Abstract
Leber’s hereditary optic neuropathy (LHON) is the most common mitochondrial disease. Mitochondrial modifiers are proposed to modify the phenotypic expression of primary LHON-associated mitochondrial DNA (mtDNA) mutations. In this study, we demonstrated that the LHON susceptibility allele (m.14502T > C, p. 58I > V) in the ND6 gene modulated the phenotypic expression of primary LHON-associated m.11778G > A mutation. Twenty-two Han Chinese pedigrees carrying m.14502T > C and m.11778G > A mutations exhibited significantly higher penetrance of optic neuropathy than those carrying only m.11778G > A mutation. We performed functional assays using the cybrid cell models, generated by fusing mtDNA-less ρo cells with enucleated cells from LHON patients carrying both m.11778G > A and m.14502T > C mutations, only m.14502T > C or m.11778G > A mutation and a control belonging to the same mtDNA haplogroup. These cybrids cell lines bearing m.14502T > C mutation exhibited mild effects on mitochondrial functions compared with those carrying only m.11778G > A mutation. However, more severe mitochondrial dysfunctions were observed in cell lines bearing both m.14502T > C and m.11778G > A mutations than those carrying only m.11778G > A or m.14502T > C mutation. In particular, the m.14502T > C mutation altered assemble of complex I, thereby aggravating the respiratory phenotypes associated with m.11778G > A mutation, resulted in a more defective complex I. Furthermore, more reductions in the levels of mitochondrial ATP and increasing production of reactive oxygen species were also observed in mutant cells bearing both m.14502T > C and m.11778G > A mutation than those carrying only 11778G > A mutation. Our findings provided new insights into the pathophysiology of LHON that were manifested by interaction between primary and secondary mtDNA mutations.
Introduction
Leber’s hereditary optic neuropathy (LHON) is the most common maternally inherited eye disease (1–4). LHON patients exhibit severe visual impairment or even blindness by death of retinal ganglion cells (3–4). Maternal transmission of LHON suggested that mutations in mitochondrial DNA (mtDNA) are the molecular bases of this disorder (5–7). More than 20 mtDNA missense mutations have been identified to contribute to LHON to varying degrees (8). These mtDNA mutations included the primary mutations that by themselves can cause LHON and the secondary mutations, which may interact with the primary mutations to increase the probability of clinical expression (1,4). In particular, ND1 m.3460G > A, ND4 m.11778G > A, and ND6 m.14484T > C, which involve genes encoding the subunits of respiratory chain complex I, were the most prevalent primary mutations (4,9,10). However, matrilineal relatives within and among families carrying the same mtDNA mutation(s) exhibited a wide range of penetrance and expressivity including severity, age-of-onset and progression in visual impairment (4,11–14). These suggest that the LHON-associated mtDNA mutation(s) is the primary causative evident, but by itself is insufficient to produce a clinical phenotype. Therefore, additional genetic or environmental determinants are necessary for the phenotypic expression of LHON (4,15–18). In particular, the genetic evidence suggest that the secondary mtDNA mutations such as m.4216T > C, m.13708G > A, m.15257G > A and m.4435A > G mutations may modulate the phenotypic manifestation of the LHON-associated primary mtDNA mutations such as m.11778G > A and m.14484T > C (4,15,19–22). However, the role of these secondary mtDNA mutations in the LHON expression remains poorly understood.
In this study, we investigated the pathophysiology of secondary ND6 m.14502T > C mutation in the LHON expression by taking advantage of a large cohort of 1281 Chinese probands with LHON (12,23,24,25). In this cohort, 438 individuals harboured the m.11778G > A (p.340R > H) mutation and 40 Chinese probands carried the ND6 14502T > C (p.58I > V) mutation (23,24,26–28). Mutational screening of other known LHON-associated mtDNA mutations showed that nine subjects carried only the m.14502T > C mutation, while 31 probands harbored the m.14502T > C and one additional mutation(s) (22 subjects carrying m.11778G > A, 6 individuals harboring m.14484T > C, 1 subject having m.3640G > A and 2 probands bearing m.3866T > C) (23,24,26–28). All nine probands carrying only m.14502T > C mutation did not have a family history of LHON (23,26). However, the average penetrances of LHON among 22 pedigrees harbouring both m.11778G > A and m.14502T > C mutations were higher than those in families carrying only m.14502T > C or m.11778G > A mutation (23,26–28). It was anticipated that m.14502T > C mutation may further deteriorate the mitochondrial dysfunction associated with m.11778G > A mutation, therefore increasing the penetrance and risk of LHON expression. To investigate the pathophysiological consequences of m.14502T > C mutation, we used the cybrid cell models, which were generated by fusing mtDNA-less ρo cells with enucleated cells from LHON patients carrying both m. 11778G > A and 14502T > C mutations, only m.14502T > C or m.11778G > A mutation and a control subject belonging to the same mtDNA haplogroup (29,30). The resultant cybrid lines, which contain the identical nuclear background and same mtDNA hapogroup, allow us to analyze directly biochemical phenotypes due to specific mtDNA mutations. Using Western blot and blue native polyacrylamide gel electrophoresis (BN-PAGE) analyses, we examined if the m.11778G > A and m.14502T > C mutations exerted effects on the stability of ND4 and ND6 subunits and assembly of complex I. These cell lines were then analyzed with the enzymatic activities of electron transport chain complexes, oxygen consumption ratio (OCR), membrane potential, ATP production and generation of reactive oxygen species (ROS).
Results
Clinical and genetic evaluation of 37 Chinese families with LHON
As a part of genetic screen program, we have recruited 1281 Han Chinese families with LHON across China (23–25). These Chinese pedigrees with LHON for this study consisted of 9 families carrying only m.14502T > C mutation, 6 families carrying only m.11778G > A mutation and 22 families carrying both m.14502T > C and m.11778G > A mutations. Ophthalmologic evaluation showed that all affected subjects exhibited the variable severity and age at onset of optic neuropathy. As shown in Table 1, the penetrance of visual impairment of nine families carrying only m.14502T > C mutation ranged from 3.9 to 16.7%, with an average of 7.9%. The affected matrilineal relatives among these families consisted of 4 males and 5 females. The penetrance of visual impairment of six families carrying only m.11778G > A mutation varied from 11.8 to 25%, with an average of 18.3%. The ratio between affected males/females was 1.47/1. Of 283 matrilineal relatives of 22 families carrying both m.14502T > C and m.11778G > A mutations, 54 males and 43 females developed the LHON phenotype. The penetrance of visual impairment of 22 families carrying both m.14502T > C and m.11778G > A mutations ranged from 9.1 to 75%, with an average of 34.3%. Thus, the average penetrance of families carrying both m.14502T > C and m.11778G > A mutations was significantly higher than those families carrying only m.14502T > C or m.11778G > A mutation.
Table 1.
Summary of clinical and molecular data for 37 Chinese families with LHON
| Pedigree | Ratio (affected male/ female) | Average age-at-onset (years) | Matrilineal relatives (affected) | Penetrancea (%) | m.14502T>C mutation | m.11778A>G mutation | Additional mutations | mtDNA haplogroup |
|---|---|---|---|---|---|---|---|---|
| WZ421 | 0:1 | 15 | 15(1) | 6.67 | Yes | No | m.7419G>A | M10a |
| WZ422 | 1:0 | 12 | 22(1) | 4.55 | Yes | No | M10a | |
| WZ423 | 0:1 | 9 | 9(1) | 11.1 | Yes | No | M10a | |
| WZ424 | 1:0 | 14 | 10(1) | 10 | Yes | No | M10a | |
| WZ425 | 0:1 | 11 | 20(1) | 5 | Yes | No | M10a | |
| WZ426 | 1:0 | 6 | 26(1) | 3.85 | Yes | No | m.4812G>A | M10a |
| WZ427 | 0:1 | 7 | 14(1) | 7.1 | Yes | No | M10a | |
| WZ402b | 1:0 | 2 | 6(1) | 16.7 | Yes | No | m.6828C>T | F1a |
| WZ403b | 0:1 | 30 | 16(1) | 6.3 | Yes | No | m.8296A>C | H2 |
| WZ428 | 1:0 | 16 | 6(1) | 16.7 | Yes | Yes | M10a | |
| WZ429 | 1:1 | 18 | 13(2) | 15.4 | Yes | Yes | M10a | |
| WZ430 | 1:2 | 13 | 17(6) | 35.3 | Yes | Yes | M10 | |
| WZ431 | 2:0 | 20 | 9(2) | 22.2 | Yes | Yes | H2 | |
| WZ432 | 1:0 | 14 | 10(1) | 10 | Yes | Yes | M10a | |
| WZ433 | 3:1 | 30 | 7(4) | 57.1 | Yes | Yes |
|
M10a |
| WZ434 | 1:0 | 35 | 15(1) | 6.7 | Yes | Yes | M10a | |
| WZ435 | 5:1 | 18 | 24(6) | 25 | Yes | Yes | M10a | |
| WZ436 | 1:0 | 19 | 11(1) | 9.1 | Yes | Yes | H2 | |
| WZ437 | 3:1 | 20 | 9(4) | 44.4 | Yes | Yes | M10a | |
| WZ438 | 2:1 | 15 | 10(3) | 30 | Yes | Yes |
|
M10a |
| WZ439 | 1:0 | 32 | 4(1) | 25 | Yes | Yes | M10a | |
| WZ440 | 1:3 | 20 | 10(4) | 40 | Yes | Yes | B4a | |
| WZ441 | 3:1 | 18 | 8(4) | 50 | Yes | Yes | M10a | |
| WZ442 | 1:2 | 22 | 4(3) | 75 | Yes | Yes | m.644A>G | M10a |
| WZ443 | 4:5 | 24 | 16(9) | 56.3 | Yes | Yes | M10a | |
| WZ444 | 4:6 | 15 | 15(10) | 66.7 | Yes | Yes | m.11204T>C | M10a |
| WZ50c | 0:4 | 17 | 12(4) | 33.3 | Yes | Yes | M10a | |
| WZ404d | 1:1 | 20 | 14(8) | 57.1 | Yes | Yes | m.3866T>C | M10a |
| WZ405d | 3:1 | 23 | 34(12) | 35.3 | Yes | Yes | M10a | |
| WZ406d | 1:3 | 25 | 15(4) | 26.7 | Yes | Yes |
|
M7c2 |
| WZ407d | 4:3 | 21 | 20(7) | 35 | Yes | Yes | M10a | |
| WZ111 | 1:1 | 27 | 17(2) | 11.8 | No | Yes | D5a | |
| WZ112 | 1:1 | 21 | 10(2) | 20 | No | Yes | D5b1 | |
| WZ113 | 0:3 | 16 | 12(3) | 25 | No | Yes | B5a | |
| WZ114 | 3:0 | 17 | 20(3) | 15 | No | Yes | m.1095T>C | M11 |
| WZ115 | 4:0 | 20 | 19(4) | 21.1 | No | Yes | D5a'b | |
| WZ116 | 1:1 | 21 | 12(2) | 16.7 | No | Yes | A |
Mitochondrial DNA analysis
In previous investigations, we performed the sequence analysis of mtDNA in four Chinese pedigrees carrying both m.11778G > A and m.14502T > C mutations and three Chinese families harbouring only m.14502T> C mutation (25,26). In this study, we performed the entire mtDNA sequence analysis in additional 7 probands carrying only m.14502T > C mutation, 6 probands harbouring only m.11778G > A mutation and 17 probands carrying both m.14502T > C and m.11778G > A mutations. In addition to the known m.14502T> C and m.11778G > A mutations, these probands, as shown in Supplementary Material, Table S1, exhibited distinct sets of mtDNA polymorphisms including 223 known and 2 novel variants. The mtDNAs from 31 Chinese families (9 carrying only m.14502T > C mutation, and 22 carrying both m.14502T > C and m.11778G > A mutations) were distributed among M10a (25 families), H2 (3 pedigrees), M7c (1 family), F1a (1 family) and B4a (1 family) haplogroups, respectively (31,32). On the other hand, six families carrying only m.11778G > A mutation belonged to the haplogroups D5, B5, M11 and A, respectively. The phylogenetic tree including 31 Chinese pedigrees carrying the m.14502T > C mutation and six Chinese families carrying the m.11778G > A mutation is shown in Supplementary Material, Figure 1.
