Abstract
Despite their economic, ecological, and experimental importance, genomic resources remain scarce for crustaceans. In lieu of genomes, many researchers have taken advantage of technological advancements to instead sequence and assemble crustacean transcriptomes de novo. However, there is little consensus on what standard operating procedures are, or should be, for the field. Here, we systematically reviewed 53 studies published during 2014–2015 that utilized transcriptomic resources from this taxonomic group in an effort to identify commonalities as well as potential weaknesses that have applicability beyond just crustaceans. In general, these studies utilized RNA-Seq data, both novel and publicly available, to characterize transcriptomes and/or identify differentially expressed genes (DEGs) between treatments. Although the software suite Trinity was popular in assembly pipelines and other programs were also commonly employed, many studies failed to report crucial details regarding bioinformatic methodologies, including read mappers and the utilized parameters in identifying and characterizing DEGs. Annotation percentages for assembled transcriptomic contigs were low, averaging 32% overall. While other metrics, such as numbers of contigs and DEGs reported, correlated with the number of sequence reads utilized per sample, these did reach apparent saturation with increasing sequencing depth. Most disturbingly, a number of studies (55%) reported DEGs based on non-replicated experimental designs and single biological replicates for each treatment. Given this, we suggest future RNA-Seq experiments targeting transcriptome characterization conduct deeper (i.e., 50–100 M reads) sequencing while those examining differential expression instead focus more on increased biological replicates at shallower (i.e., ∼10–20 M reads/sample) sequencing depths. Moreover, the community must avoid submitting for review, or accepting for publication, non-replicated differential expression studies. Finally, mining the ever growing publicly available transcriptomic data from crustaceans will allow future studies to focus on hypothesis-driven research instead of continuing to simply characterize transcriptomes. As an example of this, we utilized neurotoxin sequences from the recently described remipede venom gland transcriptome in conjunction with publicly available crustacean transcriptomic data to derive preliminary results and hypotheses regarding the evolution of venom in crustaceans.
Introduction
Crustaceans are one of the most taxonomically, ecologically, and physiologically diverse groups of animals on our planet, with >70,000 described species occupying nearly all marine and freshwater habitats (Hobbs and Hart 1982), as well as semi- and fully-terrestrial habitats (Bliss 1968), and even pools of water found in bromeliads (Anger 1995). They are also economically important, totaling 30% of the total harvest of US commercial fisheries, representing a value of $4 billion, in 2007 (Cooley and Doney 2009). Ecologically, crustaceans have invaded most major niches, ranging from the basis of marine food webs to apex predators of, or parasitic and mutualistic partners to, a wide variety of other organisms, and venom has evolved independently in at least one crustacean (von Reumont et al. 2014). Given this biological breadth, it should not be surprising that crustaceans are also physiologically diverse, having been utilized in studies elucidating effects of changing salinities, temperatures, acidification, oxygen concentrations, toxic metals, and emersion, just to name a few (Henry 1994; Morris 2002; Henry et al. 2012; McNamara and Faria 2012; Lewis et al. 2013; Harms et al. 2014; Spicer 2014).
Despite their importance, crustaceans still lack appreciable genomic resources compared with other widely-studied groups such as insects and vertebrates. As identified during the “Pancrustacean” symposium at the 2015 Annual Meeting of the Society for Integrative and Comparative Biology (SICB), there is a need for developing a model crustacean system, similar to Drosophila in insects, with one of its primary characteristics being a well-annotated genome (Mykles and Hui 2015). However, only one comprehensively assembled and annotated crustacean genome is available, that of the water flea Daphnia (Colbourne et al. 2011). While useful, the Daphnia genome does have shortcomings. Mainly, the genus is not closely related to any economically, ecologically, or physiologically important decapod crustacean species examined to date. While draft genomes are available for a few species, such as the cherry shrimp Neocaridina denticulata (Kenny et al. 2014; Sin et al. 2015), crustacean genomes are diverse, and can be large and structurally complex (Rees et al. 2007; Stillman et al. 2008), making them difficult to assemble. Furthermore, annotation of the Daphnia genome revealed that 30% of its genes lacked homologs in other animal genomes (Colbourne et al. 2011), suggesting that crustacean genomics at the functional level may not be practical for the foreseeable future.
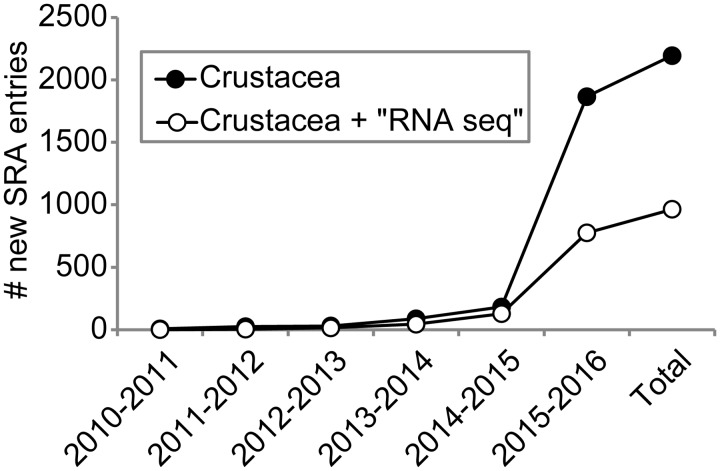
On the other hand, transcriptomics provides a viable alternative to characterizing complete genomes. For most transcriptomic approaches, the transcribed RNA population in a cell, tissue, whole organism, or pool of organisms is first sequenced using massively-parallel strategies and then computationally assembled into a series of overlapping but discrete DNA sequences (i.e., contigs). This process, termed RNA-Seq, is robust to experimental, bioinformatic, and genomic heterogeneity (Wang et al. 2009; Vijay et al. 2013). Furthermore, RNA-Seq is a powerful approach for: (1) the characterization of a transcriptome and novel transcript discovery from organisms with no available genomic resources; (2) analyses of differential gene expression that are superior to microarrays; (3) identification of alternatively spliced genes (Wang et al. 2009; Nookaew et al. 2012; Sims et al. 2014); and (4) the generation of hundreds to thousands of orthologous genetic markers for population genomic and phylogenomic studies (Meusemann et al. 2010; Regier et al. 2010; Rehm et al. 2011; Carmichael et al. 2013). With the advent of cost-affordable technologies like Illumina’s Sequencing by Synthesis chemistry, it is not surprising that transcriptomic resources are becoming widely available for a range of crustacean species, in spite of a lack of corresponding genomes. As an example, a search utilizing the term “crustacean” to the National Center for Biotechnology Information (NCBI)’s Sequence Read Archive (SRA), where many RNA-Seq datasets are deposited upon publication, reveals an exponential increase in the number of entries during the last ∼5 years (Fig. 1).
Fig. 1.
Number of entries added each year to the NCBI’s SRA database for crustaceans in general and the subset of those having a “RNA-seq” descriptor in particular (as of December 31, 2015).
