Age at diagnosis (years) | Tumor Location | Gender | Tumor Greatest Dimension (cm) | Initial Surgery | Residual Radiographic Tumor (RRT; 1 = yes, 0 = no) | Temozolomide and Radiation (1 = yes) | Other Treatments of Initial Tumor | Progression Free Survival (PFS, days) | Second Surgery | RRT After Second Surgery | PFS After Second Surgery (days) | OS After Second Surgery (days) | OS From Initial Surgery (days) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 55 | Frontal | M | 2.0 | GTR | 0 | 1 | ENZ | 190 | GTR | 1 | 62 | 134 | 346 |
| 3 | 71 | Temporal | F | ND | ND | ND | ND | HSP | ND | ND | ND | 78 | 333 | 634 |
| 4 | 53 | Parietal | F | 6.5 | NGTR | 1 | 1 | THAL | 1172 | DB | ND | 503 | 607 | 1786 |
| 5 | 40 | Frontal | M | 5.9 | ND | ND | 1 | AV | 531 | DB | 1 | ND | 117 | 673 |
| 6 | 71 | Occipital | F | 4.8 | NGTR | 1 | 1 | 854 | DB | 0 | ND | 43 | 917 | |
| 7 | 51 | Bifrontal | F | ND | ND | ND | ND | I-125 | ND | ND | ND | ND | 173 | 804 |
| 8 | 58 | Parietal | M | 3.0 | STR | 1 | 1 | 103 | DB | 1 | 90 | 347 | 495 | |
| 10 | 60 | Frontal | M | ND | ND | ND | 1 | ND | ND | ND | ND | 151 | 542 | |
| 11 | 46 | Temporal | M | 4.5 | GTR | 0 | 1 | 571 | GTR | 0 | 82 | 166 | 740 | |
| 12 | 54 | Temporal | M | 3.8 | NTR | 1 | 1 | AV+TCV | 227 | GTR | 0 | 69 | 129 | 400 |
| 13 | 46 | Temporal | F | 6.5 | GTR | 1 | 1 | 628 | STR | 1 | 153 | 290 | 940 | |
| 14 | 71 | Temporal | M | 4.1 | GTR | 0 | 1 | vIIIVAC | 315 | GTR | 1 | 119 | 177 | 497 |
| 15 | 41 | Occipital | M | 4.0 | GTR | 0 | 1 | ENZ | 1382 | GTR | 0 | 253 | 1178 | 2581 |
| 17 | 63 | Temporal | F | 4.0 | GTR | 1 | 1 | 109 | GTR | 1 | 355 | 355 a | 480 a | |
| 22 | 48 | Frontal | M | 5.9 | GTR | 0 | 1 | HSP | 875 | DB | 1 | 68 | 467 | 1406 |
| 23 | 44 | Temporal | M | 5.5 | STR | 1 | 1 | vIIIVAC | 87 | STR | 1 | 255 | 446 | 534 |
| 24 | 73 | Temporal | M | 6.5 | GTR | 1 | 1 | 221 | STR | 1 | 186 | 249 | 474 |
ND, no data; GTR, gross total resection; NTR/NGTR, near total/gross total resection; STR, subtotal resection; DB, debulking; ENZ, enzastaurine; HSP, heat shock protein (vaccine trial); THAL, thalidomide; AV, Avastin; I-125, Iodine 125 seeds; TCV, Tarceva; vIIIVAC, epidermal growth factor receptor class III variant vaccine; M, male; F, female.
a For tumor pair 17, overall survival reflects the days elapsed from initial surgery to hospice care referral. No documented date of death was available for this patient.
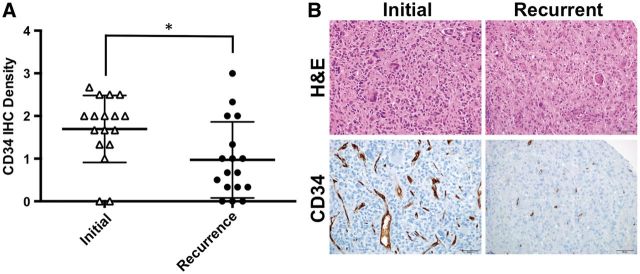
Tissue microarrays were stained with a panel of immunohistochemical markers, including markers of cell proliferation (Ki-67), tumor vascularity (CD34), microglial and macrophage infiltration (IBA1), proneural phenotype (OLIG2), mesenchymal phenotype (CD44, YKL-40, and pSTAT3, Tyr705), EGFR, and phosphorylated S6 kinase, Ser240/244 [pS6K] to assess the PI3K/AKT signaling pathway. To assess the overall stability of these markers at recurrence, the mean expression scores were compared between primary and recurrent tumors. Only CD34 showed a significant change in expression in the recurrent tumors, dropping from 1.70 ± 0.78 units in the initial resection to 0.97 ± 0.89 units in the recurrence sampling (mean ± SD, p < 0.05, Fig. 1A ). The other markers did not show a significant difference in marker expression between primary and posttreatment recurrent samplings ( Supplementary Data Fig. S3 ). Three of the initial tumors had significant nuclear staining for p53 (defined as strong nuclear staining in >10% of tumor nuclei), and the degree of staining was stable posttreatment at tumor recurrence. No tumors gained significant p53 positivity at recurrence.
