The crystal structures of (E)-4-chloro-N′-(4-chlorobenzylidene)benzenesulfonohydrazide and (E)-4-chloro-N′-(4-nitrobenzylidene)benzenesulfonohydrazide have been studied to investigate the effect of substituents on the structural parameters. The two-dimensional fingerprint plots of these two p-substituted compounds indicate that in the 4-chloro-substituted compound, the largest contribution to the Hirshfeld surface comes from the H⋯H contacts (26.6%), in contrast to the 34.8% contribution of the O⋯H/H⋯O contacts in the 4-nitro-substituted compound.
Keywords: crystal structure, hydrazones, N′-(arylidene)arylsulfonohydrazides, hydrogen bonding, graph-set motif, Hirshfeld surface analysis, fingerprint plots
Abstract
Two (E)-N′-(p-substituted benzylidene)-4-chlorobenzenesulfonohydrazides, namely, (E)-4-chloro-N′-(4-chlorobenzylidene)benzenesulfonohydrazide, C13H10Cl2N2O2S, (I), and (E)-4-chloro-N′-(4-nitrobenzylidene)benzenesulfonohydrazide, C13H10ClN3O4S, (II), have been synthesized, characterized and their crystal structures studied to explore the effect of the nature of substituents on the structural parameters. Compound (II) crystallized with two independent molecules [(IIA) and IIB)] in the asymmetric unit. In both compounds, the configuration around the C=N bond is E. The molecules are twisted at the S atom with C—S—N—N torsion angles of −62.4 (2)° in (I), and −46.8 (2)° and 56.8 (2)° in the molecules A and B of (II). The 4-chlorophenylsulfonyl and 4-substituted benzylidene rings form dihedral angles of 81.0 (1)° in (I), 75.9 (1)° in (IIA) and 73.4 (1)° in (IIB). In the crystal of (I), molecules are linked via pairs of N—H⋯O hydrogen bonds, forming inversion dimers with an R 2 2(8) ring motif. The dimers are linked by C—Cl⋯π interactions, forming a three-dimensional structure. In the crystal of (II), molecules are linked by C—H⋯π interactions and N—H⋯O hydrogen bonds, forming –A–B–A–B– chains along the c-axis direction. The chains are linked via C—H⋯O and C—H⋯π interactions, forming layers parallel to the bc plane. Two-dimensional fingerprint plots show that the most significant contacts contributing to the Hirshfeld surface for (I) are H⋯H contacts (26.6%), followed by Cl⋯H/H⋯Cl (21.3%), O⋯H/H⋯O (15.5%) and Cl⋯C/C⋯Cl (10.7%), while for (II) the O⋯H/H⋯O contacts are dominant, with a contribution of 34.8%, followed by H⋯H (15.2%), C⋯H/H⋯C (14.0%) and Cl⋯H/H⋯Cl (10.0%) contacts.
Chemical context
In the field of synthetic chemistry, hydrazones are frequently used as nucleophiles and electrophiles (Ogawa et al., 2004 ▸). They also play an important role in organic synthesis as one of the reaction intermediates due to their ring-closure reactions (Rollas & Küçükgüzel, 2007 ▸). Hydrazones have drawn considerable attention in the field of coordination chemistry (Weber et al., 2007 ▸). They also find various industrial applications (Reis et al., 2013 ▸) and exhibit a wide spectrum of biological activities (da Silva et al., 2011 ▸). Arylsulfonyl-hydrazones have shown antitumour activity in addition to their role as a versatile source of diazo compounds in many metal-catalysed and metal-free reactions (Hashemi, 2012 ▸). In a continuation of our efforts to explore the effect of site and nature of substituents on the crystal structures of 4-chloro-arylsulfonohydrazide derivatives (Salian et al., 2018 ▸), we report herein the synthesis, characterization, crystal structures and Hirshfeld surface analysis of the title compounds, (I) and (II), and compare them with those of the recently reported structures of (E)-4-chloro-N′-(benzylidene) benzenesulfonohydrazide (III), (E)-4-chloro-N′-(2-methylbenzylidene)benzenesulfonohydrazide (IV) and (E)-4-chloro-N′-(4-methylbenzylidene)benzenesulfonohydrazide (V) (Salian et al., 2018 ▸).
Structural commentary
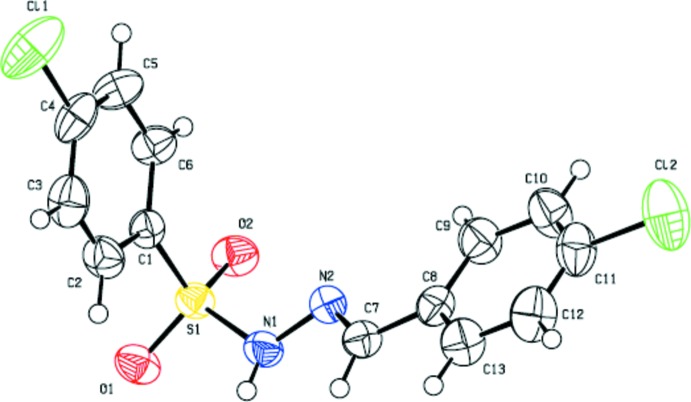
Compound (I), crystallizes in the triclinic crystal system, space group P
 , with one molecule in the asymmetric unit (Fig. 1 ▸), while compound (II) crystallizes in the monoclinic crystal system, space group P21/c, with two independent molecules [(IIA) and (IIB)] in the asymmetric unit (Fig. 2 ▸). For both the compounds, the configuration about the C=N bond is E and the conformations of the N—H and C—H bonds in the hydrazone segments are syn to each other.
, with one molecule in the asymmetric unit (Fig. 1 ▸), while compound (II) crystallizes in the monoclinic crystal system, space group P21/c, with two independent molecules [(IIA) and (IIB)] in the asymmetric unit (Fig. 2 ▸). For both the compounds, the configuration about the C=N bond is E and the conformations of the N—H and C—H bonds in the hydrazone segments are syn to each other.
Figure 1.
Molecular structure of (I), with the atom labelling and displacement ellipsoids drawn at the 50% probability level.
Figure 2.
Molecular structure of (II), with the atom labelling and displacement ellipsoids drawn at the 50% probability level.
The C=N bond lengths of 1.269 (3), 1.269 (3) and 1.269 (3) Å in (I), (IIA) and (IIB), and the N—N bond lengths of 1.388 (2) 1.397 (3) and 1.390 (2) Å in (I), (IIA) and (IIB), respectively, indicate the delocalization of the π-electron density over the hydrazone part of the molecules. The other bond lengths are in close agreement with those of the parent compound (III), and the ortho-methyl (IV) and para-methyl (V) derivatives (Salian et al., 2018 ▸). Selected geometrical parameters of compounds (I)–(V) are compared in Table 1 ▸ (Salian et al., 2018 ▸).
Table 1. Comparison of selected geometrical parameters (Å, °) of compounds (I)–(V).
The dihedral angle is that between the aromatic rings. The equivalent bond lengths and torsion angles are given for (IIB).
| Bond length | (I) | (II) Molecule A | (II) Molecule B | (III) | (IV) | (V) |
|---|---|---|---|---|---|---|
| C1—S1 | 1.763 (2) | 1.754 (2) | 1.760 (2) | 1.752 (4) | 1.751 (5) | 1.761 (2) |
| S1—N1 | 1.631 (2) | 1.645 (2) | 1.641 (2) | 1.644 (4) | 1.645 (4) | 1.625 (2) |
| N1—N2 | 1.388 (2) | 1.397 (3) | 1.390 (2) | 1.394 (5) | 1.407 (5) | 1.393 (2) |
| N2—C7 | 1.269 (3) | 1.269 (3) | 1.269 (3) | 1.258 (5) | 1.272 (5) | 1.273 (3) |
| C7—C8 | 1.463 (3) | 1.465 (3) | 1.462 (3) | 1.473 (6) | 1.461 (6) | 1.458 (3) |
| Torsion angle | ||||||
| C1—S1—N1—N2 | −62.4 (2) | −46.8 (2) | 56.8 (2) | −66.0 (3) | −66.0 (3) | −58.4 (2) |
| S1—N1—N2—C7 | 158.9 (2) | 171.4 (2) | −165.3 (2) | 166.5 (3) | 165.4 (3) | 157.9 (2) |
| N1—N2—C7—C8 | 175.0 (2) | −175.9 (2) | 178.2 (2) | 177.8 (4) | 175.8 (4) | 175.8 (2) |
| Dihedral angle | 81.0 (1) | 75.9 (1) | 73.4 (1) | 78.4 (2) | 74.8 (2) | 76.9 (1) |
In the title compounds the molecules are twisted at the S atom with C—S—N—N torsion angles of −62.4 (2)° in (I), and −46.8 (2) and 56.8 (2)° in (IIA) and (IIB), respectively. The respective S—N—N=C torsion angles of 158.9 (2)° in (I), and 171.4 (2) and −165.3 (2)° in (IIA) and (IIB), denote the non-planarity of the sulfonohydrazide parts of the molecules. However, the N—N—C—C torsion angles of 175.0 (2)° in (I), and −175.9 (2) and 178.2 (2)° in (IIA) and (IIB), indicate near coplanarity of the hydrazide units with the benzylidene rings. The dihedral angles between the 4-chloro-substituted phenylsulfonyl ring and 4-substituted benzylidene ring are 81.0 (1)° in (I), and 75.9 (1) and 73.4 (1°) in molecules A and B of compound (II). In comparison, the corresponding values in compounds (III), (IV) and (V) are 78.4 (2), 74.8 (2) and 76.9 (1)°, respectively (see Table 1 ▸). In (II) the A and B molecules are linked by a C—H⋯Cl interaction (Table 3 ▸).
