Abstract
Background
Currently, molecular xenomonitoring efforts for lymphatic filariasis rely on PCR or real-time PCR-based detection of Brugia malayi, Brugia timori and Wuchereria bancrofti in mosquito vectors. Most commonly, extraction of DNA from mosquitoes is performed using silica column-based technologies. However, such extractions are both time consuming and costly, and the diagnostic testing which follows typically requires expensive thermal cyclers or real-time PCR instruments. These expenses present significant challenges for laboratories in many endemic areas. Accordingly, in such locations, there exists a need for inexpensive, equipment-minimizing diagnostic options that can be transported to the field and implemented in minimal resource settings. Here we present a novel diagnostic approach for molecular xenomonitoring of filarial parasites in mosquitoes that uses a rapid, NaOH-based DNA extraction methodology coupled with a portable, battery powered PCR platform and a test strip-based DNA detection assay. While the research reported here serves as a proof-of-concept for the backpack PCR methodology for the detection of filarial parasites in mosquitoes, the platform should be easily adaptable to the detection of W. bancrofti and other mosquito-transmitted pathogens.
Methodology/Principal findings
Through comparisons with standard silica column-based DNA extraction techniques, we evaluated the performance of a rapid, NaOH-based methodology for the extraction of total DNA from pools of parasite-spiked vector mosquitoes. We also compared our novel test strip-based detection assay to real-time PCR and conventional PCR coupled with gel electrophoresis, and demonstrated that this method provides sensitive and genus-specific detection of parasite DNA from extracted mosquito pools. Finally, by comparing laboratory-based thermal cycling with a field-friendly miniaturized PCR approach, we have demonstrated the potential for the point-of-collection-based use of this entire diagnostic platform that is compact enough to fit into a small backpack.
Conclusions/Significance
Because this point-of-collection diagnostic platform eliminates reliance on expensive and bulky instrumentation without compromising sensitivity or specificity of detection, it provides an alternative to cost-prohibitive column-dependent DNA extractions that are typically coupled to detection methodologies requiring advanced laboratory infrastructure. In doing so, this field-ready system should increase the feasibility of molecular xenomonitoring within B. malayi-endemic locations. Of greater importance, this backpack PCR system also provides the proof-of-concept framework for the development of a parallel assay for the detection of W. bancrofti.
Author summary
Molecular xenomonitoring has demonstrated significant potential as a non-invasive means of providing reliable surveillance for the presence of lymphatic filariasis (LF)-causing parasites. Given the continuing successes of global mass drug administration efforts, the need for such non-invasive surveillance techniques is expanding. However, considering the significant infrastructural demands which such surveillance requires, the development of simplified surveillance methodologies will be fundamental to future programmatic implementation efforts. Accordingly, we have developed a novel, simplified diagnostic platform for point-of-collection-based detection of the LF-causing parasite, Brugia malayi in pools of mosquitoes. By coupling a rapid and inexpensive DNA extraction methodology with a field-friendly amplification platform and test strip-based detection assay, this backpack PCR system eliminates the need for expensive instrumentation and laboratory-based infrastructure. Furthermore, adaptation of the platform described here will allow for the straightforward and rapid development of a parallel assay for the detection of Wuchereria bancrofti, facilitating the increased use of xenomonitoring and enabling mosquito surveillance efforts in regions lacking sophisticated laboratory infrastructure.
Introduction
Lymphatic filariasis (LF) is a disfiguring and disabling tropical disease caused by parasitic filarial nematodes. It is estimated that more than 120 million people currently suffer from this mosquito-borne infection, with approximately 90% of the global LF burden caused by Wuchereria bancrofti and the remaining 10% caused by the parasites Brugia malayi and Brugia timori [1–3]. Despite this significant burden of disease, the World Health Organization has targeted LF for elimination by the year 2020 [4–7], and accordingly, the Global Programme to Eliminate Lymphatic Filariasis (GPELF) has implemented mass drug administration (MDA) programs in most endemic countries in order to interrupt disease transmission and reduce infection rates.
An important component of LF elimination efforts is monitoring changing infection and exposure rates over the course of yearly MDAs in order to evaluate programmatic success and determine when treatment can be stopped [8–11]. Following the cessation of MDA, similar monitoring is required to ensure that infection recrudescence has not occurred and is unlikely to occur going forward [12]. While human blood sampling is currently the standard procedure for monitoring these rates [13–16], PCR-based detection of parasite DNA in insect vectors, termed molecular xenomonitoring (MX), is an effective and non-invasive alternative, capable of indirectly measuring parasite burden within endemic locations [17–27]. Given this potential, the World Health Organization has championed the development of novel methodologies capable of increasing the practicality of this approach to disease surveillance [28–29], since implementation of currently available MX techniques are expensive in terms of both reagents and infrastructure requirements.
Currently, field-adaptable loop-mediated isothermal amplification (LAMP) assays exist for the detection of both W. bancrofti [30] and Brugia parasites [31], and a helicase-dependent isothermal amplification (HDA) assay exists for the detection of B. malayi DNA in human blood samples [32]. However, widespread programmatic implementation of such assays has not occurred. Limited use likely stems primarily from insufficient large-scale comparative evaluation efforts. Concerns regarding detection ambiguities and uncertainties may also contribute. When running LAMP assays detection of positive samples relies on the visualization of either sample turbidity or sample fluorescence [31]. As such, results are not “presence” or “absence”-based, but rather occur on a spectrum. This potentially raises concerns regarding susceptibility to technician bias, fatigue, or interpretation. Equally problematic, while HDA assays typically rely on agarose gel electrophoresis, a proven means of effective target detection, such techniques are prone to sample contamination [33], and are difficult to perform in a field setting. In contrast, the field-friendly, “Backpack PCR” platform we describe here utilizes test strip-based DNA detection methods. Such technology partners the advantages of standardizing assay readouts with the capacity to minimize laboratory equipment needs. Furthermore, given the ongoing work towards the development of automated card reader technologies [34], the consistency of results will likely continue to improve. For these reasons, a number of assays conducted in conjunction with the Global Programme to Eliminate Lymphatic Filariasis and other tropical disease control and elimination programs [35] make use of strip or card-based detection techniques [36–39].
Here we describe the development of a novel diagnostic platform, which we have termed “Backpack PCR”, coupling a rapid mosquito DNA extraction procedure with the test strip-based detection of B. malayi DNA. This assay amplifies a 132 base pair region of the non-coding HhaI repetitive DNA sequence element, selected as the target due to its high copy number and proven reliability in previously described molecular diagnostic assays [32, 40–45]. Following a resource-minimizing NaOH-based DNA extraction procedure, amplification occurs using a field-friendly, miniaturized PCR technology. Parasite DNA-derived amplicons are then detected using a test-strip-based methodology, enabling point-of-collection-based sample processing. While acknowledging the potential utility of backpack PCR as a tool in Brugia-endemic locations, the primary contribution of this work is likely its service as a proof-of-concept study for the parallel development of a similar assay for the detection of W. bancrofti. Such development is currently underway.
