Abstract
Background Several approaches are available for evaluating heterogeneity in meta-analysis. Sensitivity analyses are often used, but these are often implemented in various non-standardized ways.
Methods We developed and implemented sequential and combinatorial algorithms that evaluate the change in between-study heterogeneity as one or more studies are excluded from the calculations. The algorithms exclude studies aiming to achieve either the maximum or the minimum final I2 below a desired pre-set threshold. We applied these algorithms in databases of meta-analyses of binary outcome and ≥4 studies from Cochrane Database of Systematic Reviews (Issue 4, 2005, n = 1011) and meta-analyses of genetic associations (n = 50). Two I2 thresholds were used (50% and 25%).
Results Both algorithms have succeeded in achieving the pre-specified final I2 thresholds. Differences in the number of excluded studies varied from 0% to 6% depending on the database and the heterogeneity threshold, while it was common to exclude different specific studies. Among meta-analyses with initial I2 > 50%, in the large majority [19 (90.5%) and 208 (85.9%) in genetic and Cochrane meta-analyses, respectively] exclusion of one or two studies sufficed to decrease I2 < 50%. Similarly, among meta-analyses with initial I2 > 25%, in most cases [16 (57.1%) and 382 (81.3%), respectively) exclusion of one or two studies sufficed to decrease heterogeneity even <25%. The number of excluded studies correlated modestly with initial estimated I2 (correlation coefficients 0.52–0.68 depending on algorithm used).
Conclusions The proposed algorithms can be routinely applied in meta-analyses as standardized sensitivity analyses for heterogeneity. Caution is needed evaluating post hoc which specific studies are responsible for the heterogeneity.
Keywords: Heterogeneity, sensitivity analysis, sequential algorithm, meta-analysis
Introduction
Assessment of the between-study heterogeneity is an essential component of meta-analysis.1 Lack of consistency may reflect genuine differences in the design and conduct of the studies (methodological heterogeneity) or in participants, interventions, exposures or outcomes evaluated (clinical heterogeneity).2,3 Lack of consistency may also herald errors, biases or pure chance. Errors and biases may affect single studies (e.g. quality problems) or research fields at large, e.g. publication and other selective reporting biases.
Sources of heterogeneity need to be carefully examined on a case-by-case basis.4 A common approach is to perform sensitivity analyses, where one study is excluded at a time and the impact of removing each of the studies is evaluated on the summary results and the between-study heterogeneity.5 Such ‘one-out’ sensitivity analyses can tell us whether the summary effect and heterogeneity are heavily influenced by a particular study. However, it is possible that not one but several studies are primarily responsible for the between-study heterogeneity. One could generalize into sensitivity analyses where many studies are removed, according to some order. Identification of ‘outlying’ studies may also offer some post hoc hints for explaining the reasons of between-study heterogeneity.
Here, we present algorithms that evaluate the change in between-study heterogeneity as one or more studies are excluded sequentially or in combination from the meta-analysis calculations. Our aim is to show how these algorithms can be routinely adopted in meta-analyses as standardized sensitivity analyses for heterogeneity. Accordingly, we have empirically examined their performance in many meta-analyses of clinical trials and genetic epidemiology. Illustrative examples are also discussed in detail.
Methods
Several statistical tests are routinely used to assess overall the extent and statistical significance of heterogeneity. The most common test that examines the null hypothesis that all studies are evaluating the same effect is the Cochran's chi-squared test (Cochran's Q).6 The most common metric for measuring the magnitude of the between-study heterogeneity is the I2,7 the ratio of Q–df (df: degrees of freedom) over Q. I2 is easily interpretable and does not depend on the number of the studies. It ranges between 0% and 100% and is typically considered low for I2 < 25%, modest for 25–50%, and large for >50%.8
Previous methods have been proposed that examine the relative contribution to heterogeneity of each study in a meta-analysis.9,10 The standard Galbraith plot is a bivariate radial scatter plot of the inverse of the standard error of each study vs the ratio of the log of the effect size over the respective standard error.9 A variant proposed by Baujat et al. plots the contribution of each study to Q as a function of the influence on the overall effect estimate.10 Here, we have developed algorithms that are based on the I2 statistic rather than Q and that allow the removal of more than one studies, if need be.
Specifically, we implemented iterative algorithms to calculate the minimum number of studies that should be omitted to decrease the estimated between-study heterogeneity below a specific threshold  . The algorithms are applied to all meta-analyses where the initial estimate of I2 is above the
. The algorithms are applied to all meta-analyses where the initial estimate of I2 is above the  . For practical purposes, we use here the traditional threshold of large heterogeneity (I2 < 50%,
. For practical purposes, we use here the traditional threshold of large heterogeneity (I2 < 50%, 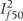 ) and of modest heterogeneity (I2 < 25%,
) and of modest heterogeneity (I2 < 25%, 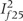 ).
).
The effect size θ and respective standard error SEθ are used as input data. Very rarely more than one study may have the same input data. Then, the algorithms randomly select one of these similar data sets to be removed first.
