Abstract
Developmental biology is among the many sub-disciplines of the life sciences being transformed by our increasing awareness of the role of coevolved microbial symbionts in health and disease. Most symbioses are horizontally acquired, i.e., begin anew each generation. In such associations, the embryonic period prepares the animal to engage with the coevolved partner(s) with fidelity following birth or hatching into the environment. Once interactions are underway, the microbial partners drive maturation of tissues that are either directly associated with or distant from the symbiont populations. Animal alliances often involve complex microbial communities, such as those in the vertebrate gastrointestinal tract. A series of simpler model systems is providing insight into the basic ‘rules’ and principles that govern the establishment and maintenance of stable animal-microbe partnerships. This review focuses upon what biologists have learned about the developmental trajectory of horizontally acquired symbioses through the study of the binary squid-vibrio model.
Keywords: Euprymna scolopes, Vibrio fischeri, morphogenesis, holobiont, horizontal transmission, colonization
INTRODUCTION – IT’S A NEW DAY FOR DEVELOPMENTAL BIOLOGY
Through recent technological advances, biologists have come to recognize the extent to which animals and plants rely on microbes for the success of their developmental programs (25; 26; 52). For example, fascinating new molecular data demonstrate that many, perhaps most, marine invertebrates require transient microbe-derived chemical cues, often associated with biofilms, to progress from one developmental stage to the next (30; 82). In such cases, it is likely that, while the animal experiences selection pressure to recognize a particular microbial cue, the fitness of the microbe may not be specifically affected. In contrast, selection pressure on the specificity of both partners is a characteristic of mutualistic symbioses. However, unlike the monospecific legume-rhizobia associations (for reviews see (36; 68)), the molecular details of such recognition are generally not well understood in animals, where microbes occur primarily as complex consortia. Only with advances in nucleic-acid technology over the last decade could biologists determine the microbial composition of such communities and study the development-inducing activities of its members (6; 26). Relevant here is the recognition that often the ‘organism’ undergoing development is not only the host, but rather a set of interdependent organisms, in persistent symbiosis, whose developmental programs have coevolved. These new findings add exciting dimensions to the field of animal developmental biology, and promise to bring an unprecedented integration of microbiology into this discipline.
As the field of host-microbe interactions grows at a fierce rate, considerable controversy about, variation in, and misuse of, terminology has understandably crept into the field of symbiosis. For clarity, I define here how I apply the key terms. I use the simple, classical definition of symbiosis, established by deBary in 1879 (14), i.e., organisms living together. The primary types of symbioses, i.e., mutualism, commensalism, and parasitism (or pathogenesis), refer to a state or outcome. In mutualistic symbioses, the fitness of both partners, i.e., their ability to contribute to the next generation, is enhanced; in commensal relationships, one partner benefits while the other is unaffected; and in parasitic associations, one partner’s fitness is positively affected while the other’s is harmed. Because these states are context specific, the terms do not refer to an organism itself, but rather the state of the interaction; whether the symbiosis is one type or another is all about outcome (72). When the effects of fitness on the partners is difficult to define, as is often true in complex systems, the most accurate usage to describe the association is as a ‘symbiosis’, a higher-level term with no designation of fitness.
Developmental biologists studying symbiosis seek to define how the holobiont, i.e., the coevolved partners in a symbiosis (99), persists through generations as a unit of selection. Microbial symbionts are either vertically transmitted, that is, passed maternally in or on the egg (e.g., insect-bacteriocyte symbionts), or horizontally transmitted, that is, acquired anew from the environment (e.g., gut consortia) (9). Terrestrial animals, such as social insects (22) and mammals (34), often have a facilitated horizontal transmission, in which symbionts are acquired through interactions with more mature conspecifics (19). No matter the mode of transmission, all symbiotic associations must alter developmental programs to accommodate the partnership, although several salient differences between vertical and horizontal transmission are noteworthy. For example, vertical transmission, by definition, ensures maintenance of the coevolved symbiont(s) with fidelity. Thus, no need exists for mechanisms that promote specificity during the initiation of symbiosis each successive generation. In contrast, during horizontal transmission, the naïve juvenile host is exposed not just to the appropriate symbiont(s), but also to an array of other environmental microbes. Thus, mechanisms must be present in the host, the symbionts, or both to facilitate the establishment of the ‘right’ partnerships to the exclusion of interlopers, and such mechanisms must be integrated into the developmental programs of all members.
In addition to mode of transmission, the patterns of development are also influenced, for one or more partners, by where along the spectrum of obligate to facultative a particular symbiosis lies. At one end of the spectrum, strong selection over evolutionary time has resulted in unequivocally obligate mutualisms. For example, in the insect-bacteriocyte associations, both partners have a strict requirement for the relationship and rarely occur independent of each other, even under laboratory conditions (18). Conversely, in obligate parasitic symbioses, such as between many insects and wolbachia, it is only the microbial partner that cannot survive without the host. Some situations, however, are more context dependent. For example, only under conditions of low nitrogen in the soil do leguminous plants entertain the development of a symbiosis with nitrogen-fixing rhizobia, a process that is tightly controlled through gene regulation in the plant and bacterium (73). In all cases examined, diverse strategies have evolved that incorporate into the host’s developmental program an accommodation to the form and function of the specific symbiosis.
In this review, through the lens of the squid-vibrio light-organ association, I discuss the developmental trajectory of a horizontally acquired symbiosis. Recent reviews(63; 83) on the biology of symbiont, Vibrio fischeri, provide a detailed up-to-date summary of what is known about V. fischeri as a species, both in symbiosis as well as in other ecological niches. Here, I will focus my efforts on a review of what is known about the development of the symbiosis, the interplay between partners that mediates this developmental program, and the near and far horizons of the system.
THE BASIC NATURE OF THE SQUID-VIBRIO SYMBIOSIS AND THE ENDPOINT OF DEVELOPMENT, THE MATURE ASSOCIATION
The symbiosis between the squid Euprymna scolopes and the luminous bacterium Vibrio fischeri appears to be obligate for the host in nature, but not for the microbial partner (see Supplemental Materials and Fig. S1 for a discussion of the phylogenetic position of the partners and the world-wide patterns of squid-vibrio symbioses). The juvenile animals are colonized shortly after hatching, and all animals examined from wild populations harbor the symbiont in their light organ. V. fischeri, however, has distinct plankton phase independent of the animal. The light produced by the symbiont population is thought to be used by the host animal as an antipredatory camouflage (35; 54), called counterillumination. In this behavioral strategy (98), the animal emits ventral luminescence to mimic down-welling moonlight and starlight. The importance of bioluminescence to the host’s biology is reflected the developmental program, which has a ‘built-in’ requirement for symbiont luminescence. This incorporation of symbiont light production into development underscores the host’s imperative to obtain and maintain a luminous microbial partner.
By beginning our discussion with the mature symbiosis, we can best understand the goal of the association’s program of development. Horizontally acquired symbioses typically undergo postembryonic differentiation in which the host tissues mature and the bacterial symbionts become stable partners (56). For example, the immune system and microbiota of humans undergo changes over the first few years of life that lead to a mature immune system and stable, resilient set of symbiont communities in various body sites (33; 50; 70). Studies of mouse development have demonstrated that the adult state results from a complex reciprocal interaction among the partners (74; 78; 88). The maturation of such symbioses can result in either morphologically elaborate regions of host-microbe interaction, such as the rumen, or more subtle yet critical relationships, such as in the human integument (29).