These mtDNA variants included 66 in the D-loop region, 10 in the 12S rRNA, 5 in the 16S rRNA, 593T > C and 644A > G mutations in the tRNAPhe, 5824G > A mutation in the tRNACys, 8296A > G mutation in the tRNALys, 14693A > G mutation in the tRNAGlu, 15900T > C, 15902A > G and 15924A > G in the tRNAThr, 9 bp deletion in the NC7 region, and 89 silent and 45 misense mutations the genes encoding polypeptides (8). These variants in RNAs and polypeptides were evaluated by phylogenetic analysis of these variants and sequences from 17 vertebrates (Supplementary Material, Table S1). These variants were further evaluated for the presence of 100 control subjects and potential structural and functional alterations. Of these, 14 variants exhibited the following criteria: 1) absent in the 100 Chinese controls and <1% in the 2704 mitochondrial genomes (33). (2) evolutional conservation among 17 vertebrates (conservation index >75% proposed by Wallace et al.) (34); (3) potential structural and functional alterations. As shown in Table 1, the known LHON-associated ND1 3866T > C and ND4 11204T > C mutations and tRNAPhe 644A > G mutation were co-segregated with both m.11778G > A and m.14502T > C mutations in the pedigrees WZ442, WZ444 and WZ404 with higher penetrance of LHON (23,24,27,35). Two or three variants were present in pedigrees carrying m.11778G > A and m.14502T > C mutations: pedigrees WZ433 (m.14596A > G, m.14617A > G and m.10844A > G), WZ438 (m.3866T > C, m.644A > G and m.8821T > G) and WZ406 (m.7598G > A and m.9957T > C). Furthermore, tRNALys 8296A > G, m.4812G > A, m.6828C > T, m.7419G > A, m.9053G > A, 8821T > G variants were present in probands carrying only m.14502T > C mutation. Moreover, 12S rRNA 1095T > C mutation coexisted with m.11778G > A mutation in the pedigree WZ114. These suggest that these mtDNA variants may play a role in the phenotypic manifestation of m.11778G > A and m.14502T > C mutations.
Generation of cell lines from three Chinese pedigrees with LHON
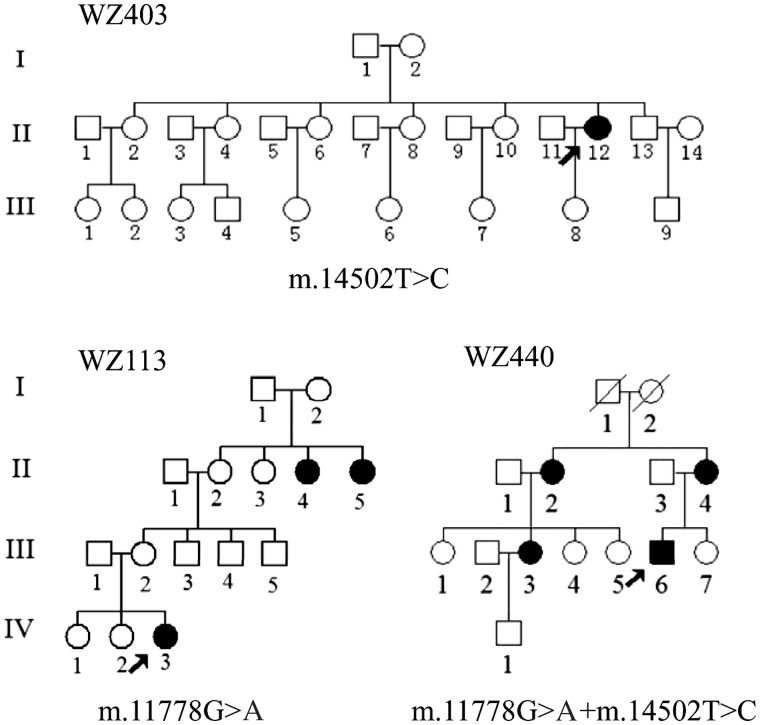
To gain the functional evidence that the m.14502T > C mutation plays a role in the phenotypic manifestation of m.11778G > A mutation, we used three representative Chinese pedigrees with LHON (WZ113 pedigree carrying only m.11778G > A mutation, WZ403 pedigree bearing only m.14502T > C mutation, WZ440 pedigree harboring both m.11778G > A and m.14502T > C mutations; and all three lacking functional significant variants) for the biochemical characterization (Fig. 1). Immortalized lymphoblastoid cell lines were derived from three probands (WZ113-IV-3 carrying only m.11778G > A mutation, WZ403-II-12 carrying only m.14502T > C mutation, WZ440-III-6 carrying both m.11778G > A and m.14502T > C mutations; all three between 30-40 years) belonging to the mtDNA haplogroups B4, B5, respectively and one genetically unrelated control individual A20 belonging to the mtDNA haplogroup B5 (male, 35 years). These lymphoblastoid cell line were enucleated, and subsequently fused to a large excess of mtDNA-less human ρ0206 cells and the cybrid clones were isolated by growing in selective DMEM medium (27,36). The cybrids derived from each donor cell line were analyzed for the presence and level of the m.11778G > A or m.14502T > C mutation and mtDNA copy numbers. The results confirmed the absence of the mtDNA mutations in the control clones and their presence in homoplasmy in all cybrids derived from the mutant cell lines (data not shown). Three cybrids derived from each donor cell line with similar mtDNA copy numbers were used for the biochemical characterization described below.
Figure 1.
Three Chinese pedigrees with LHON. Vision-impaired individuals are indicated by blackened symbols.