Sequential algorithm
In this approach, for a meta-analysis of n studies, we perform n new meta-analyses, where one study is excluded from the calculations each time. The study that is responsible for the largest decrease in I2 is dropped and a new set of n−1 studies is created. When two or more studies cause exactly the same decrease in I2 by their exclusion, we drop the study with the largest decrease in Q. We continue by successively re-analysing reduced sets of studies and applying the same rule one step before I2 decreases below the requested  . In the last step there is a chance more than one omitted studies can result in I2 dropping below the wanted threshold. We implement two variants of the algorithm: one omits the study that will result in the maximum I2 below the desired
. In the last step there is a chance more than one omitted studies can result in I2 dropping below the wanted threshold. We implement two variants of the algorithm: one omits the study that will result in the maximum I2 below the desired  , while the other omits the study that will result in the minimum possible I2 below the desired
, while the other omits the study that will result in the minimum possible I2 below the desired  . Both variants exclude eventually the same number of studies, but the last excluded study may be a different one.
. Both variants exclude eventually the same number of studies, but the last excluded study may be a different one.
Combinatorial algorithm
The combinatorial algorithm aims to identify clusters of studies whose exclusion can reduce the between-study heterogeneity below the desired threshold. When the exclusion of any single study does not suffice to drop the heterogeneity below the desired threshold, the decrease in the I2 is examined by the exclusion of all possible pairs of studies. If no pair exclusion achieves the  , we examine the decrease in the I2 by the exclusion of all possible triplets of studies, and so forth. At the last step, there are again two variants. If there are more than one set with equal number of excluded studies that decrease I2 below the desired
, we examine the decrease in the I2 by the exclusion of all possible triplets of studies, and so forth. At the last step, there are again two variants. If there are more than one set with equal number of excluded studies that decrease I2 below the desired  , one can choose to omit the set that results in the maximum or minimum I2 below
, one can choose to omit the set that results in the maximum or minimum I2 below  .
.
Sensitivity metrics
The proposed algorithms generate the number of studies that have to be omitted to decrease I2 below  (k<50 and k<25, for the 50% and 25% threshold, respectively). We can also estimate the proportions of studies that need to be excluded, l<50 = k<50/n and l<25 = k<25/n, respectively. Similarly, one can estimate the proportions based on the sample sizes of the omitted studies vs the total sample size of the meta-analysis aiming for either maximum or minimum I2 below
(k<50 and k<25, for the 50% and 25% threshold, respectively). We can also estimate the proportions of studies that need to be excluded, l<50 = k<50/n and l<25 = k<25/n, respectively. Similarly, one can estimate the proportions based on the sample sizes of the omitted studies vs the total sample size of the meta-analysis aiming for either maximum or minimum I2 below  .
.
Databases for empirical evaluation
We applied these algorithms and derived the sensitivity metrics in two datasets of meta-analyses. First, we used a previously described database of 50 meta-analyses of gene-disease associations that had found a nominally statistically significant effect (P < 0.05) by random-effects calculations (DerSimonian and Laird model).11 Large between-study heterogeneity is common in genetic epidemiology.12
Second, we used meta-analyses from the Cochrane Database of Systematic Reviews (Issue 4, 2005). We used all systematic reviews where at least one meta-analysis with four or more studies had been conducted and the outcome was binary. Among those, we kept only one meta-analysis per systematic review, the one with highest number of studies; in case of ties, we kept the one with largest sample size.
For all meta-analyses, we recorded the initial number of studies, sample size, and random effects (DerSimonian and Laird) summary effect size [natural logarithm of the odds ratio (OR)] and its standard error, I2 and Q (P-value) as well as the sensitivity metrics described earlier. We provide descriptive statistics for the sensitivity metrics for both databases and on differences between the two algorithms. Furthermore, we illustrate the concordance between the initial I2 and number and proportion of studies excluded using scatter plots and Spearman correlation coefficients.
Modules
Analyses were conducted in Intercooled STATA 8.2 (College Station, TX, USA) using the metan and hetred modules. The latter module was developed for the purposes of the study and can be downloaded from www.dhe.med.uoi.gr/software.htm and Statistical Software Components (SSC) archive.
Results
Descriptive statistics of meta-analyses for both databases can be found in the Appendix.
Sequential algorithm
In the genetic meta-analyses data set (Table 1), the median number of studies that had to be omitted from the meta-analysis to drop I2 below the requested threshold (50% and 25%) was one and two, respectively. In the majority of meta-analyses, excluding one or two studies sufficed [19 (90.5%) for the 50% threshold and 16 (57.1%) for the 25% threshold, Figure 1]. In the two variants of the algorithm (aiming for maximum or minimum I2 below  ), the excluded studies at the last step were different in 13 (61.9%) and 11 (39.3%) meta-analyses for the 50% and 25% thresholds, respectively.
), the excluded studies at the last step were different in 13 (61.9%) and 11 (39.3%) meta-analyses for the 50% and 25% thresholds, respectively.
Table 1.