Table 3. Hydrogen-bond geometry (Å, °) for (II) .
Cg3 is the centroid of the C14–C19 ring.
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1N⋯O5i | 0.85 (2) | 2.06 (2) | 2.887 (3) | 163 (2) |
| N4—H4N⋯O1ii | 0.84 (2) | 2.13 (2) | 2.918 (2) | 157 (2) |
| C10—H10⋯O7iii | 0.93 | 2.58 | 3.465 (3) | 159 |
| C16—H16⋯O4iv | 0.93 | 2.58 | 3.259 (3) | 131 |
| C25—H25⋯O5ii | 0.93 | 2.45 | 3.340 (3) | 161 |
| C12—H12⋯Cg3 | 0.93 | 2.96 | 3.843 (2) | 160 |
Symmetry codes: (i)  ; (ii)
; (ii)  ; (iii)
; (iii)  ; (iv)
; (iv)  .
.
Supramolecular features
The pattern of the hydrogen-bonding interactions in the crystal structures of (I) and (II) are different. In the crystal of (I), molecules are linked by pairs of N—H⋯O hydrogen bonds, forming inversion dimers enclosing  (8) loops (Fig. 3 ▸, Table 2 ▸). The dimers are linked by C—Cl⋯π interactions, forming a three-dimensional arrangement (Fig. 3 ▸). This is very similar to the situation observed in the crystal of compound (V) [(E)- 4-chloro-N′-(4-methylbenzylidene)benzenesulfonohydrazide; Salian et al., 2018 ▸].
(8) loops (Fig. 3 ▸, Table 2 ▸). The dimers are linked by C—Cl⋯π interactions, forming a three-dimensional arrangement (Fig. 3 ▸). This is very similar to the situation observed in the crystal of compound (V) [(E)- 4-chloro-N′-(4-methylbenzylidene)benzenesulfonohydrazide; Salian et al., 2018 ▸].
Figure 3.
Crystal packing of (I), viewed along the a axis, with hydrogen bonds (Table 2 ▸) shown as dashed lines and C—Cl⋯π interactions as blue arrows. C-bound H atoms have been omitted.
Table 2. Hydrogen-bond geometry (Å, °) for (I) .
Cg1 and Cg2 are the centroids of rings C1–C6 and C8–C13, respectively.
| D—H⋯A | D—H | H⋯A | D⋯A | D—H⋯A |
|---|---|---|---|---|
| N1—H1N⋯O1i | 0.83 (2) | 2.07 (2) | 2.903 (2) | 178 (2) |
| C4—Cl1⋯Cg2ii | 1.73 (1) | 3.41 (1) | 5.112 (2) | 166 (1) |
| C11—Cl2⋯Cg1iii | 1.74 (1) | 3.65 (1) | 5.372 (3) | 171 (1) |
Symmetry codes: (i)  ; (ii)
; (ii)  ; (iii)
; (iii)  .
.
Replacement of the 4-chloro group in (I) by the 4-nitro group to produce compound (II) introduces C—H⋯O interactions, which stabilize the crystal packing (Table 3 ▸ and Figs. 4 ▸ and 5 ▸). The N—H⋯O hydrogen bond involving the sulfonyl O atom and the amino H atom of the hydrazide segment between the A and B molecules results in the formation of –A–B–A–B– chains propagating along the c-axis direction (Fig. 4 ▸). The chains are linked by C—H⋯O interactions involving O atoms O4 in (IIA) and O5 in (IIB) and O7 of the nitro group and the aromatic hydrogen atoms ortho to the Cl or NO2 group. The sulfonyl O atom of (IIB), i.e. O5, shows bifurcated hydrogen bonding, one with the amino H atom of the hydrazide segment and the other with one of the aromatic H atoms (H25), adjacent to the nitro group. These interactions link the chains, forming layers lying parallel to the bc plane (Table 3 ▸ and Fig. 5 ▸).
Figure 4.
A partial view along the a axis of the crystal packing of (II), with hydrogen bonds (Table 3 ▸) shown as dashed lines. H atoms not involved in these interactions have been omitted. Colour code: black A molecules; red B molecules.
Figure 5.
Crystal packing of (II), viewed along the b axis, with hydrogen bonds shown as dashed lines. H atoms not involved in these interactions have been omitted.
Hirshfeld surface analysis
Hirshfeld surfaces and two-dimensional fingerprint plots were generated for the two substituted compounds (I) and (II) using CrystalExplorer (Turner et al., 2017 ▸) to visualize the intermolecular interactions, to investigate the impact of each kind of intermolecular contact on the crystal packing and to study the relative strengths of the different interactions in the two compounds. The molecular Hirshfeld surfaces were generated using a standard (high) surface resolution. d i and d e are the contact distances from the Hirshfeld surface to the nearest atom inside and outside, respectively [Fig. 6 ▸(a) for (I) and Fig. 6 ▸(b) for (II)]. The strong hydrogen bonds appear as dark-red spots and weak interactions as light-red spots on the d norm surface (McKinnon et al., 2004 ▸; Spackman & Jayatilaka, 2009 ▸).
Figure 6.
(a) View of the Hirshfeld surface mapped over d norm for (I); (b) two views of the Hirshfeld surface mapped over d norm for (II).
Comparison of fingerprint plots for various atom–atom interactions show that the percentage contributions of these interactions to the Hirshfeld surfaces vary significantly from (I) to (II). The major contribution to the Hirshfeld surface in (I) is from H⋯H contacts (26.6%), followed by Cl⋯H/H⋯Cl (21.3%), O⋯H/H⋯O (15.5%), Cl⋯C/C⋯Cl (10.7%) and C⋯H/ H⋯C (9.1%) [Fig. 7 ▸(a)], while in (II), as a result of C—H⋯O interactions, O⋯H/H⋯O contacts are dominant and serve as the major contributors (34.8%) in the crystal packing, followed by H⋯H contacts (15.2%), C⋯H/ H⋯C (14.0%) and Cl⋯H/H⋯Cl (10.0%) [Fig. 7 ▸(b)]. The Cl⋯C/C⋯Cl contribution to the d norm surface is almost negligible (0.5%) in (II). However, C⋯C, H⋯N/N⋯H and C⋯O/O⋯C contacts make very similar contributions in the two compounds, their respective contributions being 4.7, 2.8, 3.0%, in (I) and 5.3, 3.6 and 4.1% in (II). Two pairs of symmetrical, long narrow spikes are present at d i + d e ∼2.2 Å for the O⋯H/H⋯O contacts in the fingerprint plots of (I) and (II) and these values are very close to the H⋯A distances for the N—H⋯O hydrogen bonds observed in the crystal structures (Tables 2 ▸ and 3 ▸). The contributions of the other weak intermolecular contacts to the Hirshfeld surfaces are: Cl⋯N/N⋯Cl (1.0 and 1.5%), C⋯N/N⋯C (0.0 and 2.8%), O⋯O (0, 2.3%), N⋯N (0, 0.4%) in (I) and (II), respectively. The result of the quantitative analysis of all types of intermolecular contacts present in (I) and (II) is summarized in Fig. 8 ▸.
Figure 7.
Two-dimensional fingerprint plots for (a) (I) and (b) (II). d i is the closest internal distance from a given point on the Hirshfeld surface and d e is the closest external contact.
Figure 8.
Quantitative results of different intermolecular interactions contributing to the Hirshfield surfaces of (I) and (II).
Database survey
The structures reported in the literature similar to the title compounds include (E)-N′-(4-chlorobenzylidene)-p-toluenesulfonohydrazide 0.15-hydrate (Kia et al., 2009a ▸), (E)-N′-(4-chlorobenzylidene)-p-toluenesulfonohydrazide (Balaji et al., 2014 ▸), (E)-N′-(4-bromobenzylidene)-p-toluenesulfonohydrazide (Kia et al., 2009b ▸], (E)-N′-(4-nitrobenzylidene)benzenesulfonohydrazide (Hussain et al., 2017a ▸) and (E)-4-methyl-N′-(4-nitrobenzylidene)benzenesulfonohydrazide (Hussain et al., 2017b ▸). In all of these structures, intermolecular N—H⋯O hydrogen bonds link neighbouring molecules to form chains, which are linked by C—H⋯O hydrogen bonds. There are also intermolecular π–π interactions present, which further stabilize the crystal structures.
Synthesis and crystallization
Synthesis of 4-chlorobenzenesulfonohydrazide
4-Chlorobenzenesulfonohydrazide was synthesized by a recently reported procedure (Salian et al., 2018 ▸).
Synthesis of compounds (I) and (II)
A mixture of 4-chlorobenzenesulfonohydrazide (0.01 mol) and 4-chlorobenzaldehyde (0.01 mol) for (I), and 4-nitrobenzaldehyde (0.01 mol) for (II), in ethanol (30 ml) and two drops of glacial acetic acid were stirred for 4 h. The reaction mixtures were cooled to room temperature and concentrated by evaporating off the excess of solvent. The solid products obtained were washed with cold water, dried and recrystallized to constant melting points from ethanol to obtain the pure compounds. The purity of the compounds was checked by TLC.
