Abstract
Background. Sofosbuvir (SOF) exhibits a high barrier to resistance, with no S282T NS5B substitution or phenotypic resistance detected in phase 3 registration studies.
Methods. Here, emergence of the NS5B variants L159F and V321A and possible association with resistance was evaluated in 8 studies of SOF (NEUTRINO, FISSION, POSITRON, FUSION, VALENCE, PHOTON-1, PHOTON-2, and P7977-2025) and 5 studies of combination ledipasvir (LDV) and SOF (LDV/SOF; LONESTAR, ELECTRON [LDV/SOF arms], ION1, ION2, and ION3), using deep sequencing.
Results. Deep sequencing detected L159F in 15% (53 of 353) and V321A in 5% (17 of 353) of patients with virologic failure in the SOF studies. Intensification of SOF treatment with LDV reduced the emergence of L159F or V321A to 2% (1 of 50 each) at virologic failure. L159F and V321A did not influence the outcome of retreatment with SOF, ribavirin, and pegylated interferon. At baseline, L159F was detected only in genotype 1–infected patients (1%) and was only associated with increased virologic failure in patients treated for short durations with SOF and ribavirin.
Conclusions. Deep-sequencing analysis confirmed that NS5B variants L159F and V321A emerged in a subset of patients treated with SOF at virologic failure. These variants had no impact on retreatment outcome with SOF, ribavirin, and pegylated interferon. Baseline L159F in genotype 1 did not affect the treatment outcome with LDV/SOF.
Keywords: direct-acting antivirals, sofosbuvir, resistance, HCV NS5B sofosbuvir resistance–associated variants, L159F, V321A
The pangenotypic NS5B hepatitis C virus (HCV) inhibitor sofosbuvir (SOF), when used in combination with other agents, has demonstrated high efficacy in patients infected with genotypes 1–6 (GT1–6) [1–6]. SOF was approved in 2013 for treatment, in combination with ribavirin (RBV), of HCV GT2 and GT3 infection and, in combination with pegylated interferon (peginterferon) and RBV, for treatment of GT1 and GT4–6 HCV infection. Sustained virologic response (SVR) rates of ≥95% have been obtained when combining SOF with the viral NS5A inhibitor ledipasvir (LDV) for treatment of HCV GT1–infected patients [7, 8]. In 2014, the combination of SOF with LDV (LDV/SOF) was approved for treatment of GT1 in the United States and Europe.
SOF exhibits a high barrier to resistance [9, 10]. After selection, S282T in NS5B was the only substitution selected in all tested GTs, and it conferred 2.4–18.1-fold reduced susceptibility to SOF [1, 11, 12]. Other substitutions observed in replicon selection in vitro did not show reduced susceptibility to SOF [11, 12]. Even though S282T was selected in vitro, selection of S282T in SOF clinical trials was very rare [1, 4, 13, 14]. In the SOF phase 2 and 3 registrational studies, 1662 patients received SOF-containing regimens, and only 1 patient developed S282T at virologic failure [4]. Similarly, in LDV/SOF phase 2 and 3 registrational studies, >2000 patients received SOF-containing regimens, and only 1 patient developed S282T at virologic failure [5]. In comparison, the low rate of S282T development observed for patients treated with SOF is not consistent for all nucleoside inhibitors. Among patients treated with a regimen containing VX-135, 4 of 23 experienced virologic failure, of whom 3 (75%) had S282T at treatment failure [15]. For patients treated with the regimen containing mericitabine, S282T was observed in 2 of 99 (2%) at virologic failure [16].
Comprehensive analysis of all NS5B population sequences in the SOF development program identified 2 NS5B variants, L159F and V321A, which emerged at the time of virologic failure in 6 and 5 GT3-infected patients, respectively, using a standard 15% population sequencing cutoff [11]. Analyses of these variants modeled in NS5B crystal structures indicated that these substitutions are close to the SOF binding site and could possibly affect the anti-HCV activity of SOF [17]. However, the fold reduction in the 50% effective concentration (EC50) of SOF against these variants was 1.2–1.3-fold for L159F in GT1a, GT1b, and GT3a and 1.3-fold for V321A in GT3a [11, 12]. Although the substitution L320F was not observed by population sequencing in the SOF program, it has been described as a variant associated with mericitabine treatment and with potentially reduced susceptibility to SOF [18].
The prevalence of L159F, V321A, and L320F NS5B substitutions was evaluated in 13 clinical trials, using deep sequencing with a 1% assay cutoff. Moreover, the prevalence of these variants at baseline and the subsequent impact on treatment outcome were evaluated.
METHODS
Clinical Trials
Sequencing analysis of NS5B was performed on patients who were enrolled in NEUTRINO, FISSION, POSITRON, FUSION, VALENCE, PHOTON-1, PHOTON-2, liver pretransplantation study P7977-2025, LONESTAR, ELECTRON (the LDV/SOF treatment arms, with or without RBV), ION1, ION2, and ION3 clinical trials. Information regarding each clinical trial can be found online (available at: http://www.clinicaltrials.gov). All patients included in these clinical trials have provided informed consent in writing, and the study protocol conformed to the ethical guidelines of the 1975 Declaration of Helsinki, as reflected in a priori approval by the appropriate institutional review committee.
