Abstract
Among the most potent carcinogens in tobacco are the tobacco-specific nitrosamines (TSNAs), with 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) the most abundant as well as the most potent. NNK is extensively metabolized to the equally carcinogenic 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol (NNAL). Of the two NNAL enantiomers, (S)-NNAL appears to be preferentially glucuronidated and excreted in humans, but also exhibits higher stereoselective tissue retention in mice and humans and has been shown to be more carcinogenic in mice than its (R)- counterpart. Due to the differential carcinogenic potential of the two NNAL enantiomers, it is increasingly important to know which UGT enzyme targets the specific NNAL enantiomers for glucuronidation. To examine this, a chiral separation method was developed to isolate entiomerically pure (S)- and (R)-NNAL. Comparison of NNAL glucuronide (Gluc) peaks formed in reactions of UGT2B7-, UGT2B17-, UGT1A9-, and UGT2B10-over-expressing cell microsomes with pure NNAL enantiomers showed large differences in kinetics for (S)- versus (R)-NNAL, indicating varying levels of enantiomeric preference for each enzyme. UGT2B17 preferentially formed (R)-NNAL-O-Gluc and UGT2B7 preferentially formed (S)-NNAL-O-Gluc. When human liver microsomes (HLM) were independently incubated with each NNAL enantiomer, the ratio of (R)-NNAL-O-Gluc to (S)-NNAL-O-Gluc formation in HLM from subjects exhibiting the homozygous deletion UGT2B17 (*2/*2) genotype was significantly lower (p=0.012) than HLM from wild-type (*1/*1) subjects. There was a significant trend (p=0.015) towards decreased (R)-NNAL-O-Gluc:(S)-NNAL-O-Gluc ratio with increasing numbers of the UGT2B17*2 deletion allele. These data demonstrate that variations in the expression or activity of specific UGTs may affect individual susceptibility to cancers induced by specific NNAL enantiomers.
Graphical Abstract
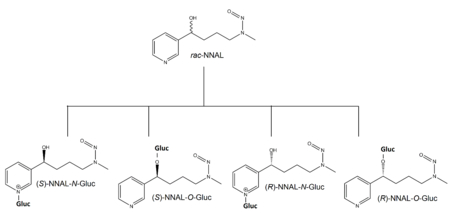
Introduction
Tobacco-specific nitrosamines (TSNAs) are an important class of carcinogens present in both tobacco smoke and smokeless tobacco products.1–6 The most abundant and potent TSNA is 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK).7–11 The major metabolic pathway for NNK (Scheme 1) is carbonyl reduction to both the (R)- and (S)- enantiomers of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol (NNAL),12, 13 which, like NNK, are very potent carcinogens in rodents.2, 14–16 By analysis of urinary metabolites, it has been estimated that 39-100% of NNK is converted to racemic NNAL (rac-NNAL) in cigarette smokers.2, 10, 12, 16, 17
Scheme 1.
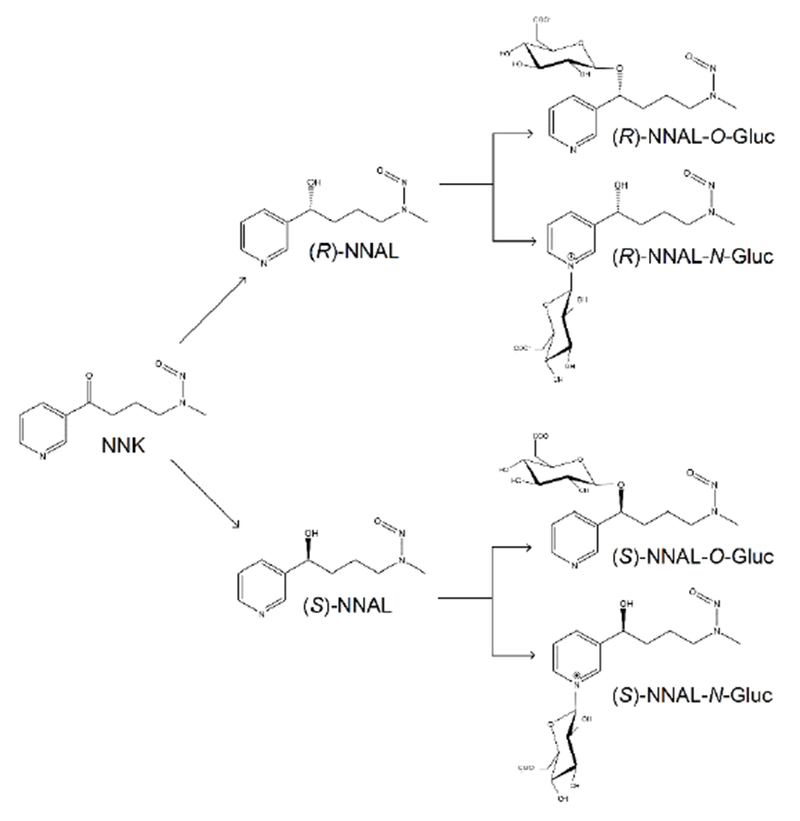
Simplified schematic of NNK metabolism to each NNAL-Gluc regioisomer and diastereomer.
The glucuronidation of NNAL is considered to be an important mechanism for NNK detoxification.2–4, 12, 18–25 In contrast to the relatively high tumorigenicity exhibited by both (R)- and (S)-NNAL, glucuronidated NNAL (NNAL-Gluc) is non-tumorigenic after subcutaneous injection into A/J mice.26 It has been shown that NNAL glucuronides are formed extensively in human liver microsomes (HLM).20, 27 The NNAL enantiomers and their glucuronides can also be detected in the urine of past and current smokers.3, 22–25, 28–32 It has been found that while (S)-NNAL is stereoselectively retained in rat lung and has a higher tumorigenicity than (R)-NNAL, (R)-NNAL exhibits a higher rate of glucuronidation in rats33–36 and the A/J mouse.26, 36 However, studies indicate that (S)-NNAL may be stereo-selectively retained in smokeless tobacco users37 but exhibits a higher rate of glucuronidation in the patas monkey.18
NNAL glucuronidation can occur at both the carbinol group (NNAL-O-Gluc)2, 12, 18–20, 38–43 and the nitrogen on the pyridine ring (NNAL-N-Gluc).27, 38, 42–44 Three enzymes, UGT1A9,20, 45 UGT2B720, 45, 46 and UGT2B17,40, 45, 47, 48 were previously found to mediate hepatic NNAL-O-Gluc formation in humans, with UGT2B17 exhibiting the lowest KM in vitro. While both UGTs 1A420, 27, 39, 45, 49 and 2B1044, 45, 49 mediate NNAL-N-Gluc formation in humans,27, 38, 39, 43, 50, 51 UGT2B10 was shown to account for ~95% of total hepatic NNAL-N-Gluc activity ex vivo.49
The variability in the urinary ratios of NNAL-Gluc:NNAL11, 32 and NNAL-O-Gluc:NNAL-N-Gluc38 from smokers is substantial, suggesting that individuals may differ greatly in their ability to detoxify NNK and form different NNAL glucuronides. In addition, variation in the levels of NNAL-O-Gluc and NNAL-N-Gluc formation was also observed in ex vivo assays performed using HLM specimens.39, 40 This variation was suggested to be, in part, mediated by genetic polymorphisms in UGT2B17 and UGT2B10. Previous studies have shown that the prevalent UGT2B17 whole-gene deletion polymorphism [31-33% allelic prevalence in Caucasians]48, 52–54 and the UGT2B10 codon 67 Asp>Tyr SNP [9.1% allelic prevalence in Caucasians]45, 55 are associated with large variability in hepatic NNAL-O-Gluc and NNAL-N-Gluc formation activities, respectively.40, 44, 45, 48, 49 In addition, the UGT2B17 gene deletion polymorphism was significantly associated with lung cancer risk in women.47 These data suggest an important role for glucuronidation-mediated detoxification of NNAL in tobacco-related cancer risk.
