The title molecule is substantially twisted, with a dihedral angle of 43.70 (2)° between the 2-(methylsulfanyl)thiophene and 4-methoxyphenyl rings. In the crystal, molecules are linked through C—H⋯O interactions, forming a bifurcated layer stacking along the b-axis direction enclosing  (10) ring motifs.
(10) ring motifs.
Keywords: crystal structure, thiophene, Hirschfeld surface
Abstract
The title compound, C13H12O2S2, crystallizes in the triclinic space group P
 . The molecular structure is substantially twisted, with a dihedral angle of 43.70 (2)° between the 2-(methylsulfanyl)thiophene and 4-methoxyphenyl rings. In the crystal, molecules are linked through C—H⋯O interactions and form a bifurcated layer stacking along the b-axis direction and enclosing R
2
2(10) ring motifs. The phenyl rings are involved in π–π interactions with a centroid–centroid separation of 3.760 (2) Å. The Hirshfeld surfaces were studied and the contributions of the various intermolecular interactions were quantified.
. The molecular structure is substantially twisted, with a dihedral angle of 43.70 (2)° between the 2-(methylsulfanyl)thiophene and 4-methoxyphenyl rings. In the crystal, molecules are linked through C—H⋯O interactions and form a bifurcated layer stacking along the b-axis direction and enclosing R
2
2(10) ring motifs. The phenyl rings are involved in π–π interactions with a centroid–centroid separation of 3.760 (2) Å. The Hirshfeld surfaces were studied and the contributions of the various intermolecular interactions were quantified.
Chemical context
Thiophenes are five-membered sulfur-containing heterocyclic compounds with important applications in areas such as agrochemistry, pharmaceuticals, molecular electronics, liquid crystalline materials and corrosion inhibition. Thiophenes are also important building blocks in organic synthesis. Their aromatic character gives enough stabilization to allow the manipulation of various substituents (Mishra et al., 2011 ▸). α-Oxoketene thioacetals are powerful building blocks for the synthesis of numerous heterocyclic scaffolds, where the carbonyl carbon generally provides hard centers and the carbon-bearing methylsulfanyl group acts as a soft electrophilic center (Junjappa et al., 1990 ▸). This synthetic building block was used for the synthesis of (4-methoxyphenyl) [2-(methylsulfanyl)thiophen-3-yl]methanone (Pradeepa Kumara et al., 2016 ▸). 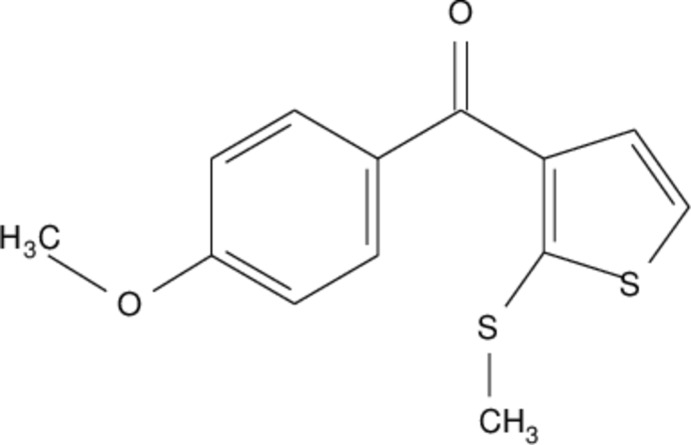
Structural commentary
In the title compound, the 2-(methylsulfanyl)thiophene and 4-methoxyphenyl aromatic rings are connected via a C(=O)—C methanone bridge (Fig. 1 ▸). The compound is substantially twisted about the methanone bridge as indicated by the dihedral angle of 43.70 (2)° between the thiophene (S1/C1/C5/C7/C10) and phenyl (C2–C6/C11/C13) rings. The methyl group at S2 is in a +syn-periplanar conformation, as indicated by the C8—S2—C10—S1 torsion angle of 6.09 (16)°. However, in the related compound (4-fluorophenyl)[2-(methylsulfanyl)thiophen-3-yl]methanone (Nagaraju et al., 2018 ▸), this group is in a -syn-periplanar conformation with a torsion angle of −1.7 (2)°. Atom C12 adopts a nearly trigonal geometry, as indicated by the bond angles C7–C12–O2 = 119.5 (2)°, O2–C12–C4 = 119.2 (2)° and C4—C12—C7 = 121.3 (2)°. The methoxy group attached at C11 is in a −anti-periplanar conformation [C3—C11—O1—C9 = −176.9 (2)°]. The bond lengths and angles are normal (Sreenatha et al., 2017 ▸; Rajni Swamy et al., 2014 ▸; Gopinath et al., 2016 ▸).
