Abstract
Effective eradication of cancer requires treatment directed against multiple targets. The p53 and nuclear factor κB (NF-κB) pathways are dysregulated in nearly all tumors, making them attractive targets for therapeutic activation and inhibition, respectively. We have isolated and structurally optimized small molecules, curaxins, that simultaneously activate p53 and inhibit NF-κB without causing detectable genotoxicity. Curaxins demonstrated anticancer activity against all tested human tumor xenografts grown in mice. We report here that the effects of curaxins on p53 and NF-κB, as well as their toxicity to cancer cells, result from “chromatin trapping” of the FACT (facilitates chromatin transcription) complex. This FACT inaccessibility leads to phosphorylation of the p53 Ser392 by casein kinase 2 and inhibition of NF-κB–dependent transcription, which requires FACT activity at the elongation stage. These results identify FACT as a prospective anticancer target enabling simultaneous modulation of several pathways frequently dysregulated in cancer without induction of DNA damage. Curaxins have the potential to be developed into effective and safe anticancer drugs.
INTRODUCTION
Because the genetic plasticity of cancer cells allows them to acquire resistance to therapies with a single molecular target, multitargeted therapies provide the best hope for effective eradication of cancer. Some conventional anticancer drugs (such as doxorubicin, cyclophosphamide, and cisplatin) do hit multiple targets, but their genotoxicity reduces their clinical value. Nongenotoxic functional analogs of these drugs could revolutionize cancer treatment; however, these analogs have not been extensively explored because induction of DNA damage has been considered an essential aspect of their mechanism of action (1–4).
Another challenge in anticancer drug development is the scarcity of “universal” targets that are important in multiple tumor types. p53 and nuclear factor κB (NF-κB) are notable in this regard, being dysregulated in the vast majority of tumors (5). Mutation or functional inactivation of p53 and/or constitutive activation of NF-κB contribute to the genomic instability, resistance to apoptosis, and unconstrained growth of tumor cells. Thus, the pathways controlled by these two transcription factors are promising anticancer targets (6).
Previously, we have isolated small molecules (such as the antimalarial drug quinacrine) capable of simultaneously activating p53 and suppressing NF-κB without inducing genotoxicity (7). Although quinacrine demonstrated antitumor efficacy in animal models, we sought to identify more potent molecules with similar properties. This led to isolation of a distinct structural class of compounds with similar effects on p53 and NF-κB, lack of genotoxicity, and tumor cell–specific cytotoxicity, which we named curaxins. Here, we show that curaxins cause functional inactivation of the FACT (facilitates chromatin transcription) complex, which results in modulation of p53 and NF-κB activities and death of tumor cells. As multitargeted nongenotoxic agents, curaxins have strong potential for development into safe, effective, and broadly applicable anticancer drugs. In addition, our investigation of FACT as the molecular target of curaxins and its role in tumors defines FACT as an anticancer target.
RESULTS
Curaxins simultaneously activate p53, inhibit NF-κB, and cause death of tumor cells
Using a diverse chemical library and a renal cell carcinoma cell line (RCC45) in which p53 cannot be effectively activated by DNA damage, we have previously identified (7, 8) several compounds, including 9-aminoacridine–like molecules [for example, quinacrine (7)] and the carbazole-like molecule CBLC000 (Fig. 1A), that were capable of simultaneously inhibiting NF-κB and activating p53 (Fig. 1B). To further optimize their properties for potential drug development, we performed a structure-activity relationship (SAR) study testing focused libraries of quinacrine and CBLC000 structural analogs in cell-based p53 and NF-κB reporter assays. The active molecules that resulted from this effort were named curaxins (Fig. 1A). Here, we present data for CBLC000 (the original hit), CBLC100 (one of the most active compounds of this class in vitro; Fig. 1A), and CBLC137 (selected for in vivo evaluation of antitumor efficacy because of its high metabolic stability and water solubility; table S1). Several inactive (in p53/NF-κB reporter assays) structural analogs were used as negative controls (CBLC101, CBLC120, and CBLC136; fig. S1A).