Deep-Sequencing Analyses
NS5B deep-sequencing analysis was performed at baseline and virologic failure time points for patients who received at least 1 dose of a SOF-containing regimen; did not achieve SVR, owing to virologic failure or early discontinuation; and had an HCV RNA level of ≥1000 IU/mL. NS5B polymerase chain reaction amplicons at baseline and posttreatment time points, generated by DDL Diagnostic Laboratory (Rijswijk, the Netherlands), were subjected to deep sequencing using the Illumina MiSeq deep-sequencing platform (Illumina, San Diego, California) at DDL or WuXi AppTec (Shanghai, China). Internally developed software was used to process and align sequencing data to identify the substitutions present at levels of >1% [11, 17]. In addition, consensus sequences were generated for each sample with inclusion of mixtures of amino acids, when present, between 15% and 85%. Deep sequencing of baseline samples was also performed for a subset of patients who achieved SVR for 12 weeks after treatment cessation (SVR12).
Site-Directed Mutagenesis, Replication Capacity, and Sofosbuvir Susceptibility Assays
Site-directed mutants of replicons were constructed by Wuxi AppTec (Shanghai, China), using a QuikChange II Site-Directed Mutagenesis Kit (Stratagene). The NS5B region containing the introduced mutant was subcloned back into a wild-type Pi-Rluc plasmid. Transient transfection assays were performed at Wuxi AppTec or Gilead Sciences, as described previously, to determine the activity of SOF against mutant replicons [17]. Briefly, 1C cells were used for transient transfection. In vitro–transcribed RNA was transfected into 1C cells by electroporation. Transfected cells were plated in 96-well microplates, and serially diluted SOF was then added to the cells. For all assays, relative light unit (RLU) signals were obtained 4 and 96 hours after transfection, and replication capacities were calculated as follows: [RLUchimera, 72 hours/RLUchimera, 4 hours]/[RLUwild type, 72 hours/RLUwild type, 4 hours)]. Seventy-two hours after compound addition, luciferase signal was measured using the Promega Renilla-GLO Luciferase Assay kit or the Promega Renilla Luciferase Assay kit according to the manufacturer's instructions (Promega). Inhibitor susceptibility was determined by evaluating 3 replicates. Intra-assay and interassay variations were each approximately 2–3 fold [19].
RESULTS
SOF Treatment–Emergent L159F and V321A Variants
The emergence of L159F and V321A NS5B substitutions was evaluated in 8 clinical studies of SOF and 5 clinical studies of LDV/SOF. The studies investigated here are described as SOF studies if the treatment regimen was SOF plus RBV (SOF +RBV) with or without peginterferon (SOF +RBV ± peginterferon), and as LDV/SOF studies if the treatment regimen was SOF plus LDV, with or without RBV. There were 1817 SOF-treated patients in the SOF studies, of whom 353 had virologic failure (19.4%). At the time of virologic failure, deep-sequencing analysis detected the L159F variant in 53 patients (15%) and the V321A variant in 17 (5%; Table 1). Four patients had both L159F and V321A detected at relapse. In the SOF studies included here, no S282T was observed. However, as reported previously, S282T was detected in 1 patient who received SOF monotherapy for 12 weeks in the ELECTRON study [20]. In a similar number of SOF-treated patients in the LDV/SOF studies, only 50 of 1807 (2.8%) experienced virologic failure. Deep-sequencing analysis showed that the L159F or V321A variants were each detected in only a single patient at the time of failure (Table 1). S282T was only detected in 1 patient, a participant in the LONESTAR study who received LDV/SOF therapy for 8 weeks [5].
Table 1.
L159F and V321A Detection by Deep Sequencing at Virologic Failure in Studies of Sofosbuvir (SOF) and Ledipasvir (LDV) Plus SOF
| Study, GT(s) | Patients Treated, No. | Patients at Virologic Failurea |
||
|---|---|---|---|---|
| Deep-Sequencing Data, No. | L159F, No. (%) | V321A, No. (%) | ||
| SOF + RBV, pretransplantation | ||||
| 1–4 | 61 | 29 | 10 (34) | 1 (3) |
| SOF + RBV, phase 3 | ||||
| 1a | 200 | 35 | 1 (3) | 3 (9) |
| 1b | 42 | 17 | 4 (22) | … |
| 2 | 388 | 17 | 1 (6) | … |
| 3 | 834 | 227 | 35 (15) | 13 (6) |
| SOF + RBV + peginterferon, phase 3 | ||||
| 1a | 226 | 18 | 1 (6) | … |
| 1b | 66 | 10 | 1 (10) | … |
| Overall | 1817 | 353 | 53 (15) | 17 (5) |
| LDV/SOF, phase 2/3 | ||||
| 1a | 1394 | 41 | 1 (2) | 1 (2) |
| 1b | 413 | 9 | … | … |
| Overall | 1807 | 50 | 1 (2) | 1 (2) |
Abbreviations: Peginterferon, pegylated interferon; RBV, ribavirin.
a Presence of L159F and V321A was counted in all cases if it was detected at virologic failure. Patients who had these variants at baseline and maintained them at the virologic failure time point, as well as patients who had these variants emerge after treatment, were counted.