Testimony to the rise of transcriptomics and RNA-Seq as formidable tools in crustacean comparative biology comes from the symposium “Tapping the Power of Crustacean Transcriptomes to Address Grand Challenges in Comparative Biology”, held in conjunction with the 2016 Annual Meeting of the SICB. However, it is unclear whether current RNA-Seq experiments of crustaceans conform to standard operating protocols (SOPs) used in model systems or even what should be considered SOPs for crustacean transcriptomic studies. We therefore surveyed 53 published studies from 2014–2015 to provide a qualitative and quantitative systematic review on the current state of crustacean transcriptomics as well as offer suggestions on ways to improve such studies in the future that we feel have applicability beyond just this taxonomic group. Our objectives were to: (1) identify the goals of current studies utilizing transcriptomic data from crustaceans, (2) describe broad trends on the methodologies being applied in analyses of crustacean transcriptomic data, and (3) suggest future directions for the field as a whole based on transcriptomic studies from more well-developed systems. Notably, we did not attempt a comprehensive review of methodologies associated with RNA-Seq in general since such reviews are published on at least a yearly basis (e.g., Fonseca et al. 2014; Hunt et al. 2014; Saliba et al. 2014; Seyednasrollah et al. 2015; Todd et al. 2016).
Methods
Data acquisition and selection of publications
Literature queries were performed against the NCBI PubMed and Thomson Reuters Web of Science databases to identify published studies meeting the following criteria: (1) describing some attribute relevant to transcriptomics (e.g., number of contigs, transcript expression levels, or identification of novel transcripts) without restriction to the level of biological organization (i.e., tissue(s), whole organism, etc.) being examined; (2) the targeted taxa must have been a crustacean; and (3) the work was published between January 1, 2014 and September 10, 2015. Notably, these criteria were selected to provide a snapshot of the methods and results of relatively recent crustacean transcriptomic studies rather than a comprehensive review of the field since its inception, as earlier work may have used drastically different methodologies due to continuing and rapid advancements in DNA sequencing technology and software for analyses. For example, many studies prior to 2014 relied on Roche 454 pyrosequencing. However, the relatively high-cost per sequenced base as well as the 2013 announcement of its close down by mid-2016 has largely led many projects to adopt and employ Illumina sequencing instead. Also of note, we did not query sequence databases such as the SRA of NCBI since we sought to examine the goals and methodologies of published transcriptomic studies, not survey projects simply generating sequence reads. Initial search terms used were combinations of the following: “Crustacea”, “transcriptome”, “RNA-Seq”, “crab”, “shrimp”, and “genome”. References of the literature identified in initial queries were also searched in order to locate other potentially relevant work (i.e., “in press” manuscripts available from publishers but not yet in either of the two databases).
Quantitative review and re-analyses of collective datasets
Along with qualitatively reviewing the studies identified from the literature queries and noting the goals of, and methodologies utilized, for each, several quantitative metrics were also summarized (Table 1; Supplementary Table S1). These included: (1) number of reads sequenced per transcriptomic sample; (2) number of sequenced samples; (3) number of contigs assembled per transcriptome; (4) percentage contigs annotated; and (5) number of differentially expressed genes (DEGs) recovered, as well as level of biological sample replication per treatment, especially when studies explicitly stated that DEGs were being identified and characterized between tissues, developmental stages, or environmental treatments. Unfortunately, given their nature, patterns and correlations drawn from the data were not amenable to examination in a formal meta-analysis statistical framework (ArchMiller et al. 2015). Specifically, much of the data we gathered lack the appropriate metrics for meta-analyses (Borenstein 2009) since they represent single counts per study without any measure of statistical deviation (e.g., number of reads sequenced). However, we feel generalized conclusions drawn from this systematic review have utility in designing future crustacean RNA-Seq experiments, even if P values were not calculated.
Table 1.
Average ± SEM for common quantitative metrics reported from the 53 crustacean transcriptomic studies reviewed here
| Metric | Average ±SEM |
|---|---|
| No. of Reads per sample | 63.0 ± 11.5 million |
| No. of Samples | 6.7 ± 1.2 |
| No. of Samples per treatment | 1.7 ± 0.2 |
| No. of Treatments | 3.8 ± 0.4 |
| % Contigs annotated | 31.8 ± 2.4 |
| No. of Contigs | 103,739 ± 13,782 |
| No. of DEGs | 4154 ± 1058 |
Note: DEGs = differentially expressed genes.
Results and Discussion
The current state of crustacean transcriptomics
Based on the search criteria, 53 studies, examining the transcriptomics of 37 crustacean species, were identified for inclusion in this review (Supplementary Table S1). In general, these studies either: (1) sequenced and characterized the transcriptome of a particular tissue or species; (2) utilized RNA-Seq to examine differential gene expression between particular tissues, treatments, or developmental stages; (3) screened transcriptomic contigs (either publicly available or ones assembled de novo) to identify novel crustacean-specific transcripts; or (4) some combination of the above. While this provides a general summary of goals associated with RNA-Seq experiments in crustaceans (additional details for each study are given in Supplementary Table S1), it likely does not represent a comprehensive list. Notably, no studies encompassing phylogenomics are included in this review, possibly due to the search criteria or terms utilized, despite the utilization of transcriptomic data in such studies (Rehm et al. 2011). Therefore, our conclusions on read depth, assembly programs, and other commonalities in crustacean transcriptomic studies may not generally apply to phylogenomic studies for the group.
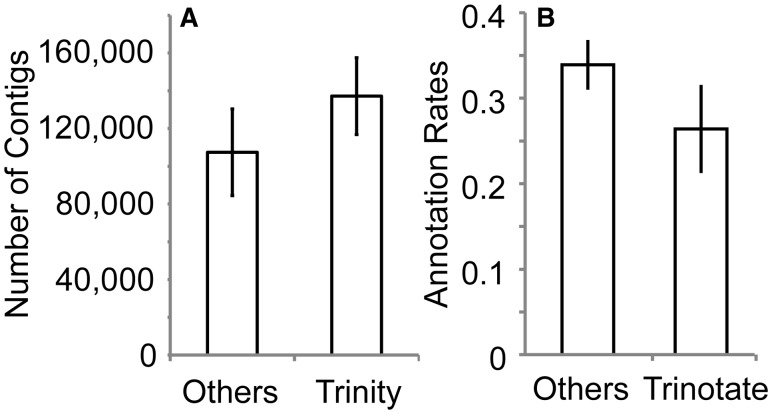
Among the 53 transcriptomic studies, the most frequently examined species was the red swamp crayfish Procambarus clarkii (used in six publications), with several others, such as the Chinese mitten crab Eriocheir sinensis and the whiteleg shrimp Litopenaeus vannamei, also represented in multiple studies. The majority employed Illumina’s Sequencing by Synthesis chemistry in generating RNA-Seq data (82% of applicable studies), with Roche 454 pyrosequencing (16%), microarrays (4%), and/or expressed sequence tags libraries (2%) also being utilized less commonly. The vast majority of studies (90%) employed some form of quality filtering of reads prior to contig assembly, with programs such as Trimmomatic (Bolger et al. 2014), SeqPrep (https://github.com/jstjohn/SeqPrep), FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/), and the FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/) being popular for such preprocessing steps. It should be noted that overzealous trimming and filtering of sequence reads can negatively impact transcriptome assembly (MacManes 2014), and we seldom perform preprocessing of reads prior to contig assembly in our own projects if overall read quality is high (e.g., Havird and Santos 2016). Contig assembly from sequence reads was most commonly (64%) accomplished with the Trinity software suite (Grabherr et al. 2011), with CLC Genomics Workbench (14%; http://www.clcbio.com), Newbler (10%; Margulies et al. 2005), SOAP (5%; Luo et al. 2012), CAP3 (2%; Huang and Madan 1999), Trans-ABySS (2%; Simpson et al. 2009), or assemblies performed at sequencing centers (2%) employed to a lesser extent. Notably, Trinity tended to generate transcriptomes with higher numbers of contigs than studies using other assembly programs, even when Roche 454 pyrosequencing data (most often assembled via Newbler) were excluded (Fig. 2(A)). This likely stems from Trinity explicitly and efficiently assembling splice variants and alternative isoforms of genes, which will necessarily produce a higher number of contigs per gene as a result. To annotate resulting contigs, the use of the Basic Local Alignment Search Tool (BLAST; Altschul et al. 1997) suite was overwhelmingly popular (95%), typically coupled with queries to Gene Ontology (GO) subject databases such as NCBI’s nr, SwissProt (Apweiler et al. 2012), or Kyoto Encyclopedia of Genes and Genomes (KEGG; Kanehisa et al. 2016). Although the number of entries in a database could potentially influence annotation success rates, most studies queried multiple databases, thus preventing identification of direct correlations between the two. A majority of studies (60%) employed a BLAST Expect (E) value criterion of 1e−5, but values ranged from 1e−10 to 1e−3, when conducting annotation, which also likely influenced success rates. A minority of studies (18%) utilized Trinity’s companion annotation suite Trinotate (https://trinotate.github.io/), which simultaneously queries the UniProt, EggNOG, and GO Pathways databases (The Gene Ontology Consortium 2000; Apweiler et al. 2012; Powell et al. 2012) using BLAST and HMMER (Mistry et al. 2013). While there was a trend for studies using Trinotate to have lower annotation rates than alternative methods, this difference was relatively minor (Fig. 2(B)).
