The molecule has mirror symmetry with the all non-H atoms, except tert-butyl groups, located on the mirror plane. An intramolecular O—H⋯N hydrogen bond forms an S(6) ring motif. In the crystal, the molecules are connected by C—H⋯π interactions
Keywords: crystal structure, Schiff bases, 3-chloro-4-methylphenylimino, Hirshfeld surface
Abstract
The title Schiff base compound, C22H28ClNO, shows mirror symmetry with all its non-H atoms, except the tert-butyl groups, located on the mirror plane. There is an intramolecular O—H⋯N hydrogen bond present forming an S(6) ring motif. In the crystal, the molecules are connected by C—H⋯π interactions, generating a three-dimensional supramolecular structure. Hirshfeld surface analysis and two dimensional fingerprint plots were used to analyse the intermolecular interactions present in the crystal, indicating that the most important contributions for the crystal packing are from H⋯H (68.9%) and C⋯H/H⋯C (11.7%) interactions.
Chemical context
In coordination chemistry, Schiff bases have found wide use as ligands (Calligaris et al., 1972 ▸; Hökelek et al., 2004 ▸; Moroz et al., 2012 ▸; Kansiz et al., 2018 ▸). Schiff bases are important for various areas of chemistry and biochemistry because of their biological activity (El-masry et al., 2000 ▸) and photochromic properties and have applications in various fields such as the measurement and control of radiation intensities in imaging systems and optical computers (Elmalı et al., 1999 ▸), electronics, optoelectronics and photonics (Iwan et al., 2007 ▸). They have been used as starting materials in the synthesis of many important medicinal substances. In the present study, a new Schiff base compound was synthesized and its crystal structure determined by X-ray diffraction. In addition, to understand the intermolecular interactions in the crystal structure, Hirshfeld surface analysis was performed.
Structural commentary
The molecular structure of the title compound is illustrated in Fig. 1 ▸. The title Schiff base compound shows mirror symmetry with all the non-H atoms, except the tert-butyl groups, located on the mirror plane. The C14—O1 bond distance is 1.349 (3) Å, the C8=N1 and C5—N1 bond lengths are 1.278 (4) and 1.412 (4) Å, respectively, and the C7—Cl1 bond distance is 1.744 (3) Å. There is an intramolecular O—H⋯N hydrogen bond present (Table 1 ▸), forming an S(6) ring motif.
Figure 1.
The molecular structure of the title compound, showing the atom labelling. Displacement ellipsoids are drawn at the 10% probability level. Symmetry code: (i) x, −y + 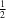 , z.
, z.
Table 1. Hydrogen-bond geometry (Å, °).
Cg is the centroid of the C9–C14 benzene ring.
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| O1—H1⋯N1 | 0.82 | 1.84 | 2.582 (3) | 149 |
| C1—H1B⋯Cg1i | 0.96 | 2.77 | 3.5072 (4) | 134 |
Symmetry code: (i)  .
.
Supramolecular features
In the crystal, the molecules are connected by C1—H1B⋯π interactions, generating a three-dimensional supramolecular structure (Table 1 ▸ and Fig. 2 ▸).
Figure 2.
A view of the crystal packing of the title compound. Dashed lines denote the intramolecular and intermolecular hydrogen bonds.
Database survey
There are no previous reports of the title structure. However, several related structure have been reported (CSD, version 5.39, update May 2018; Groom et al., 2016 ▸), including bis{(E)-1-[(3-chloro-4-methylphenylimino)methyl]naphthalen-2-olate-N,O}copper(II) (SICXOU; Toprak et al., 2018 ▸), 2-{(E)-[(3-chloro-4-methylphenyl)imino]methyl}-4-(trifluoromethoxy)phenol (TERTUI; Atalay et al., 2017 ▸), {2,2′-[4-chloro-5-methyl-o-phenylenebis(nitrilomethylidyne)]diphenolato}nickel(II) (WABDEK; Wang, 2010 ▸) and 4-[(E)-(3-chloro-4-methylphenyl)iminomethyl]-2-methoxy-3-nitrophenyl acetate (GAPPOE; Su et al., 2012 ▸). In all four compounds, the C—Cl bond lengths vary from 1.724 to 1.743 Å. In the title compound, the C7—Cl1 bond length is 1.744 (3) Å.