Sensitivity metrics for heterogeneity, median values (IQR) for the genetic meta-analyses’ database
| Sequential algorithm | Combinatorial algorithm | |
|---|---|---|
 threshold: 50% threshold: 50% |
n = 21a | |
| I 2 before exclusion of studies | 61.9% (54.1–70.8) | |
| Number of studies excluded | 1 (1–2) | 1 (1–2) |
| Proportion of studies excluded | 12.5% (7.7–16.7) | 12.5% (7.7–16.7) |
Proportion of sample size excluded, target of maximum I2 below 
|
11.2% (6.4–17.3) | 11.2% (6.4–17.3) |
Proportion of sample size excluded, target of minimum I2 below 
|
9.3% (6.4–25.8) | 9.3% (6.4–25.8) |
I
2 after exclusion of studies, target of maximum I2 below 
|
47% (39.3–49.7) | 47.0% (39.3–49.7) |
I
2 after exclusion of studies, target of minimum I2 below 
|
38.5% (23.2–47.1) | 38.5% (23.2–47.1) |
 threshold: 25% threshold: 25% |
n = 28b | |
| I 2 before exclusion of studies | 59.1% (48.9–66.9) | |
| Number of studies excluded | 2 (1–4) | 2 (1–4) |
| Proportion of studies excluded | 17.7% (10.5–24.0) | 17.7% (10.6–24.0) |
Proportion of sample size excluded, target of maximum I2 below 
|
13.8% (7.8–29.2) | 15.9% (7.8–32.3) |
Proportion of sample size excluded, target of minimum I2 below 
|
13.2% (7.1–29.2) | 11.9% (7.1–21.1) |
I
2 after exclusion of studies, target of maximum I2 below 
|
20.6% (4–23.1) | 21.5% (4.4–23.2) |
I
2 after exclusion of studies, target of minimum I2 below 
|
14.9% (0.7–23.2) | 13.7% (0.7–19.6) |
aFor two meta-analyses I2 was not able to be dropped <50%.
bFor 2 meta-analyses I2 was not able to be dropped <25%.
Figure 1.
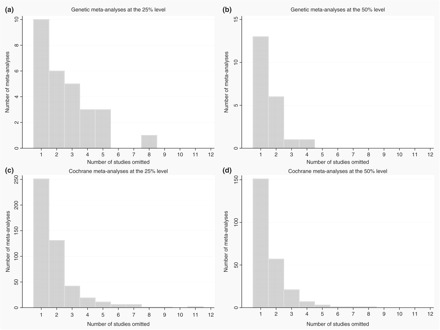
Frequency distribution of excluded studies. (a) Genetic meta-analyses at the 25% level. (b) Genetic meta-analyses at the 50% level. (c) Cochrane meta-analyses at the 25% level. (d) Cochrane meta-analyses at the 50% level
In the Cochrane meta-analyses (Table 2), the median number of studies had to be omitted from the meta-analysis to drop I2 < 50% or even 25% was one. Excluding one or two studies sufficed in the vast majority of cases [208 (85.9%) for the 50% threshold and 382 (81.3%) for the 25% threshold, Figure 1]. Different studies were selected for exclusion at the last step, when targeting for maximum or minimum I2 below the desired  , in 98 (40.5%) and 185 (39.4%) meta-analyses for the 50% and 25% threshold, respectively. Of note, heterogeneity could not be dropped below
, in 98 (40.5%) and 185 (39.4%) meta-analyses for the 50% and 25% threshold, respectively. Of note, heterogeneity could not be dropped below 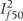 or
or 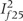 in two meta-analyses regardless of how many studies were excluded.
in two meta-analyses regardless of how many studies were excluded.
Table 2.
Sensitivity metrics for heterogeneity, median values (IQR) for the Cochrane meta-analyses’ database
| Sequential algorithm | Combinatorial algorithm | |
|---|---|---|
 threshold: 50% threshold: 50% |
n = 242a | |
| I 2 before exclusion of studies | 65.5% (56.7–74.3) | |
| Number of studies excluded | 1 (1–2) | 1 (1–2) |
| Proportion of studies excluded | 16.7% (12.0–25.0) | 16.7% (12.0–25.0) |
Proportion of sample size excluded, target of maximum I2 below 
|
17.5% (8.8–33.7) | 17.6% (8.8–37.8) |
Proportion of sample size excluded, target of minimum I2 below 
|
18.4% (10.5–35.1) | 18.4% (10.5–35.9) |
I
2 after exclusion of studies, target of maximum I2 below 
|
40.9% (15.7–47.8) | 42.4% (19.1–47.9) |
I
2 after exclusion of studies, target of minimum I2 below 
|
30.7% (4.7–43.2) | 30.0% (1.3–43.2) |
 threshold: 25% threshold: 25% |
n = 470b | n = 466b, c |
| I 2 before exclusion of studies | 51.4% (38.8–65.8) | 51.0% (38.6–65.8) |
| Number of studies excluded | 1 (1–2) | 1 (1–2) |
| Proportion of studies excluded | 20.0% (11.1–28.6) | 20.0% (11.1–28.6) |
Proportion of sample size excluded, target of maximum I2 below 
|
19.2% (9.3–33.9) | 19.5% (9.3–35.1) |
Proportion of sample size excluded, target of minimum I2 below 
|
19.5% (9.4–35.0) | 19.5% (9.4–35.3) |
I
2 after exclusion of studies, target of maximum I2 below 
|
12.3% (0.0–21.8) | 12.7% (0.0–22.4) |
I
2 after exclusion of studies, target of minimum I2 below 
|
1.4% (0.0–15.1) | 1.0% (0.0–14.7) |
aFor two meta-analyses I2 was not able to be dropped <50%.
bFor two meta-analyses I2 was not able to be dropped <25%.
cFour meta-analyses required excess amount of computational resources and were excluded from analysis.
