Abstract
The Mexican gray wolf (Canis lupus baileyi) was historically distributed throughout the southwestern United States and northern Mexico. Extensive predator removal campaigns during the early 20th century, however, resulted in its eventual extirpation by the mid 1980s. At this time, the Mexican wolf existed only in 3 separate captive lineages (McBride, Ghost Ranch, and Aragón) descended from 3, 2, and 2 founders, respectively. These lineages were merged in 1995 to increase the available genetic variation, and Mexican wolves were reintroduced into Arizona and New Mexico in 1998. Despite the ongoing management of the Mexican wolf population, it has been suggested that a proportion of the Mexican wolf ancestry may be recently derived from hybridization with domestic dogs. In this study, we genotyped 87 Mexican wolves, including individuals from all 3 captive lineages and cross-lineage wolves, for more than 172000 single nucleotide polymorphisms. We identified levels of genetic variation consistent with the pedigree record and effects of genetic rescue. To identify the potential to detect hybridization with domestic dogs, we compared our Mexican wolf genotypes with those from studies of domestic dogs and other gray wolves. The proportion of Mexican wolf ancestry assigned to domestic dogs was only between 0.06% (SD 0.23%) and 7.8% (SD 1.0%) for global and local ancestry estimates, respectively; and was consistent with simulated levels of incomplete lineage sorting. Overall, our results suggested that Mexican wolves lack biologically significant ancestry with dogs and have useful implications for the conservation and management of this endangered wolf subspecies.
Keywords: Arizona, conservation genomics, gray wolves, hybridization, New Mexico, single nucleotide polymorphisms
The Mexican gray wolf (Canis lupus baileyi; Nelson and Goldman 1929; Goldman 1937) was once distributed throughout the southwestern United States and northern Mexico. It is considered the smallest (Young and Goldman 1944; Brown 1983) and the most genetically divergent (Wayne et al. 1992; García-Moreno et al. 1996; Vila et al. 1999; vonHoldt et al. 2011; Chambers et al. 2012; Fan et al. 2016; vonHoldt et al. 2016) of North American gray wolf subspecies. As a result of extensive predator-removal campaigns during the expansion of settlements in the southwestern United States, Mexican wolf abundance declined rapidly throughout the 20th century until their eventual extirpation in the mid 1980s (McBride 1980; Brown 1983). To mitigate this decline, the Mexican wolf was listed as federally protected in 1976 (USFWS 1976) and a captive population, called the McBride population, was established. A total of 6 individuals (5 males and 1 female) were captured in Mexico, but only 3 contributed to subsequent generations and are effectively considered founders (Hedrick et al. 1997; Siminski 2011). Two additional captive lineages had also been established by this time: the Ghost Ranch lineage in the United States, and a second one at the San Juan de Aragón Zoo in Mexico. Both of these lineages were descended from 2 founders (Hedrick et al. 1997). The 3 captive populations were merged in 1995, and by 1998 the first reintroduction of Mexican wolves into a recovery zone in Arizona and New Mexico was accomplished (USFWS 2010). Currently, the population of free-ranging Mexican wolves is estimated to be ~100 individuals (Harding et al. 2016).
The recovery and reintroduction program of the Mexican wolf has been mired by multiple controversies (reviewed in Harding et al. 2016), including the designation of the Mexican wolf as a separate subspecies (Cronin et al. 2015a, 2015b; Fredrickson et al. 2015) and the potential for introgression from other canids (i.e., coyotes and domestic dogs). Both morphologic (Bogan and Mehlhop 1983) and genetic evidence (Vila et al. 1999; Wayne and Vilá 2003; vonHoldt et al. 2011; Fan et al. 2016; vonHoldt et al. 2016) support that the Mexican wolf is indeed a separate subspecies of gray wolf and likely represents one of the earliest waves of migration of Canis lupus into the New World. As a result, the endangered status of the Mexican wolf has since been amended to be an independent subspecies rather than under the larger cover of gray wolves (USFWS 2015).
The genetic purity of Mexican wolves has also been questioned, especially in the Aragón and Ghost Ranch populations (reviewed in Hedrick et al. 1997). In Ghost Ranch, the male founder was suspected to be a wolf admixed with a domestic dog, rather than of pure Mexican wolf ancestry, and consequently, the documented management of this population has been questioned (Carley 1979). In Aragón, 2 female wolves were bred with a wolf-dog hybrid, but the resulting offspring were eliminated from the population (García-Moreno et al. 1996; Hedrick et al. 1997). Furthermore, the reduced density of wild Mexican wolves before extirpation may have facilitated hybridization with other canid species (Rhymer and Simberloff 1996). For example, Leonard et al. (2005) identified a single mitochondrial haplotype among pre-extirpation Mexican wolves that differed from a Mexican coyote haplotype by a single base pair and assumed it derived from interspecific hybridization. However, this occurred before any conservation management, is considered a rare occurrence, and does not occur in extant Mexican wolves (Hailer and Leonard 2008). Reintroduced Mexican wolves have also bred with domestic dogs, but the hybrid pups were immediately identified, removed, and euthanized (USFWS 2010).
Based on skull morphometrics, there was no detectable hybridization between captive lineages of Mexican wolves and other canids (Bogan and Mehlhop 1983; Weber 1989; López and Vázquez 1991; Hedrick et al. 1997). Early genetic studies using both mitochondrial DNA (Wayne et al. 1992) and microsatellite markers (García-Moreno et al. 1996) demonstrated a lack of introgression from other canids, although the possibility of a small amount of nonwolf ancestry could not be entirely ruled out (Hedrick et al. 1997). Recent genome-wide analyses of single nucleotide polymorphisms (SNPs; vonHoldt et al. 2011; Cronin et al. 2015a) and even complete genome sequencing (Fan et al. 2016; vonHoldt et al. 2016) also did not identify relevant admixture with other canids. Fan et al. (2016) suggested the possibility of a small amount of admixture with the basenji (1.2–3.2%), but such small levels can be difficult to distinguish from incomplete lineage sorting.
Unfortunately, each of these genomic studies to date have analyzed fewer than 10 Mexican wolves—many of which are shared across studies—and have not included representatives from each of the original captive lineages (McBride, Ghost Ranch, Aragón) and extant samples from cross-lineage wolves. Despite the current genetic evidence, local laws have subsequently been passed (e.g., Arizona, Apache County ordinance 2013-07, adopted 21 May 2013) removing certain protections from the Mexican wolf claiming, “experts on predators … have legitimate concerns and suspicions about the genetic purity …” and surmising that the Mexican wolf is not a wolf but rather a “wolf-dog hybrid.” Therefore, it is imperative that a thorough interrogation of the Mexican wolf’s genome across a representative sample of the subspecies is performed to clarify the extent of introgression with domestic dogs. In this study, we genotyped Mexican wolves from each of the 3 captive lineages in addition to captive and reintroduced wild wolves of cross-lineage ancestry for >172000 SNPs. Using multiple analytical approaches, we compared our results with datasets of domestic dogs and gray wolf populations to assess the potential for admixture between these various canids. Our work is the largest genetic study, in sample size, of Mexican wolves to date and provides extensive evidence for a lack of biologically significant ancestry from domestic dogs. These results have critical legal and scientific implications for the conservation and management of this endangered wolf.
Materials and Methods
CanineHD BeadChip
In this study, we used the CanineHD BeadChip (Illumina, Inc., San Diego, CA) developed by Vaysse et al. (2011), which includes 173 662 evenly spaced SNPs (mean spacing = 13 kb). These SNPs are a combination of those identified from the existing domestic dog genome project and an additional 4353 SNPs ascertained from resequencing gaps in a panel of 4 domestic dog breeds (Irish Wolfhounds, West Highland White Terrier, Belgian Shepherds, and Shar-Pei) and a pool of wolves (Vaysse et al. 2011). The authors evaluated the BeadChip using 450 samples of domestic dogs from 26 breeds, and also demonstrated its performance in 15 wolves. The average minor allele frequency (MAF) and call rate (proportion of genotyped SNPs) across all loci and samples are 0.23, and 0.998, respectively (https://www.illumina.com/documents/products/datasheets/datasheet_caninehd.pdf).