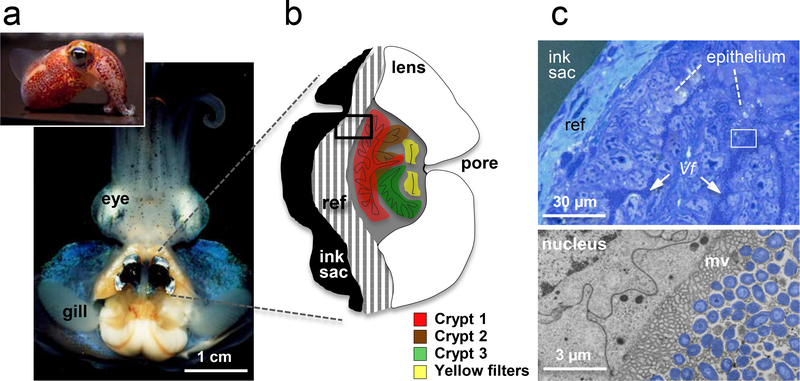
The developmental program of the squid light organ produces a dedicated, complex bilobed structure in the center of the mantle cavity (Figure 1a). These organs show strong convergence in form and function with the eye (Figure 1b; (54; 86)). Instead of the photoreceptive tissue of the retina, the interior epithelial layers of the light organ are rendered photogenic by an extracellular symbiont population that resides in crypts lined by complex microvilli (Figure 1c). To modify the incident light, each lobe of the organ has: (i) a reflective layer (or tapetum), with imbricated layers of the protein reflectin (12), which serve to direct light; (ii) a choroid/iris-like pigmented layer that controls the direction and amount of light emitted (54); and, (iii) a muscle-derived lens that achieves transparency by high concentrations of the enzyme aldehyde dehydrogenase, one of two enzyme-crystallins of the squid eye (59). (For further discussion of convergence between the light organ and eye, see Supplemental Material.) Between the lens and the photogenic tissues, just inside each lobe’s single pore, are two yellow ‘filters’. While not yet demonstrated in the squid light organ, in other bioluminescent animals, such yellow filters will red-shift the luminescence, presumably to match the color of down-welling light more effectively (16). Although features of the crypt epithelium itself are influenced by V. fischeri, studies of late postembryonic development have shown much of the maturation of the light organ is ‘hard wired’, i.e., both the organ’s adult shape and the elaboration of its light-controlling tissues are achieved, either in the presence or absence of the symbiont.
Figure 1.
The mature symbiosis. (a) A ventral dissection of the adult animal reveals the bilobed light organ in the center of the mantle (body) cavity. (b) A frontal section through one lobe of the organ illustrates the relationship of the three crypts, which house the symbionts, to the surrounding light-modulating tissues; ref, reflector; black box, region of tissue in histological section (c), upper right. (c) Upper, a low magnification view of the tissues shows each V. fischeri (Vf) cell of the symbiont population either in direct contact with host epithelial cells or only a few cell-lengths away; white box, region of the transmission electron micrograph (TEM), lower right. (d) Lower, a high magnification view shows V. fischeri cells (false color, purple) in intimate association with complex, lobate microvilli of the host cell surface.
In addition to colonizing the surface of the crypt epithelium, V. fischeri cells also interact with a small number of host blood cells found within the crypt spaces. These hemocytes are macrophage-like cells that are the only blood-cell type of squids. How the hemocytes enter the crypts is not understood, nor is their function(s) at this site. Nevertheless, histological analysis suggests that the hemocytes migrate between the cells of the epithelium (pers. obs.), much like the diapedesis of vertebrate blood cells through capillary walls (77). While it is unclear how long these migrating hemocytes remain in the crypt spaces, at most their residence time is limited by the expulsion every morning that discharges them along with most of the symbionts (28; 65) in a protein-rich matrix (79).
The vast majority of the hemocytes are found circulating in the blood stream (i.e., hemolymph), and analysis of this population has revealed significant behavioral and molecular differences depending on whether they are obtained from symbiotic or non-symbiotic adult animals (67). These differences suggest the presence of a symbiont-driven maturation process in the cellular component of the host immune system. For instance, hemocytes extracted from the hemolymph of symbiotic animals bind V. fischeri cells less well than hemocytes from non-colonized animals, while these two hemocyte populations bind other bacterial species equivalently. Interestingly, this gain of resistance is not seen in a V. fischeri mutant defective in OmpU (67), the major outer membrane protein that also mediates pathogenesis in other Vibrio spp. (20; 51). Hemocytes from symbiotic and nonsymbiotic animals also show differential gene expression, notably of genes encoding portions of the innate immune system (79). While the basis of this apparent ‘education’ of the hemocytes by its specific symbionts is unknown, determining where and how it occurs may provide clues. Because of their large numbers, as well as the daily expulsion process, it seems unlikely that each circulating hemocyte must be educated by passing through the crypt spaces and directly interacting with the symbionts. While some crypt hemocytes might reenter the blood stream, carrying information to the circulating population, a more likely mode of education would result from the uptake and transport of bacterial products into the hemolymph. Such metabolomic cues could act at a distance on the circulating hemocytes in a manner analogous to the education of T-cells in the thymus by the colonic microbiota of mammals (45). Clearly, further study of the host hemocytes, as well as other aspects of the relationship between the innate immune system and the symbiont population, offer exciting subjects for future exploration.
DAILY OSCILLATIONS IN THE ASSOCIATION
“Everything in the universe has a rhythm. Everything dances.”
This quote from Maya Angelou touches upon a feature well documented in the biological world, but poorly understood as a force governing bacterial symbioses. The members of all phylogenetic domains of the biosphere have rhythms that are entrained by natural cycles, most often those that are circadian. Circadian rhythms are synchronized in two major ways: through transcriptional and metabolic activities (4). In the former, the clock genes and their relatives regulate transcription associated with different day-to-night activities; in the later, a metabolic oscillator linked to an organism’s rhythm of nutrient utilization is driven by the cyclic oxidation and reduction of peroxiredoxin enzymes. Whereas genes controlling transcriptional oscillation are not well conserved (69), the peroxiredoxins are homologous and play a role in the metabolic rhythms of all organisms (21; 84). As biologists gain evidence of the profound influences of the microbiota on mammalian metabolism, and integrate that information with well documented daily metabolic rhythms of the host, it would not be surprising to find that host-symbiont interactions are under circadian control. Clock-gene oscillations have been documented in both the mucosal immune system and epithelium of the gut (10). Whether transcriptional activities of the associated microbiota are affected by these rhythms remains to be determined, but recent data have demonstrated that microbe-associated molecular pattern (MAMP)/toll-like receptor (TLR) interactions that maintain homeostasis of the gut epithelium require normal circadian oscillations (62).
The squid-vibrio system is the only animal-bacterial symbiosis whose daily rhythms have been studied extensively. Early on, it was discovered that the per-cell luminescence of symbiotic V. fischeri colonizing juveniles varies over the day-night cycle (5). The highest luminescence levels occur in the early evening as the animal emerges to forage in the water column, while the lowest levels are in the hours before dawn. Subsequently, the daily expulsion of most of the crypts’ contents into the surrounding seawater was observed (28).
In response to these findings, a study of the transcriptome of the adult crypt epithelium and its associated symbiont population was conducted at 6-h intervals over the 24-h day-night cycle. The data revealed a complex, daily dialogue between the partners (94). In the hours just before the dawn venting of symbionts, nearly all of the dozens of host cytoskeletal genes were up-regulated, a change that was correlated with the effacement of the epithelial microvilli into the crypt spaces; the ultrastructure of the host’s tissues at this point looks remarkably like the effacement of epithelia by enteric pathogens (80). After six hours, i.e., when most of the symbionts have been vented and the remaining population has begun to grow exponentially, the bacterial symbionts have up-regulated genes associated with the metabolism of host-membrane constituents. Specifically, the genes associated with anaerobic respiration of glycerol-phosphate are induced, and an analysis of the lipid composition of the bacterial membranes indicates that the symbionts also incorporate host-specific membrane fatty acids into their own membranes. Over this same time interval, the host down-regulates the cytoskeletal genes and begins to reconstitute a new microvillous layer on the existing crypt epithelial cells. By around noon, the symbionts have once again filled the crypt spaces and their growth is curtailed. Then, in the late afternoon and evening, the symbionts shift their metabolism to the fermentation of the polysaccharide chitin. Taken together, the data demonstrate a day-night oscillation of the symbiont’s metabolic pathways from anaerobic respiration to fermentation (94). This rhythmic switch in catabolic pathways also suggests that the crypt environment varies from a neutral to an acidic pH over the day-night cycle. Although these data provide evidence for a daily reciprocal dialogue between the partners, numerous questions about this process remain to be addressed, including: do the symbionts or their products induce effacement, and how? Do rhythmic ultrastructural and molecular changes occur in other symbioses, such as in the vertebrate gut?