The goal of the present study was to characterize the stereo-selectivity of individual UGT enzymes towards individual NNAL enantiomers and determine whether the UGT2B17 deletion polymorphism is correlated with altered levels of (R)- versus (S)-NNAL-O-Gluc formation activity in human liver microsomes (HLM). The results demonstrate that UGT2B17 and UGT2B7 exhibit high stereo-selectivity for (R)- and (S)-NNAL, respectively, and that there is a significant change in the (R)- to (S)-NNAL-O-Gluc ratio associated with the UGT2B17 null genotype in HLM.
Materials and Methods
Chemicals and materials.
rac-NNAL (#M325740) and NNK (#KIT0565) were purchased from Toronto Research Chemicals (Toronto, ON, Canada); UDP glucuronic acid (UDPGA), alamethicin, kanamycin, chloramphenicol, imidazole, methanol (MeOH) and isopropanol were purchased from Sigma (St Louis, MO); Dulbecco’s modified Eagle’s medium, fetal bovine serum (FBS), geneticin and penicillin-streptomycin were purchased from Life Technologies (Carlsbad, CA); silver stain, isopropyl β-D-1-thiogalactopyranoside (IPTG), bicinchoninic acid (BCA), ammonium acetate and formic acid were purchased from Fisher Scientific (Fair Lawn, NJ); Luria broth base and Tris-glycine gels (1.0 mm) were purchased from Invitrogen (Carlsbad, CA).
Tissues.
A description of the normal human liver tissue microsomes used for the current studies were previously described.39 Tissue samples were quick-frozen at −70°C within 2 h post-surgery. Liver microsomes were prepared through differential centrifugation as previously described56 and stored (2.5-5 mg protein/mL) at −80°C. Microsomal protein concentrations were measured using the BCA assay. The UGT2B17 gene deletion analysis was performed previously40 utilizing UGT2B17 deletion locations as determined by Wilson et al.53
Cell lines and microsomal preparation.
HEK293 cells overexpressing wild-type UGT1A9, UGT2B7, UGT2B10 and UGT2B17 have been described previously.20, 57, 58 All HEK293 cell lines were grown to 80% confluence in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum, 100 U/mL penicillin and 100 μg/mL streptomycin, and maintained in 700 μg/mL of geneticin for selection of UGT overexpression, in a humidified incubator atmosphere of 5% CO2. For the preparation of cell microsomal fractions, cells were suspended in Tris-buffered saline (25 mM Tris base, 138 mM NaCl, 2.7 mM KCl; pH 7.4) and subjected to five rounds of freeze/thaw before gentle homogenization. The cell homogenate was centrifuged at 9,000 g for 30 min at 4°C. The supernatant was then centrifuged at 105,000 g for 60 min at 4°C. The microsomal pellet was suspended in Tris-buffered saline and stored in 100 μL aliquots at −80°C. Total microsomal protein concentrations were determined using the BCA protein assay.
AKR1C1 induction and purification.
Transformation-ready expression plasmids (6X N-term His tag, pQE-T7 vector; Qiagen, Venlo, Limburg) with the AKR1C1 gene were introduced to BL21 E. coli. The transformed E. coli were grown on kanamycin selection plates for 12 h and screened for plasmid uptake by DNA sequencing. AKR1C1-expressing E. coli were incubated on a shaker for 1.75 h at 37°C in Luria broth (25 μg/μL) containing kanamycin (17 μg/μL) and chloramphenicol (8 μg/μL). Protein expression was induced with the addition of IPTG (25 mM) and incubated while shaking at 37°C for 3 h. The recombinant histidine-tagged protein was purified from cell lysate on a Ni-NTA column (Fisher Scientific, #PI-88225). Lysate was loaded onto the column in a 1:1 mixture with 10 mM imidazole and washed four times with increasing concentrations of imidazole (2 mL; 20 mM, 60 mM, 100 mM, 250 mM), then eluted with 2 mL of 500 mM imidazole. Eluted protein was then dialyzed in a Slide-A-Lyzer G2 dialysis cassette (Fisher Scientific) against PBS for a total of 8 h at 4°C. Purity (>80%) was assessed via SDS-PAGE using a 4-20% Tris-Glycine gradient gel and silver staining; protein quantity was determined using the BCA assay.
NNK reduction assay.
The NNK reduction assay was adapted from a previously determined method59 using the following conditions: AKR1C1 (1 μg) was incubated (50 μL final volume) with 1 mM NNK in buffer (0.1 M monopotassium phosphate, 0.4 mM potassium chloride, 0.2 mM magnesium chloride; pH 7.4) and NADPH regeneration system (2.5 μL solution A plus 0.5 μL solution B; Corning, Corning, NY) at 37°C for 1 h. Reactions were terminated by the addition of an equal volume of methanol on ice. The precipitate was removed by centrifugation and the supernatant was saved for LC-MS analysis.
rac-NNAL separation and collection.
NNAL enantiomer separation was achieved by liquid chromatography (LC) using the following system: an Acquity (model BSM) ultra-performance LC (Waters) equipped with an automatic injector (model SM) and a UV detector operated at 254 nm (model TUV). LC was performed using a Lux 3u Amylose-2 column (150×4.6 mm; Phenomex, #00F-4471-E0) at 23°C with an isocratic elution of 30% ultra-pure water and 70% 3:1 methanol/isopropanol at 0.3 mL/min. Peaks 1 and 2 (see Figure 1) were collected from 6.95 to 7.45 min and 7.50 to 8.20 min, respectively. This method was developed to optimize for (R)- and (S)-NNAL enantiomer separation, with the freely-interconverting E/Z NNAL isomerization contained within the (R)- and (S)-NNAL peaks.
Figure 1. rac-NNAL separation and NNAL enantiomer analysis.
