Abstract
Insects harbor a tremendous diversity of sex determining mechanisms both within and between groups. For example, in some orders such as Hymenoptera, all members are haplodiploid, whereas Diptera contain species with homomorphic as well as male and female heterogametic sex chromosome systems or paternal genome elimination. We have established a large database on karyotypes and sex chromosomes in insects, containing information on over 13000 species covering 29 orders of insects. This database constitutes a unique starting point to report phylogenetic patterns on the distribution of sex determination mechanisms, sex chromosomes, and karyotypes among insects and allows us to test general theories on the evolutionary dynamics of karyotypes, sex chromosomes, and sex determination systems in a comparative framework. Phylogenetic analysis reveals that male heterogamety is the ancestral mode of sex determination in insects, and transitions to female heterogamety are extremely rare. Many insect orders harbor species with complex sex chromosomes, and gains and losses of the sex-limited chromosome are frequent in some groups. Haplodiploidy originated several times within insects, and parthenogenesis is rare but evolves frequently. Providing a single source to electronically access data previously distributed among more than 500 articles and books will not only accelerate analyses of the assembled data, but also provide a unique resource to guide research on which taxa are likely to be informative to address specific questions, for example, for genome sequencing projects or large-scale comparative studies.
Keywords: haplodiploidy, karyotypes, insects, paternal genome elimination, sex chromosomes, sex determination
Introduction
Insects are a tremendously successful group that accounts for a great majority of animal species (Mora et al. 2011) and can be found in almost all terrestrial and freshwater habitats (Gullan and Cranston 2010). This diversity at the taxonomic level is matched by a wide variety of sex determining mechanisms, and insect model systems have provided us with important insights into the biology and mechanisms of sex determination, and how they evolve (Sánchez 2008). For example, decades of research in the genetic model species Drosophila melanogaster have allowed identification of the genes and molecular pathways involved in sex determination (Cline et al. 2010), and comparative analysis between Drosophila species have greatly increased our understanding of how sex chromosomes evolve (Carvalho et al. 2009; Zhou and Bachtrog 2012; Zhou et al. 2012, 2013). However, most of these studies are focused on just a few model systems and do not take advantage of the potential insects could offer when taking a broader phylogenetic approach to study general principals of sex determination, sex chromosomes, and karyotype evolution.
The goal of this article is to systematically characterize and catalogue sex determination mechanisms in all orders of insects, based on karyotypes of 13113 species assembled from the literature as part of the Tree of Sex Consortium (2014). All the compiled data, including references, are available at www.treeofsex.org. Here, we use this data to first describe broad-scale phylogenetic patterns on the distribution of reproductive systems, sex determination mechanisms, sex chromosomes, and karyotypes across insects. We then analyze our data to test key evolutionary models to explain the observed distributions of sex determination systems. We discuss how these insights contribute to our general understanding of sex determination across the tree of life and what questions still remain unanswered.
Diversity of Sex Determination Systems in Insects
Throughout the tree of life, sex can be determined by many different mechanisms (Bachtrog et al. 2014), and insects capture much of this diversity (Figure 1). Most insects reproduce sexually (Normark 2003) and almost all insects are gonochoristic, that is, individuals are either male or female throughout their life. Hermaphroditism, where individuals either change sex during their life, or harbor both male and female gamete-producing organs simultaneously is found in about 30% of noninsect animals but appears largely absent in insects (Jarne and Auld 2006). In other taxa with diverse sex determination systems, such as in boney fish or Crustaceans, hermaphroditism is common and much of this diversity of how sex is determined can be explained by independent origins of separate sexes. In insects, however, the absence of hermaphroditism means that different modes of sex determination evolved from a gonochoristic ancestor. Two major forms of sex determination exist in gonochoristic animals, with either the environment or the genotype of the developing embryo determining sex, and in almost all insects, sex is determined by the genotype of the zygote (genotypic sex determination) (Figure 1).
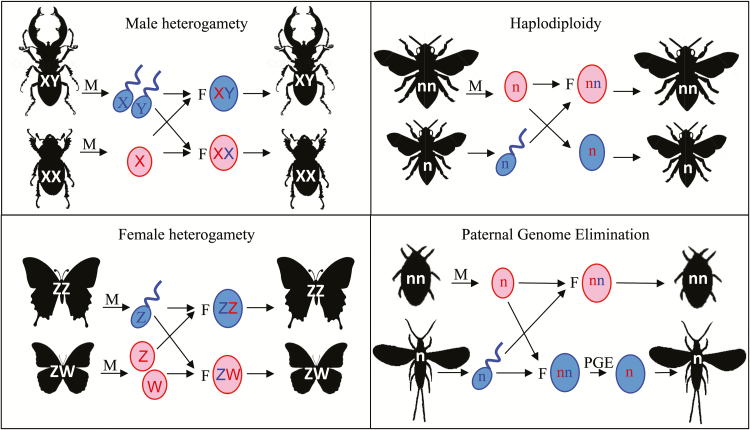
Figure 1.
Common sex determination systems in insects. With male heterogamety, males have heteromorphic sex chromosomes (XY), and females are homomorphic (XX). With female heterogamety, females have heteromorphic sex chromosomes (ZW), and males are homomorphic (ZZ). Under haplodiploid sex determination, females develop from diploid fertilized eggs (nn), and males develop from unfertilized haploid eggs (n). Under PGE, males develop from initially fertilized eggs, but eliminate their paternal genome during development (and become functionally haploid). Sperm are shown in blue, and eggs are shown in red. M indicates meiosis, F indicates fertilization, and PGE the elimination of the paternal genome.
Genotypic Sex Determination
One of the most familiar forms of sex determination is heterogametic genotypic sex determination, where either a single gene, a nonrecombining region along a chromosome, or an entire sex chromosome determines the sex of the developing embryo (also often referred to as genetic or chromosomal sex determination). In systems with heterogametic genotypic sex determination, there are 2 important distinctions between systems: whether it is the male or female that produces different gametes (male vs. female heterogamety), and whether the molecular mechanism of sex determination involves a dominant sex determining gene (dominant-Y/W) or if sex is determined by the X/Z to autosome ratio.
In male heterogametic systems, males are heterozygous for the sex determining region and carry an X and a Y chromosome, while females are homozygous carrying 2 X’s (XX/XY systems); or males carry a single X chromosome and females 2 X’s (XX/XO systems). Male heterogametic systems are found in many insects, most of the plant systems with separate sexes, and also in many vertebrates, including humans (Bachtrog et al. 2014). In female heterogamety, this situation is reversed: females are heterozygous for the sex determining region and carry a Z and a W chromosome, and males are homozygous carrying 2 Z’s (ZZ/ZW systems); or females carry a single Z chromosome and males 2 Z’s (ZZ/ZO systems). Female heterogametic systems are found in Lepidoptera (Sahara et al. 2012), Trichoptera (Marec and Novak 1998), some true fruit flies (Frías 1992), and also other invertebrates (Molluscs, crustaceans, arachnids) (Legrand et al. 1987; Tsurusaki and Cokendolpher 1990; Barsiene et al. 2000; Thiriot-Quievreux 2003) and within vertebrates (all birds and snakes, and some fish, amphibians, and lizards) (Tree of Sex Consortium 2014).
The genetic mechanism of sex determination in heterogametic systems can differ, with sex either determined by the presence of the sex-limited chromosome (i.e., a dominant-Y/W sex determining system), the ratio of X or Z chromosomes and autosomes, or the number of X or Z chromosomes. A dominant-Y system is found in Eutherian mammals and a dominant-W in silkmoth Bombyx mori, and sex is determined by the X-autosome ratio in Drosophila and Caenorhabditis elegans, or the presence of 2 Z chromosomes in birds. In systems that lack the sex-limited chromosome (XO and ZO systems), sex necessarily is determined by either the X/Z-autosome balance or the number of X/Z chromosomes present (as, e.g., is the case in many Orthoptera). Note that while the presence of sex chromosomes can often be inferred using simple cytogenetic techniques, the genetic mechanism of sex determination in species with sex chromosomes (i.e., dominant-Y/W or X/Z-autosome ratio) is in most cases unknown. The specific genes which act as the switch in the developmental pathway for sex determination have been identified in only a handful of species (Bopp et al. 2013; Bachtrog et al. 2014). In fruitflies of the genus Drosophila, the initial switch depends on the dosage of the X-linked gene sex lethal (Erickson and Quintero 2007), which regulates the down-stream sexual differentiation pathway. Interestingly, sex lethal is not involved in sex determination in other flies (Meise et al. 1998). In the mosquito Aedes aegpti, sex determination is governed by a dominant male-determining factor (the Nix protein-coding gene) located on the Y chromosome (Hall et al. 2015), while a W-linked piRNA is the primary determiner of sex in silkworm (Kiuchi et al. 2014).
Haplodiploidy and Paternal Genome Elimination
Haplodiploidy (HD) or arrhenotoky is a reproductive system where males are haploid and develop from unfertilized eggs, while females are diploid and develop from fertilized eggs. It is well known from Hymenoptera, but also found across a number of other clades of insects and other invertebrates, including nematodes, rotifers, and mites (Normark 2003; de la Filia et al. 2015). In other insects, like scale insects (Hemiptera: Coccoidea), both sexes develop as diploids from fertilized eggs, but maleness is determined by inactivation or loss of paternal chromosomes after fertilization (paternal genome elimination; PGE), leaving males functionally haploid (de la Filia et al. 2015). At which point the paternally derived chromosomes are eliminated, and from which tissues (germline-only, or germline and somatic cells) differs between species. Species with PGE display the same transmission genetics as true haplodiploid species, where males only transmit their maternal genome to their offspring. The genetic mechanisms of sex determination in haplodiploid systems can depend on complementarity of alleles at an unusually highly variable locus (as found in honey bees; Cook 1993) or genomic imprinting of paternal chromosomes, as found in Nasonia (Verhulst et al. 2010). Under complementary sex determination (CSD), a female develops if the alleles at the sex determination locus are different, and a male develops if the locus is hemizygous or homozygous (Beye et al. 2003). That is, diploid offspring develop from fertilized eggs and are normally female, whereas haploid offspring develop into males from unfertilized eggs (Cook 1993). The genomic imprinting model postulates that it is not haploidy or diploidy per se that determines sex, but the presence or absence of a paternally imprinted chromosome during early development. All eggs that only possess maternally derived chromosomes develop as males, and any egg receiving a paternal genome develops as female, regardless of ploidy. However, unlike the presence of haplodiploidy which can be relatively easily inferred using cytogenetic techniques, the genetic mechanism of sex determination in these systems is hard to study and known only in a few Hymenoptera.
Other, rare forms of sex determination exist in insects, such as monogeny, where all offspring of a particular individual female are either exclusively male or exclusively female, and is found in some Diptera and crustaceans (Stuart and Hatchett 1991). A type of temperature‐dependent sex determination is present in a species of Sciara fly with PGE. In this species, sex is determined by a temperature-sensitive maternal effect that controls X-chromosome elimination (Nigro et al. 2007). Cytoplasmic sex determination occurs if sex is under the control of cytoplasmic elements, such as endosymbionts (e.g., Wolbachia). It occurs in insects, but normally at low frequency within populations, since such sex determination systems normally result in a skew of the sex ratio within a species (Werren and Windsor 2000). Also, while the vast majority of insects reproduce sexually, parthenogenesis exists in some species where a female embryo develops from an unfertilized, diploid egg. Species that reproduce asexually are found in almost all orders of insects, but usually at low frequency (<1% of species) (Normark 2014). This is consistent with evolutionary theory that posits that asexual species are short-lived (Maynard-Smith 1978).