Materials and methods
Preparation of mosquito pools
While B. malayi-infected mosquitoes can be successfully reared in the laboratory, due to the possible failure of individual mosquitoes to ingest microfilariae when taking a blood meal, the definitive assumption cannot be made that all exposed mosquitoes actually harbor parasites. For this reason, positive mosquito pools were prepared by spiking groups of uninfected, laboratory reared Aedes aegypti mosquitoes (Oxitec Ltd., Abingdon, UK) with a single B. malayi infective-staged larva (L3) (Filarial Research Reagent Resource Center [FR3], Athens, GA). Uninfected A. aegypti mosquito pools were also prepared to serve as experimental controls. Positive pools containing 5, 10, 15, 20, and 25 uninfected mosquitoes were prepared by adding a single L3 larva to each pool. Twenty positive pools of each pool size were prepared, while two negative control pools, containing only uninfected mosquitoes with no added L3 larva, were also prepared for each pool size. Thus, 22 pools total were prepared for each of the 5 pool sizes, resulting in 110 total pools.
DNA extraction from pooled mosquitoes
Ten positive mosquito pools and 1 negative mosquito pool of each mosquito pool size (5, 10, 15, 20 and 25) were extracted using a modified version of a previously published, cost-effective, NaOH-based DNA extraction procedure [46]. Briefly, in a 1.7 ml microfuge tube, a sterile plastic micro-pestle (Axygen Scientific, Union City, CA) was used to grind each mosquito pool with 180 μl of 0.2 N NaOH for 3 min. The micro-pestle was then rinsed with an additional 180 μl of 0.2 N NaOH into the same 1.7 ml tube containing the ground mosquitoes to be sure all mosquito debris was removed from the pestle. A new, clean, sterile micro-pestle was used for each pool. Each tube was incubated at 75 °C for 10 min. 115.2 μl of 1 M Tris (pH = 8.0) and 364.8 μl of nuclease-free water were added to each sample and the tubes were thoroughly mixed using a vortex mixer for 10 sec. Samples were centrifuged at 10,000 x g for 3 min, and the supernatant, containing extracted DNA, was collected and transferred into a clean 1.7 ml microcentrifuge tube. Utilizing an additional volume of nuclease-free water, ten-fold dilutions of each sample were prepared to minimize the effects of PCR inhibitors on downstream amplification reactions. In parallel, ten positive and 1 negative mosquito pool of each mosquito pool size (5, 10, 15, 20 and 25) were also extracted utilizing the Qiagen DNeasy Blood & Tissue Kit (Qiagen, Hilden, Germany) in accordance with the previously described extraction protocol [43, 47–48].
Real-Time PCR testing of DNA extracts
Real-time PCR was used to determine whether or not each pool of mosquito DNA, extracted by either the NaOH-based or Qiagen-based method, contained B. malayi DNA. For testing purposes, the real-time PCR assay developed by Rao, et al. was considered to be the gold standard for parasite detection [43], and it was assumed that every pool containing B. malayi DNA would result in positive detection using this assay. All real-time PCR reactions were carried out in triplicate using the previously described primers, probe, reaction mixture, volume, and cycling conditions [43]. Thermal cycling was performed using the StepOnePlus Real-Time PCR System (Applied Biosystems, Foster City, CA). Results were considered to be positive when amplification occurred with a mean Ct value of less than 38.
Amplification of B. malayi DNA utilizing standard and miniPCR platforms
A comparison of amplification efficiency between a standard thermal cycler (Veriti Thermal Cycler, Applied Biosystems, Foster City, CA) and the field-friendly miniPCR platform (Amplyus LLC, Cambridge, MA), was conducted using conventional PCR-based amplification of B. malayi DNA. The reactions were carried out using the Phire Hot Start II enzyme (Thermo Fisher Scientific, Waltham, MA) with the previously described real-time PCR primer set [43], modified to contain a biotin tag on the reverse primer (Fwd: 5’—GCAATATACCGACCAGCAC—3’ / Rev: 5’—Biotin-ACATTAGACAAGGAAATTGGTT—3’). Reactions were performed in 20 μl total volumes containing 100 nM concentrations of each primer, 0.5 μl of enzyme, 5 μl of 5X reaction buffer, 0.5 μl of 10 mM dNTPs, and 1 μl of a 10-fold dilution of the DNA extraction product. Parallel amplification reactions were performed using both a standard thermal cycler, (Veriti Thermal Cycler, Applied Biosystems), and the miniPCR instrument (Amplyus LLC, Cambridge, MA). Cycling conditions consisted of an initial 30 sec hold at 95 °C, followed by 35 cycles of 30 sec at 95 °C, 40 sec at 55 °C, and 1 min at 72 °C. Following cycling, a final 5 min extension step at 72 °C was performed.
Test strip-based detection of amplification products
Labeling of the amplified product
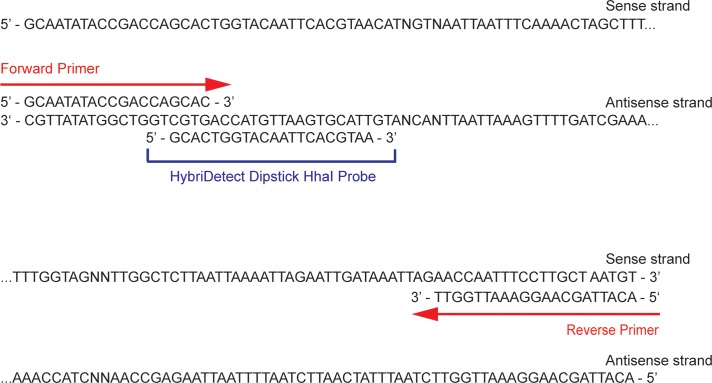
HybriDetect dipsticks (Milenia Biotec, Giessen, Germany) were used for the development of the detection assay. Detection of the amplified product with HybriDetect test strips requires that the DNA product be labeled with both a biotin tag (in order to bind to the streptavidin band of the test strip) and a FAM tag (in order to bind to the gold-labeled anti-fluorescein antibodies that result in a visible band for detection). Biotin was incorporated into the product of the PCR reaction by labeling the reverse primer as described above. FAM was incorporated into the product using a 5’ FAM-labeled sequence-specific probe that was designed using Primer Express 3.0.1 software (ThermoFisher Scientific) to be complementary to the strand of DNA tagged with the biotin label (Fig 1). Both the biotin-labeled primer and the FAM-labeled probe (5’—FAM-GCACTGGTACAATTCACGTAA—3’) were ordered from Integrated DNA Technologies (Coralville, IA).
Fig 1. B. malayi HhaI repeat DNA sequence used for test strip-based detection.
The sequence of B. malayi DNA that is amplified from the HhaI repetitive target is illustrated. Both the forward and reverse primer sites, as well as the location of the hybridization probe are indicated.