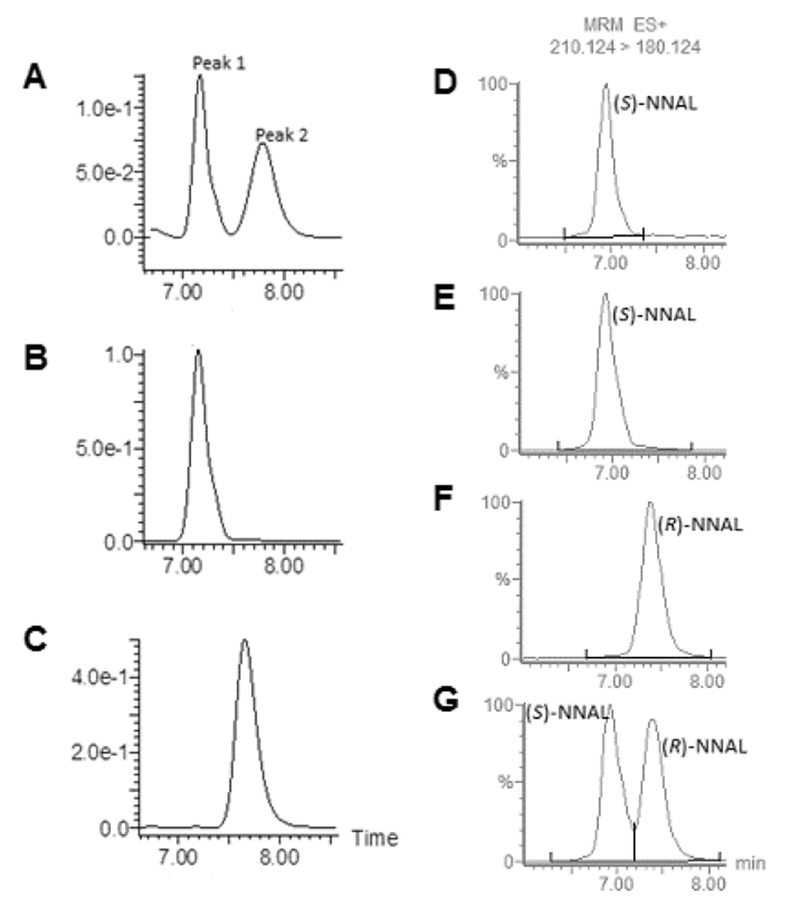
Panels A-C, rac-NNAL was separated by an isocratic method as described in the Materials and Methods and monitored by UV at 254 nm using LC (right panels). (A), rac-NNAL; (B), enantiomerically pure peak 1 collected from panel A; and (C), enantiomerically pure peak 2 collected from panel A. Panels D-G, LC-MS analysis of NNAL enantiomers. (D), AKR1C1-generated (S)-NNAL; (E), LC-collected peak 1 from panel A; (F) LC-collected peak 2 from panel A; and, (G) rac-NNAL. For panel D, AKR1C1 (1 μg total protein; >80% purity) was incubated at 37°C for 1 h with 1 mM NNK and NADPH regeneration system, and LC-MS was performed as described in the Materials and Methods. The E and Z isomers of (R)- and (S)-NNAL were not separated using this LC procedure.
NNAL glucuronidation assay.
The rate of (R)- and (S)-NNAL-O-Gluc formation by HLM and UGT over-expressing cell microsomes was determined after pre-incubation with alamethicin (50 μg/mg protein) for 10 min on ice using the following conditions: UGT over-expressing cell microsomes (15-20 μg protein) or HLM (10 μg protein) were incubated (10 μL, final volume) in 50 mM Tris-HCl (initial pH 7.4), 10 mM MgCl2, 4 mM UDPGA, and each NNAL enantiomer (UGT over-expressing cell microsomes: 0.5-16 mM; HLM: 4 mM) at 37°C for 1 h; as described previously, glucuronidation reactions were rate linear for up to 2 h incubation times.44 Reactions were terminated by the addition of an equal volume of methanol on ice. The precipitate was removed by centrifugation and the supernatant was saved for LC-MS analysis. Three individual HLM specimens from each genotype were assayed with each individual NNAL enantiomer. Each reaction was run in triplicate.
LC-MS analysis.
LC separation was achieved using an Acquity H class UPLC (Waters) equipped with an auto sampler (model FTN). NNAL peaks were analyzed with the same column and isocratic method as described above; glucuronide peaks were analyzed with a HSS T3 1.8 μm column (2.1×100 mm; Acquity, Waters, Milford, MA) at 30°C with gradient elution at 0.4 mL/min using the following conditions: 0.5 min with 99% buffer A (5 mM ammonium acetate with 0.01% formic acid) and 1% buffer B (100% MeOH), followed by a linear gradient for 3.0 min to 20% buffer B, and a subsequent linear gradient for 1.0 min to 95% buffer B. The column was washed with a 1.0 min linear gradient to 1% buffer B and regenerated for 1.0 min in 1% buffer B.
The Waters Xevo TQD tandem mass spectrometer was equipped with a Zspray electrospray ionization interface operated in the positive ion mode, with capillary voltage at 0.6 kV. Nitrogen was used as both the cone gas and desolvation gas at 50 and 800 L/hr, respectively. Ultra-pure argon was used for collision-induced dissociation. The desolvation temperature and the ion source temperature were 500°C. For the detection of NNAL enantiomers and NNAL-Glucs, the mass spectrometer was operated in the multiple reaction monitoring mode (MRM). The ion related parameters for each transition were monitored as follows: NNAL, MS transition of 210.124>180.124 with cone voltage and collision energy at 30 and 10 V, respectively; NNAL-N-Gluc, MS transition of 386.16>180.115 with cone voltage and collision energy at 15 and 20 V, respectively; NNAL-O-Gluc, MS transition of 386.16>162.115 with the cone voltage and collision energy each at 15 V. The MS transitions and LC retention times for each molecule were compared to purchased NNAL, NNAL-O-Gluc and NNAL-N-Gluc standards (Toronto Research Chemicals) for each metabolite. Reaction rates were calculated comparing the ratio of peak areas for NNAL-O-Gluc and NNAL-N-Gluc to the peak areas of deuterated NNAL-O-Gluc and NNAL-N-Gluc internal standards (kind gifts from Shantu Amin; Penn State University, Hershey, PA) and quantified against a standard curve made from purchased NNAL-O-Gluc (Toronto Research Chemicals).
Statistical analysis.
The Student’s t-test was used to compare the ratio of (R)- to (S)- NNAL-O-Gluc in subjects with UGT2B17 null (*2/*2) genotype versus subjects with the UGT2B17 (*1/*1) genotype. The linear trend test was used to examine the ratio of (R)- to (S)- NNAL-O-Gluc in HLM with decreasing copies of the UGT2B17 alleles. Kinetic constants were determined and statistical analysis were performed using Prism version 6.01 (GraphPad Softwear, San Diego, CA).
Results
To separate the individual NNAL enantiomers, a LC chiral separation method was developed using purchased (Toronto Research Chemicals) rac-NNAL (Figure 1, panel A). This method produced two distinct peaks, peak 1 (retention time of approximately 7.25 min) and peak 2 (retention time of approximately 7.85 min). Both peaks were collected and examined for purity using the same UV-monitored LC method (Figure 1, panels B and C); NNAL enantiomers corresponding to the respective peaks were collected with an enantiomeric purity of >99%.
AKR1C1 was previously shown to be selective for the formation of (S)-NNAL.59–61 To examine which of the peaks observed by LC separation corresponded to the (S)- versus (R)-NNAL enantiomers, peaks 1 and 2 were compared to the product of an AKR1C1-mediated NNK reduction assay as described in the Materials and Methods. The identity of the peaks were verified by the MRM transitions and confirmed by comparison to the purchased rac-NNAL standard (Toronto Research Chemicals). The AKR1C1 (S)-NNAL peak was observed at 6.50-7.30 min (Figure 1, panel D), and was identical to that observed for peak 1 (Figure 1, panel E), indicating that peak 1 is (S)-NNAL. Since peak 2 was observed at a different retention time (7.10-7.95 min), this suggests that peak 2 corresponds to (R)-NNAL (Figure 1, panel F).