FIGURE 1.
CD34 immunostaining in glioblastoma specimens at initial and recurrent disease. ( A ) Plots of the CD34 expression in the cohort at initial tumor resection and sampling of recurrent tumor. Whiskers represent the mean ± SD, *p < 0.05. ( B ) Representative images demonstrating the reduction in CD34 immunostaining in 1 of the paired cases. H&E, hematoxylin and eosin; IHC, immunohistochemistry.
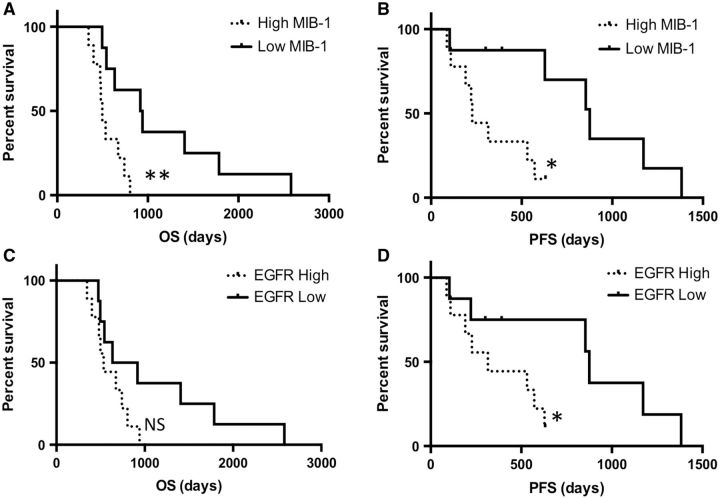
To determine whether protein expression in the initial tumor predicted OS or PFS, tumors were divided into high- and low-expressing categories based on median expression values, with tumors at the median value assigned to the high-expression group. Initial tumors at or above the median Ki-67 labeling index of 20% or greater showed a significantly shorter PFS (median PFS, 227 vs 875 days; p < 0.05) and OS (median OS, 497 vs 928.5 days; p < 0.01) compared with tumors with a low (<20%) labeling index ( Fig. 2A, B ). In addition, tumors with high EGFR expression showed a significantly shorter PFS (median PFS, 315 vs 875 days; p < 0.05; Fig. 2C ). There was a trend toward shorter OS in the EGFR-high cases, but this did not achieve statistical significance ( Fig. 2D ). No significant PFS or OS differences were observed in the other markers (OLIG2, CD44, CD34, YKL-40, IBA1, pS6K, and pSTAT3). Low IBA1 labeling index trended with longer OS; however, this difference did not achieve statistical significance (median survival, 828.5 vs 542 days; p = 0.08; Supplementary Data Fig. S4 ).
FIGURE 2.
Survival analysis for tumors with low or high Ki-67/MIB-1 labeling index. Overall survival (OS) (A) and progression-free survival (PFS) (B) ; p < 0.01 and p < 0.05, respectively. PFS (C) and OS ( D ) for tumors with low or high labeling for epidermal growth factor receptor (EGFR). *p < 0.05 for PFS.