Crystals of compounds (I) and (II), suitable for X-ray diffraction analysis, were obtained by slow evaporation of their DMF solutions at room temperature.
Both compounds were characterized by measuring their IR, 1H and 13C NMR spectra.
(E )-4-Chloro- N ′-(4-chlorobenzylidene)benzenesulfonohydrazide (I)
Colourless rod-shaped crystals; m.p. 432–433 K; IR (cm−1): 3180.6 (N—H asym. stretch), 1573.9 (C=N), 1327.0 (S=O asym. stretch) and 1166.9 (S=O sym. stretch).
1H NMR (400 MHz, DMSO-d6): δ 7.32 (d, 1H, J = 8.4Hz, Ar-H), 7.51–7.56 (m, 4H, Ar-H), 7.87–7.89 (m, 2H, Ar-H), 7.92 (s, 1H), 11.50 (s, 1H). 13C NMR (100 MHz, DMSO-d6): δ 128.38, 129.42, 130.73, 132.06, 134.96, 137.47, 138.39, 139.32, 145.66.
( E )-4-Chloro- N ′-(4-nitrobenzylidene)benzenesulfonohydrazide (II)
Yellow rod-shaped crystals; m.p. 414–415 K; IR (cm−1): 3093.8 (N—H asym. stretch), 1653.0 (C=N), 1392.6 (S=O asym. stretch) and 1153.4 (S=O sym. stretch).
1H NMR (400 MHz, DMSO-d6): δ 7.0 (d, 1H, J = 8.80, Ar-H), 7.38 (d, 1H, J = 8.52, Ar-H), 7.63 (d, 1H, J = 8.36, Ar-H), 7.64 (s, 1H), 7.79 (d, 1H, J = 8.56, Ar-H), 7.80 (d, 2H, J = 8.28, Ar-H), 7.90 (s, 1H), 11.60 (s, 1H). 13C NMR (100 MHz, DMSO-d6): δ 115.49, 124.47, 128.45, 129.63, 136.85, 137.87, 138.28, 147.97, 159.43.
Refinement
Crystal data, data collection and structure refinement details are summarized in Table 4 ▸. C-bound H atoms were positioned with idealized geometry and refined using a riding model: C—H = 0.93 Å with U iso(H) = 1.2U eq(C). The amino H atoms were located in difference-Fourier maps and refined with an N—H distance restraint of 0.86 (2) Å and U iso(H) = 1.2U eq(N). In (I), reflection 011 was masked by the beam stop and omitted from the refinement. In (II), atom O3 is disordered and was refined using a split model. The corresponding site-occupation factors were fixed at 0.55:0.45 and the corresponding N—O bond lengths in the disordered group were restrained to be equal. The U ij components of O3 and O3′ were restrained to be approximately isotropic.
Table 4. Experimental details.
| (I) | (II) | |
|---|---|---|
| Crystal data | ||
| Chemical formula | C13H10Cl2N2O2S | C13H10ClN3O4S |
| M r | 329.19 | 339.75 |
| Crystal system, space group | Triclinic, P

|
Monoclinic, P21/c |
| Temperature (K) | 293 | 293 |
| a, b, c (Å) | 5.9306 (6), 9.477 (1), 13.040 (2) | 19.903 (1), 10.2517 (7), 15.064 (1) |
| α, β, γ (°) | 98.822 (9), 96.046 (9), 92.416 (9) | 90, 103.929 (7), 90 |
| V (Å3) | 718.94 (15) | 2983.3 (3) |
| Z | 2 | 8 |
| Radiation type | Mo Kα | Mo Kα |
| μ (mm−1) | 0.60 | 0.42 |
| Crystal size (mm) | 0.48 × 0.40 × 0.36 | 0.48 × 0.40 × 0.36 |
| Data collection | ||
| Diffractometer | Oxford Diffraction Xcalibur diffractometer with Sapphire CCD | Oxford Diffraction Xcalibur diffractometer with Sapphire CCD |
| Absorption correction | Multi-scan (CrysAlis RED; Oxford Diffraction, 2009 ▸) | Multi-scan (CrysAlis RED; Oxford Diffraction, 2009 ▸) |
| T min, T max | 0.762, 0.814 | 0.825, 0.864 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 4153, 2626, 2321 | 19194, 5455, 4247 |
| R int | 0.016 | 0.025 |
| (sin θ/λ)max (Å−1) | 0.602 | 0.602 |
| Refinement | ||
| R[F 2 > 2σ(F 2)], wR(F 2), S | 0.038, 0.102, 1.08 | 0.038, 0.101, 1.02 |
| No. of reflections | 2626 | 5455 |
| No. of parameters | 185 | 414 |
| No. of restraints | 1 | 15 |
| H-atom treatment | H atoms treated by a mixture of independent and constrained refinement | H atoms treated by a mixture of independent and constrained refinement |
| Δρmax, Δρmin (e Å−3) | 0.37, −0.35 | 0.43, −0.38 |
Supplementary Material
Crystal structure: contains datablock(s) I, II, global. DOI: 10.1107/S205698901801592X/su5458sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S205698901801592X/su5458Isup2.hkl
Structure factors: contains datablock(s) II. DOI: 10.1107/S205698901801592X/su5458IIsup3.hkl
Additional supporting information: crystallographic information; 3D view; checkCIF report
Acknowledgments
The authors thank SAIF Panjab University for extending the services of their NMR facility and Mangalore University for providing all the facilities required.
supplementary crystallographic information
(E)-4-Chloro-N'-(4-chlorobenzylidene)benzenesulfonohydrazide (I) . Crystal data
| C13H10Cl2N2O2S | Z = 2 |
| Mr = 329.19 | F(000) = 336 |
| Triclinic, P1 | Dx = 1.521 Mg m−3 |
| a = 5.9306 (6) Å | Mo Kα radiation, λ = 0.71073 Å |
| b = 9.477 (1) Å | Cell parameters from 2722 reflections |
| c = 13.040 (2) Å | θ = 2.9–27.8° |
| α = 98.822 (9)° | µ = 0.60 mm−1 |
| β = 96.046 (9)° | T = 293 K |
| γ = 92.416 (9)° | Rod, colourless |
| V = 718.94 (15) Å3 | 0.48 × 0.40 × 0.36 mm |
(E)-4-Chloro-N'-(4-chlorobenzylidene)benzenesulfonohydrazide (I) . Data collection
| Oxford Diffraction Xcalibur diffractometer with Sapphire CCD | 2321 reflections with I > 2σ(I) |
| Radiation source: Enhance (Mo) X-ray Source | Rint = 0.016 |
| Rotation method data acquisition using ω scans. | θmax = 25.4°, θmin = 3.2° |
| Absorption correction: multi-scan (CrysAlis RED; Oxford Diffraction, 2009) | h = −4→7 |
| Tmin = 0.762, Tmax = 0.814 | k = −11→11 |
| 4153 measured reflections | l = −15→11 |
| 2626 independent reflections |
(E)-4-Chloro-N'-(4-chlorobenzylidene)benzenesulfonohydrazide (I) . Refinement
| Refinement on F2 | Hydrogen site location: mixed |
| Least-squares matrix: full | H atoms treated by a mixture of independent and constrained refinement |
| R[F2 > 2σ(F2)] = 0.038 | w = 1/[σ2(Fo2) + (0.0523P)2 + 0.3082P] where P = (Fo2 + 2Fc2)/3 |
| wR(F2) = 0.102 | (Δ/σ)max = 0.001 |
| S = 1.08 | Δρmax = 0.37 e Å−3 |
| 2626 reflections | Δρmin = −0.35 e Å−3 |
| 185 parameters | Extinction correction: SHELXL, Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| 1 restraint | Extinction coefficient: 0.025 (3) |
(E)-4-Chloro-N'-(4-chlorobenzylidene)benzenesulfonohydrazide (I) . Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
(E)-4-Chloro-N'-(4-chlorobenzylidene)benzenesulfonohydrazide (I) . Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | ||
| C1 | 0.3867 (3) | 0.1943 (2) | 0.08046 (14) | 0.0366 (4) | |
| C2 | 0.6099 (3) | 0.2317 (2) | 0.12163 (17) | 0.0481 (5) | |
| H2 | 0.6853 | 0.3119 | 0.1052 | 0.058* | |
| C3 | 0.7193 (4) | 0.1486 (3) | 0.18737 (19) | 0.0556 (6) | |
| H3 | 0.8683 | 0.1731 | 0.2166 | 0.067* | |
| C4 | 0.6054 (4) | 0.0289 (2) | 0.20921 (17) | 0.0526 (6) | |