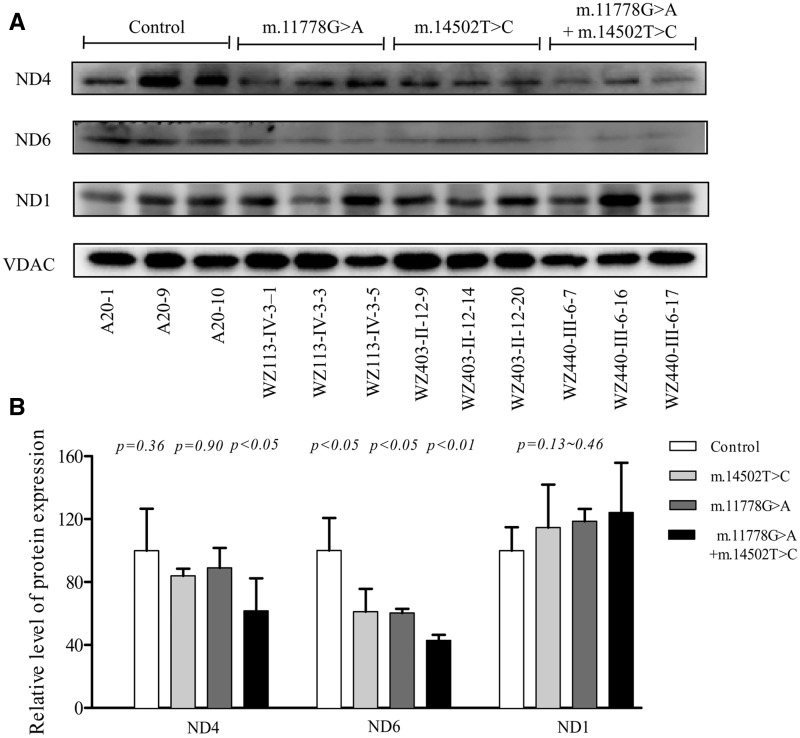
Reductions in levels of ND4 and ND6 proteins
To test the predicted effects of m.14502T > C, p.58I > V mutation on ND6 and m.11778G > A, p.340R > H mutation on ND4, we analyzed the levels of ND4, ND6, and ND1 proteins by Western blotting using total mitochondrial proteins in cell lines carrying only m.11778G > A mutation, only m.14502T > C mutation, or both m.14502T > C and m.11778G > A mutations and control cell lines with VDAC (a nuclear gene encoding mitochondrial protein) as a loading control. As shown in Figure 2, the levels of ND6 in mutant cell lines carrying only m.14502T > C, m.11778G > A, or both m.14502T > C and m.11778G > A mutations were 60.10, 60.27 and 42.83%, relative to the average control values. Furthermore, the levels of ND4 in mutant cell lines carrying only m.14502T > C, m.11778G > A, both m.14502T > C and m.11778G > A mutations were 83.7, 89 and 61.7%, relative to the average control values, respectively. However, the levels of ND1 in mutant cell lines were comparable to those in control cell lines.
Figure 2.
Western blot analysis of mitochondrial proteins. (A) 5 μg of mitochondrial proteins from various cell lines were electrophoresed through a denaturing polyacrylamide gel, electroblotted and hybridized with three respiratory complex subunits in mutant and control cell lines with VDAC as a loading control. ND1, ND4, and ND6, subunits 1, 4 and 6 of the reduced nicotinamide–adenine dinucleotide dehydrogenase. (B) Quantification of three complex I subunits. The levels of ND1, ND4, and ND6 in mutant and control cell lines were determined as described elsewhere (38). The calculations were based on three independent determinations in each cell line. The error bars indicate two standard errors of the means. p indicates the significance, according to the t-test, of the differences between mutant and control cell lines.
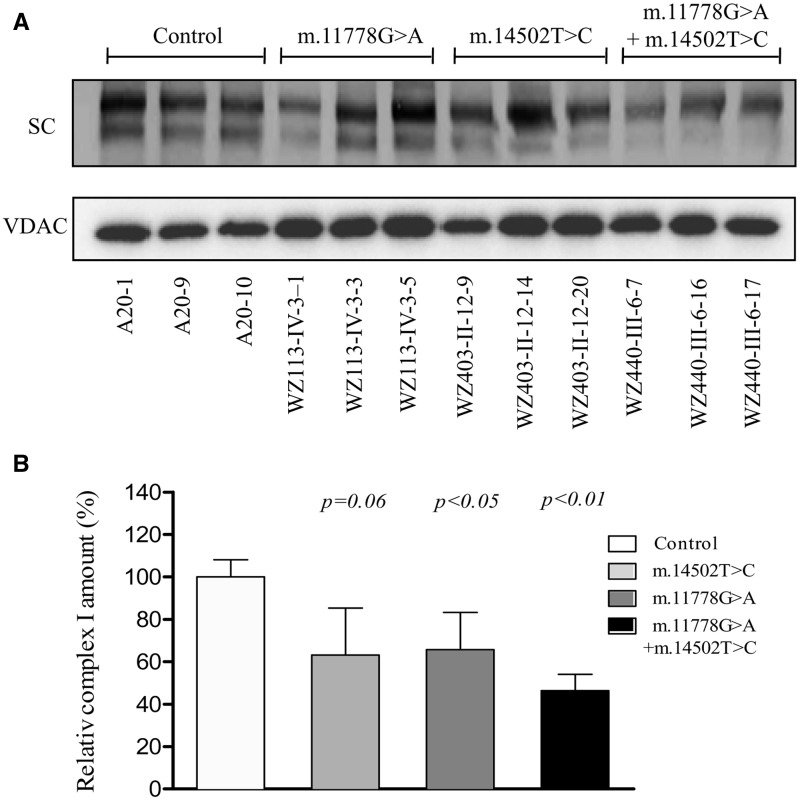
Defects in complex I assembly
To examine if m.14502T > C and m.11778G > A mutations affect the complex I assembly, mitochondrial membrane proteins isolated from mutant and control cell lines were separated by blue native polyacrylamide gel electrophoresis. Complex I was then detected with NDUFA13 antibody (a nuclear encoded complex I subunit) and VDAC was used as loading control. As shown in Figure 3, the m.14502T > C and m.11778G > A mutations altered the assembly of intact complex I. In particular, the levels of supercomplexes carrying the complex I in mutant cell lines carrying only m.11778G > A, only m.14502T > C, and both m.14502T > C and m.11778G > A mutations were 63.1, 65.8 and 46.4%, relative to the average control values. These data suggested that these cell lines carrying both m.14502T > C and m.11778G > A mutations led to more unstable complex I than those cell lines carrying the only single mutation.
Figure 3.
Analysis of complex I assembly. (A). Respiratory complex assembly and in-gel activity assay. Whole cells were solubilized with n-dodecyl-β-D-maltoside (DDM) and then subjected to BN-PAGE/immunoblot analysis. Blots were hybridized with anti-NDUFA13 antibody and VDAC as internal control. SC: supercomplex containing complex I. (B). Quantification of supercomplex containing complex I. The levels of supercomplex containing complex I in mutant and control cell lines were determined as described elsewhere (51). The calculations were based on three independent determinations in each cell line. Graph details and symbols are explained in the legend of Figure 2.
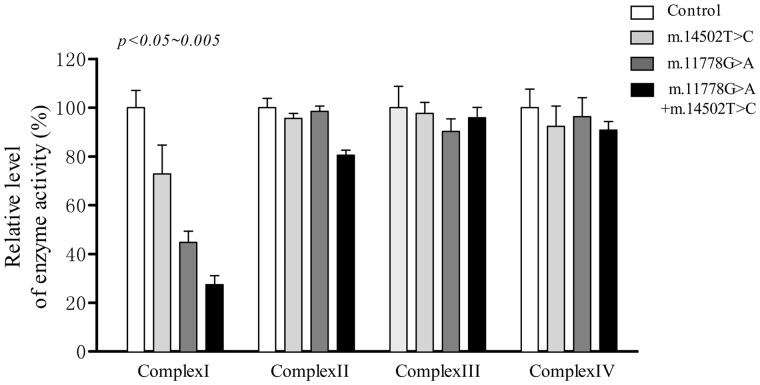
Reduced activity of complex I
To investigate the effect of the m.14502T > C and m.11778G > A mutations on oxidative phosphorylation, we measured the activities of respiratory complexes in isolating mitochondria from mutant and control cell lines. Complex I (NADH ubiquinone oxidoreductase) activity was determined by following the oxidation of NADH with ubiquinone as the electron acceptor (37). Complex III (ubiquinone cytochrome c oxidoreductase) activity was measured as the reduction of cytochrome c (III) using D-ubiquinol-2 as the electron donor. Complex IV (cytochrome c oxidase) activity was monitored by following the oxidation of cytochrome c (II). As shown in Figure 4, the activity of complex I in mutant cell lines carrying only m.14502T > C, only m.11778G > A and both m.14502T > C and m.11778G > A mutations were 72.8, 44.5 and 27.4%, relative to the average control values, respectively. However, the activities of complex II, III and IV in mutant cell lines were comparable to those of three control cell lines.
Figure 4.
Enzymatic activities of respiratory chain complexes. The activities of respiratory complexes were investigated by enzymatic assay on complexes I, II, III, and IV in mitochondria isolated from various cell lines. The calculations were based on three independent determinations. Graph details and symbols are explained in the legend to Figure 2.