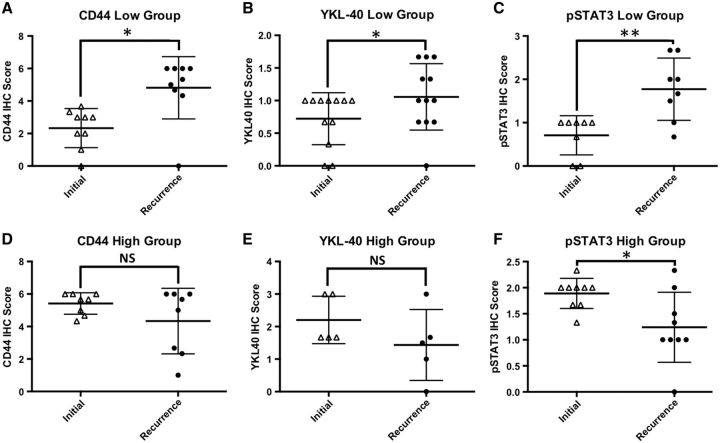
Analyses of gene expression patterns in primary and recurrent GBM have shown that tumors with a nonmesenchymal gene expression signature transition to a more mesenchymal signature at recurrence, whereas tumors with a mesenchymal gene expression signature initially tend to be more stable and only rarely shift away from the mesenchymal signature ( 4 , 8 ). These gene expression data predict that tumors with high mesenchymal protein marker expression will retain high expression at recurrence, whereas initial tumors with low mesenchymal protein marker expression will tend to have higher expression at recurrence. To determine whether protein markers of the mesenchymal subtype increased posttreatment, we examined the expression of CD44, YKL-40, and pSTAT3 (hereafter referred to as mesenchymal markers) ( 4 ). For each marker, the initial tumors were assigned to high- or low-expression groups using an IHC cutoff score of >4 for CD44, >1 for pSTAT3, and >1 for YKL-40. These groups were then examined for changes in marker expression in the matched recurrent tumors. We observed that for each marker the group of low-expressing tumors showed a significant increase in marker expression at recurrence ( Fig. 3A–C ). As predicted, the group of high-expressing tumors for CD44 and YKL-40 had stable expression levels at recurrence; the only exception to the predicted pattern was the pSTAT3 high-expressing group, which showed a significant reduction in pSTAT3 expression at recurrence ( Fig. 3D–F ). Tumors with high initial levels of OLIG2 showed a trend toward decreased OLIG2 at recurrence, but this did not achieve statistical significance (data not shown). A subset of tumors showed reciprocal changes in YKL-40 and/or CD44 and OLIG2, with the recurrent tumor showing increased mesenchymal marker expression and decreased OLIG2 ( Supplementary Data Fig. S5 ), an immunophenotypic feature of recurrent GBM that has been previously reported ( 9 ). The microglia and macrophage response, as denoted by IBA1, was not significantly increased in tumors with a more mesenchymal phenotype or in tumors that acquired a more mesenchymal phenotype at recurrence (data not shown). These data suggest that a subset of primary GBM shifts toward a more mesenchymal immunohistochemical profile at recurrence.
FIGURE 3.
Changes in immunohistochemistry (IHC) scores among high- or low-expressing groups of mesenchymal markers. Whiskers represent the mean ± SD. ( A–C ) Tumors with low initial expression of CD44, YKL-40, or phosphorylated STAT3 (pSTAT3) show increased expression of these markers on recurrence, *p < 0.05 and **p < 0.01, respectively. ( D, E) Tumors with high expression of CD44 or YKL-40 show stable levels on recurrence, p > 0.05. ( F ) pSTAT3 is reduced in recurrent tumors with high initial staining for pSTAT3, p < 0.05.
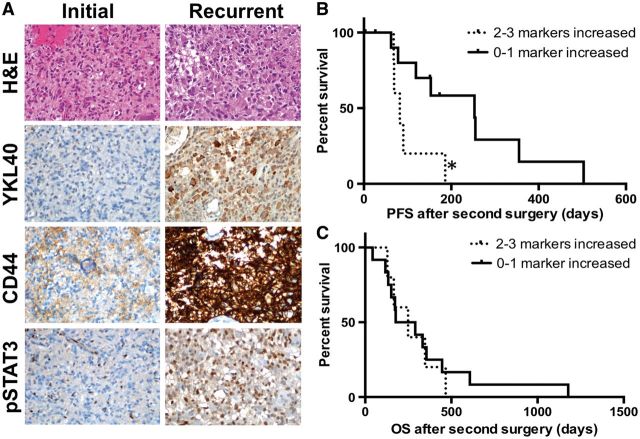
Because the mesenchymal gene expression pattern is associated with aggressive behavior, we next asked whether tumors that had the highest change in mesenchymal marker expression at recurrence showed shorter PFS after recurrence. Five tumors had increased protein expression of 2 or more mesenchymal markers ( Fig. 4A ), while 12 tumors had an increase in 1 (6 cases) or none (6 cases) of the markers. The PFS after second surgery in the group with 2 or more increased mesenchymal markers was significantly shorter (82 days) versus the group with 0-1 increased mesenchymal markers (253 days; p < 0.05; Fig. 4B ). However, the time from second surgery to death was identical between the 2 groups ( Fig. 4C ). These data suggest that a mesenchymal transition in a recurrent tumor, as defined by protein expression compared with the initial tumor sampling, may have prognostic value for time to clinical or radiographic progression but not for OS after recurrence.