Evolution of Sex Chromosomes and Complex Sex Chromosome Systems
Sex Chromosome Differentiation
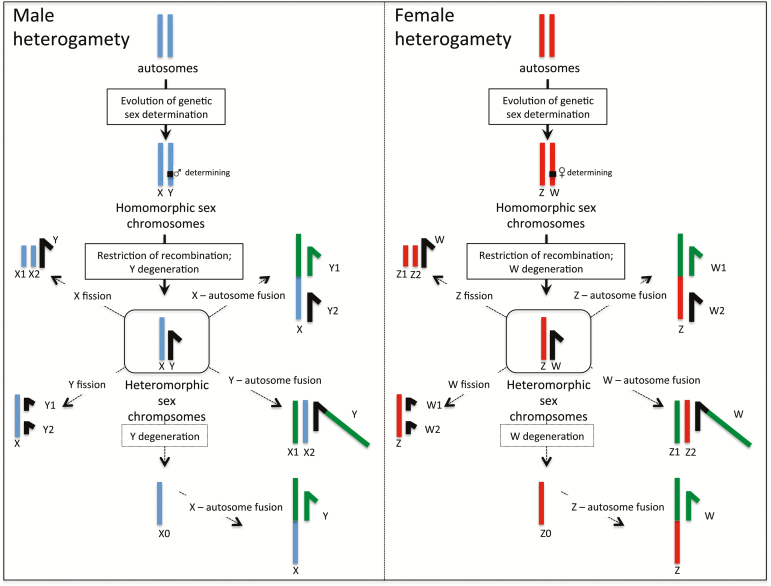
Sex chromosomes have evolved independently many times across the tree of life, including in insects. Sex chromosomes are derived from originally homologous autosomes that acquired a master-switch sex-determining gene (Bull 1983, see Figure 2). This creates sex chromosomes with a sex-determining function but an otherwise identical gene content and which recombine over most of their length (homomorphic sex chromosomes). The accumulation of sexually antagonistic mutations (i.e., mutations that are good for one sex, but bad for the other) close to the sex-determining region creates selective pressures to reduce or eliminate recombination between the proto-X/Y or proto-Z/W chromosomes, to ensure that such a sexually antagonistic allele is preferentially transmitted through the sex that it benefits. A restriction of recombination allows the sex chromosomes to diverge functionally and morphologically, and to evolve into heteromorphic sex chromosomes (Bachtrog 2013). The sex chromosome present in the homomorphic sex (the X in male heterogametic systems, the Z in female heterogametic systems) can still recombine in the homogametic sex, and typically maintains most of its ancestral gene content. The sex-limited chromosome (the Y or W chromosome), however, will be completely sheltered from recombination. The lack of recombination decreases the efficacy of natural selection on the Y/W chromosome, and may lead to the accumulation of deleterious mutations at many or most of its original genes. Over long evolutionary time periods, the Y or W chromosome might degenerate entirely, and this loss in gene function is often associated with a simultaneous accumulation of repetitive DNA on the Y or W chromosome (Bachtrog 2003). In the extreme case the Y or W chromosome may loose all essential genes and disappear entirely, leading to the evolution of XO or ZO sex determination (Blackmon and Demuth 2014, 2015b).
Figure 2.
Sex chromosome differentiation and origination of complex sex chromosomes. Sex chromosomes evolve from ordinary autosomes, after the emergence of a sex-determining locus. A restriction of recombination allows for differentiation, and Y/W chromosomes degenerate by an accumulation of deleterious mutations, and may be entirely lost (XO or ZO systems). Both fusions and fissions between sex chromosomes and autosomes can lead to the evolution of complex sex chromosomes (such as X1X2Y and XY1Y2 or Z1Z2W and ZW1W2 systems). Note that fusions are associated with a decrease in total chromosome number, while fissions increase the chromosome count.
Complex Sex Chromosomes
Complex sex chromosome systems, where a species harbors multiple X or Y or Z or W chromosomes can evolve relatively easily from an XY or ZW system by fusions between the ancestral sex chromosomes and autosomes, or fissions of the ancestral sex chromosome pair (Figure 2; Kitano and Peichel 2012; Blanco et al. 2013). For example, a fusion between an X and an autosome can lead to a system containing multiple Y chromosomes, where the second Y chromosome corresponds to the unfused homolog of the autosome that fused to the X. This second Y can undergo similar degeneration as the ancestral Y, leading to the possession of two degenerate Y’s (and an XY1Y2 sex chromosome system). On the other hand, a fusion between an autosome and a Y chromosome can result in the evolution of an X1X2Y system. Similar sex chromosome—autosome fusions in ZW systems can also produce complex sex chromosomes, with autosome-Z fusions creating a ZW1W2 karyotype, and autosome-W fusions generating a Z1Z2W karyotype. Chromosomal fusions can also lead to the gain of new Y or W chromosomes in species that had ancestrally lost them (i.e., transitions from XO to XY or ZO to ZW systems), and is believed to have occurred within Lepidoptera (Traut et al. 2008; Marec et al. 2010). Chromosomal fusions leading to complex sex chromosomes can be selected for if the fused autosome contains sexually antagonistic variation, analogous to the forces selecting for restricted recombination on the ancestral sex chromosomes, but they can also drift to fixation neutrally (Charlesworth and Charlesworth 1980), or may in fact be slightly deleterious (Pennell et al. 2015).
Complex sex chromosome systems can also derive from chromosomal fissions of the ancestral sex chromosomes (Figure 2). For example, a fission of the ancestral X chromosome will result in an X1X2Y system while a fission of the Y will generate a XY1Y2 sex chromosome system. Similar, a Z chromosome fission will create Z1Z2W sex chromosomes and a W fission will result in a ZW1W2 karyotype. Chromosomal fissions are thought to be less common than simple chromosomal fusions, since each chromosomal fragment requires a centromere for proper segregation during meiosis. Indeed, it has been suggested that the relative importance of chromosomal fissions and fusions differs among species groups that possess either monocentric chromosomes (i.e., a single centromere on each chromosome), or holocentric chromosomes (where localized centromeres are absent, and each chromosome fragment can segregate successfully during meiosis; Melters et al. 2012). Other chromosomal rearrangements, such as translocations, can also create complex sex chromosomes.
Most data that are available on sex chromosomes in insects are based on morphological differentiation from cytogenetic studies, that is, they are based on whether the X and the Y (or Z and W) appear distinct under a light microscope. Differentiation at the DNA sequence level is often accompanied by morphological differentiation, and sex chromosomes that appear morphologically similar are termed “homomorphic sex chromosomes,” and those that are distinct at the morphological level are termed “heteromorphic sex chromosomes.” While this distinction based on morphology often captures real differences in the underlying sequence divergence between sex chromosomes, systems that are classified as homomorphic may in fact be highly divergent at the DNA sequence level, yet show similar morphological features (Vicoso et al. 2013). In fact, sequencing of 37 species of Diptera showed that despite relatively homogenous karyotypes the species exhibited 12 distinct sex chromosome configurations (Vicoso and Bachtrog 2015). Thus, our classification based on morphological attributes is certainly an underestimate of sex chromosome occurrence and change, but provides an important first step in quantifying the diversity of sex chromosomes across insects, which we do below.
Evolution of Insect Sex Determination Mechanisms and Karyotypes
Our compilation of karyotype information across insects allows us to determine changes in the sex determination state across hexapods. Here, we present the results of a number of analyses aimed at understanding the evolution of sex determination across insects and to test general evolutionary theory. For some of these analyses we utilize data across all Hexapods while for others we focus on particular groups for which a sufficient amount of data is available. We highlight some major take-home points of these analyses, and discuss some mechanisms that could drive the observed patterns.
Male Heterogamety Is the Ancestral Mode of Sex Determination in Insects
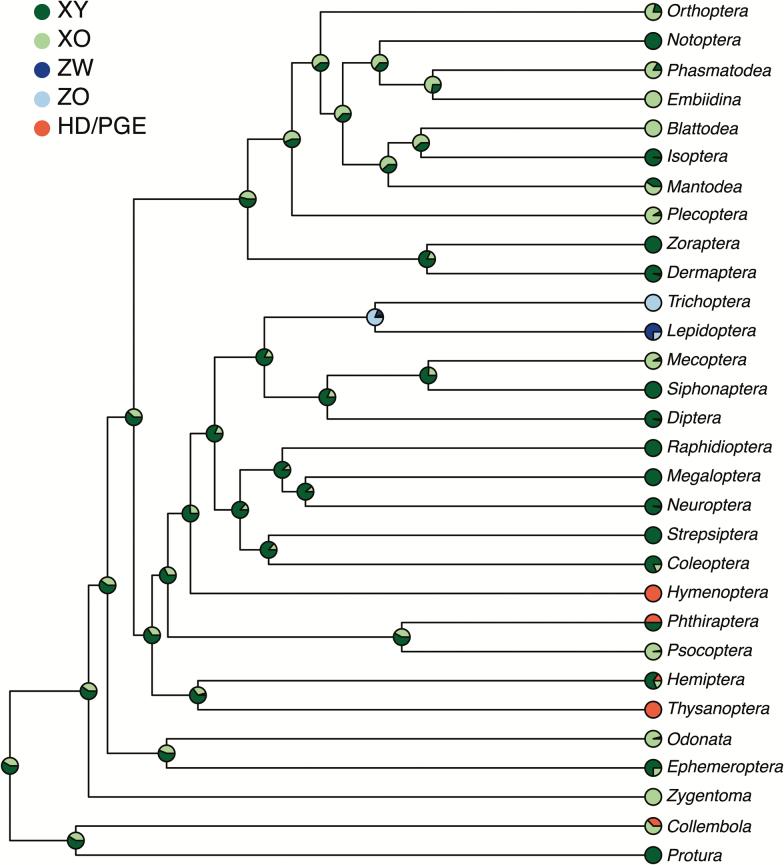
Because of the numerical dominance of male heterogamety (77% of investigated species, and within 223 of 359 families) it has long been assumed to be the ancestral state for insects (White 1977). However, to our knowledge this has never been tested quantitatively. We performed likelihood based ancestral state reconstruction to test this hypothesis in the R Phytools package (Revell 2012), using a dated phylogeny from (Misof et al. 2014). We made a number of simplifying assumptions to model the evolution of sex determination systems in insects. First, we assigned prior probabilities for sex chromosome systems for each order based on the proportion of taxa in each state in our database. We then implemented a model that treated PGE and HD as a single state and set the rate of transitions out of haplodiploidy to zero. Figure 3 shows the ancestral state reconstruction (pie charts on internal nodes of the tree) as well as the proportion of taxa in each order that exhibit each sex determination system (pie charts at tip of tree). We find strong evidence for the node leading to insects being male heterogametic (100% probability), but we have little power to distinguish between XY and XO sex chromosome systems (60% and 40% probability, respectively).
Figure 3.
Ancestral state reconstruction of sex determination systems in insects. Pie charts on nodes of the tree show the probability of the ancestral state at that node calculated from 1000 stochastic mappings. Pie charts at the tips indicate the abundance of different types of sex determination mechanisms in our dataset. The displayed values are from a model where transitions out of haplodiploidy are set to zero and the probabilities for the root state are equal to the stationary distribution.
Transitions to Female Heterogamety Are Rare
Sex determination systems where females are the heterogametic sex are rare in insects. Both parsimony and likelihood based ancestral state reconstruction suggests only 2 transitions to female heterogamety: one at the base leading to the superorder Amphiesmenoptera (Lepidoptera and Trichoptera), and a second transition within Diptera, within the family of Tephritidae. Transitions to female heterogamety from an ancestral state of male heterogamety might be difficult to achieve, as they result in offspring that are homozygous for the Y chromosome (Bull 1983; Bachtrog et al. 2014). If the X contains important genes not present on the degenerate Y, such transitions become increasingly difficult and such constraints may underlie the rarity of female heterogamety in insects. Likelihood based ancestral state reconstruction suggests that Lepidoptera and Trichoptera (female heterogametic clade) evolved from an XY rather than an XO ancestor (82% vs. 18% probability, respectively). In species groups with homomorphic sex chromosomes such transitions may be easier, and there might be several instances of undetected female heterogamety, as has been suggested for example, in Chironomidae (Thompson 1971); but see (Martin and Lee 1984).
Gain and Loss of the Sex-Limited Chromosome Is Common
As discussed, the chromosome that is limited to one sex only lacks recombination and degenerates. If the Y or W chromosome no longer contains genes that are necessary for sex determination or fitness, it may be lost entirely. Indeed, loss of the Y or W chromosome is common, both within and between families, and XO or ZO appear to be the ancestral mode of sex determination in several insect orders. In particular, ancestral state reconstruction indicates that Orthoptera, Blattodea, and Mantodea likely were ancestrally XO (with 61%, 62%, and 63% probability, respectively). The frequency at which the sex-limited chromosome is lost is probably largely driven by its gene content; that is, in groups where the Y contains genes necessary for survival or reproduction, it is unlikely to be lost unless these genes are relocated to another chromosome first. Such an event was detected within the pseudoobscura group of Drosophila: The Drosophila Y chromosome contains multiple genes necessary for male fertility, and they have become translocated to an autosome in the Drosophila pseudoobscura lineage where a new Y chromosome evolved through an X-autosome fusion (Figure 2; Carvalho and Clark 2005).