Respiration defects
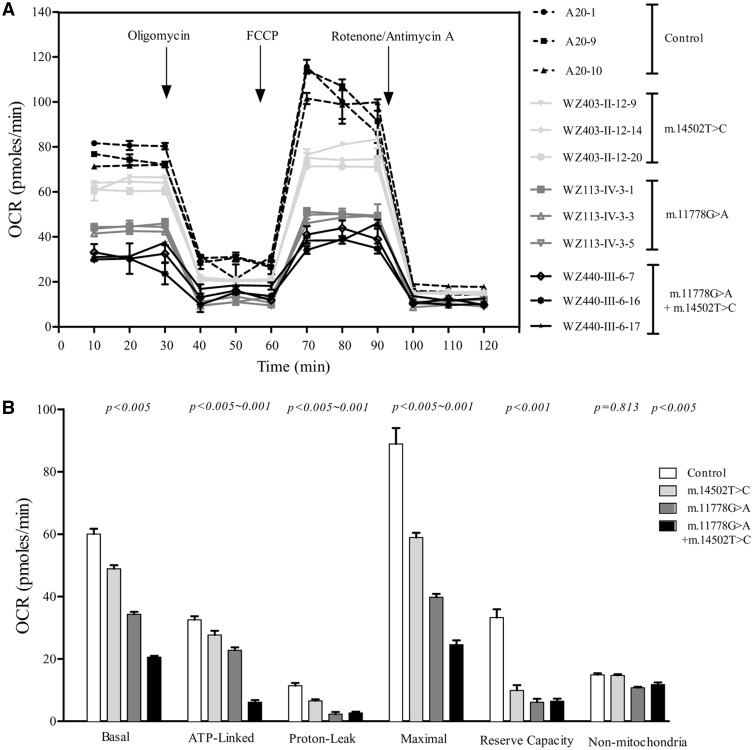
To evaluate if m.14502T > C and m.11778G > A mutations alter cellular bioenergetics, we examined the oxygen consumption rates (OCR) of mutant and control cell lines. As shown in Figure 5, the basal OCR in the mutant cell lines carrying only m.14502T > C, only m.11778G > A or both m.14502T > C and m.11778G > A mutations was 81.5, 57.2 and 34.3%, relative to the mean value measured in the control cell lines, respectively. To investigate which of the enzyme complexes of the respiratory chain was affected in the mutant cell lines, OCR was measured after the sequential additions of oligomycin (to inhibit the ATP synthase), FCCP (to allow for maximum electron flux through the ETC), rotenone (to inhibit complex I) and antimycin (to inhibit complex III). The difference between the basal OCR and the drug-insensitive OCR yielded the amount of ATP-linked OCR, proton leak OCR, maximal OCR, reserve capacity and non-mitochondrial OCR. As shown in Figure 5, we found the ATP-linked OCR, proton leak OCR, maximal OCR, reserve capacity and non-mitochondrial OCR in mutant cell lines carrying both m.14502T > C and m.11778G > A mutations were 18.8, 23.3, 27.6, 19.3 and 79.1%, those cell lines carrying only m.14502T > C were 85, 57.3, 66.35, 29.7 and 98.5%, cell lines carrying only m.11778G > A mutations were 70, 20.4, 44.8, 18.4 and 72.4%, relative to the mean value measured in the control cell lines, respectively.
Figure 5.
Respiration assays. (A) An analysis of O2 consumption in the various cell lines using different inhibitors. The rates of O2 (OCR) were first measured on 2 × 104 cells of each cell line under basal condition and then sequentially added to oligomycin (1.5µM), carbonyl cyanide p-(trifluoromethoxy) phenylhydrazone(FCCP) (0.5µM), rotenone (1 µM) and antimycin A (1μM) at indicated times to determine different parameters of mitochondrial functions. (B) Graphs presented the ATP-linked OCR, proton leak OCR, maximal OCR, reserve capacity and non-mitochondrial OCR in mutant and control cell lines. Non-mitochondrial OCR was determined as the OCR after rotenone/antimycinA treatment. Basal OCR was determined as OCR before oligomycin minus OCR after rotenone/antimycin A. ATP-lined OCR was determined as OCR before oligomycin minus OCR after oligomycin. Proton leak was determined as Basal OCR minus ATP-linked OCR. Maximal was determined as the OCR after FCCP minus non-mitochondrial OCR. Reserve Capacity was defined as the difference between Maximal OCR after FCCP minus Basal OCR. The average values of 4 determinations for each cell line were shown, the horizontal dashed lines represent the average value for each group. Graph details and symbols are explained in the legend to Figure 2.
Reductions in mitochondrial ATP production
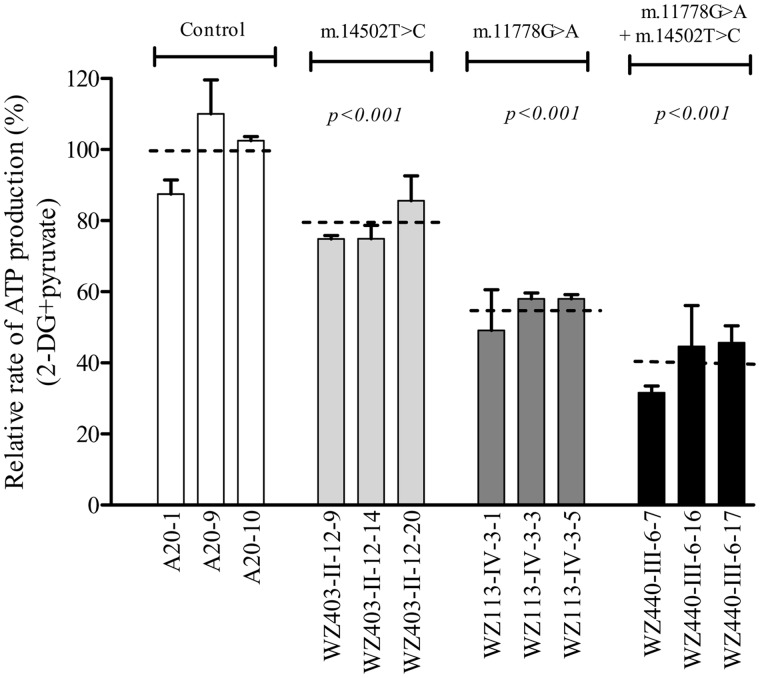
The capacity of oxidative phosphorylation in mutant and wild type cell lines was examined by measuring the levels of cellular and mitochondrial ATP using a luciferin/luciferase assay. Cells were incubated in the media in the presence of glucose, and 2-deoxy-D-glucose with pyruvate (38,39). The levels of ATP production in mutant cells in the presence of glucose (total ATP production) were comparable to those measured in the control cell lines (data now shown). As shown in Figure 6, in the presence of pyruvate and 2-deoxy-D-glucose to inhibit the glycolysis (mitochondrial ATP production), the levels of ATP production in the mutant cell lines harbouring only m.14502T > C, only m.11778G > A, both m.14502T > C and m.11778G > A mutations were 78.5, 55 and 39.3% of average value of three cybrid control cell lines, respectively.
Figure 6.
Measurement of cellular and mitochondrial ATP levels. Mutant and control cell lines were incubated with 10 mM glucose or 5 mM 2-deoxy-d-glucose plus 5 mM pyruvate to determine ATP generation under mitochondrial ATP synthesis. Average rates of ATP level per cell line in mitochondria are show. Six to seven determinations were made for each cell line. Graph details and symbols are explained in the legend to Figure 2.
Decrease in mitochondrial membrane potential
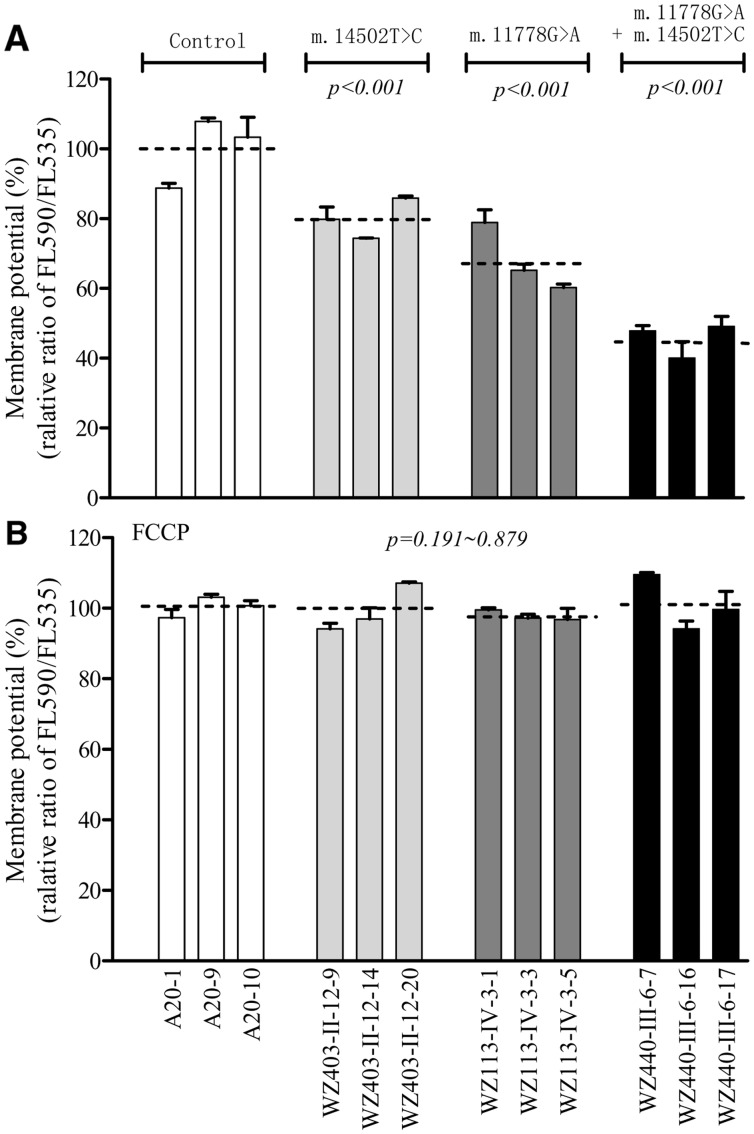
The mitochondrial membrane potential (ΔΨm) changes were measured in mutant and control cell lines using a fluorescence probe JC-10 assay system. The ratios of fluorescence intensities Ex/Em = 490/590 and 490/530 nm (FL590/FL530) were recorded to delineate the ΔΨm level of each sample. The relative ratios of FL590/FL530 geometric mean between mutant and control cell lines were calculated to represent the level of ΔΨm. As shown in Figure 7A, the levels of the ΔΨm in the mutant cell lines carrying only m.14502T > C, only m.11778G > A or both m.14502T > C and m.11778G > A mutations were 80.8, 68.2 and 46.4%, as compared with the mean value measured in the control cell lines. In contrast, as shown in Figure 7B, the levels of ΔΨm in mutant cells in the presence of FCCP were comparable to those measured in the control cell lines.