| C5 | 0.3850 (4) | −0.0103 (2) | 0.16683 (18) | 0.0549 (6) | |
| H5 | 0.3118 | −0.0925 | 0.1813 | 0.066* | |
| C6 | 0.2746 (4) | 0.0736 (2) | 0.10286 (16) | 0.0450 (5) | |
| H6 | 0.1248 | 0.0493 | 0.0747 | 0.054* | |
| C7 | 0.1757 (3) | 0.5784 (2) | 0.22864 (16) | 0.0423 (4) | |
| H7 | 0.2965 | 0.6386 | 0.2185 | 0.051* | |
| C8 | 0.0609 (3) | 0.6140 (2) | 0.32242 (16) | 0.0427 (5) | |
| C9 | −0.1405 (4) | 0.5433 (3) | 0.33612 (18) | 0.0542 (5) | |
| H9 | −0.2037 | 0.4681 | 0.2855 | 0.065* | |
| C10 | −0.2480 (4) | 0.5832 (3) | 0.4236 (2) | 0.0618 (6) | |
| H10 | −0.3837 | 0.5356 | 0.4319 | 0.074* | |
| C11 | −0.1539 (4) | 0.6935 (3) | 0.49867 (18) | 0.0575 (6) | |
| C12 | 0.0449 (4) | 0.7658 (3) | 0.48740 (19) | 0.0630 (6) | |
| H12 | 0.1068 | 0.8410 | 0.5383 | 0.076* | |
| C13 | 0.1518 (4) | 0.7252 (3) | 0.39922 (18) | 0.0548 (6) | |
| H13 | 0.2872 | 0.7734 | 0.3913 | 0.066* | |
| N1 | 0.2346 (3) | 0.45985 (18) | 0.07324 (14) | 0.0426 (4) | |
| H1N | 0.345 (3) | 0.517 (2) | 0.0743 (19) | 0.051* | |
| N2 | 0.1173 (3) | 0.46871 (18) | 0.16049 (13) | 0.0414 (4) | |
| O1 | 0.3856 (3) | 0.33553 (16) | −0.07638 (11) | 0.0487 (4) | |
| O2 | 0.0199 (2) | 0.24297 (16) | −0.03067 (12) | 0.0492 (4) | |
| Cl1 | 0.74291 (16) | −0.07477 (8) | 0.29214 (6) | 0.0886 (3) | |
| Cl2 | −0.29271 (14) | 0.74500 (11) | 0.60842 (6) | 0.0909 (3) | |
| S1 | 0.24404 (8) | 0.30435 (5) | 0.00069 (4) | 0.03746 (17) |
(E)-4-Chloro-N'-(4-chlorobenzylidene)benzenesulfonohydrazide (I) . Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| C1 | 0.0347 (9) | 0.0351 (9) | 0.0370 (9) | −0.0002 (7) | 0.0035 (7) | −0.0024 (8) |
| C2 | 0.0372 (10) | 0.0498 (12) | 0.0547 (12) | −0.0056 (9) | 0.0048 (9) | 0.0032 (10) |
| C3 | 0.0394 (11) | 0.0632 (15) | 0.0580 (13) | 0.0069 (10) | −0.0054 (10) | −0.0029 (11) |
| C4 | 0.0661 (14) | 0.0415 (11) | 0.0443 (11) | 0.0170 (10) | −0.0079 (10) | −0.0060 (9) |
| C5 | 0.0682 (15) | 0.0346 (11) | 0.0581 (13) | −0.0040 (10) | −0.0038 (11) | 0.0044 (9) |
| C6 | 0.0433 (11) | 0.0377 (10) | 0.0499 (11) | −0.0066 (8) | −0.0053 (9) | 0.0033 (9) |
| C7 | 0.0429 (11) | 0.0383 (10) | 0.0459 (11) | 0.0015 (8) | 0.0056 (9) | 0.0078 (9) |
| C8 | 0.0460 (11) | 0.0406 (10) | 0.0420 (11) | 0.0083 (8) | 0.0036 (9) | 0.0068 (8) |
| C9 | 0.0571 (13) | 0.0519 (13) | 0.0501 (12) | −0.0022 (10) | 0.0100 (10) | −0.0040 (10) |
| C10 | 0.0570 (14) | 0.0708 (16) | 0.0574 (14) | 0.0003 (12) | 0.0175 (11) | 0.0030 (12) |
| C11 | 0.0581 (14) | 0.0723 (16) | 0.0416 (11) | 0.0186 (12) | 0.0071 (10) | 0.0023 (11) |
| C12 | 0.0666 (16) | 0.0666 (16) | 0.0473 (13) | 0.0053 (12) | −0.0027 (11) | −0.0117 (11) |
| C13 | 0.0522 (13) | 0.0573 (13) | 0.0512 (12) | 0.0001 (10) | 0.0031 (10) | −0.0004 (10) |
| N1 | 0.0459 (9) | 0.0357 (9) | 0.0461 (9) | −0.0045 (7) | 0.0133 (8) | 0.0032 (7) |
| N2 | 0.0426 (9) | 0.0396 (9) | 0.0438 (9) | 0.0039 (7) | 0.0110 (7) | 0.0074 (7) |
| O1 | 0.0569 (9) | 0.0475 (8) | 0.0396 (7) | −0.0107 (7) | 0.0117 (6) | 0.0005 (6) |
| O2 | 0.0413 (8) | 0.0474 (8) | 0.0559 (9) | −0.0080 (6) | −0.0055 (6) | 0.0093 (7) |
| Cl1 | 0.1200 (7) | 0.0595 (4) | 0.0754 (5) | 0.0290 (4) | −0.0372 (4) | 0.0025 (3) |
| Cl2 | 0.0868 (5) | 0.1270 (7) | 0.0546 (4) | 0.0193 (5) | 0.0230 (4) | −0.0133 (4) |
| S1 | 0.0382 (3) | 0.0352 (3) | 0.0372 (3) | −0.00503 (18) | 0.00356 (19) | 0.00245 (19) |
(E)-4-Chloro-N'-(4-chlorobenzylidene)benzenesulfonohydrazide (I) . Geometric parameters (Å, º)
| C1—C6 | 1.384 (3) | C8—C9 | 1.386 (3) |
| C1—C2 | 1.385 (3) | C9—C10 | 1.376 (3) |
| C1—S1 | 1.763 (2) | C9—H9 | 0.9300 |
| C2—C3 | 1.382 (3) | C10—C11 | 1.373 (4) |
| C2—H2 | 0.9300 | C10—H10 | 0.9300 |
| C3—C4 | 1.377 (4) | C11—C12 | 1.371 (4) |
| C3—H3 | 0.9300 | C11—Cl2 | 1.741 (2) |
| C4—C5 | 1.377 (3) | C12—C13 | 1.382 (3) |
| C4—Cl1 | 1.734 (2) | C12—H12 | 0.9300 |
| C5—C6 | 1.374 (3) | C13—H13 | 0.9300 |
| C5—H5 | 0.9300 | N1—N2 | 1.388 (2) |
| C6—H6 | 0.9300 | N1—S1 | 1.6311 (17) |
| C7—N2 | 1.269 (3) | N1—H1N | 0.830 (16) |
| C7—C8 | 1.463 (3) | O1—S1 | 1.4316 (14) |
| C7—H7 | 0.9300 | O2—S1 | 1.4212 (15) |
| C8—C13 | 1.385 (3) | ||
| C6—C1—C2 | 120.77 (19) | C10—C9—H9 | 119.6 |
| C6—C1—S1 | 120.05 (15) | C8—C9—H9 | 119.6 |
| C2—C1—S1 | 119.18 (15) | C11—C10—C9 | 119.6 (2) |
| C3—C2—C1 | 119.3 (2) | C11—C10—H10 | 120.2 |
| C3—C2—H2 | 120.4 | C9—C10—H10 | 120.2 |
| C1—C2—H2 | 120.4 | C12—C11—C10 | 121.0 (2) |
| C4—C3—C2 | 119.3 (2) | C12—C11—Cl2 | 119.5 (2) |
| C4—C3—H3 | 120.3 | C10—C11—Cl2 | 119.4 (2) |
| C2—C3—H3 | 120.3 | C11—C12—C13 | 119.0 (2) |
| C5—C4—C3 | 121.6 (2) | C11—C12—H12 | 120.5 |
| C5—C4—Cl1 | 119.19 (19) | C13—C12—H12 | 120.5 |
| C3—C4—Cl1 | 119.22 (18) | C12—C13—C8 | 121.2 (2) |
| C6—C5—C4 | 119.2 (2) | C12—C13—H13 | 119.4 |
| C6—C5—H5 | 120.4 | C8—C13—H13 | 119.4 |
| C4—C5—H5 | 120.4 | N2—N1—S1 | 118.76 (13) |
| C5—C6—C1 | 119.84 (19) | N2—N1—H1N | 118.8 (17) |
| C5—C6—H6 | 120.1 | S1—N1—H1N | 115.6 (17) |
| C1—C6—H6 | 120.1 | C7—N2—N1 | 114.02 (16) |
| N2—C7—C8 | 122.74 (19) | O2—S1—O1 | 119.93 (9) |
| N2—C7—H7 | 118.6 | O2—S1—N1 | 109.69 (9) |
| C8—C7—H7 | 118.6 | O1—S1—N1 | 102.85 (9) |
| C13—C8—C9 | 118.4 (2) | O2—S1—C1 | 107.94 (9) |
| C13—C8—C7 | 119.19 (19) | O1—S1—C1 | 109.14 (9) |
| C9—C8—C7 | 122.40 (19) | N1—S1—C1 | 106.52 (9) |
| C10—C9—C8 | 120.8 (2) | ||
| C6—C1—C2—C3 | −1.2 (3) | C10—C11—C12—C13 | −0.6 (4) |
| S1—C1—C2—C3 | 177.57 (16) | Cl2—C11—C12—C13 | −179.08 (19) |
| C1—C2—C3—C4 | 1.1 (3) | C11—C12—C13—C8 | 0.4 (4) |
| C2—C3—C4—C5 | 0.2 (3) | C9—C8—C13—C12 | −0.3 (4) |
| C2—C3—C4—Cl1 | −179.75 (17) | C7—C8—C13—C12 | 177.7 (2) |
| C3—C4—C5—C6 | −1.3 (4) | C8—C7—N2—N1 | 175.01 (17) |
| Cl1—C4—C5—C6 | 178.64 (17) | S1—N1—N2—C7 | 158.96 (15) |
| C4—C5—C6—C1 | 1.1 (3) | N2—N1—S1—O2 | 54.13 (17) |
| C2—C1—C6—C5 | 0.1 (3) | N2—N1—S1—O1 | −177.17 (15) |
| S1—C1—C6—C5 | −178.67 (17) | N2—N1—S1—C1 | −62.44 (17) |
| N2—C7—C8—C13 | 172.1 (2) | C6—C1—S1—O2 | −0.97 (19) |
| N2—C7—C8—C9 | −10.0 (3) | C2—C1—S1—O2 | −179.78 (15) |
| C13—C8—C9—C10 | 0.3 (4) | C6—C1—S1—O1 | −132.84 (16) |
| C7—C8—C9—C10 | −177.6 (2) | C2—C1—S1—O1 | 48.35 (18) |
| C8—C9—C10—C11 | −0.5 (4) | C6—C1—S1—N1 | 116.77 (17) |
| C9—C10—C11—C12 | 0.7 (4) | C2—C1—S1—N1 | −62.04 (18) |
| C9—C10—C11—Cl2 | 179.13 (19) |
(E)-4-Chloro-N'-(4-chlorobenzylidene)benzenesulfonohydrazide (I) . Hydrogen-bond geometry (Å, º)
Cg1 and Cg2 are the centroids of rings C1–C6 and C8–C13, respectively.