FIGURE 4.
Overall (OS) and progression-free survival (PFS) after second surgery for tumors with increased mesenchymal marker expression. ( A ) Representative images from a tumor pair with increased CD44, YKL-40, and phosphorylated STAT3 (pSTAT3) immunostaining at recurrence. (B , C) PFS (B) and OS (C) after second surgery by mesenchymal marker expression, p < 0.05 and p > 0.05, respectively. H&E, hematoxylin and eosin.
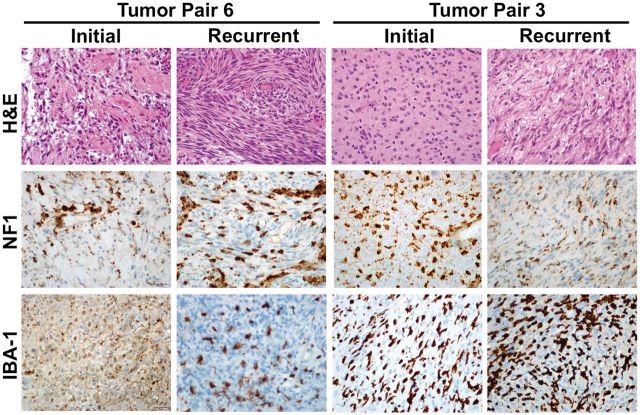
Alterations of the NF1 tumor suppressor gene are a feature of the mesenchymal molecular subgroup of GBM. Because immunohistochemical markers associated with a mesenchymal phenotype are increased in a subset of recurrent tumors, we asked whether any of the recurrent tumors also had loss of neurofibromin 1 (NF1) protein. Immunostaining of the tissue microarrays yielded neurofibromin expression data on 13 tumor pairs. Two of the tumors (15%) were negative for neurofibromin protein, showing immunoreactivity only in the microglia, inflammatory cells, and vessels, providing an internal positive control ( Fig. 5 ). Both neurofibromin-deficient tumors demonstrated a spindled or frankly sarcomatous morphology although neither tumor met criteria for a diagnosis of gliosarcoma due to focality of the sarcomatous component. The remaining 11 tumors were positive (85%). Paired samples for an additional 2 tumors that were positive for IDH1 R132H showed strong neurofibromin staining in both cases. Taken together, the neurofibromin expression was stable in initial and posttreatment recurrent samples for all 15 cases tested (13 non-IDH1 R132H and 2 IDH1 R132H cases). These results suggest that complete loss of neurofibromin, at least at the protein level, is not a common event in recurrent GBM.
FIGURE 5.
Representative images from 2 tumor pairs deficient for neurofibromin 1 (NF1) protein by immunohistochemical staining. H&E, hematoxylin and eosin; IBA1, ionized calcium-dependent binding protein 1.
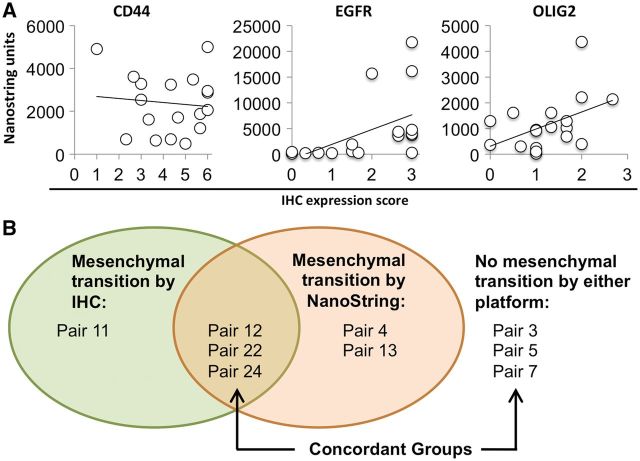
To correlate the immunohistochemical analysis with gene expression levels, we examined the expression of a panel of genes in paired tumors by NanoString. The correlation between mRNA and protein expression was low to moderate, with R2 values of 0.01 for CD44 , 0.21 for OLIG2 , and 0.25 for EGFR ( Fig. 6A ). Specimens with very high levels of EGFR mRNA (above 10 000 units) generally had a high IHC labeling index (≥2 units), but specimens with lower mRNA expression (<5000 units) showed a broad range of IHC staining (ranging from no staining to a score of 3). By protein analysis, 5 of 17 tumor pairs acquired a more mesenchymal phenotype at recurrence (i.e. increased staining for 2 or more mesenchymal markers, as described earlier). Of these 17 tumor pairs, a total of 9 were available for analysis of mRNA expression levels, including 4 of the tumors that met IHC criteria for a mesenchymal phenotype transition on recurrence ( Fig. 6B ). Based on NanoString analysis, 5 of 9 tumor pairs acquired a more mesenchymal phenotype at recurrence; however, the correlation between protein and mRNA analysis was poor, with 3 of 9 tumor pairs showing a mesenchymal transition by both platforms, 3 of 9 tumor pairs negative for a mesenchymal transition by both platforms, and 3 of 9 pairs with discordance between the 2 platforms (overall 6 of 9 concordant pairs; Pearson correlation coefficient, 0.35).
