Abstract
The X-linked reproductive homeobox (RHOX) gene cluster encodes transcription factors preferentially expressed in reproductive tissues. This gene cluster has important roles in male fertility based on phenotypic defects of Rhox-mutant mice and the finding that aberrant RHOX promoter methylation is strongly associated with abnormal human sperm parameters. However, little is known about the molecular mechanism of RHOX function in humans. Using gene expression profiling, we identified genes regulated by members of the human RHOX gene cluster. Some genes were uniquely regulated by RHOXF1 or RHOXF2/2B, while others were regulated by both of these transcription factors. Several of these regulated genes encode proteins involved in processes relevant to spermatogenesis; e.g. stress protection and cell survival. One of the target genes of RHOXF2/2B is RHOXF1, suggesting cross-regulation to enhance transcriptional responses. The potential role of RHOX in human infertility was addressed by sequencing all RHOX exons in a group of 250 patients with severe oligozoospermia. This revealed two mutations in RHOXF1 (c.515G > A and c.522C > T) and four in RHOXF2/2B (-73C > G, c.202G > A, c.411C > T and c.679G > A), of which only one (c.202G > A) was found in a control group of men with normal sperm concentration. Functional analysis demonstrated that c.202G > A and c.679G > A significantly impaired the ability of RHOXF2/2B to regulate downstream genes. Molecular modelling suggested that these mutations alter RHOXF2/F2B protein conformation. By combining clinical data with in vitro functional analysis, we demonstrate how the X-linked RHOX gene cluster may function in normal human spermatogenesis and we provide evidence that it is impaired in human male fertility.
Introduction
It is estimated that 10–15% of couples are affected by infertility, and nearly half of the cases can be attributed predominantly to a male factor (1). Some cases of male infertility are caused by known genetic defects, such as chromosomal abnormalities (e.g. Klinefelter syndrome) or Y chromosome microdeletions. However, a large proportion of infertile men (∼30%) remain diagnosed with idiopathic infertility, i.e., no known underlying cause (1–3). The X chromosome is of considerable interest to search for mutations that cause male infertility and subfertility, as this chromosome is enriched in testis-specific genes (4–7). Furthermore, mutations in X-linked genes have a complete penetrance due to their hemizygous state in men (8). Despite this potential, only few X-linked genes have been studied with respect to human male fertility. Mutations in the X-linked androgen receptor (AR) gene have been shown to be associated with disorders in male sexual differentiation and development (9). More recently, mutations in the X-linked gene, TEX11, have been identified as a cause of meiotic arrest and azoospermia (10,11).
In this report, we focus on a set of X chromosome-encoded transcription factors that previous studies have suggested are prime candidates to be involved in male infertility (12–15). These reproductive homeobox (RHOX) transcription factors contain a homeodomain, a highly conserved 60 amino-acid DNA-binding domain that interacts with DNA through its third α- helix (16). Over 200 homeobox transcription factors are present in mammalian species, where they take over a diverse set of roles, including control of key steps during embryogenesis (17). The RHOX transcription factors are a distinct subfamily that differs significantly from other homeobox family members, including those in the well-known HOX and POU subfamilies (12,18). The human RHOX cluster is composed of three genes located in Xq24: RHOXF1 (hPEPP1, OTEX), RHOXF2 (hPEPP2) and RHOXF2B (18,19). RHOXF2 and RHOXF2B are arranged in a head-to-head orientation and share high sequence similarity (>99%) and thus cannot easily be distinguished and are usually analysed as a single gene (13,14).
Compared to their human counterpart, the mouse and rat Rhox clusters are considerably larger; they are composed of at least 33 and 11 members, respectively (18). Two mouse Rhox family members—Rhox5 and Rhox8—have been shown through knockout and knockdown studies, respectively, to have essential roles in spermatogenesis and male fertility (12,20). In contrast to our considerable knowledge of rodent Rhox genes, the human RHOX genes have been poorly characterized. This is accentuated by the fact that, like most genes involved in reproduction (21,22), the RHOX cluster is rapidly evolving (23,24) and thus it is difficult to transfer information obtained in rodents to the human. Previously, it has been shown that RHOXF1 and RHOXF2/2B display highest expression levels in testis, and that the proteins are almost exclusively expressed in germ cells in a developmentally regulated manner (13). RHOXF1 is expressed in the late stages of germ cell development (pachytene spermatocytes and round spermatids), while RHOXF2/F2B protein is expressed in early stages (spermatogonia and early spermatocytes; 13). The finding that RHOXF2/F2B and RHOXF1 are expressed predominantly in early and late stages of spermatogenesis, respectively, raises the possibility that they function in these stages, and that loss-of-function mutations in these genes are good candidates to perturb male gametogenesis in humans (4). This is further supported by studies examining RHOX regulation. Both mouse and human RHOX family members have been demonstrated to be strongly regulated by DNA methylation (25–27) and there is an association between RHOX hypermethylation and abnormal sperm in idiopathic infertile patients (14).
The possibility that the X-linked RHOX cluster has roles in human fertility is intriguing. In this report, we investigate the molecular mechanism of the human RHOX transcription factors and determine whether their function is negatively impacted in male infertility patients. Our studies identified mutations that debilitate human RHOX function and they provide a foundation for understanding the transcriptional networks downstream of RHOX transcription factors in male germ cells.
Results
Identification of genes regulated by RHOX transcription factors
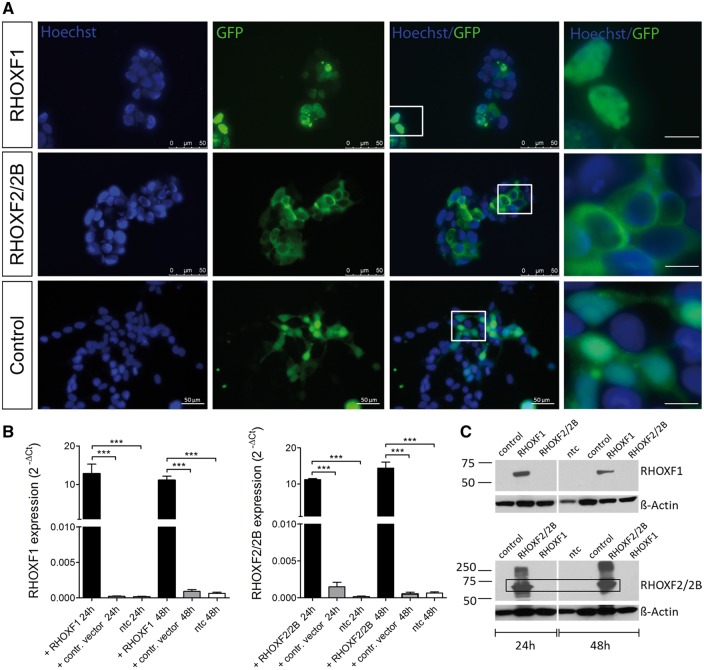
To identify genes regulated by the RHOXF1 and RHOXF2/2B transcription factors, we performed microarray analysis in transiently transfected HEK293 cells (28). These cells largely lack these two transcription factors, thus allowing us to perform gain-of-function analysis. Transfection efficiency and RHOX protein localization were monitored and evaluated using fluorescence microscopy detection of GFP (Fig. 1A). Interestingly, we found that the sub-cellular localization of RHOXF1 and RHOXF2/2B differed. While RHOXF1 expression was restricted mainly to the nucleus, RHOXF2/2B was found to be localized predominantly in the cytoplasm. Ectopic expression of RHOXF1/RHOXF2 and RHOXF1/RHOXF2 protein was validated by qRT-PCR and Western blot analyses, respectively (Fig. 1B and C).
Figure 1.
RHOX expression in transiently transfected HEK293 cells. (A) Immunofluorescent imaging of HEK293 cells 48h after transfection. GFP signal reflects RHOX expression while nuclei were counterstained with Hoechst. Scale bar: 10 µm (right row). (B) RHOX expression after 24h and 48h of transfection (n = 8). Total RNA was isolated, reverse transcribed, and amplified with specific primers. Values were normalized to endogenous GAPDH. ***P < 0.001. (C) Western Blot analysis of RHOX after 24h and 48h of transfection.