Fig. 2.
Impact of (A) assembly and (B) annotation software on number of contigs and percentage annotated, respectively, from the 53 crustacean transcriptomic studies reviewed here. Roche 454 data have been removed from (A). Error bars show ± SEM.
In the 22 studies reporting differential gene expression, Bowtie (Langmead 2010; Langmead and Salzberg 2012) was most frequently used for initial read mapping to contigs (25%), with CLC Genomics Workbench (15%) also being somewhat common. Surprisingly, the specific read mapping software employed was not explicitly stated, or readily apparent, in 35% of studies. Moreover, although the percentage of reads mapping to contigs were often provided, the mapping parameters employed (e.g., how reads mapping to multiple contigs were handled) were not stated in 79% of the studies. Similarly, the software utilized in identifying DEGs was not clearly provided in 35% of the 22 studies, despite reporting P (i.e., statistical significance cutoff) and FDR (i.e., false discovery rate) values as well as interpreting biological significance. When information on software was given, edgeR (Robinson et al. 2010) and DESeq (Anders and Huber 2010) were used most frequently to identify DEGs (35% and 25%, respectively).
Quantitative values that could be summarized from the 53 transcriptomic studies are presented in Table 1. When taken together with the most commonly utilized software (see above), this serves as a description for an “average” crustacean transcriptomic study during the 2014–2015 time period, which is consistent with RNA-Seq work in other non-model systems (Willette et al. 2014). Assembly, annotation, and differential expression analyses via Trinity were particularly widespread among the crustacean transcriptomics studies, likely due to the computational performance and speed of this software suite in assembling transcriptomes de novo (Grabherr et al. 2011) along with published, easy-to-follow protocols being available (Haas et al. 2013). Furthermore, studies using Trinity have reported transcriptome assemblies of apparently high-quality as measured by parameters such as transcript lengths (Manfrin et al. 2015) or the fraction of genes recovered in expected categories (Lenz et al. 2014). However, the percentage of contigs annotated was only 32% on average, suggesting that a significant fraction of a typical crustacean transcriptome is composed of novel or highly divergent transcripts, which is supported by a similarly low annotation rate for the Daphnia genome (Colbourne et al. 2011). Use of hidden Markov models (Yoon 2009) and/or structure-based annotation (Brylinski and Skolnick 2010) could increase annotation rates for crustacean transcriptomes or specific genes of interest in the future (see Das et al., 2016). Importantly, the bioinformatic pipelines described here and the metrics in Table 1 should not be considered the “ideal” for crustacean transcriptomic studies, as there cannot be a one-size-fits-all recommendation for RNA-Seq experiments. Rather, different sequencing depths and bioinformatic pipelines should be explored depending on the particular goal(s) of the specific study (Tarazona et al. 2011). In any case, we feel this summary provides a synopsis of the field in general as well as a useful benchmark for crustacean researchers interested in beginning to integrate RNA-Seq into their own work.
Effects of sequencing depth on RNA-Seq experiments
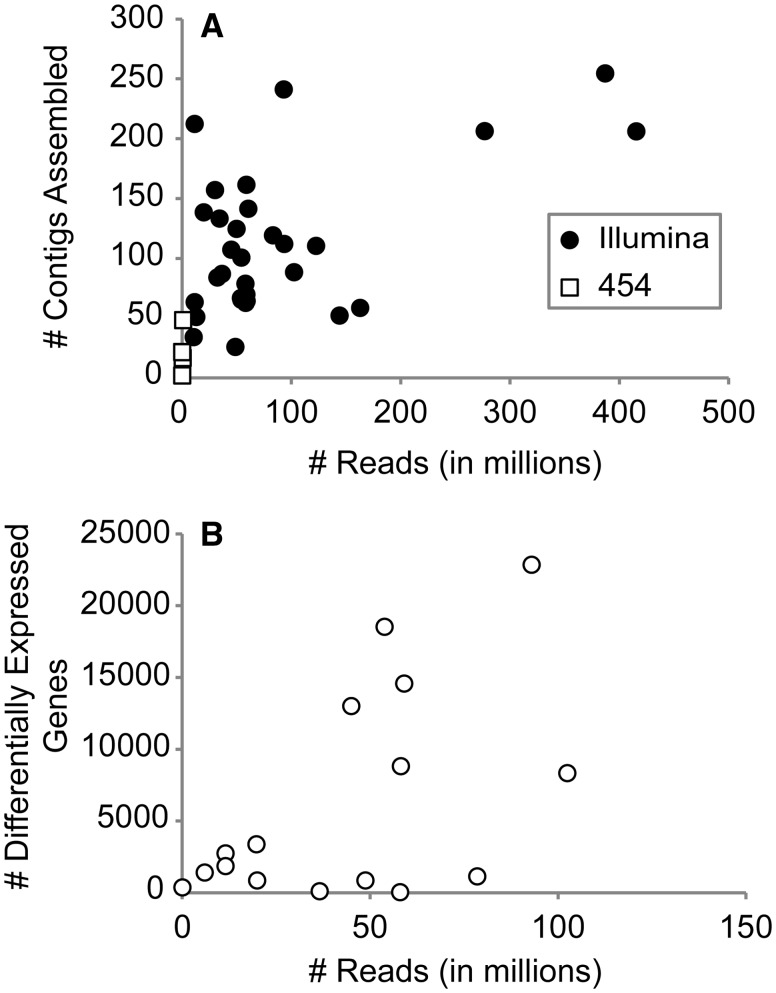
Since deciding on sequencing depth is an important consideration when designing RNA-Seq experiments (Francis et al. 2013), we examined the influence of read numbers on the resulting number of contigs in a transcriptome assembly (Fig. 3(A)) as well as the number of identified DEGs (when applicable to a study; Fig. 3(B)). Although there was a correlation between increasing read numbers and greater number of contigs and/or DEGs per sample, there were several examples where this did not turn out to be the case. This corresponds well with previous works documenting similar diminishing returns with increasing sequencing depth, both in crustaceans (Lenz et al. 2014) and other non-model systems (Francis et al. 2013; Zhang et al. 2013). Similarly, we have also observed the same trend in saturation of identified DEGs with increasing read numbers when examining salinity-induced differential expression in the gills of the blue crab Callinectes sapidus (Havird et al., unpublished data).