An indication of the variation in propensity for Y chromosome loss is seen in comparing Diptera and Coleoptera, the two orders for which we have the most data. In Coleoptera, 19% of the species (783 of 4187) are XO while in Diptera only 2% (47 of 2023) have lost their Y chromosome. We can get a rough estimate of the number of independent origins by using the taxonomy in our database to control for phylogeny. By counting only genera where a single species is XO but at least 2 species are XY, we infer a minimum of 70 Y chromosome losses in Coleoptera but only 12 in Diptera. This indicates that the fly Y chromosome may harbor more genes important for male function, relative to the beetle Y. This is consistent with a dominant male-determining role of the Y in many flies outside of Drosophila (Ullerich 1963).
Sex-limited chromosomes can be lost, but also be regained, for example, by fusions between Z or X and an autosome (Figure 2). In fact, karyotype data from orders like Lepidoptera, Orthoptera, and Odonota indicate that sex-limited chromosomes are frequently regained after their loss in an ancestral lineage. Though the sex chromosomes have been identified in only 40 species of Lepidoptera, we identify at least 4 independent origins of W chromosomes, whereas in Orthoptera and Odonata we observe 12 and 10 independent origins of Y chromosomes, respectively. To assess the importance of fusions as a source of new Y or W chromosomes, we compared the mean number of autosomes between species with and without sex-limited chromosomes within genera (Supplementary Table 1). In Coleoptera and Hemiptera, only half of the genera exhibit a pattern of reduction in chromosome number in species with a sex-limited chromosome (32 of 62 in Coleoptera, and 4 of 9 in Hemiptera). In contrast, 11 of 14 genera of Dermaptera, Lepidoptera, Orthoptera, Phasmatodea, and Plecoptera show a pattern of reduction in number of autosomes in species with a new sex-limited chromosome. This suggests a larger role for fusions between autosome and sex chromosomes as the force generating new sex-limited chromosomes in these orders.
Complex Sex Chromosomes Are Common in Some Groups
In some insect orders, such as Dermaptera, Plecoptera, and Isoptera, a majority of the taxa was found to harbor multiple X or Y chromosomes, and complex sex chromosomes are also common in Mantodea, Coleoptera, Hemiptera, and Orthoptera. Multiple sex chromosomes can originate through various chromosomal mutations, including fusions, fissions, or translocations (Figure 2). Groups with holocentric chromosomes are able to successfully segregate fragmented chromosomes (LaChance et al. 1970), which has long been assumed to account for complex sex chromosome systems in groups like Hemiptera [Ueshima (1979), but see Thomas (1987) for an alternative explanation]. To establish the origin of complex sex chromosome systems, we compared the mean number of autosomes between species with simple versus complex sex chromosomes within 81 genera that contain species with both types of sex chromosomes (Supplementary Table 1). As discussed above, fusions are expected to reduce the number of autosomes between closely related species with complex versus simple sex chromosomes while fissions and translocations should have no effect on the number of autosomes. In 41% of genera (33 of 81), species with complex sex chromosomes had fewer autosomes than species with simple sex chromosomes (consistent with fusions), while in 59% of genera (48 of 81), species with complex sex chromosome systems have equal or more autosomes (indicative of fissions/translocations). This analysis suggests that multiple processes are creating complex sex chromosomes in insects, but within orders, often one process dominates. For instance, within Coleoptera 26 genera show evidence for fission or translocations while only 13 suggest fusions. Our data include 30 genera from orders with holocentric chromosomes (25 from Hemiptera, 3 from Dermaptera, and 2 Lepidoptera). When restricting our analysis to this subset, we find roughly similar patterns as in the entire dataset: 63% of genera are consistent with fissions and 37% with fusions as the source of complex chromosomes. Thus, patterns of sex chromosome autosome fusions indicate that multiple pathways are leading to complex sex chromosomes, and we detect no significant difference in fusions vs. fissions creating complex sex chromosomes in species with monocentric versus holocentric chromosomes.
(Pseudo)-haplodiploidy Has Evolved Multiple Times in Insects, But Losses Are Rare
Haplodiploidy and PGE are thought to have evolved as a maternal adaptation that increases the reproductive value of females: A female’s haploid son will transmit her genes to future generations at twice the rate of a diploid son (Brown 1964; Bull 1979; Gardner and Ross 2014). However, this advantage is countered by the fact that haploid males will probably be less viable, at least in the early stages, which will limit the origination of haplodiploidy. Haplodiploidy has evolved at least 6 times within insects (Normark 2003). Two insect orders, Hymenoptera and Thysanoptera (thrips), are completely haplodiploid, and there are a number of smaller haplodiploid clades within Coleoptera and Hemiptera. PGE, which is often referred to as “pseudo-haplodiploidy,” has evolved at least 6 times within insects, once within Coleoptera, Hemiptera, Collembola (a sister group to the insects), Phthiraptera and twice within Diptera. The exact number of species with PGE is unclear since only a handful of species have been studied for each clade, but it may occur in up to 20000 species (2%) of insect (Table 1). Given their infrequent origin, haplodiploidy, and pseudo-haplodiploidy are remarkably widespread and estimated to be present in about 15% of invertebrate species (Normark 2003; de la Filia et al. 2015). The species-richness of haplodiploid clades could be either due to a low rate of reversal from haplodiploidy back to diplodiploid sex determination, or high rates of speciation in haplodiploid lineages (Koevoets and Beukeboom 2008; Lohse and Ross 2015; Patten et al. 2015). Haplodiploidy leads to the loss of meiotic spermatogenesis, a complex process that would be hard to re-evolve and may thus be an evolutionary trap (Bull 1983). Table 1 shows the estimated size of each haplodiploid or pseudo-haplodiploid clade as well as the number of species within each clade for which we have data. Indeed, none of the true haplodiploid clades harbors diplodiploid species, but reversions to diplodiploidy might have taken place in a clade with PGE (the scale insects). This fits the idea that PGE is less irreversible as males still have meiotic spermatogenesis (albeit a modified form). Loss of PGE could be driven by sexual conflict over the elimination of the paternal genome, which is predicted to lead to an evolutionary arms-race between maternal and paternal genomes (Herrick and Seger 1999; Ross et al. 2010). A recently published sister group analysis (Lohse and Ross 2015) does provide some support for higher speciation rates in haplodiploids, yet the effect is weak and limited to clades were haplodiploidy evolved relatively recently. Thus, the phylogenetic distribution of PGE and haplodiploidy appears to be determined mostly by their infrequent origin and rare loss.
Table 1.
Independent origins of haplodiploidy and pseudo-haplodiploidy (PGE) among insects
| Order/class | Haplodiploid clade | Type of haplodiploidy | Species number | No. of species data | Monophyletic? | Evidence of loss |
|---|---|---|---|---|---|---|
| Coleoptera | Micromalthus | Arrhenotoky | 1 | 1 | Yes | No |
| Coleoptera | Xyleborini | Arrhenotoky | 1360 | 5 | Yes | No |
| Coleoptera | Hypothenemus | PGE | 179 | 1 | Yes | ? |
| Collembola | Symphypleona | PGE | 1188 | 10 | Yes | No |
| Diptera | Sciaridae + Cecidomyiidae | PGE | 8468 | 32 | Yes | ? |
| Hemiptera | Aleyrodidae | Arrhenotoky | 1550 | 4 | Yes | No |
| Hemiptera | Iceryini | Arrhenotoky | 81 | 12 | Yes | Noa |
| Hemiptera | Neococcoidea | PGE | 7000 | 400 | Yes | Yes |
| Hymenoptera | Hymenoptera | Arrhenotoky | 115000 | 1600 | Yes | No |
| Phthirapterab | Phthiraptera | PGE | 3000 | 15b | Yes | ? |
| Thysanoptera | Thysanoptera | Arrhenotoky | 5000 | 24 | Yes | No |
Each row shows an independent origin of haplodiploidy or PGE, describing the estimated size of the clade and the number of species for which we have data in our database. The “evidence of loss” column is based on the presence or absence of diplodiploid species within these clades.
aNo reversion to diploploidy, but several origins of selfing hermaphroditism.
bPGE per se has only been described in a single species, but cytogenetic data on 14 other species shows an unusual type of spermatogenesis that might be indicative of PGE across lice families.
Parthenogenesis Evolved Often But Is Rare
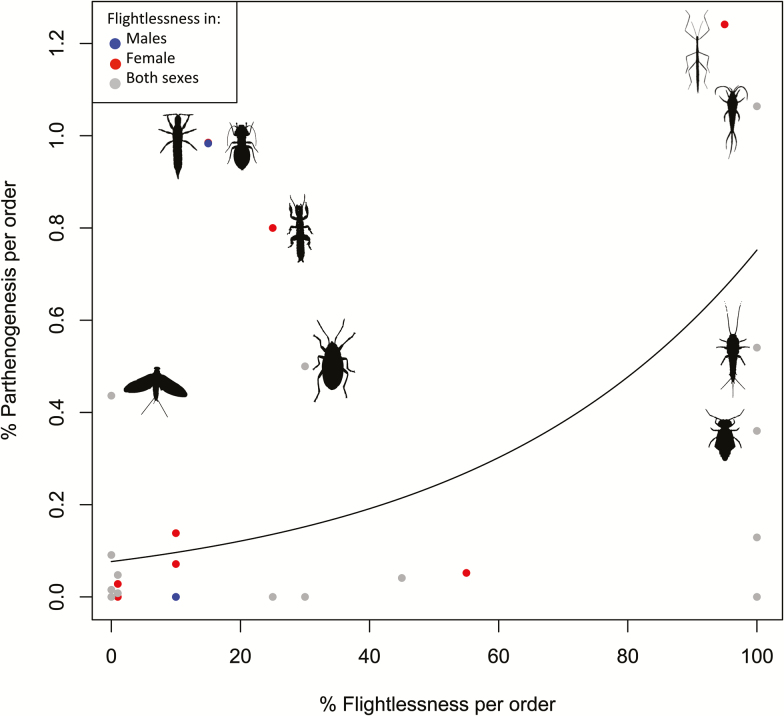
Parthenogenesis, where females develop from unfertilized eggs, has evolved many times across insects. Inclusion of a recently compiled list of parthenogenetic insect species to our database shows that parthenogenesis is found in 1169 species across 144 families (Normark 2003, 2014). Thus, while parthenogenesis is relatively rare when considering the total number of species, it has evolved independently hundreds or even thousands of times. Consistent with evolutionary predictions that asexual lineages are short-lived, there are few higher clades (genera, families, etc.) that are entirely parthenogenetic, but instead parthenogenesis is found mostly at lower taxonomic units. Hemiptera and Coleoptera harbor many of the parthenogenetic species reported (325 and 467, respectively). A wealth of theory has been published on the relative advantage of sexual versus asexual reproduction, but much less on the factors that can explain their phylogenetic distribution (Ross et al. 2012). For example, loss of flight may reduce the ability for mate search and parthenogenesis may evolve in taxa that frequently remain unmated. To test if a loss of flight in insects is associated with the evolution of parthenogenesis, we used the estimated fraction of parthenogenesis in each insect order (Normark 2014), together with order-level estimates of the frequency of flightlessness (Wagner and Liebherr 1992). We found that there is indeed higher levels of parthenogenesis in orders with a high percentage of flightlessness in either one or both sexes (F1,31 = 6.90, P = 0.013, Figure 4), but no difference between orders where flightlessness is restricted to just one of the sexes (F2,27 = 0.70, P = 0.50, Figure 4). Although this is a rather crude analysis, it is consistent with the notion that mate-search efficiency influences the mode of reproduction (Eppley and Jesson 2008).
Figure 4.
Parthenogenesis and flightlessness. The points show the percentages of parthenogenesis and flightlessness for each insect order. Grey points indicate orders where flightlessness is found in both sexes, red points those where flightlessness is restricted to females and blue points where it is restricted to males. Orders with a relatively high percentage of parthenogenesis (>0.2%) are identified with an icon of a representative species. The line plots the model prediction from a quasibinomial linear model of the relationship using the statistical package R.
Transitions to parthenogenesis in insects can also be induced by endosymbiotic bacteria (Werren and Windsor 2000). All convincing cases of parthenogenesis induction by endosymbionts come from haplodiploid taxa (Kageyama et al. 2012; Normark and Ross 2014), and parthenogenesis does appear overrepresented among haplodiploid/PGE clades relative to their frequency: haplodiploid/PGE clades account for 15% of taxa in our database, but 29% of parthenogens (Tree of Sex Consortium 2014; Table 1). Note, however, that the majority of parthenogenetic taxa (71%) are nested within diploid clades, suggesting that endosymbionts alone may be unable to explain most of the observed transitions to parthenogenesis.