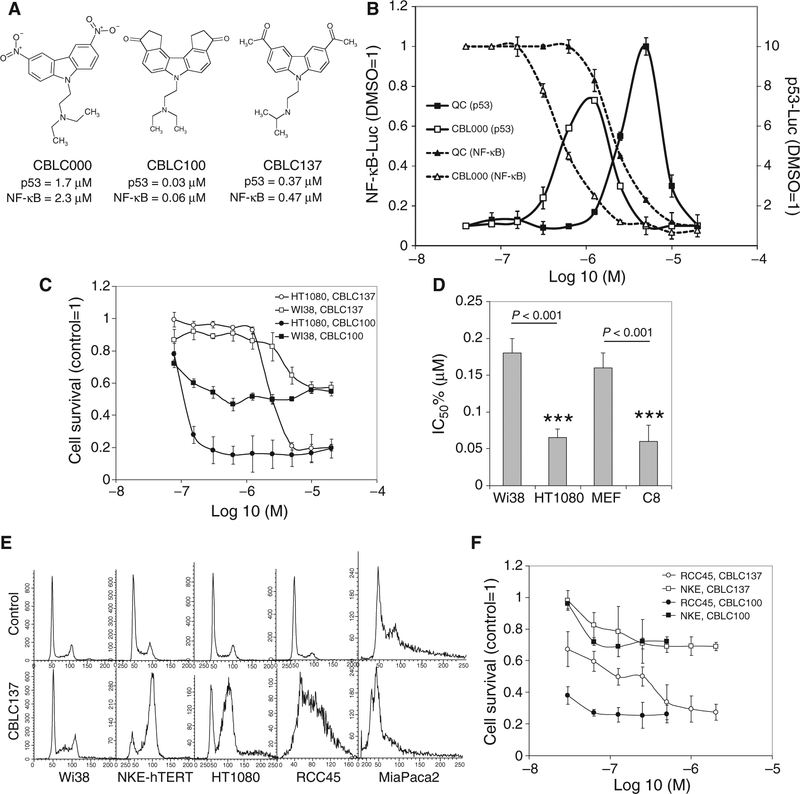
Fig. 1.
Structure and activity of curaxins. (A) Structural formulas of curaxins CBLC000, CBLC100, and CBLC137 with their EC50 in cell-based p53 and NF-κB reporter assays. (B) CBLC000 activates a p53-dependent reporter and inhibits an NF-κB–dependent reporter similarly to quinacrine (QC), but at lower concentrations (x axis). The fold change in luciferase reporter activity relative to 0.1% dimethyl sulfoxide (DMSO) treatment is shown (mean of three replicates ± SD). (C) Cytotoxicity of curaxins to cultured human diploid fibroblasts (Wi38) and fibrosarcoma (HT1080) cells (mean of three replicates ± SD). (D) Fifty percent inhibitory concentration (IC50%) of CBLC137 for human and mouse normal diploid fibroblasts (Wi38, MEF), HT1080 cells, and mouse-transformed fibroblasts [C8 (56)]. Error bars indicate 75% confidence intervals. ***P < 0.001, t test. (E) Effect of CBLC137 (2 μM for 24 hours) on cell cycle in tumor (HT1080, RCC45, MiaPaca) and normal cells (Wi38, NKE-hTERT); fluorescence-activated cell sorting (FACS) analysis of propidium iodide–stained cells. (F) Regrowth of RCC45 and NKE-hTERT cells after curaxin treatment for 3 hours. Methylene blue–stained cells were counted 5 days later. See also fig. S1.
Regression analysis of the EC50 (mean effective concentration) of compounds in the two reporter assays revealed a correlation between the p53-activating and the NF-κB–inhibiting capacities of individual molecules (fig. S1B). In addition, the EC50 of compounds in the reporter assays correlated with their toxicity [mean lethal concentration (LC50)] to cancer cell lines (fig. S1, B and C). Moreover, for any given curaxin, the effective concentrations in all three assays were similar, suggesting that the effects of curaxins on p53 and NF-κB were related to their cytotoxicity toward cancer cells (Fig. 1A and fig. S1, B and C).