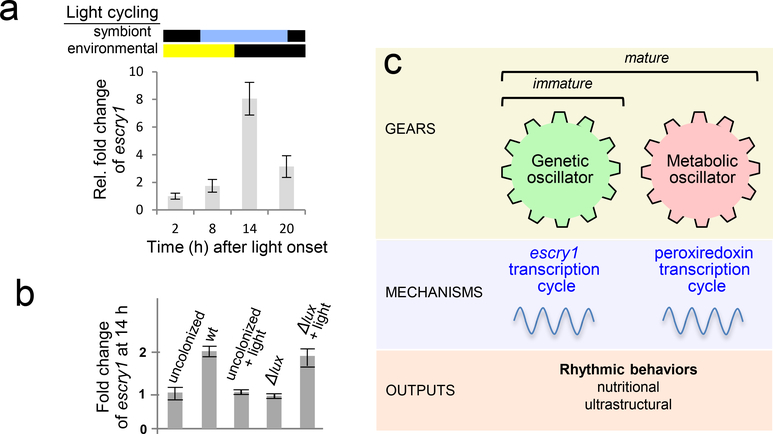
In addition to exploration of the rhythms themselves, the squid-vibrio system offers an opportunity to characterize how these rhythms are controlled. The first reported evidence for the occurrence of a bacteria-driven circadian rhythm in animals was provided by a recent study in E. scolopes (32). The transcription of genes encoding cryptochromes, proteins that are conserved regulators of animal circadian rhythms, has a daily oscillation in the host light organ. Two cryptochrome genes, escry1 and −2, have been identified in the host squid. One of these, escry2 is dominant in the eye, and cycles with environmental light, which is typical of cry genes in other animals. However, the transcriptional cycling of escry1 is offset 12 h, and corresponds to the daily rhythm of the symbiont luminescence (Figure 2a). Studies with juvenile animals demonstrated that the symbionts are required for the entrainment escry1 cycling. Further, analyses with a mutant of V. fischeri defective in light production (Δlux) demonstrated that symbiont luminescence is also required (32). While the defect in these mutants could be complemented with provision of exogenous blue light, in the absence of symbionts, illumination alone could not entrain the rhythms (Figure 2b). These data show that V. fischeri must be present to enable the host animal to respond to the daily cycle of its partner’s luminescence.
Figure 2.
Candidate mechanisms underlying molecular and biochemical control of host-symbiont rhythms. (a) escry1 gene regulation. Analyses of the regulation of one of the two host cryptochrome genes over the day/night cycle (environmental light) revealed that the escry1 gene expression peaks during peak luminescence (symbiont light). (b) A mutants defective in light production (Δlux) was unable to induce these symbiosis levels of escry1 expression. This defect could be complemented by exposing symbiotic animals to external blue light (Δlux + light), but not by exposing uncolonized animals (uncolonized + light) (32). (c) (modified from (4)). The host cryptochrome is one of a family of genetic oscillators; it begins to cycle immediately upon colonization by the symbiont. We hypothesize that daily oscillations in the mature organ are governed by both escry1, the genetic oscillator, and peroxiredoxin (PRX), the metabolic oscillator. Whereas in central clocks (the brain), the PRX genes do not cycle, in the peripheral clock (the light organ), expression of these genes does oscillate over the day/night cycle. In addition to the study of gene transcription and protein production, one can characterize these behaviors by analyzing metabolic and ultrastructural features.
Much remains to be learned about the role and mechanisms of rhythms in this association, and about how the early development of these rhythms influences maturation of the symbiosis. Analysis of a newly available RNAseq database has revealed that other key biochemical regulators of rhythms, including per and timeless, are expressed in the host light organ. Whether the cycling of these genes occurs and, if so, is similar in the eye and the light organ, remains to be determined. We also have evidence that the ‘universal’ metabolic clock proteins, peroxiredoxins, are expressed in adult E. scolopes light organs (94). These redox proteins have many functions, but may sense the day-night change in symbiont metabolism. Taken together, the data have provided enough information to construct a model of symbiosis rhythms in this system that provides testable hypotheses for future research. For instance, preliminary data suggest that the cycle between anaerobic respiration and fermentation may not be active in the juvenile symbiosis, indicating that a peroxiredoxin-based rhythm may not appear until the organ has fully developed (Figure 2c).
COLONIZATION OF THE HOST LIGHT ORGAN
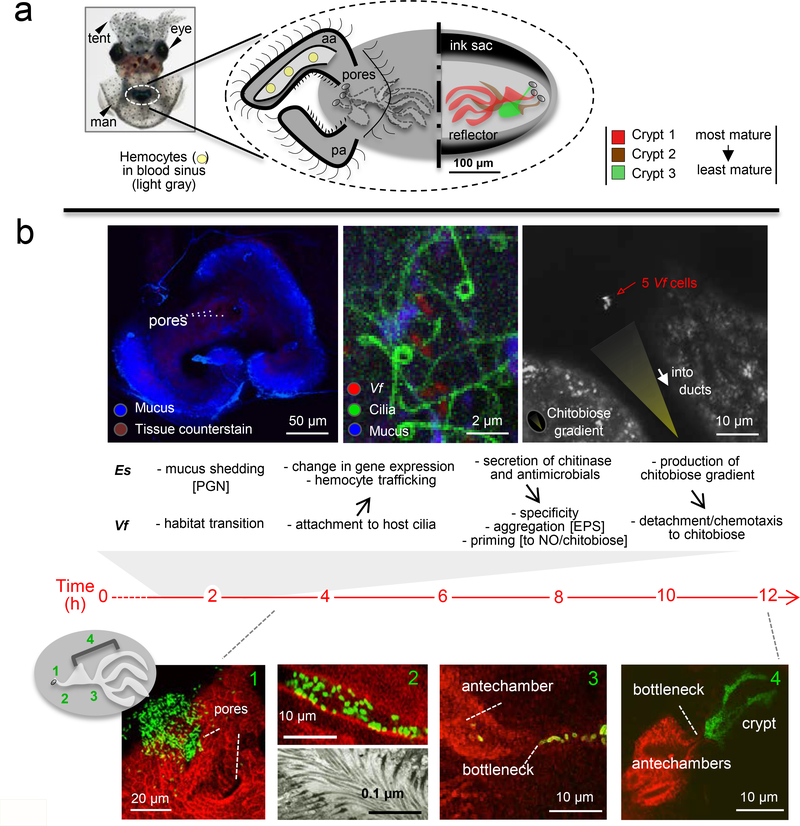
The embryonic period prepares the animal to interact with environmental V. fischeri (See Supplementary Materials and Fig. S2). At the end of embryogenesis, a complex set of differentiated cell types has developed that will promote harvesting of the symbiont (Fig. 3). The superficial ciliated epithelium consists of an anterior and posterior appendage, which covers a blood sinus, and a set of cells surrounding the pores (Figure 3a). The cells across this field vary in size, length and ciliary activity (40), but the functional significance of these variations is not well understood. By hatching, what began as invaginations of the embryonic hindgut-ink sac complex has become an elaborate set of blind-ended crypts (60; 85). In the most mature of these (crypt 1), moving lateral to medial, the pore leads to a ciliated duct, which opens into a broader ‘antechamber’. At the medial end of the antechamber is a constriction of the tissues, or bottleneck, which opens more medially into a labyrinthine crypt lined by a microvillous epithelium. Although this distance is a total of only ~100 microns, it covers a series of distinct environments to which the future symbionts must adapt during the colonization process.