Fig. 3.
Effect of sequencing depth, measured as the number of reported reads per study, on the number of (A) contigs assembled and (B) DEGs identified from the 53 transcriptomic studies reviewed here.
Given this, how does one go about determining an optimum balance between sequencing depth and sample numbers to be investigated? For example, when directly compared with a dataset possessing ∼6X more reads, results from human stem cells found that “just” ∼36 M reads per sample allowed for accurate transcript abundance estimates (Encode Project Consortium 2004; Trapnell et al. 2009), while ∼20–30 M reads have been suggested as a benchmark to describe a “complete” transcriptome (Francis et al. 2013). Along with this, if “completeness” is a priority, the generation of transcriptomes from different developmental stages, tissues, sexes, and/or under variable environmental conditions should also be considered in order to maximize gene and transcript recovery. Although sequencing depth will depend on the objectives of the study (e.g., whether identifying and estimating abundance for rare transcripts is of importance) and should be determined in the context of the question(s) being addressed (Sims et al. 2014), we can offer a few generalized recommendations based on current sequencing technologies and studies.
Future crustacean transcriptome studies should plan on sequencing ∼50–100 M reads to more-or-less adequately describe a transcriptome. Specifically, when a linear function is fitted to the Illumina data presented in Fig. 3(A), sequencing 10 M reads yields on average ∼86,000 contigs, while 50 M and 100 M reads yields 98,000 and 113,000 contigs, respectively. Given that the Daphnia genome only contains ∼31,000 genes, it is clear that many of the contigs reported from transcriptomic assemblies in Fig. 3(A) are either alternatively spliced variants or assembly artifacts, which generally are not of interest to many investigators and subsequently inflate the perceived number of reads needed. In other words, the higher number of contigs yielded from deep sequencing is likely a many-fold inaccurate estimate rather than a biological plausible one. When estimating differential expression, ∼10–20 M reads per sample is likely acceptable for identifying most DEGs. We arrive at this suggestion by utilizing a similar linear function on Fig. 3(B), where sequencing 10 M or 20 M reads per sample yields 1400 and 2500 DEGs, respectively. This syncs well with rarefication analyses in the blue crab C. sapidus, which suggested diminished returns in numbers of recovered DEGs when >10 M reads are included in analyses (Havird et al., unpublished data). Similarly to the transcriptome assembly example above, a result of ∼20,000 DEGs is likely not biologically plausible, implying again that deep sequencing can produce inaccurate estimates. While these recommendations correspond well with current practices used to characterize transcriptomes (Table 1), the current sequencing depth to identify DEGs (i.e., 44 ± 8 M reads/sample in differential expression studies) could easily be considered overkill. Moreover, this excessive level of sequencing likely prevents many differential expression analyses from incorporating sufficient, and sorely needed, biological replication (see below). In summary, sequencing more reads is not always better.
Replication and software choice in differential expression studies
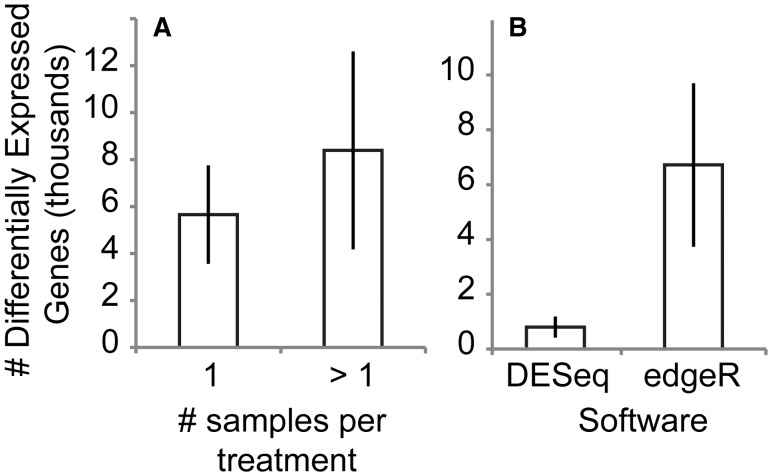
We found no significant difference in the number of identified DEGs when single or multiple biological replicates were utilized in differential expression analyses (Fig. 4(A)). However, the reported numbers of DEGs from 12 of the 22 (∼55%) studies came from examination of only a single biological replicate per treatment. In some cases, multiple sequencing or technical replicates were generated (Supplementary Table S1), but such schemes fail to quantify any variance between independent biological replicates due to practices like pooling all biological samples from a treatment before sequencing (i.e., without barcoding).
Fig. 4.
Effect of (A) biological replication and (B) software utilized on the number of DEGs identified from 22 studies reviewed here reporting numbers of DEGs. Error bars show ± SEM.
While early RNA-Seq studies commonly estimated differential expression using a non-replicated experimental design (Marioni et al. 2008), it is deeply troubling to report this is an ongoing practice. Specifically, Fisher noted the seriousness of non-replication 80 years ago (Fisher 1935) and simulations of RNA-Seq data have demonstrated the impossibility of extrapolating such non-replicated results to populations (Auer and Doerge 2010). Unfortunately, empirical studies corroborating differences in the number and identity of DEGs between replicated vs. non-replicated designs have been lacking to date. To address this, we estimated DEGs between developmental stages of the anchialine atyid shrimp Halocaridina rubra utilizing the single, non-replicated samples from transcriptome assemblies, which was done by 12 of the studies examined here, as well as from a multi-replicated design (Havird and Santos 2016). Notably, we found large differences among DEGs between non-replicated vs. replicated RNA-Seq experimental designs investigating the same developmental stages, implying results from non-replicated experiments should, at most, be used only for generating hypotheses (Havird and Santos 2016). Moreover, recent analytical tools such as Scotty allow the design of RNA-Seq experiments, including estimating the number of biological replicates needed per sample, based on such preliminary data (Busby et al. 2013).
The pitfalls of non-replicated RNA-Seq experimental designs are likely compounded by the fact that the most commonly utilized software for differential expression analyses (e.g., edgeR and DESeq) rely on Poisson models, which are inherently sensitive to biological replication (Robinson and Smyth 2007; Langmead et al. 2010). Given this, alternative statistical methods, such as those implemented in GFold (Feng et al. 2012), may be better suited for non-replicated experiments where biological replication is either extremely difficult or impossible. Until such alternative statistical methods have been thoroughly-vetted, however, we strongly recommend that the crustacean transcriptomics community, as well as those doing similar work on other taxonomic groups, avoid submitting for review, or accepting for publication, non-replicated differential expression studies (unfortunately, many will continue to be published in 2016 and potentially beyond), and only consider replicated experimental designs in the future.
