Abstract
Visceral leishmaniasis (VL) in humans is a chronic and often fatal disease if left untreated. Dogs appear to be the main reservoir host for L. infantum infection, however, in many regions other canids such as jackals, foxes, wolves and other mammals, such as hares or black rats, have been implicated as wild reservoirs. Most dogs cannot form an effective immune response against this infection, and this could be modulated by small non-coding RNAs, called microRNAs, responsible for post-transcriptional control of gene expression. Here, we evaluated the expression of miRNAs in peripheral blood mononuclear cells (PBMC) of symptomatic dogs naturally infected with Leishmania (L.) infantum (n = 10) and compared to those of healthy dogs (n = 5). Microarray analysis revealed that miR-21, miR-424, miR-194 and miR-451 had a 3-fold increase in expression, miR-192, miR-503, and miR-371 had a 2-fold increase in expression, whereas a 2-fold reduction in expression was observed for miR-150 and miR-574. Real-time PCR validated the differential expression of miR-21, miR-150, miR-451, miR-192, miR-194, and miR-371. Parasite load of PBMC was measured by real-time PCR and correlated to the differentially expressed miRNAs, showing a strong positive correlation with expression of miR-194, a regular positive correlation with miR-371 expression, and a moderate negative correlation with miR-150 expression in PBMC. These findings suggest that Leishmania infection interferes with miRNAs expression in PBMC, and their correlation with parasite load may help in the identification of therapeutic targets in Canine Visceral Leishmaniasis (CVL).
Introduction
Visceral Leishmaniasis (VL) is a zoonosis caused by the protozoan Leishmania infantum and is the most fatal form of this parasitic disease [1]. Despite occurring in 76 countries, VL is still one of the most neglected diseases in the world and, of the human cases reported in America, 95.1% are in Brazil [2]. Dogs are considered the main domestic reservoirs of L. infantum [3]. Once in the vertebrate host, the parasite may cause lesions and symptoms that are characteristic of Canine Visceral Leishmaniasis (CVL), although some infected dogs may be oligo or asymptomatic [4], others may evolve to spontaneous cure [5]. The most frequent signs of VL are lymphadenopathy, onychogryphosis, cutaneous lesions, weight loss, cachexia, fever and locomotor abnormalities [6].
Protective immunity in dogs has generally been associated with a cellular immune response manifested by a positive lymphoproliferative response to Leishmania spp antigens [7] and cytokine production, such as IFNγ and TNF-α, which are necessary for macrophage activation and parasite death [8]. The role of T cell in the induction of the cellular response is determinant for the elimination of the parasite inside the macrophages [9].
In recent years, microRNAs (miRNAs) have been shown to play a critical role in the development and function of immune responses [10]. miRNAs are a group of small, highly conserved, single-stranded non-coding RNAs that regulate gene expression at the post-transcriptional level [11].
In vitro infection of human phagocytes with Leishmania donovani showed that the parasite induces alteration on the expression of miR-21, miR-155 and miR-146b-5p and interferes with the TGF-β signaling pathway [12]. In vitro infection of J774 murine macrophages with L. infantum increases miR-155, which in turn plays a role in regulating the response to L. infantum [13]. In BALB/c mice infected with L. donovani, there is a decrease in miR-122 expression, facilitating hepatic infection, and an increase in miR-122 expression decreases parasite burden in the liver [14]. Leishmania donovani infected mice macrophages have altered expression of miR-3620, miR-6385, miR-6973a, miR-6996, miR-328, miR-8113, miR-3473f, miR-763, miR-6540 and miR-1264, that are involved in controlling macrophage effector functions [15]. An in silico study has also shown that miR-29a and miR-29b target signal transcription factors that play a role in the proliferation and differentiation of T cells in visceral leishmaniasis in human, indicating that miRNAs can regulate immune response and infection control [16]. Although there are several studies on the role of miRNAs in VL, the expression of miRNAs in infected dogs, the most important reservoir for Leishmania, has not yet been described.
In this study, we demonstrated that there are differences in the expression of miRNAs in PBMC of dogs with VL compared to healthy dogs. The parasite load showed strong positive correlation with the miR-194 expression and regular correlation with miR-371 and miR-150 expressions in PBMC.
Materials and methods
Screening of animals and collection of samples
This study was approved by the Committee of Ethics in Animal Experimental Research (COBEA), with the approval of the Institutional Animal Care and Use Committee of UNESP–Universidade Estadual Paulista "Júlio de Mesquita Filho"—Araçatuba—School of Veterinary Medicine—FMVA (process 00978/2016). The generation of the dataset analysed within this manuscript is available in a data descriptor article [17].
Five healthy animals from an endemic area (Araçatuba-SP, Brazil) with absence of parasitic DNA by PCR, upon clinical examination, complete blood count and serum biochemical profile evaluation, were selected for this study, and ten animals naturally infected with Leishmania infantum, from the Zoonosis Control Center of Araçatuba, were selected. These animals contained at least three characteristic clinical signs of the disease, including onychogriphosis, weight loss, ear-tip injuries, periocular lesions, alopecia, skin lesions and lymphadenopathy (S1 Table). Blood samples from both groups (infected and healthy) were collected in tubes without EDTA to obtain serum for biochemical profile (S2 Table) and indirect ELISA assay (S1 Table) for the detection of anti-leishmania antibodies [18], and in EDTA tubes for complete blood count (CBC) (S3 Table) and isolation of the PBMC. Based on clinical signs, CBC results and biochemical profile, infected animals were classified in the clinical stage II of the disease [19]. PCR for detection of Leishmania DNA from PBMC cells was performed for all animals. Infected animals were euthanized by barbiturate anesthesia (Tiopental, Cristália Itapira, SP), followed by intravenous injection of potassium chloride 19.1% by the same route, in compliance with local laws.
Isolation of peripheral blood mononuclear cells
PBMC were isolated by gradient Histopaque 1077 (Sigma-Aldrich) following manufacturer's instructions. They were then washed twice in phosphate buffered saline solution at pH 7.2. After isolation, these cells were counted in a Neubauer chamber for further extraction of DNA and total RNA containing the miRNAs.
DNA extraction and determination of the Leishmania species
DNA extraction from PBMC samples from the experimental dogs was performed using 5x106 cells with the commercial DNAeasy kit (Qiagen) according to the manufacturer's recommendations. Extracted DNA was quantified in spectrophotometer 260/280 (NanoDrop, Thermo Fisher Scientific) for evaluation of purity and concentration, and were then stored at -20°C until analysis.
Determination of the Leishmania species was performed by PCR-RFLP (Restriction Fragment Length Polymorphism) [20], comparing the restriction profiles of the sample with a PCR restriction profile obtained from L. infantum (IOC / L0575-MHOM / BR / 2002 / LPC-RPV), L. braziliensis (IOC / L0566-MHOM / BR / 1975 / M2903) and L. amazonensis (IOC / L0575-MHOM / BR / 1967 / PH8) as positive controls, and water as a negative control.
Quantification of the parasite load by real-time PCR
DNA extraction from PBMC samples from the experimental dogs was performed using 5x106 cells with the commercial DNAeasy kit (Qiagen) according to manufacturer's recommendation.