Sample Collection and Genotyping
We collected 87 Mexican wolf whole blood and tissue samples (dataset MW) from the Museum of Southwestern Biology’s Division of Mammals and the Division of Genomic Resources (www.msb.unm.edu) or from the Mexican Wolf Recovery Team as part of a captive breeding and monitoring program (collected by US Fish and Wildlife Service personnel as part of routine veterinary procedures under permit #TE-091551-7). Samples included individuals with pure ancestry from each of the 3 original captive lineages: McBride (MB, n = 33), Aragón (AG, n = 2), Ghost Ranch (GR, n = 7), and individuals of mixed or cross-lineage ancestry (CL, n = 45) according to the Mexican wolf pedigree (Siminski 2011). Additionally, we collected whole blood from a domestic dog of unknown breed ancestry to serve as a genotyping control. We extracted DNA from each sample using the DNeasy Blood and Tissue Kit (Qiagen, Inc., Germantown, MD) according to the manufacturer’s specifications. Purified DNA was concentrated using ethanol precipitation and quantified using the Quant-iT PicoGreen dsDNA Assay Kit (Invitrogen Corp., Carlsbad, CA). In cases where DNA concentrations were less than the requirement (~30 ng/µL) for downstream genotyping, multiple DNA extractions were performed for the sample and subsequently combined to achieve the appropriate concentrations. All samples were genotyped using the CanineHD BeadChip at the Broad Institute (www.broadinstitute.org; Cambridge, MA) according to recommended procedures. We replicated a total of 8 samples within and between different BeadChips to assess the technical reproducibility of the results. All signal intensities were imported into GenomeStudio Software (Illumina) and genotypes called according to cluster profiles for each SNP using the canineHD.egt file from Illumina.
We first excluded loci with a GenTrain score (ability to assign a genotype) less than 0.35 as recommended by Illumina and assessed reproducibility errors (genotyping disagreements among replicated samples) in GenomeStudio. Next, we removed the sample from each pair of replicates with the lowest call rate. Additional samples with a call rate less than 0.90 were excluded. Using the software PLINK v1.07 (Purcell et al. 2007), we omitted SNPs on the X- and Y-chromosomes, with a genotyping rate less than 0.90, and a MAF less than 0.05. For each locus, we calculated allele frequencies and Hardy–Weinberg equilibrium (HWE) using an exact test by Wigginton et al. (2005) in PLINK, and only retained SNPs that did not significantly deviate from HWE (P > 0.001). Using the software SNPRelate v0.9.18 (Zheng et al. 2012), we calculated linkage disequilibrium (LD) between all pairs of SNPs in a one megabase (Mb) sliding window and randomly removed a SNP if the correlation coefficient, r2, was greater than 0.5. The above SNP-cleaning procedures are standard for processing genome-wide SNP data and have been employed in other SNP-based analyses that included Mexican wolves (vonHoldt et al. 2011; Cronin et al. 2015a). The observed heterozygosity (HO) was calculated in PLINK for each individual and captive lineage. A simple linear regression was calculated in R (R Core Development Team 2017) to predict heterozygosity in both MB and CL wolves based upon birth year for wolves born post the merging of populations (1995).
The final dataset was summarized using a principle components analysis (PCA) in SNPRelate. PCA has been routinely used with genome-wide SNP genotypes to infer the clustering of individual wolves into populations (vonHoldt et al. 2011; Stronen et al. 2013; Cronin et al. 2015a). The PCA method also lacks certain assumptions used in other methods (see below) such as HWE and no LD within populations, and the SNP ascertainment scheme has been shown to have little effect on the results from PCA (Boyko et al. 2010; vonHoldt et al. 2011).
Global Ancestry
We investigated the potential for recent admixture between Mexican wolves and domestic dogs by combining our dataset with 4 additional studies that genotyped other canids on the CanineHD BeadChip (datasets are summarized in Table 1). These datasets included that of 1) Vaysse et al. (2011) who genotyped 532 dogs from 48 breeds and 15 gray wolves (LUPA, Lequarré et al. 2011; www.eurolupa.org), 2) Stronen et al. (2015a, 2015b) who genotyped 59 European gray wolves (EURO) from 4 population clusters, 3) Cronin et al. (2015a) (including Medrano et al. 2014) who genotyped 91 dogs, primarily mixed and poodle breeds, and 305 North American gray wolves that included 8 Mexican wolves (NAC), and 4) Vernau et al. (2013) who genotyped 28 Alaskan husky dogs as part of an association study (HUSK). Before merging together all the datasets, 2 Mexican wolves from NAC were removed because they overlapped with this study (studbook #1133 and #1177 with 99.6% and 99.7% genotype agreement across studies, respectively), the HUSK dataset was limited to only 10 control individuals with the lowest identity by descent as calculated in PLINK, and the strand (top vs. bottom) of each SNP was checked with the manifest (www.illumina.com) to ensure the same orientation across datasets. The datasets were subsequently merged and filtered for call rate, genotyping rate, autosomal SNPs, MAF, and LD as described above for MW.
Table 1.
Summary of the datasets used in this study
| Dataset | Mexican wolves | Gray wolves | Dogs | Total | Autosomal SNPs | XY SNPs | Total SNPs | Call rate | Citation |
|---|---|---|---|---|---|---|---|---|---|
| MW | 83 | 0 | 1 | 84a | 166582 | 5532 | 172114 | 0.989 | This study |
| MW-cleaned | 83 | 0 | 1 | 84 | 62219 | 0 | 62219 | 0.989 | This study |
| MW-cleaned-pruned | 83 | 0 | 1 | 84 | 7295 | 0 | 7268 | 0.989 | This study |
| LUPA | 0 | 15 | 532 | 547 | 169066 | 5744 | 174810 | 0.983 | Vaysse et al. (2011) |
| NAC | 8 | 297 | 91 | 396 | 120671 | 3130 | 123801 | 0.946 | Cronin et al. (2014) |
| EURO | 0 | 59 | 0 | 59 | 131118 | 3430 | 134548 | 0.982 | Stronen et al. (2013) |
| HUSK | 0 | 0 | 10 | 10 | 166583 | 5532 | 172115 | 0.987 | Vernau et al. (2013) |
| Merged | 89b | 371 | 634 | 1094 | 169066 | 0 | 169066 | 0.861 | This study |
| Merged-cleaned | 88 | 299 | 634 | 1021 | 118287 | 0 | 118287 | 0.991 | This study |
| Merged-cleaned- pruned | 88 | 299 | 634 | 1021 | 74876 | 0 | 74876 | 0.991 | This study |
The Mexican wolf (MW) and Merged datasets are shown before quality control, after removal of low quality loci and individuals (“Cleaned”), and again after removal of loci in high linkage disequilibrium (“Pruned”). “Merged” datasets include the combination of MW, LUPA, NAC, EURO, and HUSK samples.
aEighty-four individuals remained after initial removal of 8 replicates and 4 with low call rate.
bTwo Mexican wolves overlapped both the MW and NAC datasets.
We used 3 different methods to estimate global ancestry (average genome-wide proportion of ancestry from contributing populations) in Mexican wolves compared with domestic dogs and other gray wolves. We first assessed the potential for admixture by summarizing the data using a PCA as described above. The PCA provides a low-dimensional, nonparametric visualization of the variation in the dataset. If admixture is present in Mexican wolves, we expect admixed individuals to cluster closer to domestic dogs than nonadmixed ones.
Second, we used the maximum likelihood model implemented in ADMIXTURE v1.23 (Alexander et al. 2009; Alexander and Lange 2011) to estimate a priori ancestry coefficients in k different populations. The underlying model is similar to that of the popular STRUCTURE (Pritchard et al. 2000) software but uses a block relaxation strategy to accelerate the search procedure. This results in the ability to run large datasets more quickly compared with STRUCTURE while producing maximum likelihood estimates of ancestry proportions (Q values) with similar accuracy (Alexander et al. 2009; Alexander and Lange 2011). Because we are not interested in population structure within dogs or non-Mexican gray wolves, we first ran ADMIXTURE on a reduced sample set containing an equal number of Mexican wolves (n = 88) and randomly selected domestic dogs (n = 88) and gray wolves (n = 88). We ran ADMIXTURE for k = 1–10 and terminated calculations for each point estimate when the log likelihoods increased by less than 0.0001 between iterations (parameters: -C 0.0001, -c 0.0001). We assessed the most likely value of k by including a cross-validation procedure (parameter: --cv = 10), where the most probable k has the smallest cross-validation error (CVk; Alexander and Lange 2011). We repeated the above random sampling of individuals and k estimation procedure 100 times to assess confidence in the estimate of k. We then ran ADMIXTURE with the complete dataset using k = 2 through the best estimate of k. Finally, we repeated the ADMIXTURE analysis at just k = 3 with all samples using the supervised option (--supervised). The supervised method has been shown to produce less biased ancestry estimates, especially when the differentiation between ancestral populations is weak (Alexander and Lange 2011). This analysis used prior information that domestic dogs, North American gray wolves, and European grey wolves each belonged to a separate cluster. ADMIXTURE used this prior information to estimate the ancestry from these 3 clusters in Mexican wolves.
Lastly, we tested for admixture using the 3-population test (Reich et al. 2009) implemented in TREEMIX v1.12 (Pickrell and Pritchard 2012). This method tests for deviations from “treeness,” of which the test statistic f3(X;A,B) is negative when population X is a mixture of A and B, and can detect gene flow even hundreds of generations in the past (Reich et al. 2009; Patterson et al. 2012). We performed the test using each of the 3 original Mexican wolf lineages as X and compared them with all possible combinations of dog breeds and wolf populations with a sample size ≥10. The standard error of each test was estimated using a jackknife procedure and blocks of 100 SNPs—corresponding to ~3.1 Mb window length and longer than known LD in Mexican wolves (vonHoldt et al. 2011). Standard errors were Z-transformed and only Z-scores ≤−4 were examined for significant admixture (Patterson et al. 2012). As a control, we applied the f3 statistic and Z-score transformation to CL wolves using both Mexican and gray wolf populations as the parental populations. In these comparisons, Z-scores should be significantly negative when both parental populations are Mexican wolves.