Previous studies have shown that the UGTs 2B7,20, 39, 40, 45, 46, 49 2B17,40, 45, 47–49 1A920, 45, 49 and 2B10,44, 45, 49 are the major enzymes responsible for the hepatic glucuronidation of NNAL. To determine whether any or all of these enzymes exhibit stereo-selectivity against the individual NNAL enantiomers, the activity of each UGT was examined using microsomes from UGT over-expressing cells against (R)- and (S)-NNAL collected as described above. Using a LC-MS method developed to separate the O-Glucs of the (S)- versus (R)-NNAL enantiomers, UGT2B7 was shown to preferentially form the O-Gluc of (S)-NNAL while UGT2B17 preferentially forms the O-Gluc of (R)-NNAL (Figure 2). Representative plots of glucuronidation rate versus substrate concentration for individual UGT enzymes against (R)- versus (S)-NNAL are shown in Figure 3. The Vmax/KM for UGT2B7 was 30-fold higher for (S)-NNAL as compared to (R)-NNAL (Table 1) and exhibited a KM that was >1.5-fold lower than the other UGTs for (S)-NNAL. In contrast, the Vmax/KM for UGT2B17 for (S)-NNAL was 12-fold lower as compared to (R)-NNAL while exhibiting a KM > 5.6-fold lower than any other UGT for (R)-NNAL. By comparison, while UGT1A9 exhibited a relatively high KM (>8 mM) against both (S)- and (R)-NNAL, UGT1A9 does not appear to exhibit the same level of stereo-specificity as the other O-glucuronidating enzymes (Figure 2), with a Vmax/KM for (S)-NNAL that was 1.8-fold higher than that observed for (R)-NNAL (Table 1).
Figure 2. Representative traces of LC-MS analysis of (S)-NNAL-O-Gluc, (R)-NNAL-O-Gluc, and NNAL-N-Gluc formation by UGT-overexpressing cell microsomes.
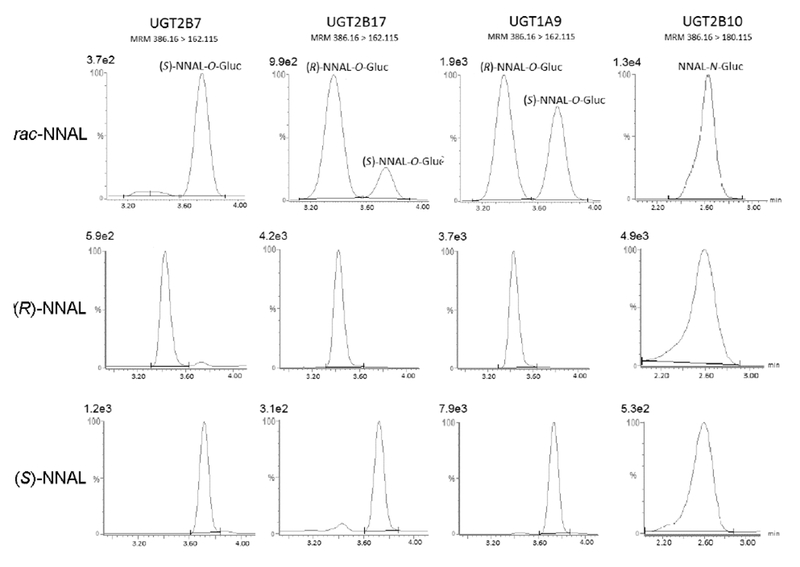
Microsomes (15-20 μg total protein) were incubated at 37 °C for 1 h with UDPGA ( 4 mM) and either rac-NNAL, (S)-NNAL, or (R)-NNAL as described in the Materials and Methods. Concentrations at or near the KM of each substrate shown as follows, listed from left to right: rac-NNAL, 4 mM, 2 mM, 16 mM, 4 mM; (R)-NNAL, 16 mM, 1 mM, 16 mM, 8 mM; (S)-NNAL, 1 mM, 8 mM, 16 mM, 4 mM . The E and Z isomers of (R)- and (S)-NNAL-O-Gluc and (R)- and (S)-NNAL-N-Gluc were not separated using this LC procedure. Absolute peak values are listed above each intensity scale on the y-axis.
Figure 3. Concentration curves and kinetic analysis for (R)-NNAL- and (S)-NNAL-Gluc formation with microsomes from UGT2B7, UGT2B17, UGT1A9 and UGT2B10 overexpressing cells.
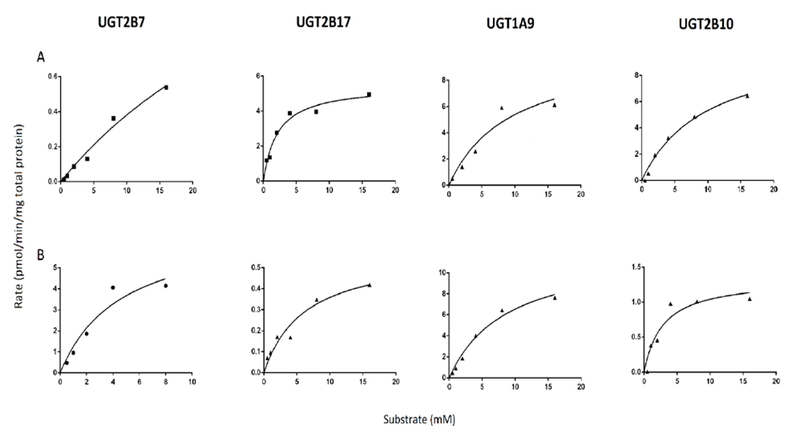
Glucuronide formation assays were performed at 37 °C for 1 h using 15-20 μg total UGT-overexpressing cell microsomal protein as described in the Materials and Methods. Rate, Vmax and Vmax/KM values are expressed per mg of total protein. Representative curves are shown and kinetic analysis was performed in three independent experiments.
Table 1.
Kinetics of UGT metabolism of (R)- versus (S)-NNAL.a
| TSNA | enzyme | Vmaxb(pmol/mg protein/min) | KM (mM) | Vmax/KMb (nL/min/mg protein) |
|---|---|---|---|---|
| (R)–NNAL | UGT2B7 | 3.1 ± 1.5 | 51 ± 9.6 | 0.06 |
| UGT2B17 | 3.0 ± 2.3 | 1.8 ± 0.47 | 1.7 | |
| UGT1A9 | 13 ± 9.8 | 13 ± 4.5 | 1.0 | |
| UGT2B10 | 8.0 ± 3.1 | 9.8 ± 4.1 | 0.81 | |
| (S)–NNAL | UGT2B7 | 4.8 ± 2.3 | 2.7 ± 1.7 | 1.8 |
| UGT2B17 | 0.54 ± 0.26 | 4.0 ± 1.9 | 0.14 | |
| UGT1A9 | 16 ± 14 | 8.7 ± 2.0 | 1.8 | |
| UGT2B10 | 1.6 ± 0.27 | 4.1 ± 1.2 | 0.40 |
Data are expressed as the mean ± SD of three independent experiments.