The requirement for replication has practical implications for crustacean transcriptomic studies, as such experiments can quickly become costly when analyzing many samples, which likely limited replication in the first place for many of the studies surveyed here. In light of decreasing sequencing costs though, we hope this will lead to non-replicated differential expression studies gradually fading from the literature. But as mentioned previously, pilot studies based on a non-replicated experimental design could be conducted in order to generate initial hypotheses. Such hypotheses could then be tested at a future time by a replicated experimental design, with the benefit of already having transcriptome assemblies in hand, which can be leveraged towards using more cost-efficient differential expression techniques. For example, by already having a composite (i.e., combined adult and larval developmental stages) transcriptome for H. rubra, we were able to take advantage of a 3′-based RNA-Seq technique to sequence 24 samples simultaneously in a single Illumina HiSeq 2000 lane (Havird and Santos 2016). Additional benefits from such an approach include: (1) eliminating length biases; (2) generating effectively deeper sequencing coverage per gene from the same number of sequence reads; (3) requiring relatively little starting RNA; and (4) correlating well with results from quantitative polymerase chain reaction (qPCR; Meyer et al. 2011). Because the other studies examined here all employed “shotgun” RNA-Seq (i.e., sequencing whole RNA populations), we could not directly compare tag-based vs. transcriptome-wide RNA-Seq methods. Lastly, possessing transcriptomic resources also allows for targeted gene expression studies by facilitating the design of qPCR primers for a smaller number of interesting genes that can be further investigated across many treatments and biological replicates (e.g., Havird et al. 2014).
As discussed above, the software utilized in differential expression analyses can have a profound effect on results and, potentially, downstream biological interpretations. For example, studies using edgeR for differential expression analyses identified ∼8X more DEGs, on average, than those utilizing DESeq (Fig. 4(B)). We have observed a similar pattern in our own RNA-Seq experiments of salinity transfer (Havird et al., unpublished data) and developmental stages (Havird and Santos 2016) in crustaceans. In this case, DESeq uses more stringent statistical parameters by default than edgeR, with manual adjustment of these parameters often reconciling results from the two software packages (Love et al. 2014). Moreover, it should be noted that different versions of these software packages differ in their default settings, with DESeq2 (Love et al. 2014), the prior iteration’s successor, potentially yielding estimates of DEGs more consistent with edgeR based on its utilization in one study included in this review. In any case, inferences based on the original DESeq, particularly under default parameters, should be considered conservative estimates of differential gene expression. Finally, like other aspects of the RNA-Seq methodologies examined here, many researchers failed to report important details of their analyses, including P and log-fold change (C) statistics employed in statistical testing of differential expression. Interestingly, 69% of differential expression studies that did report C utilized a value of 1.0, meaning DEGs had to only show a two-fold change in expression between treatments, which may not be biologically meaningful in many contexts (especially in non-replicated experiments).
The fruits of mining publicly available data: evolution of neurotoxins in crustaceans
Lastly, 7 of 53 studies reviewed here (13%) solely utilized publicly available transcriptomic data from crustaceans in order to address their hypotheses. For example, Christie and colleagues mined crustacean transcriptomes from several species to identify novel peptide-encoding transcripts as well as examine comparative peptidomes across Crustacea (Christie et al. 2013; Christie 2014a; 2014b). As such resources will continue to become available given the current pace of transcriptomics research (Fig. 1), data mining will naturally become more commonplace. This, in turn, should allow resources to be allocated to experiments targeting interesting biological questions rather than just describing transcriptomes from various crustacean species in general. In other words, generating additional transcriptomes for well-studied species such as the red swamp crayfish (P.clarkii) will likely be less fruitful than hypothesis-driven RNA-Seq experiments, such as examining differential expression of P. clarkii genes due to novel stressors.
As a further demonstration of the utility of mining publicly available transcriptomes for hypothesis driven-research, we followed on the work of von Reumont et al. (2014) and attempt to generate preliminary hypotheses for the evolution of a neurotoxin in crustaceans. As part of the description of the first venomous crustacean, the remipede Speleonectes tulumensis (von Reumont et al. 2014), a putative neurotoxin, related to those from spider venom, was identified from this remipede’s venom gland. Furthermore, von Reumont et al. (2014) identified related sequences in the Daphnia genome, implying non-toxic precursors may be widespread in crustaceans. Here, we utilized this neurotoxin sequence as a query to search for potential homologs among other crustacean transcriptomes using tBLASTx version 2.2.29 (Altschul et al. 1997). We retained all hits with a BLAST E value of 1e − 5 or less from: (1) all publicly available crustacean transcriptomes on NCBI’s Transcriptome Shotgun Assembly (TSA) database; (2) the “adult + larvae” composite transcriptome of H. rubra detailed in Havird and Santos (2016); and (3) transcriptomes generated solely from gill tissues of either C. sapidus (Havird et al., unpublished data) or publicly available sequences on the SRA from the crabs Portunus trituberculatus (Lv et al. 2013), Hyas araneus (Harms et al. 2013; Harms et al. 2014), and Carcinus aestuarii (Romiguier et al. 2014), and the giant river prawn Macrobrachium rosenbergii (Mohd-Shamsudin et al. 2013). All recovered and potentially related sequences were then aligned and analyzed via Maximum Likelihood (ML) approaches in FastTreeMP version 2.1.7 (Price et al. 2010) as well as RAxML version 8.0.23 (Stamatakis 2014; Supplementary File S4).