Parasite load quantification was performed by real-time PCR with a final reaction volume of 20μL using primers amplifying a 116bp fragment of the kinetoplast DNA (kDNA) of Leishmania spp. (5' CCTATTTTACACCAACCCCCAGT 3' and 5 'GGGTAGGGGCGTTCTGCGAAA 3'), at a concentration of 900nM [21], Power SYBR Green PCR Master Mix (Applied Biosystems) and 50ng of sample DNA. Amplification condition used was comprised of an initial heating of 95°C for 10 minutes, 40 cycles of 95°C for 15 seconds and 65°C for 60 seconds. Upon the end of amplification, a dissociation curve of the amplified fragment was determined from 60°C to 95°C with an increase of 0.5°C every 5 seconds. A standard curve with DNA from Leishmania infantum promastigotes (MHOM/BR00/MER02) with a serial dilution of 108 to 101 of parasite DNA was performed for each reaction.
Extraction and quantification of miRNA
Extraction of total RNA from 5x106 PBMC was performed on the samples with the commercial mirVana kit for isolation of miRNA with phenol (Life Technologies), following the procedure indicated by the manufacturer. Following isolation, total RNA was stored at -80°C until evaluation of quality and concentration.
Isolated RNAs were analyzed by spectrophotometry (NanoDrop, Thermo Fisher Scientific) for evaluation of their quantity and purity (260/280). Before performing microarray, samples were also analyzed for RNA quality by capillary electrophoresis (Bioanalyzer, Agilent Technologies) using the commercial Agilent RNA 6000 Nano kit, following manufacturer's instructions.
Microarray
Total RNAs with satisfactory amount (over than 30ng/μl) and quality (RNA integrity number > 8) were used to perform microarray analysis using a miRNA 4.1 Array Strip (Affymetrix) containing probes designed for the miRNAs of several species, including 291 from Canis familiaris.
MicroRNAs were biotinylated using the Affymetrix FlashTag Biotin HSR RNA Labeling Kit following manufacturer's instructions. For the microarray, GeneAtlas Hybridization, Wash, and Stain Kit for miRNA array Strips were used, following manufacturer's instructions.
Microarray data were deposited in Gene Expression Omnibus with the access number GSE105443 according to the minimum information about microarray experiment (MIAME) standards.
Microarray data analysis
Normalization and quality of microarray analysis of the miRNAs of control and infected dogs were performed in the Expression Console Software program, version 1.4.1 (Affymetrix, Thermo Fisher Scientific). Differential analysis of the miRNAs was performed in the Transcriptome Analysis Console (Affymetrix, Thermo Fisher Scientific).
The miRBase database (mirbase.org) was used to check the homology between the sequences of all differentially expressed non-canine miRNAs with known canine miRNAs.
Analysis of targets and pathways for the differentially expressed and validated miRNAs in dogs with VL were performed using the Ingenuity Pathway Analysis program (Qiagen).
Gene Ontology (GO) enrichment analysis was performed using the ENRICHR program (http://amp.pharm.mssm.edu/Enrichr/) [22,23].
Real-time PCR for miRNAs analysis
To validate the results obtained by microarray, real-time PCR (qPCR) was performed. cDNA production was performed using the miScript RT II kit (Qiagen), as recommended by the manufacturer. qPCR reactions were performed using commercially available specific primers for our Canis familiaris miRNAs of interest and the endogenous reference SNORD96A (miScript, Qiagen) using the SYBR Green system (miScript SYBR Green PCR kit, Qiagen) in real-time thermal cycler (RealPlex, Eppendorf). Amplification conditions were determined by the manufacturer. All miRNAs (including the reference) were run in separate plates. For each miRNA, a standard curve with a serial dilution of a pool of the cDNAs was performed. Absolute quantification of each miRNA was performed by converting sample cycle threshold values to a concentration (ng/μl), based on the standard curves, which were generated using 10-fold serial dilutions of the pool of cDNAs. Target amount was then divided by SNORD96A levels to obtain a normalized target value. All samples were evaluated in duplicate.
Statistical analysis
Statistical analysis were performed using the GraphPad Prism 6 software (GraphPad Software, Inc., La Jolla, CA, USA). Analysis of variance (ANOVA) was performed for treatment comparison in the microarray. Mann-Whitney test was performed for the miRNA qPCR results. Spearman correlation was done to evaluate the association between miRNA expression and blood count, biochemical molecules and parasite load. Fisher test was used by IPA for canonical pathway analysis. Results were considered significant when p<0.05.
Results
Leishmania infantum was identified in dogs with VL
PCR-RFLP was performed to identify the Leishmania species. In all dogs of the infected group, L. infantum was found as the causative agent, as shown in S1 Fig.
Differentially expressed microRNAs and validation in PBMC of dogs with VL compared to healthy dogs
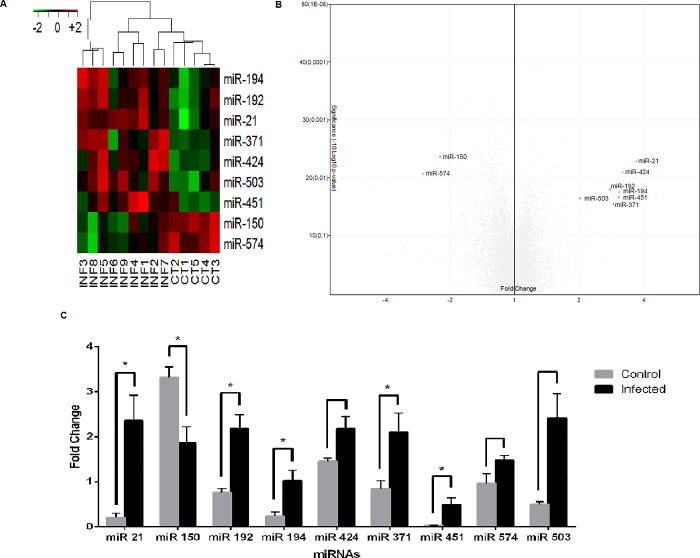
Considering that miRNAs can regulate immune response [11], microarray was employed for comparative analysis of miRNA expression in the blood of dogs naturally infected with L. infantum and healthy dogs. miRNAs miR-194, miR-192, miR-21, miR-424, miR-451, miR-503 and miR-371 showed increased expression in the blood of infected dogs (3.1, 2.8, 3.7, 3.2, 3.1, 2.0 and 2.9 fold change respectively), while miR-150 and miR-574 showed decreased blood expression (-2.2 and -2.7 fold change respectively). S4 Table shows canine differentially expressed miRNAs in dogs with VL and respective fold change and p-value. Heatmap (Fig 1A) and Volcano plot (Fig 1B) show the differentially expressed miRNAs in PBMC of dogs with VL compared to healthy ones. miRNAs of other species (mammals, birds, fish, plants) hybridized in the microarray, but none of them showed homology with currently described canine sequences. The percentage of upregulated canine miRNAs was 2,40% (7/291) and downregulated was 0,68% (2/291).
Fig 1. Differentially expressed miRNAs in PBMC of dogs infected with L. infantum.