Figure 1.
Molecular structure of the title compound, showing the atom-numbering scheme and displacement ellipsoids drawn at the 50% probability level.
Supramolecular features
The crystal structure features intermolecular hydrogen-bonding interactions of the type C9—H9A⋯O2 (Fig. 2 ▸, Table 1 ▸) and displays a bifurcated layer stacking along the b-axis direction through C6—H6⋯O2 interactions, which link inversion-related molecules into dimers enclosing an  (10) ring motif. π–π stacking interactions are also observed between the phenyl rings (centroid Cg) of adjacent molecules [Cg⋯ Cg(2 − x, −y, 1 − z) = 3.760 (2) Å]. The packing of the title compound is illustrated in Fig. 3 ▸.
(10) ring motif. π–π stacking interactions are also observed between the phenyl rings (centroid Cg) of adjacent molecules [Cg⋯ Cg(2 − x, −y, 1 − z) = 3.760 (2) Å]. The packing of the title compound is illustrated in Fig. 3 ▸.
Figure 2.
The  (10) ring motif formed via intermolecular C6—H6⋯O2 hydrogen bonds (Table 1 ▸). The π–π interactions are also shown.
(10) ring motif formed via intermolecular C6—H6⋯O2 hydrogen bonds (Table 1 ▸). The π–π interactions are also shown.
Table 1. Hydrogen-bond geometry (Å, °).
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| C6—H6⋯O2i | 0.93 | 2.48 | 3.374 (4) | 161 |
| C9—H9A⋯O2ii | 0.96 | 2.45 | 3.400 (4) | 172 |
Symmetry codes: (i)  ; (ii)
; (ii)  .
.
Figure 3.
Packing for of the title compound viewed along the b axis.
Hirshfeld surfaces and 2D fingerprint plots
Hirshfeld surface (HS) analysis and the associated fingerprint plots (FP) (CrystalExplorer; Spackman & Jayatilaka, 2009 ▸) are useful tools for visualizing the types of intermolecular interactions present in a crystal structure and quantify their percentage contributions to the crystal packing. The 3D HS mapped over d norm between −0.2106 a.u (blue) and 1.2279 a.u (red) is shown in Fig. 4 ▸. The area and volume of the HS are 287.29 Å2 and 305.24 Å3, respectively. The deep-red spots on the d norm surface are due to the presence of intermolecular C—H⋯O interactions (Sreenatha et al., 2018 ▸). The 2D FP analysis (Fig. 5 ▸) shows that the H⋯H contacts make the highest contribution (39.3%) followed by the H⋯C/C⋯H contacts (20.1%), which are seen as a pair of blunt spikes in the region 1.2 Å < (d i + d e) < 1.75 Å. The H⋯S/S⋯H contacts make a contribution of 16.9% and appear as butterfly wings in the region 1.2 Å < (d i + d e) < 1.9 Å. The pair of sharp spikes is observed in the region 1.2 Å < (d i + d e) < 1.32 Å is due to the presence of H⋯O/O⋯H contacts (15.6% contribution). The C⋯C contacts (3.4% contribution) are visible as wings in almost the same region, 1.7 Å < (d i + d e) < 1.72 Å. The relative contributions of all the contacts to the Hirshfeld surface are depicted in Fig. 6 ▸.
Figure 4.
Hirshfeld surface for the title compound mapped over d norm in the range −0.2106 to 1.2279 a.u. highlighting the C—H⋯O intermolecular interactions.
Figure 5.