Figure 3:
The symbiont’s journey. (a) The anatomy of the host organ - The light organ (dashed circle) as seen through the ventral surface of an anesthetized hatchling animal. A diagram of the external (left) and internal (right) features of this organ; aa, anterior appendage; pa, posterior appendage. (b) Colonization events – Upper, early host (Es, Euprymna scolopes) engagement of the symbiont (Vf, Vibrio fischeri) showing events, critical partner interactions, and biomolecules involved, where known. Left, Confocal image of the organ’s ciliated field, which is shedding abundant mucus in the regions around the pores in response to the MAMPs in environmental seawater; PGN, peptidoglycan (64). Middle, High-magnification confocal image of a living specimen, showing symbiont cells attaching to the cilia (2). Experimental manipulations provide evidence that the host’s perception of symbiont MAMPs, induces both cellular (41) and transcriptomic (43) changes in the host tissues. Right, Colonization in response to a host-generated chitobiose gradient. Following attachment, the symbionts aggregate using an exopolysaccharide (EPS) matrix on their surfaces (90). The observed priming to NO (92) and chitobiose (43) likely occurs in these aggregated cells. Once primed, the V. fischeri cells perceive the host gradient, and chemotax into the organ. Lower, with large numbers of GFP-labeled V. fischeri cells, their transit through tissues can be visualized by confocal microscopy (85); host tissues (red). After entering the pores (Step 1), V. fischeri cells move through ducts (Step 2) lined by outward-beating cilia (bottom TEM), which requires that the symbionts move counter-current. The symbionts then proceed across the antechamber to a bottleneck (Step 3). Once in the crypts, symbiont cells proliferate to fill each crypt by ~12 h post-inoculation (Step 4) (96).
To the author’s knowledge, the only horizontally transmitted symbioses in which the initiation of the partnership has been extensively studied are the legume-rhizobia and squid-vibrio associations. Establishment of a specific partnership during initiation of symbiosis (i.e., engagement of the coevolved partner to the exclusion of other environmental bacteria) is resolved in both systems over the first minutes to hours. In contrast, whereas nitrogenase activity characteristic of a functional root nodule takes days to weeks to develop (89), the juvenile squid has a fully colonized, bioluminescent organ within hours of hatching (76). Nevertheless, in both these and many other horizontally transmitted symbioses, the initiation of an association represents a dramatic niche change for the symbionts, from a diverse, diffuse microbial community, to a densely packed, monospecific population.
How does this niche transition progress? The initiation process proceeds through a series of stages that includes host-symbiont interactions both at the ciliated surfaces and deep within the tissues (Figure 3b). Within seconds of hatching, the animal begins to ventilate environmental seawater through its body cavity. The host accomplishes two interrelated processes during the subsequent 3- to 4-h period: it harvests its V. fischeri inoculum from the seawater, and excludes all other bacterial species from successful engagement. This harvesting is an impressive feat considering that, in near-shore Hawaiian seawater, V. fischeri typically occurs at no more than a few hundreds to thousands within communities of millions of other bacteria per ml (36; 46). The initiation events, which begin along the ciliated epithelia, can be observed directly by confocal microscopy. The V. fischeri cells first attach to the cilia on the epithelial surface and then form small aggregations of 5–10 cells, when the bacteria are present at a typical seawater concentration (2). While, under experimental conditions of the laboratory, aggregates of hundreds of cells can be obtained with large inocula (66), changing the inoculum size does not influence the timing of colonization. After 3–4 h, the aggregated cells undergo a tortuous migration into deep tissues (Fig. 3b) (85). On the last leg of this journey, the symbionts proceed single file through bottlenecks just outside of the crypts. Even with a large inoculum, apparently only a few inoculating V. fischeri cells will proliferate to fill any given crypt (96). Remarkably, even in the absence of V. fischeri, while other species initially associate with the epithelial surface, none will colonize the crypts.
Several studies have investigated the biochemical underpinnings of this process. The few hours of residence as an aggregate on the organ surface not only select for the specific symbiont, but also prepare them for the journey. Shortly after hatching, in response to the peptidoglycan present in natural seawater, the epithelial fields shed copious mucus (Figure 3b), which may serve both biophysical and biochemical functions during initiation. As mentioned above, the mucus contains antimicrobial molecules, including nitric oxide (NO) that is produced by host NO synthases (NOS) in the ciliated epithelia and transported into the mucus in vesicles (13). While this NO could discourage aggregation by some bacterial species, its main role appears to be in priming V. fischeri cells to resist their subsequent encounter with more intense stresses (92). Specifically, while NO first appears in the mucus, it is in the duct and antechambers that NOS/NO are most abundant (13; 85). Genetic studies of V. fischeri have determined that induction of NO resistance during the attachment and/or aggregation stages is essential for completion of colonization (93).
Association of V. fischeri cells with the cilia (Figure 3b) induces hemocyte trafficking into the blood sinuses of the organ. Although the precise function of this cell migration is not known, these data demonstrate that the host detects the presence of as few as 4–5 cells and respond with a robust cellular phenotype (43). Host hemocyte trafficking results from the sensing of symbiont peptidoglycan (PGN) monomer, or tracheal cytotoxin (TCT), which is constitutively exported by V. fischeri cells (41). The response to so few cells attests to the high specific activity of the TCT molecule. Sensitivity to the few attaching V. fischeri cells, against a background of other, non-specific bacterial cells in the ambient seawater, also results in a robust change in host gene expression. Genes up-regulated include several antimicrobial proteins, peptidases and a chitotriosidase. From the symbiont’s side, the bacterium-bacterium aggregation that follows ciliary attachment requires the release of a constitutively produced exopolysaccharide (EPS) (49; 97). The genetic control of the pathway responsible for EPS release has been extensively studied (61; 90). Nevertheless, the function of EPS-mediated aggregation has not been determined, although it may be essential for critical cell-cell communication, detachment from the cilia, or protection from the harsh environment of host tissues. Interestingly symbionts defective in the production of a secreted aminopeptidase-N aggregated poorly and colonized slowly (23), perhaps because the peptidase activity is critical for interaction with either the cilia (attachment or detachment) or adjacent V. fischeri cells prior to migration.
Aggregated V. fischeri cells are poised to enter the nascent light organ (Figure 3b), but require a number of additional traits to complete colonization, including flagellar motility and chemotaxis (8; 15; 27). Remarkably, the up-regulation of the host chitotriosidase mentioned above (43) plays a critical role in this migration. Chitotriosidase is endochitolytic, cleaving chitin polymers into dimeric chitobiose, and V. fischeri chemotaxes into the pores on the light organ surface in response to a chitobiose gradient (48). Culture studies of V. fischeri demonstrated that they require priming by chitobiose prior to migrating toward this molecule. Because the host chitotriosidase is secreted into the host’s mucus, which contains chitin, it catalyzes the production of chitobiose that primes the aggregated V. fischeri cells (43). A more concentrated chitotriosidase activity in the ducts apparently produces the gradient to which V. fischeri responds as it enters the light-organ pores (43; 48).
As with the legume-rhizobium symbiosis, initial interactions between E. scolopes and V. fischeri require a complex host-symbiont dialogue. Clearly, many questions remain unanswered, including some that must await further tool development. Application of bacterial molecular genetics has been a powerful approach revealing critical symbiont features (75); however, genome-wide transcriptomic analyses of the small number of aggregating V. fischeri cells have yet not been possible. Future development of single bacterial-cell transcriptomics will provide a window into the genetic regulation involved in a symbiont’s physiological transition from the planktonic to the symbiotic state. Conversely, while transcriptomic approaches are possible in the host (11; 43), development of methods for genetic knock-down of host genes, while ongoing, have not yet been achieved. Together, these advances should provide insight into why V. fischeri is the only bacterium that can leave its seawater habitat and colonize the E. scolopes light organ.
SYMBIONT TRIGGERING OF LIGHT ORGAN MORPHOGENESIS: LOCATION, LOCATION, LOCATION!