Probe hybridization and test strip detection
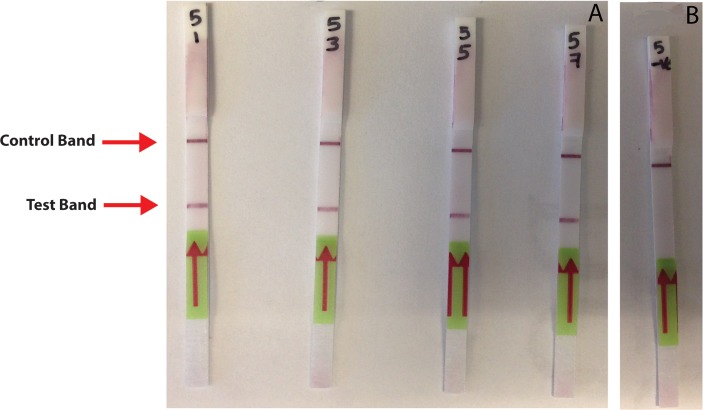
Following conventional PCR amplification of each sample using both the standard PCR and the miniPCR instruments in parallel, the 5’ FAM-labeled probe was hybridized to each resulting amplicon creating a double-stranded DNA molecule containing both a primer-derived biotin tag and a probe-derived FAM label. This hybridization reaction was performed in accordance with the manufacturer’s published protocol. After hybridization, the full volume of the reaction mixture was added to 50 μl of HybriDetect Assay Buffer (Tris-buffered saline), and a HybriDetect dipstick was placed into each sample tube and allowed to incubate at room temperature for 15 min. Following incubation, the dipstick was removed and visually inspected for the presence of a control band and a positive test band. A positive test band appears as a result of the biotin-labeled PCR product binding to a horizontal streptavidin test band as the sample travels up the strip by capillary action. Polyclonal (rabbit) anti-FITC antibodies, each conjugated to a gold particle, then bind to the FAM-labeled product, and the congregation of gold particles at the streptavidin test line causes a visible accumulation of color that is indicative of a positive result. A control band of anti-rabbit antibodies located above the test band also becomes visible as the sample travels along the length of the dipstick. The presence of this band is used to confirm the proper function of the test strip (Fig 2).
Fig 2. Test strip-based detection of amplification products generated by miniPCR.
A dark purple control band is visible near the top of each test strip, indicating the proper functioning of the dipsticks. (A) Only samples containing PCR amplification products generated from B. malayi DNA produce a visible band (test band) below the location of the control band. (B) Negative control reactions do not display visible accumulation of tagged amplification product at the location of the test band, indicating that parasite DNA was not present in these samples.
Comparative assay sensitivity testing
Analysis of the DNA extracted from spiked mosquito pools prepared as described above was used to assess the sensitivity of the test strip-based detection technique. For all tested samples, test strip results were compared with results of both the real-time PCR and the conventional PCR coupled with agarose gel electrophoresis. All conventional PCR reactions were performed using both the standard PCR and the miniPCR instruments.
Assay specificity testing
To verify the specificity of the test strip-based detection platform, genomic DNA samples isolated from Dirofilaria immitis, Mansonella perstans, Wuchereria bancrofti, Brugia pahangi, Acanthocheilonema viteae, and Loa loa, were used to measure off-target PCR amplification and strip test product detection. In order to confirm the presence of filarial-derived DNA in each sample, all gDNA isolates were first tested by conventional PCR using the previously published pan-filarial DIDR primer set [49]. All reactions were performed in 20 μl total volumes, utilizing 1 μl of gDNA template and 100 nM concentrations of both the forward and reverse DIDR primers. All remaining reaction components and cycling conditions were identical to those described above for the conventional PCR amplification of target DNA from B. malayi. Once the presence of PCR-amplifiable genomic DNA was verified, all DNA samples were then tested by real-time PCR utilizing the Brugia-specific HhaI assay [43]. The genomic DNA samples were also tested by conventional PCR for B. malayi, using the abovementioned real-time PCR assay primers and employing the same reaction recipes and conditions as described above. Detection of conventional PCR amplification products occurred using both test strips and agarose gel electrophoresis.
Blind test for assay validation
Fifty pools of mosquitoes were prepared which contained either zero or one B. malayi-L3-staged larva in pools of 5, 10, 15, 20, or 25 uninfected mosquitoes. Pools were then coded blind to the technician performing the assay. All mosquito pools underwent DNA extraction using the NaOH-based method described above, and extracts underwent amplification using the miniPCR platform, a standard conventional PCR platform, and the StepOnePlus Real-Time PCR System. All conventional PCR products were visualized using both test strip-based detection and agarose gel electrophoresis. Real-time PCR results were used to confirm the presence or absence of parasite DNA within each sample. After recording all results, the samples were un-blinded, and results were assessed.
Results
Testing the efficiency of the rapid DNA NaOH extraction method
The efficiency of the rapid NaOH-based DNA extraction methodology was examined through comparative real-time PCR testing of NaOH and Qiagen column-extracted sample panels. Qiagen extractions resulted in positive parasite detection in 50/50 samples, while NaOH-based extractions resulted in positive parasite detection in 49/50 samples. The NaOH-based extraction method was sufficiently efficient to give consistent detection of a single B. malayi L3-staged worm in all pools of up to 20 uninfected mosquitoes and in 9/10 pools containing 25 mosquitoes (Table 1). All mosquito pools not containing an L3-staged parasite were negative by both methods.
Table 1. Comparative performance of NaOH-based and Qiagen column-based DNA extraction procedures for the real-time PCR-based detection of B. malayi in pools of mosquitoes.
| Number of Samples | Pool Size (# of mosquitoes per pool) | Column-based Extraction Mean Ct Value (Ct range) | NaOH-based Extraction Mean Ct Value (Ct range) |
|---|---|---|---|
| 10 | 5 | 32.6* (27.7–37.0) | 31.2 (29.3–32.9) |
| 1 Uninfected | 5 | Negative | Negative |
| 10 | 10 | 31.0 (28.1–38.3) | 31.6 (30.0–34.2) |
| 1 Uninfected | 10 | Negative | Negative |
| 10 | 15 | 30.9 (27.9–33.0) | 32.6^ (28.0–38.6) |
| 1 Uninfected | 15 | Negative | Negative |
| 10 | 20 | 31.7 (26.7–35.6) | 32.5 (28.8–36.6) |
| 1 Uninfected | 20 | Negative | Negative |
| 10 | 25 | 35.0 (31.7–39.4) | 31.6# (28.8–35.6) |
| 1 Uninfected | 25 | Negative | Negative |
| 1 B. malayi + | Positive Control | 6.5 | 11.4 |
| 1 NTC | No Template Control | Negative | Negative |
* Positive detection of one sample from this set occurred in 2 out of 3 replicates tested
^ Positive detection of one sample from this set occurred in 1 out of 3 replicates tested
# One sample in this set failed to allow for detection of parasite signal
Sensitivity Testing of the miniPCR and Test Strip-Based Detection Methods
Following re-blinding of the panel of NaOH-extracted samples described above, conventional PCR-based amplification of all samples was conducted using both the miniPCR and the standard Veriti PCR instruments. Results were compared with those previously obtained by real-time PCR. For all samples tested, real-time PCR results agreed with the miniPCR results using both test strip-based detection and electrophoresis-based detection (Table 2).