Hirshfeld surface analysis
The Hirshfeld surface analysis was performed using CrystalExplorer (Turner et al., 2017 ▸). The Hirshfeld surfaces and their associated two-dimensional fingerprint plots were used to quantify the various intermolecular interactions in the synthesized complex. The Hirshfeld surfaces mapped over d norm, d e and d i are illustrated in Figs. 3 ▸ and 4 ▸. The red spots on the surfaces indicate the intermolecular contacts involved in strong hydrogen bonds and interatomic contacts (Şen et al., 2017 ▸; Kansız & Dege, 2018 ▸; Sen et al., 2018 ▸; Gumus et al., 2018 ▸). The Hirshfeld surfaces were calculated using a standard (high) surface resolution with the three-dimensional d norm surfaces mapped over a fixed colour scale of −0.031 (red) to 2.139 (blue) a.u. The red spots identified in Fig. 3 ▸ correspond to the near-type H⋯π contacts resulting from the C—H⋯π interactions (Table 1 ▸).
Figure 3.
The Hirshfeld surface of the title compound mapped over d norm, d i and d e.
Figure 4.
Hirshfeld surface mapped over d norm for visualizing the intermolecular interactions of the title compound.
Fig. 5 ▸ shows the two-dimensional fingerprint of the sum of the contacts contributing to the Hirshfeld surface represented in normal mode. The graph shown in Fig. 6 ▸(a) (H⋯H) shows the two-dimensional fingerprint of the (d i, d e) points associated with hydrogen atoms. It is characterized by an end point that points to the origin and corresponds to d i = d e = 1.08 Å, which indicates the presence of the H⋯H contacts in this study (68.9%). The graph shown in Fig. 6 ▸(b) shows the (C⋯H/H⋯C) contacts between the carbon atoms inside the surface and the hydrogen atoms outside the Hirshfeld surface and vice versa, which contribute 11.7%. There are two symmetrical wings on the left and right sides. Furthermore, there are also Cl⋯H/H⋯Cl (11%), C⋯C (4.5%), C⋯N/N⋯C (2.2%), O⋯H/H⋯O (1.3%) and N⋯H/H⋯N (0.4%) contacts.
Figure 5.
Overall fingerprint plot for the title compound.
Figure 6.
Two-dimensional fingerprint plots with a d norm view of the (a) H⋯H (68.9%), (b) C⋯H/H⋯C (11.7%), (c) Cl⋯H/H⋯Cl (11%) and (d) C⋯C (4.5%) contacts in the title compound.
A view of the three-dimensional Hirshfeld surface of the title compound plotted over electrostatic potential energy in the range −0.030 to 0.044 a.u. using the STO-3G basis set at the Hartree–Fock level of theory is shown in Fig. 7 ▸ where the C—H⋯π donors and acceptors are shown as blue and red areas around the atoms related with positive (hydrogen-bond donors) and negative (hydrogen-bond acceptors) electrostatic potentials, respectively.
Figure 7.
A view of the three-dimensional Hirshfeld surface of the title compound plotted over electrostatic potential energy.
Synthesis and crystallization
The title compound was prepared by refluxing a mixture of a solution containing 3,5-di-tert-butyl-2-hydroxybenzaldehyde (46.8 mg, 0.2 mmol) in ethanol (30 mL) and a solution containing 3-chloro-4-methylaniline (28.32 mg, 0.2 mmol) in ethanol (20 mL). The reaction mixture was stirred for 4 h under reflux. Crystals suitable for X-ray analysis were obtained by slow evaporation of an ethanol solution (m.p. 417–419 K; yield 84%).