Hermaphrodites Are Rare/Absent in Insects
Hermaphrodites, where both male and female sexual function exist within the same individual (either simultaneously or sequentially), are found in diverse animal groups, including both invertebrates (such as corals, gastropods, earthworms) and vertebrates (many fish; Bachtrog et al. 2014; Tree of Sex Consortium 2014). Insects, however, generally lack this form of reproduction, with the exception of 3 species of Iceryini scale insects, where hermaphroditism appears to have evolved from haplodiploidy (Gardner and Ross 2011). In noninsect invertebrates, hermaphroditism is often found in animals with low mobility (Eppley and Jesson 2008), and may thus be absent in insects where many species can fly. However, many primitive insect clades lack flight, and flight has been lost secondarily across nearly all orders of winged insects (Figure 4), yet hermaphroditism is not present in these clades. Our analysis instead shows that flightlessness in insects is more likely to be associated with parthenogenesis than with hermaphroditism. The reason for this is currently unclear, but may be due to the way sexual differentiation is achieved: In most insects sex determination is cell-autonomous, that is, each cell expresses the full sex determining cascade (Beukeboom and Perrin 2014). In contrast, many crustacean lineages that are frequently hermaphrodites regulate sexual differentiation by circulation of an androgenic hormone that causes male sex differentiation in embryos, and can override genetic sex determination. This might make sex a more flexible trait in crustaceans and allow for more frequent transitions between hermaphroditism and separate sexes (Cordaux et al. 2011).
Chromosome Numbers Differ Dramatically Among Insect Groups
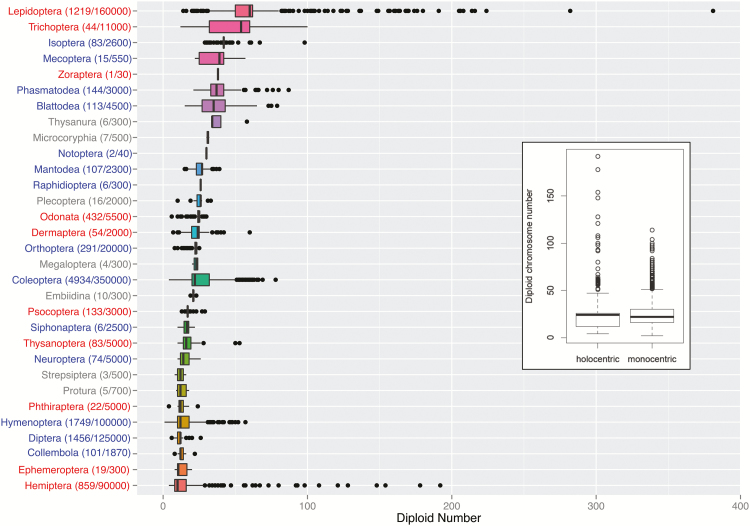
In addition to harboring diverse sex determining mechanisms, insects also vary enormously in chromosome number. Figure 5 shows variation in diploid chromosome numbers across orders of insects, and both the mean as well as the variance in chromosome number differs dramatically among insect orders. Lepidoptera, for example, have an average of 30 chromosomes, ranging from 7 to 190. Diptera, on the other hand, only have 11 chromosomes on average, and reported chromosome numbers vary from 6 to 26. Differences in chromosomal mutation rates could contribute to this diversity in chromosome numbers across taxa, and genome rearrangement rates are an order of magnitude higher in Lepidoptera than in Diptera (d’Alencon et al. 2010). Another factor that might influence chromosome numbers is the presence or absence of localized centromeres. While the majority of insects have either acrocentric or metacentric chromosomes, a significant fraction, including for example, all Lepidoptera and Hemiptera, have holocentric chromosomes. Here, localized centromeres are absent and even highly fragmented chromosomes can segregate successfully during meiosis and might enable more flexible karyotypes (Melters et al., 2012). We found that while there is no absolute difference in chromosome number between holocentric and monocentric species (Figure 5, PMCMC= 0.86, Bayesian GLM with order as a random effect), those orders with the highest variance in chromosome number tend to have holocentric chromosomes (Figure 5). However, without lower level phylogenies the significance of holo- versus monocentric chromosomes is difficult to assess, as differences in the ages of groups could also lead to differences in the variance in chromosome number among species groups. Other factors might also be important in determining variation in chromosome number, including differences in meiosis that might allow some groups to segregate rearranged chromosomes more reliably. Chromosome number, sex determination mechanisms, and sex chromosome systems are all intrinsically linked. For instance, a recent analysis suggests that lower chromosome number can increase the probability of transitioning to haplodiploidy, and certain sex chromosome systems may favor fusions more strongly than others (Blackmon and Demuth 2015b; Blackmon et al. 2015; Pennell et al. 2015; Ross et al. 2015).
Figure 5.
Distribution of chromosome numbers across hexapods. Diploid chromosome numbers are reported. Boxes represent the range of 25th to 75th percentile. Outliers are plotted as individual points. The color of the order names indicates whether chromosomes are holocentric (red), monocentric (blue), or unknown (gray). The number of species for which data is available and the size of each order is indicated in parentheses. The insert shows diploid chromosome numbers for holocentric versus monocentric species (Melters et al. 2012), where boxes represent the range of 25th to 75th percentile, and individual points the outliers.
Summary of Karyotype Data Across Orders of Insects
Sex in most insects is determined genetically, and Table 2 provides an overview of the phylogenetic distribution of sex determination mechanisms across insects. Below we give a short description of karyotype and sex chromosome composition within insect orders. We also include a short discussion on asexual species in the various groups, taken from the compilation of (Normark 2003; Normark and Ross 2014). For completeness, we include the limited data available for Entognatha (Collembola, Diplura, and Protura) that are wingless arthropods, which, together with insects, make up the subphylum Hexapoda.
Table 2.
Sex determination systems across insects
| XO | XY | C XOa | C XYb | ZO | ZW | C ZWc | Homd | HD/PGEe | Parthf | CNg | Taxa | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Orthoptera | 223 | 49 | — | 9 | — | — | — | — | — | 10 | — | 291 |
| Notoptera | — | 2 | — | — | — | — | — | — | — | — | — | 2 |
| Phasmatodea | 69 | 14 | — | — | — | — | — | — | — | 37 | 25 | 144 |
| Embiidina | 8 | — | — | — | — | — | — | — | — | 2 | — | 10 |
| Blattodea | 108 | — | — | — | — | — | — | — | — | 2 | 3 | 113 |
| Isoptera | 1 | 2 | — | 61 | — | — | — | 62 | — | — | 18 | 83 |
| Mantodea | 60 | 1 | — | 40 | — | — | — | — | — | 2 | 4 | 107 |
| Plecoptera | 3 | 1 | 8 | — | — | — | — | — | — | — | 4 | 16 |
| Zoraptera | — | 1 | — | — | — | — | — | — | — | — | — | 1 |
| Dermaptera | 3 | 22 | — | 27 | — | — | — | — | — | — | 2 | 54 |
| Trichoptera | — | — | — | — | 15 | — | — | — | — | 6 | 23 | 44 |
| Lepidoptera | — | — | — | — | 10 | 18 | 12 | — | — | 16 | 1163 | 1219 |
| Mecoptera | 13 | — | — | 1 | — | — | — | — | — | — | 1 | 15 |
| Siphonaptera | — | 2 | — | 4 | — | — | — | — | — | — | — | 6 |
| Diptera | 48 | 1893 | — | 10 | — | 7 | — | 93 | — | 46 | 97 | 1456 |
| Raphidioptera | — | 6 | — | — | — | — | — | — | — | — | — | 6 |
| Megaloptera | — | 4 | — | — | — | — | — | — | — | — | — | 4 |
| Neuroptera | 2 | 70 | — | 2 | — | — | — | — | — | — | — | 74 |
| Strepsiptera | — | 1 | — | — | — | — | — | — | — | 1 | 1 | 3 |
| Coleoptera | 770 | 3198 | 12 | 207 | — | — | — | — | 10 | 326 | 484 | 4934 |
| Hymenoptera | — | — | — | — | — | — | — | — | 1591 | 158 | — | 1749 |
| Phthiraptera | — | 1 | — | — | — | — | — | — | 1 | 4 | 16 | 22 |
| Psocoptera | 91 | 2 | — | — | — | — | — | — | — | 39 | 1 | 133 |
| Hemiptera | 155 | 284 | 1 | — | — | — | — | 3 | 255 | 467 | 114 | 1313 |
| Thysanoptera | — | — | — | — | — | — | — | — | 24 | 59 | — | 83 |
| Odonata | 403 | 20 | — | — | — | — | — | — | — | 1 | 11 | 432 |
| Ephemeroptera | 2 | 6 | — | — | — | — | — | — | — | 11 | — | 19 |
| Zygentoma | 3 | — | — | — | — | — | — | — | — | 2 | 1 | 6 |
| Archaeognatha | — | — | — | — | — | — | — | — | — | 5 | 2 | 7 |
| Diplura | — | — | — | — | — | — | — | — | — | — | — | — |
| Collembola | 17 | — | — | — | — | — | — | — | 10 | 21 | 53 | 101 |
| Protura | — | 3 | — | — | — | — | — | 1 | — | — | 2 | 5 |
| Totals | 1979 | 5582 | 21 | 361 | 25 | 25 | 12 | 159 | 1891 | 1215 | 2025 | 12452 |
The number of taxa reported for each type of sex determination system, the number of asexual species, and the number of taxa for which only chromosome number is available is indicated for each order of insects. The order of taxa matches the phylogeny in Figure 3.
aComplex XO.
bComplex XY.
cComplex ZW.
dHomomorphic.
eHaplodiploid.
fParthenogenetic.
gChromosome number only.
Collembola
There are 8000 described species of springtail (Cicconardi et al. 2013) across 4 separate orders (considered as suborders by some authors). Reproduction is sexual in most species with males depositing spermatophores that are picked up by females, although parthenogenesis is described from 21 species. Species from 3 out of the 4 orders of Collembola are male heterogametic with either XY or XO sex chromosome karyotypes. Members of the order Symphypleona, however, are characterized by PGE: Here both sexes develop from fertilized eggs with a X1X2/X1X2 sex chromosome karyotype. During early development, however, 2 X chromosomes are eliminated in males rendering them X1X2OO, which is followed by the elimination of the rest of the paternal genome from the male germline later in development (Dallai et al. 1999, 2000). Diploid chromosome numbers across the 4 orders range from 8 to 22.
Diplura
Two-pronged bristletails contain approximately 1000 described species, and ecological studies have revealed that many species reproduce sexually, and in some groups females even guard their eggs. Unfortunately, we have been unable to find any cytological investigations revealing the presence or absence of sex chromosomes or chromosome numbers in this lineage.
Protura
Approximately 700 species of these small primarily soil dwelling hexapods, known as coneheads, have been described. Chromosomal sex determination of the XY type has been identified in 3 Italian taxa from the families Acerentomidae and Eosentomidae (Fratello and Sabatini 1989). Records for the species Eosentomon transitorium indicate chromosome numbers ranging from 12 to 20 and both homomorphic and heteromorphic XY sex chromosomes, which likely is due to multiple cryptic species.
Archaeognatha (Microcoryphia)
Jumping bristletails are among the least evolutionarily changed insects, and approximately 500 species have been described. The method of sex determination has not been identified and cytological data is limited to diploid chromosome numbers in 2 species (32 in Machilis noctis and 30 in Dilta littoralis; (Bach and Petitpierre 1978). Reproduction by parthenogenesis has been identified in 5 taxa.
Zygentoma
Silverfish are the sister group of all other insects and have approximately 300 extant species. Cytogenetic data is available for 4 species and reveal an XO sex chromosome system with diploid chromosome number of 34 in 3 taxa and 58 in one (Makino 1951). Two additional species have been reported to reproduce parthenogenetically which belong to 2 separate families (Ateluridae and Nicoletiidae; Molero-Baltanás et al. 1998).
Ephemeroptera
Approximately 3000 species of mayflies have been described, and cytogenetic data is available for 19 species. XY sex chromosomes have been found in 6 species from 5 genera, and 2 species have XO sex chromosomes (Kiauta and Mol 1977). Eleven species belonging to 8 genera reproduce parthenogenetically (Gibbs 1977). Diploid chromosome number in this group ranges from 10 (which is found in 3 species of the family Baetidae) to 20 in Ecdyonurus dispar.