The full two-dimensional fingerprint plots for the title compound, showing (a) all interactions, and delineated into (b) H⋯H, (c) H⋯C/C⋯H, (d) H⋯S/S⋯H, (e) H⋯O/O⋯H, (f) C⋯C, (g) S⋯C/C⋯S and (h) S⋯S interactions. The d i and d e values are the closest internal and external distances (in Å) from given points on the Hirshfeld surface.
Figure 6.
The relative contributions (%) to the Hirshfeld surface for the various contacts.
Database survey
A search for thiophene derivatives was carried out in the Cambridge Structural Database (CSD, Version 5.39, update of February 2018; Groom et al., 2016 ▸). The most relevant compounds are 5-[bis(4-ethoxyphenyl)amino]thiophene-2-carbaldehyde (HOJCIU; Tan et al., 2014 ▸) and 2-[4-(benzyloxy)phenyl]-5-(3,4-dimethoxyphenyl)-3, 4-dimethylthiophene (ACETEI; Shi et al., 2004 ▸), which are both non-planar. In ethyl 4-acetyl-5-anilino-3-methylthiophene-2-carboxylate (AFIGIH; Mabkhot et al., 2013 ▸), the thiophene and phenyl rings make a dihedral angle of 36.81 (10)°.
Synthesis and crystallization
To α-oxoketene dithioacetal (0.1 mol) and 1,4-dithiane-2,5-diol (0.05 mol) in dry ethanol (10 mL), anhydrous potassium carbonate (0.12 mol) was added. The reaction mixture was refluxed on a water bath for 30 minutes (the condenser being protected by a calcium chloride guard tube). After completion of the reaction (monitored by TLC), the catalyst was filtered off and washed with fresh ethanol. The combined ethanol solution was removed on a rotary evaporator to obtain a viscous liquid. The crude product was purified by column chromatography using silica gel with 5% ethyl acetate and petroleum ether to yield the title compound as a yellow solid product, which was recrystallized from dichloromethane solution. M.p. 489–493 K. IR (KBr) νmax = 3449, 3079, 2923, 2841, 1772, 1600, 1493, 1253, 1167, 1015, 842, 694, 550 cm−1. 1H NMR (300 MHz, CDCl3): 7.79–7.77 (m, 2 H), 7.27–7.25 (m, 1H), 7.16–7.14 (m, 1H), 6.9–6.93 (m, 2H), 3.86 (s, 3H), 2.58 (s, 3H) ppm. 13 C NMR (75 MHz, CDCl3): 188.86, 162.73, 151.33, 135.36, 131.60, 131.47, 130.24, 130.59, 122.02, 113.44, 55.37, 18.06. HRMS (ESI): calculated for C13H12O2S2 [M + H]+ 265.0312; found 265.0407.
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 2 ▸. All hydrogen atoms were placed at calculated positions and refined using a riding model with C—H = 0.93 Å and U iso(H) = 1.2U eq(C) for aromatic ring atoms and with C—H = 0.96 Å with U iso(H) = 1.5U eq(C) for methyl groups.
Table 2. Experimental details.