Although p53 activation was used to screen for curaxins, curaxin toxicity was not limited to cells expressing wild-type p53 (fig. S1C). Using isogenic cell pairs, we observed that p53 wild-type cells were only slightly more susceptible to curaxin-induced death than p53-null cells (fig. S1D). The greater sensitivity of p53 wild-type cells is likely due to induction of apoptosis after curaxin treatment, whereas death of p53-deficient cells occurred without biochemical signs of apoptosis (fig. S1F). Consistently, ectopic expression of the apoptosis inhibitor Bcl-2 resulted in only a slight shift in curaxin LC50 in p53-positive RCC45 cells (fig. S1E), indicating that p53-dependent apoptosis is not the only mechanism of curaxin-mediated cell killing.
Curaxins were found to be more toxic to tumor than to normal cells. Mouse and human normal diploid fibroblasts were less sensitive to curaxin treatment in vitro than their transformed variants or fibrosarcoma cells (Fig. 1, C and D). Furthermore, human tumor cells died after curaxin treatment in vitro, whereas normal cells (human fibroblasts and immortalized kidney epithelial cells) displayed growth arrest (Fig. 1E) and resumed growth upon curaxin withdrawal (Fig. 1F).
Curaxins have broad anticancer activity in mice
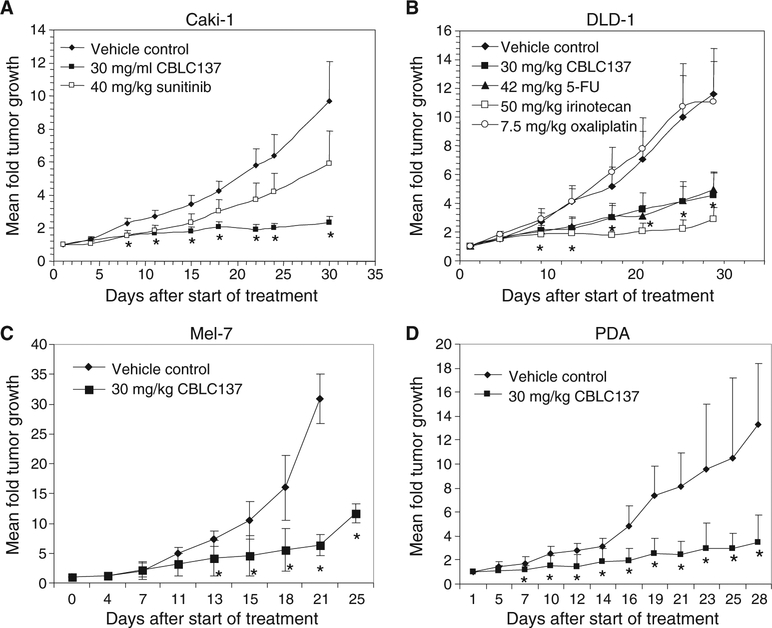
CBLC137, given by oral gavage at a nontoxic dose of 30 mg/kg per day on a 5 days on/2 days off schedule, suppressed tumor growth in xenografts of colon (DLD-1), renal cell carcinoma (Caki-1), and melanoma (Mel-7) tumor cell lines (Fig. 2, A to C) and transplanted surgical samples from patients with pancreatic ductal adenocarcinoma (Fig. 2D). CBLC137 was comparable or superior in efficacy to current standard-of-care chemotherapeutic drugs, 5-fluorouacil, irinotecan, and oxaliplatin, and a new targeted therapy used in the clinic, sunitinib (Fig. 2, A and B). Thus, curaxins display antitumor activity in mice at doses that do not cause any systemic toxicity as judged by extensive toxicological testing and comprehensive histopathological examination (fig. S2).
Fig. 2.
Antitumor effect of curaxin CBLC137 in xenograft mouse models of cancer. (A to D) Renal cell carcinoma Caki-1 (A), colon carcinoma DLD-1 (B), melanoma Mel-7 (C), and pancreatic ductal adeno-carcinoma (PDA) (D). Data are mean fold change in tumor volume (5 to 10 mice per group) relative to day 1 of treatment ± SD. *P < 0.005 for comparison of CBLC137 and vehicle, analysis of variance (ANOVA) test. See also fig. S2.