(A) Heatmap of differentially expressed miRNAs in PBMC of dogs infected with L. infantum compared to control group. Heatmap shows the average signal of miRNAs. Upregulated miRNAs are plotted in red and downregulated miRNAs are plotted in green. Fold change cut-off was lower than -2 and greater than 2. Analysis of variances (ANOVA) was performed between groups. (B) To the left, in green, are illustrated the miRNAs with negative expression and to the right in red the miRNAs with positive expression compared to control. miRNAs illustrated in grey hybridized, but did not show homology with known canine miRNAs. (C) miRNAs validated by real-time PCR. Expression of microRNAs in infected and healthy groups was quantified by real-time PCR in PBMC of infected and healthy dogs. Data represent the mean values of miRNA expression +/- standard error of the mean, and the asterisks represent statistically significant data following Mann-Whitney test. Results was considered significant when p<0.05.
To confirm the differential expression of miRNAs in the blood of dogs with VL in the microarray, real-time PCR validation was performed. Reactions showed efficiency from 0.84 to 1.42, slope from -2.603 to 3.761 and R2 from 0.933 to 0.998 (S5 Table).
miR-21, miR-451, miR-192, miR-194 and miR-371 were increased similarly to the results seen for the microarray, and miR-150 decreased in the PBMC (Fig 1C).
Canonical pathways regulated by differentially expressed miRNAs in PBMC of infected animals
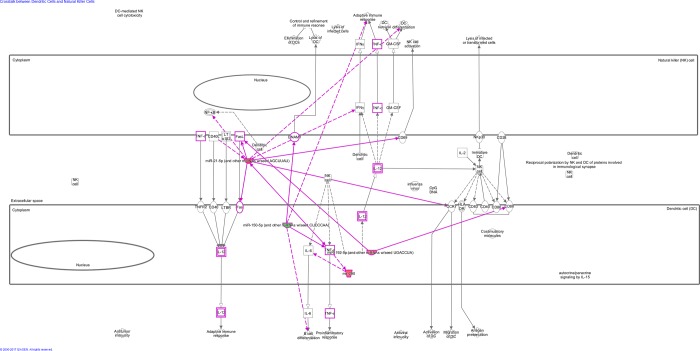
IPA's microRNA Target Filter (employing highly predicted and experimentally observed only) showed target genes and demonstrated 63 canonical pathways regulated by these miRNAs/targets (S6 Table). The top 20 canonical pathways and target genes are presented in Table 1, including p53 signaling, antiproliferative role of TOB in T cell signaling, STAT3 pathway, PTEN signaling, death receptor signaling and crosstalk between dendritic cells and natural killer cells, that can regulate the immune response in CVL. We further showed the canonical pathway “crosstalk between dendritic cells and natural killer cells” (Fig 2), which targets important genes involved in immunopathogenesis in CVL, such as NFκB, TNF-α, CD80, IFN-γ and DNAM-1.
Table 1. Top twenty canonical pathways predicted for the differentially regulated miRNAs in CVL.
| Ingenuity Canonical Pathways | P value | miRNAs in the Pathway | Target genes |
|---|---|---|---|
| Aryl Hydrocarbon Receptor Signaling | 0.00014 | miR-150 | CDKN1B, CHEK2, HSP90B1, IL1A, TNF, TP53 |
| miR-192 | ATM, DHFR, FASLG, GSTA2, RB1 | ||
| miR-194 | ALDH4A1, GSTT2, IL1A, MAPK1 | ||
| miR-21 | ALDH1A1, APAF1, CDK6, CDKN1A, FAS, FASLG, IL1B, NFIA, NFIB, TNF | ||
| p53 Signaling | 0.00015 | miR-150 | CHEK2, MDM4, PERP, TP53 |
| miR-192 | ATM, PERP, RB1, STAG1 | ||
| miR-194 | CCNG1, CSNK1D, SERPINE2, THBS1 | ||
| miR-21 | APAF1, CCNG1, CDKN1A, FAS, FRS2, PIK3R1, PTEN, SERPINB5, TNFRSF10B | ||
| Tumoricidal Function of Hepatic Natural Killer Cells | 0.00030 | miR-150 | GZMB |
| miR-192 | CYCS, FASLG | ||
| miR-194 | CASP7, SRGN | ||
| miR-21 | APAF1, CASP8, FAS, FASLG | ||
| Osteoarthritis Pathway | 0.00068 | miR-150 | CEBPB, CREB1, MMP13, TNF, VEGFA |
| miR-192 | ADIPOQ, FGF2, HTRA1, IL1RAP, SDC4, TGFBR1 | ||
| miR-194 | CASP7, FZD6, RAC1 | ||
| miR-21 | BMPR2, CASP8, FGF18, FZD6, GLIS2, IL1B, JAG1, RBPJ, SMAD7, TGFBR2, TIMP3, TNF, VEGFC | ||
| Hepatic Fibrosis / Hepatic Stellate Cell Activation | 0.00098 | miR-150 | IL1A, MMP13, PDGFB, STAT1, TNF, VEGFA |
| miR-192 | CCR5, CSF1, FASLG, FGF2, FLT1, IL1RAP, SERPINE1, TGFBR1 | ||
| miR-194 | CXCL3, EDN1, IL1A | ||
| miR-21 | ACTA2, CCR7, FAS, FASLG, IL1B, IL6R, KLF6, SMAD7, TGFBR2, TNF, TNFRSF11B, VEGFC | ||
| Crosstalk between Dendritic Cells and Natural Killer Cells | 0.00141 | miR-150 | CD226, TNF |
| miR-192 | CD80, FASLG | ||
| miR-194 | ACTG2, CD83, FSCN3 | ||
| miR-21 | ACTA2, CCR7, CD69, FAS, FASLG, IL12A, TNF | ||
| dTMP De Novo Biosynthesis | 0.00182 | miR-192 | DHFR, DHFR2, TYMS |
| Cell Cycle: G1/S Checkpoint Regulation | 0.00186 | miR-150 | CDKN1B, HDAC8, SKP1, TP53 |
| miR-192 | ATM, RB1 | ||
| miR-194 | BMI1, E2F6, HDAC8 | ||
| miR-21 | CDC25A, CDK6, CDKN1A, SKP2 | ||
| STAT3 Pathway | 0.00347 | miR-192 | FLT1, PIM1, SOCS6, TGFBR1 |
| miR-194 | MAPK1, RAC1, RRAS2, SOCS2 | ||
| miR-21 | BMPR2, CDC25A, CDKN1A, MAPK10, SOCS5, SOCS6, STAT3, TGFBR2 | ||
| Human Embryonic Stem Cell Pluripotency | 0.00363 | miR-150 | FOXD3, PDGFB |
| miR-192 | ATM, FGF2, LEFTY2, TGFBR1 | ||
| miR-194 | ACVR1, FZD6, H2BFM | ||
| miR-21 | BMPR2, FRS2, FZD6, NTF3, PIK3R1, SMAD7, SOX2, TGFBR2 | ||
| Estrogen-mediated S-phase Entry | 0.00417 | miR-150 | CDKN1B |