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 2 ▸. C-bound H atoms were positioned geometrically with C—H distances of 0.93–0.97 Å and refined as riding, with U iso(H) = 1.2U eq(C).
Table 2. Experimental details.
| Crystal data | |
| Chemical formula | C22H28ClNO |
| M r | 357.90 |
| Crystal system, space group | Monoclinic, P21/m |
| Temperature (K) | 296 |
| a, b, c (Å) | 9.6753 (10), 7.0072 (6), 15.3749 (13) |
| β (°) | 93.425 (7) |
| V (Å3) | 1040.51 (17) |
| Z | 2 |
| Radiation type | Mo Kα |
| μ (mm−1) | 0.19 |
| Crystal size (mm) | 0.74 × 0.65 × 0.48 |
| Data collection | |
| Diffractometer | Stoe IPDS 2 |
| Absorption correction | Integration (X-RED32; Stoe & Cie, 2002 ▸) |
| T min, T max | 0.879, 0.940 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 5895, 2300, 1298 |
| R int | 0.028 |
| (sin θ/λ)max (Å−1) | 0.628 |
| Refinement | |
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.053, 0.168, 1.00 |
| No. of reflections | 2300 |
| No. of parameters | 145 |
| H-atom treatment | H-atom parameters constrained |
| Δρmax, Δρmin (e Å−3) | 0.28, −0.22 |
Supplementary Material
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989018016377/xu5948sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989018016377/xu5948Isup2.hkl
CCDC reference: 1873612
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
The authors acknowledge the Faculty of Arts and Sciences, Ondokuz Mayıs University, Turkey, for the use of the Stoe IPDS 2 diffractometer (purchased under grant F.279 of the University Research Fund).
supplementary crystallographic information
Crystal data
| C22H28ClNO | F(000) = 384 |
| Mr = 357.90 | Dx = 1.142 Mg m−3 |
| Monoclinic, P21/m | Mo Kα radiation, λ = 0.71073 Å |
| a = 9.6753 (10) Å | Cell parameters from 5548 reflections |
| b = 7.0072 (6) Å | θ = 2.1–30.7° |
| c = 15.3749 (13) Å | µ = 0.19 mm−1 |
| β = 93.425 (7)° | T = 296 K |
| V = 1040.51 (17) Å3 | Prism, orange |
| Z = 2 | 0.74 × 0.65 × 0.48 mm |
Data collection
| Stoe IPDS 2 diffractometer | 2300 independent reflections |
| Radiation source: sealed X-ray tube, 12 x 0.4 mm long-fine focus | 1298 reflections with I > 2σ(I) |
| Detector resolution: 6.67 pixels mm-1 | Rint = 0.028 |
| rotation method scans | θmax = 26.5°, θmin = 2.1° |
| Absorption correction: integration (X-RED32; Stoe & Cie, 2002) | h = −12→12 |