Table 2. Comparison of the standard PCR-based amplification and miniPCR-based amplification of B. malayi DNA from mosquito pools coupled with real-time PCR-based detection.
| Sample ID | Pool Size | Real-Time PCR Result (Ct Value) | Standard PCR Gel Result | miniPCR Gel Result | miniPCR Test Strip Result |
|---|---|---|---|---|---|
| 1 | 5 | 30.7 | + | + | + |
| 2 | 5 | 32.2 | + | + | + |
| 3 | 5 | 29.3 | + | + | + |
| 4 | 5 | 32.9 | + | + | + |
| 5 | 5 | 30.4 | + | + | + |
| 6 | 5 | 31.9 | + | + | + |
| 7 | 5 | 32.3 | + | + | + |
| 8 | 5 | 29.5 | + | + | + |
| 9 | 5 | 30.9 | + | + | + |
| 10 | 5 | 31.6 | + | + | + |
| 11 Uninfected | 5 | - | - | - | - |
| 12 | 10 | 30.2 | + | + | + |
| 13 | 10 | 31.2 | + | + | + |
| 14 | 10 | 31.2 | + | + | + |
| 15 | 10 | 34.2 | + | + | + |
| 16 | 10 | 31.3 | + | + | + |
| 17 | 10 | 31.2 | + | + | + |
| 18 | 10 | 30.0 | + | + | + |
| 19 | 10 | 32.4 | + | + | + |
| 20 | 10 | 32.9 | + | + | + |
| 21 | 10 | 31.3 | + | + | + |
| 22 Uninfected | 10 | - | - | - | - |
| 23 | 15 | 33.9 | + | + | + |
| 24 | 15 | 31.7 | + | + | + |
| 25 | 15 | 35.0 | + | + | + |
| 26 | 15 | 30.8 | + | + | + |
| 27 | 15 | 31.9 | + | + | + |
| 28 | 15 | 34.1 | + | + | + |
| 29 | 15 | 28.0 | + | + | + |
| 30 | 15 | 33.9 | + | + | + |
| 31 | 15 | 28.1 | + | + | + |
| 32 | 15 | 38.6 | + | + | + |
| 33 Uninfected | 15 | - | - | - | - |
| 34 | 20 | 31.0 | + | + | + |
| 35 | 20 | 30.5 | + | + | + |
| 36 | 20 | 36.6 | + | + | + |
| 37 | 20 | 35.8 | + | + | + |
| 38 | 20 | 30.0 | + | + | + |
| 39 | 20 | 28.8 | + | + | + |
| 40 | 20 | 31.9 | + | + | + |
| 41 | 20 | 34.5 | + | + | + |
| 42 | 20 | 36.6 | + | + | + |
| 43 | 20 | 29.3 | + | + | + |
| 44 Uninfected | 20 | - | - | - | - |
| 45 | 25 | 35.6 | + | + | + |
| 46 | 25 | 30.6 | + | + | + |
| 47 | 25 | 32.5 | + | + | + |
| 48 | 25 | 30.0 | + | + | + |
| 49 | 25 | 33.2 | + | + | + |
| 50 | 25 | 34.3 | + | + | + |
| 51 | 25 | - | - | - | - |
| 52 | 25 | 28.8 | + | + | + |
| 53 | 25 | 30.3 | + | + | + |
| 54 | 25 | 29.0 | + | + | + |
| 55 Uninfected | 25 | - | - | - | - |
| 56 B. malayi + | Positive Control | 11.4 | + | + | + |
| 57 NTC | No Template Control | - | - | - | - |
Assay specificity testing
In order to verify the specificity of the “Backpack PCR”-based assay for the detection of B. malayi DNA, gDNA samples from D. immitis, M. perstans, W. bancrofti, B. pahangi, A. viteae, and L. loa were assayed along with B. malayi DNA as a positive control. To verify that these samples contained amplifiable filarial parasite DNA, extracts from all species were subjected to amplification utilizing the previously described pan-filarial primer set (DIDR) [49], and all samples amplified successfully (S1 Fig). When tested using the B. malayi field-friendly PCR platform described above, all pools containing non-Brugian filarial DNA tested negative, regardless of whether results were visualized by gel electrophoresis or test strip-based detection. However, B. pahangi genomic DNA produced positive results, indicating cross-reactivity, when examined by both visualization techniques (S1 Fig). Although the primers and probe selected for use with this assay were designed using the HhaI repeat DNA sequence from B. malayi (GenBank Accession No. AF499129.1), these results indicate that this assay also detects the closely related animal parasite B. pahangi. This is not surprising since the HhaI repeat found in B. malayi has a sequence that is 89% identical to the HhaI repeat found in B. pahangi [40]. Thus, the assay we describe here is Brugia genus-specific, but not B. malayi species-specific.
Blind test for assay validation
To further validate the use of our field-friendly platform and to further demonstrate its comparable sensitivity to real-time PCR, a series of 50 mosquito samples were prepared and coded blind to the processing technician. Following NaOH-based DNA extraction, each sample underwent cPCR-based amplification using both the standard and miniPCR instruments, followed by analysis using both gel electrophoresis and test strip-based detection methodologies. Quantitative real-time PCR was also performed for comparison. For 48 out of 50 samples, results for all assays were in agreement (Table 3). Examination of the two discordant samples revealed that in one instance (Sample #13) all assays yielded negative results with the exception of standard amplification coupled with test strip-based detection. In the second instance of discordance (Sample #46), real-time PCR and test strip-based detection assays returned positive results, while the gel electrophoresis results were negative. Thus, real-time PCR and the test strip-based detection agreed on 49 of 50 samples.