| miR-192 | RB1 | ||
| miR-194 | E2F6 | ||
| miR-21 | CDC25A | ||
| Actin Cytoskeleton Signaling | 0.00427 | miR-150 | ARPC3, FGF16, PDGFB, TMSB4Y |
| miR-192 | ATM, BRK1, CRK, FGF14, FGF2, FGF7, MSN, MYLK, PIP4K2B | ||
| miR-194 | ACTG2, CFL2, GNA13, LIMK2, MAPK1, PFN2, PIP4K2C, RAC1, RRAS2, TMSB4Y | ||
| miR-21 | ACTA2, ARHGAP24, ARHGEF12, CFL2, FGF18, FRS2, PFN2, PIK3R1, SOS2, TIAM1, TMSB4Y | ||
| Type I Diabetes Mellitus Signaling | 0.00427 | miR-150 | GZMB, STAT1, TNF |
| miR-192 | CD80, CYCS, FASLG, GAD1, HLA-DOB, IL1RAP, SOCS6 | ||
| miR-194 | GAD1, MAPK1, SOCS2, TRAF6 | ||
| miR-21 | APAF1, CASP8, FAS, FASLG, IL12A, IL1B, MAP2K3, MAPK10, SOCS5, SOCS6, TNF, TNFRSF11B | ||
| Cyclins and Cell Cycle Regulation | 0.00437 | miR-150 | CDKN1B, HDAC8, SKP1, TP53 |
| miR-192 | ATM, PPP2CB | ||
| miR-194 | E2F6, HDAC8 | ||
| miR-21 | CDC25A, CDK6, CDKN1A, SKP2 | ||
| Regulation of the Epithelial-Mesenchymal Transition Pathway | 0.00468 | miR-150 | FGF16, ZEB1 |
| miR-192 | ATM, FGF14, FGF2, FGF7, TGFBR1, ZEB1, ZEB2 | ||
| miR-194 | FZD6, MAPK1, RRAS2 | ||
| miR-21 | FGF18, FRS2, FZD6, JAG1, MAP2K3, PIK3R1, RBPJ, SOS2, STAT3, TGFBR2 | ||
| Antiproliferative Role of TOB in T Cell Signaling | 0.00525 | miR-150 | CDKN1B, SKP1 |
| miR-192 | PABPC4, RB1, TGFBR1 | ||
| miR-194 | MAPK1 | ||
| miR-21 | SKP2, TGFBR2 | ||
| Molecular Mechanisms of Cancer | 0.00603 | miR-150 | CDKN1B, CHEK2, RAPGEF3, TP53 |
| miR-192 | ATM, CRK, CYCS, FASLG, PRKAR1A, PRKCQ, RALB, RB1, TGFBR1, XIAP | ||
| miR-194 | ADCY7, CASP7, CDK14, E2F6, FNBP1, FZD6, GNA13, H2BFM, MAPK1, PRKAR1A, RAC1, RAP2B, RRAS2 | ||
| miR-21 | APAF1, ARHGEF12, BMPR2, CASP8, CDC25A, CDK6, CDKN1A, FAS, FASLG, FRS2, FZD6, PIK3R1, SMAD7, TGFBR2 | ||
| PTEN Signaling | 0.00603 | miR-150 | CDKN1B |
| miR-192 | FASLG, FLT1, TGFBR1 | ||
| miR-194 | H2BFM, MAPK1, RAC1, RRAS2 | ||
| miR-21 | BMPR2, CDKN1A, FASLG, PIK3R1, PREX2, PTEN, SOS2, TGFBR2 | ||
| Small Cell Lung Cancer Signaling | 0.00631 | miR-150 | CDKN1B, CKS1B, TP53 |
| miR-192 | ATM, CYCS, RB1, TRAF5 | ||
| miR-194 | CKS1B, MAPK1, RRAS2, TRAF6 | ||
| miR-21 | APAF1, CDK6, FRS2, PIK3R1, PTEN, SKP2, SOS2 | ||
| Death Receptor Signaling | 0.00912 | miR-150 | MAP4K4, TNF |
| miR-192 | CYCS, FASLG | ||
| miR-194 | XIAP, ACTG2, CASP7, MAP4K4, PARP11 | ||
| miR-21 | ACTA2, APAF1, CASP8, FAS, FASLG, TNF |
Fig 2. IPA’s Canonical Pathway “crosstalk between dendritic cells and natural killer cells”.
Upregulated miRNAs are in red, and downregulated are in green. Indirect relationships are represented by dotted lines and direct relationships by solid lines. miR-290 is a synonym of miR-371 in IPA’s analysis. miR-192 targets CD80 and FASL. miR-371 targets IL-6 and TNF-α. miR-150 targets TNF-α and DNAM-1. miR-21 targets FAS, FASL, TNFα, CD40L, NFκB, IFNγ, CD69 and CCR7. This figure was obtained with Ingenuity Pathway Analysis (Qiagen).
To understand the functional networks of the target genes, GO analysis was performed. GO analysis showed that these target genes were involved in a large number of physiological processes at the levels of biological processes, cellular component and molecular function (S7–S9 Tables).
Blood count, biochemical molecules and correlation with the expression of microRNAs miR-194 and miR-371 in dogs naturally infected with L. infantum
There was a strong positive correlation between miR-194 expression and serum urea (r = 0.73 p = 0.031) (S2A Fig). There was a strong negative correlation between miR-194 expression and hemoglobin concentration (r = -0.70 p = 0.037) (S2B Fig) and a strong negative correlation between miR-371 expression and globular volume (S2C Fig) (r = -0.76 p = 0.019).
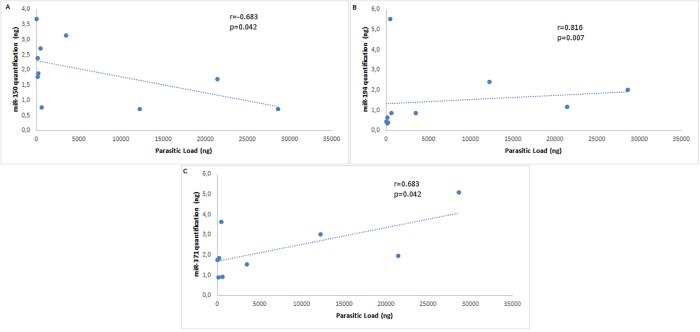
Parasite load on PBMC and correlation with expression of microRNAs miR-194, miR-371 and miR-150 in dogs naturally infected with L. infantum
Real-time PCR reaction for quantification of parasite load presented efficiency values of 0.97, slope -3.405 and R2 0.958. Parasite load ranged from 10.1ng to 28633ng.
There was a strong positive correlation between parasite load and miR-194 expression (r = 0.816 p = 0.007) and a regular positive correlation between parasite load and miR-371 expression (r = 0.683 p = 0.042) in PBMC from infected dogs compared to healthy dogs. There was a regular negative correlation between parasite load and the expression of miR-150 (r = -0.683 p = 0.042) in these cells. Fig 3 illustrates the correlations between these miRNAs and parasite load.
Fig 3. Correlation on miRNAs validated by RT-PCR and parasite load in PBMC of dogs infected by L. infantum.
Data represents a negative correlation of miR-150 (A) and positive correlation of miR-194 (B) and miR-371 (C).