On average, exclusion of 13% and 17% of the studies in genetic and Cochrane meta-analyses, respectively, sufficed to reach below 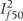 ; the respective percentages were 18% and 20% for reaching below
; the respective percentages were 18% and 20% for reaching below 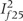 (Tables 1 and 2). There was however some diversity across meta-analyses (Tables 1 and 2). The respective percentages were similar, when based on sample size rather than number of studies (Tables 1 and 2).
(Tables 1 and 2). There was however some diversity across meta-analyses (Tables 1 and 2). The respective percentages were similar, when based on sample size rather than number of studies (Tables 1 and 2).
Differences with the combinatorial algorithm
Results were always very similar and often identical with the use of the combinatorial algorithm (Tables 1 and 2). We will thus focus only on describing the relatively uncommon differences between the two algorithms (Table 3). Differences in the number of studies that had to be excluded occurred in only 0–6% of the meta-analyses, depending on the dataset and desired heterogeneity threshold. Differences in the specific studies to be excluded were somewhat more common and ranged from 11% to 17% when aiming for minimum I2 below  , and 0% to 29% when aiming for the maximum I2 below
, and 0% to 29% when aiming for the maximum I2 below  in the last step.
in the last step.
Table 3.
Differences between sequential and combinatorial algorithm (meta-analyses with two or more excluded studies)
| Different number of studies excluded (%) | Different studies excluded, aiming for maximum I2 below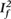 (%) (%) |
Different studies excluded, aiming for minimum I2 below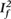 (%) (%) |
|
|---|---|---|---|
| Genetic meta-analyses | |||
| 50% threshold (n = 8) | 0 (0) | 0 (0) | 1 (11.1) |
| 25% threshold (n = 18) | 1 (5.6) | 5 (27.8) | 2 (11.1) |
| Cochrane meta-analyses | |||
| 50% threshold (n = 91) | 5 (5.6) | 26 (28.6) | 15 (16.5) |
| 25% threshold (n = 215) | 10 (4.7) | 52 (24.2) | 33 (15.3) |
The combinatorial algorithm was also time-consuming when several studies had to be excluded. A meta-analysis with 38 studies of which seven had to be excluded eventually (requiring almost 16 000 000 meta-analyses), required 86 h in a Intel(R) Pentium(R) 4 CPU 550 3.40 GHz with 1 GB RAM.
Concordance between initial I2 estimate and sensitivity metrics
Figure 2 shows the correlation between the estimated initial I2 of each meta-analysis and the number of studies that had to be removed to reduce this <50 and 25% (panels A and B, respectively; Cochrane and genetics meta-analyses are combined in both panels) using the sequential algorithm (the results with the combinatorial algorithm are almost identical, as earlier). The Spearman correlation coefficients were 0.65 [(95% confidence interval (CI): 0.58–0.72, n = 263)] and 0.65 (95% CI: 0.60–0.70, n = 498), respectively. The correlation coefficients between the estimated initial I2 and the proportion of studies excluded were 0.52 (95% CI: 0.42–0.60, n = 263) and 0.68 (95% CI: 0.62–0.72, n = 498), respectively (Figures 2C and D for 50% and 25% threshold, respectively). When the proportion of sample size was considered, the correlation coefficients were 0.52 (95% CI: 0.42–0.60, n = 263) aiming for minimum and 0.63 (95% CI: 0.55–0.70, n = 263) aiming for maximum I2 below  of 50%. Targeting for I2 below
of 50%. Targeting for I2 below  of 25%, the correlation coefficient was 0.58 (95% CI: 0.52–0.64, n = 498) for both minimum and maximum variants.
of 25%, the correlation coefficient was 0.58 (95% CI: 0.52–0.64, n = 498) for both minimum and maximum variants.
Figure 2.
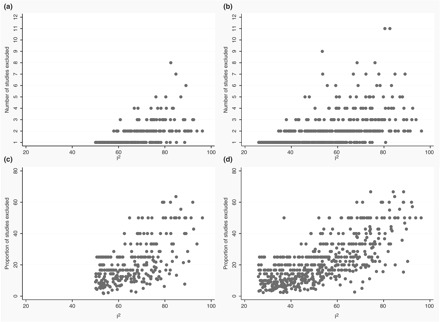
Scatter plots of the number and proportion of excluded studies vs the initial I2 The (a) and (b) represent analyses for number of excluded studies using the target threshold of 50 and 25%, respectively. The (c) and (d) represent analyses for proportion of excluded studies using the target threshold of 50 and 25%, respectively
While the average correlation was modestly high, there was considerable variability in the range of k<50 and k<25 for a given amount of initial heterogeneity. For example, for initial I2 of > 60% the range of k<50 and k<25 was 1 to 8 and 1 to 11, respectively. For initial I2 between 50% and 60%, the range of k<50 and k<25 was 1–2 and 1–9, respectively.