| Tmin = 0.879, Tmax = 0.940 | k = −8→8 |
| 5895 measured reflections | l = −18→19 |
Refinement
| Refinement on F2 | 0 restraints |
| Least-squares matrix: full | Hydrogen site location: mixed |
| R[F2 > 2σ(F2)] = 0.053 | H-atom parameters constrained |
| wR(F2) = 0.168 | w = 1/[σ2(Fo2) + (0.0964P)2] where P = (Fo2 + 2Fc2)/3 |
| S = 1.00 | (Δ/σ)max < 0.001 |
| 2300 reflections | Δρmax = 0.28 e Å−3 |
| 145 parameters | Δρmin = −0.22 e Å−3 |
Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 0.2129 (4) | 0.250000 | 0.2502 (2) | 0.0799 (9) | |
| H1A | 0.122313 | 0.250000 | 0.272431 | 0.120* | |
| H1B | 0.223846 | 0.361862 | 0.215178 | 0.120* | |
| C2 | 0.3181 (3) | 0.250000 | 0.3253 (2) | 0.0633 (8) | |
| C3 | 0.2795 (3) | 0.250000 | 0.4107 (2) | 0.0703 (9) | |
| H3 | 0.185649 | 0.250000 | 0.420637 | 0.084* | |
| C4 | 0.3723 (3) | 0.250000 | 0.4806 (2) | 0.0700 (9) | |
| H4 | 0.340566 | 0.250000 | 0.536544 | 0.084* | |
| C5 | 0.5133 (3) | 0.250000 | 0.46997 (18) | 0.0556 (7) | |
| C6 | 0.5560 (3) | 0.250000 | 0.38600 (19) | 0.0600 (7) | |
| H6 | 0.649961 | 0.250000 | 0.376348 | 0.072* | |
| C7 | 0.4596 (3) | 0.250000 | 0.3165 (2) | 0.0639 (8) | |
| C8 | 0.5942 (3) | 0.250000 | 0.61807 (19) | 0.0568 (7) | |
| H8 | 0.502444 | 0.250000 | 0.632881 | 0.068* | |
| C9 | 0.7006 (3) | 0.250000 | 0.68668 (18) | 0.0521 (7) | |
| C10 | 0.6637 (3) | 0.250000 | 0.77307 (19) | 0.0566 (7) | |
| H10 | 0.570247 | 0.250000 | 0.784241 | 0.068* | |
| C11 | 0.7595 (3) | 0.250000 | 0.84146 (18) | 0.0571 (7) | |
| C12 | 0.8995 (3) | 0.250000 | 0.82148 (19) | 0.0620 (8) | |
| H12 | 0.966318 | 0.250000 | 0.867572 | 0.074* | |
| C13 | 0.9445 (3) | 0.250000 | 0.7374 (2) | 0.0590 (7) | |
| C14 | 0.8423 (3) | 0.250000 | 0.66939 (18) | 0.0551 (7) | |
| C15 | 1.0991 (3) | 0.250000 | 0.7198 (2) | 0.0722 (9) | |
| C16 | 1.1330 (2) | 0.0710 (4) | 0.6677 (2) | 0.0980 (10) | |
| H16A | 1.076649 | 0.068596 | 0.614163 | 0.147* | |
| H16B | 1.228999 | 0.073060 | 0.655013 | 0.147* | |
| H16C | 1.114737 | −0.040681 | 0.701290 | 0.147* | |
| C17 | 1.1899 (4) | 0.250000 | 0.8041 (3) | 0.1274 (18) | |
| H17A | 1.285592 | 0.250000 | 0.790824 | 0.191* | |
| H17C | 1.170713 | 0.138138 | 0.837395 | 0.191* | |
| C18 | 0.7224 (3) | 0.250000 | 0.9368 (2) | 0.0753 (9) | |
| C19 | 0.7730 (5) | 0.4303 (8) | 0.9793 (2) | 0.176 (2) | |