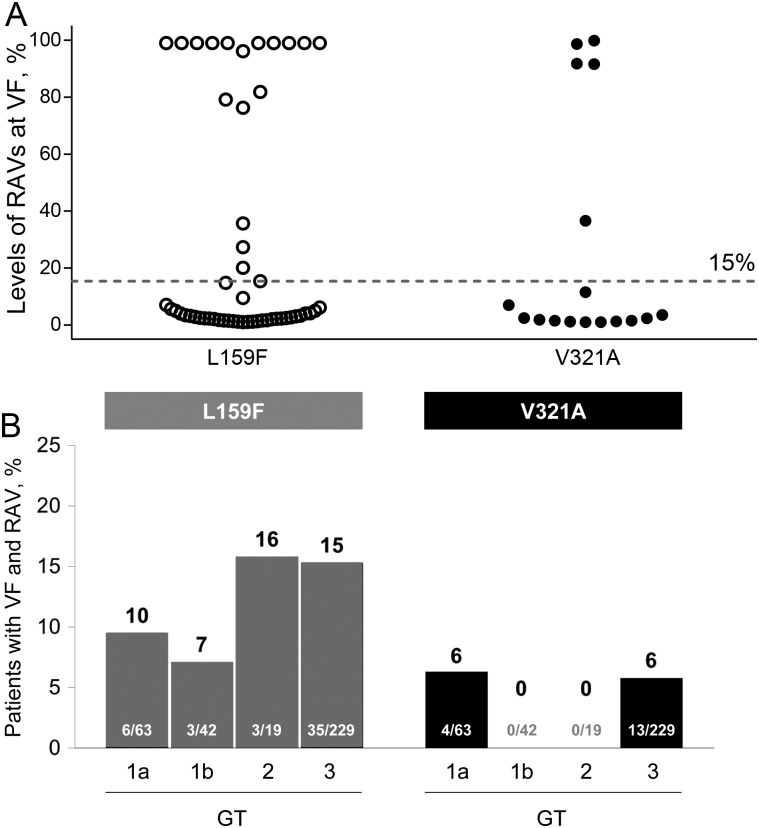
In the SOF studies, the frequency of the L159F and V321A variants at the time of virologic failure was evaluated on the basis of the number of reads containing these variants divided by the total number of reads analyzed at these positions. L159F and V321A variants were detected as a minor viral population in 35 of 53 patients (66%) and 12 of 17 patients (70%), respectively (Figure 1A). A minor population was defined as <15% of the total virus population, which is below the detection limit for standard population sequencing. Of these patients with minor variants, 19 of 35 (54%) and 9 of 12 (75%) had only 1%–2% of the viral quasispecies containing L159F and V321A, respectively. At subsequent posttreatment follow-up time points, the frequency of the variant rapidly declined in the majority of the patients. L159F and V321A declined in frequency in 18 of 24 and 12 of 13 patients, respectively, with longitudinal samples obtained between 4 and 20 weeks after initial detection (Supplementary Figure 1).
Figure 1.
Levels of L159F and V321A detected in sofosbuvir (SOF) studies by deep-sequencing analysis. A, The levels of emergent L159F and V321A in samples obtained from patients after SOF treatment are shown on the y-axis. The cutoff of 15% for standard population sequencing is shown by the dotted line indicating that, for most patients, the level of L159F and V321A was <15% and only detectable by deep sequencing. B, Emergence of L159F and V321A in SOF studies across hepatitis C virus genotypes (GTs). The number of patients with emergent L159F or V321A detected at virologic relapse divided by the total number of patients with virologic relapse is shown inside the bars for each GT. The resulting percentages are shown on top of the bars. Abbreviations: RAV, resistance-associated variant; VF, virologic failure.
The emergence of L159F and V321A was compared in GT-1a, GT1b, GT2, and GT3 (Figure 1B). Emergence of these variants following SOF treatment was not restricted to any particular GT, with similar rates observed across GT1, GT2, and GT3. Lack of emergence of V321A in GT1b and GT2 was possibly related to the low number of patients who experienced virologic failure with these GTs in the SOF studies. There was a limited number of patients infected with GT4, GT5, and GT6 (n = 51) who were treated in these SOF studies, with only a single patient experiencing virologic failure, in whom neither variant was detected.
Another nucleotide inhibitor resistance–associated variant, L320F, was observed in 5 cases (1.4%) at relapse in the SOF studies, with 4 of these cases observed in the liver pretransplantation study. The level of L320F at the time of relapse in these patients ranged from 1% to 7%. In vitro analyses of L320F showed a low fold reduction in SOF susceptibility (1.8-fold and 1.7-fold for GT1a and GT1b, respectively) [21]. L320F was not observed in any patient in the LDV/SOF studies.
Retreatment of Patients With Emergent L159F and V321A
To investigate whether the emergence of L159F and V321A variants affected retreatment outcome with SOF regimens, 23 of the virologic failures across the SOF studies with either L159F or V321A variants were retreated with either SOF + RBV +peginterferon for 12 weeks or with SOF + RBV for 24 weeks. Of these 23 patients, 18 (78%) achieved SVR following retreatment, which is similar the SVR rate of 78% (382 of 490 patients) observed in patients without L159F or V321A in the retreatment study, GS-US-334-0109. All 5 patients from the retreatment study who experienced relapse had GT3a infection and L159F, and 1 also had emergent V321A from the parental study. At the time of retreatment failure, L159F was no longer detectable in any patient, and V321A was detected in 1 patient. In this patient, V321A was detected as a minor viral population, and the level was not enriched following retreatment.