Local Ancestry
In contrast with global ancestry estimation, which aims to measure the ancestral proportions of population averaged across the entire genome of an individual, local ancestry methods infer the identity of distinct chromosomal segments within a genome (Liu et al. 2013). We estimated local ancestry in Mexican wolves using the software LAMP-LD v1.3 (Baran et al. 2012). This program uses hidden Markov models to infer the haplotype structure in reference, or ancestral, populations and incorporates this structure into inferring the local ancestry in an admixed population. Results from LAMP-LD increase in accuracy with reference sample size, and larger reference panels can compensate for the loss of accuracy as divergence between the ancestral populations increases (Baran et al. 2012). LAMP-LD also natively infers the local recombination rate and consistently outperforms other methods in accuracy and computational speed (Baran et al. 2012; Zhang 2013; Brown and Pasaniuc 2014; Thornton and Bermejo 2014). Similar to the supervised analyses above in ADMIXTURE, we assigned local ancestry in Mexican wolves to either North American gray wolf, European gray wolf, or domestic dog reference populations. This analysis required phased reference haplotypes that were generated from the filtered genotypes before LD pruning using the software BEAGLE v3.3.2 (Browning and Browning 2007) with the “-nsamples” parameter set to 20 to improve accuracy. We ran LAMP-LD separately for each chromosome with a window size of 100 SNPs and 50 states. A window size of 50–100 SNPs and number of states >10 have been shown to provide the highest levels of accuracy (Baran et al. 2012).
Admixture Simulations
To assess the theoretical distribution of local ancestry fragments assigned to domestic dog in Mexican wolves, we simulated coalescent histories of canid populations using MACS v0.4 (Chen et al. 2009). The simulated histories we performed according to a subset of the model presented by Fan et al. (2016) using 4 extant populations: Mexican wolves, North American (Yellowstone) gray wolves, European gray wolves, and domestic dogs. The model, including effective population sizes and divergence times, is shown in Supplementary Figure S1. Using this model, we simulated 1 null (no migration) and 12 recent migration schemes between domestic dogs and Mexican wolves (Table 2). The migration schemes included either 5% or 15% admixture into the Mexican wolf population lasting either 1 or 5 generations and occurring 2, 20, or 200 generations in the past. The simulations included a mutation rate of 1 × 10−8 changes · site−1 · generation−1 and a generation time of 3 years (Fan et al. 2016). We simulated each of the 38 autosomes independently according to their length with recombination rates per chromosome as reported for domestic dogs (Wong et al. 2010). All rates were scaled to a reference effective population size of 10000. At the end of each simulation, we sampled the number of chromosomes matching that of our observed dataset (176, 460, 138, and 1268 chromosomes from Mexican wolves, Yellowstone wolves, European wolves, and domestic dogs, respectively). We first removed loci with a MAF < 0.1 among domestic dogs to mimic the ascertainment bias of SNPs, then filtered SNPs differing from HWE as described above (Wigginton et al. 2005) and randomly thinning the number of SNPs per chromosome to match the number and distribution in our observed dataset. Local admixture from wolves and domestic dogs into Mexican wolves was subsequently inferred as described above using LAMP-LD. Simulations were repeated 10 times for each migration scheme, resulting in a total of 880 simulated Mexican wolf genomes per scheme. A full description of the simulations and accompanying computer code can be found in the Data Accessibility section below.
Table 2.
The different migration schemes used in the simulations
| Scheme | m | T | g |
|---|---|---|---|
| 1 | 0 | — | — |
| 2 | 0.05 | 2 | 1 |
| 3 | 0.05 | 20 | 1 |
| 4 | 0.05 | 200 | 1 |
| 5 | 0.05 | 2 | 5 |
| 6 | 0.05 | 20 | 5 |
| 7 | 0.05 | 200 | 5 |
| 8 | 0.15 | 2 | 1 |
| 9 | 0.15 | 20 | 1 |
| 10 | 0.15 | 200 | 1 |
| 11 | 0.15 | 2 | 5 |
| 12 | 0.15 | 20 | 5 |
| 13 | 0.15 | 200 | 5 |
Each scheme varied in either migration rate from domestic dogs into Mexican wolves (m), the time (in generations past, T), or duration of migration (in generations, g).
Results
We genotyped a total of 88 samples (87 Mexican wolves and 1 domestic dog) on the CanineHD array for 173662 SNPs. We refer to all Mexican wolves hereafter by their official studbook number (Siminski 2011) and provide sample information in Supplementary Table S1. The average call rate across all samples was 0.982 (SD 0.029) and the mean reproducibility was 0.992 (SD 0.023) (Supplementary Tables S1 and S2). The SNPs removed included 1548 (0.89%) for a low GenTrain score, 5532 (3.2%) found on the X or Y chromosomes, 101742 (58.6%) monomorphic, and 2621 (1.5%) for a low genotyping rate. We removed 4 wolves (GR = 1 and CL = 3) with a genotyping rate <0.90. After excluding SNPs that failed our quality criteria and pruning for LD, a total of 7295 SNPs remained (Table 1).
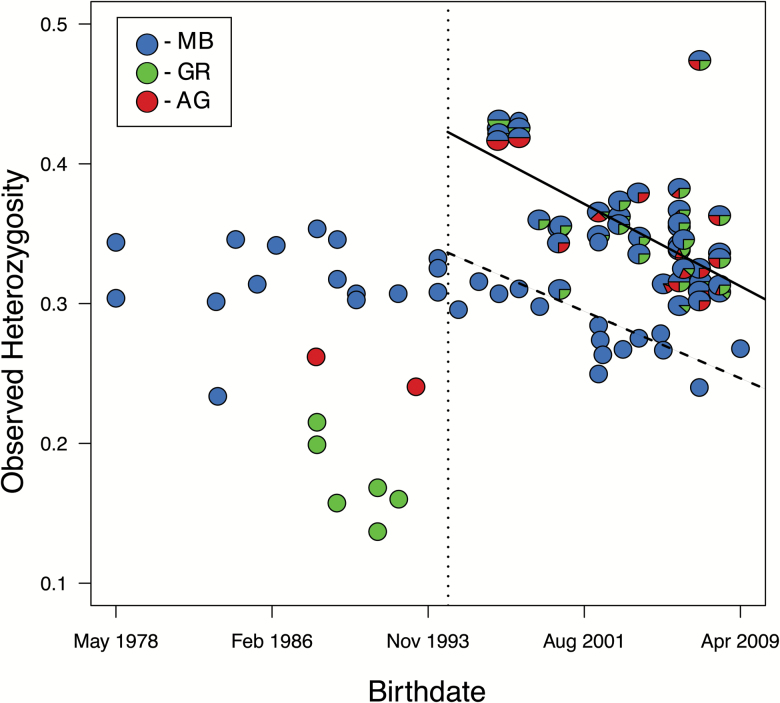
The mean HO across all Mexican wolves was 0.32 (SD 0.063). Before the merging of the 3 captive lineages, HO was higher in MB wolves (0.30 SD 0.039) than both AG (0.25 SD 0.015) and GR (0.17 SD 0.029) wolves (Figure 1). CL wolves consistently had the highest HO (0.35 SD 0.041), although after the initial merging of lineages, heterozygosity has been continuously decreasing in both MB and CL (Figure 1). Birth year significantly predicted HO in both MB [F(1, 15) = 6.4; P = .024; R2 = 0.30] and CL [F(1, 39) = 50.0; P = 1.7 × 10−8; R2 = 0.56); with HO being lost at rates of 0.62% and 0.76% per decade in MB and CL, respectively.
Figure 1.
Observed heterozygosity in Mexican wolves (Canis lupus baileyi) from 7295 unlinked SNPs. Individual Mexican wolves are shown as a pie chart with slices proportional to each wolf’s predicted ancestry in the 3 captive lineages and ordered by birthdate according to the official studbook (Siminski 2011). The linear regressions that describe the relationship between heterozygosity and birthdate for McBride (dashed line) and cross-lineage (solid line) wolves since the time when the 3 captive populations were merged (vertical dotted line) are shown. MB = McBride, AG = Aragón, GR = Ghost Ranch. See online version for full colors.