Odonata
The dragonflies and damselflies contain approximately 5500 described species, and have a long history of cytogenetic studies. Data is available for over 400 species representing 149 genera. The majority of taxa (400 species) have XO sex determination and this is believed to be the ancestral state for the order (Kiauta and Mol 1977). XY sex chromosome systems have been observed in 20 species. Of the 7 genera having both XO and XY taxa, 4 genera show a reduction in the number of autosomes in XY species compared with closely related XO species, consistent with X-autosome fusions creating new Y chromosomes. Ishnura hastate, a North American damselfly that has colonized the Azores is the only documented instance of parthenogenetic reproduction in the order (Rivera et al. 2005). Chromosome number ranges from 6 in the dragonfly Macrothemis hemichlora to 30 in an unidentified damselfly Mecistogaster species.
Psocoptera
Psocoptera or book lice are sister to the sucking lice (Phthiraptera) and contain approximately 3000 species. Karyotypes are known for species in 23 families, all of which display XO sex determination, with the exceptions of XY systems (probably created by X-autosome fusions) in Amphipsocus japonicas and Kolbia quisquiliarum (Golub and Nokkala 2009). Parthenogenesis has been reported in about 30 species. Like Phthirapteran and Hemipteran insects, all Psocopteran have holocentric chromosomes. Diploid chromosome number ranges from 14 in genera Elipsocus and Loensia to 30 in the family Psyllipsocidae.
Phthiraptera
All approximately 5000 species of lice are obligate ectoparasites on birds or mammals that survive poorly away from their host. The sex determining system of lice is poorly understood but it is likely that many have mating systems characterized by limited dispersal and frequent sibmating. There are published karyotypes for 18 species, which show that both sexes are diploid, but in most species, no sex chromosomes can be distinguished (Tombesi and Papeschi 1993). Only one species, Bovicola limbata, was found to have a XY male heterogametic sex determination system (Golub and Nokkala 2004). A recent molecular analysis in the human body louse suggested that this species may have PGE where males, although diploid, only transmit their maternal chromosomes to their offspring. It is unclear if PGE is found in other species of lice but their unusual spermatogenesis—where haploid sperm cells undergo several rounds of mitotic division after meiosis with an elimination of half of all sperm during the last division—is found across the order and might be a signature of PGE. Parthenogenesis has been described in 4 species. Diploid chromosome number in lice range from 10 in several species to 16 in the genera Hoplopleura and Polyplax.
Thysanoptera
There are approximately 5000 described species of thrips. It is generally assumed that all thrips species are haplodiploid, which would make them the only other haplodiploid insect order besides Hymenoptera. However, only a small percentage of thrips (24 species) have been studied by cytogenetic methods and as a result, haplodiploidy has been confirmed only in 2 out of the 8 families (Brito et al. 2010). There is currently no data on the molecular mechanism of sex determination in thrips. Like other groups of insects with haplodiploidy, some thrips display mating systems with high levels of sib-mating and females appear to have control over the sex ratios they produce (Choe and Crespi 1997). Thysanoptera are part of the superorder Paraneoptera, together with the Hemiptera and the Psocodea. Although haplodiploidy occurs in some Hemipteran insects, the condition there is clearly derived and originated independently from thrips. Parthenogenesis occurs frequently and is described in 59 species. Unlike members of the other two orders of the Paraneoptera that display holocentric chromosomes, all species of thrips have metacentric or acrocentric chromosomes. Diploid chromosome number in thrips ranges from 20 (reported in 2 families) to 106 in Aptinothrips rutua.
Hemiptera
There are approximately 90000 described species of hemipterans, and they are among the most diverse in terms of sex determination systems. Hemipteran insects are divided into 4 suborders. In 3 of these the sex determining systems are relatively homogeneous, mostly of the XY and XO type (890 and 299, respectively) and frequent complex karyotypes (171) and origins of new Y chromosomes. The suborder Sternorrhyncha, which includes white flies, aphids and scale insects is much more diverse though. Each of these 3 clades displays a different and unique set of sex determining systems: White flies are haplodiploid, with males developing from unfertilized eggs. Most aphids reproduce through cyclic parthenogenesis, where a species goes through several rounds of parthenogenesis followed by a single generation of sexual reproduction. All parthenogenetic and sexually produced offspring have a XX sex chromosome karyotype, and males are produced by random elimination of 1 of the 2 X chromosomes during early development, resulting in XO males (Wilson et al. 1997). Some aphid species have lost their sexual life cycle, and reproduce exclusively through parthenogenesis. Scale insects display the most diverse array of sex determination systems. Sex determination in a number of basal clades is of the XO type, but there have been at least 2 independent transitions to systems with haploid males within scale insects. Haplodiploidy evolved in the tribe Iceryiini, while PGE evolved in the neococcids and is the most common mode of reproduction among scale insects (found in approximately 6000 species; (Gavrilov 2007). In some species, the paternal genome is silenced (heterochromatinized) in somatic cells and eliminated from the germline, while in others the paternal genome is lost entirely from all cells during early development (Ross et al. 2010). Finally, a number of species in the tribe Iceryiini have evolved true hermaphroditism, where individuals produce both male and female gametes and reproduce through self fertilization (Ross et al. 2010). This is the only confirmed case of hermaphroditism in insects, and it evolved from haplodiploidy. Diploid chromosome number varies widely among hemipteran insects, ranging from 4 in some scale insect species of the family Monophlebidae to 192 in the scale insect Apiomorpha macqueeni.
Blattodea
About 4500 species of roaches have been described. Cytological data for over 100 species were available, and all of the sexual species have chromosomal sex determination with XO sex chromosomes. The overwhelming majority of roaches possess a metacentric X chromosome which cannot easily form centric fusions with autosomes, and might explain the rarity of complex sex chromosomes in this order (White 1976). Two species that reproduce parthenogenetically are reported. Chromosome number ranges from 16 in Lophoblatta fissa to 80 in Macropanesthia rhinoceros.
Isoptera
Long considered an independent order, recent molecular studies indicate that the termites are actually a highly derived clade that nests within Blattodea as sister to the genus Cryptocercus. Approximately 2600 species of termites have been described. Termites are eusocial with overlapping generations and contain multiple castes including soldiers and sterile workers that care for the young. Unlike eusocial Hymenoptera, termites are diplodiploid. Cytogenetic data is available for 83 species representing 4 families and 41 genera. Sex chromosomes have been identified in 63 taxa, and the most frequently observed sex chromosome system found in 51 species is X1X2Y1Y2 (Bergamaschi et al. 2007). One species (Stolotermes victoriensis) has XO sex determination (Luykx 1990). Chromosome number in Isoptera ranges from a high of 98 in Mastotermes darwiniensis to a low of 30 in Cryptotermes domesticus.
Mantodea
With about 2300 described species, mantids are the sister group of Blattodea and Isoptera, and are the most basally branching group of dictyopterans. With the exception of one parthenogen all species exhibit male heterogamety. XO sex chromosomes are found in approximately 60% of the studied species, but complex sex chromosome complements are also common. Specifically, a X1X2Y sex chromosome system has been documented in approximately 40 species, and only a single XY species has been found. The X1X2Y species were suggested to form a monophyletic group whose sex chromosomes derived from a reciprocal translocation between a metacentric autosome and a metacentric X chromosome. This would result in 2 X chromosomes, both of which have a single arm that chiasmatically pairs with one of the arms of the autosomes that became the Y chromosome. Achiasmatic male meiosis has evolved multiple times within mantids (White 1976). Chromosome numbers in mantids range from a low of 16 found in several groups to a high of 40 in Leptomantis parva and an unidentified species in the genus Humbertiella.
Zoraptera
Approximately 30 species of zorapterans have been described. These primarily tropical insects live in small colonies of less than 200 individuals. The colonies exhibit a polygynous mating system with the dominant males responsible for the majority of successful mating attempts. Zorotypus hubbardi is the only species that has been studied cytogenetically and it exhibits XY sex determination with a diploid chromosome number of 38 (Kuznetsova et al. 2002).
Orthoptera
The order Orthoptera contains over 20000 species that are distributed world-wide. The large size and low number of chromosomes have made Orthoptera an important group for our general understanding of chromosome biology and cytogenetics. XO sex chromosomes are found in about 80% of the species, and is considered the ancestral mode of sex determination in this clade. However, many species within Saltatoria have XY and X1X2Y sex chromosomes (Castillo et al. 2010). Parthenogenesis was found in 10 species (Lehmann et al. 2011). Extensive cytogenetic work on natural populations has revealed many examples of chromosomal variation within and between species, including inversions, translocations, centric fusions and fissions, sex chromosome rearrangements and supernumerary B-chromosomes (Karamysheva et al. 2011). Diploid chromosome number in Orthoptera ranges from a low of 8 in Dichroplus silveiraguidoi to a high of 26 in Conometopus sulcaticollis.
Phasmatodea
There are approximately 3000 species of stick insects. Data is available for 144 taxa, 37 of which reproduce parthenogenetically, and 83 species that have sex chromosomes. The majority of stick insects have XO sex chromosomes which likely is the ancestral system (68 species, present in 36 of 46 studied genera), and a minority exhibits XY sex chromosomes (13 species, 8 genera; White 1976). One species, Didymuria violescens, has males with both XO and XY sex chromosome complements. Chromosome number in this group is highly variable and polyploidy is well documented in parthenogenetic taxa. Mean chromosome number for all species that have identified sex chromosomes is 36.5, while parthenogenetic species have a mean chromosome number of 49.1. Diploid chromosome number in stick insects ranges from a low of 22 found in several species to 80 in Sipyloidea sipylus.
Embioptera (Embiidina)
The order of webspinners contains approximately 300 described species. Data on sex determination is available for 4 taxa from the family Embiidae and 5 from the family Oligotomidae (White 1976). All studied sexually reproducing Embiidina have XO sex determination, and female diploid chromosome numbers range from 20 to 24. Two parthenogenetic species have been identified, one of which, Haploembia solieri, occurs as both a diploid and triploid race.
Notoptera
The order Notoptera unites 2 small insect groups, Grylloblattodea and Mantophasmatodea, which have at times been considered independent orders, and have approximately 40 extant species. Cytogenetic data is available only for 2 species of the family Grylloblattodea, Grylloblatta campodeioformis and Galloisiana nipponensis, and both species have XY sex determination (White 1976). The diploid chromosome number of G. nipponensis is 30.
Plecoptera
There are approximately 2000 species of stoneflies. Cytogenetic data is available for 16 species from the families Perlidae and Perlodidae, both members of the suborder Systellognatha. The sex chromosome system has been identified in 11 taxa, and 7 species have X1X2O and 3 have XO sex chromosome complements, whereas only one has XY chromosomes. Multiple sex chromosome systems are often formed through the fusion of autosomes and sex chromosomes, but the available cytogenetic evidence indicates that the complex sex chromosomes in Plecoptera are the result of fission rather than fusion. For instance, in the genus Perla, species with multiple sex chromosomes have more chromosomes than Perla species with either XY or XO sex chromosomes (22 and 26 vs. 10, 19, and 21; Matthey and Aubert 1947); this together with the absence of Y chromosomes suggests that multiple sex chromosome systems originated from fissions of the ancestral X chromosome. Diploid chromosome number in this group ranges from 10 to 33.
Dermaptera
Approximately 2000 species of earwigs have been described. Cytogenetic data from over 50 species are available (White 1976), and all are male heterogametic, with about half having XY sex chromosomes, and the other half having complex sex chromosomes (X1X2Y and X1X2X3Y). Two earwig species are XO. The chromosomes of Dermaptera appear to be holocentric. Sex chromosome polymorphism has been documented in Forficula auricularia, where XY and X1X2Y males coexist within populations, and X1X2Y males have an additional chromosome, whose origin is unclear (Henderson 1970). Chromosome number varies from a low of 4 in Hemimerus bouvieri to a high of 30 in Arixenia esau.