Baseline Prevalence of L159F and V321A
To evaluate pretreatment prevalence of L159F and V321A, baseline samples were deep sequenced from 1611 and 1470 patients who were subsequently treated with SOF-containing regimens in the SOF and LDV/SOF studies, respectively (Table 2). The L159F variant was detected in only 0.6% of patients in the SOF studies and in 1.6% of patients in the LDV/SOF studies, while V321A was not detected in any patient at baseline. Of the patients with baseline L159F, the majority (32 of 33) had GT1b HCV, and 1 had GT1a HCV. The prevalence of baseline L159F was 7% in GT1b and <0.01% in GT1a, respectively. No baseline L159F was detected in patients with HCV GT2 or GT3 infection. In the SOF and LDV/SOF studies, 2 of 6 and 0 of 23 patients with L159F experienced virologic failure, respectively. The 6 patients with baseline L159F in the SOF studies were treated with SOF + RBV ± peginterferon for 12 or 24 weeks (Table 3). The SVR rate for GT1b-infected patients in the NEUTRINO study who were treated with SOF + RBV +peginterferon and those in the PHOTON-1 study who were treated with SOF + RBV without baseline L159F was 84% and 54%, respectively. Thus, baseline L159F was not associated with treatment failure in these studies.
Table 2.
Baseline L159F and V321A in Studies of Sofosbuvir (SOF) and Ledipasvir (LDV) Plus SOF
| Study, GT | Baseline Sequencing Data, No.a | Baseline L159F, No. (%) | Baseline L159F and VF, Proportion |
|---|---|---|---|
| SOF + RBV, pretransplantation | |||
| 1–4 | 60 | 4 (all GT1b) | 4/4 |
| SOF + RBV, phase 3 | |||
| 1a | 128 | 0 | … |
| 1b | 33 | 2 | 1/2 |
| 2 | 402 | 0 | … |
| 3 | 699 | 0 | … |
| SOF + RBV + peginterferon, phase 3 | |||
| 1a | 224 | 0 | … |
| 1b | 65 | 4 | 1/4 |
| Overall | 1611 | 10 (0.6) | 6/10 |
| LDV/SOF, phase 2/3 | |||
| 1a | 1150 | 1 | 0/1 |
| 1b | 320 | 22 | 0/22 |
| Overall | 1470 | 23 (1.6) | 0/23 |
Abbreviations: GT1b, genotype 1b; peginterferon, pegylated interferon; RBV, ribavirin; VF, virologic failure.
a No V321A was detected in any patient at baseline.
Table 3.
Characteristics of Patients with Baseline L159F in Studies of Sofosbuvir (SOF) and Their Treatment Outcome
| Study | Patient | GT | Treatment | Treatment Duration, wk | IL28 | TN/TE | AGRE | NS5B RAVs at BL (Population Sequencing) | HCV RNA Load, IU/mL | Relapse |
|---|---|---|---|---|---|---|---|---|---|---|
| NEUTRINO | A | 1b | SOF + peginterferon + RBV | 12 | CC | TN | 63/M/WH/H | L159F C316N | 4 770 000 | No |
| NEUTRINO | B | 1b | SOF + peginterferon + RBV | 12 | CC | TN | 63/F/WH/NH | L159F C316N S556G | 12 100 000 | No |
| NEUTRINO | C | 1b | SOF + peginterferon + RBV | 12 | TT | TN | 65/F/WH/NH | L159F C316N | 1 440 000 | Yes |
| NEUTRINO | D | 1b | SOF + peginterferon + RBV | 12 | CC | TN | 52/M/WH/NH | L159F C316N | 1 220 000 | No |
| PHOTON-1 | E | 1b | SOF + RBV | 24 | TT | TN | 55/M/WH/NH | L159F C316N | 6 450 000 | Yes |
| PHOTON-1 | F | 1b | SOF + RBV | 24 | CT | TN | 52/M/WH/NH | L159F C316N | 692 000 | No |
Abbreviations: AGRE, age, sex, race, ethnicity; BL, baseline; GT, genotype; H, Hispanic; HCV, hepatitis C virus; NH, non-Hispanic; peginterferon, pegylated interferon; RAV, resistance-associated variant; RBV, ribavirin; TE, treatment experienced; TN, treatment naive; WH, white.
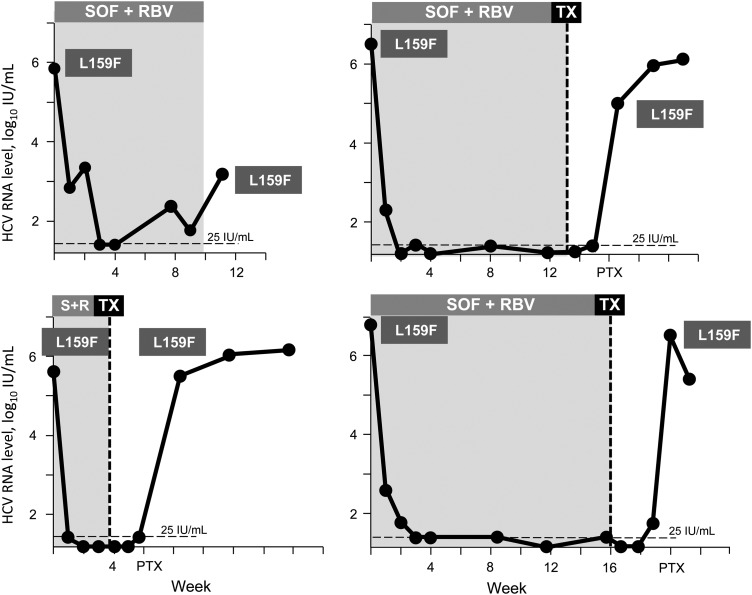
In comparison, in the liver pretransplantation study, 4 of 4 patients with pretreatment L159F experienced virologic failure following a short duration of treatment with SOF + RBV (4–16 weeks). The individual viral kinetics of these 4 GT1b HCV–infected patients is shown in Figure 2. L159F was the dominant variant within the viral population at baseline in all patients (>99%). During SOF + RBV treatment, the HCV RNA level was suppressed, and 1 patient had virologic breakthrough with L159F at week 8 of treatment. Treatment of the other 3 patients was stopped after only 4–16 weeks, when liver transplantation was performed. All had HCV recurrence soon after transplantation, with L159F as the major species (>99%). At the time of SOF + RBV treatment, the patients had advanced liver disease, which could have contributed to failure. Moreover, the treatment duration of 4–16 weeks for these GT1-infected patients was shorter than the anticipated 24 weeks.