Table 3. Comparative testing of five methods for the detection of B. malayi PCR amplification products.
| Sample # | Sample Content (# of A. aegypti mosquitoes / number of L3 larvae) | Real-Time PCR Result (Ct value) | Gel Result–Standard PCR | Test-Strip Result–Standard PCR | Gel Result -miniPCR | Test-Strip Result–miniPCR |
|---|---|---|---|---|---|---|
| 22 | 5 / 1 | 32 | + | + | + | + |
| 32 | 5 / 1 | 27.7 | + | + | + | + |
| 38 | 5 / 1 | 30.2 | + | + | + | + |
| 47 | 5 / 1 | 30.5 | + | + | + | + |
| 2 | 10 / 1 | 29.9 | + | + | + | + |
| 17 | 10 / 1 | 29 | + | + | + | + |
| 29 | 10 / 1 | 30.6 | + | + | + | + |
| 37 | 10 / 1 | 29.7 | + | + | + | + |
| 49 | 10 / 1 | 32.2 | + | + | + | + |
| 9 | 15 / 1 | 28.9 | + | + | + | + |
| 11 | 15 / 1 | 26.9 | + | + | + | + |
| 18 | 15 / 1 | 32.9 | + | + | + | + |
| 23 | 15 / 1 | 29.4 | + | + | + | + |
| 1 | 20 / 1 | 26.8 | + | + | + | + |
| 4 | 20 / 1 | 28 | + | + | + | + |
| 6 | 20 / 1 | 34.8 | + | + | + | + |
| 25 | 20 / 1 | 32.4 | + | + | + | + |
| 43 | 20 / 1 | 36.5 | + | + | + | + |
| 45 | 20 / 1 | 28.6 | + | + | + | + |
| 46* | 20 / 1 | 39.8 | - | + | - | + |
| 13* | 25 / 1 | - | - | + | - | - |
| 21 | 25 / 1 | 36.5 | + | + | + | + |
| 30 | 25 / 1 | 28.6 | + | + | + | + |
| 33 | 25 / 1 | 27.5 | + | + | + | + |
| 35 | 25 / 1 | 31.4 | + | + | + | + |
| 40 | 25 / 1 | 30.5 | + | + | + | + |
| 41 | 25 / 1 | 27.5 | + | + | + | + |
| 44 | 25 / 1 | 29.9 | + | + | + | + |
| 3 | 5 / 0 | - | - | - | - | - |
| 5 | 5 / 0 | - | - | - | - | - |
| 7 | 5 / 0 | - | - | - | - | - |
| 8 | 5 / 0 | - | - | - | - | - |
| 10 | 5 / 0 | - | - | - | - | - |
| 42 | 5 / 0 | - | - | - | - | - |
| 12 | 10 / 0 | - | - | - | - | - |
| 19 | 10 / 0 | - | - | - | - | - |
| 20 | 10 / 0 | - | - | - | - | - |
| 28 | 10 / 0 | - | - | - | - | - |
| 50 | 10 / 0 | - | - | - | - | - |
| 14 | 15 / 0 | - | - | - | - | - |
| 15 | 15 / 0 | - | - | - | - | - |
| 24 | 15 / 0 | - | - | - | - | - |
| 27 | 15 / 0 | - | - | - | - | - |
| 39 | 15 / 0 | - | - | - | - | - |
| 48 | 15 / 0 | - | - | - | - | - |
| 26 | 20 / 0 | - | - | - | - | - |
| 34 | 20 / 0 | - | - | - | - | - |
| 36 | 20 / 0 | - | - | - | - | - |
| 16 | 25 / 0 | - | - | - | - | - |
| 31 | 25 / 0 | - | - | - | - | - |
* Testing of sample produced a false negative result by one or more detection methods.
Discussion
As a platform for filarial parasite detection, the coupling of a simple DNA extraction method with a field-friendly amplification platform and test strip-based detection technology has the potential to greatly expand the reach of MX efforts. Due to the reduced need for infrastructure and expensive, highly technical equipment, “Backpack PCR” is ideally suited for use in endemic locations currently lacking the capacity to perform real-time PCR reactions for MX purposes. We have demonstrated that this “Backpack PCR” platform has the capacity to reliably detect a single B. malayi L3 infective larva in pools of up to 25 uninfected A. aegypti mosquitoes. "Backpack PCR” minimizes equipment needs since it requires only a dry bath, a low-speed microcentrifuge, and a portable, battery-powered and smartphone-controlled thermal cycler (miniPCR) which weighs less than 1 lb. All of the equipment and materials needed for the assay can be easily transported in a backpack (Fig 3) and sample analysis can be carried out at, or near the point-of-collection in remote locations with limited resources. Building upon the proof-of-concept work described here, the development of a parallel assay for the detection of W. bancrofti will further empower local scientists by enabling their independent use of MX as a tool for the mapping of filarial parasite prevalence and for post-MDA surveillance.
Fig 3. The contents of the “Backpack PCR” platform.
The entire "Backpack PCR" platform weighs less than 10 lbs and can be easily transported by a single person. The “Backpack PCR” platform, which couples a simple and inexpensive NaOH-based extraction method with miniPCR amplification and test strip detection, will facilitate the use of molecular xenomonitoring in resource-limited settings. Furthermore, as this platform is based upon commercially available technologies, it will prove readily adaptable to the detection of W. bancrofti and other mosquito-borne pathogens. Research to develop this platform for the detection of other parasite and viral pathogens found in mosquitos is ongoing in our laboratory.
Despite the many advantages of this point-of-collection platform, the current design does present some challenges. Foremost, at a cost of approximately $7.50 per pool of mosquitoes, extraction and detection using this assay requires a significantly greater reagent investment than does extraction coupled with real-time PCR testing, estimated to cost $5.45 per equally-sized pool. Much of the cost of this system is due to the expensive test-strips, so we are actively researching less expensive alternatives that could reduce the cost to be competitive with real-time PCR. Utilization of this field-friendly platform will eliminate the need to maintain and service the sophisticated thermal cyclers required to perform real-time PCR diagnostics in a laboratory setting. In addition, the large capital commitments required for the initial purchase of such real-time PCR instruments would be eliminated, substantially lowering both initial overhead costs, and the recurring costs that arise from maintenance contracts and service fees. Furthermore, in many settings, increased reagent costs could be offset by eliminating the expenses associated with transporting and/or shipping samples to a reference laboratory for analysis. Eliminating the need for shipment, oftentimes out of country, has added benefits, as government regulations in many endemic countries require samples to be tested in their country of origin.
Although not unique to MX assays [48], the capacity of this assay to amplify DNA from both the human parasite B. malayi and the animal parasite B. pahangi presents an additional limitation. While sufficient data does not exist to reliably estimate the prevalence of B. pahangi within most locations co-endemic for both Brugian parasites, this shortcoming certainly merits additional investigation in such co-endemic areas.
Effectively trapping large numbers of Anopheles and Mansonia mosquitoes, primary vectors of B. malayi [50], presents the most substantial obstacle for all Brugia-based xenomonitoring efforts. Historically, such trapping difficulties have greatly restricted xenomonitoring efforts in Brugia-endemic locations, resulting in very few published examples of implementation [47, 51]. However, novel trap designs and improved trapping techniques continue to emerge [52–56] and it is imperative that the appropriate molecular tools be available to most effectively capitalize upon trap improvements as they occur. More importantly, the successful development of the “Backpack PCR” platform described here provides proof-of-principle for the development of future MX assays for the detection of other mosquito-borne infections. Accordingly, the development of a similar “Backpack PCR”-based assay for the detection of W. bancrofti, the parasite responsible for approximately 90% of the global LF burden, would be of significant use to the research community, and efforts to create such a platform in our laboratory are currently underway. Similarly, this method could also be extended to the detection of pathogens causing other mosquito-borne infections such as malaria, Zika, dengue fever, Chikungunya and others.