| Crystal data | |
| Chemical formula | C13H12O2S2 |
| M r | 264.35 |
| Crystal system, space group | Triclinic, P

|
| Temperature (K) | 293 |
| a, b, c (Å) | 7.806 (4), 8.263 (3), 10.414 (6) |
| α, β, γ (°) | 97.260 (11), 109.65 (2), 93.79 (2) |
| V (Å3) | 623.3 (5) |
| Z | 2 |
| Radiation type | Mo Kα |
| μ (mm−1) | 0.41 |
| Crystal size (mm) | 0.30 × 0.26 × 0.20 |
| Data collection | |
| Diffractometer | Bruker APEX |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 2924, 2165, 1899 |
| R int | 0.109 |
| (sin θ/λ)max (Å−1) | 0.595 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.046, 0.128, 1.09 |
| No. of reflections | 2165 |
| No. of parameters | 157 |
| H-atom treatment | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 0.37, −0.33 |
Supplementary Material
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S2056989018016043/vm2213sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989018016043/vm2213Isup2.hkl
CCDC reference: 1871776
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
The authors are thankful to the Department of Physics, University of Mysore, and Adichuchanagiri Institute of Technology, Chikkamagaluru, Karnataka for support
supplementary crystallographic information
Crystal data
| C13H12O2S2 | Z = 2 |
| Mr = 264.35 | F(000) = 276 |
| Triclinic, P1 | Dx = 1.409 Mg m−3 |
| a = 7.806 (4) Å | Mo Kα radiation, λ = 0.71073 Å |
| b = 8.263 (3) Å | Cell parameters from 2924 reflections |
| c = 10.414 (6) Å | θ = 3.5–25.0° |
| α = 97.260 (11)° | µ = 0.41 mm−1 |
| β = 109.65 (2)° | T = 293 K |
| γ = 93.79 (2)° | Block, colourless |
| V = 623.3 (5) Å3 | 0.30 × 0.26 × 0.20 mm |
Data collection
| Bruker APEX diffractometer | 1899 reflections with I > 2σ(I) |
| Radiation source: graphite | Rint = 0.109 |
| Detector resolution: 0.894 pixels mm-1 | θmax = 25.0°, θmin = 3.5° |
| SAINT (Bruker, 2006) [not correct; type of scans needed] | h = −9→9 |
| 2924 measured reflections | k = −9→9 |
| 2165 independent reflections | l = −12→11 |
Refinement
| Refinement on F2 | Hydrogen site location: inferred from neighbouring sites |
| Least-squares matrix: full | H-atom parameters constrained |
| R[F2 > 2σ(F2)] = 0.046 | w = 1/[σ2(Fo2) + (0.072P)2 + 0.1135P] where P = (Fo2 + 2Fc2)/3 |
| wR(F2) = 0.128 | (Δ/σ)max < 0.001 |
| S = 1.09 | Δρmax = 0.37 e Å−3 |
| 2165 reflections | Δρmin = −0.33 e Å−3 |
| 157 parameters | Extinction correction: SHELXL2018 (Sheldrick, 2015), Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| 0 restraints | Extinction coefficient: 0.060 (18) |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| S1 | 0.77364 (8) | 0.32143 (7) | −0.09953 (6) | 0.0412 (3) | |
| S2 | 0.73606 (8) | 0.60555 (7) | 0.10012 (6) | 0.0422 (3) | |
| O1 | 0.7606 (3) | −0.1498 (2) | 0.62854 (18) | 0.0582 (5) | |
| O2 | 0.7950 (3) | 0.4642 (2) | 0.33504 (18) | 0.0601 (5) | |
| C1 | 0.8084 (3) | 0.1302 (3) | −0.0527 (3) | 0.0450 (6) | |