Figure 2.
Viral response to sofosbuvir plus ribavirin (SOF + RBV) in patients with L159F at baseline in the P7977-2025 Pretransplantation Study. The SOF + RBV treatment duration for each patient is shaded in the figure. The limit of the detection of the viral load assay at 25 IU/mL is shown as a black dotted line. Abbreviation: TX, liver transplantation.
Baseline L159F commonly coexisted with C316N, where 31 of 33 patients harboring L159F also had C316N in NS5B. No emergent C316N variant was observed in studies investigated here, and therefore it was not considered to be associated with SOF resistance.
Site-Directed Mutagenesis and Phenotypic Analyses
To understand the role of L159F, V321A, and L320F or a combination of these substitutions in the susceptibility to SOF and viral replication capacity, a panel of mutant NS5B replicons was constructed and tested in a transient transfection replicon assay (Table 4).
Table 4.
Activity of Sofosbuvir (SOF) Against Replicons Encoding NS5B Variants
| NS5B variant | Replication Capacity, %, Mean±SDa | SOF EC50 Fold Change, Mean±SDb |
|---|---|---|
| GT1a | ||
| L159F | 8.9 ± 1.0 | 1.2 ± 0.014 |
| C316N | 191.4 ± 57.4 | 1.6 ± 0.41 |
| C316H | 100.0 ± 0.99 | 0.9 ± 0.05 |
| C316F | 41.2 ± 4.0 | 1.6 ± 0.36 |
| L320F | 39.3 ± 3.5 | 1.8 ± 0.2 |
| V321A | 17.6 ± 11.5 | 1.2 ± 0.07 |
| S282R | NR | NA |
| D61G | NR | NA |
| L159F/C316N | 114.6 ± 34.8 | 1.8 ± 0.10 |
| L159F/L320F | 12.5 ± 10.2 | 2.2 ± 0.2 |
| S282R/L320F | NR | NA |
| D61G/S62H | NR | NA |
| D61G/S62D | NR | NA |
| D61G/S62N | NR | NA |
| S282T | 1.3 ± 0.3 | 8.4 ± 2.0 |
| GT1b | ||
| L159F | 24.1 ± 6.3 | 1.3 ± 0.3 |
| C316N | 90.4 ± 18.4 | 1.0 ± 0.21 |
| C316H | 7.4 ± 2.4 | 0.9 ± 0.05 |
| C316F | 28.2 ± 14.2 | 1.4 ± 0.04 |
| L320F | 22.9 ± 3.4 | 1.7 ± 0.17 |
| V321A | 6.8 ± 3.6 | 1.4 ± 0.07 |
| E440G | 137.2 ± 9.7 | 0.9 ± 0.002 |
| L159F/C316N | 65.3 ± 3.5 | 1.6 ± 0.60 |
| L159F/L320F | 9.3 ± 1.5 | 2.2 ± 0.47 |
| S282T | 8.4 ± 1.6 | 8.8 ± 1.9 |
| GT2b | ||
| L159F | 10.1 ± 2.9 | 1.6 ± 0.5 |
| L159F/C316N | 6.4 ± 2.4 | 0.9 ± 0.17 |
| L159F/L320F | 6.5 ± 5.5 | 1.2c |
| S282T | 11.3 ± 1.5 | 16.2 ± 0.8 |
| GT3a | ||
| L159F | 23.0 ± 6.5 | 1.3 ± 0.02 |
| V321A | 20.0 ± 7.1 | 1.3 ± 0.3 |
| K211R | 152.7 ± 116.5 | 1.1 ± 0.18 |
| P540L | 6.8 ± 5.2 | 1.2 ± 0.35 |
| T542A | 67.0 ± 17.0 | 1.4 ± 0.35 |
| L159F/C316N | 6.7 ± 0.66 | 0.7 ± 0.05 |
| L159F/L320F | 0.85 ± 0.09 | 1.9 ± 0.41 |
| S282T | 11.3 ± 1.2 | 3.5 ± 0.3 |
GT1a, GT1b, and GT3a transient replicons consisted of full-length sequences, while the chimeric GT1b replicon carrying the NS5B gene from GT2b was used for transient evaluation.
Abbreviations: EC50, 50% effective concentration; GT, genotype; NA, not applicable (replication was too low to calculate EC50 and fold change); NR, no replication was observed.
a Percentage replication capacity relative to wild type.
b Fold change from corresponding wild type.
c Fold change obtained from only 1 experiment; in 2 more experiments, replication was too low to calculate EC50.