Practical examples
Figure 3A displays a Cochrane meta-analysis with large heterogeneity where the exclusion of a single outlying study decreased significantly the estimated between-study heterogeneity and where a specific plausible explanation for the heterogeneity could be raised. The systematic review13 (CD000173) assessed anti-epileptic drugs for the prevention of seizures following acute traumatic brain injury. The specific meta-analysis used in our dataset had six studies and compared anti-epileptic drugs vs standard care for late seizures (Comparison: 02, Outcome: 02 in the specific Cochrane review). The review authors had found significant heterogeneity and stated they should not synthesize the data, nevertheless a figure with an overall estimate was provided. Using OR as the metric of risk, I2 was originally 63% (95% CI: 0–83%). Application of our algorithms resulted in omission of one study, resulting in an I2 of 19% (95% CI: 0–64%). This study (Pechadre 1991) demonstrated a very large effect of the anti-epileptic treatment (OR 0.09, 95% CI, 0.02–0.39), while the remaining studies showed no benefit, either each one in isolation, or all of them combined (summary OR 1.14, 95% CI, 0.80–1.62). The excluded study was the only one that was not blinded. Lack of blinding could have affected the outcome ascertainment or could have been a marker for other quality deficiencies.14
Figure 3.
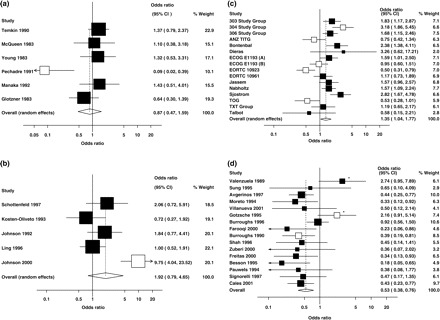
Examples of meta-analyses with very large initial estimate of between-study heterogeneity that required omission of one or several studies for the heterogeneity estimate to decrease <25%. Open boxes represent excluded studies. (a) Cochrane Database ID: CD000173 (one study omitted). (b) Cochrane Database ID: CD002208 (one study omitted). (c) Cochrane Database ID: CD003366 (five studies omitted). (d) Cochrane Database ID: CD000193 (two studies omitted); open boxes represent studies excluded using the sequential algorithm, whereas asterisks denote studies excluded using the combinatorial one
Figure 3B shows an example of a Cochrane meta-analysis with very large initial I2, where the exclusion of a single study could also reduce the I2 estimate <25%, but where it was more difficult to tell what exactly was different or wrong with the excluded study. The systematic review15 (CD002208) was about methadone maintenance at different dosages for opioid dependence. The specific meta-analysis used in our dataset had five studies and compared high (60–109 mg/day) vs low (1–39 mg/day) methadone maintenance doses for 17–26 weeks and the outcome was retention rate (Comparison: 01, Outcome: 01, Subgroup: 01 in the specific Cochrane review). In the OR scale, the initial I2 was 81% (95% CI: 40–90%). The omission of one study (Johnson 2000) dropped the estimated heterogeneity to 8% (95% CI: 0–70%). This study had shown a very large and statistically significant difference in retention rate (10-fold larger odds with the high vs low dose), while the other four studies had shown no significant difference. The outlier study was the newest one and was the only one that used methadone doses >80 mg/day. However, the review authors also noted that ‘missing samples were considered positive for purposes of analysis’, while this was not mentioned for any of the four other studies. It is difficult to decide whether any of these reasons, or others still not captured and/or not reported by the reviewers, may explain the results of this outlier study vs the others.
Figure 3C shows an example of a meta-analysis with equally large I2 that could not be reduced to <25% unless many studies were removed. This Cochrane review16 (CD003366) compared taxane containing regimens for metastatic breast cancer. The comparison used in this specific dataset was overall effect of taxanes and the outcome overall response among randomized patients (Comparison: 01 Outcome: 03 in the specific Cochrane review). The meta-analysis had 16 studies and I2 estimated at 75% (95% CI: 57–84%). The omission of five studies (open boxes in the graph) decreased I2 to 17% (95% CI: 0–52%). The same studies were selected for removal in all variants of the sequential or combinatorial algorithm. The reviewers had separated the dataset in subgroups based on the comparison regimen used. This was thought to explain some of the statistical heterogeneity, but actually the largest subgroup (10 studies, single agent taxane vs another agent) had an even larger point estimate of between-study heterogeneity (I2 = 83%) than the overall analysis. All five excluded studies were from this subgroup. The reviewers commented also about the low reporting quality of all studies and the differences in the definition of response across trials. Poor reporting may be associated with other flaws and biases that may have affected the estimates of the treatment effect, introducing substantial heterogeneity, but these additional flaws or biases are probably occult and difficult to decipher.