| H1 | 0.820956 | 0.039871 | −0.110524 | 0.054* | |
| C2 | 0.6715 (3) | 0.0359 (3) | 0.3139 (2) | 0.0419 (6) | |
| H2 | 0.607492 | 0.011357 | 0.219380 | 0.050* | |
| C3 | 0.8621 (3) | 0.1072 (3) | 0.5930 (2) | 0.0437 (6) | |
| H3 | 0.927331 | 0.131023 | 0.687398 | 0.052* | |
| C4 | 0.7766 (3) | 0.1880 (3) | 0.3679 (2) | 0.0361 (5) | |
| C5 | 0.8153 (3) | 0.1275 (3) | 0.0773 (2) | 0.0402 (5) | |
| H5 | 0.834586 | 0.034541 | 0.119774 | 0.048* | |
| C6 | 0.8701 (3) | 0.2215 (3) | 0.5107 (2) | 0.0407 (5) | |
| H6 | 0.938609 | 0.323029 | 0.550038 | 0.049* | |
| C7 | 0.7900 (3) | 0.2813 (3) | 0.1446 (2) | 0.0352 (5) | |
| C8 | 0.6913 (4) | 0.6721 (3) | −0.0642 (3) | 0.0525 (6) | |
| H8A | 0.594501 | 0.598579 | −0.133310 | 0.079* | |
| H8B | 0.800026 | 0.672480 | −0.087893 | 0.079* | |
| H8C | 0.655487 | 0.781032 | −0.059371 | 0.079* | |
| C9 | 0.6614 (5) | −0.3123 (4) | 0.5746 (3) | 0.0726 (9) | |
| H9A | 0.704580 | −0.365458 | 0.505805 | 0.109* | |
| H9B | 0.533040 | −0.303439 | 0.533903 | 0.109* | |
| H9C | 0.681017 | −0.375822 | 0.648323 | 0.109* | |
| C10 | 0.7680 (3) | 0.4010 (3) | 0.0595 (2) | 0.0341 (5) | |
| C11 | 0.7571 (3) | −0.0450 (3) | 0.5371 (2) | 0.0426 (5) | |
| C12 | 0.7881 (3) | 0.3201 (3) | 0.2846 (2) | 0.0397 (5) | |
| C13 | 0.6597 (3) | −0.0792 (3) | 0.3964 (2) | 0.0459 (6) | |
| H13 | 0.586967 | −0.179094 | 0.358019 | 0.055* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| S1 | 0.0427 (4) | 0.0471 (4) | 0.0361 (4) | 0.0025 (3) | 0.0176 (3) | 0.0050 (3) |
| S2 | 0.0402 (4) | 0.0346 (4) | 0.0488 (4) | −0.0026 (2) | 0.0148 (3) | 0.0010 (2) |
| O1 | 0.0729 (13) | 0.0652 (12) | 0.0423 (10) | 0.0013 (10) | 0.0272 (9) | 0.0124 (8) |
| O2 | 0.0870 (15) | 0.0460 (10) | 0.0429 (10) | 0.0040 (9) | 0.0214 (9) | −0.0041 (8) |
| C1 | 0.0512 (14) | 0.0421 (13) | 0.0453 (13) | 0.0072 (10) | 0.0235 (11) | 0.0008 (10) |
| C2 | 0.0356 (12) | 0.0535 (14) | 0.0302 (10) | −0.0047 (10) | 0.0073 (9) | −0.0002 (9) |
| C3 | 0.0400 (12) | 0.0615 (15) | 0.0258 (10) | 0.0005 (11) | 0.0106 (9) | −0.0013 (10) |
| C4 | 0.0325 (11) | 0.0448 (12) | 0.0296 (10) | −0.0001 (9) | 0.0115 (8) | 0.0009 (9) |
| C5 | 0.0409 (12) | 0.0393 (12) | 0.0421 (12) | 0.0079 (9) | 0.0157 (10) | 0.0067 (9) |
| C6 | 0.0375 (12) | 0.0484 (13) | 0.0311 (11) | −0.0036 (10) | 0.0110 (9) | −0.0048 (9) |
| C7 | 0.0290 (10) | 0.0398 (11) | 0.0335 (11) | 0.0008 (8) | 0.0089 (8) | 0.0007 (9) |
| C8 | 0.0504 (15) | 0.0444 (13) | 0.0623 (16) | −0.0009 (11) | 0.0170 (12) | 0.0173 (12) |
| C9 | 0.108 (3) | 0.0627 (18) | 0.0627 (18) | −0.0042 (17) | 0.0516 (18) | 0.0122 (14) |
| C10 | 0.0243 (10) | 0.0389 (11) | 0.0356 (11) | −0.0033 (8) | 0.0096 (8) | −0.0005 (9) |
| C11 | 0.0407 (13) | 0.0548 (14) | 0.0372 (11) | 0.0036 (11) | 0.0204 (9) | 0.0068 (10) |
| C12 | 0.0351 (12) | 0.0443 (12) | 0.0336 (11) | −0.0001 (9) | 0.0080 (9) | −0.0026 (9) |
| C13 | 0.0409 (13) | 0.0528 (14) | 0.0387 (12) | −0.0097 (11) | 0.0129 (10) | −0.0014 (10) |