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1N···O1i | 0.83 (2) | 2.07 (2) | 2.903 (2) | 178 (2) |
| C4—Cl1···Cg2ii | 1.73 (1) | 3.41 (1) | 5.112 (2) | 166 (1) |
| C11—Cl2···Cg1iii | 1.74 (1) | 3.65 (1) | 5.372 (3) | 171 (1) |
Symmetry codes: (i) −x+1, −y+1, −z; (ii) x+1, y−1, z; (iii) −x, −y+1, −z+1.
(E)-4-chloro-N'-(4-Nitrobenzylidene)benzenesulfonohydrazide (II) . Crystal data
| C13H10ClN3O4S | F(000) = 1392 |
| Mr = 339.75 | Dx = 1.513 Mg m−3 |
| Monoclinic, P21/c | Mo Kα radiation, λ = 0.71073 Å |
| a = 19.903 (1) Å | Cell parameters from 7054 reflections |
| b = 10.2517 (7) Å | θ = 2.8–27.8° |
| c = 15.064 (1) Å | µ = 0.42 mm−1 |
| β = 103.929 (7)° | T = 293 K |
| V = 2983.3 (3) Å3 | Rod, yellow |
| Z = 8 | 0.48 × 0.40 × 0.36 mm |
(E)-4-chloro-N'-(4-Nitrobenzylidene)benzenesulfonohydrazide (II) . Data collection
| Oxford Diffraction Xcalibur diffractometer with Sapphire CCD | 4247 reflections with I > 2σ(I) |
| Radiation source: Enhance (Mo) X-ray Source | Rint = 0.025 |
| Rotation method data acquisition using ω scans. | θmax = 25.4°, θmin = 2.8° |
| Absorption correction: multi-scan (CrysAlis RED; Oxford Diffraction, 2009) | h = −23→23 |
| Tmin = 0.825, Tmax = 0.864 | k = −12→12 |
| 19194 measured reflections | l = −18→18 |
| 5455 independent reflections |
(E)-4-chloro-N'-(4-Nitrobenzylidene)benzenesulfonohydrazide (II) . Refinement
| Refinement on F2 | Hydrogen site location: mixed |
| Least-squares matrix: full | H atoms treated by a mixture of independent and constrained refinement |
| R[F2 > 2σ(F2)] = 0.038 | w = 1/[σ2(Fo2) + (0.0435P)2 + 1.8451P] where P = (Fo2 + 2Fc2)/3 |
| wR(F2) = 0.101 | (Δ/σ)max = 0.001 |
| S = 1.02 | Δρmax = 0.43 e Å−3 |
| 5455 reflections | Δρmin = −0.38 e Å−3 |
| 414 parameters | Extinction correction: SHELXL, Fc*=kFc[1+0.001xFc2λ3/sin(2θ)]-1/4 |
| 15 restraints | Extinction coefficient: 0.0034 (3) |
(E)-4-chloro-N'-(4-Nitrobenzylidene)benzenesulfonohydrazide (II) . Special details
| Geometry. All esds (except the esd in the dihedral angle between two l.s. planes) are estimated using the full covariance matrix. The cell esds are taken into account individually in the estimation of esds in distances, angles and torsion angles; correlations between esds in cell parameters are only used when they are defined by crystal symmetry. An approximate (isotropic) treatment of cell esds is used for estimating esds involving l.s. planes. |
(E)-4-chloro-N'-(4-Nitrobenzylidene)benzenesulfonohydrazide (II) . Fractional atomic coordinates and isotropic or equivalent isotropic displacement parameters (Å2)
| x | y | z | Uiso*/Ueq | Occ. (<1) | |
| Cl1 | 0.44715 (4) | 0.44299 (7) | 0.63321 (5) | 0.0692 (2) | |
| S1 | 0.41281 (3) | 1.03308 (5) | 0.53686 (4) | 0.03843 (15) | |
| O1 | 0.42255 (8) | 1.04883 (15) | 0.44626 (10) | 0.0447 (4) | |
| O2 | 0.44990 (8) | 1.11288 (16) | 0.60973 (11) | 0.0529 (4) | |
| O3 | 0.0285 (6) | 0.5036 (8) | 0.3381 (6) | 0.081 (3) | 0.55 |
| O3' | 0.0324 (8) | 0.5149 (10) | 0.3044 (7) | 0.075 (3) | 0.45 |
| O4 | −0.04648 (9) | 0.6576 (2) | 0.30599 (13) | 0.0681 (5) | |
| N1 | 0.33087 (10) | 1.06413 (19) | 0.53180 (14) | 0.0449 (5) | |
| H1N | 0.3271 (13) | 1.086 (2) | 0.5849 (12) | 0.054* | |
| N2 | 0.28443 (9) | 0.97344 (18) | 0.48183 (12) | 0.0426 (4) | |
| N3 | 0.01310 (11) | 0.6206 (2) | 0.32944 (15) | 0.0562 (5) | |
| C1 | 0.42530 (10) | 0.8671 (2) | 0.56436 (14) | 0.0367 (5) | |
| C2 | 0.44115 (12) | 0.8290 (2) | 0.65534 (15) | 0.0465 (6) | |
| H2 | 0.4461 | 0.8912 | 0.7014 | 0.056* | |
| C3 | 0.44953 (12) | 0.6982 (3) | 0.67705 (16) | 0.0519 (6) | |
| H3 | 0.4610 | 0.6714 | 0.7378 | 0.062* | |
| C4 | 0.44062 (11) | 0.6085 (2) | 0.60760 (17) | 0.0457 (6) | |
| C5 | 0.42596 (13) | 0.6447 (2) | 0.51720 (17) | 0.0534 (6) | |
| H5 | 0.4209 | 0.5821 | 0.4714 | 0.064* | |
| C6 | 0.41891 (13) | 0.7760 (2) | 0.49545 (15) | 0.0484 (6) | |
| H6 | 0.4099 | 0.8027 | 0.4347 | 0.058* | |
| C7 | 0.22211 (11) | 0.9867 (2) | 0.48701 (16) | 0.0438 (5) | |
| H7 | 0.2100 | 1.0569 | 0.5192 | 0.053* | |
| C8 | 0.16896 (11) | 0.8929 (2) | 0.44293 (15) | 0.0406 (5) | |
| C9 | 0.09969 (12) | 0.9292 (2) | 0.42131 (17) | 0.0499 (6) | |
| H9 | 0.0877 | 1.0142 | 0.4327 | 0.060* | |
| C10 | 0.04837 (12) | 0.8412 (2) | 0.38318 (17) | 0.0497 (6) | |
| H10 | 0.0021 | 0.8661 | 0.3679 | 0.060* | |
| C11 | 0.06736 (11) | 0.7159 (2) | 0.36837 (16) | 0.0443 (5) | |
| C12 | 0.13570 (12) | 0.6763 (2) | 0.38937 (18) | 0.0508 (6) | |
| H12 | 0.1472 | 0.5907 | 0.3787 | 0.061* | |
| C13 | 0.18635 (12) | 0.7649 (2) | 0.42624 (17) | 0.0476 (6) | |
| H13 | 0.2326 | 0.7395 | 0.4402 | 0.057* | |
| Cl2 | 0.11071 (4) | 0.30845 (9) | 0.53034 (6) | 0.0814 (3) | |
| S2 | 0.32436 (3) | 0.34333 (5) | 0.28991 (4) | 0.04203 (16) | |
| O5 | 0.28924 (9) | 0.38026 (17) | 0.19868 (11) | 0.0542 (4) | |
| O6 | 0.38459 (9) | 0.41231 (17) | 0.33703 (13) | 0.0610 (5) | |
| O7 | 0.10708 (11) | −0.49600 (19) | 0.13632 (16) | 0.0778 (6) | |
| O8 | 0.03109 (10) | −0.3435 (2) | 0.12357 (18) | 0.0896 (7) | |
| N4 | 0.34961 (10) | 0.19182 (18) | 0.28409 (13) | 0.0413 (4) | |
| H4N | 0.3793 (11) | 0.167 (2) | 0.3306 (13) | 0.050* | |