| H19A | 0.734855 | 0.537623 | 0.947277 | 0.264* | |
| H19B | 0.744536 | 0.434730 | 1.037976 | 0.264* | |
| H19C | 0.872234 | 0.434730 | 0.979833 | 0.264* | |
| C20 | 0.5646 (4) | 0.250000 | 0.9437 (3) | 0.1131 (15) | |
| H20A | 0.525969 | 0.136290 | 0.917256 | 0.170* | |
| H20B | 0.543785 | 0.250000 | 1.003968 | 0.170* | |
| Cl1 | 0.52181 (11) | 0.250000 | 0.21237 (6) | 0.1032 (4) | |
| N1 | 0.6191 (2) | 0.250000 | 0.53735 (15) | 0.0599 (6) | |
| O1 | 0.8791 (2) | 0.250000 | 0.58613 (13) | 0.0748 (6) | |
| H1 | 0.809294 | 0.250000 | 0.553116 | 0.112* |
Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.088 (2) | 0.066 (2) | 0.083 (2) | 0.000 | −0.0157 (18) | 0.000 |
| C2 | 0.0734 (18) | 0.0410 (17) | 0.0749 (19) | 0.000 | −0.0011 (15) | 0.000 |
| C3 | 0.0559 (16) | 0.077 (2) | 0.077 (2) | 0.000 | 0.0003 (16) | 0.000 |
| C4 | 0.0621 (17) | 0.086 (2) | 0.0633 (18) | 0.000 | 0.0113 (15) | 0.000 |
| C5 | 0.0611 (16) | 0.0480 (17) | 0.0576 (16) | 0.000 | 0.0032 (13) | 0.000 |
| C6 | 0.0630 (16) | 0.0543 (18) | 0.0631 (17) | 0.000 | 0.0085 (14) | 0.000 |
| C7 | 0.0786 (19) | 0.0547 (19) | 0.0587 (16) | 0.000 | 0.0068 (15) | 0.000 |
| C8 | 0.0531 (14) | 0.0530 (18) | 0.0649 (17) | 0.000 | 0.0079 (13) | 0.000 |
| C9 | 0.0527 (14) | 0.0458 (16) | 0.0582 (15) | 0.000 | 0.0074 (13) | 0.000 |
| C10 | 0.0546 (15) | 0.0551 (18) | 0.0616 (16) | 0.000 | 0.0165 (14) | 0.000 |
| C11 | 0.0642 (16) | 0.0530 (18) | 0.0550 (15) | 0.000 | 0.0112 (14) | 0.000 |
| C12 | 0.0595 (16) | 0.065 (2) | 0.0611 (17) | 0.000 | 0.0016 (13) | 0.000 |
| C13 | 0.0554 (15) | 0.0556 (18) | 0.0669 (17) | 0.000 | 0.0113 (14) | 0.000 |
| C14 | 0.0574 (15) | 0.0524 (17) | 0.0566 (16) | 0.000 | 0.0128 (13) | 0.000 |
| C15 | 0.0519 (16) | 0.082 (2) | 0.083 (2) | 0.000 | 0.0124 (15) | 0.000 |
| C16 | 0.0684 (14) | 0.090 (2) | 0.139 (2) | 0.0156 (13) | 0.0353 (16) | −0.0004 (17) |
| C17 | 0.0520 (18) | 0.219 (6) | 0.110 (3) | 0.000 | −0.002 (2) | 0.000 |
| C18 | 0.079 (2) | 0.091 (3) | 0.0563 (17) | 0.000 | 0.0149 (16) | 0.000 |
| C19 | 0.204 (4) | 0.235 (5) | 0.095 (2) | −0.106 (4) | 0.061 (3) | −0.087 (3) |
| C20 | 0.102 (3) | 0.168 (4) | 0.073 (2) | 0.000 | 0.036 (2) | 0.000 |
| Cl1 | 0.1117 (8) | 0.1390 (10) | 0.0595 (5) | 0.000 | 0.0104 (5) | 0.000 |
| N1 | 0.0608 (13) | 0.0617 (16) | 0.0578 (14) | 0.000 | 0.0075 (11) | 0.000 |