Units are expressed per mg total microsomal protein.
While the individual (R)- and (S)-NNAL-N-Glucs could not be separated using this LC-MS method, NNAL-N-Gluc (retention time = 2.30-2.90 min) was separated from the NNAL-O-Gluc peaks (retention times = 3.20 – 4.00 min; Figure 2). Similar to that observed for UGT1A9 (which forms NNAL-O-Gluc), the NNAL-N-Gluc forming UGT2B10 does not appear to exhibit the same level of stereo-specificity as the O-Gluc forming UGTs 2B7 and 2B17 against the (R)- and (S)-NNAL enantiomers, with a Vmax/KM ratio for (S)-NNAL:(R)-NNAL of 0.49 (Table 1).
The UGT2B17 deletion polymorphism has previously been shown to be significantly associated with decreased NNAL-O-Gluc formation in HLM.40, 47, 48 To determine whether UGT2B17 genotype affects the stereo-selectivity of HLM glucuronidation activities against (R) versus (S)-NNAL enantiomers, HLMs from subjects exhibiting either the (*1/*1), (*1/*2) and (*2/*2) genotypes were examined. While there was some individual differences in the total levels of NNAL-O-Gluc formation in the individual HLMs within genotype groups (Table 2), the ratio of (R)-NNAL-O-Gluc to (S)-NNAL-O-Gluc formation in HLM from (*2/*2) subjects was significantly lower (p=0.012) than HLM from (*1/*1) subjects (Figure 4). There was a significant trend (p=0.015) towards decreased (R)-NNAL-O-Gluc:(S)-NNAL-O-Gluc ratio with increasing numbers of the UGT2B17*2 allele.
Table 2.
Rate of NNAL-O-Gluc formation stratified by UGT2B17 deletion genotype.a
| *1/*1 |
*1/*2 |
*2/*2 |
||||
|---|---|---|---|---|---|---|
| Subject | (R)-NNAL-O-Gluc | (S)-NNAL-O-Gluc | (R)-NNAL-O-Gluc | (S)-NNAL-O-Gluc | (R)-NNAL-O-Gluc | (S)-NNAL-O-Gluc |
| 1 | 360 | 1106 | 316 | 855 | 333 | 2784 |
| 2 | 422 | 747 | 496 | 928 | 4.8 | 174 |
| 3 | 360 | 593 | 140 | 1007 | 144 | 1181 |
| Mean ± SD | 380 ± 36 | 815 ± 263 | 317 ± 178 | 930 ± 76 | 161 ± 165 | 1380 ± 1317 |
Rate units are expressed as pmol/min/mg total HLM protein. Values shown for each HLM specimen are the mean of three independent assays.
Figure 4. The (R)-NNAL-O-Gluc to (S)-NNAL-O-Gluc ratio in HLM stratified by UGT2B17 genotype.
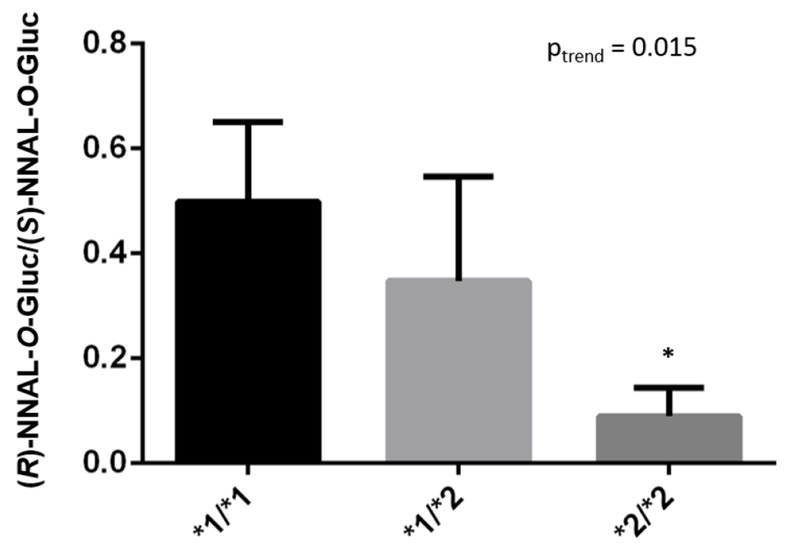
HLM (10 μg total protein) were incubated at 37 °C for 1 h with UDPGA and 4 mM (R)- and (S)-NNAL as described in the Materials and Methods. Columns represent the mean ± SD of three HLM specimens from different subjects for each of the three UGT2B17 genotype groups (indicated within the bars within the figure). *1 refers to the wild-type UGT2B17 allele, *2 refers to the UGT2B17 gene deletion allele. * p=0.012.
Discussion
In the present study, UGT2B7 exhibited the highest stereo-specificity of all of the NNAL glucuronidating UGTs, with a Vmax/KM that was 30-fold higher for (S)-NNAL as compared to (R)-NNAL, suggesting that UGT2B7 is relatively selective for the O-Gluc formation of (S)-NNAL. No detectable (R)-NNAL-O-Gluc formation was observed in assays with rac-NNAL for UGT2B7 until the substrate concentration approached the KM towards rac-NNAL,20 and the KM of UGT2B7 against enantiomerically pure (R)-NNAL was >50 mM, suggesting that UGT2B7 is unlikely to contribute to (R)-NNAL-O -Gluc formation in vivo. The KM exhibited for UGT2B7 against (S)-NNAL was similar to that observed for UGTs 2B17 and 2B10 and 3.2-fold lower than that observed for UGT1A9, suggesting that multiple UGTs may be involved in (S)-NNAL-Gluc formation in different human tissues. However, the hepatic expression of UGT2B7 appears to be higher than other NNAL-glucuronidating UGTs,62, 63 this suggests that hepatic (S)-NNAL-Gluc formation is largely mediated by UGT2B7.
UGT2B17 exhibited high O-glucuronidation activity towards (R)-NNAL, with a KM that was >5.6-fold lower than any other UGT. In addition, the ratio of (R)- to (S)-NNAL-O-Gluc formation significantly decreased in HLM from subjects with increasing numbers of the UGT2B17 gene deletion allele. The ratio of (R)- to (S)-NNAL-O-Gluc formation decreased in HLM from subjects with the UGT2B17 (*2/*2) genotype by 5.5-fold as compared to HLM from subjects with the wild-type UGT2B17 (*1/*1) genotype, suggesting that UGT2B17 is the major enzyme involved in hepatic (R)-NNAL-O-Gluc formation.
While stereo-specificity was not extensively observed for UGT2B10 against NNAL enantiomers, the data are consistent with previous studies indicating that hepatic NNAL-N-Gluc formation is generally less prominent than hepatic NNAL-O-Gluc formation:27, 39, 44 (1) (S)-NNAL-O-Gluc was the major NNAL-O-Gluc form in HLM irrespective of UGT2B17 genotype in the present study; (2) while the KM observed for UGT2B10 against (S)-NNAL in the present study was comparable to that observed for UGT2B7 (1.5-fold higher), previous studies indicate that its expression in human liver appears to be 4- to 15-fold lower than UGT2B7;62, 63 and (3) previous studies have consistently demonstrated that the mean levels of urinary NNAL-O-Gluc is higher than urinary NNAL-N-Gluc in smokers.21, 27, 32 Given the low KM’s observed for UGT1A9 forming the O-Gluc of either the (S) or (R)-NNAL enantiomers, the present studies are also consistent with previous studies20 demonstrating that UGT1A9 plays only a minor role in NNAL glucuronidation.