Figure 3D shows an example of a meta-analysis with large I2 that both algorithms exclude the same number of studies but different ones. The systematic review (CD000193)17 assessed somatostatin analogues for acute bleeding oesophageal varices. The comparison we have included in our analysis was ‘somatostatin analogues vs placebo or no treatment’ and the respective outcome ‘number of failing initial haemostasis’ (01/04). This outcome included 16 studies with an I2 of 53% (95% CI: 1–72%, using OR as effect estimate). The reviewers stratified the studies as high quality (those with allocation concealment and double-blinding) vs others trial. The high quality trials had less impressive treatment effects for other outcomes such as units of blood transfused and re-bleeding risk and showed no improvement in mortality. The sequential algorithm excluded two studies (first Gøtzshe 1995 and then Burroughs 1996), one of which was a study of high quality. The combinatorial algorithm excluded also two studies (Burroughs 1996 and Valenzuela 1989) neither of which was considered to be of high quality.
Discussion
We have developed and implemented algorithms for sensitivity analyses of between-study heterogeneity in meta-analyses. The different algorithms almost always excluded the same number of studies to reach below a desired heterogeneity threshold. The proposed sensitivity metrics add another useful dimension to the routine examination of between-study heterogeneity, besides the testing of statistical significance and the estimation of the amount of heterogeneity beyond chance (I2). While the number of studies that need to be excluded correlates with the I2, the correlation is only modest, thus there is some independent insight to be gained.
Several methods have already been described for exploration of heterogeneity in meta-analysis, as previously reviewed.18 Some of these methods, such as the traditional Galbraith plot and variants thereof, use graphic presentations to indicate the influence of individual studies.9,10,19 When only one study causes the extreme heterogeneity, our algorithms pinpoint to the same study as these other methods suggest. However, in situations where the heterogeneity is attributed to several studies, the above methods are either impractical or may yield considerably different inferences. Our algorithms complement the existing methods, given the differences in the analytical approach and objectives. In general, the contribution of a study in the overall heterogeneity is analogous to the squared difference of the study's effect from the overall effect. The exclusion of the first study results in a new overall effect and a new set of outlier studies is created. This is not necessarily the same as before, especially if the exclusion of the first study modifies also substantially the summary effect. Our proposed algorithms overcome this problem: each time a study is omitted, the influence of the remaining studies in overall heterogeneity is automatically reappraised.
Although the different variants of our algorithms exclude almost always the same number of studies to reach below a specific desired heterogeneity threshold, discrepancies in which are the specific studies to be excluded are common. Inferences on which are the specific studies that cause the between-study heterogeneity should be cautious and there is a considerable risk of over interpretation. In one extreme situation, two studies may have identical effects and uncertainty thereof, in which case our algorithms randomly select one of them to be excluded first. In the far more common situation, studies may have minor differences in their contribution to heterogeneity, and the key to understanding heterogeneity may not lie in the one that has the largest estimated contribution based on the observed data. Random noise may change the order of outliers. Moreover, when heterogeneity is explained by patient-level rather than study-level characteristics20–22 any algorithm that excludes whole studies may not reveal the true reasons for the heterogeneity. Overall, in-depth examination of the characteristics of the excluded studies may be useful in some circumstances. However, this should be seen as an exploratory post hoc evaluation of the sources of heterogeneity.
We should acknowledge that in general exploration of the reasons of heterogeneity is a difficult task in meta-analysis. Sensitivity analyses where studies are subgrouped based on various characteristics are often performed.23 Subgroups may be selected based on clinical expertise, prior knowledge on the scientific field, or other factors.4 Meta-regression for one or multiple factors may serve similar purposes.24 All of these exploratory analyses may suffer from both lack of power and high false-positive rate.20–22Post hoc selection of the covariates based on subjective interpretation of the already available data can be misleading. The proposed algorithms offer at least an objective approach that is ‘agnostic’, i.e. is not influenced initially by consideration of known specific study characteristics. The important characteristics that contribute to the heterogeneity may be unknown or unrecorded in the examined studies. Meta-analysis of individual-level information may offer a better handle for examining some characteristics, especially patient-level covariates.25 However, such analyses are more difficult, more uncommonly conducted, and often there is insufficient information on covariates of interest.25
We should also acknowledge that heterogeneity in a meta-analysis may vary depending to the effect estimate used, especially for binary outcomes.26 One should have this in mind before investigating any source of inconsistency. Selection of a different metric of effect may result in different studies being excluded.
Another limitation stems from the unavoidable uncertainty in the estimates of I2. This metric has some advantages over the Q statistic, in that it can be compared across different meta-analyses with different metrics and different number of studies. However, in the typical meta-analysis where there are only a few studies, poor power may cause substantial uncertainty in the inferences that are based on either Q or I2.27 Reducing the amount of heterogeneity below a specific level or even aiming for residual I2 = 0% is not reassuring that the remaining studies will be homogeneous. CIs for I2 are generally wide, unless many studies exist.28 Our practical examples also demonstrate this uncertainty. Apparent lack of heterogeneity based on the point estimates of I2 is not proof of homogeneity. We have recently proposed that presentation of confidence intervals around I2 would be useful to promote in the reporting of meta-analyses.28 In this regard, also the selection of the 25% and 50% thresholds should only be seen as a standardized convenience.