Microarray analysis of HEK293 cells 24h and 48h after transfection with the RHOXF1 and RHOXF2 expression vectors identified several genes that were significantly differentially expressed. In total, 17 (24h) and 37 (48h) transcripts were determined as differentially regulated in response to the RHOXF1 transfection vector compared to control vector transfection (fold change [FC] ≥1.5, uncorrected p-value ≤0.05, Supple mentary Material, Table S1), while 35 (24h) and 85 (48h) transcripts were significantly differentially expressed in response to the RHOXF2/2B transfection vector (Supplementary Material, Table S2). Hierarchical clustering of the differentially expressed genes (DEGs) revealed two clusters showing a homogeneous expression pattern across biological triplicates, indicating high reproducibility of the microarray data (Fig. 2A and B). By combining results from both time points analysed, a deduced list of 8 RHOXF1 and 30 RHOXF2/2B unique protein coding DEGs was defined (Table 1). Among these genes, RHOXF1 was found to be regulated by RHOXF2/2B, which would explain why these two RHOX transcription factors regulate some common genes (Supplementary Material, Table S3).
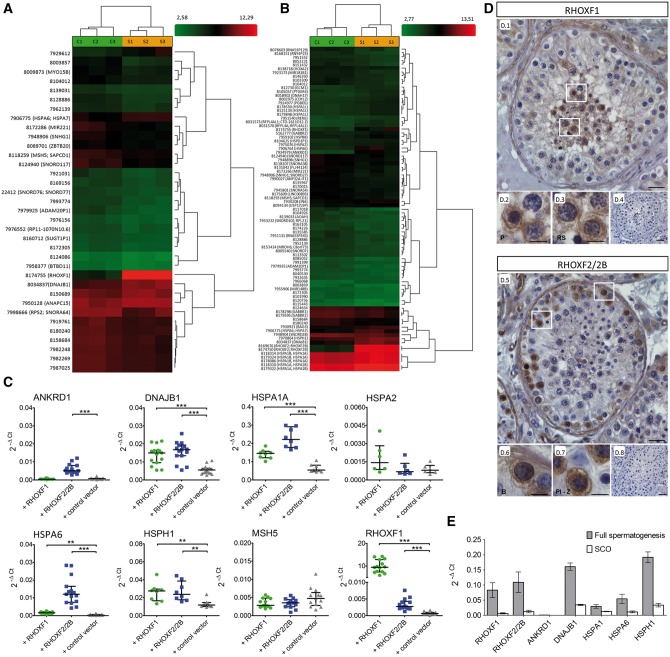
Figure 2.
Identification of genes regulated by the RHOX gene cluster. Heat map of RHOXF1 (A) and RHOXF2/2B (B) differentially expressed genes (DEGs) after 48h of transfection. Each column represents individual samples (S: RHOX transfected samples, C: control vector transfected samples), each row shows a single gene and expression levels were indicated by a color scale (red: high expression, green: low expression). (C) Analysis of DEGs by qRT-PCR. After different transfections with RHOX, mRNA expression levels of MSH5 (n = 15), ANKRD1 (n = 15), DNAJB1 (n = 15), HSPA1A (n = 8), HSPA2 (n = 6), HSPA6 (n = 15), HSPH1 (n = 8) and RHOXF1 (n = 15) were measured and normalized to GAPDH. The data are represented as median and interquartile range. **P < 0.01, ***P < 0.001. (D) Immunohistological analysis of RHOX in men with normal spermatogenesis. Pachytene spermatocytes and round spermatids showed positive staining for RHOXF1 (D.1–D.3), while type B spermatogonia and preleptotene – zygotene spermatocytes were predominantly stained with RHOXF2/2B specific antibody (D.5–D.7). IgG controls were negative (D.4, D.8). B, type B spermatogonia; P, pachytene spermatocytes; Pl-Z, preleptotene – zygotene spermatocytes; RS, round spermatids. Scale bar: 50 μm (D.1, D.4, D.5, D.8) and 25 μm (D.2, D.3, D.6, D.7). (E) Expression of RHOX regulated genes in testis with normal spermatogenesis (grey, n = 3) and in testis with SCO (white; n = 3). Data are presented as mean with SEM.
Table 1.
Protein coding differently expressed genes (DEGs) in RHOX transfected cells compared to control vector transfected cells. A threshold of fold change (FC) ≥1.5 and P-value ≤0.05 was used to identify DEGs.
| RHOXF1 |
RHOXF2/2B |
||||||
|---|---|---|---|---|---|---|---|
| Gene | Description | FC | P-value | Gene | Description | FC | P-value |
| DNAJB1 | DnaJ (Hsp40) homolog, subfamily B, member 1 | 1,97 | 0,003801 | ANKRD1 | ankyrin repeat domain 1 | 5,63 | 0,001485 |
| HSPA6/A7 | heat shock 70kDa protein 6 | 1,81 | 0,017932 | HSPA6 | heat shock 70kDa protein 6 | 4,64 | 0,018291 |
| ANAPC15 | anaphase promoting complex subunit 15 | 1,55 | 0,037242 | RHOXF1 | Rhox homeobox family, member 1 | 3,31 | 0,049385 |
| SLITRK3 | SLIT and NTRK-like family, member 3 | 1,52 | 0,021905 | DNAJB1 | DnaJ (Hsp40) homolog, subfamily B, member 1 | 2,73 | 0,002394 |
| TCL6f4 | T-cell leukaemia/lymphoma 6 ORF105 | −1,5 | 0,02937 | HSPH1 | heat shock 105kDa/110kDa protein 1 | 2,03 | 0,002656 |
| BTBD11 | BTB (POZ) domain containing 11 | −1,5 | 0,019466 | RFPL4A | ret finger protein-like 4A | 1,94 | 0,002071 |
| MSH5 | mutS homolog 5 | −1,5 | 0,009626 | HSPA1L | heat shock 70kDa protein 1-like | 1,84 | 0,046973 |
| ZBTB20 | zinc finger and BTB domain containing 20 | −1,6 | 0,009686 | INA | internexin neuronal intermediate filament protein, alpha | 1,74 | 0,031778 |
| HSPA1A | heat shock 70kDa protein 1A | 1,7 | 0,002545 | ||||
| DNAH17 | dynein, axonemal, heavy chain 17 | 1,68 | 0,027365 | ||||
| HSPA1B | heat shock 70kDa protein 1B | 1,68 | 0,002698 | ||||
| GABBR2 | gamma-aminobutyric acid (GABA) B receptor, 2 | 1,67 | 0,044556 | ||||
| BAG3 | BCL2-associated athanogene 3 | 1,64 | 0,019302 | ||||
| PGBD5 | piggyBac transposable element derived 5 | 1,63 | 0,006543 | ||||
| HSPB8 | heat shock 22kDa protein 8 | 1,61 | 0,030736 | ||||
| GCM1 | glial cells missing homolog 1 | 1,6 | 0,006529 | ||||
| RERG | RAS-like, oestrogen-regulated, growth inhibitor | 1,57 | 0,046276 | ||||
| USP17L | ubiquitin specific peptidase 17-like | 1,57 | 0,049222 | ||||
| PTGER4 | prostaglandin E receptor 4 (subtype EP4) | 1,55 | 0,042114 | ||||
| HSPA2 | heat shock 70kDa protein 2 | 1,54 | 0,018599 | ||||
| CDYL2 | chromodomain protein, Y-like 2 | 1,52 | 0,024896 | ||||
| HOXA2 | homeobox A2 | −1,5 | 0,047907 | ||||
| ZFP92 | ZFP92 zinc finger protein | −1,6 | 0,030135 | ||||
| SPRR2D | small proline-rich protein 2D | −1,6 | 0,032626 | ||||
| LCE1B | late cornified envelope 1B | −1,6 | 0,01887 | ||||
| GABBR1 | gamma-aminobutyric acid (GABA) B receptor, 1 | −1,6 | 0,031487 | ||||
| MSH5 | mutS homolog 5 | −1,6 | 0,031154 | ||||
| CES1 | carboxylesterase 1 | −1,6 | 0,00629 | ||||
| MROH6 | maestro heat-like repeat family member 6 | −1,6 | 0,012475 | ||||
| AOAH | acyloxyacyl hydrolase (neutrophil) | −1,8 | 0,031603 | ||||
A subset of eight candidate genes, composed of the five most strongly regulated genes (ANKRD1, DNAJB1, HSPA6, HSPH1, RHOXF1) and three regulated genes that have been previously associated with human fertilization (HSPA1A; (29,30) and abnormal spermatogenesis (HSPA2, MSH5; (31,32), were selected for validation by qRT-PCR analysis. This analysis confirmed that DNAJB1, HSPA1A, HSPA6 and HSPH1 were significantly upregulated by both RHOXF1 and RHOXF2/2B, whereas ANKRD1 was selectively upregulated in response to RHOXF2/2B overexpression (all P < 0.01, Fig. 2C). Also confirmed was that RHOXF2 regulates RHOXF1 (Fig. 2C). This is of possible physiological relevance given that the former is mainly expressed in early stage germ cells (type-B spermatogonia and early spermatocytes), while the latter is predominantly expressed in late stage germ cells (pachytene spermatocytes and round spermatids; Fig. 2D; 13). RHOXF1 and RHOXF2/2B are not exclusively expressed by germ cells, since we also noticed the very faint expression in somatic cells, e.g. Sertoli cells, Leydig cells and peritubular cells (Fig. 2D).