(S)-NNAL-O-Gluc comprised at least 60% of the total NNAL-O-Gluc in HLM in the present study. These data are consistent with previous studies demonstrating that (S)-NNAL-Gluc is the major excreted form of NNAL-Gluc in humans, accounting for 68% of the NNAL-Gluc formed in current smokers23 and was nearly 3 times the amount of the (R)-NNAL-Gluc metabolite in smokeless tobacco product users.37
Previous studies indicate that the expression of UGT2B17 is roughly 10-fold higher in lung and significantly higher in tissues of the aerodigestive tract including larynx, tonsil, tongue and esophagus62 as compared to the expression of UGTs 2B7 and 2B10. Therefore, the glucuronidation of (R)-NNAL may play a relatively more important role in NNAL detoxification in tobacco target tissues than what might be observed hepatically. This possibility is consistent with the fact that (R)-NNAL is the major enantiomer of NNAL formed in both lung microsomes and homogenates in vitro (unpublished results), a pattern not observed for hepatic fractions. Direct assessment of (R)-NNAL vs (S)-NNAL glucuronidation rates in lung tissue and aerodigestive tract tissues will be required to better assess this important possibility.
In summary, this study is the first to identify the UGTs responsible for the glucuronidation for individual NNAL enantiomers, with both UGTs 2B7 and 2B17 exhibiting high stereo-specificity. More comprehensive studies examining how changes in the expression or activity of these enzymes affect the production of NNAL glucuronide enantiomers will be required to better determine the potential impact of such changes on cancer susceptibility.
Acknowledgements
We thank Amity Platt for her outstanding work and for providing purified AKR1C1 protein. We also thank the MS Core Facility at Washington State University Spokane for their help with LC-MS.
Funding Support
This work was supported in part by the National Institutes of Health, National Institutes of Environmental Health Sciences [Grant R01-ES025460, to P Lazarus] and the Health Sciences and Services Authority of Spokane, WA.
Abbreviations
- TSNAs
Tobacco-specific nitrosamines
- NNK
4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone
- NNAL
4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol
- rac-NNAL
racemic NNAL
- NNAL-Gluc
glucuronidated NNAL
- HLM
human liver microsomes
References
- (1).Hecht SS, and Hoffmann D (1989) The relevance of tobacco-specific nitrosamines to human cancer. Cancer Surv. 8, 273–294. [PubMed] [Google Scholar]
- (2).Hecht SS (1998) Biochemistry, biology, and carcinogenicity of tobacco-specific N-nitrosamines. Chem. Res. Toxicol. 11, 559–603. [DOI] [PubMed] [Google Scholar]
- (3).Wei B, Blount BC, Xia B, and Wang L (2015) Assessing exposure to tobacco-specific carcinogen NNK using its urinary metabolite NNAL measured in US population: 2011–2012. J. Expo. Sci. Environ. Epidemiol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Czoli CD, and Hammond D (2015) TSNA Exposure: Levels of NNAL Among Canadian Tobacco Users. Nicotine Tob. Res. 17, 825–830. [DOI] [PubMed] [Google Scholar]
- (5).Inaba Y, Ohkubo T, Uchiyama S, and Kunugita N (2014) Determination of amounts of tar, nicotine, carbon monoxide, and tobacco-specific nitrosamines in the fillers of and mainstream smoke from privately imported cigarettes. Nihon Eiseigaku Zasshi 69, 205–210. [DOI] [PubMed] [Google Scholar]
- (6).Hatsukami DK, Stepanov I, Severson H, Jensen JA, Lindgren BR, Horn K, Khariwala SS, Martin J, Carmella SG, Murphy SE, and Hecht SS (2015) Evidence supporting product standards for carcinogens in smokeless tobacco products. Cancer Prev. Res. (Phila.) 8, 20–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Appleton S, Olegario RM, and Lipowicz PJ (2013) TSNA levels in machine-generated mainstream cigarette smoke: 35 years of data. Regul. Toxicol. Pharmacol. 66, 197–207. [DOI] [PubMed] [Google Scholar]
- (8).Digard H, Gale N, Errington G, Peters N, and McAdam K (2013) Multi-analyte approach for determining the extraction of tobacco constituents from pouched snus by consumers during use. Chem. Cent. J. 7, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Hearn BA, Renner CC, Ding YS, Vaughan-Watson C, Stanfill SB, Zhang L, Polzin GM, Ashley DL, and Watson CH (2013) Chemical analysis of Alaskan Iq’mik smokeless tobacco. Nicotine Tob. Res. 15, 1283–1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Appleton S, Olegario RM, and Lipowicz PJ (2014) TSNA exposure from cigarette smoking: 18 years of urinary NNAL excretion data. Regul. Toxicol. Pharmacol. 68, 269–274. [DOI] [PubMed] [Google Scholar]
- (11).Carmella SG, Akerkar SA, Richie JP Jr., and Hecht SS (1995) Intraindividual and interindividual differences in metabolites of the tobacco-specific lung carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) in smokers’ urine. Cancer Epidemiol. Biomarkers Prev. 4, 635–642. [PubMed] [Google Scholar]
- (12).Carmella SG, Akerkar S, and Hecht SS (1993) Metabolites of the tobacco-specific nitrosamine 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone in smokers’ urine. Cancer Res. 53, 721–724. [PubMed] [Google Scholar]
- (13).Hecht SS, Spratt TE, and Trushin N (1997) Absolute configuration of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol formed metabolically from 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone. Carcinogenesis 18, 1851–1854. [DOI] [PubMed] [Google Scholar]
- (14).Rivenson A, Hoffmann D, Prokopczyk B, Amin S, and Hecht SS (1988) Induction of lung and exocrine pancreas tumors in F344 rats by tobacco-specific and Areca-derived N-nitrosamines. Cancer Res. 48, 6912–6917. [PubMed] [Google Scholar]
- (15).Upadhyaya P, Carmella SG, Guengerich FP, and Hecht SS (2000) Formation and metabolism of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol enantiomers in vitro in mouse, rat and human tissues. Carcinogenesis 21, 1233–1238. [PubMed] [Google Scholar]
- (16).Richter E, Engl J, Friesenegger S, and Tricker AR (2009) Biotransformation of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone in lung tissue from mouse, rat, hamster, and man. Chem. Res. Toxicol. 22, 1008–1017. [DOI] [PubMed] [Google Scholar]
- (17).Hecht SS, Carmella SG, Stepanov I, Jensen J, Anderson A, and Hatsukami DK (2008) Metabolism of the tobacco-specific carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone to its biomarker total NNAL in smokeless tobacco users. Cancer Epidemiol. Biomarkers Prev. 17, 732–735. [DOI] [PubMed] [Google Scholar]