| O1 | 0.0613 (11) | 0.1072 (18) | 0.0574 (12) | 0.000 | 0.0166 (10) | 0.000 |
Geometric parameters (Å, º)
| C1—C2 | 1.494 (5) | C12—C13 | 1.389 (4) |
| C1—H1A | 0.9600 | C12—H12 | 0.9300 |
| C1—H1B | 0.9600 | C13—C14 | 1.395 (4) |
| C1—H1Bi | 0.9600 | C13—C15 | 1.536 (4) |
| C2—C7 | 1.383 (4) | C14—O1 | 1.349 (3) |
| C2—C3 | 1.387 (4) | C15—C17 | 1.523 (5) |
| C3—C4 | 1.359 (4) | C15—C16 | 1.534 (3) |
| C3—H3 | 0.9300 | C15—C16i | 1.534 (3) |
| C4—C5 | 1.383 (4) | C16—H16A | 0.9600 |
| C4—H4 | 0.9300 | C16—H16B | 0.9600 |
| C5—C6 | 1.379 (4) | C16—H16C | 0.9600 |
| C5—N1 | 1.412 (4) | C17—H17A | 0.9600 |
| C6—C7 | 1.376 (4) | C17—H17C | 0.9600 |
| C6—H6 | 0.9300 | C17—H17Ci | 0.9600 |
| C7—Cl1 | 1.744 (3) | C18—C19i | 1.491 (4) |
| C8—N1 | 1.278 (4) | C18—C19 | 1.491 (4) |
| C8—C9 | 1.429 (4) | C18—C20 | 1.537 (5) |
| C8—H8 | 0.9300 | C19—H19A | 0.9600 |
| C9—C10 | 1.396 (4) | C19—H19B | 0.9600 |
| C9—C14 | 1.412 (3) | C19—H19C | 0.9600 |
| C10—C11 | 1.360 (4) | C20—H20A | 0.9600 |
| C10—H10 | 0.9300 | C20—H20B | 0.9595 |
| C11—C12 | 1.407 (4) | C20—H20Ai | 0.9600 |
| C11—C18 | 1.530 (4) | O1—H1 | 0.8200 |
| C2—C1—H1A | 108.6 | O1—C14—C13 | 119.7 (2) |
| C2—C1—H1B | 109.9 | O1—C14—C9 | 119.5 (3) |
| H1A—C1—H1B | 109.5 | C13—C14—C9 | 120.8 (2) |
| C2—C1—H1Bi | 109.91 (9) | C17—C15—C16 | 108.31 (19) |
| H1A—C1—H1Bi | 109.5 | C17—C15—C16i | 108.3 (2) |
| H1B—C1—H1Bi | 109.5 | C16—C15—C16i | 109.7 (3) |
| C7—C2—C3 | 114.6 (3) | C17—C15—C13 | 111.6 (3) |
| C7—C2—C1 | 123.8 (3) | C16—C15—C13 | 109.45 (18) |
| C3—C2—C1 | 121.5 (3) | C16i—C15—C13 | 109.45 (18) |
| C4—C3—C2 | 123.1 (3) | C15—C16—H16A | 109.5 |
| C4—C3—H3 | 118.4 | C15—C16—H16B | 109.5 |
| C2—C3—H3 | 118.4 | H16A—C16—H16B | 109.5 |
| C3—C4—C5 | 121.1 (3) | C15—C16—H16C | 109.5 |
| C3—C4—H4 | 119.5 | H16A—C16—H16C | 109.5 |
| C5—C4—H4 | 119.5 | H16B—C16—H16C | 109.5 |
| C6—C5—C4 | 117.6 (3) | C15—C17—H17A | 109.4 |
| C6—C5—N1 | 116.2 (2) | C15—C17—H17C | 109.5 |
| C4—C5—N1 | 126.1 (2) | H17A—C17—H17C | 109.5 |
| C7—C6—C5 | 120.0 (3) | C15—C17—H17Ci | 109.48 (8) |
| C7—C6—H6 | 120.0 | H17A—C17—H17Ci | 109.5 |
| C5—C6—H6 | 120.0 | H17C—C17—H17Ci | 109.5 |
| C6—C7—C2 | 123.6 (3) | C19i—C18—C19 | 115.8 (5) |
| C6—C7—Cl1 | 117.2 (2) | C19i—C18—C11 | 109.23 (18) |
| C2—C7—Cl1 | 119.2 (3) | C19—C18—C11 | 109.23 (18) |