FIGURE 6.
( A ) Correlation between immunohistochemistry (IHC) scores and gene expression levels. R2 = 0.0103 for CD44, 0.250 for epidermal growth factor receptor (EGFR), and 0.208 for oligodendrocyte transcription factor 2 (OLIG2). ( B ) Diagrammatic representation of the concordance between tumors that underwent a mesenchymal transition at recurrence by IHC and/or NanoString analysis.
DISCUSSION
GBM are highly aggressive tumors that invariably recur despite current therapies. Knowledge about tumor properties at recurrence is essential for a better understanding of tumor evolution and to improve GBM therapies. The analysis of paired GBM from the same patient at initial diagnosis and after treatment at recurrence provides a unique opportunity to examine this evolution. Transcriptional profiling of GBM suggests tumors acquire a more mesenchymal phenotype at recurrence. How this corresponds to protein level changes is unclear ( 17 , 18 ). To begin to address this question, we generated a platform for the analysis of protein and gene expression changes upon posttreatment tumor recurrence using paired GBM specimens. Significant changes in protein expression, as demonstrated by IHC, were seen in a subset of GBM at tumor recurrence, providing a strong rationale for retesting these emerging immunophenotypic markers in recurrent tumors. In particular, markers associated with a mesenchymal gene expression pattern increased markedly in some recurrent tumors, which showed a more aggressive course (i.e. shortened PFS). Our data support an emerging model in which tumors can shift toward a more mesenchymal phenotype upon posttreatment recurrence. Our data add to and expand upon the growing data from paired primary and recurrent GBM by providing an in-depth analysis of mesenchymal marker expression and correlation with gene expression patterns ( 4 , 8 , 19–27 ). In support of our findings, a recent publication on the clonal evolution of posttreatment GBM also identified frequent gene expression subtype switching in recurrent tumors with the mesenchymal subtype being the most stable upon recurrence ( 28 ). This study also demonstrated a worse OS in tumors with a mesenchymal subtype at recurrence.
In aggregate, paired samples did not show a consistent pattern of protein expression changes in OLIG2, CD44, or YKL-40 at recurrence. Analysis of tumor subsets, however, revealed increased protein expression of mesenchymal markers specifically in the subset of tumors with initial low expression. Moreover, a few tumor pairs showed increased YKL-40 and reduced OLIG2 at recurrence, consistent with prior reports demonstrating a shift toward a more mesenchymal phenotype at recurrence with increased YKL-40 and/or CD44 expression and decreased OLIG2 expression ( 9 ). These data indicate that although a mesenchymal immunophenotypic transition can occur in recurrent posttreatment GBM, only a subset of tumors demonstrate such a transition, and knowledge of the protein expression level in the initial tumor is required as a point of reference to assess the changes at recurrence. Although GBM subtypes have been defined based on transcriptional analysis, several studies suggest protein level data can also be used to subtype tumors, at least at initial resection ( 17 , 18 ). Our data demonstrate that the correlation between mRNA and protein levels may be poor in some tumors. Several reasons may explain this discrepancy, including posttranscriptional regulation of protein levels, sensitivity of IHC, and mRNA or protein stability. Unexpectedly, we observed a drop in pSTAT3 in tumors that initially had high expression, which did not match the pattern seen for YKL-40 and CD44 ( Fig. 3D–F ). The significance of this finding is uncertain. STAT3 can act as an oncogene or a tumor suppressor depending on the mutational profile of a tumor ( 29 ). This phenomenon may account for some of the discrepancy, i.e. if a subset of tumors have a survival advantage by downregulating, rather than increasing, STAT3 activation upon recurrence ( 29 , 30 ).