- (18).Hecht SS, Trushin N, Reid-Quinn CA, Burak ES, Jones AB, Southers JL, Gombar CT, Carmella SG, Anderson LM, and Rice JM (1993) Metabolism of the tobacco-specific nitrosamine 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone in the patas monkey: pharmacokinetics and characterization of glucuronide metabolites. Carcinogenesis 14, 229–236. [DOI] [PubMed] [Google Scholar]
- (19).Morse MA, Eklind KI, Toussaint M, Amin SG, and Chung FL (1990) Characterization of a glucuronide metabolite of 4-(methyl-nitrosamino)-1-(3-pyridyl)-1-butanone (NNK) and its dose-dependent excretion in the urine of mice and rats. Carcinogenesis 11, 1819–1823. [DOI] [PubMed] [Google Scholar]
- (20).Ren Q, Murphy SE, Zheng Z, and Lazarus P (2000) O-Glucuronidation of the lung carcinogen 4-(methylnitrosamino)-1- (3-pyridyl)-1-butanol (NNAL) by human UDP-glucuronosyltransferases 2B7 and 1A9. Drug Metab. Dispos. 28, 1352–1360. [PubMed] [Google Scholar]
- (21).Modesto JL, Hull A, Angstadt AY, Berg A, Gallagher CJ, Lazarus P, and Muscat JE (2014) NNK reduction pathway gene polymorphisms and risk of lung cancer. Mol. Carcinog. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Hu CW, Hsu YW, Chen JL, Tam LM, and Chao MR (2014) Direct analysis of tobacco-specific nitrosamine NNK and its metabolite NNAL in human urine by LC-MS/MS: evidence of linkage to methylated DNA lesions. Arch. Toxicol. 88, 291–299. [DOI] [PubMed] [Google Scholar]
- (23).Hecht SS, Carmella SG, Chen M, Dor Koch JF, Miller AT, Murphy SE, Jensen JA, Zimmerman CL, and Hatsukami DK (1999) Quantitation of urinary metabolites of a tobacco-specific lung carcinogen after smoking cessation. Cancer Res. 59, 590–596. [PubMed] [Google Scholar]
- (24).Yuan JM, Butler LM, Stepanov I, and Hecht SS (2014) Urinary tobacco smoke-constituent biomarkers for assessing risk of lung cancer. Cancer Res. 74, 401–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Park SL, Carmella SG, Ming X, Vielguth E, Stram DO, Le Marchand L, and Hecht SS (2015) Variation in levels of the lung carcinogen NNAL and its glucuronides in the urine of cigarette smokers from five ethnic groups with differing risks for lung cancer. Cancer Epidemiol. Biomarkers Prev. 24, 561–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Upadhyaya P, Kenney PM, Hochalter JB, Wang M, and Hecht SS (1999) Tumorigenicity and metabolism of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol enantiomers and metabolites in the A/J mouse. Carcinogenesis 20, 1577–1582. [DOI] [PubMed] [Google Scholar]
- (27).Wiener D, Doerge DR, Fang JL, Upadhyaya P, and Lazarus P (2004) Characterization of N-glucuronidation of the lung carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol (NNAL) in human liver: importance of UDP-glucuronosyltransferase 1A4. Drug Metab. Dispos. 32, 72–79. [DOI] [PubMed] [Google Scholar]
- (28).Hwang SH, Ryu HJ, Kang SJ, Yun EH, Lim MK, Kim HT, Lee JS, and Lee DH (2014) Levels of tobacco-specific metabolites among non-smoking lung cancer cases at diagnosis: case-control findings. Asian Pac. J. Cancer Prev. 14, 6591–6593. [DOI] [PubMed] [Google Scholar]
- (29).Xia Y, Wong LY, Bunker BC, and Bernert JT (2014) Comparison of creatinine and specific gravity for hydration corrections on measurement of the tobacco-specific nitrosamine 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol (NNAL) in urine. J. Clin. Lab. Anal. 28, 353–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Kotandeniya D, Carmella SG, Ming X, Murphy SE, and Hecht SS (2015) Combined analysis of the tobacco metabolites cotinine and 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol in human urine. Anal. Chem. 87, 1514–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Yang JY, Ahn HK, Lee SW, Han YJ, Oh YJ, Velazquez-Armenta EY, and Nava-Ocampo AA (2015) Simple high-throughput analytical method using ultra-performance liquid chromatography coupled with tandem mass spectrometry to quantify total 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol in urine. Clin. Chem. Lab. Med. [DOI] [PubMed] [Google Scholar]
- (32).Richie JP Jr., Carmella SG, Muscat JE, Scott DG, Akerkar SA, and Hecht SS (1997) Differences in the urinary metabolites of the tobacco-specific lung carcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone in black and white smokers. Cancer Epidemiol. Biomarkers Prev. 6, 783–790. [PubMed] [Google Scholar]
- (33).Zimmerman CL, Wu Z, Upadhyaya P, and Hecht SS (2004) Stereoselective metabolism and tissue retention in rats of the individual enantiomers of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol (NNAL), metabolites of the tobacco-specific nitrosamine, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK). Carcinogenesis 25, 1237–1242. [DOI] [PubMed] [Google Scholar]
- (34).Lao Y, Yu N, Kassie F, Villalta PW, and Hecht SS (2007) Formation and accumulation of pyridyloxobutyl DNA adducts in F344 rats chronically treated with 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone and enantiomers of its metabolite, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol. Chem. Res. Toxicol. 20, 235–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Zhang S, Wang M, Villalta PW, Lindgren BR, Upadhyaya P, Lao Y, and Hecht SS (2009) Analysis of pyridyloxobutyl and pyridylhydroxybutyl DNA adducts in extrahepatic tissues of F344 rats treated chronically with 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone and enantiomers of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol. Chem. Res. Toxicol. 22, 926–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Hecht SS, Spratt TE, and Trushin N (2000) Absolute configuration of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol formed metabolically from 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone. Carcinogenesis 21, 850. [PubMed] [Google Scholar]
- (37).Hecht SS, Carmella SG, Ye M, Le KA, Jensen JA, Zimmerman CL, and Hatsukami DK (2002) Quantitation of metabolites of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone after cessation of smokeless tobacco use. Cancer Res. 62, 129–134. [PubMed] [Google Scholar]
- (38).Carmella SG, Le Ka KA, Upadhyaya P, and Hecht SS (2002) Analysis of N- and O-glucuronides of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol (NNAL) in human urine. Chem. Res. Toxicol. 15, 545–550. [DOI] [PubMed] [Google Scholar]
- (39).Wiener D, Fang JL, Dossett N, and Lazarus P (2004) Correlation between UDP-glucuronosyltransferase genotypes and 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone glucuronidation phenotype in human liver microsomes. Cancer Res. 64, 1190–1196. [DOI] [PubMed] [Google Scholar]