Discussion
Leishmania infantum was the species observed in dogs with VL in the endemic area studied. Identification of the infecting species is important because differences in miRNA expression by L. donovani and L. major have already been observed in infected human dendritic cells in vitro [12]. Also, L. amazonensis has been identified in dogs with visceral leishmaniasis in the nearby city of Bauru-SP, Brazil [20], thus, the determination of L. infantum in the samples was important to ensure the subsequent results of the research. We demonstrated by microarray that there was altered expression of nine miRNAs in PBMC of animals with CVL compared to healthy animals; the miRNAs miR-21, miR-192, miR-194, miR-371, miR-503, miR-424 and miR-451 were upregulated, and miR-574 and miR-150 were downregulated in CVL samples. Differentially expressed miRNAs in the microarray in the Canis familiaris species were also evaluated by real-time PCR. miR-21, miR-150, miR-451, miR-192, miR-194 and miR-371 qPCR results confirmed the microarray data in PBMC. miR-424, miR-574 and miR-503 showed similar patterns to microarray results, but statistical significance was not achieved, likely due to the small sample size (10 infected animals and 5 healthy animals).
The IPA program was efficient in identifying gene targets and demonstrated 63 canonical pathways regulated by these miRNAs, including p53 signaling, antiproliferative role of TOB in T cell signaling, STAT3 pathway, PTEN signaling, death receptor signaling and crosstalk between dendritic cells and natural killer cells involved in immune response. GO analysis were performed for the predicted target genes of the differentially expressed miRNAs in PBMC from dogs with visceral leishmaniasis. GO analysis results showed that the predicted target genes participated in numerous biological processes, suggesting that the six miRNAs could play important roles in regulation of a variety of biological processes. The results for IPA and GO enrichment analyses can therefore provide direction for future research in canine visceral leishmaniasis.
Canonical pathway “crosstalk between dendritic cells and natural killer cells” involved several target genes of differentially expressed miRNAs and their products, between them, TNF-α and IFN-γ are important cytokines in CVL and are related with disease resistance [8], The expression of TNF-α and IFN-γ is downregulated by miR 21 [24–26], which would favor parasitic growth [8].
Canonical pathway analysis also showed relation with apoptosis signaling. Fas and FasL, important molecules in apoptosis, were downregulated by miR-21 [27,28]. Expression levels of Fas and FasL in CD4+ cells from blood and spleen have already been shown to be lower in infected dogs when compared to healthy dogs [29], and the soluble levels Fas (sFas) from spleen leukocytes were lower in dogs with VL compared to controls [30]. Thus, miRNAs are acting in apoptosis pathway that occurs in CVL.
miR-21 showed a significant increase in PBMC of animals with CVL (3.7 fold change). This miRNA is involved in immune regulation, negatively regulating the activation of T lymphocytes [31]. Also, it shows a negative correlation with IL-6 and TNF-α production [32]. TNF-α is associated with resistance in CVL, suggesting that the higher levels of miR-21 may be decreasing TNF-α, which in turn could increase parasite load. In addition, miR-21 negatively regulates proteins involved with adaptative immune response generation, CD40L [33], NFKB [34], CD69 [31], CCR7 [35] and a decreased of NFKB [36], CD40L [37], CCR7 [38] were previously observed in Leishmania infection in mice, thus suggesting that miR-21 could be modulation these immune regulatory proteins in CVL.
Likewise, miR-192 showed a significant increase in PBMC of animals with CVL, and it has been shown to down regulate immune response in human [39]. Leishmania infection in canine monocyte-derived macrophages decreased expression of costimulatory B7 molecules, and the co-culture with B7-transfected cells resulted in the restoration of cell proliferation and IFN-γ production by a Leishmania specific T-cell line [9]. Therefore, it is tempting to suggest that inhibition of T-cell activation and cytokine production are modulated by the expression of miR-192 in dogs with VL.
On the other hand, miR-150 was decreased in PBMC of dogs with VL and had a regular negative correlation with parasitic load in the blood. miR-150 was first described as a key miRNA in the development of B cells [40]. Knockout mice and mutants for miR-150 had an overproduction of immunoglobulins (Igs) of all classes, due to an increase in the number of B1 cells through increased c-Myb gene expression [41]. In dogs with symptomatic VL, there is an increase in the serum level of immunoglobulins of all classes (IgG, IgA IgE, and IgM) [42] and regulatory B cells from L. infantum–infected dogs co-cultured with enriched T cells, were sufficient to suppress T cell function through IL-10 and PD1 [43]. It is possible that miR-150 is acting in hypergammaglobulin, and in the development of regulatory B-cells, by increasing the parasite burden due to T-cell suppression in CVL.
Increased parasite load associated with decreased miR-150 could also act in other effector pathways of anti-parasitic immunity. Mutant mice lacking miR-150 have a decrease in the number of mature natural killer (NK) cells, whereas mice with increased expression of this miRNA have enhanced development of NK cells [44]. NK cells may play a role in controlling the burden of Leishmania spp. parasites during early stages of infection by their ability to respond rapidly to IFN-γ production, which activates macrophages to kill the parasite. In vitro, human NK cells were shown to be directly activated by Leishmania promastigotes or their lipophosphoglycan (LPG) to produce IFN-γ [45–47]. Thus, the decrease in NK cells modulated by miR-150 expression could be related to a higher parasite load in the CVL animals.
Unlike miR-150, an increase in miR-194 expression was observed in PBMC in dogs with VL and a positive and strong correlation with parasite blood load was observed. In human THP-1 monocytes, an increase in miR-194 expression directly decreases the expression of the TRAF6 gene, decreasing the release of the proinflammatory cytokine TNF-α [48]. In dogs with asymptomatic VL there is a high expression of TNF-α in lymph nodes with low parasitism, and this expression is lower in symptomatic dogs with a higher parasite load, suggesting that this cytokine has a protective function in L. infantum infection [8]. A miR-194 increase in infected dogs could be regulating the secretion of inflammatory cytokines, such as TNF-α, modulating parasite load in these animals.
Moreover, expression of miR-371 is increased in PBMC of dogs with VL and has a positive correlation with parasite load in PBMC. In patients with asthma, miR-371 targets the Runx3 gene and is related to the Th1 and Th2 response balance, and its increased expression is related to a higher Th2 response [49]. In CVL, the response is both Th1 and Th2, but predominantly Th2, with high production of cytokines by these cells, such as IL-10 and TGF-β, and decreased production of Th1-type cytokines [50,51]. Thus, miR-371 could be involved with pathways that regulate Th2 immune response polarization in CVL.
miR-194 showed a strong positive correlation with serum urea of dogs with VL. This miRNA was increased in plasma of rats with renal ischemia-reperfusion injury [52], dogs with VL can present renal failure [53], suggesting that miR-194 could be explored as an early plasma biomarker in renal lesion in dogs with VL.
Further, miR-194 showed a strong negative correlation with hemoglobin concentration and miR-371 showed a strong negative correlation with erythrocyte globular volume. In CVL hemoglobin and globular volume are decreased [54], as observed in our results, and anemia is a common clinical sign observed in complete blood count of dogs with VL [55]. Other miRNAs have been associated with anemia (miR-150-5p, miR-146b-5p and miR-1) [56] and erythroid homeostasis (miR-144 and miR-451) [57], however miR-194 and miR-371 were not associated with anemia, therefore more studies are needed to confirm our results.