Geometric parameters (Å, º)
| S1—C10 | 1.719 (2) | C4—C6 | 1.400 (3) |
| S1—C1 | 1.724 (3) | C4—C12 | 1.494 (3) |
| S2—C10 | 1.744 (2) | C5—C7 | 1.430 (3) |
| S2—C8 | 1.793 (3) | C5—H5 | 0.9300 |
| O1—C11 | 1.360 (3) | C6—H6 | 0.9300 |
| O1—C9 | 1.448 (4) | C7—C10 | 1.391 (3) |
| O2—C12 | 1.230 (3) | C7—C12 | 1.458 (3) |
| C1—C5 | 1.340 (4) | C8—H8A | 0.9600 |
| C1—H1 | 0.9300 | C8—H8B | 0.9600 |
| C2—C13 | 1.379 (3) | C8—H8C | 0.9600 |
| C2—C4 | 1.394 (3) | C9—H9A | 0.9600 |
| C2—H2 | 0.9300 | C9—H9B | 0.9600 |
| C3—C6 | 1.365 (3) | C9—H9C | 0.9600 |
| C3—C11 | 1.397 (4) | C11—C13 | 1.387 (3) |
| C3—H3 | 0.9300 | C13—H13 | 0.9300 |
| C10—S1—C1 | 92.19 (11) | S2—C8—H8A | 109.5 |
| C10—S2—C8 | 100.76 (12) | S2—C8—H8B | 109.5 |
| C11—O1—C9 | 117.8 (2) | H8A—C8—H8B | 109.5 |
| C5—C1—S1 | 111.72 (18) | S2—C8—H8C | 109.5 |
| C5—C1—H1 | 124.1 | H8A—C8—H8C | 109.5 |
| S1—C1—H1 | 124.1 | H8B—C8—H8C | 109.5 |
| C13—C2—C4 | 121.9 (2) | O1—C9—H9A | 109.5 |
| C13—C2—H2 | 119.0 | O1—C9—H9B | 109.5 |
| C4—C2—H2 | 119.0 | H9A—C9—H9B | 109.5 |
| C6—C3—C11 | 120.7 (2) | O1—C9—H9C | 109.5 |
| C6—C3—H3 | 119.6 | H9A—C9—H9C | 109.5 |
| C11—C3—H3 | 119.6 | H9B—C9—H9C | 109.5 |
| C2—C4—C6 | 117.6 (2) | C7—C10—S1 | 110.79 (16) |
| C2—C4—C12 | 124.3 (2) | C7—C10—S2 | 127.08 (17) |
| C6—C4—C12 | 118.1 (2) | S1—C10—S2 | 122.13 (14) |
| C1—C5—C7 | 113.6 (2) | O1—C11—C13 | 125.0 (2) |
| C1—C5—H5 | 123.2 | O1—C11—C3 | 115.7 (2) |
| C7—C5—H5 | 123.2 | C13—C11—C3 | 119.3 (2) |
| C3—C6—C4 | 121.0 (2) | O2—C12—C7 | 119.5 (2) |
| C3—C6—H6 | 119.5 | O2—C12—C4 | 119.2 (2) |
| C4—C6—H6 | 119.5 | C7—C12—C4 | 121.33 (19) |
| C10—C7—C5 | 111.7 (2) | C2—C13—C11 | 119.5 (2) |
| C10—C7—C12 | 120.6 (2) | C2—C13—H13 | 120.3 |
| C5—C7—C12 | 127.7 (2) | C11—C13—H13 | 120.3 |
| C10—S1—C1—C5 | 0.0 (2) | C8—S2—C10—S1 | 6.09 (16) |
| C13—C2—C4—C6 | −0.5 (4) | C9—O1—C11—C13 | 2.7 (4) |
| C13—C2—C4—C12 | −177.3 (2) | C9—O1—C11—C3 | −176.9 (2) |
| S1—C1—C5—C7 | 0.8 (3) | C6—C3—C11—O1 | 179.4 (2) |
| C11—C3—C6—C4 | −1.4 (4) | C6—C3—C11—C13 | −0.2 (4) |
| C2—C4—C6—C3 | 1.7 (3) | C10—C7—C12—O2 | −11.1 (3) |
| C12—C4—C6—C3 | 178.7 (2) | C5—C7—C12—O2 | 168.1 (2) |
| C1—C5—C7—C10 | −1.4 (3) | C10—C7—C12—C4 | 168.49 (19) |
| C1—C5—C7—C12 | 179.4 (2) | C5—C7—C12—C4 | −12.4 (4) |
| C5—C7—C10—S1 | 1.3 (2) | C2—C4—C12—O2 | 141.9 (3) |
| C12—C7—C10—S1 | −179.42 (16) | C6—C4—C12—O2 | −34.8 (3) |
| C5—C7—C10—S2 | −179.26 (16) | C2—C4—C12—C7 | −37.6 (3) |
| C12—C7—C10—S2 | 0.0 (3) | C6—C4—C12—C7 | 145.6 (2) |
| C1—S1—C10—C7 | −0.78 (18) | C4—C2—C13—C11 | −1.1 (4) |
| C1—S1—C10—S2 | 179.77 (14) | O1—C11—C13—C2 | −178.2 (2) |
| C8—S2—C10—C7 | −173.3 (2) | C3—C11—C13—C2 | 1.4 (4) |
Hydrogen-bond geometry (Å, º)
| D—H···A | D—H | H···A | D···A | D—H···A |
| C6—H6···O2i | 0.93 | 2.48 | 3.374 (4) | 161 |
| C9—H9A···O2ii | 0.96 | 2.45 | 3.400 (4) | 172 |
Symmetry codes: (i) −x+2, −y+1, −z+1; (ii) x, y−1, z.