| N5 | 0.29454 (9) | 0.10878 (17) | 0.24828 (12) | 0.0386 (4) | |
| N6 | 0.09076 (11) | −0.3814 (2) | 0.14089 (16) | 0.0582 (6) | |
| C14 | 0.26262 (11) | 0.3420 (2) | 0.35620 (15) | 0.0388 (5) | |
| C15 | 0.19424 (12) | 0.3099 (2) | 0.31583 (15) | 0.0432 (5) | |
| H15 | 0.1803 | 0.2948 | 0.2532 | 0.052* | |
| C16 | 0.14730 (12) | 0.3006 (2) | 0.36964 (17) | 0.0479 (6) | |
| H16 | 0.1014 | 0.2790 | 0.3437 | 0.058* | |
| C17 | 0.16929 (13) | 0.3237 (2) | 0.46229 (17) | 0.0483 (6) | |
| C18 | 0.23655 (13) | 0.3573 (3) | 0.50252 (16) | 0.0534 (6) | |
| H18 | 0.2500 | 0.3742 | 0.5650 | 0.064* | |
| C19 | 0.28388 (12) | 0.3655 (2) | 0.44925 (16) | 0.0489 (6) | |
| H19 | 0.3298 | 0.3868 | 0.4757 | 0.059* | |
| C20 | 0.30626 (12) | −0.0126 (2) | 0.25909 (15) | 0.0413 (5) | |
| H20 | 0.3503 | −0.0419 | 0.2881 | 0.050* | |
| C21 | 0.25043 (11) | −0.1061 (2) | 0.22580 (15) | 0.0395 (5) | |
| C22 | 0.18148 (12) | −0.0665 (2) | 0.20002 (18) | 0.0496 (6) | |
| H22 | 0.1707 | 0.0215 | 0.2026 | 0.060* | |
| C23 | 0.12929 (12) | −0.1552 (2) | 0.17098 (18) | 0.0511 (6) | |
| H23 | 0.0834 | −0.1283 | 0.1534 | 0.061* | |
| C24 | 0.14638 (12) | −0.2851 (2) | 0.16848 (16) | 0.0436 (5) | |
| C25 | 0.21363 (12) | −0.3280 (2) | 0.19346 (17) | 0.0498 (6) | |
| H25 | 0.2238 | −0.4163 | 0.1907 | 0.060* | |
| C26 | 0.26565 (12) | −0.2384 (2) | 0.22262 (17) | 0.0482 (6) | |
| H26 | 0.3114 | −0.2663 | 0.2404 | 0.058* |
(E)-4-chloro-N'-(4-Nitrobenzylidene)benzenesulfonohydrazide (II) . Atomic displacement parameters (Å2)
| U11 | U22 | U33 | U12 | U13 | U23 | |
| Cl1 | 0.0651 (4) | 0.0455 (4) | 0.0853 (5) | −0.0022 (3) | −0.0048 (4) | 0.0214 (3) |
| S1 | 0.0359 (3) | 0.0367 (3) | 0.0404 (3) | −0.0048 (2) | 0.0046 (2) | −0.0003 (2) |
| O1 | 0.0414 (8) | 0.0469 (9) | 0.0454 (9) | −0.0022 (7) | 0.0096 (7) | 0.0086 (7) |
| O2 | 0.0544 (10) | 0.0450 (10) | 0.0524 (10) | −0.0110 (8) | −0.0006 (8) | −0.0087 (8) |
| O3 | 0.058 (3) | 0.050 (3) | 0.129 (7) | −0.007 (2) | 0.010 (5) | −0.002 (4) |
| O3' | 0.069 (4) | 0.062 (4) | 0.087 (5) | −0.005 (4) | 0.005 (4) | −0.018 (4) |
| O4 | 0.0382 (10) | 0.0845 (15) | 0.0791 (13) | −0.0053 (9) | 0.0093 (9) | −0.0026 (11) |
| N1 | 0.0406 (10) | 0.0446 (11) | 0.0488 (11) | −0.0001 (9) | 0.0094 (9) | −0.0083 (9) |
| N2 | 0.0380 (10) | 0.0443 (11) | 0.0438 (10) | −0.0011 (9) | 0.0065 (8) | −0.0028 (9) |
| N3 | 0.0421 (12) | 0.0600 (15) | 0.0649 (14) | −0.0050 (11) | 0.0102 (10) | −0.0023 (12) |
| C1 | 0.0310 (10) | 0.0404 (12) | 0.0371 (11) | −0.0048 (9) | 0.0051 (8) | 0.0017 (9) |
| C2 | 0.0483 (13) | 0.0506 (14) | 0.0360 (12) | −0.0052 (11) | 0.0014 (10) | −0.0009 (10) |
| C3 | 0.0507 (14) | 0.0594 (16) | 0.0393 (13) | −0.0033 (12) | −0.0015 (11) | 0.0136 (12) |
| C4 | 0.0367 (12) | 0.0401 (13) | 0.0557 (14) | −0.0023 (10) | 0.0021 (10) | 0.0126 (11) |
| C5 | 0.0664 (16) | 0.0405 (13) | 0.0511 (14) | −0.0042 (12) | 0.0097 (12) | −0.0031 (11) |
| C6 | 0.0650 (15) | 0.0447 (14) | 0.0337 (12) | −0.0035 (12) | 0.0085 (11) | 0.0028 (10) |
| C7 | 0.0400 (12) | 0.0392 (12) | 0.0527 (14) | 0.0061 (10) | 0.0119 (10) | 0.0010 (10) |
| C8 | 0.0355 (11) | 0.0437 (13) | 0.0434 (12) | 0.0033 (10) | 0.0113 (9) | 0.0039 (10) |
| C9 | 0.0397 (12) | 0.0435 (13) | 0.0667 (16) | 0.0103 (11) | 0.0131 (11) | −0.0002 (12) |
| C10 | 0.0324 (11) | 0.0535 (15) | 0.0614 (15) | 0.0076 (11) | 0.0081 (11) | 0.0050 (12) |
| C11 | 0.0383 (12) | 0.0475 (14) | 0.0472 (13) | −0.0010 (10) | 0.0105 (10) | 0.0034 (11) |
| C12 | 0.0419 (13) | 0.0418 (13) | 0.0677 (16) | 0.0059 (11) | 0.0113 (11) | −0.0033 (12) |
| C13 | 0.0327 (11) | 0.0481 (14) | 0.0616 (15) | 0.0079 (10) | 0.0106 (10) | 0.0033 (12) |
| Cl2 | 0.0824 (5) | 0.0998 (6) | 0.0759 (5) | −0.0171 (5) | 0.0459 (4) | −0.0163 (4) |
| S2 | 0.0445 (3) | 0.0341 (3) | 0.0487 (3) | −0.0011 (2) | 0.0137 (2) | 0.0007 (2) |
| O5 | 0.0642 (11) | 0.0510 (10) | 0.0514 (10) | 0.0132 (9) | 0.0218 (8) | 0.0169 (8) |
| O6 | 0.0524 (10) | 0.0512 (10) | 0.0824 (13) | −0.0141 (8) | 0.0219 (9) | −0.0147 (9) |
| O7 | 0.0715 (13) | 0.0450 (11) | 0.1165 (18) | −0.0122 (10) | 0.0217 (12) | −0.0072 (11) |
| O8 | 0.0452 (11) | 0.0735 (15) | 0.143 (2) | −0.0107 (10) | 0.0096 (12) | −0.0044 (14) |
| N4 | 0.0401 (10) | 0.0353 (10) | 0.0446 (11) | 0.0022 (8) | 0.0027 (8) | −0.0006 (8) |
| N5 | 0.0395 (10) | 0.0349 (10) | 0.0397 (10) | −0.0002 (8) | 0.0064 (8) | 0.0001 (8) |
| N6 | 0.0521 (13) | 0.0532 (14) | 0.0700 (15) | −0.0095 (11) | 0.0160 (11) | −0.0011 (11) |
| C14 | 0.0439 (12) | 0.0315 (11) | 0.0407 (12) | 0.0031 (9) | 0.0094 (9) | −0.0009 (9) |
| C15 | 0.0451 (13) | 0.0432 (13) | 0.0388 (12) | 0.0024 (10) | 0.0050 (10) | −0.0056 (10) |
| C16 | 0.0425 (13) | 0.0468 (14) | 0.0533 (14) | −0.0016 (11) | 0.0093 (11) | −0.0062 (11) |
| C17 | 0.0542 (14) | 0.0443 (13) | 0.0511 (14) | −0.0032 (11) | 0.0216 (11) | −0.0061 (11) |
| C18 | 0.0642 (16) | 0.0565 (16) | 0.0388 (13) | −0.0021 (13) | 0.0110 (11) | −0.0084 (11) |
| C19 | 0.0462 (13) | 0.0501 (14) | 0.0474 (14) | −0.0060 (11) | 0.0051 (11) | −0.0078 (11) |
| C20 | 0.0411 (12) | 0.0405 (13) | 0.0419 (12) | 0.0052 (10) | 0.0092 (10) | 0.0001 (10) |