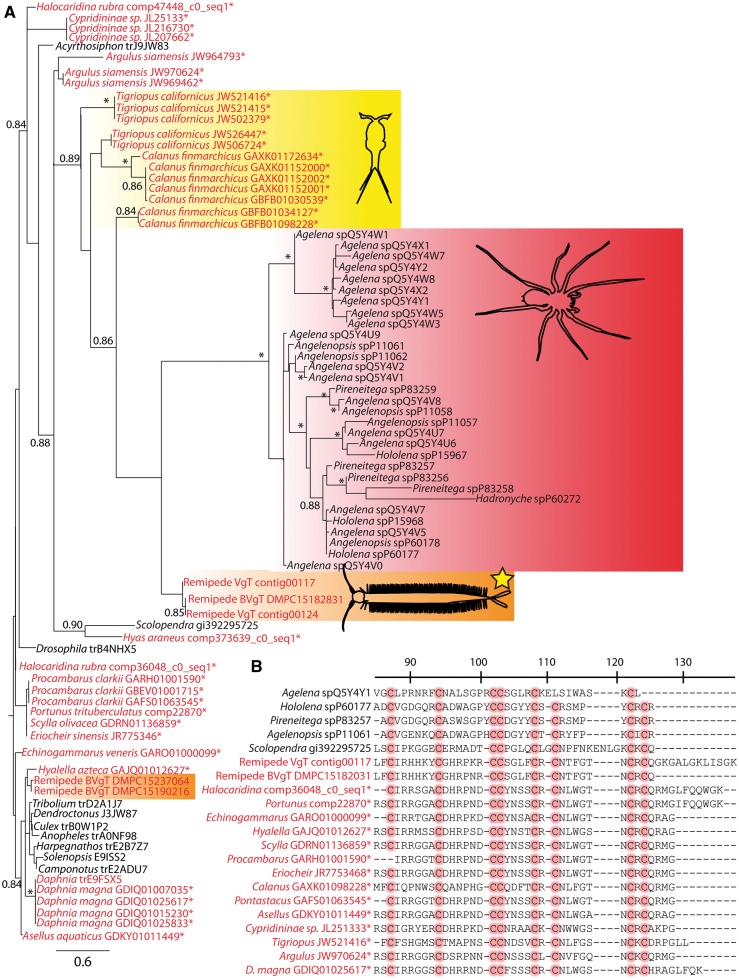
Several interesting hypotheses on the evolution of this neurotoxin in crustaceans can be generated from our inferred, but preliminary, phylogeny (Fig. 5(A)). For example, it is clear that precursor proteins for this neurotoxin exist across crustaceans, and are expressed at appreciable enough levels to be detected in obviously non-venomous tissues such as gills. Moreover, all these novel sequences possessed the multiple conserved cysteine residues which form disulfide bonds characteristic of β/δ agatoxins (Fig. 5(B)). Other notable features of the phylogeny include neurotoxin homologs from the copepods Tigriopus and Calanus forming a monophyletic clade with recognized neurotoxins from remipedes and spiders as well as the known functional neurotoxin from the venom gland of the centipede Scolopendra falling outside of this clade (Fig. 5(A)). When coupled with a relatively long branch leading to the copepod homologs, which was used to infer functional evolution in the original remipede neurotoxin (von Reumont et al. 2014), this suggests a functional neurotoxin may also be expressed in at least some copepods. Obviously, the preliminary nature of these results must be stressed and additional, more directed studies need to be undertaken to further elaborate on neurotoxin and venom evolution in crustaceans. However, the primary point of this exercise was to demonstrate how publicly available transcriptomic data can be utilized in a relatively short time frame (i.e., ∼hours) to generate preliminary hypotheses on crustacean biology that are of potential interest to the field or funding agencies.
Fig. 5.
(A) Phylogenetic hypothesis of the evolution of the β/δ agatoxin-like domain from putative neurotoxins in crustaceans, based on the approximate ML methods implemented in FastTreeMP (a qualitatively similar tree was generated using RAxML —see Supplementary File S4). Crustacean taxa names are in red; other sequences are in black; new crustacean sequences from the TSA database of NCBI or from unpublished transcriptomic assemblies are indicated with an asterisk (e.g., Hyas and Portunus sequences are from gill tissue). The clade forming known nuerotoxins from spiders is outlined in red, the described neurotoxin from the remipede venom gland is outlined in orange and highlighted with a yellow star, while the “nontoxin” paralog expressed in other remipede tissues is outlined in orange only (no star). The copepod sequences closely related to known neurotoxins are outlined in yellow. Scale bar indicates amino acid replacements per site. Numbers at nodes indicate “SH-like local support values” based on 1000 resamples (Price et al. 2010), with those < 0.8 removed and those > 0.9 indicated with an asterisk. The alignment used in generating this phylogeny and the resulting tree in Newick format are available as Supplementary Files S2 and S3. (B) Representative alignment of the β/δ agatoxin-like domain from putative neurotoxin-like proteins in crustaceans, with functionally important and conserved cysteines highlighted in red. Numbering and other details follow von Reumont et al. (2014).
Conclusions
Here, we systematically reviewed 53 studies developing or utilizing transcriptomic resources in crustaceans and published in the last ∼2 years to provide a current snapshot of the field. Importantly, crustaceans were targeted due to their roles as models in ecological and physiological studies as well as the likelihood that functional genomic tools for the group will be lacking through the near future. However, we postulate that the trends and recommendations identified here for crustaceans easily extend to transcriptomic studies in other non-model organisms. Based on this review, it is clear that RNA-Seq has become a popular tool in furthering understanding of crustacean biology, with a handful of fairly standard methodologies and software, common to similar work in other non-model systems, dominating such studies. However, some identified trends were worrisome, including the omission of critical methodological details from many analyses of DEGs. Particularly, over half of the examined studies describing DEGs from RNA-Seq data were based on non-replicated experimental designs. Coupled with diminishing returns due to saturation as more sequence reads are generated, this suggests future RNA-Seq experiments examining differential expression should focus more on increasing the number of biological replicates and experimental treatments rather than sequencing a smaller number of samples to greater depths. Finally, the relatively unexplored aspects of small, regulatory RNAs as well as alternative splicing in crustaceans represent areas of study that could benefit from further utilization of RNA-Seq experiments.
Supplementary Material
Acknowledgments
We thank five anonymous reviewers for comments that improved the manuscript. We are also grateful to Donald Mykles, Karen Burnett, David Durica, and Jonathon Stillman for organizing the “Tapping the Power of Crustacean Transcriptomes to Address Grand Challenges in Comparative Biology” symposium at the 2016 meeting of the Society for Integrative and Comparative Biology and inviting us to participate. This represents contributions No. 142 and No. 50 to the Auburn University (AU) Marine Biology Program and Molette Biology Laboratory for Environmental and Climate Change Studies, respectively.
Funding
This work was supported in part by the National Science Foundation (NSF) [DEB #0949855 to SRS] and its Doctoral Dissertation Improvement Grant program [DEB #1311500 to JCH]. Funding support to JCH also came from the Auburn University Cellular and Molecular Biosciences Peaks of Excellence Graduate Fellowship program, a Professional Association of Diving Instructors (PADI) Foundation Research Award (#5089), a Fellowship in Graduate Studies for Genetics/Physiology from The Crustacean Society, and a Doctoral Graduate Research Fellowship from the Alabama Council on Higher Education (ACHE). Travel support for JCH to the Society for Integrative and Comparative (SICB) Annual Meeting 2016 came from NSF [IOS #1551003], The Crustacean Society, and the Division of Comparative Physiology & Biochemistry.