To investigate the expression pattern of RHOX-regulated genes in the human testis, we performed qRT-PCR analysis on testis samples with either complete spermatogenesis or Sertoli cell only (SCO) syndrome (i.e. without germ cells). With the exception of ANKRD1, all RHOX-regulated genes we tested were expressed in the testis, with HSPH1 (0.1917 ± 0.018) and DNAJB1 (0.1606 ± 0.012) showing the highest expression levels (Fig. 2E). All these genes exhibited decreased expression levels in SCO testes compared to normal testes, suggesting that they are predominantly expressed in germ cells. For controls, we examined the expression of RHOXF1 (0.083 ± 0.025) and RHOXF2/2B (0.109 ± 0.034) and found they both exhibited dramatically decreased expression in SCO testes, consistent with the previous direct evidence that they are primarily expressed in germ cells in the human testis (13).
Identification of RHOX sequence variations in infertile men
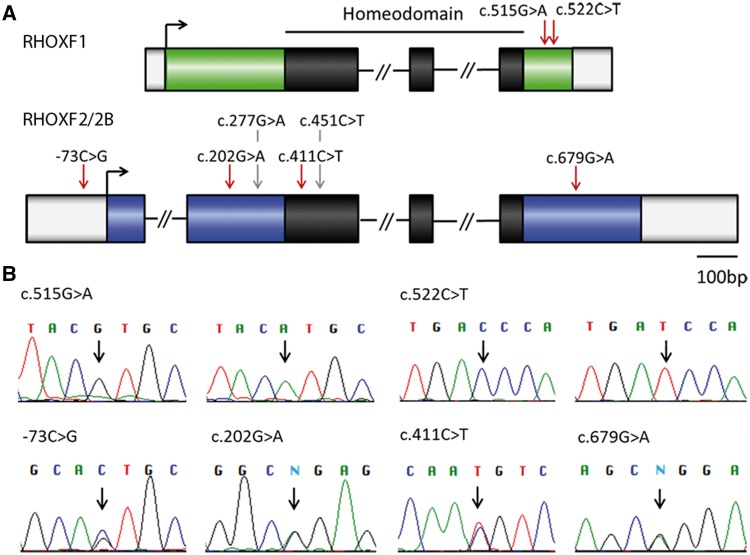
We next determined whether there are genetic alterations in RHOX genes in infertile men as a precursor to determining whether these mutations affect RHOX transcription factor function and thus are candidates to cause human spermatogenic defects. Towards this end, the coding region of RHOX was sequenced in 250 idiopathic infertile patients suffering from severe oligozoospermia (total sperm count ≤ 10×106, concentration ≤ 5×106/ml) to identify Single Nucleotide Polymorphisms (SNPs, minor allele frequency [MAF]>0.01) and rare genetic variants (also termed mutations, MAF < 0.01). This analysis revealed a non-synonymous mutation c.515G > A (p.Arg172His) and a synonymous mutation c.522C > T (p.Asp174Asp) in RHOXF1 (Table 2), both located downstream of the homeodomain (Fig. 3A). We also identified four rare variants in RHOXF2/2B: a novel mutation in the 5’UTR (-73C > G), one in exon 4 (c.679G > A, p.Gly227Arg), and two known mutations in Exon 2 (c.202G > A, p.Gly68Arg, rs148604152 and c.411C > T, p.Asn137Asn, rs142963365; Table 2, Fig. 3A), all of which were confirmed by Sanger sequencing (Fig. 3B). Clinical parameters of the patients carrying these variants are shown in the Supplementary Material, Table S4. Analysis of 174 normozoospermic controls did not identify any of these variants with the exception of c.202G > A (Table 2). Mutations c.202G > A and c.411C > T in RHOXF2/2B were present in the ExAC database (http://exac.broadinstitute.org; date last accessed August 31, 2016), whereas -73C > G and c.679G > A were not present in either dbSNP, ClinVar, 1000 Genome project or the ExAC dataset, and thus are novel variants. In silico prediction of the effect of the c.202G > A RHOXF2/2B mutation on protein function (using the tool PolyPhen-2 [http://genetics.bwh.harvard.edu/pph2/]) rated it as likely damaging (0.956). In contrast, c.679G > A RHOXF2/2B and c.515G > A RHOXF1 were predicted by both PolyPhen-2 and SIFT (http://sift.bii.a-star.edu.sg/; date last accessed February 26, 2016) to be benign. In addition to rare variants, several known non-synonymous SNPs were found for RHOXF2/2B (Supplementary Material, Table S5). Two of these SNPs (c.277G > A, p.Asp93Asn and c.451C > T, p.Arg151Cys) were further analysed because they affect the homeodomain and adjacent regions and one (c.451C > T) has a high PolyPhen-2 score (0.945), suggesting it impacts function.
Table 2.
RHOX mutations identified in 250 severe oligozoospermic patients. A list of all mutations (MAF < 0.01) identified in RHOXF1 and RHOXF2/2B in men with severe oligozoospermia. Mutation frequencies are given for the patient group, controls with normal sperm concentrations and data available at the ExAC database. NA: data not available, nt: not tested.
| mRNA | Type | Protein | Frequency (MAF) | Previously described | MAF ExAC database (European/Total) | Prediction (PolyPhen-2/SIFT) | Effect on | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Patients | Controls | target genes | protein structure | |||||||
| RHOXF1 | c. 515G>A | Non-synonymous | p.Arg172His | 1/250(0.004) | 0/174 | rs2301977 | 0.0001/0.0002 | benign (0.001)/tolerated (0.025) | no | yes (weak) |
| c.522C>T | Synonymous | p.Asp174Asp | 1/250(0.004) | 0/174 | ExAC | 0.003/0.008 | NA | no | nt | |
| RHOXF2/2B | c.-73C>G | 5‘UTR | 1/250(0.004) | no | NA | NA | nt | nt | ||
| c.202G>A | Non-synonymous | p.Gly68Arg | 2/250(0.008) | 2/174 (0.01) | rs148604152 | 0.008/0.005 | probably damaging (0.956)/tolerated (0.08) | yes | yes (strong) | |
| c.411C>T | Synonymous | p.Asn137Asn | 1/250(0.004) | 0/174 | rs142963365 | 0.002/0.001 | NA | no | nt | |
| c.679G>A | Non-synonymous | p.Gly227Arg | 1/250(0.004) | 0/269 | no | NA | benign (0.012)/NA | yes | Yes (strong) | |
Figure 3.
Genetic variants identified in RHOX genes in men with severe oligozoospermia. (A) Location of identified variants within RHOX gene structure. The homeodomain is shown in black, mutations are marked with red arrows and SNPs with grey arrows. (B) Chromatograms obtained from Sanger sequencing validate the mutations found by IonTorrent sequencing. For RHOXF1 c.515G > A and c.522C > T wild type sequence (left) and mutated sequence (right) are shown. In case of mutations -73C > G, c.202G > A, c.411C > T, and c.679G > A only one of the two RHOXF2 copies is affected resulting in a double peak at these positions. N = ambiguous call.