Figure 7.
Mitochondrial membrane potential analysis. The mitochondrial membrane potential (ΔΨm) was measured in mutant and control cell lines using a fluorescence probe JC-10 assay system. The ratio of fluorescence intensities Ex/Em = 490/590 nm and 490/530 nm (FL590/FL530) were recorded to delineate the ΔΨm level of each sample. The relative ratios of FL590/FL530 geometric mean between mutant and control cell lines were calculated to reflect the level of ΔΨm. Relative ratio of JC-10 fluorescence intensities at Ex/Em = 490/530 nm and 490/590 nm in absence (A) and presence (B) of 10µM of carbonyl cyanide 3-chlorophenylhydrazone (FCCP). The average of 3-5 determinations for each cell line is shown. Graph details and symbols are explained in the legend to Figure 2.
The increase of ROS production
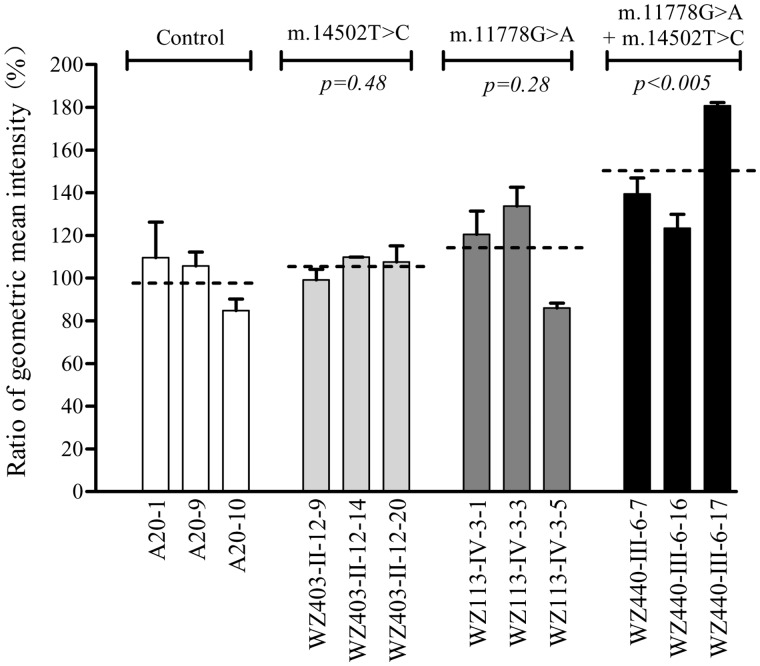
The levels of the ROS generation in the cells derived from control and mutant cell lines were measured with flow cytometry under normal and H2O2 stimulation (40). Geometric mean intensity was recorded to measure the rate of ROS of each sample. The ratio of geometric mean intensity between unstimulated and stimulated with H2O2 in each cell line was calculated to delineate the reaction upon increasing levels of ROS under oxidative stress. As shown in Figure 8, the levels of ROS generation in the mutant cell lines carrying only m.14502T > C, only m.11778G > A mutation, both m.11778G > A and m.14502T > C mutations were 105.5, 113.5 and 147.9% of three control cell lines.
Figure 8.
Ratio of geometric mean intensity between levels of the ROS generation in the vital cells with or without H2O2 stimulation. The rates of production in ROS from mutant and control cell lines were analyzed by BD-LSR II flow cytometer system with or without H2O2 stimulation. The relative ratio of intensity (stimulated versus unstimulated with H2O2) was calculated. The average of three determinations for each cell line is shown. Graph details and symbols are explained in the legend to Figure 2.
Discussion
The secondary LHON-associated mtDNA mutations were proposed to increase the susceptibility to LHON-associated mtDNA mutations such as m.11778G > A mutation (4,7,13,20,22,41). Using genetic and molecular approaches, in combination with functional assays, we demonstrated that the LHON susceptibility allele (m.14502T > C, p.58I > V) in the ND6 gene modulates for the phenotypic expression of the primary m.11778G > A mutation. The m.14502T > C mutation resulted in the substitution of a conserved isoleucine for valine (p.58Ile > Val) at amino acid position 58 in ND6, which is an essential subunit of complex I (37). The occurrence of m.14502T > C mutation in some genetically unrelated pedigrees affected by LHON and differing considerably in their mtDNA haplotypes (M10, B4, B5, H2, M7) suggested that this mutation is involved in the pathogenesis of the disorder (23,26–28,42–44). In fact, the average penetrance of optic neuropathy in all nine Chinese families carrying only m.14502T > C mutation were much lower than those in these pedigrees bearing only m.11778G > A mutation (4,14,22,45,46). Strikingly, the average penetrance of visual impairment of 22 Chinese families carrying both m.14502T > C and m.11778G > A mutations exhibited significantly higher than average penetrance of optic neuropathy in those families (European and Asian origins) carrying only m.11778G > A mutation (4,11,14,24,46). However, nuclear modifier genes or other mtDNA mutations such as m.3866T > C may also contribute to the phenotypic manifestation of LHON-associated mtDNA mutations in some of these families (17,22,47). Furthermore, the other Chinese families bearing m.14502T > C mutation with the m.3640G > A, m.3635G > A, m.3866T > C or m.14484T > C mutation had relatively higher penetrance of optic neuropathy, as compared with those families carrying only one of primary mtDNA mutations (13,23,28,36,42–44,48). These observations strongly suggested that the m.14502T > C mutation may increase the expression of optic neuropathy in these families carrying the primary mtDNA mutations, as in the cases of m.4216T > C and m.13708G > A mutations in European families (16,21) and m.4435A > G and m.15951A > G mutations in other Chinese pedigrees (22,47).
It was anticipated that the m.14502T > C mutation altered the structure and function of NADH dehydrogenase (complex I), thereby worsening mitochondrial dysfunction associated with m.11778G > A mutation. In the present investigation, functional significance of m.14502T > C mutation was evaluated by using cybrid cell lines derived mutant and control subjects belonging to very similar mtDNA haplotypes B4 and B5. Cybrids cell lines bearing only m.14502T > C mutation exhibited only mild effect on mitochondrial function as in the cases of other LHON-associated mtDNA mutations (17,35,49,50). However, more severe mitochondrial dysfunctions were observed in cybrids cell lines bearing both m.14502T > C and m.11778G > A mutations than those carrying only m.11778G > A or m.14502T > C mutation. The primary defects of m.11778G > A and m.14502T > C mutations altered the structure and function of NADH dehydrogenase (complex I). The instability of mutated ND6 and ND4 was evidenced by the reduced levels of ND6 and ND4 observed in cell lines carrying only m.14502T > C or m.11778G > A and both m.14502T > C and m.11778G > A mutations. The defects in assembly of complex I caused by the mutated ND6 and ND4 were consistent within the previous observations that the primary mtDNA mutation(s) affected the assemble of OXPHOS complexes (20,51). Notably, the cell lines carrying both m.14502T > C and m.11778G > A mutations exhibited more unstable complex I than those bearing only m.14502T > C or m.11778G > A mutation. The defects in the assembly of complex I were apparently responsible for the respiratory phenotypes. As a result, the m.14502T > C mutation worsens the respiratory phenotypes associated with m.11778G > A mutation. In this study, ∼27 and 55% decrease in the activity of complex I were observed in these mutant cell lines carrying only m.14502T > C or m.11778G > A mutation, respectively. By contrast, 73% decreases of complex I activity was observed in cell lines harbouring both m.11778G > A and m.14502T > C mutations. Furthermore, the m.14502T > C mutation further deteriorated the reduced rates in the basal OCR, or ATP-linked OCR, reserve capacity and maximal OCR in mutant cell lines carrying only m.11778G > A mutation.
The respiratory deficiency then affected the efficiency of mitochondrial ATP synthesis. In this investigation, ∼21.5 and 45% decrease in mitochondrial ATP production were observed in these mutant cell lines carrying only m.14502T > C or m.11778G > A mutation, respectively. However, 60.6% reduction of mitochondrial ATP production in mutant cell lines harbouring both m.11778G > A and m.14502T > C mutations were comparable with those in lymphoblastoid cell lines bearing both m.11778G > A and homozygous YARS2 p.191Gly > Val mutations (17) and lymphoblastoid cell lines derived from a large Chinese pedigree with high penetrance of LHON (39). As a result, retinal ganglion cells harbouring both m.11778G > A and m.14502T > C mutations may be particularly sensitive to increased ATP demand. Furthermore, the deficient activities of respiratory chain complexes often alter mitochondrial membrane potentials, which is a key indicator of cellular viability (52). Indeed, mitochondrial membrane potentials reflect the pumping of hydrogen ions across the inner membrane during the process of electron transport and oxidative phosphorylation (53). In this study, 54, 32 and 19% reductions in mitochondrial membrane potential was observed in mutant cell lines bearing both m.11778G > A and m.14502T > C mutations, only m.11778G > A or m.14502T > C mutation. The defects in mitochondrial membrane potential may be due to significantly decreased efficiency of respiratory chain-mediated proto extrusion for the matrix (38,54,55). The impairment of both oxidative phosphorylation and mitochondrial membrane potential would elevate the production of reactive oxygen species (ROS) in mutant cells carrying mtDNA mutations. Here, a relative mild reduction of ROS production in the cybrid cell lines carrying only m.14502T > C or m.11778G > A mutation was consistent with the previous observation in cybrid cell lines harbouring only m.11778G > A mutation (56,57). However, a 48% increase of ROS production in cells harbouring both m.11778G > A and m.14502T > C mutations was comparable to 47% increase of ROS production in cells carrying both m.11778G > A and homozygous YARS2 p.191Gly > Val mutations (17). However, 2.5 fold more cellular hydroperoxide were detected in neuronal NT2 cells carrying the m.11778G > A mutation (58). This discrepancy is likely due to differentiation-specific effects. The overproduction of ROS can establish a vicious cycle of oxidative stress in the mitochondria, thereby damaging mitochondrial and cellular proteins, lipids and nuclear acids (59). The retinal ganglion cells may be preferentially involved because they are somehow exquisitely sensitive to subtle imbalance in cellular redox state or increased level of free radicals (4,58,60). This would lead to dysfunction or apoptosis of retinal ganglion cells carrying both m.11778G > A and m.14502T > C mutations, thereby producing a phenotype of optic neuropathy (4,60).