Supplementary data
Supplementary Data available at ICB online.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nuc Aci Res 25:3389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, Huber W. 2010. Differential expression analysis for sequence count data. Gen Biol 11:R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anger K. 1995. The conquest of freshwater and land by marine crabs: Adaptations in life-history patterns and larval bioenergetics. J Exp Mar Biol Ecol 193:119–45. [Google Scholar]
- Apweiler R, Jesus Martin M, O'onovan C, Magrane M. 2012. Reorganizing the protein space at the Universal Protein Resource (UniProt). Nuc Aci Res 40:D71–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ArchMiller AA, Bauer EF, Koch RE, Wijayawardena BK, Anil A, Kottwitz JJ, Munsterman AS, Wilson AE. 2015. Formalizing the definition of meta-analysis in Molecular Ecology. Mol Ecol 24:4042–51. [DOI] [PubMed] [Google Scholar]
- Auer PL, Doerge RW. 2010. Statistical design and analysis of RNA sequencing data. Genetics 185:405–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bliss DE. 1968. Transition from water to land in decapod crustaceans. Amer Zool 8:355–92. [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borenstein M. 2009. Introduction to meta-analysis. Chichester, UK: John Wiley & Sons. [Google Scholar]
- Busby MA, Stewart C, Miller CA, Grzeda KR, Marth GT. 2013. Scotty: a web tool for designing RNA-Seq experiments to measure differential gene expression. Bioinformatics 29:656–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brylinski M, Skolnick J. 2010. Comparison of structure-based and threading-based approaches to protein functional annotation. Pro Struc Func Bioinf 78:118–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmichael SN, Bekaert M, Taggart JB, Christie HR, Bassett DI, Bron JE, Skuce PJ, Gharbi K, Skern-Mauritzen R, Sturm A. 2013. Identification of a sex-linked SNP marker in the salmon louse (Lepeophtheirus salmonis) using RAD sequencing. PLoS One 8:e77832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie AE. 2014a. Expansion of the Litopenaeus vannamei and Penaeus monodon peptidomes using transcriptome shotgun assembly sequence data. Gen Comp Endocrinol 206:235–54. [DOI] [PubMed] [Google Scholar]
- Christie AE. 2014b. Prediction of the peptidomes of Tigriopus californicus and Lepeophtheirus salmonis (Copepoda, Crustacea). Gen Comp Endocrinol 201:87–106. [DOI] [PubMed] [Google Scholar]
- Christie AE, Roncalli V, Wu LS, Ganote CL, Doak T, Lenz PH. 2013. Peptidergic signaling in Calanus finmarchicus (Crustacea, Copepoda): In silico identification of putative peptide hormones and their receptors using a de novo assembled transcriptome. Gen Comp Endocrinol 187:117–35. [DOI] [PubMed] [Google Scholar]
- Colbourne JK, Pfrender ME, Gilbert D, Thomas WK, Tucker A, Oakley TH, Tokishita S, Aerts A, Arnold GJ, Basu MK. et al. 2011. The ecoresponsive genome of Daphnia pulex. Science 331:555–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooley SR, Doney SC. 2009. Anticipating ocean acidification’s economic consequences for commercial fisheries. Environ Res Lett 4:15–9. [Google Scholar]
- Das S, Shyamal SD, Durica DS. 2016b. Analysis of annotation and differential expression methods used in RNA-seq studies in crustaceans systems. Integr Comp Biol 56:1067–69. [DOI] [PubMed] [Google Scholar]
- Encode Project Consortium 2004. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science 306:636–40. [DOI] [PubMed] [Google Scholar]
- Feng J, Meyer CA, Wang Q, Liu JS, Shirley Liu X, Zhang Y. 2012. GFOLD: a generalized fold change for ranking differentially expressed genes from RNA-Seq data. Bioinformatics 28:2782–8. [DOI] [PubMed] [Google Scholar]
- Fisher RA. 1935. The design of experiments. Edinburgh, London: Oliver and Boyde. [Google Scholar]
- Fonseca NA, Marioni J, Brazma A. 2014. RNA-Seq gene profiling–a systematic empirical comparison. PLoS One 9:e107026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis WR, Christianson LM, Kiko R, Powers ML, Shaner NC, Haddock SH. 2013. A comparison across non-model animals suggests an optimal sequencing depth for de novo transcriptome assembly. BMC Genom 14:167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q. et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29:644–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M. et al. 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc 8:1494–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms L, Frickenhaus S, Schiffer M, Mark FC, Storch D, Held C, Portner HO, Lucassen M. 2014. Gene expression profiling in gills of the great spider crab Hyas araneus in response to ocean acidification and warming. BMC Genom 15:789.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms L, Frickenhaus S, Schiffer M, Mark FC, Storch D, Portner HO, Held C, Lucassen M. 2013. Characterization and analysis of a transcriptome from the boreal spider crab Hyas araneus. Comp Biochem Physiol Part D Genom Prot 8:344–51. [DOI] [PubMed] [Google Scholar]
- Havird JC, Mitchell RT, Henry RP, Santos SR. Salinity-induced changes in gene expression from anterior and posterior gills of Callinectes sapidus (Crustacea: Portunidae) with implications for crustacean ecological genomics. In press at Comparative Biochemistry and Physiology Part D: Genomics and Proteomics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havird JC, Santos SR, Henry RP. 2014. Osmoregulation in the Hawaiian anchialine shrimp Halocaridina rubra (Crustacea: Atyidae): expression of ion transporters, mitochondria-rich cell proliferation and hemolymph osmolality during salinity transfers. Journal of Experimental Biology 217(13):2309–2320. [DOI] [PubMed] [Google Scholar]
- Havird JC, Santos SR. 2016. Developmental transcriptomics of the hawaiian anchialine shrimp halocaridina rubra holthuis, 1963 (Crustacea: Atyidae). Integr Comp Biol 56:1170–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry RP. 1994. Morphological, behavioral, and physiological characterization of bimodal breathing crustaceans. Amer Zool 34:205–15. [Google Scholar]
- Henry RP, Lucu C, Onken H, Weihrauch D. 2012. Multiple functions of the crustacean gill: osmotic/ionic regulation, acid-base balance, ammonia excretion, and bioaccumulation of toxic metals. Front Physiol 3:431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobbs HH, Hart CW. 1982. The shrimp genus Atya (Decapoda:Atyidae). Washington: Smithsonian Institution Press. [Google Scholar]
- Huang X, Madan A. 1999. CAP3: A DNA sequence assembly program. Gen Res 9:868–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt M, Newbold C, Berriman M, Otto TD. 2014. A comprehensive evaluation of assembly scaffolding tools. Gen Biol 15:R42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. 2016. KEGG as a reference resource for gene and protein annotation. Nuc Aci Res 44:D457–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenny NJ, Sin YW, Shen X, Zhe Q, Wang W, Chan TF, Tobe SS, Shimeld SM, Chu KH, Hui JH. 2014. Genomic sequence and experimental tractability of a new decapod shrimp model, Neocaridina denticulata. Mar Drugs 12:1419–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B. 2010. Aligning short sequencing reads with Bowtie. Curr Protoc Bioinfor Chapter 11, Unit 7. [DOI] [PMC free article] [PubMed]