The effect of RHOX variants on protein function
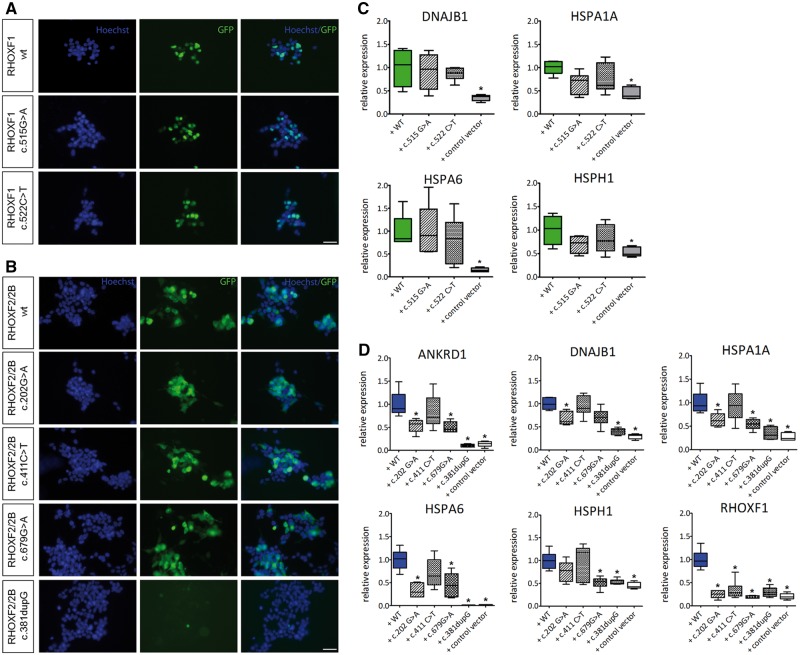
To determine the impact of the sequence variations on RHOX function, expression vectors with the identified modifications were generated by site-directed mutagenesis. A frame-shift mutation (c.381dupG) in RHOXF2/2B leading to a premature stop codon and therefore to a truncated protein without the functional homeodomain (p.L128Afs*34) was used as a positive control. Qualitative microscopic evaluation of the transfected cells revealed that all the mutant proteins exhibited normal localization (Fig. 4A and B), except for the truncated mutant, which lacks GFP expression due to the premature stop codon. Figure 4C and D shows the ability of these mutants to upregulate RHOX target genes that were identified in our microarray analysis (Fig. 2C). The RHOXF2/F2B truncation mutant, c.381dupG, failed to upregulate the RHOX target genes, as it exhibited comparable expression values to empty control vector transfection. Futhermore, the c.202G > A and c.679G > A mutations in RHOXF2/2B caused a significant reduction in the expression levels of the ANKRD1, DNAJB1, HSPA1A, HSPA6 and RHOXF1 genes compared to ectopic expression of wild-type RHOXF2/2B (P < 0.01; Fig. 4D). This indicated that these two mutations compromised the ability of RHOXF2/F2B to regulate the transcription of its targets. In contrast, the RHOXF1 mutations had no effect on target gene regulation (Fig. 4C).
Figure 4.
Functional analysis of RHOX mutations. (A) Immunofluorescent staining of HEK293 cells after 48h of transfection with RHOXF1 c.515G > A, c.522C > T and (B) RHOXF2/2B c.202G > A, c.411C > T, c.679G > A, and c.381dupG expression vectors. GFP signal reflects RHOX expression, nuclei were counterstained with Hoechst. Scale bar: 50 μm. (C) Gene expression levels of RHOXF1 DEGs after transfection with c.515G > A and c.522C > T expression vectors. (D) Gene expression levels of RHOXF2/2B DEGs after transfection with c.202G > A, c.411C > T, c.679G > A, and c.381dupG expression vectors. Transformed data are represented as whiskers with 5–95 percentiles. *Results are significant after Bonferroni correction for multiple testing (P < 0.01).
To study the impact of two common RHOXF2/2B SNPs, c.277G > A and c.451C > T (Fig. 3), the same experimental setup was used. This analysis revealed that the single-mutants, c.277G > A and c.451C > T, as well as the double-mutant, c.277G > A/c.451C > T, did not exhibit significantly altered expression of most of the RHOXF2/2B-regulated genes. In marked contrast, the RHOXF2/F2B target, RHOXF1, was no longer regulated by most of these mutants (Supplementary Material, Fig. S2A), possibly due to a direct interaction between RHOXF1 and RHOXF2/2B. We investigated the potential clinical impact of c.277G > A and c.451C > T in patients diagnosed with idiopathic infertility and found an association between SNPs with ejaculate volume, sperm count and morphology in a discovery cohort (n = 444, Supplementary Material, Fig. S2B). However, we could not confirm these results in a subsequently analysed confirmation cohort (n = 451, Supplementary Material, Fig. S2C).
Modelling of RHOX protein structure
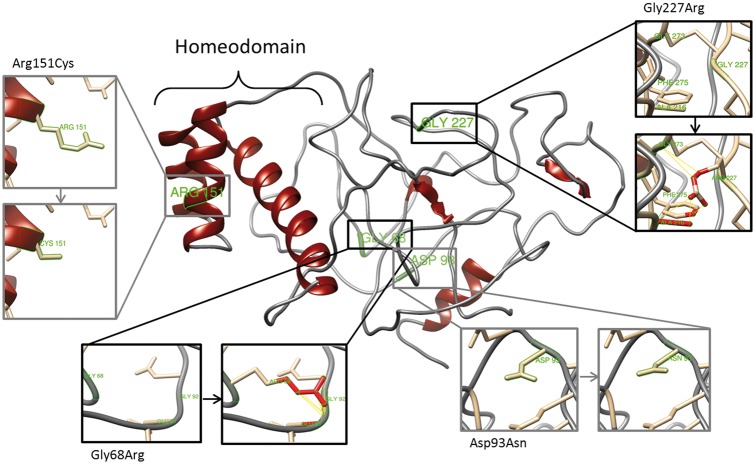
To determine the effect of the identified mutations on RHOX protein conformation we used three-dimensional homology modelling to predict the protein structure of RHOXF1 and RHOXF2/2B. Using Phyre2, the homeodomain region was modelled based on templates belonging to the homeodomain superfamily with high confidence (99%; Supplementary Materials, Fig. S3 and S4A). As a control, we used the p.L128Afs*34 RHOXF2/2B truncation, as this removes most of the homeodomain. Although the truncated protein had a low-confidence score (3%; Supplementary Material, Fig. S3), all models share the fact that c.381dupG leads to a frame shift mutation eight amino acids upstream of the homeodomain, which severely affects the mutual protein conformation (Supplementary Material, Fig. S4B and C). Using the full length RHOX protein, we analysed the structural consequences of the identified non-synonymous mutations and SNPs in this study. It was investigated whether the substituted amino acids lead to potential contacts (proximity) or clashes (direct interactions) with their surrounding amino acids, which could therefore result in an altered protein conformation. For RHOXF2/2B c.202G > A (p.Gly68Arg) and c.679G > A (p.Gly227Arg) we identified 14 and 5 potential clashes and contacts by substituting Glycine by Arginine, respectively (Fig. 5). Therefore, both mutations are expected to affect the position of the surrounding amino acids and thereby might affect the correct folding of this region. Furthermore, substitution of an uncharged amino acid by a charged amino acid, as is the case for Gly68Arg and Gly227Arg, will presumably change the surface charge of the protein and thereby lead to different binding properties. In contrast, no clashes and contacts were identified for the non-synonymous SNPs c.277G > A (p.Asp93Asn) and c.451C > T (p.Arg151Cys), making them unlikely to change RHOXF2/2B conformation. Analysing the structural effect of RHOXF1 mutations, c.515G > A (p.Arg172His) was predicted to lead to one potential contact with Pro175, which is directly located on the opposite side of Arg172 and therefore might change a loop formation (Supplementary Material, Fig. S4D). This approach corroborated experimental data, in which predicted conformational changes induced by mutations lead to altered transcription of target genes caused by changed RHOX function.
Figure 5.
Effect of non-synonymous mutations on RHOXF2/2B protein structure. Diagram of the top predicted tertiary structure model of RHOXF2/2B using Phyre2. The homeodomain region was modeled with template structures of homeodomain containing proteins and the remaining residues ab initio. The model was colored by secondary structure using UCSF Chimera software: helix structures are shown in red and coiled structures are shown in grey. Each insert shows the identified non-synonymous mutations (grey) and SNPs (black) as sticks. Predicted clashes/contacts for c.202G > A (p.Gly68Arg) and c.679G > A (p.Gly227Arg) are marked with yellow lines (14 and 5, respectively) and atoms involved in the potential contacts are highlighted in red (10 and 8, respectively). No clashes/contacts were predicted for c.277G > A (p.Asp93Asn) and c.451C > T (p.Arg151Cys).