| N1—C8—C9 | 123.2 (2) | C19i—C18—C20 | 105.8 (2) |
| N1—C8—H8 | 118.4 | C19—C18—C20 | 105.8 (2) |
| C9—C8—H8 | 118.4 | C11—C18—C20 | 110.9 (3) |
| C10—C9—C14 | 119.1 (3) | C18—C19—H19A | 109.5 |
| C10—C9—C8 | 119.2 (2) | C18—C19—H19B | 109.5 |
| C14—C9—C8 | 121.7 (2) | H19A—C19—H19B | 109.5 |
| C11—C10—C9 | 122.3 (2) | C18—C19—H19C | 109.5 |
| C11—C10—H10 | 118.9 | H19A—C19—H19C | 109.5 |
| C9—C10—H10 | 118.9 | H19B—C19—H19C | 109.5 |
| C10—C11—C12 | 116.9 (2) | C18—C20—H20A | 109.5 |
| C10—C11—C18 | 123.5 (2) | C18—C20—H20B | 109.4 |
| C12—C11—C18 | 119.6 (3) | H20A—C20—H20B | 108.1 |
| C13—C12—C11 | 124.2 (3) | C18—C20—H20Ai | 109.49 (9) |
| C13—C12—H12 | 117.9 | H20A—C20—H20Ai | 112.2 |
| C11—C12—H12 | 117.9 | H20B—C20—H20Ai | 108.1 |
| C12—C13—C14 | 116.8 (2) | C8—N1—C5 | 122.8 (2) |
| C12—C13—C15 | 121.8 (3) | C14—O1—H1 | 109.5 |
| C14—C13—C15 | 121.5 (3) | ||
| C7—C2—C3—C4 | 0.000 (1) | C12—C13—C14—O1 | 180.000 (1) |
| C1—C2—C3—C4 | 180.000 (1) | C15—C13—C14—O1 | 0.000 (1) |
| C2—C3—C4—C5 | 0.000 (1) | C12—C13—C14—C9 | 0.000 (1) |
| C3—C4—C5—C6 | 0.000 (1) | C15—C13—C14—C9 | 180.000 (1) |
| C3—C4—C5—N1 | 180.000 (1) | C10—C9—C14—O1 | 180.000 (1) |
| C4—C5—C6—C7 | 0.000 (1) | C8—C9—C14—O1 | 0.000 (1) |
| N1—C5—C6—C7 | 180.000 (1) | C10—C9—C14—C13 | 0.000 (1) |
| C5—C6—C7—C2 | 0.000 (1) | C8—C9—C14—C13 | 180.000 (1) |
| C5—C6—C7—Cl1 | 180.000 (1) | C12—C13—C15—C17 | 0.000 (1) |
| C3—C2—C7—C6 | 0.000 (1) | C14—C13—C15—C17 | 180.000 (1) |
| C1—C2—C7—C6 | 180.000 (1) | C12—C13—C15—C16 | 119.9 (2) |
| C3—C2—C7—Cl1 | 180.000 (1) | C14—C13—C15—C16 | −60.1 (2) |
| C1—C2—C7—Cl1 | 0.000 (1) | C12—C13—C15—C16i | −119.9 (2) |
| N1—C8—C9—C10 | 180.000 (1) | C14—C13—C15—C16i | 60.1 (2) |
| N1—C8—C9—C14 | 0.000 (1) | C10—C11—C18—C19i | 116.2 (3) |
| C14—C9—C10—C11 | 0.000 (1) | C12—C11—C18—C19i | −63.8 (3) |
| C8—C9—C10—C11 | 180.000 (1) | C10—C11—C18—C19 | −116.2 (3) |
| C9—C10—C11—C12 | 0.000 (1) | C12—C11—C18—C19 | 63.8 (3) |
| C9—C10—C11—C18 | 180.000 (1) | C10—C11—C18—C20 | 0.000 (2) |
| C10—C11—C12—C13 | 0.000 (2) | C12—C11—C18—C20 | 180.000 (1) |
| C18—C11—C12—C13 | 180.000 (1) | C9—C8—N1—C5 | 180.000 (1) |
| C11—C12—C13—C14 | 0.000 (1) | C6—C5—N1—C8 | 180.000 (1) |
| C11—C12—C13—C15 | 180.000 (1) | C4—C5—N1—C8 | 0.000 (1) |
Symmetry code: (i) x, −y+1/2, z.