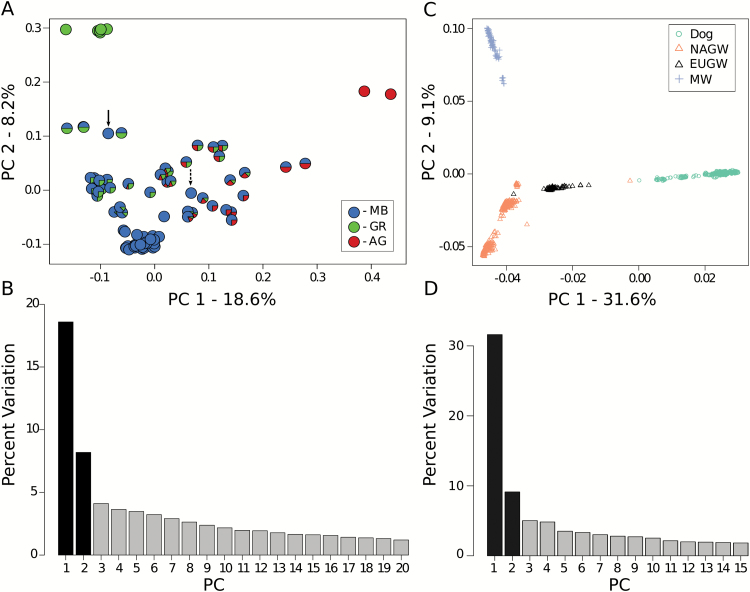
The distribution of genetic variation among Mexican wolves was summarized using PCA and shown in Figure 2A. The first 2 principle components combined to explain 26.8% of the variation (Figure 2B) in the dataset and easily differentiated the 3 captive lineages. The first component accounted for 18.6% of variation and primarily distinguished between AG and non-AG wolves, whereas the second component (8.2%) separated MB from GR wolves. Wolves with ancestry from multiple captive populations were intermediate between the 3 lineage clusters, corresponding approximately with the proportion of each population predicted by the pedigree. Two wolves (Studbook #547 and #858) are of particular interest because the pedigree predicts them to be pure MB wolves, yet the PCA suggests that #547 is an F1 hybrid between pure GR and MB parents and #858 may have ancestry from all 3 lineages.
Figure 2.
Principle component analyses of Mexican wolves (Canis lupus baileyi) and other canids (Canis lupus ssp). The first 2 principle components (PC) are shown (A, B) within Mexican wolves and (C, D) among Mexican wolves (MW), domestic dogs (Canis lupus familiaris), North American gray wolves (NAGW), and European gray wolves (EUGW). The proportion of variation explained by each component is shown for (B) Mexican wolves and (D) all canids. Individual Mexican wolves are shown as a pie chart with slices proportional to each wolf’s predicted ancestry in the 3 captive lineages according to the official studbook (Siminski 2011). The solid and dashed arrows in (A) depict Mexican wolves (studbook #547 and #858, respectively) whose predicted ancestries don’t coincide with the clustering in the plot. MB = McBride, AG = Aragón, GR = Ghost Ranch. See online version for full colors.
Global Ancestry
We combined our Mexican wolf genotypes with those of domestic dogs and gray wolves and repeated the filtering criteria as described above to produce a dataset containing 74876 unlinked SNPs (Table 1). Using PCA, the largest component of variation separated wolves from domestic dogs (Figure 2C) and accounted for 31.6% of variation in the dataset (Figure 2D). The second component distinguished between Mexican wolves and other gray wolves (Figure 2C) and explained 9.1% of variation (Figure 2D).
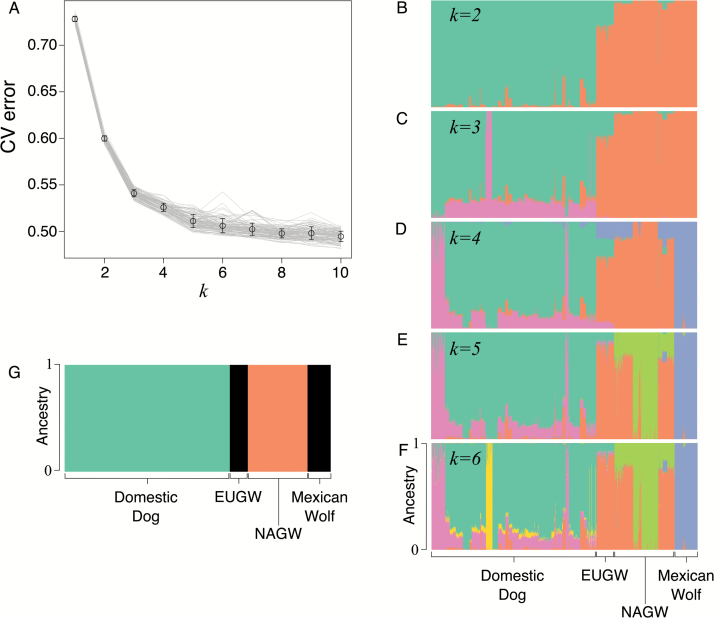
The software ADMIXTURE consistently estimated an optimal k ≥ 5 (mean CV5 = 0.51, SD = 0.007) among 100 resampled datasets, with very little decrease in CV error for k ≥ 6 (mean CV6 = 0.51, SD = 0.008, Figure 3A). Because we wanted to avoid detection of the extensive structuring within breeds of domestic dogs, we ran ADMIXTURE on the complete dataset for k = 2–6 since larger values of k provide little improvement in describing the population structure. At k = 2 and k = 3, domestic dogs and gray wolves formed distinct clusters, with Mexican wolves sharing only a mean of 0.06% (SD 0.23%) ancestry with domestic dogs (Figure 3B,C). Mexican wolves appear as a distinct cluster for k ≥ 4, and only a small amount of shared ancestry with clusters of gray wolves in both AG (16%) and GR (0.08%) (Figure 3D-F) existed. The mean proportion of ancestry in Mexican wolves assigned to the Mexican wolf cluster was 98.9% (SD = 3.2%) at k = 5, and individual-level values for k = 4–6 are provided in Supplementary Table S1. No other gray wolf ancestry was observed in MB or CL Mexican wolves. Using the “supervised” method to assign Mexican wolf ancestry a priori to 3 population clusters, all Mexican wolves assign completely to the European gray wolf cluster and no evidence of introgression with domestic dogs was observed (Figure 3G).
Figure 3.
Results of the population clustering analysis from ADMIXTURE. (A) Cross-validation error for different potential numbers of clusters, k, between 1 and 10. Each line represents a different subsample of an equal number (n = 88) of Mexican wolves, gray wolves, and domestic dogs. (B–F) Ancestry plot across all individuals for k = 2–6. Each vertical bar represents an individual with colors corresponding to the proportion of ancestry in the predefined number of clusters, k. (G) Ancestry plot for the supervised analysis, where Mexican wolves were assigned ancestry to either domestic dog, European gray wolf (EUGW) or North American gray wolf (NAGW) parental populations. See online version for full colors.
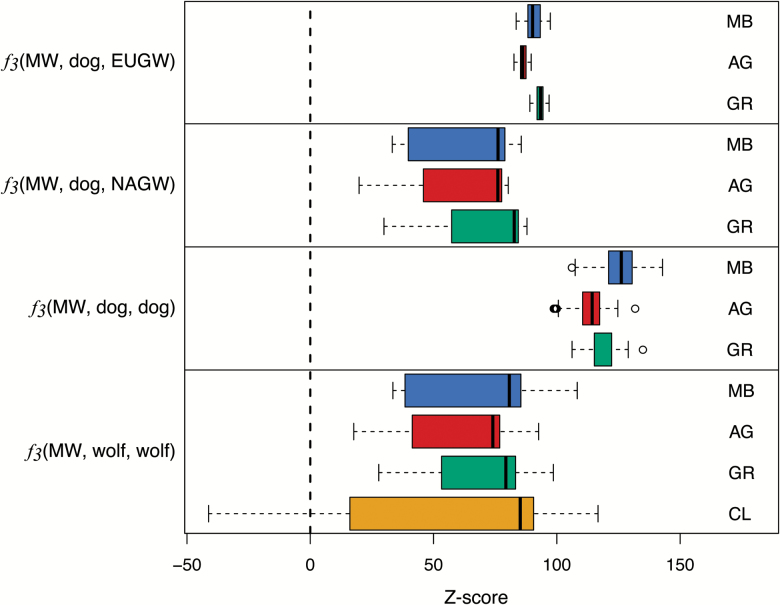
Using the 3-population test, we found no evidence for admixture (Z-score ≤ −4) in each of the 3 original captive lineages of Mexican wolf for all population comparisons (Figure 4). As expected, we only found evidence for admixture in CL between MB and either AG [f3(CL; MB, AG) = −0.015, Z-score = −31.0] or GR [f3(CL; MB, GR) = −0.020, Z-score = −41.3] and not for admixture in CL between AG and GR [f3(CL; GR, AG) = 0.027, Z-score = 21.4].
Figure 4.
Boxplots of the Z-scores of the f3 statistic for Mexican wolves (MW) when compared with either domestic dog breeds (dog), European gray wolves (EUGW), North American gray wolves (NAGW), or all populations of wolves (wolf). The statistic is in the form f3(X;A,B), where significantly negative values (Z-score ≤ −4) indicate that X is a result of admixture between A and B. Because CL wolves are known to be admixed between the Mexican wolf captive populations, the significantly negative scores for CL wolves are as expected. MB = McBride, AR = Aragón, GR = Ghost Ranch, CL = cross-lineage. See online version for full colors.