Supporting information
(A) Amplification reactions utilizing the previously published pan-filarial DIDR primer set were conducted to demonstrate the integrity and amplifiable-nature of the isolated DNA templates from various filarial parasites. 100 bp ladder (lane 1), B. malayi (lane 2), D. immitis (lane 3), M. perstans (lane 4), W. bancrofti (lane 5), B. pahangi (lane 6), A. viteae (lane 7), L. loa (lane 8), No Template Control (lane 9), 100 bp ladder (lane 10).
(B) These same genomic DNA samples were included as template in amplification reactions utilizing the Brugia spp.-specific primer pair employed for test strip-based detection and amplification products were visualized on an agarose gel. 100 bp ladder (lane 1), B. malayi (lane 2), D. immitis (lane 3), M. perstans (lane 4), W. bancrofti (lane 5), B. pahangi (lane 6), A. viteae (lane 7), L. loa (lane 8), No Template Control (lane 9), 100 bp ladder (lane 10). (C) With the exception of A. viteae (product volume was exhausted) amplification products were also visualized using test strip-based detection. B. malayi (strip 1), D. immitis (strip 2), M. perstans (strip 3), W. bancrofti (strip 4), B. pahangi (strip 5), L. loa (strip 6), No template control (strip 7).
(TIF)
Acknowledgments
We would like to thank Dr. Eric Ottesen and Dr. Patrick Lammie (NTD Support Center, Task Force for Global Health, Decatur, GA), for their continued support, encouragement, and advice. We would also like to thank the Filarial Research Reagent Resource Center (FR3) for parasites and molecular reagents, and Oxitec Ltd. for the provision of mosquitoes. Our grateful appreciation also goes to Ryan R. Hemme, for providing us with the preliminary protocol for NaOH-based DNA isolation that we adapted for use with this project. We also wish to thank Susan Haynes for logistical support.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was principally funded by a Filarial Research Reagent Resource Center sub-award to SAW (RR211-508/4787366) funded by NIH contract HHSN272200001 (Animal Models of Infectious Diseases). Additional support was provided by the Blakeslee Fund for Genetics Research at Smith College. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Scott AL. Lymphatic-dwelling filariae In: Nutman T, editor. Lymphatic Filariasis. London: Imperial College Press; 2000. pp. 5–39. [Google Scholar]
- 2.Mendoza N, Li A, Gill A, Tyring S. Filariasis: diagnosis and treatment. Dermatol Ther. 2009; 22(6): 475–490. 10.1111/j.1529-8019.2009.01271.x [DOI] [PubMed] [Google Scholar]
- 3.Taylor MJ, Hoerauf A, Bockarie M. Lymphatic filariasis and onchocerciasis. Lancet. 2010; 376(9747): 1175–1185. 10.1016/S0140-6736(10)60586-7 [DOI] [PubMed] [Google Scholar]
- 4.Molyneux DH, Bradley M, Hoerauf A, Kyelem D, Taylor MJ. Mass drug treatment for lymphatic filariasis and onchocerciasis. Trends Parasitol. 2003; 19(11): 516–523. [DOI] [PubMed] [Google Scholar]
- 5.Rebollo MP, Bockarie MJ. Toward the elimination of lymphatic filariasis by 2020: treatment update and impact assessment for the endgame. Expert Rev Anti Infect Ther. 2013; 11(7): 723–731. 10.1586/14787210.2013.811841 [DOI] [PubMed] [Google Scholar]
- 6.Rebollo MP, Bockarie MJ. Can lymphatic filariasis be eliminated by 2020? Trends Parasitol. 2017; 33(2): 83–92. 10.1016/j.pt.2016.09.009 [DOI] [PubMed] [Google Scholar]
- 7.Shamsuzzaman AK, Haq R, Karim MJ, Azad MB, Mahmood AS, Khair A, et al. The significant scale up and success of Transmission Assessment Surveys ‘TAS’ for endgame surveillance of lymphatic filariasis in Bangladesh: One step closer to the elimination goal of 2020. PLoS Negl Trop Dis. 2017; 11(1): e0005340 10.1371/journal.pntd.0005340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Helmy H, Weil GJ, Ellethy AS, Ahmed ES, Setouhy ME, Ramzy RM. Bancroftian filariasis: effect of repeated treatment with diethylcarbamazine and albendazole on microfilaraemia, antigenaemia and antifilarial antibodies. Trans R Soc Trop Med Hyg. 2006; 100(7): 656–662. 10.1016/j.trstmh.2005.08.015 [DOI] [PubMed] [Google Scholar]
- 9.Weil GJ, Kastens W, Susapu M, Laney SJ, Williams SA, King CL, et al. The impact of repeated rounds of mass drug administration with diethylcarbamazine plus albendazole on bancroftian filariasis in Paupau New Guinea. PLoS Negl Trop Dis. 2008; 2(12): e344 10.1371/journal.pntd.0000344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Swaminathan S, Perumal V, Adinarayanan S, Kaliannagounder K, Rengachari R, Purushothaman J. Epidemiological assessment of eight rounds of mass drug administration for lymphatic filariasis in India: implications for monitoring and evaluation. PLoS Negl Trop Dis. 2012; 6(11): e1926 10.1371/journal.pntd.0001926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simonsen PE, Derua YA, Magesa SM, Pedersen EM, Stensgaard AS, Malecela MN, et al. Lymphatic filariasis control in Tanga Region, Tanzania: status after eight rounds of mass drug administration. Parasit Vectors. 2014; 12(7): 507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Owusu IO, de Souza DK, Anto F, Wilson MD, Boakye DA, Bockarie MJ, et al. Evaluation of human and mosquito based diagnostic tools for defining endpoints for elimination of Anopheles transmitted lymphatic filariasis in Ghana. Trans R Soc Trop Med Hyg. 2015; 109(10): 628–635. 10.1093/trstmh/trv070 [DOI] [PubMed] [Google Scholar]
- 13.Weil GJ, Lammie PJ, Weiss N. The ICT Filariasis Test: A rapid-format antigen test for diagnosis of bancroftian filariasis. Parasitol Today. 1997; 13(10): 401–404. [DOI] [PubMed] [Google Scholar]
- 14.Schuetz A, Addiss DG, Eberhard ML, Lammie PJ. Evaluation of the whole blood filariasis ICT test for short-term monitoring after antifilarial treatment. Am J Trop Med Hyg. 2000; 62(4): 502–503. [DOI] [PubMed] [Google Scholar]
- 15.Weil GJ, Ramzy RM. Diagnostic tools for filariasis elimination programs. Trends Parasitol. 2007; 23(2): 78–82. 10.1016/j.pt.2006.12.001 [DOI] [PubMed] [Google Scholar]