In summary, our results provide the strongest evidence that the m.14502T > C mutation modulates the phenotypic manifestation of LHON-associated m.11778G > A mutation. The m.14502T > C mutation can further deteriorate the defects in the structure and function of complex I, associated with the primary m.11778G > A mutation. The resultant biochemical defects worsen the mitochondrial dysfunction associated with m.11778G > A mutation, thereby leading to the higher penetrance and occurrence of optic neuropathy in these Chinese families carrying both m.14502T > C and 11778G > A mutations. Thus, our findings may provide new insights into the pathophysiology of LHON that were manifested by interaction between primary and secondary mtDNA mutations.
Materials and Methods
Families and subjects
These Chinese pedigrees with LHON for this study consisted of 9 families carrying only m.14502T > C mutation, 6 families carrying only m.11778G > A mutation and 22 families carrying m.14502T > C and m.11778G > A mutations. A total of 100 control DNA samples lacking the m.11778G > A and m.14502T > C mutations were obtained from adult Han Chinese from the same area. The ophthalmic examinations and other clinical evaluations of probands, other members of these families and 100 control subjects were conducted as detailed elsewhere (13,14,23). Informed consent, blood samples, and clinical evaluations were obtained from all participants and families, under protocols approved by Ethic Committees of Zhejiang University and Wenzhou Medical University and the Institutional Review Board of Cincinnati Children’s Hospital.
Mitochondrial DNA analysis
The entire mitochondrial genomes of 7 probands carrying only m.14502T > C mutation, 6 probands harbouring only m.11778G > A mutation and 17 probands carrying both m.14502T > C and m.11778G > A mutations were analyzed as described elsewhere (61). The resulting sequence data were compared with the updated consensus Cambridge sequence (GenBank accession number: NC_012920) (62). The entire mtDNA sequences of 37 pedigrees and 1 control subject were assigned to the mitochondrial haplogroups using the nomenclature as described previously (31,32). An analysis for the presence and levels of m.11778G > A and m.14502T > C mutations in mutant and control cell lines were carried out as described previously (22,26). The quantification of mtDNA copy numbers from different cell lines was performed as detailed elsewhere (63).
Cell lines and culture conditions
Lymphoblastoid cell lines derived from three probands (WZ113-IV-3 carrying only m.11778G > A mutation, WZ403-II-12 carrying only m.14502T > C mutation, WZ440-III-6 carrying both m.11778G > A and m.14502T > C mutations) and one control individual A20 were immortalized by transformation with the Epstein-Barr virus, as described previously (64). Lymphoblastoid cell lines were grown in RPMI 1640 medium (Invitrogen), supplemented with 10% fetal bovine serum. The 143B.TK– cell line was grown in DMEM (containing 4.5 mg of glucose and 0.11 mg pyruvate per ml), supplemented with 100 µg of BrdU per ml and 5% FBS. The mtDNA-less ρ0206 cell line, derived from 143B.TK– (29,36) was grown under the same conditions as the parental line, except for the addition of 50 µg of uridine/ml. Transformation by cytoplasts of mtDNA less ρ0206 cells was performed by using four immortalized lymphoblastoid cell lines, as detailed elsewhere (36,63). All cybrid cell lines were maintained in the same medium as the 143B.TK– cell line.
Western blot analysis
Mitochondria were isolated from cell lines according to procedures previously described (65). Western blotting analysis was performed by using total mitochondrial proteins, as detailed previously (38,55). The antibodies used for this investigation were from Abcam [anti VDAC (ab14734), ND1 (ab74257), Santa Cruz Biotechnology [ND4 (sc-20499-R) and ND6 (sc-20667)]. Peroxidase Affini Pure goat anti-mouse IgG and goat anti-rabbit IgG (Jackson) were used as a secondary antibody and protein signals were detected using the ECL system (CWBIO). Quantification of density in each band was performed as detailed previously (34).
Blue native gel electrophoresis
Blue native gel electrophoresis were carried out using mitochondrial proteins isolated from mutant and control cell lines, as detailed elsewhere (51,66). The antibodies used for this investigation were from Sigma-Aldrich [anti-NDUFA13 (SAB1101649) and Abcam [anti-VDAC (ab14734). Peroxidase Affini Pure goat anti-mouse IgG and goat anti-rabbit IgG (Cell Signalling) were used as a secondary antibody. Protein signals were detected using Super Signal West Pico Chemiluminescent Substrate (Thermo Scientific, Waltham, USA) or 5-bromo-4-chloro-3-indolyl-phosphate/nitro blue tetrazolium (BCIP/NBT) substrate (Promega, Madison, USA). Quantification of density in each band was performed as detailed previously (51,66).
Enzymatic assays
The enzymatic activities of complex I, II, III and IV were assayed as detailed elsewhere (30,55,67).
Measurements of oxygen consumption
The rates of oxygen consumption in cybrid cell lines were measured with a Seahorse Bioscience XF-96 extracellular flux analyzer (Seahorse Bioscience), as detailed previously (38,68).
ATP measurements
The Cell Titer-Glo® Luminescent Cell Viability Assay kit (Promega) was used for the measurement of cellular and mitochondrial ATP levels, according to the modified manufacturer’s instructions (38,55).
Assessment of mitochondrial membrane potential
Mitochondrial membrane potential was assessed with JC-10 Assay Kit-Microplate (Abcam) following general manufacturer’s recommendations with some modifications, as detailed elsewhere (38).
ROS measurements
ROS measurements were performed as detailed previously (38–39).
Computer analysis
Statistical analysis was carried out using the unpaired, two-tailed Student’s t-test contained in the Microsoft-Excel program or Macintosh (version 2007). Differences were considered significant at a P < 0.05.
Supplementary Material
Supplementary Material is available at HMG online.
Supplementary Material
Acknowledgements
We are grateful to patients and their family members for their participation. This work was supported by the National Key Technologies R&D Program grant 2012BAI09B03 from the Ministry of Science and Technology of China to MXG and PJ, grants 31471191, 81200724 and 81400434 from the National Natural Science Foundation of China to MXG, JZ and YJ, respectively.
Conflict of Interest Statement. None declared.
Funding
This work was supported by the National Key Technologies R&D Program grant 2012BAI09B03 from the Ministry of Science and Technology of China to M.-X.G. and P.J., and grants 31471191, 81200724 and 81400434 from the National Natural Science Foundation of China to M.-X.G., J.Z. and Y.J., respectively.
Footnotes
†The authors wish it to be known that, in their opinion, these authors contributed equally to this work.