Local Ancestry
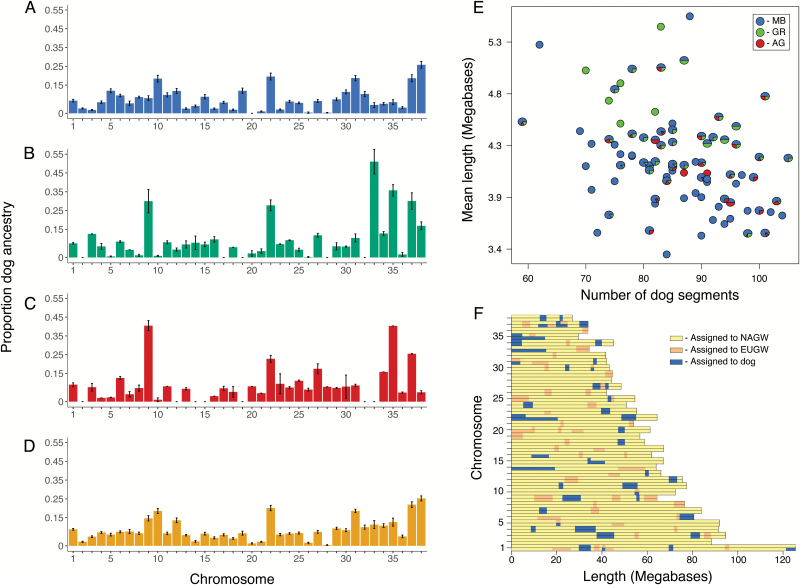
We assigned the ancestry of chromosomal segments of Mexican wolves to either gray wolf or domestic dog parental populations. Across all 88 Mexican wolves, an average of 7.8% (SD 1.0%) and 6.7% (SD 0.8%) of the genome contained fragments that could have originated from domestic dogs and European gray wolves, respectively (see Discussion below; Supplementary Figures S2 and S3 and Supplementary Table S1). Among chromosomes within populations, more variation existed, with most chromosomes containing <5% assignment to domestic dog ancestry (Figure 5A-D). However, a few exceptions were present. In both GR and AG chromosome 9 had a mean of 30.0% (SD 12.6%) and 40.5% (SD 3.8%) domestic dog ancestry, respectively, chromosome 33 in GR had a mean of 50.1% (SD 16.0%), and chromosome 35 in AG had 40.3% (SD 0%) domestic dog ancestry (Figure 5B,C). All chromosomes in both MB and CL wolves assigned ≤26% to domestic dog ancestry (Figure 5A,D).
Figure 5.
Local ancestry assignment in Mexican wolves. The proportion of each chromosome assigned to domestic dog ancestry in (A) MB, (B) GR, (C) AG, and (D) CL Mexican wolves. (E) The relationship between the number and length of local ancestry fragments assigned to domestic dogs in Mexican wolves. Mexican wolves are shown as pie charts with slices proportional to each wolf’s predicted ancestry in the 3 captive lineages according to the official studbook (Siminski 2011). (F) Locations of local ancestry fragments in an example Mexican wolf (#431, 100% predicted GR ancestry). Ancestry plots for all individual Mexican wolves can be found in Supplementary Figure S2. See online version for full colors. MB = McBride, AR = Aragón, GR = Ghost Ranch, CL = cross-lineage, NAGW = North American gray wolves, EUGW = European gray wolves.
On average, the number of segments assigned to domestic dog ancestry was 85.8 (SD 9.8) per individual Mexican wolf, with a mean segment length of 4.2 Mb (SD 0.44). No apparent pattern between each wolf’s pedigree ancestry and the length and number of domestic dog segments was evident, although GR wolves had slightly more admixed segments than other wolves despite no difference in mean length of these segments (Figure 5E). The fragments of domestic dog and European gray wolf ancestry were scattered more or less randomly across Mexican wolf genomes with no differences across populations (Figure 5F; Supplementary Figure S3).
Admixture Simulations
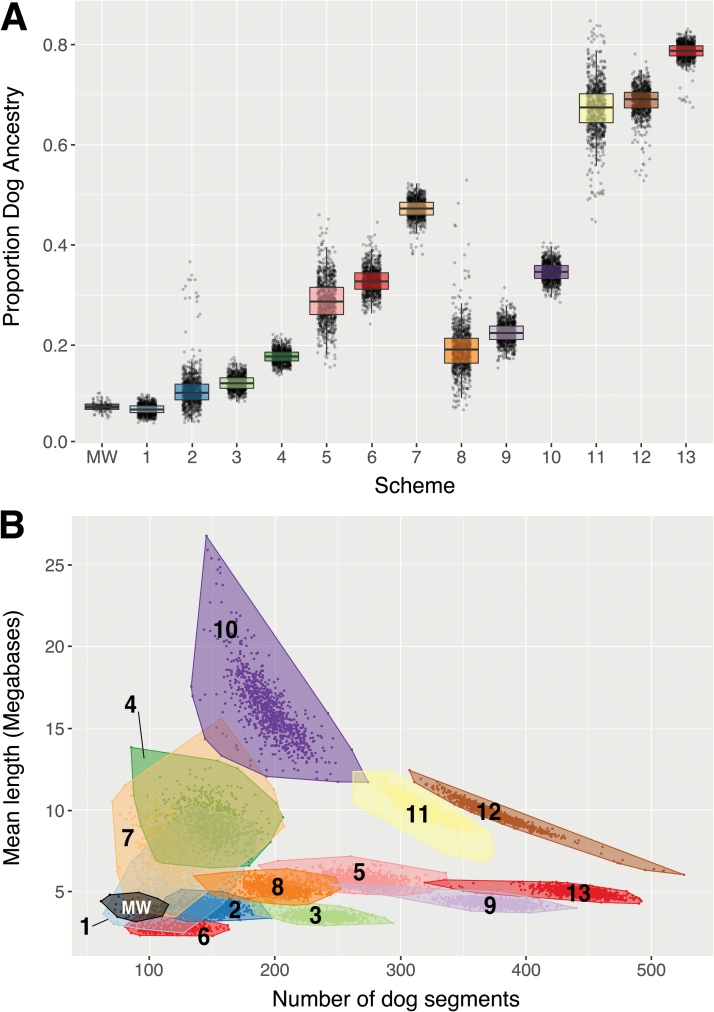
We simulated SNPs in 11440 Mexican wolf genomes to assess the theoretical expectation of local ancestry segments assigned to domestic dogs under 13 migration schemes (Table 2). The genome-wide proportion of ancestry summed across local segments assigned to domestic dogs is summarized in Figure 6A. Without migration, simulated Mexican wolves on average had 7.2% (SD = 1.0%) of their ancestry assigned to domestic dogs. The highest average proportion of domestic dog ancestry (79%, SD = 1.7%) was observed in scheme 13 which experienced 15% admixture from domestic dogs into Mexican wolves for 5 generations beginning 200 generations ago. The average admixture in schemes 2 (11%), 3 (13%), and 4 (18%) were most similar, albeit larger, than that observed in Mexican wolves (7.8%). When comparing the mean length of domestic dog fragments, schemes 1 (2.9 Mb), 3 (4.0 Mb), and 4 (3.5 Mb) were most similar to that observed in Mexican wolves (4.2 Mb, Figure 6B). The mean number of domestic dog fragments in schemes 1 (116), 2 (112), and 8 (117) were also similar but consistently larger than those in our observed Mexican wolf dataset (85.8, Figure 6B)
Figure 6.
Local ancestry assignment to domestic dogs in simulated Mexican wolves. (A) The proportion of local ancestry assigned per individual to domestic dogs summed across the genome. (B) Scatterplot of the number and mean length of local segments assigned to domestic dog ancestry per individual. In both panels, points represent individual observed Mexican wolves (MW) or individuals from simulated migration schemes (1–13, see Table 2). In (B), groups (MW or schemes 1–13) are bounded by the minimum convex polygon and labeled accordingly. See online version for full colors.
Discussion
Analyses within Mexican wolves
We were able to generate high-quality SNP genotypes for Mexican wolves using the CanineHD BeadChip. The genotype concordance across replicates was also markedly high (>99%) and consistent with technical replicates of human samples using the Illumina BeadChip platform (Hong et al. 2012). A majority of SNPs (58.6%) were monomorphic across all samples; a result expected based upon the small number of founders, extensive inbreeding, and possibility of historical small population size in Mexican wolves. The observed heterozygosity in each of the 3 original captive populations is consistent with pedigree expectations and microsatellite analyses (García-Moreno et al. 1996; Hedrick et al. 1997; Hedrick and Fredrickson 2008). For example, GR is the most inbred population (F = 0.61), and the mean heterozygosity observed for the SNP loci (HO = 0.17) was similar to that estimated from microsatellites (HO = 0.13). Of noticeable interest is the increase in genetic variation (genetic rescue) after merging the captive lineages in 1995 (Figure 1). The HO of first-generation CL wolves was >0.4, nearly a 33% increase over levels in the MB population—the most outbred lineage—during the same period. Unfortunately, it appears that despite this rescue of heterozygosity, genetic variation continues to deteriorate around a loss of ~0.6–0.7% HO per year, even in CL wolves. This is possibly the result of most individuals having a majority of ancestry from MB, whose loss of heterozygosity due to inbreeding and drift may be the dominating component.