Hymenoptera
There are approximately 100000 species of Hymenoptera and it is assumed that all of them have a haplodiploid sex determining system, which has been confirmed in all of the 1300 species for which karyotype data is available. Many hymenopteran insects have CSD, where a single locus (or a small number of loci) controls female development when heterozygous and male development when hemizygous (in haploids developing from unfertilized eggs; Cook 1993). CSD appears to be the ancestral state in the Hymenoptera with subsequent evolution of either CSD systems based on multiple loci or a complete loss of CSD in a number of taxa (Heimpel and de Boer 2007). Loss of CSD might be an adaptation to mating systems with high levels of inbreeding that would generate a high percentage of diploid males (homozygous for the CSD loci), which tend to be sterile. The molecular mechanisms of sex determination have been studied in detail in a number of species. In honeybees, a species with CSD, the complementary sex determining switch gene is highly polymorphic in populations, and in the jewel wasp Nasonia vitripennis, sex is determined by maternal imprinting (Verhulst et al. 2010). Chromosome number varies from 1 in Myrmecia croslandi while Dinoponera lucida has 57 chromosomes.
Coleoptera
Beetles, with 350000 described species, have been the focus of intense cytogenetic investigation, and karyotypes have often been used to identify cryptic species and to resolve phylogenetic relationships (Smith and Virkki 1978). Cytogenetic data for over 4797 taxa exist, and the vast majority of sexually reproducing beetles are male heterogametic with 3197 species possessing heteromorphic XY sex chromosomes. Of the remaining species, 771 are XO and more than 100 are asexual (Blackmon and Demuth 2015a). Despite the wealth of cytogenetic data, the genetic mechanism for sex determination has not been identified for beetles; however the widespread loss of the Y chromosome (e.g., 24 of 59 studied families have XO species) suggests that sex is determined by the X-autosome ratio, at least in some families. There are likely 2 origins of haplodiploidy and at least one origin of PGE in beetles (Brun et al. 1995; Jordal et al. 2000; Normark 2013). Coleoptera consists of 4 extant suborders. Archostemata has only 42 extant species, and Distocupes varians has 9 autosomes and XO sex determination, while the other archostematan species studied, Micromalthus debilis, has a diploid chromosome number of 20 and cyclic parthenogenesis, paedogenesis (reproduction by sexually mature larvae), and haplodiploidy (Normark 2013). The suborder Myxophaga has approximately 65 species but only one has been studied cytogenetically. Adephaga, the second largest order of beetles, contains approximately 40000 described species, and data is available for 1273 species from 7 families. Chromosome number in Adephaga is lowest in Graphipterus serrator, which has a diploid chromosome number of 8, and highest in Dixus capito obscuroides, which has 70 chromosomes. XO and XY sex chromosome systems are both common in this suborder (39% and 46%, respectively). Complex sex chromosome systems with multiple X chromosomes are present in 111 species. Polyphaga is the largest suborder of beetles and contains over 300000 described species, with XY sex chromosomes by far the most common (over 1938 species from 43 families). XO sex chromosomes are generally less prevalent but have been recorded in 18 out of 53 families, and complex sex chromosome systems have been found in 12 families. Chromosome number ranges from 4 in Chalcolepidius zonatus to 66 in Disonycha bicarinata. Polyploidy is frequent in parthenogenetic species, and parthenogenesis has been identified in 16 families. True haplodiploidy has evolved at least once in the subfamily Scolytinae, and it is thought that all Xylobrini (>1200 taxa) are haplodiploid but this has been investigated only in a handful of species. Another scolitine not closely related to the tribe Xylobrini, Hypothenemus hamperi, exhibits functional haplodiploidy in the form of PGE (Brun et al. 1995).
Strepsiptera
Twisted-wing insects are a highly derived and enigmatic group of endoparasitic insects and contain over 500 species. Their phylogenetic placement was debated for some time, but most recent studies indicate a close relationship with Coleoptera. Cytogenetic data for this group exists for just 2 species. The diploid number of Xenos peckii was identified as 16 and in an unidentified species of Xenos from Brazil, 3 autosomes and an XY sex chromosome system was observed (Ferreira et al. 1984). There are scattered reports of parthenogenesis in this family based on collecting only females, but the only convincing case of parthenogenesis is in Stichotrema dallatorreanum, a species that is facultative parthenogenetic with isolated females reproducing for multiple generations (Kathirithamby et al. 2001).
Neuroptera
With over 5000 described species, this group has cytogenetic information for 72 taxa belonging to 5 families. XY sex determination is dominant in the group and is found in 70 taxa and in all studied families. The Y chromosome has been lost at least twice, once in the Sisyrid Climacia areolaris and again in the Mantispid Plega dactyloya (Hughes-Schrader 1975b). The family Mantispidae also contains a species, Entanoneura phithisica that has a X1X2X3Y1Y2Y3 sex chromosome system (Hughes-Schrader 1969). Entanoneura phithisica has 7 autosomes, a reduction of 2 when compared to Entanoneura limbata, which indicates that it was a conversion of 2 of the dot like autosomes into sex chromosomes to produce this multiple sex chromosome system. Diploid chromosome number in the group ranges from 10 to 26.
Megaloptera
This group has approximately 300 described species, and along with Raphidioptera was formerly considered part of Neuroptera. Four of these species have been examined cytogenetically and all have XY sex chromosomes, and diploid chromosome numbers range from 9 to 11 (Takeuchi et al. 2002).
Raphidioptera
There are approximately 300 species of extant described snakeflies. Cytogenetic data is limited to 6 species from the genera Agulla and Rhaphidia, all with XY sex chromosomes (Hughes-Schrader 1975a), and all studied species have 12 autosomes.
Trichoptera
Caddisflies contain approximately 11000 extant species, and are the sister group of Lepidoptera. Sex determination data is limited to 15 taxa, all of which have a ZO system (Lukhtanov 2000). An additional 6 taxa are likely parthenogenetic (Corbet 1966). Diploid chromosome number has been identified for 44 species in this group and varies from 12 in Limnephilus affinis to 100 in Agrypnetes crassicornis.
Lepidoptera
There are over 160000 described butterfly species. Lepidoptera are one of the few orders that are female heterogametic. While there have been many cytogenetic studies in butterflies, testis squashes cannot reveal the karyotype of the heterogametic sex and most cytogenetic studies have only reported chromosome number. The sex chromosome system has been identified in only 40 of the 1219 studied species, and Lepidoptera have both ZW and ZO systems. Based on the distribution of ZO within Lepidoptera as well as its presence in its sister group Trichoptera, it is believed that butterflies were ancestrally ZO (Lukhtanov 2000). ZO sex chromosomes have been identified in 6 families of Lepidoptera, 3 of which (Arctiidae, Gelechiidae, and Saturniidae) also have species with W chromosomes. These ZW sex chromosome systems are usually found with matching reductions in the number of autosomes, indicating that fusions between autosomes and the ancestral Z chromosome are the source of new W chromosomes or even multiple ZW chromosome systems. The genus Samia within Saturniidae offers a particularly striking example with species that have 13, 12, and 11 autosomes having ZO, ZW, and Z1Z2W sex chromosome systems (Yoshido et al. 2005). Some of these transitions may have also coincided with a transition of the sex determining mechanism, from an ancestral Z counting state to a dominant feminizing allele on the W, as is found in B. mori (Fujii and Shimada 2007). Chromosome number in Lepidoptera is highly variable with diploid numbers ranging from 14 to 382. Parthenogenesis has been identified in 16 taxa, 10 of which belong to the family Psychidae.
Diptera
The 125000 plus species of Diptera have been subject to intense research and cytogenetic data for over 2123 taxa are available (White 1949). The majority of species (1866) have XY sex chromosomes, but the family Tephritidae has evolved ZW sex chromosome systems and several lineages lack heteromorphic sex chromosomes all together. (White 1949) proposed a classification of Diptera based on cytological grounds. Lower Diptera (Nematocera) were divided into 4 groups. The most primitive Diptera (superfamily Tipuloidea) are characterized by the presence of chiasmata in both sexes, and cytologically distinguishable XY sex chromosomes. A second assemblage of families (including Culicidae, Chironomidae, and Simuliidae) generally lack cytologically distinguishable XY sex chromosomes, but have retained chiasmata in males. The third group, which includes Bibionidae and Thaumaleidae, is characterized by heteromorphic XY sex chromosomes but a lack of chiasmata in males. A fourth cytological group of Nematocera is characterized by a highly specialized chromosome cycle, the loss of the Y chromosome, and sex is determined by elimination of the paternal X chromosome, and includes the families Sciaridae and Cecidomyidae. Most families of Diptera, including Drosophila, fall within the suborder Brachycera (higher Diptera). All Brachycera appear to lack chiasmata in males, and most species have heteromorphic XY chromosomes. At least 9 families have evolved parthenogenetic species, with the greatest concentration of parthenogens in the family Chironomidae (where 25 of the 52 studied taxa are parthenogenetic). In contrast to the lability of their sex determination systems, chromosome number varies less in this group. A number of families have genomes with just 3 autosomes, while Tabanidae has the highest with 12 autosomes. Flies such as D. melanogaster and A. gambiae are important model organisms for studies in genetics and development, and the sex determination pathway in Drosophila has been worked out, with the X-autosome ratio determining gender, while other species, such as Lucilia cuprina and Ceratitis capitata harbor Y-dominant sex determination (Willhoeft and Franz 1996). A recent study has demonstrated that while many Diptera species have male heterogamety, the chromosome that is sex-linked can differ among families. In particular, the sex chromosome of Drosophila was found to be autosomal in several outgroup species from different families, which shared a sex chromosome that is autosomal in Drosophila (Vicoso and Bachtrog 2013). Furthermore, genome sequence analysis of 37 species of Diptera showed that species investigated exhibited 12 distinct sex chromosome configurations despite relatively homogenous karyotypes (Vicoso and Bachtrog 2015).
Mecoptera
Scorpion or hang flies, with approximately 550 extant species, are the sister group of Diptera, but have not been widely studied cytogenetically. 13 of the 14 taxa in which sex chromosomes have been identified exhibited XO sex determination, and one species, Boreus brumalis, has a X1X2Y sex chromosome complement (Xu et al. 2013). Achiasmatic male meiosis has evolved independently in different species.
Siphonaptera
Fleas have an estimated 2500 species, and recent molecular evidence indicates that fleas are actually highly derived Mecopterans most closely related to the family Borediae (Whiting 2002). Despite their abundance and medical importance, cytogenetic data is available for only 7 taxa (Thomas 1990). The sex chromosome system has been identified in 6 taxa, 2 of which have XY sex chromosomes and 4 have multiple sex chromosomes, a characteristic that would lend support to their close association with Borediae. The number of autosomes in this group ranges from 3 in Xenopsylla prasadi to 10 in Leptopsylla musculi.
Conclusions
Our synthesis of karyotype data provides a first step in understanding the evolution of sex determination systems and genome organization in one of the most abundant and economically important groups of organisms on the planet. The taxonomic breadth of our analysis allows us to make broader and more general inferences than earlier syntheses of insect sex determination (e.g., Cook 2002; Kaiser and Bachtrog 2010; Verhulst et al. 2010). In general, we find strong support for male heterogamety as the ancestral state for insects, while female heterogamety has evolved only twice. Our data indicate that sex-limited chromosomes are lost and gained more readily in some clades than others, and that the importance of fusions and fissions varies among orders. Our database reinforces that some groups (i.e., Dermaptera, Plecoptera, Mantodea, and Isoptera) exhibit a propensity for complex sex chromosomes not seen in closely related clades. We find that the phylogenetic distribution of haplodiploidy and related systems is shaped by infrequent origins and even fewer reversions back to diploidy. Finally our data show a correlation between the loss of flight and the frequency of parthenogenesis, suggesting that ineffective mate search ability might have contributed to drive transitions to asexually.
Our data set also highlights a number of unanswered questions: Why does variance in chromosome number differ so greatly between orders? Why is hermaphroditism largely absent in insects? Do some sex determination systems allow for more frequent transitions to asexuality? Do some sex determination systems lead to higher diversification? The data described in this manuscript provide an ideal starting point to answer these and related questions. We have made this data publicly available through the Tree of Sex project (www.treeofsex.org).
Supplementary Materials
Supplementary material can be found at http://www.jhered.oxfordjournals.org/.
Funding
This work was supported by NESCent as part of the “Tree of Sex” working group.
Supplementary Material
Acknowledgments
We thank Benjamin Normark for sharing his data set on the occurrence of parthenogenesis. We thank Leo Beukeboom for comments on an earlier version of this manuscript.