References
- 1. Newman N.J. (1993) Leber’s hereditary optic neuropathy. Ophthalmol.Clin. N.A., 50, 540–548. [DOI] [PubMed] [Google Scholar]
- 2. Nikoskelainen E.K. (1994) Clinical Picture of LHON. Clin. Neurosci., 2,115–120. [Google Scholar]
- 3. Yu-Wai-Man P., Griffiths P.G., Hudson G., Chinnery P.F. (2009) Inherited mitochondrial optic neuropathies. J. Med. Genet., 46, 145–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Carelli V., La Morgia C., Valentino M.L., Barboni P., Ross-Cisneros F.N., Sadun A.A. (2009) Retinal ganglion cell neurodegeneration in mitochondrial inherited disorders. Biochim. Biophys. Acta., 1787, 518–528. [DOI] [PubMed] [Google Scholar]
- 5. Wallace D.C., Singh G., Lott M.T., Hodge J.A., Schurr T.G., Lezza A.M., Elsas L.J., Nikoskelainen E.K. (1998) Mitochondrial DNA mutation associated with Leber’s hereditary optic neuropathy. Science., 242, 1427–1430. [DOI] [PubMed] [Google Scholar]
- 6. Mashima Y., Yamada K., Wakakura M., Kigasawa K., Kudoh J., Shimizu N., Oguchi Y. (1998) Spectrum of pathogenic mitochondrial DNA mutations and clinical features in Japanese families with Leber’s hereditary optic neuropathy. Curr. Eye Res., 17, 403–408. [DOI] [PubMed] [Google Scholar]
- 7. Brown M.D., Torroni A., Reckord C.L., Wallace D.C. (1995) Phylogenetic analysis of Leber’s hereditary optic neuropathy mitochondrial DNA’s indicates multiple independent occurrences of the common mutations. Hum.Mutat., 6, 311–325. [DOI] [PubMed] [Google Scholar]
- 8. Ruiz-Pesini E., Lott M.T., Procaccio V., Poole J.C., Brandon M.C., Mishmar D., Yi C., Kreuziger J., Baldi P., Wallace D.C. (2007) An enhanced mitomap with a global mtDNA mutational phylogeny. Nucleic Acids Res., 35, D823–D828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mackey D.A., Oostra R.J., Rosenberg T., Nikoskelainen E., Bronte-Stewart J., Poulton J., Harding A.E., Govan G., Bolhuis P.A., Norby S. (1996) Primary pathogenic mtDNA mutations in multigeneration pedigrees with Leber hereditary optic neuropathy. Am. J. Hum.Genet., 59, 481–485., [PMC free article] [PubMed] [Google Scholar]
- 10. Kirches E. (2011) LHON: Mitochondrial Mutations and More. Curr. Genom., 212, 44–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Riordan-Eva P., Sanders M.D., Govan G.G., Sweeney M.G., Da Costa J., Harding A.E. (1995) The clinical features of Leber’s hereditary optic neuropathy defined by the presence of a pathogenic mitochondrial DNA mutation. Brain, 118, 319–337. [DOI] [PubMed] [Google Scholar]
- 12. Newman N.J., Lott M.T., Wallace D.C. (1991) The clinical characteristics of pedigrees of Leber’s hereditary optic neuropathy with the 11778 mutation. Am. J.Ophthalmol., 111, 750–762. [DOI] [PubMed] [Google Scholar]
- 13. Zhang J., Zhao F., Fu Q., Liang M., Tong Y., Liu X., Lin B., Mi H., Zhang M., Wei Q.P., et al. (2013) Mitochondrial haplotypes may modulate the phenotypic manifestation of the LHON-associated m.14484T>C (MT-ND6) mutation in Chinese families. Mitochondrion., 13, 772–781. [DOI] [PubMed] [Google Scholar]
- 14. Qu J., Zhou X., Zhang J., Zhao F., Sun Y.H., Tong Y., Wei Q.P., Cai W., Yang L., West C.E., Guan M.X. (2009) Extremely low penetrance of Leber’s hereditary optic neuropathy in 8 Han Chinese families carrying the ND4 G11778A mutation. Ophthalmology, 116, 558–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Torroni A., Petrozzi M., D'Urbano L., Sellitto D., Zeviani M., Carrara F., Carducci C., Leuzzi V., Carelli V., Barboni P., et al. (1997) Haplotype and phylogenetic analyses suggest that one European-specific mtDNA background plays a role in the expression of Leber hereditary optic neuropathy by increasing the penetrance of the primary mutations 11778 and 14484. Am. J. Hum. Genet., 60, 1107–1121. [PMC free article] [PubMed] [Google Scholar]
- 16. Hudson G., Keers S., Yu Wai Man P., Griffiths P., Huoponen K., Savontaus M.L., Nikoskelainen E., Zeviani M., Carrara F., Horvath R., et al. (2005) Identification of an X-chromosomal locus and haplotype modulating the phenotype of a mitochondrial DNA disorder. Am. J. Hum. Genet., 77, 1086–1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jiang P., Jin X., Peng Y., Wang M., Liu H., Liu X., Zhang Z., Ji Y., Zhang J., Liang M., et al. (2016) The exome sequencing identified the mutation in YARS2 encoding the mitochondrial tyrosyl-tRNA synthetase as a nuclear modifier for the phenotypic manifestation of Leber’s hereditary optic neuropathy-associated mitochondrial DNA mutation. Hum. Mol. Genet., 25, 584–596. [DOI] [PubMed] [Google Scholar]
- 18. Giordano L., Deceglie S., d'Adamo P., Valentino M.L., La Morgia C., Fracasso F., Roberti M., Cappellari M., Petrosillo G., Ciaravolo S., et al. (2016) Cigarette toxicity triggers Leber’s hereditary optic neuropathy by affecting mtDNA copy number, oxidative phosphorylation and ROS detoxification pathways. Cell Death Dis, 6, e2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Johns D.R., Berman J. (1991) Alternative, simultaneous complex I mitochondrial DNA mutations in Leber’s hereditary optic neuropathy. Biochem. Biophys. Res. Commun., 174, 1324–1330. [DOI] [PubMed] [Google Scholar]
- 20. Pello R., Martin M.A., Carelli V., Nijtmans L.G., Achilli A., Pala M., Torroni A., Gomez-Duran A., Ruiz-Pesini E., Martinuzzi A., et al. (2008) Mitochondrial DNA background modulates the assembly kinetics of OXPHOS complexes in a cellular model of mitochondrial disease. Hum. Mol.Genet., 17, 4001–4011. [DOI] [PubMed] [Google Scholar]
- 21. Howell N., Kubacka I., Halvorson S., Howell B., McCullough D.A., Mackey D. (1995) Phylogenetic analysis of the mitochondrial genomes from Leber hereditary optic neuropathy pedigrees. Genetics, 140, 285–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Qu J., Li R., Zhou X., Tong Y., Lu F., Qian Y., Hu Y., Mo J.Q., West C.E., Guan M.X. (2006) The novel A4435G mutation in the mitochondrial tRNAMet may modulate the phenotypic expression of the LHON-associated ND4 G11778A mutation in a Chinese family. Invest. Ophthalmol. Vis. Sci., 47, 475–483. [DOI] [PubMed] [Google Scholar]
- 23. Liang M., Jiang P., Li F., Zhang J., Ji Y., He Y., Xu M., Zhu J., Meng X., Zhao F., et al. (2014) Frequency and spectrum of mitochondrial ND6 mutations in 1218 Han Chinese subjects with Leber’s hereditary optic neuropathy. Invest. Ophthalmol. Vis. Sci., 55, 1321–1331. [DOI] [PubMed] [Google Scholar]
- 24. Jiang P., Liang M., Zhang J., Gao Y., He Z., Yu H., Zhao F., Ji Y., Liu X., Zhang M, et al. (2015) Prevalence of mitochondrial ND4 mutations in 1281 Han Chinese subjects with Leber’s hereditary optic neuropathy. Invest. Ophthalmol. Vis. Sci., 56, 4778–4788. [DOI] [PubMed] [Google Scholar]
- 25. Ji Y., Liang M., Zhang J., Zhu L., Zhang Z., Fu R., Liu X., Zhang M., Fu Q., Zhao F., et al. (2016) Mitochondrial ND1 Variants in 1281 Chinese Subjects With Leber’s Hereditary Optic Neuropathy. Invest. Ophthalmol. Vis. Sci., 57, 2377–2389. [DOI] [PubMed] [Google Scholar]
- 26. Zhao F., Guan M., Zhou X., Yuan M., Liang M., Liu Q., Liu Y., Zhang Y., Yang L., Tong Y., et al. (2009) Leber’s hereditary optic neuropathy is associated with mitochondrial ND6 T14502C mutation. Biochem.Biophys. Res.Commun., 389, 466–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhang J., Zhou X., Zhou J., Li C., Zhao F., Wang Y., Meng Y., Wang J., Yuan M., Cai W, et al. (2010) Mitochondrial ND6 T14502C variant may modulate the phenotypic expression of LHON-associated G11778A mutation in four Chinese families. Biochem.Biophys. Res.Commun., 399, 647–653. [DOI] [PubMed] [Google Scholar]
- 28. Qu J., Wang Y., Tong Y., Zhou X., Zhao F., Yang L., Zhang S., Zhang J., West C.E., Guan M.X. (2010) Leber’s hereditary optic neuropathy affects only female matrilineal relatives in two Chinese families. Invest. Ophthalmol. Vis. Sci., 51, 4906–4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. King M.P., Attardi G. (1989) Human cells lacking mtDNA: repopulation with exogenous mitochondria by complementation. Science, 246, 500–503. [DOI] [PubMed] [Google Scholar]
- 30. Zhang J., Jiang P., Jin X., Liu X., Zhang M., Xie S., Gao M., Zhang S., Sun Y.H., Zhu J., et al. (2014) Leber’s hereditary optic neuropathy caused by the homoplasmic ND1 m.3635G>A mutation in nine Han Chinese families. Mitochondrion, 18, 18–26. [DOI] [PubMed] [Google Scholar]
- 31. Tanaka M., Cabrera V.M., Gonzalez A.M., Larruga J.M., Takeyasu T., Fuku N., Guo L.J., Hirose R., Fujita Y., Kurata M., et al. (2004) Mitochondrial genome variation in eastern Asia and the peopling of Japan. Genom. Res., 14, 1832–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kong Q.P., Bandelt H.J., Sun C., Yao Y.G., Salas A., Achilli A., Wang C.Y., Zhong L., Zhu C.L., Wu S.F., et al. (2006) Updating the East Asian mtDNA phylogeny: a prerequisite for the identification of pathogenic mutations. Hum. Mol. Genet., 15, 2076–2086. [DOI] [PubMed] [Google Scholar]