L159F and V321A displayed diminished replication capacity of 6.8%–24.1% in GT1a, GT1b, and GT3a replicons, with a small effect on susceptibility to SOF (1.2–1.4-fold change). When L159F was combined with C316N, replication capacity was restored to 65.3%–114.6% of the wild-type replicon in GT1a and GT1b. No compensatory effect of C316N on the replication defect of L159F was observed in GT2b and 3a replicons. Similarly, the replication capacity of the L320F mutant was also diminished (range, 22.9%–39.3% of the wild-type replicon), and the effect on susceptibility to SOF was low (1.7–1.8-fold change) in GT1a and GT1b replicons. For comparison, the fold-change of S282T is 2.4–18.1 in GT1-6 replicons [5, 10], with a replication capacity of 3.2%–11.3% of the corresponding wild-type replicon in GT1-6 replicons [12].
DISCUSSION
SOF exhibits a high barrier to resistance, with no S282T NS5B substitution or phenotypic resistance detected in the phase 3 registration studies that included >990 SOF-treated patients. S282T is the only variant known to reduce in vitro susceptibility to SOF [1], and it has been rarely detected in patients treated with SOF [5, 20]. In vitro phenotypic analysis of L159F and V321A variants demonstrates a low fold reduction in the SOF EC50. However, since virologic failure in general is associated with resistance, additional analysis beyond in vitro selection is necessary to fully exclude selection or the presence of resistance-associated variants (RAVs). Here, the prevalence and frequencies of these mutations in pretreatment and posttreatment samples obtained in 13 phase 2/3 studies of SOF and LDV/SOF were investigated using a deep-sequencing assay to enable detection of low-frequency variants. Moreover, the pretreatment prevalence and the impact of L159F and V321A on treatment outcome were evaluated.
Of 1817 patients in the SOF studies, 353 experienced virologic failure, among whom L159F and V321A were detected in 15% and 5%, respectively. No other emergent variants were identified at relapse. Donaldson et al described C316N and S282R as possibly associated with SOF failure [17]. However, in our data set, with 13 clinical trials, C316N did not emerge at virologic failure in any patient and commonly coexisted with L159F at baseline in patients with GT1b infection. Minor levels of S282R were also observed very rarely in the SOF studies, with 2 patients developing this mutant at relapse to date. The site-directed mutant S282R failed to replicate in vitro, and the effect of this mutant on SOF susceptibility could not be established [21]. L320F was previously described as a variant associated with mericitabine treatment and with potentially reduced susceptibility to SOF [18]. In our data set, L320F was observed in 5 cases (1.4%) at relapse in the SOF studies, with 4 of these 5 cases observed in the liver pretransplantation study. L320F was not observed in any patient in the LDV/SOF studies. The rare occurrence of this substitution and the low fold change in the SOF EC50 [21] indicates that further studies are needed to investigate any possible association of L320F with SOF resistance.
When SOF treatment was intensified through addition of peginterferon or LDV, the emergence of L159F and V321A was dramatically reduced. In the LDV/SOF studies, which included 1807 patients, the number of virologic failures was lower, and only a single patient developed L159F and 1 developed V321A, indicating a decreased selection of these variants during LDV/SOF combination therapy.
To study the impact of L159F and V321A on treatment outcome, the prevalence of these variants at baseline was investigated in the SOF and LDV/SOF studies. At baseline, the prevalence of L159F was 7% and <0.1% in GT1b and GT1a, respectively, and it was not detected in any other GTs. V321A was not detected in the HCV of any patient at baseline. The high cure rate of LDV/SOF in phase 2/3 studies indicates that this treatment is highly effective for GT1-infected patients. Indeed, baseline L159F did not preclude an SVR in the LDV/SOF and in most SOF studies, and thus no association with treatment failure was observed. Baseline L159F was only associated with treatment failure in the liver pretransplantation study and in 1 study conducted in Russia, where GT1-infected patients received SOF + RBV for 16 weeks (Isakov V, Zhdanov K, Kersey K, et al, unpublished data) [22]. In the liver transplant study, 4 GT1b-infected patients with pretreatment L159F experienced virologic failure. L159F was present as the major variant within the viral population at baseline in these patients. It is important to note that virologic failure in all 4 patients was possibly associated with the short treatment duration of SOF +RBV (<24 weeks in GT1-infected patients) in combination with advanced liver disease. Any independent effect of L159F on the response to SOF-based treatment is not clear. However, in a recent study conducted in Russia, L159F was detected in 22 of 65 patients (34%) at baseline (Isakov V, Zhdanov K, Kersey K, et al, unpublished data). Of these 22 patients, 12 received SOF + RBV for 16 weeks, and 10 patients received SOF + RBV for 24 weeks. The resulting rates of SVR 12 for these studies were 25% (3 of 12 patients) in the 16-week arm and 80% (8 of 10) in the 24-week arm. These results and those from the pretransplantation study suggest that the L159F variant is associated with treatment failure following shortened duration of SOF + RBV therapy. However, any impact of baseline L159F appears to be obviated by either the prolongation of SOF + RBV treatment to 24 weeks or the addition of peginterferon or LDV.