Discussion
Spermatogenesis is a highly complex process that requires the concerted action of a large number of genes. Mutations in genes located on the X chromosome have a complete penetrance in the male population due to their hemizygous state and are therefore of primary interest to cause spermatogenic failure. One set of X-linked genes that are strong candidates to have a role in human infertility are those in the RHOX gene cluster. They encode transcription factors that are selectively expressed in the reproductive tract; in humans this gene cluster is mainly expressed in male and female germ cells (12–15). Mice with loss or depletion of Rhox genes exhibit male reproductive defects (12,20) and human RHOX gene promoters are hypermethylated in sperm from men with abnormal semen parameters (14). Here, we report the identification of specific genes regulated by the human RHOX transcription factors and we identify specific mutations in RHOX genes that disrupt their ability to regulate transcription.
One of the most striking findings of our investigation was that several RHOX-regulated genes encode chaperons belonging to the HSP70 family, including HSPA1A, HSPA2, HSPA6 and HSPH1 (Table 1, Fig. 2A–C). Such factors are critical for adaptive responses to stress and thus cell survival (33). Several of these RHOX-regulated genes have been shown to have crucial roles in human male reproduction. For example, idiopathic infertile men who completely lack HSPA2 in their sperm exhibit impaired fertilizing ability (31,34) and reduced expression of HSPA1A in ejaculated sperm has been implicated in the pathogenesis of male infertility (35). Very recently, the aberrant methylation pattern of two closely related genes of HSPA1A, HSPA1B and HSPA1L, was associated with reduced fecundity (30). DNAJB1 belongs to the HSP40 family and encodes a protein in human spermatozoa (29) that has also been localized to the sperm head and tail of rodent spermatozoa (36). Given that HSP70 family members and other heat shock-induced proteins protect cells from heat stress, it is tempting to speculate that RHOX transcription factors regulate these genes to protect germ cells from heat stress. In support of this notion, spermatogenesis normally occurs at temperatures well below body temperature and thus deviations above this would require protection mechanisms. This is underscored by the fact that germ cells (particularly spermatocytes and spermatids) are exquisitely sensitive to elevated temperatures (37). Our experimental design, using a control consisting of cells transfected with an empty vector, excludes that the heat shock proteins might be upregulated due to the transfection procedure.
Interestingly, we discovered that RHOXF1 is regulated by RHOXF2/2B (Table 1, Fig. 2A–C) and thus it is likely that RHOXF1 is a RHOXF2/F2B target gene. As evidence that this regulatory relationship is physiologically significant, RHOXF2 protein is expressed in earlier stages of germ cell development than RHOXF1 (Fig. 2D; 13). We suggest that these two transcription factors function in ‘a cascade regulatory circuit’ in which RHOXF2/F2B not only regulates genes critical for early stages of spermatogenesis (e.g. in spermatogonia and leptotene and pachytene spermatocytes) but is also responsible for driving the expression of RHOXF1, which serves as the next transcription factor in a temporal development pathway by regulating genes critical for later stages of spermatogenesis (e.g. in spermatids). In support of the notion that RHOXF2/F2B regulates RHOXF1, we identified several genes co-regulated by these two transcription factors (Supplementary Material, Table S3). This regulative scenario also enables RHOXF1 and RHOXF2/2B to maintain some common functions despite major sequence differences and distinct expression pattern. We note that even though RHOXF1 was not detectable after the round spermatid stage (13), its influence may extend beyond this stage, as many mRNAs transcribed in the round spermatid stage are translated and function at later stages (38).
Another major finding of our study was the discovery that human idiopathic infertile patients have RHOX mutations. We identified two mutations in RHOXF1 and four in RHOXF2/2B in patients with severe oligozoospermia, of which only one (c.202G > A) was also detected in the control group with normal sperm concentrations (Table 2). We note that a similar mutation frequency was observed in the ExAC database (exomes from 60,706 individuals derived from different studies [http://exac.broadinstitute.org/about]) which can be explained by the fact the ExAC dataset includes exomes from infertile men as well. Interestingly, we observed more genetic variants for RHOXF2/2B than RHOXF1, consistent with a previous study showing that there was a more rapid evolution of RHOXF2/2B than RHOXF1 in primates (24). Most of the RHOXF2/2B variants we identified were located in exon 2 (Fig. 3A). This encodes the homeodomain N-terminal region, which is of interest given that this region is involved in protein-protein interactions that modulate homebox transcription factor target gene specificity and the transcription rate (19,23). We suggest that mutations in this region might disrupt functional interactions of RHOX proteins with other proteins (such as heterodimeric transcription factor partners) and might even allow RHOX proteins to gain new functional properties.
Our functional analysis showed that both the c.202G > A and c.679G > A mutations significantly impaired the ability of RHOXF2/2B to upregulate the expression of several of its downstream genes (Fig. 4D). This critical data indicated that the c.202G > A and c.679G > A mutations impair the ability of RHOXF2/2B to function. Therefore, these mutations have the potential to impair the in vivo function of RHOXF2/F2B and thus perturb human spermatogenesis. A caveat is that we used a cell line for these studies and thus our results do not necessarily reflect the in vivo situation. Nevertheless, we could show that the identified target genes were also expressed within the testis (Fig. 2E) and data available at the human protein atlas repository (39) display their germ cell specific expression at the protein level. Thus, the identified RHOX network is presumably not restricted to the HEK293 cell system. Moreover, our protein modelling indicated that both the c.202G > A and c.679G > A mutations are predicted to alter the tertiary structure of RHOXF2/2B protein (Fig. 5), providing further weight to the argument that these mutations cause deleterious effects. Both mutations lead to a Glycine being replaced by an Arginine, whose larger amino acid side chain could lead to altered protein folding as a result of steric interactions with the surrounding molecules. Furthermore, the mutated amino acid side chains change from a neutral charge to a positive charge. These alterations, in turn, can lead to modified binding properties of already existing target genes or enable novel interactions with different cofactors to regulate a new set of targets. The c.202G > A mutation impacts the region upstream of the homeodomain, a region that is known to be a protein-protein interaction domain in other homeobox proteins (19,23). Our finding that the c.202G > A mutation is present with a similar frequency in the control group indicates that this mutation alone is not sufficient to cause severe oligozoospermia. Instead, this mutation might predispose individuals to infertility and/or cause a more modest (subfertile) phenotype. In contrast to c.202G > A, we observed that c.679G > A only occurred in our patient group which indicates that this mutation might cause severe oligozoospermia. As a limitation of this study, we could not distinguish whether mutations occurred in RHOXF2 or RHOXF2B, as the two gene paralogs are almost identical (13,14). In the future, it will be interesting to identify men who are double hemizygous (i.e. have loss-of-function mutations in both RHOXF2 and RHOXF2B), as this would allow one to exclude compensatory mechanisms that might mask a more severe effect of complete loss of RHOXF2 function.
A relatively unusual feature of RHOX proteins is their cytoplasmatic accumulation. In contrast, most homeobox proteins and other transcription factors are localized in the nucleus, where they bind DNA and modulate transcription. Only in exceptional cases are transcription factors retained in the cytoplasm or exported from the nucleus to the cytoplasm, where they are either sequestered from regulating transcription or confer different activities in the cytoplasm, such as modulating translation (40–42). RHOXF1 and RHOXF2 differ in their subcellular localization. In the foetal testes, RHOXF1 is predominantly expressed in the nucleus, while RHOXF2/2B is mostly restricted to the cytoplasm (13). We observed the same differential localization pattern of these two proteins in transfected HEK293 cells (Fig. 1A). In contrast, RHOXF1 and RHOXF2/2B are expressed in both the nucleus and the cytoplasm of adult testes (Fig. 2D;13). Because RHOX proteins are relatively small (RHOXF1: 20.5 kDa, RHOXF2/2B: 31.6 kDa; 13), they do not require active mechanisms to pass through the nuclear pore into the cytoplasm and thus their presence in the cytoplasm may simply reflect excess proteins that are not actively regulating translation.
In this work, we coupled genetic and transcriptional analyses with functional validation and in silico modelling to uncover molecular mechanisms by which RHOX transcription factors function in the context of male fertility. We suggest that the RHOX transcription factors and some of their downstream genes represent new potential targets to be use for the diagnostics and treatment of male infertility.