All major light-organ regions through which V. fischeri cells pass during colonization (Figure 3b) undergo symbiont-induced developmental changes. These modifications in morphology and function appear to transform the organ from a form that promotes colonization to one that fosters a life-long relationship with the symbiont. Once V. fischeri cells have populated the crypt spaces, their presence induces alterations in the tissues, only some of which can be reversed by antibiotic treatments that eliminate the symbionts (17). The most dramatic of the changes is the loss of the superficial ciliated epithelium (Figure 4a). This process, which results from an apoptosis of the host cells, takes ~4–5 d, but has already been irreversibly triggered by ~12 h post inoculation (17), at which time the symbiont population has colonized the crypts (76). Although some symbiont activities that trigger this process have been defined, how they signal the apoptotic events from the crypts, located several cell layers away, is not known. As such, this process is reminiscent of the triggering of cortical cell division in the roots of legumes in response to rhizobia interacting with the root hairs (68). In the juvenile squid, apoptosis of the superficial epithelial layer is accompanied by breakdown of the supporting basement membrane through the activity of matrix metalloproteinases (40). Cells detach from the basement membrane into the mantle cavity, and the cytoskeleton of the surrounding cells coordinately mobilizes to ‘zip’ closed the space created by the leaving cell. In addition, within 24–48 h of colonization, the symbionts shut down mucus shedding from the surface epithelium (64). Unlike the apoptotic morphogenesis, this developmental change is reversible; if the crypts are cured of the symbionts, the surface cells once again initiate mucus shedding, and symbionts will again begin to aggregate, although at lower numbers than previously (64).
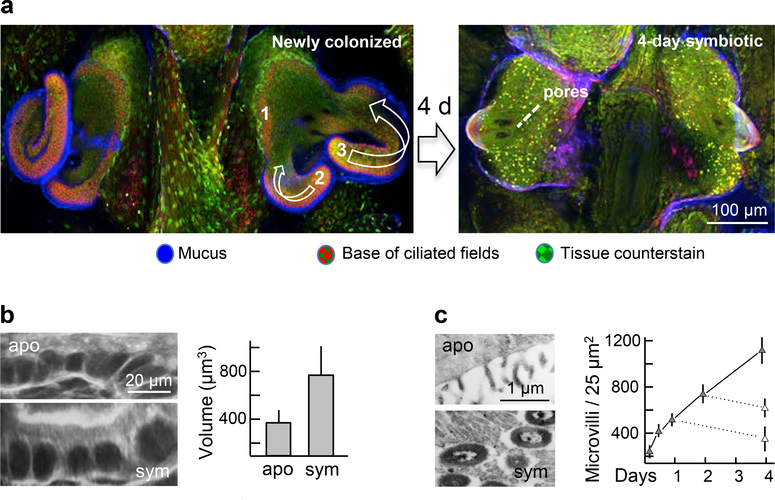
Figure 4.
Symbiont-induced light-organ morphogenesis that follows crypt colonization. (a) The 4-d program of regression of the ciliated epithelial fields on the organ surface occurs through apoptosis, which is induced by symbiont populations deep within the crypts. Left, the process begins with the loss of the ridge of ciliated cells just medial to the pores (Step 1), followed by shortening and loss of the posterior (Step 2) and anterior (Step 3) appendages. (b,c) Changes of the crypt epithelia that result from direct interactions between host and symbiont cells; swelling of crypt epithelial cells (b), and increase in the density of the microvilli on the their surfaces (c). Eliminating V. fischeri cells at 1 or 2 days with antibiotic treatment (dashed lines in graph, (c)) causes a return to the aposymbiotic morphological state of this tissue.
The interior tissues undergo more subtle changes, which are most often reversible. Although V. fischeri cells migrate through the ducts and antechambers of the organ, they do not reside there, i.e., they interact with these tissues only during the initial colonization event and as they are released during the daily expulsions (85). The timing of developmental changes in these tissues in response to colonization suggests that the effects of V. fischeri are also not the result of direct host cell/bacterial cell interactions. By 12 h, the ducts have constricted ~50%, (i.e., from ~30 down to 15 μm) due to a decrease in the number of cells surrounding the duct lumen (37). This reversible change is correlated with an increase in actin synthesis and abundance of filamentous actin in the terminal web of the duct cells. By ~7 d, the three pores on either side of the organ (Figure 3a) begin to coalesce into a single orifice, characteristic of the adult condition (Figure 1b). Similarly, the bottleneck narrows by 48 h from ~9 to ~2 μm (85), slightly wider than a single V. fischeri cell. The underlying mechanisms of bottleneck constriction, and whether this event is reversible, have not been explored. In contrast, direct interaction with V. fischeri cells induces modifications of the crypt epithelia with which the symbionts associate, and these changes can also be reversed by curing. Specifically, by 48 h following colonization, the symbiont population has induced a ~2-fold increase in cytoplasmic volume (58; 85; 91) and a ~4-fold increase in the density of the microvilli (44) of the crypt epithelial cells (Figure 4b,c). These modifications of result in a greater intimacy between host cells and symbionts.
Numerous studies have investigated symbiont features that induce these morphological and ultrastructural changes, and are the subject of recent reviews (53; 55); thus, this topic will not be covered in detail here. However, the surprising finding was that conserved MAMPs, a term that was first used in a publication on development of squid-vibrio symbiosis (41), are key inducers of symbiont-mediated host development. Specifically, lipopolysaccharide (LPS) and PGN derivatives are potent morphogens, and work synergistically to induce much of the regression of the superficial epithelium (1; 41; 87). Their likely involvement in other developmental events occurring within internal host tissues is the subject of ongoing studies. Interestingly, these MAMPs, similar to their role in pathogenesis, appear to modulate immune responses. For example, beyond triggering apoptosis, they induce hemocyte trafficking into the light organ (41). In addition, LPS and PGN derivatives work synergistically to attenuate NOS activity and NO production (3), which is the opposite of how these molecules function in pathogenesis, where MAMPs typically increase nitrosylative stress (see, e.g., (24; 38)).
Evidence is accumulating that light production by the symbiont synergizes with these chemical MAMPs, and together drive much, if not all, of the developmental program (53). Although the basis of light perception by host tissues are not well understood, two possible mechanisms have been discovered. The above-mentioned cryptochromes, which are blue light receptors, may perceive and respond to the blue luminescence of the symbiont. Cryptochromes have also been recently implicated in induction of plant development (47). The second possible receptor is rhodopsin. In addition to having much the same structure as the eye (86), the light organ produces all of the protein components of the eye’s visual transduction cascade, including the visual pigment rhodopsin, and exhibits physiological sensitivity to light much like the eye (86). Whatever the receptor, the mechanism of action is still unknown. V. fischeri mutants in light production are also defective in normal transcription of the light organ’s eye-specification genes, which may be involved in developmental induction by luminescence (71). Further, a microarray study demonstrated that bacteria defective in luminescence are not capable of inducing normal transcriptional levels of conserved host pattern-recognition receptors (PRRs) (11), which may be the receptors for the MAMPs that are critical for inducing normal development. Finally, transcription of key genes of the NF-kB pathway occurs in the light organ. As the principal response mechanism to MAMPs, this pathway presents a likely link between symbiont signals and host responses. Obviously these various jigsaw-puzzle pieces, while suggesting an underlying image, do not yet provide a coherent picture. Much research will be required to fully understand how light and MAMPs function together to drive developmental processes.
FRONTIERS
New horizons in the E. scolopes-V. fischeri symbiosis are strongly linked to the ongoing development of tools for dissecting and revealing the events underlying the initiation and maintenance of the system. Three emerging technologies deserve mention here. For much of the first 25 years that the squid-vibrio symbiosis has been studied, research has focused on the association’s first few days. While this period of onset and early development offers a rich landscape for experimentation, it had been delimited by the technical difficulty in raising the juvenile host beyond about one week (31). With intensive efforts over the last few years, robust methods for raising the animal to maturity have been developed. Our initial experiments with this new capability indicate that the association is surprisingly sensitive to the presence of nonluminous V. fischeri, and over the course of 4 weeks is able to essentially clear a population of such ‘cheaters’, even when co-colonized with a normal luminous symbiont (39). While uncolonized animals continue to be susceptible to symbiotic infection for at least several weeks after hatching, colonization triggers a critical change in the symbiosis’ subsequent receptivity and resilience (39). Specifically, animals colonized for 3 days can be cured and successfully recolonized; however, after 5 days they cannot. The capability to experimentally manipulate the host through all stages of ontogeny opens a whole new arena for research into the squid-vibrio symbiosis.