NF1 mutations have been reported in both primary GBM and in IDH1 mutant (secondary) GBM arising from a lower-grade infiltrating glioma precursor ( 27 , 31 ). In the latter group, the sequence alterations in the NF1 gene suggest that the mutation may be a consequence of temozolomide therapy. Based on the association of NF1 alterations with the mesenchymal subtype (defined by expression profiling) and the possibility that NF1 alterations may contribute to tumor progression, we hypothesized that NF1 alterations could be a driving factor in the mesenchymal shift in recurrent GBM ( 5 ). However, we observed that none of the tumors in this series showed loss of neurofibromin at recurrence. Possible explanations for this could be the presence of newly acquired heterozygous mutations that may be functionally significant without altering protein expression levels. A recent study of genetic alterations in matched initial and posttreatment GBM identified loss of heterozygosity at the NF1 locus in recurrent tumors in 3 of 78 paired samples (3.8%), suggesting that the small sample size of our study was a limiting factor in detecting neurofibromin loss in recurrent tumors ( 28 ).
In agreement with our study, the frequency of homozygous NF1 disruption in primary GBM has been estimated at approximately 3%–5% ( 32 ). A recent clinicopathologic correlation study identified chromosomal losses at the NF1 locus in 6% of GBM, and NF1 loss correlated with high expression of the mesenchymal IHC marker podoplanin ( 33 ). The role of neurofibromin in mesenchymal GBM is currently unknown. In NF-associated neurofibromas, Schwann cells, and breast cancer cells in vitro, NF1 loss is associated with upregulation of 1 or more epithelial–mesenchymal transition (EMT)-related transcription factors including SNAI1, SNAI2, TWIST1, TWIST2, ZEB1, and ZEB2, providing a potential mechanism for NF1 involvement in a mesenchymal phenotype ( 34 ). Our data show a similar frequency of neurofibromin loss in recurrent GBM, and further demonstrate stability of this marker, at least at the protein level, across initial and recurrent posttreatment tumor pairs. To our knowledge, this study is the first application of the NFC antibody to GBM specimens and validates this tool as a useful marker for neurofibromin loss in GBM. The NFC antibody is positive in background endothelial and inflammatory cells. Some tumors showed a high degree of microglia/macrophage infiltration as determined by IBA1 immunostaining. Because inflammatory cells are positive for NF1 by IHC, dense inflammatory infiltrates could mask NF1 loss in the tumor cells Application of a macrophage/microglial immunohistochemical marker such as IBA1 can therefore be very helpful in interpreting the NF1 immunostain by highlighting the background macrophage/microglial population. The correlation of neurofibromin loss and sarcomatous/spindled morphology is intriguing but requires further study for validation. EMT-related transcription factors are upregulated in the sarcomatous component of gliosarcoma, which raises the intriguing possibility that this finding is driven by neurofibromin loss ( 35 ).
Several studies have evaluated Ki-67 labeling index as a prognostic marker in GBM, with variable and sometimes conflicting results. Most studies identified an increasing Ki-67 labeling index with increasing tumor grade, and a poorer prognosis in tumors with higher Ki-67 labeling. However, studies differ on the cutoff value, with ranges from 1.5% to 20% ( 36 ). In this cohort, a Ki-67 labeling index at initial diagnosis of ≥20% was associated with shorter PFS and OS. This cutoff was selected a priori to the survival analysis because it was the median labeling index value in the cohort. Using this cutoff, our data are consistent with the study of Reavey-Cantwell et al, which showed a 2.2-fold higher risk of death in GBM patients whose tumors had a labeling index of >20% ( 37 ). Our data support the use of Ki-67 labeling index in the context of other clinical parameters such as performance status, extent of resection, and radiographic evaluation, as was advocated in a review on this subject ( 36 ).
As for any retrospective study, this analysis has limitations. First, our cohort of paired samples was relatively small and tumor features associated with differences in survival need to be interpreted with caution until they are evaluated in a prospective manner. Second, GBM are heterogeneous tumors and the examined tissue cores may not be representative of the entire lesion. We partially mitigated this by examining up to 3 cores per specimen when possible. Third, the mesenchymal subtype of GBM has been defined by gene expression profiling. Thus, although our data support a shift to a more mesenchymal phenotype, we cannot conclude they fit within the mesenchymal transcriptional subtype of GBM ( 4 , 5 ). Finally, we detected 3 IDH1 R132H mutant tumors by IHC but did not perform sequencing studies to exclude less common IDH1 mutations, or mutations in IDH2 , in the IHC-negative cohort. Therefore, we cannot exclude the possibility that some of our IDH1 R132H -negative tumors harbored less common mutations in IDH1 or IDH2 .