Hydrogen-bond geometry (Å, º)
Cg is the centroid of the C9–C14 benzene ring.
| D—H···A | D—H | H···A | D···A | D—H···A |
| O1—H1···N1 | 0.82 | 1.84 | 2.582 (3) | 149 |
| C1—H1B···Cg1ii | 0.96 | 2.77 | 3.5072 (4) | 134 |
Symmetry code: (ii) −x+1, y+1/2, −z+1.
References
- Atalay, Ş., Gerçeker, S., Meral, S. & Bülbül, H. (2017). IUCrData, 2, x171725.
- Calligaris, M., Nardin, G. M. J. & Randaccio, C. (1972). Coord. Chem. Rev. 7, 385–403.
- Elmalı, A., Kabak, M., Kavlakoğlu, E. & Elerman, Y. (1999). J. Mol. Struct. 510, 207–214.
- El-masry, A. H., Fahmy, H. H. & Ali Abdelwahed, S. (2000). Molecules, 5, 1429–1438.
- Farrugia, L. J. (2012). J. Appl. Cryst. 45, 849–854.
- Groom, C. R., Bruno, I. J., Lightfoot, M. P. & Ward, S. C. (2016). Acta Cryst. B72, 171–179. [DOI] [PMC free article] [PubMed]
- Gumus, M. K., Kansiz, S., Dege, N. & Kalibabchuk, V. A. (2018). Acta Cryst. E74, 1211–1214. [DOI] [PMC free article] [PubMed]
- Hökelek, T., Bilge, S., Demiriz, Ş., Özgüç, B. & Kılıç, Z. (2004). Acta Cryst. C60, o803–o805. [DOI] [PubMed]
- Iwan, A., Kaczmarczyk, B., Janeczek, H., Sek, D. & Ostrowski, S. (2007). Spectrochim. Acta A Mol. Biomol. Spectrosc. 66, 1030–1041. [DOI] [PubMed]
- Kansız, S. & Dege, N. (2018). J. Mol. Struct. 1173, 42–51.
- Kansiz, S., Macit, M., Dege, N. & Tsapyuk, G. G. (2018). Acta Cryst. E74, 1513–1516. [DOI] [PMC free article] [PubMed]
- Moroz, Y. S., Demeshko, S., Haukka, M., Mokhir, A., Mitra, U., Stocker, M., Müller, P., Meyer, F. & Fritsky, I. O. (2012). Inorg. Chem. 51, 7445–7447. [DOI] [PubMed]
- Sen, P., Kansiz, S., Golenya, I. A. & Dege, N. (2018). Acta Cryst. E74, 1147–1150. [DOI] [PMC free article] [PubMed]
- Şen, F., Kansiz, S. & Uçar, İ. (2017). Acta Cryst. C73, 517–524. [DOI] [PubMed]
- Sheldrick, G. M. (2015a). Acta Cryst. A71, 3–8.
- Sheldrick, G. M. (2015b). Acta Cryst. C71, 3–8.
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Stoe & Cie (2002). X-AREA and X-RED32. Stoe & Cie GmbH, Darmstadt, Germany.
- Su, D.-C., Wang, F.-T., Mao, C.-G. & Qian, S.-S. (2012). Acta Cryst. E68, o303. [DOI] [PMC free article] [PubMed]
- Toprak, Ş., Tanak, H., Macit, M., Dege, N. & Orbay, M. (2018). J. Mol. Struct. 1174, 184–191.
- Turner, M. J., MacKinnon, J. J., Wolff, S. K., Grimwood, D. J., Spackman, P. R., Jayatilaka, D. & Spackman, M. A. (2017). CrystalExplorer17.5. University of Western Australia, Perth.
- Wang, H. (2010). Acta Cryst. E66, m1571. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I. DOI: 10.1107/S2056989018016377/xu5948sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S2056989018016377/xu5948Isup2.hkl
CCDC reference: 1873612
Additional supporting information: crystallographic information; 3D view; checkCIF report