Materials and Methods
Study population
All subjects in this study were attending the Department of Clinical Andrology at the Centre of Reproductive Medicine and Andrology (Münster, Germany) for a couple infertility. Semen analysis was performed according to the 2010 criteria of the World Health Organization (WHO; 43) and all men underwent a complete physical examination including scrotal ultrasonography. In order to detect rare sequence variations in RHOX genes, a cohort of 250 idiopathic infertile men with severe oligozoospermia (total sperm count ≤10×106, concentration ≤5x106 sperm per millilitre) was retrospectively selected using the Androbase database (44). Patients with known clinical (e.g. maldescended testes, testicular tumours, infections of the genital tract) or genetic (karyotype anomalies, Y-chromosomal deletions) causes of infertility were excluded. Genotyping of SNPs c.277G > A and c.451C > T in RHOXF2/2B was performed in a discovery cohort of 444 idiopathic infertile patients (45). The subsequent confirmation cohort comprised 451 prospectively screened patients (46) and 174 men with normal sperm concentrations (>39 × 106 sperm per millilitre) served as a control group (11). For RHOXF2/2B c.679G > A, 95 additional controls were genotyped.
All participants provided informed written consent for evaluation of their clinical data and agreed to genetic analysis as approved by the Ethics Committee of the University and the state medical board (Az4INie).
DNA isolation, library preparation and sequencing
Genomic DNA was isolated from 1ml EDTA-preserved blood using the FlexiGene DNA extraction kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The quantity and quality of DNA was evaluated using NanoDrop1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and all samples were diluted to 50 ng/µl and pooled (N = 20). Prior to sequencing, RHOXF1 (NM_139282.2) and RHOXF2/2B (NM_032498.1/NM_001099685.1) exons (including exon-intron boundaries) were amplified via polymerase chain reaction (PCR). Primers are listed in Supplementary Material, Table S6. Libraries were constructed using the Ion Xpress™ Plus Fragment Library Kit according to the manufacturer’s instructions and sequenced on the Ion Torrent Personal Genome Machine (PGM™, Thermo Fisher Scientific). Data analysis was performed using GensearchNGS (PhenoSystems SA Wallonia, Belgium) using hg19 as reference genome (https://genome.ucsc.edu/; date last accessed March 16, 2016). Gene fragments were sequenced with an average coverage of at least 3041 reads each, and the results were filtered to ≥80% quality, 2–10% frequency and ≥20% balance. Frequencies of already known genetic variants were obtained from the (ExAC) database (Cambridge, MA, URL: http://exac.broadinstitute.org) and the sequences (with the novel mutations annotated) were submitted to the ENA (LT601603, LT601604).
SNP genotyping
Genomic DNA was isolated using DNA Extract All Reagents Kit (Applied Biosystems, Carlsbad, CA, USA) according to the product protocol and samples were stored at 4 °C over night before use. TaqMan GTXpress Master Mix and a customized TaqMan genotyping assay mix (Applied Biosystems, Assay no. AHOI4J0, AHLJSWI) were used for genotyping RHOXF2/2B c.277G > A (rs146311958), c.451C > T (rs40785) and c.679G > A with a StepOnePlus Real-Time PCR-System. Primer and reporter sequence of the designed TaqMan Assays are shown in the Supplementary Material Table S6. For each sample, the reaction mix consisted of 2 µl isolated DNA was added to 5 µl Master Mix, 0,3 µl Assaymix and 2,7 µl sterile water. Fast run thermal cycling conditions were as follows: 20 s at 25 °C followed by 20 s at 95 °C and 40 cycles of 3 s at 95 °C plus 20 s at 60 °C.
Site-directed mutagenesis
Mutations and SNPs identified in RHOXF1 (c.515G > A [rs2301977], c.522C > T) and RHOXF2/2B (c.202G > A [rs14860 4152], c.277G > A [rs146311958], c.411C > T [rs142963365], c.451C > T [rs40785], c.679G > A and c.381dupG) were introduced into expression vectors using site-directed mutagenesis (QuickChangeII Site-directed mutagenesis kit; Stratagene, Heidelberg, Germany) according to the manufacturer’s instructions. Primers were designed with Agilent’s online tool QuikChange Primer Design (www.genomics.agilent.com) and are listed in Supplementary Material, Table S6. Successful mutagenesis was confirmed by Sanger sequencing.
Cell culture and transient transfection of RHOX
Human Embryonic Kidney 293 (HEK293) cells (CLS, Eppelheim, Germany) were cultured in DMEM/F12 1:1 (Gibco, Carlsbad, CA, USA) containing 10% (v/v) FCS (PAA Laboratories, Pasching, Austria), 1% penicillin/streptomycin (Gibco) and 2 mM L-Glutamine (Gibco) at 37 °C and 5% CO2 (v/v). Twenty four hours prior to transfection, HEK293 cells were seeded in 24-well plates (9×105) containing round coverslips (Ø 10mm, Medishop, Markgröningen, Germany). For transient transfection, wild type contructs (RHOXF1: RG217618, RHOXF2: RG205957, pCMV6-AC-GFP: PS100010, OriGene, Rockville, MD, USA) and mutated constructs (see site-directed mutagenesis) were mixed with Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) in a 1:2.5 ratio. Cells were covered with 475 ml serum-free medium (DMEM/F12) and 25 µl of the prepared DNA-lipofectamine complex was added. Each DNA construct was transfected in quadruplicates and the medium was replaced after 5h with medium containing 10% (v/v) FCS. After 24h and 48h, cells were washed with PBS and two wells per construct were suspended in 300 µl Lysis Buffer (Total RNA Isolation Kit; Agilent, Santa Clara, CA, USA) and stored at −80 °C for RNA isolation. Cells of the remaining 2 wells were fixed in 4% paraformaldehyde for 20 min at 37 °C and nuclei were counterstained with Hoechst 33258 (1:1000, Sigma Aldrich, St. Louis, MO, USA). Images were obtained with inverted microscope Axio Observer.Z1 and Axio Vision Viewer 4.8 (Zeiss, Oberkochen, Germany).
Microarray analysis
Total RNA was isolated using Total RNA Isolation Kit (Agilent) according to the protocol provided by the manufacturer. The quality of the RNA was assessed with the Bioanalyzer 2100 (Agilent) and only samples with an RNA integrity number (RIN) > 7 were included in the analysis. The Affymetrix GeneChip Human Exon 1.0 ST array (Affymetrix, Santa Clara, CA, USA) was used to evaluate changes in mRNA expression in RHOX transfected samples compared to control transfected samples (24h and 48h after transfection, analysed in biological triplicates). Samples were processed as previously described (47). Normalization, quality control, and output file (CHP format) generation were performed with Affymetrix Gene Expression Console v.3.1 using the Robust Multi-Chip Analysis (RMA) algorithm. DEGs were identified with Affymetrix transcriptome analysis console (TAC) v.2.0 (Affymetrix). Data were obtained using one-way analysis of variance (ANOVA) and p-value correction according to Benjamini and Hochberg False Discovery Rate (FDR) for multiple testing. Genes with a fold change in expression of at least 1.5 and uncorrected P-values ≤ 0.05 were considered as statistically significant DEGs. Gene ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis were performed using DAVID (https://david.ncifcrf.gov; date last accessed April 14, 2016; 48). The P-value < 0.05 and count > 2 were applied as cut-off criteria to identify over-represented biological categories. For RHOXF1 no significant GO could be determined due to low numbers of DEGs Ingenuity pathway analysis (IPA, Qiagen, www.qiagen.com/ingenuity) software was used to evaluate molecule connectivity. Genes of interest were assigned to specific biologically significant networks based on known associations in the databases and ranked as to their biological relevance. Microarray data obtained in this study were submitted to the GEO database (accession number: GSE84463).
qRT-PCR
RNA was isolated as described above. A total of 500 ng of mRNA were transcribed into cDNA using iScript cDNA synthesis kit according to the manufacturer’s protocol (Bio-Rad, Hercules, CA, USA). Quantitative real-time PCR analyses were conducted using SYBR®-Green reagents on a StepOnePlus PCR system (Applied Biosystems). All samples were measured in duplicates and data analysis was performed using the StepOnePlus software 2.2.1. (Applied Biosystems). Primer sequences are listed in Supplementary Material, Table S6. The results were normalized using GAPDH as internal control and shown as 2-ΔCt values (49).
Western blot analysis
Western blotting analysis was performed as previously described (13). Briefly, 50µg of total cell lysates was electrophoresed in 12% SDS-PAGE transferred to Immobilon-P PVDF membrane (Millipore) and probed with custom generated antibodies against RHOXF1 or RHOXF2/2B (ProteinTech Group Inc., Chicago, IL; 13) and β -ACTIN (Sigma Chemical Co., St. Louis, MO). The membranes were washed 3×10 min with 0.1% Tween-20 in phosphate-buffered saline and then incubated for 1h at room temperature with the secondary antibody (ECL anti-rabbit or anti-mouse; GE Healthcare UK). Afterwards, membranes were developed using the Super Signal West Pico Chemiluminescent substrate (Thermo Scientific, Rockford, IL).