References
- Bach C, Petitpierre E. 1978. Notas preliminares sobre cromosomas de. Misc Zool. 4:43–46. [Google Scholar]
- Bachtrog D. 2003. Adaptation shapes patterns of genome evolution in sexual and asexual genomes in Drosophila. Nat Genet. 34:215–219. [DOI] [PubMed] [Google Scholar]
- Bachtrog D. 2013. Y-chromosome evolution: emerging insights into processes of Y-chromosome degeneration. Nat Rev Genet. 14:113–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog D, Mank JE, Peichel CL, Kirkpatrick M, Otto SP, Ashman TL. 2014. Sex determination: why so many ways of doing it? PLoS Biol. 12:e1001899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barsiene J, Ribi G, Barsyte D. 2000. Comparative karyological analysis of five species of Viviparus (Gastropoda: Prosobranchia). J Mollus Stud. 66:259–271. [Google Scholar]
- Bergamaschi S, Dawes-Gromadzki TZ, Scali V, Marini M, Mantovani B. 2007. Karyology, mitochondrial DNA and the phylogeny of Australian termites. Chromosome Res. 15:735–753. [DOI] [PubMed] [Google Scholar]
- Beukeboom LW, Perrin N. 2014. The evolution of sex determination. Oxford (UK): Oxford University Press. [Google Scholar]
- Beye M, Hasselmann M, Fondrk MK, Page RE, Omholt SW. 2003. The gene csd is the primary signal for sexual development in the honeybee and encodes an SR-type protein. Cell. 114:419–429. [DOI] [PubMed] [Google Scholar]
- Blackmon H, Demuth JP. 2014. Estimating tempo and mode of Y chromosome turnover: explaining Y chromosome loss with the fragile Y hypothesis. Genetics. 197:561–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackmon H, Demuth JP. 2015. a. Coleoptera Karyotype Database. Coleopts Bull. 69:174–175. [Google Scholar]
- Blackmon H, Demuth JP. 2015. b. The fragile Y hypothesis: Y chromosome aneuploidy as a selective pressure in sex chromosome and meiotic mechanism evolution. Bioessays. 37:942–950. [DOI] [PubMed] [Google Scholar]
- Blackmon H, Hardy NB, Ross L. 2015. The evolutionary dynamics of haplodiploidy: genome architecture and haploid viability. Evolution. 69:2971–2978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco DR, Vicari MR, Lui RL, Bertollo LAC, Traldi JB, Moreira-Filho O. 2013. The role of the Robertsonian rearrangements in the origin of the XX/XY1Y2 sex chromosome system and in the chromosomal differentiation in Harttia species (Siluriformes, Loricariidae). Rev Fish Biol Fisher. 23:127–134. [Google Scholar]
- Bopp D, Saccone G, Beye M. 2013. Sex determination in insects: variations on a common theme. Sex Dev. 8:20–28. [DOI] [PubMed] [Google Scholar]
- Brito RO, Affonso PRAM, Silva JC., Jr 2010. Chromosomal diversity and phylogenetic inferences concerning thrips (Insecta, Thysanoptera) in a semi-arid region of Brazil. Genet Mol Res. 9:2230–2238. [DOI] [PubMed] [Google Scholar]
- Brown SW. 1964. Automatic frequency response in evolution of male haploidy + other coccid chromosome systems. Genetics. 49:797–817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brun LO, Stuart J, Gaudichon V, Aronstein K, French-Constant RH. 1995. Functional haplodiploidy: a mechanism for the spread of insecticide resistance in an important international insect pest. Proc Natl Acad Sci USA. 92:9861–9865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull JJ. 1979. An advantage for the evolution of male haploidy and systems with similar genetic transmission. Heredity. 43:361–381. [Google Scholar]
- Bull JJ. 1983. The evolution of sex determining mechanisms. Menlo Park (CA): Benjamin/Cummings Publishing Company. [Google Scholar]
- Carvalho AB, Clark AG. 2005. Y chromosome of D. pseudoobscura is not homologous to the ancestral Drosophila Y. Science. 307:108–110. [DOI] [PubMed] [Google Scholar]
- Carvalho AB, Koerich LB, Clark AG. 2009. Origin and evolution of Y chromosomes: Drosophila tales. Trends Genet. 25:270–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo ER, Marti DA, Bidau CJ. 2010. Sex and neo-sex chromosomes in Orthoptera: a review. J Orthoptera Res. 19:213–231. [Google Scholar]
- Charlesworth D, Charlesworth B. 1980. Sex differences in fitness and selection for centric fusions between sex-chromosomes and autosomes. Genet Res. 35:205–214. [DOI] [PubMed] [Google Scholar]
- Choe JC, Crespi BJ. 1997. The evolution of social behaviour in insects and arachnids. Cambridge: Cambridge University Press. [Google Scholar]
- Cicconardi F, Fanciulli PP, Emerson BC. 2013. Collembola, the biological species concept and the underestimation of global species richness. Mol Ecol. 22:5382–5396. [DOI] [PubMed] [Google Scholar]
- Cline TW, Dorsett M, Sun S, Harrison MM, Dines J, Sefton L, Megna L. 2010. Evolution of the Drosophila feminizing switch gene Sex-lethal. Genetics. 186:1321–1336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook JM. 1993. Sex determination in the Hymenoptera: a review of models and evidence. Heredity. 71:421–435. [Google Scholar]
- Cook JM. 2002. Sex determination in invertebrates. In: Hardy ICW, editor. Sex ratios: concepts and research methods. Cambridge (UK): Cambridge University Press. p. 178–194. [Google Scholar]
- Corbet PS. 1966. Parthenogenesis in caddisflies (Trichoptera). Can J Zool. 44:981–982. [Google Scholar]
- Cordaux R, Bouchon D, Grève P. 2011. The impact of endosymbionts on the evolution of host sex-determination mechanisms. Trends Genet. 27:332–341. [DOI] [PubMed] [Google Scholar]
- d’Alencon E, Sezutsu H, Legeai F, Permal E, Bernard-Samain S, Gimenez S, Gagneur C, Cousserans F, Shimomura M, Brun-Barale A. 2010. Extensive synteny conservation of holocentric chromosomes in Lepidoptera despite high rates of local genome rearrangements. Proc Natl Acad Sci USA. 107:7680–7685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dallai R, Fanciulli PP, Frati F. 1999. Chromosome elimination and sex determination in springtails (Insecta, Collembola). J Exp Zool. 285:215–225. [DOI] [PubMed] [Google Scholar]
- Dallai R, Fanciulli PP, Frati F. 2000. Aberrant spermatogenesis and the peculiar mechanism of sex determination in Symphypleonan Collembola (Insecta). J Hered. 91:351–358. [DOI] [PubMed] [Google Scholar]
- de la Filia AG, Bain SA, Ross L. 2015. Haplodiploidy and the reproductive ecology of Arthropods. Curr Opin Insect Sci. 9:36–43. [DOI] [PubMed] [Google Scholar]
- Eppley S, Jesson L. 2008. Moving to mate: the evolution of separate and combined sexes in multicellular organisms. J Evol Biol. 21:727–736. [DOI] [PubMed] [Google Scholar]
- Erickson JW, Quintero JJ. 2007. Indirect effects of ploidy suggest X chromosome dose, not the X:A ratio, signals sex in Drosophila. PLoS Biol. 5:e332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira A, Cella D, Mesa A, Virkki N. 1984. Cytology and systematical position of Stylopids (= Strepsiptera). Hereditas. 100:51–52. [Google Scholar]
- Fratello B, Sabatini MA. 1989. Chromosome studies in Protura Eosentomoidea. 3rd International Seminar on Apterigota, Siena, Italy. [Google Scholar]
- Frías D. 1992. Aspectos de la biología evolutiva de especies de Tephritidae (Diptera) de distribución chilena. Acta Entomol Chil. 17:69–79. [Google Scholar]
- Fujii T, Shimada T. 2007. Sex determination in the silkworm, Bombyx mori: a female determinant on the W chromosome and the sex-determining gene cascade. Semin Cell Dev Biol. 18:379–388. [DOI] [PubMed] [Google Scholar]
- Gardner A, Ross L. 2011. The evolution of hermaphroditism by an infectious male-derived cell lineage: an inclusive-fitness analysis. Am Nat. 178:191–201. [DOI] [PubMed] [Google Scholar]
- Gardner A, Ross L. 2014. Mating ecology explains patterns of genome elimination. Ecol Lett. 17:1602–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrilov I. 2007. A catalog of chromosome numbers and genetic systems of scale insects (Homoptera: Coccinea) of the world. Isr J Entomol. 37:1–45. [Google Scholar]
- Gibbs EK. 1977. Evidence for obligatory parthenogenesis and its possible effect on the emergence period of Cloeon triangulifer (Ephemeroptera: Baetidae). Can Entomol. 109:337–340. [Google Scholar]
- Golub N, Nokkala S. 2004. Chromosome numbers of two sucking louse species (Insecta, Phthiraptera, Anoplura). Hereditas. 141:94–96. [DOI] [PubMed] [Google Scholar]
- Golub N, Nokkala S. 2009. Chromosome numbers in eight species of Palaearctic Psocoptera (Insecta). Comp Cytogenet. 3:33–41. [Google Scholar]
- Gullan PJ, Cranston PS. 2010. The insects: an outline of entomology. West Sussex (UK): Wiley-Blackwell. [Google Scholar]
- Hall AB, Basu S, Jiang X, Qi Y, Timoshevskiy VA, Biedler JK, Sharakhova MV, Elahi R, Anderson MA, Chen X-G. 2015. A male-determining factor in the mosquito Aedes aegypti. Science. 348:1268–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heimpel G, de Boer J. 2007. Sex determination in the Hymenoptera. Annu Rev. 53:209–230. [DOI] [PubMed] [Google Scholar]
- Henderson SA. 1970. Sex chromosomal polymorphism in the earwig Forficula. Chromosoma. 31:139–164. [DOI] [PubMed] [Google Scholar]
- Herrick G, Seger J. 1999. Imprinting and paternal genome elimination in insects. In: Ohlsson R, editor. Genomic imprinting: an interdisciplinary approach. New York: Springer-Verlag; p. 41–71. [DOI] [PubMed] [Google Scholar]
- Hughes-Schrader S. 1969. Distance segregation and compound sex chromosomes in mantispids (Neuroptera:Mantispidae). Chromosoma. 27:109–129. [DOI] [PubMed] [Google Scholar]
- Hughes-Schrader S. 1975. a. Male meiosis in camel-flies (Raphidioptera; Neuropteroidea). Chromosoma. 51:99–110. [DOI] [PubMed] [Google Scholar]
- Hughes-Schrader S. 1975. b. Segregational mechanisms of sex chromosomes in Spongilla-flies (Neuroptera: Sisyridae). Chromosoma. 52:1–10. [DOI] [PubMed] [Google Scholar]
- Jarne P, Auld JR. 2006. Animals mix it up too: the distribution of self-fertilization among hermaphroditic animals. Evolution. 60:1816–1824. [DOI] [PubMed] [Google Scholar]
- Jordal BH, Normark BB, Farrell BD. 2000. Evolutionary radiation of an inbreeding haplodiploid beetle lineage (Curculionidae, Scolytinae). Biol J Linn Soc. 71:483–499. [Google Scholar]
- Kageyama D, Narita S, Watanabe M. 2012. Insect sex determination manipulated by their endosymbionts: incidences, mechanisms and implications. Insects. 3:161–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser VB, Bachtrog D. 2010. Evolution of sex chromosomes in insects. Annu Rev Genet. 44:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karamysheva T, Lehmann AW, Lehmann GU, Heller KG. 2011. Changes in the numbers of chromosomes and sex determination system in bushcrickets of the genus Odontura (Orthoptera: Tettigoniidae: Phaneropterinae). Eur J Entomol. 108:183–195. [Google Scholar]
- Kathirithamby J, Solulu T, Caudwell R. 2001. Descriptions of female Myrmecolacidae (Strepsiptera) parasitic in Orthoptera (Tettigoniidae) in Papua New Guinea. Tijdschrift voor Entomologie. 144:187–196. [Google Scholar]
- Kiauta B, Mol AWM. 1977. Behaviour of the spermatocyte chromosomes of the Mayfly, Cloeon dipterum (Linnaeus, 1761) s.l. (Ephemeroptera:Baetidae), with a note on the cytology of the order. Genen Phaenen. 19:31–39. [Google Scholar]