The continuing development of new molecular genetic technology has opened exciting directions for both host and symbiont. For example, sequencing of the genomes of dozens of V. fischeri strains, both symbiotic and strictly environmental, is underway. Comparative analysis of the resulting data has begun to provide insight into that portion of the flexible genome of V. fischeri that is required for the establishment and maintenance of a horizontally transmitted animal symbiosis (81; 95). The sequencing of the host genome is nearly complete, and will pave the way for the study of such exciting questions as the nature of gene regulation during the developmental trajectory of the association. In addition, attempts at genetic manipulation of the host are continuing, and refinement of the methods for the transcriptomic, proteomic, and metabolomic analyses of small amounts of tissue will provide an opportunity to unravel the temporal and spatial details of symbiosis development. The ultimate goal is to understand the complex dialogue between symbiotic partners from a biochemical to an ecological level.
Finally, the squid-vibrio association also offers rare opportunities for merging the fields of biophysics and mathematics into the investigation of symbiosis, an exciting new synergy that is currently under development. For example, the initial interaction between E. scolopes and V. fischeri offers the rare chance to visualize and model the capture and selection of bacterial cells along ciliated surfaces in an intact animal. The vision is that a thorough understanding of this process will inform many areas of biomedical research about how animals establish healthy interactions along their ciliated epithelial surfaces, while dissuading pathogenic ones.
In conclusion, the squid-vibrio system is just one of a number of genetic models of animal-bacterial symbiosis that have been pioneered over the last few years (7; 42; 57; 75). Evolution has done remarkable ‘experiments’ with this phenomenon, providing biologists with a rich palette for research. Just as models have been invaluable for deciphering how animal development programs themselves are executed, together these symbiosis models, and those to be generated in the future, promise to provide a coherent picture of both evolutionarily conserved features and the mechanisms driving diversification that underlie the development of host-microbe interactions.
Supplementary Material
ACKNOWLEDGMENTS
I thank Ned Ruby for helpful discussions and editing of the manuscript. I would also like to express my appreciation to the members of my laboratory, past and present, and other squid-vibrio researchers for their contributions. I thank Dennis Kunkel Microscopy, Inc., Kailua, HI, for providing the image of V. fischeri. This work was supported by grants from the National Institutes of Health, National Science Foundation, Office of Naval Research, WM Keck Foundation, Gordon and Betty Moore Foundation, and the John Simon Guggenheim Foundation.
Footnotes
DISCLOSURE STATEMENT
The author is not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Adin DM, Engle JT, Goldman WE, McFall-Ngai MJ, Stabb EV. 2009. Mutations in ampG and lytic transglycosylase genes affect the net release of peptidoglycan monomers from Vibrio fischeri. J Bacteriol 191:2012–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altura MA, Heath-Heckman EA, Gillette A, Kremer N, Krachler AM, et al. 2013. The first engagement of partners in the Euprymna scolopes-Vibrio fischeri symbiosis is a two-step process initiated by a few environmental symbiont cells. Environ Microbiol 15:2937–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Altura MA, Stabb E, Goldman W, Apicella M, McFall-Ngai MJ. 2011. Attenuation of host NO production by MAMPs potentiates development of the host in the squid-vibrio symbiosis. Cell Microbiol 13:527–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bass J, Takahashi JS. 2011. Circadian rhythms: Redox redux. Nature 469:476–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boettcher KJ, Ruby EG, McFall-Ngai MJ. 1996. Bioluminescence in the symbiotic squid Euprymna scolopes is controlled by a daily biological rhythm. J Comp Physiol 179:65–73 [Google Scholar]
- 6.Bosch TC, McFall-Ngai MJ. 2011. Metaorganisms as the new frontier. Zoology (Jena) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bosch TCG. 2013. Cnidarian-microbe interactions and the origin of innate immunity in metazoans. Annu Rev Microbiol 67:499–518 [DOI] [PubMed] [Google Scholar]
- 8.Brennan CA, Mandel MJ, Gyllborg MC, Thomasgard KA, Ruby EG. 2013. Genetic determinants of swimming motility in the squid light-organ symbiont Vibrio fischeri. MicrobiologyOpen 2:576–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bright M, Bulgheresi S. 2010. A complex journey: transmission of microbial symbionts. Nat Rev Microbiol 8:218–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bron R, Furness JB. 2009. Rhythm of digestion: keeping time in the gastrointestinal tract. Clin Exp Pharmacol Physiol 36:1041–8 [DOI] [PubMed] [Google Scholar]
- 11.Chun CK, Troll JV, Koroleva I, Brown B, Manzella L, et al. 2008. Effects of colonization, luminescence, and autoinducer on host transcription during development of the squid-vibrio association. Proc Natl Acad Sci U S A 105:11323–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Crookes WJ, Ding LL, Huang QL, Kimbell JR, Horwitz J, McFall-Ngai MJ. 2004. Reflectins: the unusual proteins of squid reflective tissues. Science 303:235–8 [DOI] [PubMed] [Google Scholar]
- 13.Davidson SK, Koropatnick TA, Kossmehl R, Sycuro L, McFall-Ngai MJ. 2004. NO means ‘yes’ in the squid-vibrio symbiosis: nitric oxide (NO) during the initial stages of a beneficial association. Cell Microbiol 6:1139–51 [DOI] [PubMed] [Google Scholar]
- 14.deBary HA. 1879. Die Erscheinung der Symbiose, p. 30 Strasburg [Google Scholar]
- 15.Deloney-Marino CR, Visick KL. 2012. Role for cheR of Vibrio fischeri in the Vibrio-squid symbiosis. Can J Microbiol 58:29–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Denton EJ, Herring PJ, Widder EA, Latz MF, Case JF. 1985. The roles of filters in the photophores of oceanic animals and their relation to vision in the oceanic environment. Proc Roy Soc Lond B 225:63–97 [Google Scholar]
- 17.Doino JA, McFall-Ngai MJ. 1995. A transient exposure to symbiosis competent bacteria induces light organ morphogenesis in the host squid. Biol Bull 189:347–55 [DOI] [PubMed] [Google Scholar]
- 18.Douglas AE. 1989. Mycetocyte symbiosis in insects. Biological reviews of the Cambridge Philosophical Society 64:409–34 [DOI] [PubMed] [Google Scholar]
- 19.Douglas AE. 2010. The symbiotic habit. Princeton, NJ: Princeton Univ Press; 216 pp. [Google Scholar]
- 20.Duperthuy M, Binesse J, Le Roux F, Romestand B, Caro A, et al. 2010. The major outer membrane protein OmpU of Vibrio splendidus contributes to host antimicrobial peptide resistance and is required for virulence in the oyster Crassostrea gigas. Environ Microbiol 12:951–63 [DOI] [PubMed] [Google Scholar]
- 21.Edgar RS, Green EW, Zhao Y, van Ooijen G, Olmedo M, et al. 2012. Peroxiredoxins are conserved markers of circadian rhythms. Nature 485:459–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Engel P, Moran NA. 2013. The gut microbiota of insects - diversity in structure and function. FEMS Microbiol Rev 37:699–735 [DOI] [PubMed] [Google Scholar]
- 23.Fidopiastis PM, Rader BA, Gerling DG, Gutierrez NA, Watkins KH, et al. 2012. Characterization of a Vibrio fischeri aminopeptidase and evidence for its influence on an early stage of squid colonization. J Bacteriol 194:3995–4002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flak TA, Heiss LN, Engle JT, Goldman WE. 2000. Synergistic epithelial responses to endotoxin and a naturally occurring muramyl peptide. Infect Immun 68:1235–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gilbert SF, Sapp J, Tauber AI. 2012. A symbiotic view of life: we have never been individuals. The Quarterly review of biology 87:325–41 [DOI] [PubMed] [Google Scholar]