Immunohistochemical analysis
Immunohistochemical analysis was performed on paraffin-embedded testis tissue that were fixed in Bouin’s solution. Sections (5 µm) were deparaffinized two times in Paraclear before rehydration in serial dilutions of ethanol. Heat-induced antigen retrieval was performed in citrate buffer (pH 6) in a microwave. Internal peroxidase activity was quenched by incubation with H2O2 for 15 min and incubated with 20% goat serum diluted in 5% bovine serum albumin (BSA) for 30 min at room temperature (RT). Sections were blocked with Avidin and Biotin for 15 min each (SP-2001 Vector Laboratory, Inc.) and incubated over night at 4 °C against primary antibodies for RHOXF1 (1:200 dilution) or RHOXF2/2B (1:300 dilution). Immunoglobulin G (IgG; 1:1000 dilution) and blocking serum (without antibody) served as negative controls. Primary antibodies were detected using a secondary antibody from the ABC kit (PK-4001, Vector Laboratory, Inc.) following the manufacturers’ instructions and incubation with 3,3′-diaminobenzidine tetrahydrochloride (DAB, Sigma D-4168) substrate. Nuclei were counterstained with hematoxylin (Sigma, St. Louis, USA) and sections were dehydrated in a rising ethanol row, followed by incubation in Paraclear. Finally, sections were mounted with Merckoglas® (Merck 1.03973.0001), viewed with an Olympus BX61 microscope (Olympus Corporation, Tokio, Japan) and images were recorded with a Retiga 4000R camera (QImaging, Surrey, Canada). Expression data of RHOX target genes are available in the protein atlas (http://www.proteinatlas.org; date last accessed July 15, 2016).
Protein modelling and visualization
RHOXF1 (NP_644811.1) and RHOXF2 (NP_115887.1) amino acid sequence was submitted to the I-TASSER (50) and Phyre2 (51) servers for the prediction of the three dimensional structure of RHOX protein. A list of templates used for homology based model generation by Phyre2 is given in Supplementary Material, Fig. 3. In total, 149 residues (81%) and 95 residues (33%) were modelled at >90% accuracy for RHOXF1 and RHOXF2/2B, respectively. The altered amino acid sequence of truncated RHOXF2/2B L128Afs*34 was determined (Supplementary Material, Fig. S4C) and the tertiary structure modelled as for full length RHOX. The best predicted models were selected and visualized using UCSF Chimera v.1.10.2. Mutated amino acids for the identified non-synonymous mutations and SNPs were introduced with the chimera rotamere tool. Highest ranked rotamers were chosen for each variation and used to identify interatomic clashes and contacts of the substituted amino acid with the surrounding atoms.
Statistical analysis
Significant differences between RHOX expression and RHOX target gene expression levels were obtained using Mann-Whitney test with a 95% confidence level. Adjustment of p-values for multiple testing was performed using the Bonferroni post hoc test. All statistical calculations were performed using GraphPad Prism v.5.00 (GraphPad Software, La Jolla, CA, USA), and the results were presented as mean ± SEM unless stated otherwise.
Supplementary Material
Supplementary Material is available at HMG online.
Supplementary Material
Acknowledgements
The authors would like to thank the patients and attending clinicians involved in this study, Elisabeth Lahrmann, Reinhild Sandhowe, Nicole Terwort, Sylvia Fleige-Menzen and Laura Hankamp for their excellent technical assistance and Andreas Huge, PhD, for expert assistance and advice concerning bioinformatic analysis.
Conflict of Interest statement. None declared.
Funding
The study was supported by the German Research Foundation (DFG) collaborative grant (GE1547/15-1) and by Innovative Medizinische Forschung (IMF, I-LA121315).
References
- 1. Tüttelmann F., Gromoll J., Kliesch S. (2008) Genetics of male infertility. Der. Urol., 47, 1561–1562. 1564–1567. [DOI] [PubMed] [Google Scholar]
- 2. Tüttelmann F., Gromoll J. (2010) Novel genetic aspects of Klinefelter’s syndrome. Mol. Hum. Reprod., 16, 386–395. [DOI] [PubMed] [Google Scholar]
- 3. Bansal S.K., Jaiswal D., Gupta N., Singh K., Dada R., Sankhwar S.N., Gupta G., Rajender S. (2016) Gr/gr deletions on Y-chromosome correlate with male infertility: an original study, meta-analyses, and trial sequential analyses. Sci. Rep., 6, 19798.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang P.J., McCarrey J.R., Yang F., Page D.C. (2001) An abundance of X-linked genes expressed in spermatogonia. Nat. Genet., 27, 422–426. [DOI] [PubMed] [Google Scholar]
- 5. Mueller J.L., Skaletsky H., Brown L.G., Zaghlul S., Rock S., Graves T., Auger K., Warren W.C., Wilson R.K., Page D.C. (2013) Independent specialization of the human and mouse X chromosomes for the male germ line. Nat. Genet., 45, 1083–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Krausz C., Giachini C., Giacco D.L., Daguin F., Chianese C., Ars E., Ruiz-Castane E., Forti G., Rossi E. (2012) High resolution X chromosome-specific array-CGH detects new CNVs in infertile males. PLoS One, 7, e44887.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mou L., Xie N., Yang L., Liu Y., Diao R., Cai Z., Li H., Gui Y. (2015) A Novel Mutation of DAX-1 Associated with Secretory Azoospermia. PLoS One, 10, e0133997.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Stouffs K., Lissens W. (2012) X chromosomal mutations and spermatogenic failure. Biochim. Biophys. Acta, 1822, 1864–1872. [DOI] [PubMed] [Google Scholar]
- 9. O’Hara L., Smith L.B. (2015) Androgen receptor roles in spermatogenesis and infertility. Best Pract. Res. Endocrinol. Metab., 29, 595–605. [DOI] [PubMed] [Google Scholar]
- 10. Yang F., Silber S., Leu N.A., Oates R.D., Marszalek J.D., Skaletsky H., Brown L.G., Rozen S., Page D.C., Wang P.J. (2015) TEX11 is mutated in infertile men with azoospermia and regulates genome-wide recombination rates in mouse. EMBO Mol. Med., 7, 1198–1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yatsenko A.N., Georgiadis A.P., Ropke A., Berman A.J., Jaffe T., Olszewska M., Westernstroer B., Sanfilippo J., Kurpisz M., Rajkovic A., et al. (2015) X-linked TEX11 mutations, meiotic arrest, and azoospermia in infertile men. N. Engl. J. Med., 372, 2097–2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. 2nd J.A.M., Chen M.A., Wayne C.M., Bruce S.R., Rao M., Meistrich M.L., Macleod C., Wilkinson M.F. (2005) Rhox: a new homeobox gene cluster. Cell, 120, 369–382. [DOI] [PubMed] [Google Scholar]
- 13. Song H.W., Anderson R.A., Bayne R.A., Gromoll J., Shimasaki S., Chang R.J., Parast M.M., Laurent L.C., de Rooij D.G., Hsieh T.C., et al. (2013) The RHOX homeobox gene cluster is selectively expressed in human oocytes and male germ cells. Hum. Reprod., 28, 1635–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Richardson M.E., Bleiziffer A., Tüttelmann F., Gromoll J., Wilkinson M.F. (2014) Epigenetic regulation of the RHOX homeobox gene cluster and its association with human male infertility. Hum. Mol. Genet., 23, 12–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Frainais C., Kannengiesser C., Albert M., Molina-Gomes D., Boitrelle F., Bailly M., Grandchamp B., Selva J., Vialard F. (2014) RHOXF2 gene, a new candidate gene for spermatogenesis failure. Basic Clin. Androl., 24, 3–4190. 24–3. eCollection 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. McGinnis W., Krumlauf R. (1992) Homeobox genes and axial patterning. Cell, 68, 283–302. [DOI] [PubMed] [Google Scholar]