- Kitano J, Peichel CL. 2012. Turnover of sex chromosomes and speciation in fishes. Environ Biol Fishes 94:549–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiuchi T, Koga H, Kawamoto M, Shoji K, Sakai H, Arai Y, Ishihara G, Kawaoka S, Sugano S, Shimada T, et al. 2014. A single female-specific piRNA is the primary determiner of sex in the silkworm. Nature. 509:633–636. [DOI] [PubMed] [Google Scholar]
- Koevoets T, Beukeboom LW. 2008. Genetics of postzygotic isolation and Haldane’s rule in haplodiploids. Heredity. 102:16–23. [DOI] [PubMed] [Google Scholar]
- Kuznetsova VG, Nokkala S, Shcherbakov DE. 2002. Karyotype, reproductive organs, and pattern of gametogenesis in Zorotypus hubbardi Caudell (Insecta: Zoraptera, Zorotypidae), with discussion on relationships of the order. Can J Zool. 80:1047–1054. [Google Scholar]
- LaChance LE, Degrugillier M, Leverich AP. 1970. Cytogenetics of inherited partial sterility in three generations of the large milkweed bug as related to holokinetic chromosomes. Chromosoma. 29:20–41. [DOI] [PubMed] [Google Scholar]
- Legrand J, Legrand‐Hamelin E, Juchault P. 1987. Sex determination in Crustacea. Biol Rev. 62:439–470. [Google Scholar]
- Lehmann GU, Siozios S, Bourtzis K, Reinhold K, Lehmann AW. 2011. Thelytokous parthenogenesis and the heterogeneous decay of mating behaviours in a bushcricket (Orthopterida). J Zool Syst Evol Res. 49:102–109. [Google Scholar]
- Lohse K, Ross L. 2015. What haplodiploids can teach us about hybridization and speciation. Mol Ecol. 24:5075–5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukhtanov VA. 2000. Sex chromatin and sex chromosome systems in nonditrysian Lepidoptera (Insecta). J Zool Syst Evol Res. 38:73–79. [Google Scholar]
- Luykx P. 1990. A cytogenetic survey of 25 species of lower termites from Australia. Genome. 33:80–88. [Google Scholar]
- Makino S. 1951. An atlas of the chromosome numbers in animals. Ames (IA): Iowa State College Press. [Google Scholar]
- Marec F, Novak K. 1998. Absence of sex chromatin corresponds with a sex-chromosome univalent in females of Trichoptera. Eur J Entomol. 95:197–210. [Google Scholar]
- Marec F, Sahara K, Traut W. 2010. Rise and fall of the W chromosome in Lepidoptera. Mol Biol Genet Lepidoptera. 3:49–63. [Google Scholar]
- Martin J, Lee BT. 1984. Are there female heterogametic strains of Chironomus tentans Fabricius? Can J Genet Cytol. 26:743–747. [DOI] [PubMed] [Google Scholar]
- Matthey R, Aubert J. 1947. Les chromosomes des plecopteres. Bull Biol Fr Bel. 81:202–246. [Google Scholar]
- Maynard-Smith J. 1978. The evolution of sex. Cambridge: Cambridge University Press. [Google Scholar]
- Meise M, Hilfiker-Kleiner D, Dübendorfer A, Brunner C, Nöthiger R, Bopp D. 1998. Sex-lethal, the master sex-determining gene in Drosophila, is not sex-specifically regulated in Musca domestica. Development. 125:1487–1494. [DOI] [PubMed] [Google Scholar]
- Melters DP, Paliulis LV, Korf IF, Chan SWL. 2012. Holocentric chromosomes: convergent evolution, meiotic adaptations, and genomic analysis. Chromosome Res. 20:579–593. [DOI] [PubMed] [Google Scholar]
- Misof B, Liu S, Meusemann K, Peters RS, Donath A, Mayer C, Frandsen PB, Ware J, Flouri T, Beutel RG, et al. 2014. Phylogenomics resolves the timing and pattern of insect evolution. Science. 346:763–767. [DOI] [PubMed] [Google Scholar]
- Molero-Baltanás R, Gaju-Ricart M, de Roca CB. 1998. Description of Atelura valenciana n. sp. (Insecta, Zygentoma) and distribution and myrmecophilic relationships of Proatelurina pseudolepisma in the Iberian peninsula. Misc Zool. 21:101–117. [Google Scholar]
- Mora C, Tittensor DP, Adl S, Simpson AG, Worm B. 2011. How many species are there on Earth and in the ocean? PLoS Biol. 9:e1001127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigro RG, Campos MCC, Perondini AP. 2007. Temperature and the progeny sex-ratio in Sciara ocellaris (Diptera, Sciaridae). Genet Mol Biol. 30:152–158. [Google Scholar]
- Normark BB. 2003. The evolution of alternative genetic systems in insects. Annu Rev Entomol. 48:397–423. [DOI] [PubMed] [Google Scholar]
- Normark BB. 2013. Micromalthus debilis Curr Biol. 23:R430–R431. [DOI] [PubMed] [Google Scholar]
- Normark BB. 2014. Modes of reproduction. In Shuker D, Simmons L, editors. The evolution of insect mating systems. Oxford (UK): University Press Oxford. [Google Scholar]
- Normark BB, Ross L. 2014. Genetic conflict, kin and the origins of novel genetic systems. Philos Trans R Soc B Biol Sci. 369:20130364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patten MM, Carioscia SA, Linnen CR. 2015. Biased introgression of mitochondrial and nuclear genes: a comparison of diploid and haplodiploid systems. Mol Ecol. 24:5200–5210. [DOI] [PubMed] [Google Scholar]
- Pennell MW, Kirkpatrick M, Otto SP, Vamosi JC, Peichel CL, Valenzuela N, Kitano J. 2015. Y fuse? Sex chromosome fusions in fishes and reptiles. PLoS Genet. 11:e1005237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revell LJ. 2012. phytools: an R package for phylogenetic comparative biology (and other things). Methods Ecol Evol. 3:217–223. [Google Scholar]
- Rivera AC, Carballa ML, Utzeri C, Vieira V. 2005. Parthenogenetic Ischnura hastata (Say), widespread in the Azores (Zygoptera: Coenagrionidae). Odonatologica. 34:11–26. [Google Scholar]
- Ross L, Blackmon H, Lorite P, Gokhman V, Hardy N. 2015. Recombination, chromosome number and eusociality in the Hymenoptera. J Evol Biol. 28:105–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross L, Hardy NB, Okusu A, Normark BB. 2012. Large population size predicts the distribution of asexuality in scale insects. Evolution. 67:196–206. [DOI] [PubMed] [Google Scholar]
- Ross L, Pen I, Shuker DM. 2010. Genomic conflict in scale insects: the causes and consequences of bizarre genetic systems. Biol Rev. 85:807–828. [DOI] [PubMed] [Google Scholar]
- Sahara K, Yoshido A, Traut W. 2012. Sex chromosome evolution in moths and butterflies. Chromosome Res. 20:83–94. [DOI] [PubMed] [Google Scholar]
- Sánchez L. 2008. Sex-determining mechanisms in insects. Int J Dev Biol. 52:837–856. [DOI] [PubMed] [Google Scholar]
- Smith SG, Virkki N. 1978. Coleoptera. Animal cytogenetics Vol. 3: Insecta. Berlin-Stuttgart: Gebruder Borntraeger. [Google Scholar]
- Stuart JJ, Hatchett JH. 1991. Genetics of sex determination in the Hessian fly, Mayetiola destructor. J Hered. 81:43–52. [Google Scholar]
- Takeuchi Y, Iizuka K, Yamada T. 2002. Chromosomes of the Japanese dobsonfly Protohermes grandis (Megaloptera: Corydalidae). Chromosome Sci. 62:49–51. [Google Scholar]
- Thiriot-Quievreux C. 2003. Advances in chromosomal studies of gastropod molluscs. J Molluscan Stud. 69:187–202. [Google Scholar]
- Thomas C. 1990. Cytogenetics of fleas (Siphonaptera: Pulicidae). 1. Rat fleas of the genus Xenopsylla. Cytobios. 67:29–43. [PubMed] [Google Scholar]
- Thomas DB. 1987. Chromosome evolution in the Heteroptera (Hemiptera): agmatoploidy versus aneuploidy. Ann Entomol Soc Am. 80:720–730. [Google Scholar]
- Thompson PE. 1971. Male and female heterogamety in populations of Chironomus tentans (Diptera: Chironomidae). Can Entomol. 103:369–372. [Google Scholar]
- Tombesi ML, Papeschi AG. 1993. Meiosis in Haematopinus suis and Menacanthus stramineus (Phthiraptera, Insecta). Hereditas. 119:31–38. [Google Scholar]
- Traut W, Sahara K, Marec F. 2008. Sex chromosomes and sex determination in Lepidoptera. Sex Dev. 1:332–346. [DOI] [PubMed] [Google Scholar]
- Tree of Sex Consortium 2014. Tree of Sex: a database of sexual systems. Sci Data. 1:140015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsurusaki N, Cokendolpher JC. 1990. Chromosomes of sixteen species of harvestmen (Arachnida, Opiliones, Caddidae and Phalangiidae). J Arachnol. 18:151–166. [Google Scholar]
- Ueshima N. 1979. Animal cytogenetics: Hemiptera II: Heteroptera. Berlin: Gebr. Borntraeger. [Google Scholar]
- Ullerich F-H. 1963. Geschlechtschromosomen und geschlechtsbestimmung bei einigen Calliphorinen (Calliphoridae, Diptera). Chromosoma. 14:45–110. [Google Scholar]
- Verhulst EC, Beukeboom LW, van de Zande L. 2010. Maternal control of haplodiploid sex determination in the wasp Nasonia. Science. 328:620–623. [DOI] [PubMed] [Google Scholar]
- Vicoso B, Bachtrog D. 2013. Reversal of an ancient sex chromosome to an autosome in Drosophila. Nature. 499:332–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicoso B, Bachtrog D. 2015. Numerous transitions of sex chromosomes in Diptera. PLoS Biol. 13:e1002078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicoso B, Emerson JJ, Zektser Y, Mahajan S, Bachtrog D. 2013. Comparative sex chromosome genomics in snakes: differentiation, evolutionary strata, and lack of global dosage compensation. PLoS Biol. 11:e1001643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner DL, Liebherr JK. 1992. Flightlessness in insects. Trends Ecol Evol. 7:216–220. [DOI] [PubMed] [Google Scholar]
- Werren JH, Windsor DM. 2000. Wolbachia infection frequencies in insects: evidence of a global equilibrium? Proc R Soc Lond B Biol Sci. 267:1277–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White MJ. 1949. Cytological evidence on the phylogeny and classification of the Diptera. Evolution. 3:252–261. [DOI] [PubMed] [Google Scholar]
- White MJD. 1976. Blattodea, Mantodea, Isoptera, Grylloblattodea, Phasmatodea, Dermaptera and Embioptera. Berlin-Stuttgart: Gebrüder Borntraeger. [Google Scholar]
- White MJD. 1977. Animal cytology & evolution. Cambridge: Cambridge University Press. [Google Scholar]
- Whiting MF. 2002. Mecoptera is paraphyletic: multiple genes and phylogeny of Mecoptera and Siphonaptera. Zool Scripta. 31:93–104. [Google Scholar]
- Willhoeft U, Franz G. 1996. Identification of the sex-determining region of the Ceratitis capitata Y chromosome by deletion mapping. Genetics. 144:737–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson ACC, Sunnucks P, Hales DF. 1997. Random loss of X chromosome at male determination in an aphid, Sitobion near fragariae, detected using an X-linked polymorphic microsatellite marker. Genet Res. 69:233–236. [Google Scholar]
- Xu B, Li Y, Hua B. 2013. A chromosomal investigation of four species of Chinese Panorpidae (Insecta, Mecoptera). Comp Cytogenet. 7:229–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshido A, Marec F, Sahara K. 2005. Resolution of sex chromosome constitution by genomic in situ hybridization and fluorescence in situ hybridization with (TTAGG)(n) telomeric probe in some species of Lepidoptera. Chromosoma. 114:193–202. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Bachtrog D. 2012. Sex-specific adaptation drives early sex chromosome evolution in Drosophila. Science. 337:341–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Ellison CE, Kaiser VB, Alekseyenko AA, Gorchakov AA, Bachtrog D. 2013. The epigenome of evolving Drosophila neo-sex chromosomes: dosage compensation and heterochromatin formation. PLoS Biol. 11:e1001711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Zhu H-m, Huang Q-f, Zhao L, Zhang G-j, Roy SW, Vicoso B, Xuan Z-l, Ruan J, Zhang Y, et al. 2012. Deciphering neo-sex and B chromosome evolution by the draft genome of Drosophila albomicans. BMC Genomics 13:109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.