| C21 | 0.0423 (12) | 0.0346 (12) | 0.0413 (12) | 0.0043 (10) | 0.0094 (10) | 0.0013 (10) |
| C22 | 0.0452 (13) | 0.0356 (12) | 0.0669 (16) | 0.0070 (10) | 0.0112 (12) | 0.0032 (11) |
| C23 | 0.0389 (12) | 0.0443 (14) | 0.0683 (16) | 0.0054 (11) | 0.0089 (11) | 0.0055 (12) |
| C24 | 0.0441 (13) | 0.0399 (13) | 0.0486 (13) | −0.0041 (10) | 0.0149 (10) | −0.0008 (10) |
| C25 | 0.0517 (14) | 0.0334 (12) | 0.0660 (16) | 0.0041 (11) | 0.0173 (12) | −0.0051 (11) |
| C26 | 0.0404 (12) | 0.0403 (13) | 0.0631 (15) | 0.0058 (10) | 0.0107 (11) | −0.0031 (11) |
(E)-4-chloro-N'-(4-Nitrobenzylidene)benzenesulfonohydrazide (II) . Geometric parameters (Å, º)
| Cl1—C4 | 1.738 (2) | Cl2—C17 | 1.735 (2) |
| S1—O2 | 1.4245 (16) | S2—O6 | 1.4252 (17) |
| S1—O1 | 1.4331 (16) | S2—O5 | 1.4346 (17) |
| S1—N1 | 1.6452 (19) | S2—N4 | 1.6414 (19) |
| S1—C1 | 1.754 (2) | S2—C14 | 1.760 (2) |
| O3—N3 | 1.237 (8) | O7—N6 | 1.226 (3) |
| O3'—N3 | 1.239 (10) | O8—N6 | 1.217 (3) |
| O4—N3 | 1.213 (3) | N4—N5 | 1.390 (2) |
| N1—N2 | 1.397 (3) | N4—H4N | 0.839 (16) |
| N1—H1N | 0.849 (16) | N5—C20 | 1.269 (3) |
| N2—C7 | 1.269 (3) | N6—C24 | 1.467 (3) |
| N3—C11 | 1.469 (3) | C14—C19 | 1.384 (3) |
| C1—C6 | 1.379 (3) | C14—C15 | 1.390 (3) |
| C1—C2 | 1.386 (3) | C15—C16 | 1.380 (3) |
| C2—C3 | 1.381 (3) | C15—H15 | 0.9300 |
| C2—H2 | 0.9300 | C16—C17 | 1.379 (3) |
| C3—C4 | 1.372 (4) | C16—H16 | 0.9300 |
| C3—H3 | 0.9300 | C17—C18 | 1.374 (3) |
| C4—C5 | 1.373 (3) | C18—C19 | 1.379 (3) |
| C5—C6 | 1.385 (3) | C18—H18 | 0.9300 |
| C5—H5 | 0.9300 | C19—H19 | 0.9300 |
| C6—H6 | 0.9300 | C20—C21 | 1.462 (3) |
| C7—C8 | 1.465 (3) | C20—H20 | 0.9300 |
| C7—H7 | 0.9300 | C21—C26 | 1.393 (3) |
| C8—C9 | 1.389 (3) | C21—C22 | 1.394 (3) |
| C8—C13 | 1.396 (3) | C22—C23 | 1.370 (3) |
| C9—C10 | 1.380 (3) | C22—H22 | 0.9300 |
| C9—H9 | 0.9300 | C23—C24 | 1.377 (3) |
| C10—C11 | 1.372 (3) | C23—H23 | 0.9300 |
| C10—H10 | 0.9300 | C24—C25 | 1.373 (3) |
| C11—C12 | 1.381 (3) | C25—C26 | 1.375 (3) |
| C12—C13 | 1.371 (3) | C25—H25 | 0.9300 |
| C12—H12 | 0.9300 | C26—H26 | 0.9300 |
| C13—H13 | 0.9300 | ||
| O2—S1—O1 | 120.35 (10) | C8—C13—H13 | 119.8 |
| O2—S1—N1 | 104.40 (10) | O6—S2—O5 | 120.24 (11) |
| O1—S1—N1 | 107.10 (10) | O6—S2—N4 | 105.33 (10) |
| O2—S1—C1 | 110.93 (10) | O5—S2—N4 | 106.46 (10) |
| O1—S1—C1 | 106.87 (10) | O6—S2—C14 | 109.87 (10) |
| N1—S1—C1 | 106.33 (10) | O5—S2—C14 | 107.46 (10) |
| N2—N1—S1 | 114.59 (15) | N4—S2—C14 | 106.70 (10) |
| N2—N1—H1N | 118.9 (18) | N5—N4—S2 | 112.16 (14) |
| S1—N1—H1N | 108.5 (17) | N5—N4—H4N | 118.0 (17) |
| C7—N2—N1 | 115.17 (19) | S2—N4—H4N | 113.8 (17) |
| O4—N3—O3 | 122.2 (6) | C20—N5—N4 | 116.56 (18) |
| O4—N3—O3' | 122.5 (8) | O8—N6—O7 | 123.5 (2) |
| O4—N3—C11 | 118.8 (2) | O8—N6—C24 | 118.5 (2) |
| O3—N3—C11 | 117.5 (6) | O7—N6—C24 | 118.0 (2) |
| O3'—N3—C11 | 116.8 (8) | C19—C14—C15 | 120.8 (2) |
| C6—C1—C2 | 120.7 (2) | C19—C14—S2 | 119.21 (17) |
| C6—C1—S1 | 119.78 (17) | C15—C14—S2 | 119.83 (17) |
| C2—C1—S1 | 119.49 (17) | C16—C15—C14 | 119.3 (2) |
| C3—C2—C1 | 119.6 (2) | C16—C15—H15 | 120.3 |
| C3—C2—H2 | 120.2 | C14—C15—H15 | 120.3 |
| C1—C2—H2 | 120.2 | C17—C16—C15 | 119.2 (2) |
| C4—C3—C2 | 119.0 (2) | C17—C16—H16 | 120.4 |
| C4—C3—H3 | 120.5 | C15—C16—H16 | 120.4 |
| C2—C3—H3 | 120.5 | C18—C17—C16 | 121.9 (2) |
| C3—C4—C5 | 122.2 (2) | C18—C17—Cl2 | 118.95 (19) |
| C3—C4—Cl1 | 119.75 (19) | C16—C17—Cl2 | 119.19 (19) |
| C5—C4—Cl1 | 118.1 (2) | C17—C18—C19 | 119.2 (2) |
| C4—C5—C6 | 118.8 (2) | C17—C18—H18 | 120.4 |
| C4—C5—H5 | 120.6 | C19—C18—H18 | 120.4 |
| C6—C5—H5 | 120.6 | C18—C19—C14 | 119.6 (2) |
| C1—C6—C5 | 119.6 (2) | C18—C19—H19 | 120.2 |
| C1—C6—H6 | 120.2 | C14—C19—H19 | 120.2 |
| C5—C6—H6 | 120.2 | N5—C20—C21 | 119.9 (2) |
| N2—C7—C8 | 120.8 (2) | N5—C20—H20 | 120.1 |
| N2—C7—H7 | 119.6 | C21—C20—H20 | 120.1 |
| C8—C7—H7 | 119.6 | C26—C21—C22 | 118.8 (2) |
| C9—C8—C13 | 119.0 (2) | C26—C21—C20 | 119.8 (2) |
| C9—C8—C7 | 119.9 (2) | C22—C21—C20 | 121.3 (2) |
| C13—C8—C7 | 121.0 (2) | C23—C22—C21 | 121.0 (2) |
| C10—C9—C8 | 121.0 (2) | C23—C22—H22 | 119.5 |
| C10—C9—H9 | 119.5 | C21—C22—H22 | 119.5 |
| C8—C9—H9 | 119.5 | C22—C23—C24 | 118.5 (2) |
| C11—C10—C9 | 118.3 (2) | C22—C23—H23 | 120.8 |
| C11—C10—H10 | 120.8 | C24—C23—H23 | 120.8 |
| C9—C10—H10 | 120.8 | C25—C24—C23 | 122.3 (2) |
| C10—C11—C12 | 122.2 (2) | C25—C24—N6 | 118.8 (2) |
| C10—C11—N3 | 118.8 (2) | C23—C24—N6 | 118.9 (2) |
| C12—C11—N3 | 119.0 (2) | C24—C25—C26 | 118.9 (2) |
| C13—C12—C11 | 119.0 (2) | C24—C25—H25 | 120.6 |
| C13—C12—H12 | 120.5 | C26—C25—H25 | 120.6 |
| C11—C12—H12 | 120.5 | C25—C26—C21 | 120.6 (2) |
| C12—C13—C8 | 120.4 (2) | C25—C26—H26 | 119.7 |
| C12—C13—H13 | 119.8 | C21—C26—H26 | 119.7 |
| O2—S1—N1—N2 | −164.14 (15) | C7—C8—C13—C12 | 176.5 (2) |
| O1—S1—N1—N2 | 67.20 (18) | O6—S2—N4—N5 | 173.52 (15) |
| C1—S1—N1—N2 | −46.79 (18) | O5—S2—N4—N5 | −57.76 (17) |
| S1—N1—N2—C7 | 171.43 (17) | C14—S2—N4—N5 | 56.77 (17) |
| O2—S1—C1—C6 | −153.93 (18) | S2—N4—N5—C20 | −165.33 (16) |
| O1—S1—C1—C6 | −21.0 (2) | O6—S2—C14—C19 | −19.8 (2) |
| N1—S1—C1—C6 | 93.2 (2) | O5—S2—C14—C19 | −152.28 (18) |
| O2—S1—C1—C2 | 26.4 (2) | N4—S2—C14—C19 | 93.9 (2) |