- 26.Gordon JI. 2012. Honor thy gut symbionts redux. Science 336:1251–3 [DOI] [PubMed] [Google Scholar]
- 27.Graf J, Dunlap PV, Ruby EG. 1994. Effect of transposon-induced motility mutations on colonization of the host light organ by Vibrio fischeri. J Bacteriol 176:6986–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Graf J, Ruby EG. 1998. Host-derived amino acids support the proliferation of symbiotic bacteria. Proc Natl Acad Sci U S A 95:1818–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grice EA, Segre JA. 2011. The skin microbiome. Nat Rev Microbiol 9:244–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hadfield MG. 2011. Biofilms and marine invertebrate larvae: what bacteria produce that larvae use to choose settlement sites. Ann Rev Mar Sci 3:453–70 [DOI] [PubMed] [Google Scholar]
- 31.Hanlon RT, Claes MF, Ashcraft SE, Dunlap PV. 1997. Laboratory culture of the sepiold squid Euprymna scolopes; a model system for bacteria-animal symbiosis. Biol Bull 192:364–74 [DOI] [PubMed] [Google Scholar]
- 32.Heath-Heckman EA, Peyer SM, Whistler CA, Apicella MA, Goldman WE, McFall-Ngai MJ. 2013. Bacterial bioluminescence regulates expression of a host cryptochrome gene in the squid-vibrio symbiosis. MBio 4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hooper LV, Littman DR, Macpherson AJ. 2012. Interactions between the microbiota and the immune system. Science 336:1268–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hungate RE. 1966. The rumen and its microbes. New York: Academic Press; 533 pp. [Google Scholar]
- 35.Jones BW, Nishiguchi MK. 2004. Counterillumination in the bobtail squid, Euprymna scolopes (Mollusca: Cephalopoda). Mar Biol 144:1151–5 [Google Scholar]
- 36.Jones KM, Kobayashi H, Davies BW, Taga ME, Walker GC. 2007. How rhizobial symbionts invade plants: the Sinorhizobium-Medicago model. Nat Rev Microbiol 5:619–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kimbell JR, McFall-Ngai MJ. 2004. Symbiont-induced changes in host actin during the onset of a beneficial animal-bacterial association. Appl Environ Microbiol 70:1434–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kleinert H, Schwarz PM, Forstermann U. 2003. Regulation of the expression of inducible nitric oxide synthase. Biol Chem 384:1343–64 [DOI] [PubMed] [Google Scholar]
- 39.Koch EJ, Miyashiro T, McFall-Ngai MJ, Ruby EG. 2013. Features governing symbiont persistence in the squid-vibrio association. Molecular ecology [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koropatnick T, Goodson MS, Heath-Heckman EAC, McFall-Ngai M. 2014. Identifying the cellular mechanisms of symbiont induced epithelial morphogenesis in the squid-vibrio association. Biol Bull (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koropatnick TA, Engle JT, Apicella MA, Stabb EV, Goldman WE, McFall-Ngai MJ. 2004. Microbial factor-mediated development in a host-bacterial mutualism. Science 306:1186–8 [DOI] [PubMed] [Google Scholar]
- 42.Kostic AD, Howitt MR, Garrett WS. 2013. Exploring host-microbiota interactions in animal models and humans. Genes Dev 27:701–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kremer N, Philipp EE, Carpentier MC, Brennan CA, Kraemer L, et al. 2013. Initial symbiont contact orchestrates host-organ-wide transcriptional changes that prime tissue colonization. Cell Host Microbe 14:183–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lamarcq LH, McFall-Ngai MJ. 1998. Induction of a gradual, reversible morphogenesis of its host’s epithelial brush border by Vibrio fischeri. Infect Immun 66:777–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lathrop SK, Bloom SM, Rao SM, Nutsch K, Lio CW, et al. 2011. Peripheral education of the immune system by colonic commensal microbiota. Nature 478:250–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee K, Ruby EG. 1995. Symbiotic role of the viable but nonculturable state of Vibrio fischeri in Hawaiian coastal seawater. Appl Environ Microbiol 61:278–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu CX, Yin QQ, Zhou HC, Wu YL, Pu JX, et al. 2012. Adenanthin targets peroxiredoxin I and II to induce differentiation of leukemic cells. Nat Chem Biol 8:486–93 [DOI] [PubMed] [Google Scholar]
- 48.Mandel MJ, Schaefer AL, Brennan CA, Heath-Heckman EA, Deloney-Marino CR, et al. 2012. Squid-derived chitin oligosaccharides are a chemotactic signal during colonization by Vibrio fischeri. Appl Environ Microbiol 78:4620–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mandel MJ, Wollenberg MS, Stabb EV, Visick KL, Ruby EG. 2009. A single regulatory gene is sufficient to alter bacterial host range. Nature 458:215–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Matamoros S, Gras-Leguen C, Le Vacon F, Potel G, de La Cochetiere MF. 2013. Development of intestinal microbiota in infants and its impact on health. Trends Microbiol 21:167–73 [DOI] [PubMed] [Google Scholar]
- 51.Mathur J, Davis BM, Waldor MK. 2007. Antimicrobial peptides activate the Vibrio cholerae sigmaE regulon through an OmpU-dependent signalling pathway. Mol Microbiol 63:848–58 [DOI] [PubMed] [Google Scholar]
- 52.McFall-Ngai M, Hadfield MG, Bosch TC, Carey HV, Domazet-Loso T, et al. 2013. Animals in a bacterial world, a new imperative for the life sciences. Proc Natl Acad Sci U S A 110:3229–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McFall-Ngai M, Heath-Heckman EA, Gillette AA, Peyer SM, Harvie EA. 2012. The secret languages of coevolved symbioses: insights from the Euprymna scolopes-Vibrio fischeri symbiosis. Semin Immunol 24:3–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McFall-Ngai M, Montgomery MK. 1990. The anatomy and morphology of the sdult bacterial light organ of Euprymna scolopes (Cephalopoda: Sepiolidae). Biol Bull 179:332–9 [DOI] [PubMed] [Google Scholar]
- 55.McFall-Ngai M, Nyholm SV, Castillo MG. 2010. The role of the immune system in the initiation and persistence of the Euprymna scolopes--Vibrio fischeri symbiosis. Semin Immunol 22:48–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McFall-Ngai MJ. 2002. Unseen forces: the influence of bacteria on animal development. Dev Biol 242:1–14 [DOI] [PubMed] [Google Scholar]
- 57.Milligan-Myhre K, Charette JR, Phennicie RT, Stephens WZ, Rawls JF, et al. 2011. Study of host-microbe interactions in zebrafish. Methods in cell biology 105:87–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Montgomery MK, McFall-Ngai M. 1994. Bacterial symbionts induce host organ morphogenesis during early postembryonic development of the squid Euprymna scolopes. Development 120:1719–29 [DOI] [PubMed] [Google Scholar]
- 59.Montgomery MK, McFall-Ngai MJ. 1992. The muscle-derived lens of a squid bioluminescent organ is biochemically convergent with the ocular lens. Evidence for recruitment of aldehyde dehydrogenase as a predominant structural protein. J Biol Chem 267:20999–1003 [PubMed] [Google Scholar]
- 60.Montgomery MK, McFall-Ngai MJ. 1993. Embryonic development of the light organ of the sepiolid squid Euprymna scolopes Berry. Biol Bull 184:296–308 [DOI] [PubMed] [Google Scholar]
- 61.Morris AR, Visick KL. 2013. Inhibition of SypG-induced biofilms and host colonization by the negative regulator SypE in Vibrio fischeri. PLoS One 8:e60076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mukherji A, Kobiita A, Ye T, Chambon P. 2013. Homeostasis in intestinal epithelium is orchestrated by the circadian clock and microbiota cues transduced by TLRs. Cell 153:812–27 [DOI] [PubMed] [Google Scholar]