Overall, we suggest that our algorithms can be routinely used in meta-analyses with large or moderate estimated heterogeneity to complement existing heterogeneity metrics. Compared with existing methods, the proposed algorithms cater routinely to the possibility of excluding more than one study. The excluded studies offer a starting point for exploring heterogeneity and their selection is made based on purely statistical criteria, potentially avoiding post hoc subjective interpretation of the data. Either the sequential or the combinatorial algorithm can be used. The latter is computationally more demanding when many studies have to be excluded, but in general both algorithms almost always agree on how many studies should be removed. Conversely, great caution is needed in avoiding over-interpreting the data on which specific studies cause heterogeneity and why. The algorithms make this obvious when the two versions exclude different studies, but caution should be applied even when both algorithm versions exclude the same studies.
ACKNOWLEDGEMENTS
N.A.P. and E.E. are funded by a PENED training grant from the European Union Social Fund and the Greek Ministry of Development, General Secretariat of Research and Technology.
Conflict of interest: None declared.
Appendix
Descriptive of meta-analyses
I 2 was estimated to be above 25% in 28 of the 50 (56.0%) genetic meta-analyses and in 472 of the 1011 (46.7%) Cochrane meta-analyses, while values of 50% or higher were seen in 21 (42.0%) and 244 (24.1%), respectively. Appendix Table 1 shows baseline characteristics of the two analysed sets of meta-analyses. Genetic meta-analyses were more heterogeneous, had more studies and larger sample sizes.
Appendix Table 1.
Characteristics of meta-analyses
| Characteristic | Genetics (n = 50) | Cochrane (n = 1011) |
|---|---|---|
| Number of studies, median (IQR) | 13 (8–20) | 7 (5–11) |
| Sample size, median (IQR) | 4670 (2823–8761) | 1112 (512–2691) |
| Effect size, median (IQR)a | 0.360 (0.257–0.499) | 0.371 (0.161–0.827) |
| Variance, median (IQR) | 0.0127 (0.0070–0.0281) | 0.0516 (0.0228–0.1246) |
| I 2, median (IQR) | 37.6% (4.6–59.5) | 21.1% (0.0–49.7) |
| 0–25%: n (%) | 22 (44.0) | 539 (53.3) |
| 25–50%: n (%) | 7 (14.0) | 228 (22.6) |
| 50–75%: n (%) | 19 (38.0) | 187 (18.5) |
| 75–100%: n (%) | 2 (4.0) | 57 (5.6) |
| Q P < 0.10: n (%) | 27 (54.0) | 350 (34.6) |
aAll effect sizes (presented as natural logarithm of OR) have been coined to be ≥0.
REFERENCES
- 1.Lau J, Ioannidis JP, Schmid CH. Summing up evidence: one answer is not always enough. Lancet. 1998;351:123–27. doi: 10.1016/S0140-6736(97)08468-7. [DOI] [PubMed] [Google Scholar]
- 2.Thompson SG. Why sources of heterogeneity in meta-analysis should be investigated. Br Med J. 1994;309:1351–55. doi: 10.1136/bmj.309.6965.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Egger M, Smith GD, Altman DG. Systematic Reviews in Health care: Meta-analysis in Context. London, UK: BMJ Publishing Group; 2001. [Google Scholar]
- 4.Glasziou PP, Sanders SL. Investigating causes of heterogeneity in systematic reviews. Stat Med. 2002;21:1503–11. doi: 10.1002/sim.1183. [DOI] [PubMed] [Google Scholar]
- 5.Sutton A, Abrams K, Jones D, Sheldon T, Song F. Methods for Meta-analysis in Medical Research. Chichester, UK: Wiley; 2000. [Google Scholar]
- 6.Cochran WG. The combination of estimates from different experiments. Biometrics. 1954;10:101–29. [Google Scholar]
- 7.Higgins JPT, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539–58. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 8.Higgins JPT, Thompson SG, Deeks J, Altman DG. Measuring inconsistency in meta-analyses. Br Med J. 2003;327:557–60. doi: 10.1136/bmj.327.7414.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galbraith RF. A note on graphical presentation of estimated odds ratios from several clinical trials. Stat Med. 1988;7:889–94. doi: 10.1002/sim.4780070807. [DOI] [PubMed] [Google Scholar]
- 10.Baujat B, Mahe C, Pignon JP, Hill C. A graphical method for exploring heterogeneity in meta-analyses: application to a meta-analysis of 65 trials. Stat Med. 2002;21:2641–52. doi: 10.1002/sim.1221. [DOI] [PubMed] [Google Scholar]
- 11.Ioannidis JP, Trikalinos TA, Khoury MJ. Implications of small effect sizes of individual genetic variants on the design and interpretation of genetic association studies of complex diseases. Am J Epidemiol. 2006;164:609. doi: 10.1093/aje/kwj259. [DOI] [PubMed] [Google Scholar]