The nonrandom mating between closely related individuals is a concern when inferring population structure, as it may lead to false inference of structure when it does not exist (Anderson and Dunham 2008). Our results suggested that extensive inbreeding within each of the captive lineages resulted in rapid and substantial subpopulation structure visible using PCA (Figure 2A). However, it remains possible that this substructure may have existed in Mexican wolves before their extirpation. For instance, the founders of MB were from the states of Chihuahua and Durango, Mexico, whereas the GR founders were from Tumacacori, Arizona, United States, and Sonora, Mexico. The founders of AR were collected from an unknown location (Siminski 2011).
As a result of this substructure the use of clustering algorithms, like PCA, may be valuable for estimating the contribution from different captive lineages in wild-born or otherwise unknown individuals. For example, the location of 2 Mexican wolves (#547 and #858) in the PCA (Figure 2A) did not correspond with their predicted contribution from the original lineages. It is possible that pedigree errors are responsible for these disagreements. The pedigree predicted that wolf #547 has 100% ancestry from MB; whereas the PCA predicted that this wolf most likely derived 50% ancestry from MB and 50% from GR. This is quite possible considering the particular facility where wolf #547 was born had several pure MB and GR individuals present at that time (Siminski 2011). Wolf #858 is also predicted to be of 100% MB ancestry, but the PCA indicated possible contribution from all 3 lineages. This wolf was actually born wild in the recovery area, and 2 of its grandparents were kept at one point at the AG facility—suggesting that this wolf may have incorrectly identified ancestors. However, we cannot exclude the possibility that an error had occurred in the proper identification of the tissue sample collected. Nevertheless, it is important to identify and minimize pedigree errors through proper documentation and the addition of genetic data because even a modest number of pedigree errors (i.e., >15%) can reduce the benefit of commonly used captive breeding strategies in conserving genetic variation (Oliehoek and Bijma 2009). Future work examining additional Mexican wolves needs to be performed to evaluate the pedigree error rate. Once corrected, an accurate pedigree will improve the selection of individuals and mating pairs to release into the wild, and will be useful for investigating quantitative traits, inbreeding depression, and inbreeding avoidance in the wild Mexican wolf population (Pemberton 2008).
Admixture with Domestic Dogs
As reported elsewhere for other genetic markers, including SNPs (Wayne et al. 1992; García-Moreno et al. 1996; Vila et al. 1999; vonHoldt et al. 2011; Fan et al. 2016), our analyses were consistent with a lack of biologically significant ancestry from domestic dogs. Although intensive management practices make ongoing hybridization between Mexican wolves and dogs of minimal concern in the extant population (USFWS 2010), the potential for historical admixture between Mexican wolves and domestic dogs has been of concern (Carley 1979; Hedrick et al. 1997). Unlike previous studies, our study was the first to investigate admixture in a comprehensive set of samples that included each of the 3 original captive lineages in addition to cross-lineage wolves. Estimates of global ancestry from the PCA, ADMIXTURE (both a priori and supervised), and the 3-population test each supported a lack of admixture with domestic dogs.
In addition to global ancestry analyses discussed above, estimates of local ancestry summed across the genome were consistent with no admixture (mean = 7.8%) with domestic dogs. Interestingly, the observed number and length of the segments assigned to domestic dog ancestry were relatively consistent across all samples analyzed. These putatively “admixed” segments were shorter and fewer in number than segments in Mexican wolves with simulated admixture with domestic dogs (scheme 2–13), and nearly identical to simulated Mexican wolves without admixture with dogs (scheme 1). If admixture was recent (e.g., last few generations), we would expect long, admixed regions distributed through the subset of individuals sharing recent dog ancestry as observed in simulated schemes 4, 7, and 10 (Figure 6B). However, the consistent distribution of short, admixed segments among all individual Mexican wolves suggests that these admixed regions were shared among all founders prior to the origin of the captive populations.
There are 2 general mechanisms that could theoretically generate this result. First, before extirpation, Mexican wolves might have interbred on rare occasion with domestic dogs belonging to local humans. This type of situation has been observed in gray wolves from many parts of the Old World (Fan et al. 2016). Second, and more likely, is that the common ancestor of Mexican wolves and domestic dogs shared these admixed fragments. In other words, this is the result of these genetic regions failing to coalesce within Mexican wolf evolutionary history, a process known as incomplete lineage sorting. Indeed, incomplete lineage sorting has already been reported as a major concern in genomic analyses of canines as a result of the large population size estimated in the wolf-dog ancestor, where 32.0% of variants were shared across wolves and dogs, and only 0.5% were fixed between them (Freedman et al. 2014). Recent phylogenetic work from complete genomes also showed that North American gray wolves (including the Mexican wolf) share a common ancestor, or equal amount of divergence, with Old World gray wolves and dogs (Fan et al. 2016). Since our panel of phased parental populations included 1) domestic dogs and 2) the combination of Old World and North American gray wolves, even in the absence of admixture we would expect to match a majority of haplotypes with North American gray wolves. A smaller, but equal, number of haplotypes would then be expected to assign to both domestic dogs and Old World gray wolves. Our simulated model without migration indicated that 7.2% of local ancestry with domestic dogs could be attributed to incomplete lineage sorting, a finding nearly identical to that in our observed dataset (7.8%). The small differences could be a result of the recent demographic processes not modeled by our simulations, such as bottlenecks and inbreeding, occurring within dog breeds and extant Mexican wolves. Furthermore, our model did not include ancient admixture between the ancestors of North American wolves and the domestic dog-European gray wolf ancestral population. It is likely that this kind of ancient admixture would increase the amount of local ancestry shared between North American gray wolves (including the Mexican wolf) and domestic dogs. Some methods that infer local ancestry, including LAMP-LD, can also utilize additional parental populations (e.g., Brisbin et al. 2012; Omberg et al. 2012; Guan 2014), albeit with decreasing accuracy (Padhukasahasram 2014). In our study, we limited the analyses to 3-way admixture since the primary focus was on introgression with domestic dogs, but additional, multi-way admixture using specific breeds of domestic dogs or populations of gray wolves may be of interest. Future management practices that track the distribution of local ancestry fragments from the 3 original captive lineages in CL wolves may also be practical for ensuring that the maximum amount of genetic variation is being maintained. Furthermore, haplotypes from the original captive lineages may provide a powerful tool for detecting beneficial and/or detrimental variation using admixture mapping requiring orders of magnitude fewer samples than conventional genome-wide association studies (Smith and O’Brien 2005).
It is possible that when comparing wolves and dogs, some of the differences between populations could be inflated due to ascertainment bias (Albrechtsen et al. 2010). Ascertainment bias refers to systematic deviations in allele frequencies between the SNP discovery panel and the genotyping panel. In our case, a majority of the SNP loci were discovered from a panel of domestic dogs, potentially biasing analyses that compare parameters sensitive to the allele frequency spectrum (e.g., FST) between dogs and wolves. To avoid this issue, we did not calculate FST values or compare heterozygosity between dogs and wolves. Analyses such as ADMIXTURE and the 3-population test, however, may be influenced by ascertainment bias. Therefore, results from the PCA, which is generally unaffected by ascertainment bias (Albrechtsen et al. 2010; vonHoldt et al. 2011; McTavish and Hillis 2015), should carry additional weight when inferring potential admixture (at least the relative differences between components are unaffected, McTavish and Hillis 2015). Furthermore, haplotype-based approaches, such as local ancestry inference using LAMP-LD, also are minimally affected by ascertainment bias (Lachance and Tishkoff 2013). Although future studies using markers devoid of this bias are needed (e.g., whole genome sequencing, RAD sequencing), all analyses in this study supported minimal admixture between Mexican wolves and domestic dogs.
Conclusions
In this study, we reported the potential for dense SNP datasets to improve the genetic management of the endangered Mexican wolf and the lack of biologically significant introgression from domestic dogs—a result shared across other studies and genetic markers in Mexican wolves. It remains possible that the >46 breeds of domestic dog (including mixed-breed individuals) used for comparison did not include specific breeds or populations that may have hybridized recently with Mexican wolves. The likelihood of this is rather low, considering that 1) the dogs used to design the CanineHD array were selected to represent a majority of the genetic variation in dogs, 2) “ancient” breeds of Native American origin were included (i.e., Alaskan husky, Greenland sledge dog; vonHoldt et al. 2010), and 3) a substantial majority of local ancestry was still assigned to gray wolf populations. We did not investigate admixture with coyotes (Canis latrans), because although possible, it is of little concern in the management program (Leonard et al. 2005; USFWS 2010). Additionally, we restricted the analyses to autosomal loci, and future studies targeting examination of the X and Y chromosomes may provide additional insight into both neutral and adaptive processes not manifested by the autosomes (Johnson and Lachance 2012). We encourage continued and expanded monitoring of Mexican wolves using a genome-wide set of SNPs to conserve the integrity of the Mexican wolf’s genome and maintain sufficient genetic variation necessary for a population with future evolutionary potential.