- 33. Ingman M., Gyllensten U. (2006) mtDB: Human Mitochondrial Genome Database, a resource for population genetics and medical sciences. Nucleic Acids Res., 34, D749–D751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ruiz-Pesini E., Wallace D.C. (2006) Evidence for adaptive selection acting on the tRNA and rRNA genes of human mitochondrial DNA. Hum. Mutat., 27, 1072–1081. [DOI] [PubMed] [Google Scholar]
- 35. Zhou X., Qian Y., Zhang J., Tong Y., Jiang P., Liang M., Dai X., Zhou H., Zhao F., Ji Y., et al. (2012) Leber’s hereditary optic neuropathy is associated with the T3866C mutation in mitochondrial ND1 gene in three Han Chinese Families. Invest. Ophthalmol. Vis. Sci., 53, 4586–4594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. King M.P., Attadi G. (1996) Mitochondria-mediated transformation of human rho(0) cells. Methods Enzymol., 264, 313–334. [DOI] [PubMed] [Google Scholar]
- 37. Scheffler I.E. (2015) Mitochondrial disease associated with complex I (NADH-CoQ oxidoreductase) deficiency. J. Inherit.Metab. Dis., 38, 405–415. [DOI] [PubMed] [Google Scholar]
- 38. Gong S., Peng Y., Jiang P., Wang M., Fan M., Wang X., Zhou H., Li H., Yan Q., Huang T., Guan M.X. (2014) A deafness-associated tRNAHis mutation alters the mitochondrial function, ROS production and membrane potential. Nucleic Acids Res., 42, 8039–8048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Qian Y., Zhou X., Liang M., Qu J., Guan M.X. (2011) The altered activity of complex III may contribute to the high penetrance of Leber’s hereditary optic neuropathy in a Chinese family carrying the ND4 G11778A mutation. Mitochondrion., 11, 871–877. [DOI] [PubMed] [Google Scholar]
- 40. Mahfouz R., Sharma R., Lackner J., Aziz N., Agarwal A. (2009) Evaluation of chemiluminescence and flow cytometry as tools in assessing production of hydrogen peroxide and superoxide anion in human spermatozoa. Fertil.Steril., 92, 819–827. [DOI] [PubMed] [Google Scholar]
- 41. Chen C., Chen Y., Guan M.X. (2015) A peep into mitochondrial disorder: multifaceted from mitochondrial DNA mutations to nuclear gene modulation. Protein Cell., 6, 862–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang S., Wang L., Hao Y., Wang P., Hao P., Yin K., Wang Q.K., Liu M. (2008) T14484C and T14502C in the mitochondrial ND6 gene are associated with Leber’s hereditary optic neuropathy in a Chinese family. Mitochondrion., 8, 205–210. [DOI] [PubMed] [Google Scholar]
- 43. Shu L., Zhang Y.M., Huang X.X., Chen C.Y., Zhang X.N. (2012) Complete mitochondrial DNA sequence analysis in two southern Chinese pedigrees with Leber hereditary optic neuropathy revealed secondary mutations along with the primary mutation. Int. J. Ophthalmol.,5, 28–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jin X., Wang L., Gong Y., Chen B., Wang Y., Chen T., Wei S. (2015) Leber’s hereditary optic neuropathy is associated with compound primary mutations of mitochondrial ND1 m.3635G > A and ND6 m.14502 T > C. Ophthalmic. Genet., 36, 291–298. [DOI] [PubMed] [Google Scholar]
- 45. Zhou X., Zhang H., Zhao F., Ji Y., Tong Y., Zhang J., Zhang Y., Yang L., Qian Y, Lu F., et al. (2010) Very high penetrance and occurrence of Leber’s hereditary optic neuropathy in a large Han Chinese pedigree carrying the ND4 G11778A mutation. Mol. Genet. Metab., 100, 379–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Harding A.E., Sweeney M.G., Govan G.G., Riordan-Eva P. (1995) Pedigree analysis in Leber hereditary optic neuropathy families with a pathogenic mtDNA mutation. Am. J. Hum. Genet., 57, 77–86. [PMC free article] [PubMed] [Google Scholar]
- 47. Li R., Qu J., Zhou X., Tong Y., Hu Y., Qian Y., Lu F., Mo J.Q., West C.E., Guan M.X. (2006) The mitochondrial tRNAThr A15951G mutation may influence the phenotypic expression of the LHON-associated ND4 G11778A mutation in a Chinese family. Gene, 376, 79–86. [DOI] [PubMed] [Google Scholar]
- 48. Ji Y., Liang M., Zhang J., Zhang M., Zhu J., Meng X., Zhang S., Gao M., Zhao F., Wei Q.P., et al. (2014) Mitochondrial haplotypes may modulate the phenotypic manifestation of the LHON-associated ND1 G3460A mutation in Chinese families. J. Hum.Genet., 59, 134–140. [DOI] [PubMed] [Google Scholar]
- 49. Brown M.D., Trounce I.A., Jun A.S., Allen J.C., Wallace D.C. (2000) Functional analysis of lymphoblast and cybrid mitochondria containing the 3460, 11778, or 14484 Leber’s hereditary optic neuropathy mitochondrial DNA mutation. J. Biol. Chem., 275, 39831–39836. [DOI] [PubMed] [Google Scholar]
- 50. Hofhaus G., Johns D.R., Hurkoi O., Attardi G., Chomyn A. (1996) Respiration and growth defects in transmitochondrial cell lines carrying the 11778 mutation associated with Leber’s hereditary optic neuropathy. J. Biol. Chem., 271, 13155–13161. [DOI] [PubMed] [Google Scholar]
- 51. Li Y., D'Aurelio M., Deng J.H., Park J.S., Manfredi G., Hu P., Lu J., Bai Y. (2007) An assembled complex IV maintains the stability and activity of complex I in mammalian mitochondria. J. Biol. Chem., 282, 17557–17562. [DOI] [PubMed] [Google Scholar]
- 52. Szczepanowska J., Malinska D., Wieckowski M.R., Duszynski J. (2012) Effect of mtDNA point mutations on cellular bioenergetics. Biochim. Biophys. Acta, 1817, 1740–1746. [DOI] [PubMed] [Google Scholar]
- 53. Chen Y.B., Aon M.A., Hsu Y.T., Soane L., Teng X., McCaffery J.M., Cheng W.C., Qi B., Li H., Alavian K.N., et al. (2011) Bcl-xL regulates mitochondrial energetics by stabilizing the inner membrane potential. J. Cell Biol., 195, 263–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. de Andrade P.B., Rubi B., Frigerio F., van den Ouweland J.M., Maassen J.A., Maechler P. (2006) Diabetes-associated mitochondrial DNA mutation A3243G impairs cellular metabolic pathways necessary for beta cell function. Diabetologia, 49, 1816–1826. [DOI] [PubMed] [Google Scholar]
- 55. Jiang P., Wang M., Xue L., Xiao Y., Yu J., Wang H., Yao J., Liu H., Peng Y., Liu H., et al. (2016) A hypertension-associated tRNAAla mutation alters the tRNA metabolism and mitochondrial function. Mol. Cell. Biol., 36, 1920–1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Beretta S., Mattavelli L., Sala G., Tremolizzo L., Schapira A.H., Martinuzzi A., Carelli V., Ferrarese C. (2004) Leber hereditary optic neuropathy mtDNA mutations disrupt glutamate transport in cybrid cell lines. Brain., 127, 2183–2192. [DOI] [PubMed] [Google Scholar]
- 57. Porcelli A.M., Angelin A., Ghelli A., Mariani E., Martinuzzi A., Carelli V., Petronilli V., Bernardi P., Rugolo M. (2009) Respiratory complex I dysfunction due to mitochondrial DNA mutations shifts the voltage threshold for opening of the permeability transition pore toward resting levels. J. Biol. Chem., 284, 2045–2052. [DOI] [PubMed] [Google Scholar]
- 58. Wong A., Cavelier L., Collins-Schramm H.E., Seldin M.F., McGrogan M., Savontaus M.L., Cortopassi G.A. (2002) Differentiation-specific effects of LHON mutations introduced into neuronal NT2 cells. Hum. Mol. Genet., 11, 431–438. [DOI] [PubMed] [Google Scholar]
- 59. Hayashi G., Cortopassi G. (2015) Oxidative stress in inherited mitochondrial diseases. Free Radic. Biol. Med., 88A, 10–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ghelli A., Zanna C., Porcelli A.M., Schapira A.H., Martinuzzi A., Carelli V., Rugolo M. (2003) Leber’s hereditary optic neuropathy (LHON) pathogenic mutations induce mitochondrial-dependent apoptotic death in transmitochondrial cells incubated with galactose medium. J. Biol. Chem., 278, 4145–4150. [DOI] [PubMed] [Google Scholar]
- 61. Rieder M.J., Taylor S.L., Tobe V.O., Nickerson D.A. (1998) Automating the identification of DNA variations using quality-based fluorescence re-sequencing: analysis of the human mitochondrial genome. Nucleic Acids Res., 26, 967–973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Andrews R.M., Kubacka I., Chinnery P.F., Lightowlers R.N., Turnbull D.M., Howell N. (1999) Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat. Genet., 23, 147.. [DOI] [PubMed] [Google Scholar]
- 63. Guan M.X., Fischel-Ghodsian N., Attardi G. (2001) Nuclear background determines biochemical phenotype in the deafness-associated mitochondrial 12S rRNA mutation. Hum. Mol. Genet., 10, 573–580. [DOI] [PubMed] [Google Scholar]
- 64. Miller G., Lipman M. (1973) Release of infectious Epstein-Barr virus by transformed marmoset leukocytes. Proc. Natl. Acad. Sci. USA, 70, 190–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ausenda C., Chomyn A. (1996) Purification of mitochondrial DNA from human cell cultures and placenta. Methods Enzymol., 264, 122–128. [DOI] [PubMed] [Google Scholar]
- 66. Wittig I., Braun H.P., Schagger H. (2006) Blue native PAGE. Nat. Protoc., 1, 418–428. [DOI] [PubMed] [Google Scholar]
- 67. Birch-Machin M.A., Turnbull D.M. (2001) Assaying mitochondrial respiratory complex activity in mitochondria isolated from human cells and tissues. Methods Cell. Biol., 65, 97–117. [DOI] [PubMed] [Google Scholar]
- 68. Dranka B.P., Benavides G.A., Diers A.R., Giordano S., Zelickson B.R., Reily C., Zou L., Chatham J.C., Hill B.G., Zhang J., et al. (2011) Assessing bioenergetic function in response to oxidative stress by metabolic profiling. Free Radic. Biol. Med., 51, 1621–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.