- 17. 2nd J.A.M., Wilkinson M.F. (2010) The Rhox genes. Reproduction, 140, 195–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Geserick C., Weiss B., Schleuning W.D., Haendler B. (2002) OTEX, an androgen-regulated human member of the paired-like class of homeobox genes. Biochem. J., 366, 367–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wayne C.M., MacLean J.A., Cornwall G., Wilkinson M.F. (2002) Two novel human X-linked homeobox genes, hPEPP1 and hPEPP2, selectively expressed in the testis. Gene, 301, 1–11. [DOI] [PubMed] [Google Scholar]
- 20. Welborn J.P., Davis M.G., Ebers S.D., Stodden G.R., Hayashi K., Cheatwood J.L., Rao M.K. and 2nd ,J.A.M. (2015) Rhox8 Ablation in the Sertoli Cells Using a Tissue-Specific RNAi Approach Results in Impaired Male Fertility in Mice. Biol. Reprod., 93, 8.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Swanson W.J., Vacquier V.D. (2002) The rapid evolution of reproductive proteins. Nat. Rev, 3, 137–144. [DOI] [PubMed] [Google Scholar]
- 22. Torgerson D.G., Singh R.S. (2003) Sex-linked mammalian sperm proteins evolve faster than autosomal ones. Mol. Biol. Evol., 20, 1705–1709. [DOI] [PubMed] [Google Scholar]
- 23. Sutton K.A., Wilkinson M.F. (1997) Rapid evolution of a homeodomain: evidence for positive selection. J. Mol. Evol., 45, 579–588. [DOI] [PubMed] [Google Scholar]
- 24. Niu A.L., Wang Y.Q., Zhang H., Liao C.H., Wang J.K., Zhang R., Che J., Su B. (2011) Rapid evolution and copy number variation of primate RHOXF2, an X-linked homeobox gene involved in male reproduction and possibly brain function. BMC Evol. Biol., 11, 298.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Oda M., Yamagiwa A., Yamamoto S., Nakayama T., Tsumura A., Sasaki H., Nakao K., Li E., Okano M. (2006) DNA methylation regulates long-range gene silencing of an X-linked homeobox gene cluster in a lineage-specific manner. Genes Dev., 20, 3382–3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Maclean J.A., Bettegowda A., Kim B.J., Lou C.H., Yang S.M., Bhardwaj A., Shanker S., Hu Z., Fan Y., Eckardt S., et al. (2011) The rhox homeobox gene cluster is imprinted and selectively targeted for regulation by histone h1 and DNA methylation. Mol. Cell. Biol., 31, 1275–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bhardwaj A., Song H.W., Beildeck M., Kerkhofs S., Castoro R., Shanker S., Gendt K.D., Suzuki K., Claessens F., Issa J.P., et al. (2012) DNA demethylation-dependent AR recruitment and GATA factors drive Rhox5 homeobox gene transcription in the epididymis. Mol. Endocrinol., 26, 538–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Graham F.L., Smiley J., Russell W.C., Nairn R. (1977) Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol., 36, 59–74. [DOI] [PubMed] [Google Scholar]
- 29. Mitchell L.A., Nixon B., Aitken R.J. (2007) Analysis of chaperone proteins associated with human spermatozoa during capacitation. Mol. Hum. Reprod., 13, 605–613. [DOI] [PubMed] [Google Scholar]
- 30. Jenkins T.G., Aston K.I., Meyer T.D., Hotaling J.M., Shamsi M.B., Johnstone E.B., Cox K.J., Stanford J.B., Porucznik C.A., Carrell D.T. (2016) Decreased fecundity and sperm DNA methylation patterns. Fertil. Steril., 105, 51–57e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Scieglinska D., Krawczyk Z. (2015) Expression, function, and regulation of the testis-enriched heat shock HSPA2 gene in rodents and humans. Cell Stress Chaperones, 20, 221–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ji G., Long Y., Zhou Y., Huang C., Gu A., Wang X. (2012) Common variants in mismatch repair genes associated with increased risk of sperm DNA damage and male infertility. BMC Med., 10, 49.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Noonan E.J., Place R.F., Giardina C., Hightower L.E. (2007) Hsp70B’ regulation and function. Cell Stress Chaperones, 12, 393–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Redgrove K.A., Nixon B., Baker M.A., Hetherington L., Baker G., Liu D.Y., Aitken R.J. (2012) The molecular chaperone HSPA2 plays a key role in regulating the expression of sperm surface receptors that mediate sperm-egg recognition. PLoS One, 7, e50851.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Cedenho A.P., Lima S.B., Cenedeze M.A., Spaine D.M., Ortiz V., Oehninger S. (2006) Oligozoospermia and heat-shock protein expression in ejaculated spermatozoa. Hum. Reprod., 21, 1791–1794. [DOI] [PubMed] [Google Scholar]
- 36. Doiguchi M., Kaneko T., Urasoko A., Nishitani H., Iida H. (2007) Identification of a heat-shock protein Hsp40, DjB1, as an acrosome- and a tail-associated component in rodent spermatozoa. Mol. Reprod. Dev., 74, 223–232. [DOI] [PubMed] [Google Scholar]
- 37. Gau B.H., Chu I.M., Huang M.C., Yang K.T., Chiou S.H., Fan Y.H., Chen M.Y., Lin J.H., Chuang C.K., Huang S.Y., et al. (2008) Transcripts of enriched germ cells responding to heat shock as potential markers for porcine semen quality. Theriogenology, 69, 758–766. [DOI] [PubMed] [Google Scholar]
- 38. Chalmel F., Rolland A.D. (2015) Linking transcriptomics and proteomics in spermatogenesis. Reproduction, 150, R149–R157. [DOI] [PubMed] [Google Scholar]
- 39. Uhlen M., Fagerberg L., Hallstrom B.M., Lindskog C., Oksvold P., Mardinoglu A., Sivertsson A., Kampf C., Sjostedt E., Asplund A., et al. (2015) Proteomics. Tissue-based map of the human proteome. Science, 347, 1260419.. [DOI] [PubMed] [Google Scholar]
- 40. Gorlich D., Kutay U. (1999) Transport between the cell nucleus and the cytoplasm. Annu. Rev. Cell Dev. Biol., 15, 607–660. [DOI] [PubMed] [Google Scholar]
- 41. Niessing D., Blanke S., Jackle H. (2002) Bicoid associates with the 5’-cap-bound complex of caudal mRNA and represses translation. Genes Dev., 16, 2576–2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cho P.F., Poulin F., Cho-Park Y.A., Cho-Park I.B., Chicoine J.D., Lasko P., Sonenberg N. (2005) A new paradigm for translational control: inhibition via 5’-3' mRNA tethering by Bicoid and the eIF4E cognate 4EHP. Cell, 121, 411–423. [DOI] [PubMed] [Google Scholar]
- 43. Organization,W.H. (2010) WHO Laboratory Manual for the Examination and Processing of Human Semen. [PubMed]
- 44. Tüttelmann F., Luetjens C.M., Nieschlag E. (2006) Optimising workflow in andrology: a new electronic patient record and database. Asian J. Androl., 8, 235–241. [DOI] [PubMed] [Google Scholar]
- 45. Tüttelmann F., Laan M., Grigorova M., Punab M., Sober S., Gromoll J. (2012) Combined effects of the variants FSHB -211G>T and FSHR 2039A>G on male reproductive parameters. J. Clin. Endocrinol. Metab., 97, 3639–3647. [DOI] [PubMed] [Google Scholar]
- 46. Busch A.S., Kliesch S., Tüttelmann F., Gromoll J. (2015) FSHB -211G>T stratification for follicle-stimulating hormone treatment of male infertility patients: making the case for a pharmacogenetic approach in genetic functional secondary hypogonadism. Andrology, 3, 1050–1053. [DOI] [PubMed] [Google Scholar]
- 47. Zitzmann M., Bongers R., Werler S., Bogdanova N., Wistuba J., Kliesch S., Gromoll J., Tüttelmann F. (2015) Gene expression patterns in relation to the clinical phenotype in Klinefelter syndrome. J. Clin. Endocrinol. Metab., 100, E518–E523. [DOI] [PubMed] [Google Scholar]
- 48. Dennis G., Jr, Sherman B.T., Hosack D.A., Yang J., Gao W., Lane H.C., Lempicki R.A. (2003) DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol., 4, P3. [PubMed] [Google Scholar]
- 49. Schmittgen T.D., Livak K.J. (2008) Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc., 3, 1101–1108. [DOI] [PubMed] [Google Scholar]
- 50. Zhang Y. (2008) I-TASSER server for protein 3D structure prediction. BMC Bioinformatics, 9, 40.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J. (2015) The Phyre2 web portal for protein modelling, prediction and analysis. Nat. Protoc., 10, 845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.