Expression of miR-150, miR-194 and miR-371 showed correlation with parasitic load, indicating that target proteins and pathways should be investigated in future studies. Similarly, human macrophages infected with L. donovani showed increased expression of miR-30A-3p, and transitory transfection with inhibitor, followed by infection by L. donovani, promotes autophagy and decrease parasitic load in these cells [58]. These results indicate that differential expression of miRNA is dependent on the parasite species and infected host, thus emphasizing the need for studies in the canine model, the most important disease reservoir.
These findings suggest that L. infantum may modulate miRNAs in naturally infected dogs. We suggest that their role in immune regulation and their correlation to parasite load may help in the identification of therapeutic targets in Canine Visceral Leishmaniasis.
Supporting information
Restriction fragment length polymorphism (RFLP) analysis of ITS1-PCR fragments amplified from DNA samples, by using Hae III. M: molecular marker (123 bp); La: Leishmania amazonensis (IOC / L0575-MHOM / BR / 1967 / PH8); Lb: Leishmania braziliensis (IOC / L0566-MHOM / BR / 1975 / M2903); Li: Leishmania infantum (IOC / L0575-MHOM / BR / 2002 / LPC-RPV);1–10: samples profile identical to Leishmania infantum. The restriction fragment length polymorphism (RFLP) are indicated by arrows.
(TIFF)
Data shows a positive strong correlation of miR-194 and Urea (A), negative strong correlation of miR-194 and Hemoglobin (B) and negative strong correlation of miR-371 and Globular Volume (C).
(TIF)
Optical density on ELISA and clinical signs of naturally infected and healthy animals (control group).
(DOCX)
Biochemical profile of animals from infected and control group. Abbreviations: ALT (alanine aminotransferase) AST (aspartate aminotransferase) ALP (alkaline phosphatase) GGT (gamma glutamyl transferase).
(DOCX)
Blood cells of both infected and control groups. Abbreviations: RBC (red blood cells) GV (globular volume) MCHC (Mean corpuscular hemoglobin concentration) MCV (Mean corpuscular volume) TPP (total plasma protein).
(DOCX)
Differentially expressed miRNAs of Canis familiaris species and respective fold change and p value.
(DOCX)
Values of efficiency and R2 on real time PCR to validate differentially expressed miRNAs.
(DOCX)
Canonical pathways predicted for the differentially regulated miRNAs in CVL.
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
The authors would like to thank Flavia Mari Yamamoto for her help with the experiments.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by Fundação de Amparo a Pesquisa do Estado de São Paulo (www.fapesp.br), grants 2015/16972-7, 2016/02359-4, and 2015/16101-6. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Murray HW, Berman JD, Davies CR, Saravia NG. Advances in leishmaniasis. Lancet. 2005;366: 1561–1577. 10.1016/S0140-6736(05)67629-5 [DOI] [PubMed] [Google Scholar]
- 2.Pan American Health Organization (PAHO). Epidemiological Report of the Americas US11. PAHO; 2017; 2–5. Available: http://iris.paho.org/xmlui/handle/123456789/34112 [Google Scholar]
- 3.Moreno J, Alvar J. Canine leishmaniasis: Epidemiological risk and the experimental model. Trends Parasitol. 2002;18: 399–405. 10.1016/S1471-4922(02)02347-4 [DOI] [PubMed] [Google Scholar]
- 4.Mancianti F, Gramiccia M, Gradoni L, Pieri S. Studies on canine leishmaniasis control. 1. Evolution of infection of different clinical forms of canine leishmaniasis following antimonial treatment. Trans R Soc Trop Med Hyg. 1988;82: 566–7. 10.1016/0035-9203(88)90510-X [DOI] [PubMed] [Google Scholar]
- 5.Fisa R, Gállego M, Castillejo S, Aisa MJ, Serra T, Riera C, et al. Epidemiology of canine leishmaniosis in Catalonia (Spain) The example of the Priorat focus. Vet Parasitol. 1999;83: 87–97. 10.1016/S0304-4017(99)00074-6 [DOI] [PubMed] [Google Scholar]
- 6.Baneth G, Koutinas AF, Solano-Gallego L, Bourdeau P, Ferrer L. Canine leishmaniosis–new concepts and insights on an expanding zoonosis: part one. Trends Parasitol. 2008;24: 324–330. 10.1016/j.pt.2008.04.001 [DOI] [PubMed] [Google Scholar]
- 7.Cabral M, O’Grady J, Alexander J. Demonstration of Leishmania specific cell mediated and humoral immunity in asymptomatic dogs. Parasite Immunol. 1992;14: 531–9. 10.1111/j.1365-3024.1992.tb00026.x [DOI] [PubMed] [Google Scholar]
- 8.Alves CF, de Amorim IFG, Moura EP, Ribeiro RR, Alves CF, Michalick MS, et al. Expression of IFN-γ, TNF-α, IL-10 and TGF-β in lymph nodes associates with parasite load and clinical form of disease in dogs naturally infected with Leishmania (Leishmania) chagasi. Vet Immunol Immunopathol. 2009;128: 349–358. 10.1016/j.vetimm.2008.11.020 [DOI] [PubMed] [Google Scholar]
- 9.Pinelli E, Rutten VP, Bruysters M, Moore PF, Ruitenberg EJ. Compensation for decreased expression of B7 molecules on Leishmania infantum-infected canine macrophages results in restoration of parasite-specific T-cell proliferation and gamma interferon production. Infect Immun. 1999;67: 237–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu G, Abraham E. MicroRNAs in Immune Response and Macrophage Polarization. Arterioscler Thromb Vasc Biol. 2013;33: 170–177. 10.1161/ATVBAHA.112.300068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bartel DP. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell. 2004;116(2): 281–97. 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- 12.Geraci NS, Tan JC, Mcdowell MA. Characterization of microRNA expression profiles in Leishmania-infected human phagocytes. Parasite Immunol. 2015;37: 43–51. 10.1111/pim.12156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Silva SC, Silva DF, Almeida TC, Perasoli FB, da Silva ATP, da Silva GN, et al. Behavior of two Leishmania infantum strains-evaluation of susceptibility to antimonials and expression of microRNAs in experimentally infected J774 macrophages and in BALB/c mice. Parasitol Res. 2018;117: 2881–2893. 10.1007/s00436-018-5979-3 [DOI] [PubMed] [Google Scholar]