| O1—S1—C1—C2 | 159.36 (17) | O6—S2—C14—C15 | 163.92 (18) |
| N1—S1—C1—C2 | −86.50 (19) | O5—S2—C14—C15 | 31.5 (2) |
| C6—C1—C2—C3 | −1.0 (3) | N4—S2—C14—C15 | −82.38 (19) |
| S1—C1—C2—C3 | 178.61 (18) | C19—C14—C15—C16 | −0.5 (3) |
| C1—C2—C3—C4 | −1.2 (4) | S2—C14—C15—C16 | 175.69 (17) |
| C2—C3—C4—C5 | 2.3 (4) | C14—C15—C16—C17 | 0.2 (3) |
| C2—C3—C4—Cl1 | −177.17 (18) | C15—C16—C17—C18 | 0.8 (4) |
| C3—C4—C5—C6 | −1.0 (4) | C15—C16—C17—Cl2 | −178.57 (18) |
| Cl1—C4—C5—C6 | 178.45 (19) | C16—C17—C18—C19 | −1.3 (4) |
| C2—C1—C6—C5 | 2.3 (4) | Cl2—C17—C18—C19 | 178.0 (2) |
| S1—C1—C6—C5 | −177.32 (19) | C17—C18—C19—C14 | 1.0 (4) |
| C4—C5—C6—C1 | −1.3 (4) | C15—C14—C19—C18 | −0.1 (4) |
| N1—N2—C7—C8 | −175.87 (19) | S2—C14—C19—C18 | −176.29 (19) |
| N2—C7—C8—C9 | −157.7 (2) | N4—N5—C20—C21 | 178.17 (18) |
| N2—C7—C8—C13 | 25.5 (3) | N5—C20—C21—C26 | 168.5 (2) |
| C13—C8—C9—C10 | −0.5 (4) | N5—C20—C21—C22 | −13.9 (3) |
| C7—C8—C9—C10 | −177.3 (2) | C26—C21—C22—C23 | −0.7 (4) |
| C8—C9—C10—C11 | 1.0 (4) | C20—C21—C22—C23 | −178.4 (2) |
| C9—C10—C11—C12 | −0.7 (4) | C21—C22—C23—C24 | 0.5 (4) |
| C9—C10—C11—N3 | 178.8 (2) | C22—C23—C24—C25 | −0.4 (4) |
| O4—N3—C11—C10 | 4.8 (3) | C22—C23—C24—N6 | 177.7 (2) |
| O3—N3—C11—C10 | −161.9 (5) | O8—N6—C24—C25 | 176.1 (2) |
| O3'—N3—C11—C10 | 169.5 (6) | O7—N6—C24—C25 | −4.1 (4) |
| O4—N3—C11—C12 | −175.6 (2) | O8—N6—C24—C23 | −2.1 (4) |
| O3—N3—C11—C12 | 17.7 (6) | O7—N6—C24—C23 | 177.8 (2) |
| O3'—N3—C11—C12 | −10.9 (6) | C23—C24—C25—C26 | 0.5 (4) |
| C10—C11—C12—C13 | 0.0 (4) | N6—C24—C25—C26 | −177.6 (2) |
| N3—C11—C12—C13 | −179.6 (2) | C24—C25—C26—C21 | −0.7 (4) |
| C11—C12—C13—C8 | 0.5 (4) | C22—C21—C26—C25 | 0.8 (4) |
| C9—C8—C13—C12 | −0.2 (4) | C20—C21—C26—C25 | 178.5 (2) |
(E)-4-chloro-N'-(4-Nitrobenzylidene)benzenesulfonohydrazide (II) . Hydrogen-bond geometry (Å, º)
Cg3 is the centroid of the C14–C19 ring.
| D—H···A | D—H | H···A | D···A | D—H···A |
| N1—H1N···O5i | 0.85 (2) | 2.06 (2) | 2.887 (3) | 163 (2) |
| N4—H4N···O1ii | 0.84 (2) | 2.13 (2) | 2.918 (2) | 157 (2) |
| C10—H10···O7iii | 0.93 | 2.58 | 3.465 (3) | 159 |
| C16—H16···O4iv | 0.93 | 2.58 | 3.259 (3) | 131 |
| C25—H25···O5ii | 0.93 | 2.45 | 3.340 (3) | 161 |
| C12—H12···Cg3 | 0.93 | 2.96 | 3.843 (2) | 160 |
Symmetry codes: (i) x, −y+3/2, z+1/2; (ii) x, y−1, z; (iii) −x, y+3/2, −z+1/2; (iv) −x, y−1/2, −z+1/2.
Funding Statement
This work was funded by Department of Science and Technology, Government of India grant DST-PURSE to Akshatha R. Salian. University Grants Commission, Government of India grant UGC-BSR one-time grant to faculty to B. Thimme Gowda.
References
- Balaji, J., John Francis Xavier, J., Prabu, S. & Srinivasan, P. (2014). Acta Cryst. E70, o1250–o1251. [DOI] [PMC free article] [PubMed]
- Hashemi, S. A. (2012). Tetrahedron Lett. 53, 5141–5143.
- Hussain, M. M., Rahman, M. M., Arshad, M. N. & Asiri, A. M. (2017a). ACS Omega, 2, 420–431. [DOI] [PMC free article] [PubMed]
- Hussain, M. M., Rahman, M. M., Arshad, M. N. & Asiri, A. M. (2017b). Sci. Rep. 7, 5832. [DOI] [PMC free article] [PubMed]
- Kia, R., Etemadi, B., Fun, H.-K. & Kargar, H. (2009b). Acta Cryst. E65, o821–o822. [DOI] [PMC free article] [PubMed]
- Kia, R., Fun, H.-K. & Kargar, H. (2009a). Acta Cryst. E65, o1119–o1120. [DOI] [PMC free article] [PubMed]
- McKinnon, J. J., Spackman, M. A. & Mitchell, A. S. (2004). Acta Cryst. B60, 627–668. [DOI] [PubMed]
- Ogawa, C., Konishi, H., Sugiura, M. & Kobayashi, S. (2004). Org. Biomol. Chem. 2, 446–448. [DOI] [PubMed]
- Oxford Diffraction (2009). CrysAlis CCD and CrysAlis RED. Oxford Diffraction Ltd., Abingdon, England.
- Reis, D. C., Despaigne, A. A. R., Da Silva, J. G., Silva, N. F., Vilela, C. F., Mendes, I. C., Takahashi, J. A. & Beraldo, H. (2013). Molecules, 18, 12645–12662. [DOI] [PMC free article] [PubMed]
- Rollas, S. & Küçükgüzel, S. G. (2007). Molecules, 12, 1910–1939. [DOI] [PMC free article] [PubMed]
- Salian, A. R., Foro, S. & Gowda, B. T. (2018). Acta Cryst. E74, 1613–1618. [DOI] [PMC free article] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sheldrick, G. M. (2015). Acta Cryst. C71, 3–8.
- Silva, C. M. da, da Silva, D. L., Modolo, L. V., Alves, R. B., de Resende, M. A., Martins, C. V. B. & de Fátima, A. (2011). J. Adv. Res. 2, 1–8.
- Spackman, M. A. & Jayatilaka, D. (2009). CrystEngComm, 11, 19–32.
- Spek, A. L. (2009). Acta Cryst. D65, 148–155. [DOI] [PMC free article] [PubMed]
- Turner, M. J., McKinnon, J. J., Wolff, S. K., Grimwood, D. J., Spackman, P. R., Jayatilaka, D. & Spackman, M. A. (2017). Crystal Explorer. The University of Western Australia.
- Weber, B., Tandon, R. & Himsl, D. (2007). Z. Anorg. Allg. Chem. 633, 1159–1162.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Crystal structure: contains datablock(s) I, II, global. DOI: 10.1107/S205698901801592X/su5458sup1.cif
Structure factors: contains datablock(s) I. DOI: 10.1107/S205698901801592X/su5458Isup2.hkl
Structure factors: contains datablock(s) II. DOI: 10.1107/S205698901801592X/su5458IIsup3.hkl
Additional supporting information: crystallographic information; 3D view; checkCIF report