References
- Bruker (2006). APEX2 and SAINT. Bruker AXS Inc., Madison, Wisconsin, USA.
- Gopinath, S., Sethusankar, K., Stoeckli-Evans, H., Rafiq, M. & Mohanakrishnan, A. K. (2016). Acta Cryst. E72, 1310–1314. [DOI] [PMC free article] [PubMed]
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Junjappa, H., Ila, H. & Asokan, C. V. (1990). Tetrahedron, 46, 5423–5506.
- Mabkhot, Y. N., Alatibi, F., Barakat, A., Choudhary, M. I. & Yousuf, S. (2013). Acta Cryst. E69, o1049. [DOI] [PMC free article] [PubMed]
- Macrae, C. F., Bruno, I. J., Chisholm, J. A., Edgington, P. R., McCabe, P., Pidcock, E., Rodriguez-Monge, L., Taylor, R., van de Streek, J. & Wood, P. A. (2008). J. Appl. Cryst. 41, 466–470.
- Mishra, R., Jha, K. K., Kumar, S. & Isha, T. (2011). Pharma Chem 3, 38–54.
- Nagaraju, S., Sridhar, M. A., Sreenatha, N. R., Pradeepa Kumara, C. S. & Sadashiva, M. P. (2018). X-ray Struct. Anal. Online, 34, 13–14.
- Pradeepa Kumara, C. S., Byre Gowda, G., Vinay Kumar, K. S., Ramesh, N., Sadashiva, M. P. & Junjappa, H. (2016). Tetrahedron Lett. 57, 4302–4305.
- Rajni Swamy, V., Gunasekaran, P., Krishnakumar, R. V., Srinivasan, N. & Müller, P. (2014). Acta Cryst. E70, o974–o975. [DOI] [PMC free article] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sheldrick, G. M. (2015). Acta Cryst C71, 3–8.
- Shi, J.-X., Zheng, X.-F., Zhu, K., Lei, Y.-J. & Shi, J.-G. (2004). Acta Cryst. E60, o1977–o1978.
- Spackman, M. A. & Jayatilaka, D. (2009). CrystEngComm, 11, 19–32.
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Sreenatha, N. R., Lakshminarayana, B. N., Ganesha, D. P., Vijayshankar, S. & Nagaraju, S. (2018). X-ray Struct. Anal. Online, 34, 23–24.
- Sreenatha, N. R., Lakshminarayana, B. N., Madan Kumar, S., Mahadeva Prasad, T. N. K. S., Kiran, D., Vijayshankar, S. & Byrappa, K. (2017). Chem. Data Collections, 11, 131–138.
- Tan, J.-Y., Kong, M. & Wu, J.-Y. (2014). Acta Cryst. E70, o1075–o1076. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) global, I. DOI: 10.1107/S2056989018016043/vm2213sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989018016043/vm2213Isup2.hkl
CCDC reference: 1871776
Additional supporting information: crystallographic information; 3D view; checkCIF report