Taken together, the data reported here provide evidence for a transition toward a mesenchymal phenotype in recurrent posttreatment GBM that is detectable by immunohistochemical staining of paired initial and posttreatment recurrent tumor specimens. This transition, defined in our study by protein expression as determined by IHC, was only partially captured by gene expression analysis on a panel of 8 proneural and mesenchymal genes. Given the significant degree of intratumoral transcriptional heterogeneity in GBM ( 38 ), protein expression data may provide some advantages as GBM subtyping continues to be refined. Furthermore, the examination of paired primary and posttreatment recurrent samples is critical to assess the stability of molecular subtype and response to therapy. These findings expand upon the growing body of literature on GBM evolution after treatment.
This study was supported by the University of California San Francisco Resident Research Fund, NIH/NINDS R01NS081117, and The Children’s Tumor Foundation NF1 LGG Synodos.
Conflict of interest: The authors have no duality or conflicts of interest to declare.
Supplementary Data can be found at http://www.jnen.oxfordjournals.org .
Supplementary Material
REFERENCES
- 1. Ostrom QT, Gittleman H, Fulop J , et al. . CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2008-2012 . Neuro-Oncology 2015. ; 17(suppl 4) : iv1 – iv62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ohgaki H, Kleihues P. Population-based studies on incidence, survival rates, and genetic alterations in astrocytic and oligodendroglial gliomas . J Neuropathol Exp Neurol 2005. ; 64 : 479 – 89 [DOI] [PubMed] [Google Scholar]
- 3. Furnari FB, Fenton T, Bachoo RM , et al. . Malignant astrocytic glioma: genetics, biology, and paths to treatment . Genes Dev 2007. ; 21 : 2683 – 710 [DOI] [PubMed] [Google Scholar]
- 4. Phillips HS, Kharbanda S, Chen R , et al. . Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis . Cancer Cell 2006. ; 9 : 157 – 73 [DOI] [PubMed] [Google Scholar]
- 5. Verhaak RGW, Hoadley KA, Purdom E , et al. . Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1 . Cancer Cell 2010. ; 17 : 98 – 110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brennan CW, Verhaak RGW, McKenna A , et al. . The somatic genomic landscape of glioblastoma . Cell 2013. ; 155 : 462 – 77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ceccarelli M, Barthel FP, Malta TM , et al. . Molecular profiling reveals biologically discrete subsets and pathways of progression in diffuse glioma . Cell 2016. ; 164 : 550 – 63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lai A, Kharbanda S, Pope WB , et al. . Evidence for sequenced molecular evolution of IDH1 mutant glioblastoma from a distinct cell of origin . J Clin Oncol 2011. ; 29 : 4482 – 90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bhat KPL, Balasubramaniyan V, Vaillant B , et al. . Mesenchymal differentiation mediated by NF-κB promotes radiation resistance in glioblastoma . Cancer Cell 2013. ; 24 : 331 – 46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kim S-H, Ezhilarasan R, Phillips E , et al. . Serine/threonine kinase MLK4 determines mesenchymal identity in glioma stem cells in an NF-κB-dependent manner . Cancer Cell 2016. ; 29 : 201 – 13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Patel AP, Tirosh I, Trombetta JJ , et al. . Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma . Science 2014. ; 344 : 1396 – 401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yan P, Seelentag W, Bachmann A , et al. . An agarose matrix facilitates sectioning of tissue microarray blocks . J Histochem Cytochem 2007. ; 55 : 21 – 4 [DOI] [PubMed] [Google Scholar]
- 13. Takami H, Yoshida A, Fukushima S , et al. . Revisiting TP53 mutations and immunohistochemistry–a comparative study in 157 diffuse gliomas . Brain Pathol 2015. ; 25 : 256 – 65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Reuss DE, Habel A, Hagenlocher C , et al. . Neurofibromin specific antibody differentiates malignant peripheral nerve sheath tumors (MPNST) from other spindle cell neoplasms . Acta Neuropathol 2014. ; 127 : 565 – 72 [DOI] [PubMed] [Google Scholar]