- 12.Ioannidis JP, Ntzani EE, Trikalinos TA, Contopoulos-Ioannidis DG. Replication validity of genetic association studies. Nat Genet. 2001;29:306–9. doi: 10.1038/ng749. [DOI] [PubMed] [Google Scholar]
- 13.Schierhout G, Roberts I. Anti-epileptic drugs for preventing seizures following acute traumatic brain injury. The Cochrane Database of Systematic Reviews. 2001;(4) doi: 10.1002/14651858.CD000173. Art. No.: CD000173, doi: 10.1002/14651858.CD000173. [DOI] [PubMed] [Google Scholar]
- 14.Juni P, Altman DG, Egger M. Systematic reviews in health care: assessing the quality of controlled clinical trials. Br Med J. 2001;323:42–46. doi: 10.1136/bmj.323.7303.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Faggiano F, Vigna-Taglianti F, Versino E, Lemma P. Methadone maintenance at different dosages for opioid dependence. The Cochrane Database of Systematic Reviews. 2003;(3) doi: 10.1002/14651858.CD002208. Art. No.: CD002208, doi: 10.1002/14651858.CD002208. [DOI] [PubMed] [Google Scholar]
- 16.Ghersi D, Wilcken N, Simes J, Donoghue E. Taxane containing regimens for metastatic breast cancer. The Cochrane Database of Systematic Reviews. 2005;(2) doi: 10.1002/14651858.CD003366.pub2. Art. No.: CD003366.pub2, doi: 10.1002/14651858.CD003366.pub2. [DOI] [PubMed] [Google Scholar]
- 17.Gøtzsche PC, Hróbjartsson A. Somatostatin analogues for acute bleeding oesophageal varices. The Cochrane Database of Systematic Reviews. 2005;(1) doi: 10.1002/14651858.CD000193.pub2. Art. No.: CD000193.pub2, doi: 10.1002/14651858.CD000193.pub2. [DOI] [PubMed] [Google Scholar]
- 18.Song F, Sheldon TA, Sutton AJ, Abrams KR, Jones DR. Methods for exploring heterogeneity in meta-analysis. Eval Health Prof. 2001;24:126–51. doi: 10.1177/016327870102400203. [DOI] [PubMed] [Google Scholar]
- 19.Hardy RJ, Thompson SG. Detecting and describing heterogeneity in meta-analysis. Stat Med. 1998;17:841–56. doi: 10.1002/(sici)1097-0258(19980430)17:8<841::aid-sim781>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 20.Schmid CH, Stark PC, Berlin JA, Landais P, Lau J. Meta-regression detected associations between heterogeneous treatment effects and study-level, but not patient-level, factors. J Clin Epidemiol. 2004;57:683–97. doi: 10.1016/j.jclinepi.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 21.Lambert PC, Sutton AJ, Abrams KR, Jones DR. A comparison of summary patient-level covariates in meta-regression with individual patient data meta-analysis. J Clin Epidemiol. 2002;55:86–94. doi: 10.1016/s0895-4356(01)00414-0. [DOI] [PubMed] [Google Scholar]
- 22.Berlin JA, Santanna J, Schmid CH, Szczech LA, Feldman HI. Anti-lymphocyte antibody induction therapy study group. Individual patient- versus group-level data meta-regressions for the investigation of treatment effect modifiers: ecological bias rears its ugly head. Stat Med. 2002;21:371–87. doi: 10.1002/sim.1023. [DOI] [PubMed] [Google Scholar]
- 23.Petiti DB. Approaches to heterogeneity in meta-analysis. Stat Med. 2001;20:3625–33. doi: 10.1002/sim.1091. [DOI] [PubMed] [Google Scholar]
- 24.Higgins JP, Thompson SG. Controlling the risk of spurious findings from meta-regression. Stat Med. 2004;23:1663–82. doi: 10.1002/sim.1752. [DOI] [PubMed] [Google Scholar]
- 25.Simmonds MC, Higgins JP, Stewart LA, Tierney JF, Clarke MJ, Thompson SG. Meta-analysis of individual patient data from randomized trials: a review of methods used in practice. Clin Trials. 2005;2:209–17. doi: 10.1191/1740774505cn087oa. [DOI] [PubMed] [Google Scholar]
- 26.Engels EA, Schmid CH, Terrin N, Olkin I, Lau J. Heterogeneity and statistical significance in meta-analysis: an empirical study of 125 meta-analyses. Stat. Med. 2000;19:1707–28. doi: 10.1002/1097-0258(20000715)19:13<1707::aid-sim491>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 27.Huedo-Medina TB, Sanchez-Meca J, Marin-Martinez F, Botella J. Assessing heterogeneity in meta-analysis: Q statistic or I2 index? Psychol Methods. 2006;11:193–206. doi: 10.1037/1082-989X.11.2.193. [DOI] [PubMed] [Google Scholar]
- 28.Ioannidis JP, Patsopoulos NA, Evangelou E. Uncertainty in heterogeneity estimates in meta-analyses. Br Med J. 2007;335:914–16. doi: 10.1136/bmj.39343.408449.80. [DOI] [PMC free article] [PubMed] [Google Scholar]