- Langmead B, Hansen KD, Leek JT. 2010. Cloud-scale RNA-sequencing differential expression analysis with Myrna. Gen Biol 11:R83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Meth 9:357–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz PH, Roncalli V, Hassett RP, Wu LS, Cieslak MC, Hartline DK, Christie AE. 2014. De novo assembly of a transcriptome for Calanus finmarchicus (Crustacea, Copepoda)–the dominant zooplankter of the North Atlantic Ocean. PLoS One 9:e88589.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis CN, Brown KA, Edwards LA, Cooper G, Findlay HS. 2013. Sensitivity to ocean acidification parallels natural pCO(2) gradients experienced by Arctic copepods under winter sea ice. Proc Nat Acad Sci USA 110:E4960–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Gen Biol 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo R, Liu B, Xie Y, Li Z, Huang W, Yuan J, He G, Chen Y, Pan Q, Liu Y. et al. 2012. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience 1:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv J, Liu P, Wang Y, Gao B, Chen P, Li J. 2013. Transcriptome analysis of Portunus trituberculatus in response to salinity stress provides insights into the molecular basis of osmoregulation. PLoS One 8:e82155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacManes MD. 2014. On the optimal trimming of high-throughput mRNA sequence data. Front Genet 5:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manfrin C, Tom M, De Moro G, Gerdol M, Giulianini PG, Pallavicini A. 2015. The eyestalk transcriptome of red swamp crayfish Procambarus clarkii. Gene 557:28–34. [DOI] [PubMed] [Google Scholar]
- Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen YJ, Chen Z. et al. 2005. Genome sequencing in microfabricated high-density picolitre reactors. Nature 437:376–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. 2008. RNA-seq: an assessment of technical reproducibility and comparison with gene expression arrays. Gen Res 18:1509–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara JC, Faria SC. 2012. Evolution of osmoregulatory patterns and gill ion transport mechanisms in the decapod Crustacea: a review. J Comp Physiol B Biochem Syst Environ Physiol 182:997–1014. [DOI] [PubMed] [Google Scholar]
- Meusemann K, von Reumont BM, Simon S, Roeding F, Strauss S, Kuck P, Ebersberger I, Walzl M, Pass G, Breuers S. et al. 2010. A phylogenomic approach to resolve the arthropod tree of life. Mol Biol E 27:2451–64. [DOI] [PubMed] [Google Scholar]
- Meyer E, Aglyamova GV, Matz MV. 2011. Profiling gene expression responses of coral larvae (Acropora millepora) to elevated temperature and settlement inducers using a novel RNA-Seq procedure. Mol Ecol 20(17):3599–616. [DOI] [PubMed] [Google Scholar]
- Mistry J, Finn RD, Eddy SR, Bateman M, Punta M. 2013. Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nuc Aci Res 41:e121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohd-Shamsudin MI, Kang Y, Lili Z, Tan TT, Kwong QB, Liu H, Zhang G, Othman RY, Bhassu S. 2013. In-depth tanscriptomic analysis on giant freshwater prawns. PLoS One 8:e60839.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris S. 2002. The ecophysiology of air-breathing in crabs with special reference to Gecarcoidea natalis. Comp Biochem Physiol B Biochem Mol Biol 131:559–70. [DOI] [PubMed] [Google Scholar]
- Mykles DL, Hui JH. 2015. Neocaridina denticulata: A decapod crustacean model for functional genomics. Integr Comp Biol 891–7. [DOI] [PubMed] [Google Scholar]
- Nookaew I, Papini M, Pornputtapong N, Scalcinati G, Fagerberg L, Uhlen M, Nielsen J. 2012. A comprehensive comparison of RNA-Seq-based transcriptome analysis from reads to differential gene expression and cross-comparison with microarrays: a case study in Saccharomyces cerevisiae. Nuc Aci Res 40:10084–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell S, Szklarczyk D, Trachana K, Roth A, Kuhn M, Muller J, Arnold R, Rattei T, Letunic I, Doerks T. et al. 2012. eggNOG v3.0: orthologous groups covering 1133 organisms at 41 different taxonomic ranges. Nucleic Acids Res 40(Database issue):D284–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. 2010. FastTree 2–approximately maximum-likelihood trees for large alignments. Plos One 5:e9490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees DJ, Dufresne F, Glemet H, Belzile C. 2007. Amphipod genome sizes: first estimates for Arctic species reveal genomic giants. Genome 50:151–8. [DOI] [PubMed] [Google Scholar]
- Regier JC, Shultz JW, Zwick A, Hussey A, Ball B, Wetzer R, Martin JW, Cunningham CW. 2010. Arthropod relationships revealed by phylogenomic analysis of nuclear protein-coding sequences. Nature 463:1079–83. [DOI] [PubMed] [Google Scholar]
- Rehm P, Borner J, Meusemann K, von Reumont BM, Simon S, Hadrys H, Misof B, Burmester T. 2011. Dating the arthropod tree based on large-scale transcriptome data. Mol Phylogenet E 61:880–7. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, Smyth GK. 2007. Moderated statistical tests for assessing differences in tag abundance. Bioinformatics 23:2881–7. [DOI] [PubMed] [Google Scholar]
- Romiguier J, Gayral P, Ballenghien M, Bernard A, Cahais V, Chenuil A, Chiari Y, Dernat R, Duret L, Faivre N. et al. 2014. Comparative population genomics in animals uncovers the determinants of genetic diversity. Nature 515:261–3. [DOI] [PubMed] [Google Scholar]
- Saliba AE, Westermann AJ, Gorski SA, Vogel J. 2014. Single-cell RNA-seq: advances and future challenges. Nuc Aci Res 42:8845–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyednasrollah F, Laiho A, Elo LL. 2015. Comparison of software packages for detecting differential expression in RNA-seq studies. Bri Bioinform 16:59–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJ, Birol I. 2009. ABySS: a parallel assembler for short read sequence data. Genome Res 19:1117–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims D, Sudbery I, Ilott NE, Heger A, Ponting CP. 2014. Sequencing depth and coverage: key considerations in genomic analyses. Nat Rev Genet 15:121–32. [DOI] [PubMed] [Google Scholar]
- Sin YW, Kenny NJ, Qu Z, Chan KW, Cheong SP, Leung RW, Chan TF, Bendena WG, Chu KH, Tobe SS. et al. 2015. Identification of putative ecdysteroid and juvenile hormone pathway genes in the shrimp Neocaridina denticulata. Gen Comp Endocrinol 214:167–76. [DOI] [PubMed] [Google Scholar]
- Spicer JI. 2014. What can an ecophysiological approach tell us about the physiological responses of marine invertebrates to hypoxia? J Exp Biol 217:46–56. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30:1312–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stillman JH, Colbourne JK, Lee CE, Patel NH, Phillips MR, Towle DW, Eads BD, Gelembuik GW, Henry RP, Johnson EA. et al. 2008. Recent advances in crustacean genomics. Integr Comp Biol 48:852–68. [DOI] [PubMed] [Google Scholar]
- Tarazona S, Garcia-Alcalde F, Dopazo J, Ferrer A, Conesa A. 2011. Differential expression in RNA-seq: a matter of depth. Gen Res 21:2213–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd EV, Black MA, Gemmell NJ. 2016. The power and promise of RNA-seq in ecology and evolution. Mol Ecol 1224–41. [DOI] [PubMed] [Google Scholar]
- The Gene Ontology Consortium. (2000) Gene Ontology: tool for the unification of biology. Nature Genetics 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25:1105–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijay N, Poelstra JW, Kunstner A, Wolf JB. 2013. Challenges and strategies in transcriptome assembly and differential gene expression quantification. A comprehensive in silico assessment of RNA-seq experiments. Mol Ecol 22:620–34. [DOI] [PubMed] [Google Scholar]
- von Reumont BM, Blanke A, Richter S, Alvarez F, Bleidorn C, Jenner RA. 2014. The first venomous crustacean revealed by transcriptomics and functional morphology: remipede venom glands express a unique toxin cocktail dominated by enzymes and a neurotoxin. Mol Biol E 31:48–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M. 2009. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet 10:57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willette DA, Allendorf FW, Barber PH, Barshis DJ, Carpenter KE, Crandall ED, Cresko WA, Fernandez-Silva I, Matz MV, Meyer E. et al. 2014. So, you want to use next-generation sequencing in marine systems? Insight from the Pan-Pacific Advanced Studies Institute. Bull Mar Sci 90:79–122. [Google Scholar]
- Yoon BJ. 2009. Hidden markov models and their applications in biological sequence analysis. Curr Gen 10:402–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Ruhlman TA, Mower JP, Jansen RK. 2013. Comparative analyses of two Geraniaceae transcriptomes using next-generation sequencing. BMC Plant Biol 13:228.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.