- 63.Norsworthy AN, Visick KL. 2013. Gimme shelter: how Vibrio fischeri successfully navigates an animal’s multiple environments. Front Microbiol 4:356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nyholm SV, Deplancke B, Gaskins HR, Apicella MA, McFall-Ngai MJ. 2002. Roles of Vibrio fischeri and nonsymbiotic bacteria in the dynamics of mucus secretion during symbiont colonization of the Euprymna scolopes light organ. Appl Environ Microbiol 68:5113–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nyholm SV, McFall-Ngai MJ. 1998. Sampling the light-organ microenvironment of Euprymna scolopes: description of a population of host cells in association with the bacterial symbiont Vibrio fischeri. Biol Bull 195:89–97 [DOI] [PubMed] [Google Scholar]
- 66.Nyholm SV, Stabb EV, Ruby EG, McFall-Ngai MJ. 2000. Establishment of an animal-bacterial association: recruiting symbiotic vibrios from the environment. Proc Natl Acad Sci U S A 97:10231–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nyholm SV, Stewart JJ, Ruby EG, McFall-Ngai MJ. 2009. Recognition between symbiotic Vibrio fischeri and the haemocytes of Euprymna scolopes. Environ Microbiol 11:483–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Oldroyd GE, Downie JA. 2008. Coordinating nodule morphogenesis with rhizobial infection in legumes. Annual review of plant biology 59:519–46 [DOI] [PubMed] [Google Scholar]
- 69.Ozturk N, Song SH, Ozgur S, Selby CP, Morrison L, et al. 2007. Structure and function of animal cryptochromes. Cold Spring Harb Symp Quant Biol 72:119–31 [DOI] [PubMed] [Google Scholar]
- 70.Parfrey LW, Knight R. 2012. Spatial and temporal variability of the human microbiota. Clinical microbiology and infection : the official publication of the European Society of Clinical Microbiology and Infectious Diseases 18 Suppl 4:8–11 [DOI] [PubMed] [Google Scholar]
- 71.Peyer SM, Pankey MS, Oakley TH, McFall-Ngai MJ. 2013. Eye-specification genes in the bacterial light organ of the bobtail squid Euprymna scolopes, and their expression in response to symbiont cues. Mechanisms of development [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Pirofski LA, Casadevall A. 2008. The damage-response framework of microbial pathogenesis and infectious diseases. Adv Exp Med Biol 635:135–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Reid DE, Ferguson BJ, Hayashi S, Lin YH, Gresshoff PM. 2011. Molecular mechanisms controlling legume autoregulation of nodulation. Annals of botany 108:789–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Round JL, Lee SM, Li J, Tran G, Jabri B, et al. 2011. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science 332:974–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ruby EG. 2008. Symbiotic conversations are revealed under genetic interrogation. Nat Rev Microbiol 6:752–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ruby EG, Asato LM. 1993. Growth and flagellation of Vibrio fischeri during initiation of the sepiolid squid light organ symbiosis. Arch Microbiol 159:160–7 [DOI] [PubMed] [Google Scholar]
- 77.Sage PT, Carman CV. 2009. Settings and mechanisms for trans-cellular diapedesis. Frontiers in bioscience 14:5066–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sawa S, Lochner M, Satoh-Takayama N, Dulauroy S, Berard M, et al. 2011. RORgammat+ innate lymphoid cells regulate intestinal homeostasis by integrating negative signals from the symbiotic microbiota. Nat Immunol 12:320–6 [DOI] [PubMed] [Google Scholar]
- 79.Schleicher TR, Nyholm SV. 2011. Characterizing the host and symbiont proteomes in the association between the bobtail squid, Euprymna scolopes, and the bacterium, Vibrio fischeri. PLoS One 6:e25649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schmidt MA. 2010. LEEways: tales of EPEC, ATEC and EHEC. Cell Microbiol 12:1544–52 [DOI] [PubMed] [Google Scholar]
- 81.Shapiro BJ, Friedman J, Cordero OX, Preheim SP, Timberlake SC, et al. 2012. Population genomics of early events in the ecological differentiation of bacteria. Science 336:48–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shikuma NJ, Pilhofer M, Weiss GL, Hadfield MG, Jensen GJ, Newman DK. 2014. Marine tubeworm metamorphosis induced by arrays of bacterial phage tail-like structures. Science (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Stabb EV, Visick KL. 2013. Vibrio fischeri: a bioluminescent light-organ symbiont of the bobtail squid Euprymna scolopes In The Prokaryotes, ed. Rosenberg E, DeLong EF, Stackebrandt E, Lory S, Thompson F:497–532. Berlin: Springer-Verlag; Number of 497–532 pp. [Google Scholar]
- 84.Stangherlin A, Reddy AB. 2013. Regulation of circadian clocks by redox homeostasis. J Biol Chem 288:26505–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sycuro LK, Ruby EG, McFall-Ngai M. 2006. Confocal microscopy of the light organ crypts in juvenile Euprymna scolopes reveals their morphological complexity and dynamic function in symbiosis. J Morphol 267:555–68 [DOI] [PubMed] [Google Scholar]
- 86.Tong D, Rozas NS, Oakley TH, Mitchell J, Colley NJ, McFall-Ngai MJ. 2009. Evidence for light perception in a bioluminescent organ. Proc Natl Acad Sci U S A 106:9836–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Troll JV, Adin DM, Wier AM, Paquette N, Silverman N, et al. 2009. Peptidoglycan induces loss of a nuclear peptidoglycan recognition protein during host tissue development in a beneficial animal-bacterial symbiosis. Cell Microbiol 11:1114–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Vaishnava S, Yamamoto M, Severson KM, Ruhn KA, Yu X, et al. 2011. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science 334:255–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vedam V, Haynes JG, Kannenberg EL, Carlson RW, Sherrier DJ. 2004. A Rhizobium leguminosarum lipopolysaccharide lipid-A mutant induces nitrogen-fixing nodules with delayed and defective bacteroid formation. Molecular plant-microbe interactions : MPMI 17:283–91 [DOI] [PubMed] [Google Scholar]
- 90.Visick KL. 2009. An intricate network of regulators controls biofilm formation and colonization by Vibrio fischeri. Mol Microbiol 74:782–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Visick KL, Foster J, Doino J, McFall-Ngai M, Ruby EG. 2000. Vibrio fischeri lux genes play an important role in colonization and development of the host light organ. J Bacteriol 182:4578–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang Y, Dunn AK, Wilneff J, McFall-Ngai MJ, Spiro S, Ruby EG. 2010. Vibrio fischeri flavohaemoglobin protects against nitric oxide during initiation of the squid-Vibrio symbiosis. Mol Microbiol 78:903–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wang Y, Ruby EG. 2011. The roles of NO in microbial symbioses. Cell Microbiol 13:518–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wier AM, Nyholm SV, Mandel MJ, Massengo-Tiasse RP, Schaefer AL, et al. 2010. Transcriptional patterns in both host and bacterium underlie a daily rhythm of anatomical and metabolic change in a beneficial symbiosis. Proc Natl Acad Sci U S A 107:2259–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wollenberg MS, Preheim SP, Polz MF, Ruby EG. 2012. Polyphyly of non-bioluminescent Vibrio fischeri sharing a lux-locus deletion. Environ Microbiol 14:655–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wollenberg MS, Ruby EG. 2009. Population structure of Vibrio fischeri within the light organs of Euprymna scolopes squid from two Oahu (Hawaii) populations. Appl Environ Microbiol 75:193–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yip ES, Geszvain K, DeLoney-Marino CR, Visick KL. 2006. The symbiosis regulator rscS controls the syp gene locus, biofilm formation and symbiotic aggregation by Vibrio fischeri. Mol Microbiol 62:1586–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Young RE, Roper CF. 1976. Bioluminescent countershading in midwater animals: evidence from living squid. Science 191:1046–8 [DOI] [PubMed] [Google Scholar]
- 99.Zilber-Rosenberg I, Rosenberg E. 2008. Role of microorganisms in the evolution of animals and plants: the hologenome theory of evolution. FEMS Microbiol Rev 32:723–35 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.