- 16.Chu BK, Deming M, Biritwum N-K, Bougma WR, Dorkenoo AM, El-Setouhy M, et al. Transmission assessment surveys (TAS) to define endpoints for lymphatic filariasis mass drug administration: A multicenter evaluation. PLoS Negl Trop Dis. 2013; 7(12): e2584 10.1371/journal.pntd.0002584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chanteau S, Luguiaud P, Failloux AB, Williams SA. Detection of Wuchereria bancrofti larvae in pools of mosquitoes by the polymerase chain reaction. Trans R Soc Trop Med Hyg. 1994; 88(6): 665–666. [DOI] [PubMed] [Google Scholar]
- 18.Williams SA, Laney SJ, Bierwert LA, Saunders LJ, Boakye DA, Fischer P, et al. Development and standardization of a rapid, PCR-based method for the detection of Wuchereria bancrofti in mosquitoes, for xenomonitoring the human prevalence of bancroftian filariasis. Ann Trop Med Parasitol. 2002; 96 Suppl 2: S41–S46. [DOI] [PubMed] [Google Scholar]
- 19.Goodman DS, Orelus JN, Roberts JM, Lammie PJ, Streit TG. PCR and mosquito dissection as tools to monitor filarial infection levels following mass treatment. Filaria J. 2003; 2(1): 11 10.1186/1475-2883-2-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bockarie MJ. Molecular xenomonitoring of lymphatic filariasis. Am J Trop Med Hyg. 2007; 77(4): 591–592. [PubMed] [Google Scholar]
- 21.Farid HA, Morsy ZS, Helmy H, Ramzy RM, El Setouhy M, A critical appraisal of molecular xenomonitoring as a tool for assessing progress toward elimination of Lymphatic Filariasis. Am J Trop Med Hyg. 2007; 77(4): 593–600. [PMC free article] [PubMed] [Google Scholar]
- 22.Schmaedick MA, Koppel AL, Pilotte N, Torres M, Williams SA, Dobson SL, et al. Molecular xenomonitoring using mosquitoes to map lymphatic filariasis after mass drug administration in American Samoa. PLoS Negl Trop Dis. 2014; 8(8): e3087 10.1371/journal.pntd.0003087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rao RU, Samarasekera SD, Nagodavithana KC, Punchihewa MW, Dassanayaka TDM, Gamini PKD, et al. Programmatic use of molecular xenomonitoring at the level of evaluation units to assess persistence of lymphatic filariasis in Sri Lanka. PLoS Negl Trop Dis. 2016; 10(5): e0004722 10.1371/journal.pntd.0004722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lau CL, Won KY, Lammie PJ, Graves PM. Lymphatic Filariasis Elimination in American Samoa: Evaluation of Molecular Xenomonitoring as a Surveillance Tool in the Endgame. PLoS Negl Trop Dis. 2016; 10(11): e0005108 10.1371/journal.pntd.0005108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pilotte N, Zaky WI, Abrams BP, Chadee DD, Williams SA. A Novel Xenomonitoring Technique Using Mosquito Excreta/Feces for the Detection of Filarial Parasites and Malaria. PLoS Negl Trop Dis. 2016; 10(4): e0004641 10.1371/journal.pntd.0004641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Subramanian S, Jambulingam P, Chu BK, Sadanandane C, Vasuki V, Srividya A, et al. PLoS Negl Trop Dis. 2017; 11(4): e0005519 10.1371/journal.pntd.0005519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pilotte N, Unnasch TR, Williams SA. The Current Status of Molecular Xenomonitoring for Lymphatic Filariasis and Onchocerciasis. Trends Parasitol. 2017; 33(10): 788–798. 10.1016/j.pt.2017.06.008 [DOI] [PubMed] [Google Scholar]
- 28.Ichimori K. Lymphatic filariasis: A handbook of practical entomology for national lymphatic filariasis elimination programmes Geneva: WHO Press; 2013. [Google Scholar]
- 29.Ichimori K. Monitoring and epidemiological assessment of mass drug administration in the global programme to eliminate lymphatic filariasis: A manual for national elimination programmes Geneva: WHO Press; 2011. [Google Scholar]
- 30.Takaqi H, Itoh M, Kasai S, Yahathugoda TC, Weerasooriya MV, Kimura E. Development of loop-mediated isothermal amplification method for detecting Wuchereria bancrofti DNA in human blood and vector mosquitoes. Parasitol Int. 2011; 60(4): 493–497. 10.1016/j.parint.2011.08.018 [DOI] [PubMed] [Google Scholar]
- 31.Poole CB, Tanner NA, Zhang Y, Evans Jr, TC, Carlow CK. Diagnosis of brugian filariasis by loop-mediated isothermal amplification. PLoS Negl Trop Dis. 2012; 6(12): e1948 10.1371/journal.pntd.0001948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vincent M, Xu Y, Kong H. Helicase-dependent isothermal DNA amplification. EMBO Reports. 2004; 5(8): 795–800. 10.1038/sj.embor.7400200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Adey NB, Emery DB, Bosh DD, Parry RJ. A gel electrophoresis loading system to prevent laboratory contamination by amplification products. Biotechniques. 2014; 57(4): 199–200. 10.2144/000114216 [DOI] [PubMed] [Google Scholar]
- 34.Chesnais CB, Vlaminck J, Kunyu-Shako B, Pion SD, Awaca-Uvon N- P, Weil GJ, et al. Measurement of circulating filarial antigen levels in human blood with a point-of-care test strip and a portable spectrodensitometer. Am J Trop Med Hyg. 2016; 94(6): 1324–1329. 10.4269/ajtmh.15-0916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu Y, Gonzales G, Chen SH, Li H, Cai YC, Chu YH, Ai L, Chen MX, Chen HN, Chen JX. Urgent needs in fostering neglected tropical diseases (NTDs) laboratory capacity in WHO Western Pacific Region: results from the external quality assessment on NTDs diagnosis in 2012–2015. Infect Dis Poverty. 2017; 6(1): 106 10.1186/s40249-017-0319-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weil GJ, Lammie PJ, Weiss N. The ICT Filariasis Test: A rapid-format antigen test for diagnosis of bancroftian filariasis. Parasitol Today. 1997; 13(10): 401–404. [DOI] [PubMed] [Google Scholar]
- 37.Weil GJ, Steel C, Liftis F, Li BW, Mearns G, Lobos E, et al. A rapid-format antibody card test for diagnosis of onchocerciasis. J Infect Dis. 2000; 182(6): 1796–1799. 10.1086/317629 [DOI] [PubMed] [Google Scholar]
- 38.Supali T, Rahmah N, Djuardi Y, Sartono E, Rückert P, Fischer P. Detetion of filarial-specific IgG4 antibodies using Brugia Rapid test in individuals from an area highly endemic for Brugia timori. Acta Trop. 2004; 90(3): 255–261. 10.1016/j.actatropica.2004.02.001 [DOI] [PubMed] [Google Scholar]