Supplementary Material
Supplementary data are available at Journal of Heredity online.
Funding
This work was supported by a National Science Foundation Integrative Graduate Education and Research Traineeship (#0654435) to R.R.F. and United States Geological Survey Quick Response Program (#G12AC20256) and Science Support Partnership (#10-R2-08) grants.
Data Availability
We have deposited the primary data underlying these analyses as follows:
• Mexican wolf sample information is available in Supplementary Table S1.
• Computer code and scripts for processing the data are available at GitHub (https://github.com/rfitak/Mexican_Wolf_SNPs)
• Mexican wolf SNP genotypes along with the merged genotypes: Dryad (https://doi.org/10.5061/dryad.g68k008)
• Other canid genotypes were downloaded from existing databases:
o Medrano et al. (2014); Cronin et al. (2015a): Dryad (https://doi.org/10.5061/dryad.p6598)
o Stronen et al. (2015a, 2015b): Dryad (https://doi.org/10.5061/dryad.284tf)
o Vernau et al. (2013): Please contact the listed corresponding author of this study.
o Vaysse et al. (2011): http://dogs.genouest.org/SWEEP.dir/Supplemental.html
Acknowledgments
We would like to thank Joseph Cook at the Museum of Southwestern Biology for providing many of the archived samples used in this study and Peter Siminski for discussions regarding the official Mexican wolf pedigree. We also thank the Mexican Wolf Recovery Team of the US Fish and Wildlife Service, especially Steven Chambers, Sherry Barrett, and Maggie Dwire, for additional support in sample collection and useful dialogue regarding the Mexican wolf program. We are grateful to Alex Ochoa, Phil Morin, and Phil Hedrick for comments on earlier drafts of this article. All Mexican wolf samples were collected and held under permit #TE-091551-7. Any use of trade, product, or firm names is for descriptive purposes only and does not imply endorsement by the US Government.
References
- Albrechtsen A, Nielsen FC, Nielsen R. 2010. Ascertainment biases in SNP chips affect measures of population divergence. Mol Biol Evol. 27:2534–2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander DH, Lange K. 2011. Enhancements to the ADMIXTURE algorithm for individual ancestry estimation. BMC Bioinformatics. 12:246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander DH, Novembre J, Lange K. 2009. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19:1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson EC, Dunham KK. 2008. The influence of family groups on inferences made with the program structure. Mol Ecol Resour. 8:1219–1229. [DOI] [PubMed] [Google Scholar]
- Baran Y, Pasaniuc B, Sankararaman S, Torgerson DG, Gignoux C, Eng C, Rodriguez-Cintron W, Chapela R, Ford JG, Avila PC et al. 2012. Fast and accurate inference of local ancestry in Latino populations. Bioinformatics. 28:1359–1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogan MA, Mehlhop P. 1983. Systematic relationships of gray wolves (Canis lupus) in southwestern North America. Occ Pap Mus Southwest Biol. 1:1–24. [Google Scholar]
- Boyko AR, Quignon P, Li L, Schoenebeck JJ, Degenhardt JD, Lohmueller KE, Zhao K, Brisbin A, Parker HG, vonHoldt BM et al. 2010. A simple genetic architecture underlies morphological variation in dogs. PLoS Biol. 8:e1000451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brisbin A, Bryc K, Byrnes J, Zakharia F, Omberg L, Degenhardt J, Reynolds A, Ostrer H, Mezey JG, Bustamante CD. 2012. PCAdmix: principal components-based assignment of ancestry along each chromosome in individuals with admixed ancestry from two or more populations. Hum Biol. 84:343–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DE. 1983. The wolf in the Southwest: the making of an endangered species. Tucson: The University of Arizona Press. [Google Scholar]
- Brown R, Pasaniuc B. 2014. Enhanced methods for local ancestry assignment in sequenced admixed individuals. PLoS Comput Biol. 10:e1003555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning SR, Browning BL. 2007. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet. 81:1084–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carley CJ. 1979. Narrative report on alleged Mexican gray wolf (Canis lupus baileyi) lineages held in captivity. Albuquerque: US Fish and Wildlife Service. [Google Scholar]
- Chambers SM, Fain SR, Fazio B, Amaral M. 2012. An account of the taxonomy of North American wolves from morphological and genetic analyses. North Am Fauna. 77:1–67. [Google Scholar]
- Chen GK, Marjoram P, Wall JD. 2009. Fast and flexible simulation of DNA sequence data. Genome Res. 19:136–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronin MA, Cánovas A, Bannasch DL, Oberbauer AM, Medrano JF. 2015a. Single nucleotide polymorphism (SNP) variation of wolves (Canis lupus) in Southeast Alaska and comparison with wolves, dogs, and coyotes in North America. J Hered. 106:26–36. [DOI] [PubMed] [Google Scholar]
- Cronin MA, Cánovas A, Bannasch DL, Oberbauer AM, Medrano JF. 2015b. Wolf subspecies: reply to Weckworth et al. and Fredrickson et al. J Hered. 106:417–419. [DOI] [PubMed] [Google Scholar]
- Fan Z, Silva P, Gronau I, Wang S, Armero AS, Schweizer RM, Ramirez O, Pollinger J, Galaverni M, Ortega Del-Vecchyo D et al. 2016. Worldwide patterns of genomic variation and admixture in gray wolves. Genome Res. 26:163–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredrickson RJ, Hedrick PW, Wayne RK, vonHoldt BM, Phillips MK. 2015. Mexican wolves are a valid subspecies and an appropriate conservation target. J Hered. 106:415–416. [DOI] [PubMed] [Google Scholar]
- Freedman AH, Gronau I, Schweizer RM, Ortega-Del Vecchyo D, Han E, Silva PM, Galaverni M, Fan Z, Marx P, Lorente-Galdos B et al. 2014. Genome sequencing highlights the dynamic early history of dogs. PLoS Genet. 10:e1004016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Moreno J, Matocq MD, Roy MS, Geffen E, Wayne RK. 1996. Relationships and genetic purity of the endangered Mexican wolf based on analysis of microsatellite loci. Conserv Biol. 10:376–389. [Google Scholar]
- Goldman EA. 1937. The wolves of North America. J Mammal. 18:37–45. [Google Scholar]
- Guan Y. 2014. Detecting structure of haplotypes and local ancestry. Genetics. 196:625–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hailer F, Leonard JA. 2008. Hybridization among three native North American Canis species in a region of natural sympatry. PLoS One. 3:e3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding LE, Heffelfinger J, Paetkau D, Rubin E, Dolphin J, Aoude A. 2016. Genetic management and setting recovery goals for Mexican wolves (Canis lupus baileyi) in the wild. Biol Conserv. 203:151–159. [Google Scholar]
- Hedrick PW, Fredrickson RJ. 2008. Captive breeding and the reintroduction of Mexican and red wolves. Mol Ecol. 17:344–350. [DOI] [PubMed] [Google Scholar]
- Hedrick PW, Miller PS, Geffen E, Wayne R. 1997. Genetic evaluation of the three captive Mexican wolf lineages. Zoo Biol. 16:47–69. [Google Scholar]
- Hong HX, Xu L, Liu J, Jones WD, Su ZQ, Ning BT, Perkins R, Ge WG, Miclaus K, Zhang L. 2012. Technical reproducibility of genotyping SNP arrays ised in genome-wide association studies. PLoS One. 7:e44483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson NA, Lachance J. 2012. The genetics of sex chromosomes: evolution and implications for hybrid incompatibility. Ann N Y Acad Sci. 1256:E1–E22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachance J, Tishkoff SA. 2013. SNP ascertainment bias in population genetic analyses: why it is important, and how to correct it. Bioessays. 35:780–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard JA, Vilà C, Wayne RK. 2005. Legacy lost: genetic variability and population size of extirpated US grey wolves (Canis lupus). Mol Ecol. 14:9–17. [DOI] [PubMed] [Google Scholar]
- Lequarré AS, Andersson L, André C, Fredholm M, Hitte C, Leeb T, Lohi H, Lindblad-Toh K, Georges M. 2011. LUPA: a European initiative taking advantage of the canine genome architecture for unravelling complex disorders in both human and dogs. Vet J. 189:155–159. [DOI] [PubMed] [Google Scholar]
- Liu Y, Nyunoya T, Leng S, Belinsky SA, Tesfaigzi Y, Bruse S. 2013. Softwares and methods for estimating genetic ancestry in human populations. Hum Genomics. 7:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López G, Vázquez CB. 1991. Linaje de lobos Mexicanos “San Juan de Aragón”: historia, evidencia de su autenticidad y posibilidad de certificación. Mexico City: Zoologico San Juan de Aragon. [Google Scholar]
- McBride RT. 1980. The Mexican wolf (Canis lupus baileyi): a historical review and observations on it status and distribution. Albuquerque: US Fish and Wildlife Service Southwest Region. [Google Scholar]