Owing to the close proximity of L159 to the S282 position and of V321 to the NS5B catalytic triad (D220, D318, and D319), these variants have been predicted to enable conformational alterations of the active site of the polymerase and thus to have a possible association with SOF resistance [17]. However, in vitro phenotypic analyses of L159F and V321A demonstrated a low change in susceptibility to SOF (1.2–1.6-fold). In agreement, a similar fold change has been described for L159F to SOF [18]. In contrast, the S282T mutant has about a 10-fold reduction in susceptibility to SOF and is the primary mutation selected in vitro in all GTs by SOF, whereas L159F or V321A have not been observed in in vitro selection studies [1, 18]. However, despite the low phenotypic susceptibility shift of L159F and V321A, these substitutions are still of clinical relevance. In in vitro replicon systems, L159F are associated with a fitness cost, and interestingly the replication capacity was restored when L159F was combined with C316N, suggesting that C316N may act as a compensatory mutation and restore replication defects associated with L159F specifically in GT1 HCV. Since L159F was shown to be associated with virologic failure in GT1b-infected patients in the liver pretransplantation study and in the 16-week SOF + RBV study conducted in Russia, it indicates that these resistance-associated variants that lack a strong fold change might still have clinical significance in a subset of patients.
At postrelapse follow-up, the levels of L159F and V321A declined over time in the majority of patients for whom longitudinal samples after initial detection were available, indicating a reduced viral fitness of these variants. Moreover, at the time of failure, L159F and V321A were detected as a minor species in most patients (<15% of the viral population), suggesting that levels might already have been reduced at the time of sampling. Based on these results, we propose a hypothetical mechanism underlying L159F and V321A emergence. Owing to the high genetic variation of HCV, L159F and V321A likely preexist in the viral population before treatment initiation at low abundance, owing to reduced replicative capacity. During treatment, both wild-type and mutant virus are suppressed by SOF, but the small reduced susceptibility of L159F and V321A to SOF enriches these variants relative to wild type. After cessation of treatment, L159F and/or V321A are rapidly replaced by wild-type virus with a greater replication efficiency due to its higher replicative capacity, compared with L159F and V321A, and may only represent a minority of the viral quasispecies. However, depending on the sampling time, the mutants may still be detectable by deep sequencing. The addition of either LDV or peginterferon reduces the emergence of these variants owing to further suppression of L159F and V321A mutants. Encouragingly, 23 patients with L159F and/or V321A detected at relapse were retreated with either SOF +peginterferon/RBV (n = 13) or with SOF + RBV for 24 weeks (n = 8), and 78% achieved an SVR. This SVR rate was similar to that for patients who did not harbor L159F and/or V321A. In the few patients who experienced relapse after retreatment, L159F was not detectable at the time of retreatment failure, and V321A remained detectable in one patient but was not enriched at the time of retreatment failure. This suggests that the presence of these resistance-associated variants does not impact retreatment with a SOF-based treatment.
In summary, deep-sequencing analysis showed that NS5B variants L159F and V321A emerged in a subset of patients treated with SOF at virologic failure but were rare in patients treated with LDV/SOF. The emergence of these variants did not impact the outcome of retreatment with SOF + RBV ± peginterferon. Moreover, baseline L159F was associated with virologic failure in a subset of GT1b-infected patients treated for a shorter duration with SOF + RBV but not in patients who received LDV/SOF.
Supplementary Material
Notes
Acknowledgments. We thank the patients who participated in the phase 2/3 clinical studies of sofosbuvir, the research staff in the clinical virology department at Gilead, and the clinical investigators and resistance testing companies.
Financial support. This work was supported by Gilead Sciences.
Potential conflicts of interest. E. S. S., H. D.-S., R. M., B. D., D. B., J. G. M., M. D. M., and H. M. are Gilead employees and stockholders. E. L. consults for, and received grants from AbbVie, Achillion Pharmaceuticals, Bristol-Myers Squibb; Enanta, Gilead Sciences, Janssen, Merck & Co., Novartis, Santaris Pharmaceuticals, Regulus,Theravance. He is on the speakers bureaus for, and received grants from AbbVie, Bristol-Myers Squibb, Gilead, Janssen. E. G. is a board member for, and received grants from Abbvie, Janssen, Gilead Sciences and Merck. He is on the speakers bureaus for, and received grants from Abbvie, Gilead Sciences and Merck. I. M. J. is on the speaker bureaus for, and received grants from Abbvie, Bistrol Myers Squibb, Gilead Sciences and Janssen. He consults for, and received grants from Achillion, Enanta, Merck and Tobira. D. R. N. has received grants from Abbvie, Bristol-Myers Squibb, Gilead Sciences, Janssen and Merck. All other authors report no potential conflicts. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Lam AM, Espiritu C, Bansal S et al. Genotype and subtype profiling of PSI-7977 as a nucleotide inhibitor of hepatitis C virus. Antimicrob Agents Chemother 2012; 56:3359–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kowdley KV, Lawitz E, Crespo I et al. Sofosbuvir with pegylated interferon alfa-2a and ribavirin for treatment-naive patients with hepatitis C genotype-1 infection (ATOMIC): an open-label, randomised, multicentre phase 2 trial. Lancet 2013; 381:2100–7. [DOI] [PubMed] [Google Scholar]