- 14.Ghosh J, Bose M, Roy S, Bhattacharyya SN. Leishmania donovani targets dicer1 to downregulate miR-122, lower serum cholesterol, and facilitate murine liver infection. Cell Host Microbe. 2013;13: 277–288. 10.1016/j.chom.2013.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tiwari N, Kumar V, Gedda MR, Singh AK, Singh VK, Gannavaram S, et al. Identification and Characterization of miRNAs in Response to Leishmania donovani Infection: Delineation of Their Roles in Macrophage Dysfunction. Front Microbiol. 2017;8: 314 10.3389/fmicb.2017.00314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pandey RK, Sundar S, Prajapati VK. Differential expression of miRNA regulates T cell differentiation and plasticity during visceral leishmaniasis infection. Front Microbiol. 2016;7: 1–9. 10.3389/fmicb.2016.00001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bragato JP, Melo LM, Venturin GL, Rebech GT, Garcia LE, Lopes FL, et al. Data on differentially expressed miRNAs in dogs infected with Leishmania infantum. Data Br. Elsevier; 2018;17: 218–225. 10.1016/J.DIB.2018.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lima VMF, Gonçalves ME, Ikeda FA, Luvizotto MCR, Feitosa MM. Anti-leishmania antibodies in cerebrospinal fluid from dogs with visceral leishmaniasis. Braz J Med Biol Res. 2003;36: 485–489. 10.1590/S0100-879X2003000400010 [DOI] [PubMed] [Google Scholar]
- 19.Solano-Gallego L, Koutinas A, Miró G, Cardoso L, Pennisi MG, Ferrer L, et al. Directions for the diagnosis, clinical staging, treatment and prevention of canine leishmaniosis. Vet Parasitol. 2009;165: 1–18. 10.1016/j.vetpar.2009.05.022 [DOI] [PubMed] [Google Scholar]
- 20.Sanches L da C, Martini CC de, Nakamura AA, Santiago MEB, Dolabela de Lima B, Lima VMF de. Natural canine infection by Leishmania infantum and Leishmania amazonensis and their implications for disease control. Rev Bras Parasitol Veterinária. 2016;25: 465–469. 10.1590/s1984-29612016071 [DOI] [PubMed] [Google Scholar]
- 21.Ranasinghe S, Rogers ME, Hamilton JGC, Bates PA, Maingon RDC. A real-time PCR assay to estimate Leishmania chagasi load in its natural sand fly vector Lutzomyia longipalpis. Trans R Soc Trop Med Hyg. 2008;102: 875–882. 10.1016/j.trstmh.2008.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles G, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14: 128 10.1186/1471-2105-14-128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuleshov M V., Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44: W90–W97. 10.1093/nar/gkw377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang X, Ng W-L, Wang P, Tian L, Werner E, Wang H, et al. MicroRNA-21 modulates the levels of reactive oxygen species by targeting SOD3 and TNFα. Cancer Res. 2012;72: 4707–13. 10.1158/0008-5472.CAN-12-0639 [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 25.Liu F, Liu C, Hu X, Shang Y, Wu L. Microrna-21: A positive regulator for optimal production of type i and type iii interferon by plasmacytoid dendritic cells. Front Immunol. 2017;8: 1–14. 10.3389/fimmu.2017.00001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang N, Li Y, Wang G, Ding Y, Jin Y, Xu Y. Tumor necrosis factor-α suppresses adipogenic and osteogenic differentiation of human periodontal ligament stem cell by inhibiting miR-21/Spry1 functional axis. Differentiation. 2017;97: 33–43. 10.1016/j.diff.2017.08.004 [DOI] [PubMed] [Google Scholar]
- 27.Sayed D, He M, Hong C, Gao S, Rane S, Yang Z, et al. MicroRNA-21 is a downstream effector of AKT that mediates its antiapoptotic effects via suppression of fas ligand. J Biol Chem. 2010;285: 20281–20290. 10.1074/jbc.M110.109207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang K, Li PF. Foxo3a regulates apoptosis by negatively targeting miR-21. J Biol Chem. 2010;285: 16958–16966. 10.1074/jbc.M109.093005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Silva KLO, Melo LM, Perosso J, Oliveira BB, Santos PSP dos, Eugênio F de R, et al. CD95 (FAS) and CD178 (FASL) induce the apoptosis of CD4+ and CD8+ cells isolated from the peripheral blood and spleen of dogs naturally infected with Leishmania spp. Vet Parasitol. 2013;197: 470–476. 10.1016/j.vetpar.2013.07.012 [DOI] [PubMed] [Google Scholar]
- 30.Perosso J, Silva KLO, Ferreira SÍ de S, Avanço SV, dos Santos PSP, Eugênio F de R, et al. Alteration of sFAS and sFAS ligand expression during canine visceral leishmaniosis. Vet Parasitol. 2014;205: 417–423. 10.1016/j.vetpar.2014.09.006 [DOI] [PubMed] [Google Scholar]
- 31.Carissimi C, Carucci N, Colombo T, Piconese S, Azzalin G, Cipolletta E, et al. miR-21 is a negative modulator of T-cell activation. Biochimie. 2014;107: 319–326. 10.1016/j.biochi.2014.09.021 [DOI] [PubMed] [Google Scholar]
- 32.Mazloom H, Alizadeh S, Esfahani EN, Razi F, Meshkani R. Decreased expression of microRNA-21 is associated with increased cytokine production in peripheral blood mononuclear cells (PBMCs) of obese type 2 diabetic and non-diabetic subjects. Mol Cell Biochem. 2016;419: 11–17. 10.1007/s11010-016-2743-9 [DOI] [PubMed] [Google Scholar]
- 33.Garchow B, Kiriakidou M. MicroRNA-21 deficiency protects from lupus-like autoimmunity in the chronic graft-versus-host disease model of systemic lupus erythematosus. Clin Immunol. 2016;162: 100–6. 10.1016/j.clim.2015.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang Q, Liu S, Tang Y, Liu Q, Yao Y. MPT64 protein from Mycobacterium tuberculosis inhibits apoptosis of macrophages through NF-kB-miRNA21-Bcl-2 pathway. PLoS One. 2014;9: 1–8. 10.1371/journal.pone.0100949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smigielska-Czepiel K, van den Berg A, Jellema P, Slezak-Prochazka I, Maat H, van den Bos H, et al. Dual Role of miR-21 in CD4+ T-Cells: Activation-Induced miR-21 Supports Survival of Memory T-Cells and Regulates CCR7 Expression in Naive T-Cells. PLoS One. 2013;8: 1–10. 10.1371/journal.pone.0076217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Buates S, Matlashewski G. General suppression of macrophage gene expression during Leishmania donovani infection. J Immunol. 2001;166: 3416–22. 10.4049/jimmunol.166.5.3416 [DOI] [PubMed] [Google Scholar]
- 37.Chamekh M. CD40-CD40L Interaction in Immunity Against Protozoan Infections. J Biomed Biotechnol. 2007;2007: 1–6. 10.1155/2007/59430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Förster R, Schubel A, Breitfeld D, Kremmer E, Renner-Müller I, Wolf E, et al. CCR7 coordinates the primary immune response by establishing functional microenvironments in secondary lymphoid organs. Cell. 1999;99: 23–33. 10.1016/S0092-8674(00)80059-8 [DOI] [PubMed] [Google Scholar]
- 39.Zhou X, Mao Y, Zhu J, Meng F, Chen Q, Tao L, et al. TGF-beta promotes colorectal cancer immune escape by elevating B7-H3 and B7-H4 via the miR-155/miR-143 axis. Oncotarget. 2016;7: 67196–67211. doi: 10.18632/oncotarget.11950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou B, Wang S, Mayr C, Bartel DP, Lodish HF. miR-150, a microRNA expressed in mature B and T cells, blocks early B cell development when expressed prematurely. Proc Natl Acad Sci. 2007;104: 7080–7085. 10.1073/pnas.0702409104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xiao C, Calado DP, Galler G, Thai T-H, Patterson HC, Wang J, et al. MiR-150 Controls B Cell Differentiation by Targeting the Transcription Factor c-Myb. Cell. 2007;131: 146–159. 10.1016/j.cell.2007.07.021 [DOI] [PubMed] [Google Scholar]