- 15. Chen L, Voronovich Z, Clark K , et al. . Predicting the likelihood of an isocitrate dehydrogenase 1 or 2 mutation in diagnoses of infiltrative glioma . Neurooncology 2014. ; 16 : 1478 – 83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Parsons DW, Jones S, Zhang X , et al. . An integrated genomic analysis of human glioblastoma multiforme . Science 2008. ; 321 : 1807 – 12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Conroy S, Kruyt FAE, Joseph JV , et al. . Subclassification of newly diagnosed glioblastomas through an immunohistochemical approach . PLoS ONE 2014. ; 9 : e115687 – 21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Le Mercier M, Hastir D, Moles Lopez X , et al. . A simplified approach for the molecular classification of glioblastomas . PLoS ONE 2012. ; 7 : e45475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kim J-H, Bae Kim Y, Han JH , et al. . Pathologic diagnosis of recurrent glioblastoma: morphologic, immunohistochemical, and molecular analysis of 20 paired cases . Am J Surg Pathol 2012. ; 36 : 620 – 8 [DOI] [PubMed] [Google Scholar]
- 20. Stark AM, Witzel P, Strege RJ , et al. . p53, mdm2, EGFR, and msh2 expression in paired initial and recurrent glioblastoma multiforme . J Neurol Neurosurg Psychiatr 2003. ; 74 : 779 – 83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Martinez R, Rohde V, Schackert G. Different molecular patterns in glioblastoma multiforme subtypes upon recurrence . J Neurooncol 2010. ; 96 : 321 – 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hulsebos TJM. Molecular-genetic characterisation of gliomas that recur as same grade or higher grade tumours . J Neurol Neurosurg Psychiatr 2004. ; 75 : 723 – 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Spiegl-Kreinecker S, Pirker C, Marosi C , et al. . Dynamics of chemosensitivity and chromosomal instability in recurrent glioblastoma . Br J Cancer 2007. ; 96 : 960 – 9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wiewrodt D, Nagel G, Dreimüller N , et al. . MGMT in primary and recurrent human glioblastomas after radiation and chemotherapy and comparison with p53 status and clinical outcome . Int J Cancer 2008. ; 122 : 1391 – 9 [DOI] [PubMed] [Google Scholar]
- 25. Kubelt C, Hattermann K, Sebens S , et al. . Epithelial-to-mesenchymal transition in paired human primary and recurrent glioblastomas . Int J Oncol 2015. ; 46 : 2515 – 25 [DOI] [PubMed] [Google Scholar]
- 26. Mahabir R, Tanino M, Elmansuri A , et al. . Sustained elevation of Snail promotes glial-mesenchymal transition after irradiation in malignant glioma . Neurooncology 2014. ; 16 : 671 – 85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Virk SM, Gibson RM, Quinones-Mateu ME , et al. . Identification of variants in primary and recurrent glioblastoma using a cancer-specific gene panel and whole exome sequencing . PLoS ONE 2015. ; 10 : e0124178 – 10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wang J, Cazzato E, Ladewig E , et al. . Clonal evolution of glioblastoma under therapy . Nat Genet 2016. ; 48 : 768 – 76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. la Iglesia de N, Konopka G, Puram SV , et al. . Identification of a PTEN-regulated STAT3 brain tumor suppressor pathway . Genes Dev 2008. ; 22 : 449 – 62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Luwor RB, Stylli SS, Kaye AH. The role of Stat3 in glioblastoma multiforme . J Clin Neurosci 2013. ; 20 : 907 – 11 [DOI] [PubMed] [Google Scholar]
- 31. Johnson BE, Mazor T, Hong C , et al. . Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma . Science 2014. ; 343 : 189 – 93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McGillicuddy LT, Fromm JA, Hollstein PE , et al. . Proteasomal and genetic inactivation of the NF1 tumor suppressor in gliomagenesis . Cancer Cell 2009. ; 16 : 44 – 54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vizcaíno MA, Shah S, Eberhart CG , et al. . Clinicopathologic implications of NF1 gene alterations in diffuse gliomas . Hum Pathol 2015. ; 46 : 1323 – 30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Arima Y, Hayashi H, Kamata K , et al. . Decreased expression of neurofibromin contributes to epithelial-mesenchymal transition in neurofibromatosis type 1 . Exp Derm 2009. ; 19 : e136 – 41 [DOI] [PubMed] [Google Scholar]
- 35. Nagaishi M, Paulus W, Brokinkel B , et al. . Transcriptional factors for epithelial-mesenchymal transition are associated with mesenchymal differentiation in gliosarcoma . Brain Pathol 2012. ; 22 : 670 – 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Johannessen AL, Torp SH. The clinical value of Ki-67/MIB-1 labeling index in human astrocytomas . Pathol Oncol Res 2006. ; 12 : 143 – 7 [DOI] [PubMed] [Google Scholar]
- 37. Reavey-Cantwell JF, Haroun RI, Zahurak M , et al. . The prognostic value of tumor markers in patients with glioblastoma multiforme: analysis of 32 patients and review of the literature . J Neurooncol 2001. ; 55 : 195 – 204 [DOI] [PubMed] [Google Scholar]
- 38. Patel AP, Tirosh I, Trombetta JJ , et al. . Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma . Science 2014. ; 344 : 1396 – 401 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.