- 3. Rodriguez-Torres M, Lawitz E, Kowdley KV et al. Sofosbuvir (GS-7977) plus peginterferon/ribavirin in treatment-naive patients with HCV genotype 1: a randomized, 28-day, dose-ranging trial. J Hepatol 2013; 58:663–8. [DOI] [PubMed] [Google Scholar]
- 4. Lawitz E, Lalezari JP, Hassanein T et al. Sofosbuvir in combination with peginterferon alfa-2a and ribavirin for non-cirrhotic, treatment-naive patients with genotypes 1, 2, and 3 hepatitis C infection: a randomised, double-blind, phase 2 trial. Lancet Infect Dis 2013; 13:401–8. [DOI] [PubMed] [Google Scholar]
- 5. Lawitz E, Poordad FF, Pang PS et al. Sofosbuvir and ledipasvir fixed-dose combination with and without ribavirin in treatment-naive and previously treated patients with genotype 1 hepatitis C virus infection (LONESTAR): an open-label, randomised, phase 2 trial. Lancet 2014; 383:515–23. [DOI] [PubMed] [Google Scholar]
- 6. Zeuzem S, Dusheiko GM, Salupere R et al. Sofosbuvir and ribavirin in HCV genotypes 2 and 3. N Engl J Med 2014; 370:1993–2001. [DOI] [PubMed] [Google Scholar]
- 7. Afdhal N, Zeuzem S, Kwo P et al. Ledipasvir and sofosbuvir for untreated HCV genotype 1 infection. N Engl J Med 2014; 370:1889–98. [DOI] [PubMed] [Google Scholar]
- 8. Afdhal N, Reddy KR, Nelson DR et al. Ledipasvir and sofosbuvir for previously treated HCV genotype 1 infection. N Engl J Med 2014; 370:1483–93. [DOI] [PubMed] [Google Scholar]
- 9. Lam AM, Murakami E, Espiritu C et al. PSI-7851, a pronucleotide of beta-D-2′-deoxy-2′-fluoro-2′-C-methyluridine monophosphate, is a potent and pan-genotype inhibitor of hepatitis C virus replication. Antimicrob Agents Chemother 2010; 54:3187–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sofia MJ, Bao D, Chang W et al. Discovery of a beta-d-2′-deoxy-2′-alpha-fluoro-2′-beta-C-methyluridine nucleotide prodrug (PSI-7977) for the treatment of hepatitis C virus. J Med Chem 2010; 53:7202–18. [DOI] [PubMed] [Google Scholar]
- 11. Svarovskaia ES, Dvory-Sobol H, Parkin N et al. Infrequent development of resistance in genotype 1–6 hepatitis C virus-infected subjects treated with sofosbuvir in phase 2 and 3 clinical trials. Clin Infect Dis 2014; 59:1666–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Xu S, Rajyaguru S, Chiu S et al. Sofosbuvir selects the NS5B S282T mutation in vitro in genotype 1‒6 Replicons and is not cross-resistant to resistance-associated variants selected by other classes of antiviral inhibitors [poster 1094]. Presented at: 64th Annual Meeting of the American Association for the Study of Liver Diseases: The Liver Meeting, Washington, DC,1–5 November 2013. [Google Scholar]
- 13. Jacobson IM, Gordon SC, Kowdley KV et al. Sofosbuvir for hepatitis C genotype 2 or 3 in patients without treatment options. N Engl J Med 2013; 368:1867–77. [DOI] [PubMed] [Google Scholar]
- 14. Lawitz E, Mangia A, Wyles D et al. Sofosbuvir for previously untreated chronic hepatitis C infection. N Engl J Med 2013; 368:1878–87. [DOI] [PubMed] [Google Scholar]
- 15. Gane E, Stedman C, Garg V et al. An interferon- and ribavirin-free 12-week regimen of once-daily VX-135 and daclatasvir in treatment-naïve patients with genotype 1 HCV infection. Presented at: 49th Annual Meeting of the European Association for the Study of the Liver, London, United Kingdom, 9–13 April 2014. [Google Scholar]
- 16. Gane EJ, Pockros PJ, Zeuzem S et al. Mericitabine and ritonavir-boosted danoprevir with or without ribavirin in treatment-naive HCV genotype 1 patients: INFORM-SVR study. Liver Int 2015; 35:79–89. [DOI] [PubMed] [Google Scholar]
- 17. Donaldson EF, Harrington PR, O'Rear JJ, Naeger LK. Clinical evidence and bioinformatics characterization of potential hepatitis C virus resistance pathways for sofosbuvir. Hepatology 2015; 61:56–65. [DOI] [PubMed] [Google Scholar]
- 18. Tong X, Le Pogam S, Li L et al. In vivo emergence of a novel mutant L159F/L320F in the NS5B polymerase confers low-level resistance to the HCV polymerase inhibitors mericitabine and sofosbuvir. J Infect Dis 2014; 209:668–75. [DOI] [PubMed] [Google Scholar]
- 19. Han D, Strommen K, Anton ED et al. Analytical validation of an HCV replicon-based phenotypic assay for evaluating replication capacity and susceptibility of genotype 1a/1b patient viruses to polymerase inhibitors. Presented at: International Workshop on HIV & Hepatitis Virus Drug Resistance and Curative Strategies, Los Cabos, Mexico, 7–11 June 2011. [Google Scholar]
- 20. Gane EJ, Stedman CA, Hyland RH et al. Nucleotide polymerase inhibitor sofosbuvir plus ribavirin for hepatitis C. N Engl J Med 2013; 368:34–44. [DOI] [PubMed] [Google Scholar]
- 21. Dvory-Sobol H, Han B, Ouyang W et al. Susceptibility of HCV genotype 1, 2, 3 and 4 baseline clinical isolates to sofosbuvir. Presented at: 21st International Symposium on Hepatitis C and Related Viruses, Banff, Canada, 7–11 September 2014. [Google Scholar]
- 22. Chulanov V, Zhdanov K, Kersey K et al. Sofosbuvir plus ribavirin for the treatment of Russian patients with chronic HCV genotype 1 or 3 infection. In: 65th Annual Meeting of the American Association for the Study of Liver Diseases, Boston, Massachusetts, 7–11 November 2014. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.