- McTavish EJ, Hillis DM. 2015. How do SNP ascertainment schemes and population demographics affect inferences about population history?BMC Genomics. 16:266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medrano J, Cronin M, Cánovas A, Oberbauer AM, Bannasch DL. 2014. Data from: single nucleotide polymorphism (SNP) variation of wolves (Canis lupus) in Southeast Alaska and comparison with wolves, dogs, and coyotes in North America. Dryad Digital Repository. doi: 10.5061/dryad.284tf [DOI] [PubMed] [Google Scholar]
- Nelson EW, Goldman EA. 1929. A new wolf from Mexico. J Mammal. 10:165–166. [Google Scholar]
- Oliehoek PA, Bijma P. 2009. Effects of pedigree errors on the efficiency of conservation decisions. Genet Sel Evol. 41:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omberg L, Salit J, Hackett N, Fuller J, Matthew R, Chouchane L, Rodriguez-Flores JL, Bustamante C, Crystal RG, Mezey JG. 2012. Inferring genome-wide patterns of admixture in Qataris using fifty-five ancestral populations. BMC Genet. 13:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padhukasahasram B. 2014. Inferring ancestry from population genomic data and its applications. Front Genet. 5:204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N, Moorjani P, Luo Y, Mallick S, Rohland N, Zhan Y, Genschoreck T, Webster T, Reich D. 2012. Ancient admixture in human history. Genetics. 192:1065–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pemberton JM. 2008. Wild pedigrees: the way forward. Proc R Soc Lond B Biol Sci. 275:613–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell JK, Pritchard JK. 2012. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 8:e1002967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. 2000. Inference of population structure using multilocus genotype data. Genetics. 155:945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ et al. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 81:559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Development Team 2017. R: a language and environment for statistical computing. Vienna (Austria): R Foundation for Statistical Computing. [Google Scholar]
- Reich D, Thangaraj K, Patterson N, Price AL, Singh L. 2009. Reconstructing Indian population history. Nature. 461:489–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhymer JM, Simberloff D. 1996. Extinction by hybridization and introgression. Annu Rev Ecol Evol Syst. 27:83–109. [Google Scholar]
- Siminski DP. 2011. Mexican wolf, Canis lupus baileyi, international studbook, 2011. Palm Desert: The Living Desert. [Google Scholar]
- Smith MW, O’Brien SJ. 2005. Mapping by admixture linkage disequilibrium: advances, limitations and guidelines. Nat Rev Genet. 6:623–632. [DOI] [PubMed] [Google Scholar]
- Stronen AV, Jędrzejewska B, Pertoldi C,Demontis D, Randi E, Niedziałkowska M, Borowik T, Sidorovich VE, Kusak J, Kojola I. 2015a. Data from: genome-wide analyses suggest parallel selection for universal traits may eclipse local environmental selection in a highly mobile carnivore. Dryad Digital Repository. doi: 10.5061/dryad.p6598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stronen AV, Jędrzejewska B, Pertoldi C, Demontis D, Randi E, Niedziałkowska M, Borowik T, Sidorovich VE, Kusak J, Kojola I et al. 2015b. Genome-wide analyses suggest parallel selection for universal traits may eclipse local environmental selection in a highly mobile carnivore. Ecol Evol. 5:4410–4425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stronen AV, Jędrzejewska B, Pertoldi C, Demontis D, Randi E, Niedziałkowska M, Pilot M, Sidorovich VE, Dykyy I, Kusak J et al. 2013. North-South differentiation and a region of high diversity in European wolves (Canis lupus). PLoS One. 8:e76454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton TA, Bermejo JL. 2014. Local and global ancestry inference and applications to genetic association analysis for admixed populations. Genet Epidemiol. 38(Suppl 1):S5–S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- USFWS 1976. Determination that two species of butterflies are threatened species and two species of mammals are endangered species. Fed Regist. 41:17736–17740. [Google Scholar]
- USFWS 2010. Mexican wolf conservation assessment. Albuquerque (NM): U.S. Fish and Wildlife Service Southwest Region. [Google Scholar]
- USFWS 2015. Endangered and threatened wildlife and plants; endangered status for the Mexican wolf. Fed Regist. 80:2488–2512. [Google Scholar]
- Vaysse A, Ratnakumar A, Derrien T, Axelsson E, Rosengren Pielberg G, Sigurdsson S, Fall T, Seppälä EH, Hansen MS, Lawley CT et al. ; LUPA Consortium. 2011. Identification of genomic regions associated with phenotypic variation between dog breeds using selection mapping. PLoS Genet. 7:e1002316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernau KM, Runstadler JA, Brown EA, Cameron JM, Huson HJ, Higgins RJ, Ackerley C, Sturges BK, Dickinson PJ, Puschner B et al. 2013. Genome-wide association analysis identifies a mutation in the thiamine transporter 2 (SLC19A3) gene associated with Alaskan Husky encephalopathy. PLoS One. 8:e57195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vila C, Amorim IR, Leonard JA, Posada D, Castroviejo J, Petrucci-Fonseca F, Crandall KA, Ellegren H, Wayne RK. 1999. Mitochondrial DNA phylogeography and population history of the grey wolf Canis lupus. Mol Ecol. 8:2089–2103. [DOI] [PubMed] [Google Scholar]
- vonHoldt BM, Cahill JA, Fan ZX, Gronau I, Robinson J, Pollinger JP, Shapiro B, Wall J, Wayne RK. 2016. Whole-genome sequence analysis shows that two endemic species of North American wolf are admixtures of the coyote and gray wolf. Sci Adv. 2:e1501714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- vonHoldt BM, Pollinger JP, Earl DA, Knowles JC, Boyko AR, Parker H, Geffen E, Pilot M, Jedrzejewski W, Jedrzejewska B et al. 2011. A genome-wide perspective on the evolutionary history of enigmatic wolf-like canids. Genome Res. 21:1294–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonholdt BM, Pollinger JP, Lohmueller KE, Han E, Parker HG, Quignon P, Degenhardt JD, Boyko AR, Earl DA, Auton A et al. 2010. Genome-wide SNP and haplotype analyses reveal a rich history underlying dog domestication. Nature. 464:898–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne RK, Lehman N, Allard MW, Honeycutt RL. 1992. Mitochondrial DNA variability of the gray wolf—genetic consequences of population decline and habitat fragmentation. Conserv Biol. 6:559–569. [Google Scholar]
- Wayne RK, Vilá C. 2003. Molecular genetic studies of wolves. In: Mech LD, Boitani L, editors. Wolves: behavior, ecology, and conservation. Chicago: The University of Chicago Press. p. 218–238. [Google Scholar]
- Weber M. 1989. La pureza racial del lobo gris Mexicano (Canis lupus baileyi) en cautiverio en Mexico: estudios preliminares. Memoria del VI Simposio Sobre Fauna Silvestre; F.M.V.Z Mexico: UNAM. [Google Scholar]
- Wigginton JE, Cutler DJ, Abecasis GR. 2005. A note on exact tests of Hardy–Weinberg equilibrium. Am J Hum Genet. 76:887–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong AK, Ruhe AL, Dumont BL, Robertson KR, Guerrero G, Shull SM, Ziegle JS, Millon LV, Broman KW, Payseur BA et al. 2010. A comprehensive linkage map of the dog genome. Genetics. 184:595–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young SP, Goldman EA. 1944. The wolves of North America: Part I. New York: Dover Publications, Inc. [Google Scholar]
- Zhang Y. 2013. De novo inference of stratification and local admixture in sequencing studies. BMC Bioinformatics. 14(Suppl 5):S17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X, Levine D, Shen J, Gogarten SM, Laurie C, Weir BS. 2012. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinformatics. 28:3326–3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
We have deposited the primary data underlying these analyses as follows:
• Mexican wolf sample information is available in Supplementary Table S1.
• Computer code and scripts for processing the data are available at GitHub (https://github.com/rfitak/Mexican_Wolf_SNPs)
• Mexican wolf SNP genotypes along with the merged genotypes: Dryad (https://doi.org/10.5061/dryad.g68k008)
• Other canid genotypes were downloaded from existing databases:
o Medrano et al. (2014); Cronin et al. (2015a): Dryad (https://doi.org/10.5061/dryad.p6598)
o Stronen et al. (2015a, 2015b): Dryad (https://doi.org/10.5061/dryad.284tf)
o Vernau et al. (2013): Please contact the listed corresponding author of this study.
o Vaysse et al. (2011): http://dogs.genouest.org/SWEEP.dir/Supplemental.html