- 39.Weil GJ, Curtis KC, Fakoli L, Fischer K, Galkpala L, Lammie PJ, et al. Laboratory and field evaluation of a new rapid test for detecting Wuchereria bancrofti antigen in human blood. Am J Trop Med Hyg. 2013; 89(1): 11–15. 10.4269/ajtmh.13-0089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Williams SA, DeSimone SM, McReynolds LA. Species-specific oligonucleotide probes for the identification of human filarial parasites. Mol Biochem Parasitol. 1988; 28(2): 163–169. [DOI] [PubMed] [Google Scholar]
- 41.Poole CB, Williams SA. A rapid DNA assay for the species-specific detection and quantification of Brugia in blood samples. Mol Biochem Parasitol. 1990; 40(1): 129–136. [DOI] [PubMed] [Google Scholar]
- 42.Lizotte MR, Supali T, Partono F, Williams SA. A polymerase chain reaction assay for the detection of Brugia malayi in blood. Am J Trop Med Hyg. 1994; 51(3): 314–321. [DOI] [PubMed] [Google Scholar]
- 43.Rao RU, Weil GJ, Fischer K, Supali T, Fischer P. Detection of Brugia parasite DNA in human blood by real-time PCR. J Clin Microbiol. 2006; 44(11): 3887–3893. 10.1128/JCM.00969-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thanchomnang T, Intapan PM, Lulitanond V, Choochote W, Manjai A, Prasongdee TK, et al. Rapid detetion of Brugia malayi in mosquito vectors using a real-time fluorescence resonance energy transfer PCR and melting curve analysis. Am J Trop Med Hyg. 2008; 78(3): 509–513. [PubMed] [Google Scholar]
- 45.Pilotte N, Torres M, Tomaino FR, Laney SJ, Williams SA. A TaqMan-based multiplex real-time PCR assay for the simultaneous detection of Wuchereria bancrofti and Brugia malayi. Mol Biochem Parasitol. 2013; 189(1–2): 33–37. 10.1016/j.molbiopara.2013.05.001 [DOI] [PubMed] [Google Scholar]
- 46.Martinez-de la Puente J, Ruiz S, Soriguer R, Figuerola J. Effect of blood meal digestion and DNA extraction protocol on the success of blood meal source determination in the malaria vector Anopheles atroparvus. Malar J. 2013; 12:109 10.1186/1475-2875-12-109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fischer P, Wibowo H, Pischke S, Rückert P, Liebau E, Ismid IS, et al. PCR-based detection and identification of the filarial parasite Brugia timori from Alor Island, Indonesia. Ann Trop Med Parasitol. 2002; 96(8): 809–821. 10.1179/000349802125002239 [DOI] [PubMed] [Google Scholar]
- 48.Laney SJ, Buttaro CJ, Visconti S, Pilotte N, Ramzy RM, Weil GJ, et al. A reverse transcriptase-PCR assay for detecting filarial infective larvae in mosquitoes. PLoS Negl Trop Dis. 2008; 2(6): e251 10.1371/journal.pntd.0000251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rishniw M, Barr SC, Simpson KW, Frongillo MF, Franz M, Alpizar JLD. Discrimination between six species of canine microfilariae by a single polymerase chain reaction. Vet Parasitol. 2006; 135(3–4): 303–314. 10.1016/j.vetpar.2005.10.013 [DOI] [PubMed] [Google Scholar]
- 50.Saeung A, Hempolchom C, Baimai V, Thongsahuan S, Taai K, Jariyapan N, et al. Susceptibility of eight species members in the Anopheles hyrcanus group to nocturnally subperiodic Brugia malayi. Parasit Vectors. 2013; 6:5 10.1186/1756-3305-6-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Beng TS, Ahmad R, Hisam RSR, Heng SK, Leaburi J, Ismail Z, et al. Molecular xenomonitoring of filarial infection in Malaysian mosquitoes under the National Program for Elimination of Lymphatic Filariasis. Southeast Asian J Trop Med Public Health. 2016; 47(4): 617–624. [Google Scholar]
- 52.Govella NJ, Chaki PP, Geissbuhler Y, Kannady K, Okumu F, Charlwood JD, et al. A new tent trap for sampling exophagic and endophagic members of the Anopheles gambiae complex. Malar J. 2009; 8:157 10.1186/1475-2875-8-157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sikulu M, Govella NJ, Ogoma SB, Mpangile J, Kambi SH, Kannady K, et al. Comparative evaluation of the Ifakara tent trap-B, the standardized resting boxes and the human landing catch for sampling malaria vectors and other mosquitoes in urban Dar es Salaam, Tanzania. Malar J. 2009; 8:197 10.1186/1475-2875-8-197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Okumu FO, Madumla EP, John AN, Lwetoijera DW, Sumaye RD. Attracting, trapping and killing disease-transmitting mosquitoes using odor-bated stations–The Ifakara Odor-Baited Stations. Parasit Vectors. 2010; 3:12 10.1186/1756-3305-3-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dugassa S, Lindh JM, Oyieke F, Mukabana WR, Lindsay SW, Fillinger U. Development of a Gravid Trap for Collecting Live Malaria Vectors Anopheles gambiae s.l. PLoS One. 2013; 8(7): e68948 10.1371/journal.pone.0068948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dugassa S, Lindh JM, Lindsay SW, Fillinger U. Field evaluation of two novel sampling devices for collecting wild oviposition site seeking malaria vector mosquitoes: OviART gravid traps and squares of electrocuting nets. Parasit Vectors. 2016; 9(1): 272 10.1186/s13071-016-1557-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(A) Amplification reactions utilizing the previously published pan-filarial DIDR primer set were conducted to demonstrate the integrity and amplifiable-nature of the isolated DNA templates from various filarial parasites. 100 bp ladder (lane 1), B. malayi (lane 2), D. immitis (lane 3), M. perstans (lane 4), W. bancrofti (lane 5), B. pahangi (lane 6), A. viteae (lane 7), L. loa (lane 8), No Template Control (lane 9), 100 bp ladder (lane 10).
(B) These same genomic DNA samples were included as template in amplification reactions utilizing the Brugia spp.-specific primer pair employed for test strip-based detection and amplification products were visualized on an agarose gel. 100 bp ladder (lane 1), B. malayi (lane 2), D. immitis (lane 3), M. perstans (lane 4), W. bancrofti (lane 5), B. pahangi (lane 6), A. viteae (lane 7), L. loa (lane 8), No Template Control (lane 9), 100 bp ladder (lane 10). (C) With the exception of A. viteae (product volume was exhausted) amplification products were also visualized using test strip-based detection. B. malayi (strip 1), D. immitis (strip 2), M. perstans (strip 3), W. bancrofti (strip 4), B. pahangi (strip 5), L. loa (strip 6), No template control (strip 7).
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.