- 42.Reis AB, Teixeira-Carvalho A, Vale AM, Marques MJ, Giunchetti RC, Mayrink W, et al. Isotype patterns of immunoglobulins: Hallmarks for clinical status and tissue parasite density in brazilian dogs naturally infected by Leishmania (Leishmania) chagasi. Vet Immunol Immunopathol. 2006;112: 102–116. 10.1016/j.vetimm.2006.02.001 [DOI] [PubMed] [Google Scholar]
- 43.Schaut RG, Lamb IM, Toepp AJ, Scott B, Mendes-Aguiar CO, Coutinho JF V, et al. Regulatory IgDhi B Cells Suppress T Cell Function via IL-10 and PD-L1 during Progressive Visceral Leishmaniasis. J Immunol. 2016;196: 4100–9. 10.4049/jimmunol.1502678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bezman NA, Chakraborty T, Bender T, Lanier LL. miR-150 regulates the development of NK and iNKT cells. J Exp Med. 2011;208: 2717–2731. 10.1084/jem.20111386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Becker I, Salaiza N, Aguirre M, Delgado J, Carrillo-Carrasco N, Kobeh LG, et al. Leishmania lipophosphoglycan (LPG) activates NK cells through toll-like receptor-2. Mol Biochem Parasitol. 2003;130: 65–74. 10.1016/S0166-6851(03)00160-9 [DOI] [PubMed] [Google Scholar]
- 46.Nylén S, Maasho K, Söderstrom K, Ilg T, Akuffo H. Live Leishmania promastigotes can directly activate primary human natural killer cells to produce interferon-gamma. Clin Exp Immunol. 2003;131: 457–67. 10.1046/j.1365-2249.2003.02096.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nylén S, Gautam S. Immunological perspectives of leishmaniasis. J Glob Infect Dis. Wolters Kluwer—Medknow Publications; 2010;2: 135–46. 10.4103/0974-777X.62876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tian H, Liu C, Zou X, Wu W, Zhang C, Yuan D. MiRNA-194 Regulates Palmitic Acid-Induced Toll-Like Receptor 4 Inflammatory Responses in THP-1 Cells. Nutrients. 2015;7: 3483–3496. 10.3390/nu7053483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Qiu Y-Y, Zhang Y-W, Qian X-F, Bian T. miR-371, miR-138, miR-544, miR-145, and miR-214 could modulate Th1/Th2 balance in asthma through the combinatorial regulation of Runx3. Am J Transl Res. 2017;9: 3184–3199. [PMC free article] [PubMed] [Google Scholar]
- 50.Corrêa APFL Dossi ACS, de Oliveira Vasconcelos R, Munari DP, de Lima VMF. Evaluation of transformation growth factor β1, interleukin-10, and interferon-γ in male symptomatic and asymptomatic dogs naturally infected by Leishmania (Leishmania) chagasi. Vet Parasitol. 2007;143: 267–274. 10.1016/j.vetpar.2006.08.023 [DOI] [PubMed] [Google Scholar]
- 51.Lage RS, Oliveira GC, Busek SU, Guerra LL, Giunchetti RC, Corrêa-Oliveira R, et al. Analysis of the cytokine profile in spleen cells from dogs naturally infected by Leishmania chagasi. Vet Immunol Immunopathol. 2007;115: 135–145. 10.1016/j.vetimm.2006.10.001 [DOI] [PubMed] [Google Scholar]
- 52.Yu G, Zha Y, Li H, Wang F, Bian Q, Lai X, et al. Screening plasma miRNAs as biomarkers for renal ischemia-reperfusion injury in rats. Med Sci Monit. 2014;20: 283–289. doi: 10.12659/MSM.889937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Esch KJ, Schaut RG, Lamb IM, Clay G, Morais Lima ÁL, do Nascimento PRP, et al. Activation of Autophagy and Nucleotide-Binding Domain Leucine-Rich Repeat–Containing-Like Receptor Family, Pyrin Domain–Containing 3 Inflammasome during Leishmania infantum–Associated Glomerulonephritis. Am J Pathol. 2015;185: 2105–2117. 10.1016/j.ajpath.2015.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nicolato R de C, de Abreu RT, Roatt BM, Aguiar-Soares RD de O, Reis LES, Carvalho M das G, et al. Clinical forms of canine visceral Leishmaniasis in naturally Leishmania infantum-infected dogs and related myelogram and hemogram changes. Arez AP, editor. PLoS One. 2013;8: e82947 10.1371/journal.pone.0082947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.De Luna R, Ferrante M, Severino L, Ambrosio R, Piantedosi D, Gradoni L, et al. Decreased lipid fluidity of the erythrocyte membrane in dogs with leishmaniasis-associated anaemia. J Comp Pathol. 2000;122: 213–6. 10.1053/jcpa.1999.0357 [DOI] [PubMed] [Google Scholar]
- 56.Hosokawa K, Kajigaya S, Feng X, Desierto MJ, Fernandez Ibanez MDP, Rios O, et al. A plasma microRNA signature as a biomarker for acquired aplastic anemia. Haematologica. 2017;102: 69–78. 10.3324/haematol.2016.151076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rasmussen KD, Simmini S, Abreu-Goodger C, Bartonicek N, Di Giacomo M, Bilbao-Cortes D, et al. The miR-144/451 locus is required for erythroid homeostasis. J Exp Med. 2010;207: 1351–1358. 10.1084/jem.20100458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Singh AK, Pandey RK, Shaha C, Madhubala R. MicroRNA expression profiling of Leishmania donovani-infected host cells uncovers the regulatory role of MIR30A-3p in host autophagy. Autophagy. 2016; 1–15. 10.1080/15548627.2015.1100356 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Restriction fragment length polymorphism (RFLP) analysis of ITS1-PCR fragments amplified from DNA samples, by using Hae III. M: molecular marker (123 bp); La: Leishmania amazonensis (IOC / L0575-MHOM / BR / 1967 / PH8); Lb: Leishmania braziliensis (IOC / L0566-MHOM / BR / 1975 / M2903); Li: Leishmania infantum (IOC / L0575-MHOM / BR / 2002 / LPC-RPV);1–10: samples profile identical to Leishmania infantum. The restriction fragment length polymorphism (RFLP) are indicated by arrows.
(TIFF)
Data shows a positive strong correlation of miR-194 and Urea (A), negative strong correlation of miR-194 and Hemoglobin (B) and negative strong correlation of miR-371 and Globular Volume (C).
(TIF)
Optical density on ELISA and clinical signs of naturally infected and healthy animals (control group).
(DOCX)
Biochemical profile of animals from infected and control group. Abbreviations: ALT (alanine aminotransferase) AST (aspartate aminotransferase) ALP (alkaline phosphatase) GGT (gamma glutamyl transferase).
(DOCX)
Blood cells of both infected and control groups. Abbreviations: RBC (red blood cells) GV (globular volume) MCHC (Mean corpuscular hemoglobin concentration) MCV (Mean corpuscular volume) TPP (total plasma protein).
(DOCX)
Differentially expressed miRNAs of Canis familiaris species and respective fold change and p value.
(DOCX)
Values of efficiency and R2 on real time PCR to validate differentially expressed miRNAs.
(DOCX)
Canonical pathways predicted for the differentially regulated miRNAs in CVL.
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.