- (40).Lazarus P, Zheng Y, Runkle EA, Muscat JE, and Wiener D (2005) Genotype-phenotype correlation between the polymorphic UGT2B17 gene deletion and NNAL glucuronidation activities in human liver microsomes. Pharmacogenet. Genomics 15, 769–778. [DOI] [PubMed] [Google Scholar]
- (41).Muscat JE, Chen G, Knipe A, Stellman SD, Lazarus P, and Richie JP Jr. (2009) Effects of menthol on tobacco smoke exposure, nicotine dependence, and NNAL glucuronidation. Cancer Epidemiol. Biomarkers Prev. 18, 35–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Carmella SG, Ming X, Olvera N, Brookmeyer C, Yoder A, and Hecht SS (2013) High throughput liquid and gas chromatography-tandem mass spectrometry assays for tobacco-specific nitrosamine and polycyclic aromatic hydrocarbon metabolites associated with lung cancer in smokers. Chem. Res. Toxicol. 26, 1209–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Yao L, Zheng S, Guan Y, Yang J, Liu B, Wang W, and Zhu X (2013) Development of a rapid method for the simultaneous separation and determination of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol and its N- and O-glucuronides in human urine by liquid chromatography-tandem mass spectrometry. Anal. Chim. Acta 788, 61–67. [DOI] [PubMed] [Google Scholar]
- (44).Chen G, Dellinger RW, Gallagher CJ, Sun D, and Lazarus P (2008) Identification of a prevalent functional missense polymorphism in the UGT2B10 gene and its association with UGT2B10 inactivation against tobacco-specific nitrosamines. Pharmacogenet. Genomics 18, 181–191. [DOI] [PubMed] [Google Scholar]
- (45).Wassenaar CA, Conti DV, Das S, Chen P, Cook EH, Ratain MJ, Benowitz NL, and Tyndale RF (2015) UGT1A and UGT2B genetic variation alters nicotine and nitrosamine glucuronidation in european and african american smokers. Cancer Epidemiol. Biomarkers Prev. 24, 94–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Chung CJ, Pu YS, Shiue HS, Lee HL, Lin P, Yang HY, Su CT, and Hsueh YM (2013) 4-(Methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) metabolism-related enzymes gene polymorphisms, NNK metabolites levels and urothelial carcinoma. Toxicol. Lett. 216, 16–22. [DOI] [PubMed] [Google Scholar]
- (47).Gallagher CJ, Muscat JE, Hicks AN, Zheng Y, Dyer AM, Chase GA, Richie J, and Lazarus P (2007) The UDP-glucuronosyltransferase 2B17 gene deletion polymorphism: sex-specific association with urinary 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol glucuronidation phenotype and risk for lung cancer. Cancer Epidemiol. Biomarkers Prev. 16, 823–828. [DOI] [PubMed] [Google Scholar]
- (48).Gruber M, Le T, Filipits M, Gsur A, Mannhalter C, Jager U, and Vanura K (2013) UDP-glucuronosyltransferase 2B17 genotype and the risk of lung cancer among Austrian Caucasians. Cancer Epidemiol. 37, 625–628. [DOI] [PubMed] [Google Scholar]
- (49).Chen G, Dellinger RW, Sun D, Spratt TE, and Lazarus P (2008) Glucuronidation of tobacco-specific nitrosamines by UGT2B10. Drug Metab. Dispos. 36, 824–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Upadhyaya P, McIntee EJ, and Hecht SS (2001) Preparation of pyridine-N-glucuronides of tobacco-specific nitrosamines. Chem. Res. Toxicol. 14, 555–561. [DOI] [PubMed] [Google Scholar]
- (51).Carmella SG, Han S, Fristad A, Yang Y, and Hecht SS (2003) Analysis of total 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol (NNAL) in human urine. Cancer Epidemiol. Biomarkers Prev. 12, 1257–1261. [PubMed] [Google Scholar]
- (52).Murata M, Warren EH, and Riddell SR (2003) A human minor histocompatibility antigen resulting from differential expression due to a gene deletion. J. Exp. Med. 197, 1279–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Wilson W 3rd, Pardo-Manuel de Villena F, Lyn-Cook BD, Chatterjee PK, Bell TA, Detwiler DA, Gilmore RC, Valladeras IC, Wright CC, Threadgill DW, and Grant DJ (2004) Characterization of a common deletion polymorphism of the UGT2B17 gene linked to UGT2B15. Genomics 84, 707–714. [DOI] [PubMed] [Google Scholar]
- (54).Gallagher CJ, Kadlubar FF, Muscat JE, Ambrosone CB, Lang NP, and Lazarus P (2007) The UGT2B17 gene deletion polymorphism and risk of prostate cancer. A case-control study in Caucasians. Cancer Detect. Prev. 31, 310–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (55).Chen G, Blevins-Primeau AS, Dellinger RW, Muscat JE, and Lazarus P (2007) Glucuronidation of nicotine and cotinine by UGT2B10: loss of function by the UGT2B10 Codon 67 (Asp>Tyr) polymorphism. Cancer Res. 67, 9024–9029. [DOI] [PubMed] [Google Scholar]
- (56).Coughtrie MW, Burchell B, and Bend JR (1987) Purification and properties of rat kidney UDP-glucuronosyltransferase. Biochem. Pharmacol. 36, 245–251. [DOI] [PubMed] [Google Scholar]
- (57).Dellinger RW, Fang JL, Chen G, Weinberg R, and Lazarus P (2006) Importance of UDP-glucuronosyltransferase 1A10 (UGT1A10) in the detoxification of polycyclic aromatic hydrocarbons: decreased glucuronidative activity of the UGT1A10139Lys isoform. Drug Metab. Dispos. 34, 943–949. [DOI] [PubMed] [Google Scholar]
- (58).Sun D, Chen G, Dellinger RW, Duncan K, Fang JL, and Lazarus P (2006) Characterization of tamoxifen and 4-hydroxytamoxifen glucuronidation by human UGT1A4 variants. Breast Cancer Res. 8, R50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Breyer-Pfaff U, Martin HJ, Ernst M, and Maser E (2004) Enantioselectivity of carbonyl reduction of 4-methylnitrosamino-1-(3-pyridyl)-1-butanone by tissue fractions from human and rat and by enzymes isolated from human liver. Drug Metab. Dispos. 32, 915–922. [PubMed] [Google Scholar]
- (60).Atalla A, Breyer-Pfaff U, and Maser E (2000) Purification and characterization of oxidoreductases-catalyzing carbonyl reduction of the tobacco-specific nitrosamine 4-methylnitrosamino-1-(3-pyridyl)-1-butanone (NNK) in human liver cytosol. Xenobiotica 30, 755–769. [DOI] [PubMed] [Google Scholar]
- (61).Atalla A, and Maser E (2001) Characterization of enzymes participating in carbonyl reduction of 4-methylnitrosamino-1-(3-pyridyl)-1-butanone (NNK) in human placenta. Chem. Biol. Interact. 130–132, 737–748. [DOI] [PubMed] [Google Scholar]
- (62).Jones NR, and Lazarus P (2014) UGT2B gene expression analysis in multiple tobacco carcinogen-targeted tissues. Drug Metab. Dispos. 42, 529–536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (63).Fallon JK, Neubert H, Hyland R, Goosen TC, and Smith PC (2013) Targeted quantitative proteomics for the analysis of 14 UGT1As and -2Bs in human liver using NanoUPLC-MS/MS with selected reaction monitoring. J. Proteome Res. 12, 4402–4413. [DOI] [PubMed] [Google Scholar]