Curaxins activate p53 through casein kinase 2 associated with the FACT complex
Although death of curaxin-treated tumor cells was found to be only partially p53-dependent, we investigated curaxin-induced p53 activation as a starting point to decipher the mechanism of action of curaxins. p53 posttranslational modifications induced by curaxins were different from those induced by DNA damage: Ser15 phosphorylation was significantly weaker and did not correlate with the scale of specific activity of curaxins (Fig. 3A). In addition, acetylation of p53 on Lys382, which is usually associated with DNA damage (9–11), was not induced by curaxins (fig. S3A). On the other hand, phosphorylation of Ser392 of p53 was induced but not with structurally related inactive curaxin compounds (Fig. 3A). p53 with alanine substitution of Ser392 (S392A) was not effectively activated by curaxins, whereas p53 with alanine substitution of Ser15 (S15A) was activated to the same extent as wild-type p53 (Fig. 3, B and C, and fig. S3B). Thus, the mode of p53 activation by curaxins is different from that of DNA-damaging agents and it involves Ser392 phosphorylation.
Fig. 3.
Dependence of p53 activation by curaxin on CK2 and FACT. (A) Western analysis of HT1080 cells treated with quinacrine (6 μM), CBLC000 (2 μM), CBLC137 (0.8 μM), “inactive” curaxin-like molecules (10 μM), or doxorubicin (DX, 1 μM) for 8 hours with phosphospecific or “total” (D01) p53 antibodies. C, untreated control; arrow, p53 phosphorylated on Ser392; asterisk, nonspecific band used as loading control. (B and C) Effect of p53 Ser392 to alanine substitution (S392A) on curaxin-induced p53 activation. (B) Western analysis (anti-p53 D01 antibody) of HCT116-p53 null cells transduced with wild-type p53 (wt) or S15A or S392A mutant p53 and treated with CBLC137, quinacrine, or doxorubicin (1 μM) for 8 hours. (C) Quantification of data in (B) with ImageJ software. (D) Effect of chemical inhibitors of CK2 (CK2i, 50 μM) and DNA-PK (DNA-PKi, 25 μM) on CBLC000-induced p53-Luc reporter activation. Cells were treated with CBLC000 (1 μM) and inhibitors for 16 hours. Data are mean fold change ± SD. ***P < 0.001 (t test) compared with treatment with CBLC000. (E) Effect of CK2 inhibition on CBLC000-induced p53 Ser392 phosphorylation (Western blotting, three upper panels) and DNA binding (EMSA, lower panel). Extracts were prepared from HT1080 cells treated with CBLC000 (1 μM) and CK2i (in μM) for 16 hours. NS, nonspecific bands used as loading controls; p53BE, specific p53-binding element probe. (F) Phosphorylation of the C terminus of p53 by CK2 induced by curaxins and UV treatment. CK2β immunoprecipitated from lysates of HT1080 cells treated with quinacrine, CBLC100, or UV was used in in vitro kinase assays with a peptide substrate corresponding to the C terminus of p53 (amino acids 311 to 393). (G) Effect of siRNA-mediated knockdown of SSRP1 or CK2 on CBLC137-induced(0.6 μM, 8 hours) p53 activation in HT1080 cells. Western blots were probed with antibodies against the proteins indicated above each panel. See also fig. S3.
Ser392 of p53 can be phosphorylated by several kinases, including ataxia-telangiectasia mutated (ATM) (12), ATM- and Rad3-related (ATR) (13), DNA-dependent protein kinase (DNA-PK) (14), protein kinase R (PKR) (15), and casein kinase 2 (CK2) (16). Quinacrine does not activate ATM (7), and ATM and PKR were not activated in response to curaxins (fig. S3C). ATR and downstream target, Chk1, were weakly activated by CBLC137, but not quinacrine, which suggests that these kinases do not play critical roles in the mechanism of action of curaxins (fig. S3C). Inhibition of DNA-PK did not affect curaxin activity (Fig. 3D). In contrast, inhibition of CK2 resulted in reduced curaxin-induced p53 reporter activity (Fig. 3D), Ser392 phosphorylation, and p53-DNA binding (Fig. 3E). Moreover, a peptide corresponding to p53 amino acids 311 to 393 was phosphorylated in vitro by CK2 immunoprecipitated from curaxin-treated cells to a greater extent than by CK2 from control cells (Fig. 3F).
It has been reported that CK2 phosphorylates p53 on Ser392 when the kinase is bound to FACT (16, 17). FACT consists of two subunits: structure-specific recognition protein 1 (SSRP1) and suppressor of Ty16 (SPT16). FACT binds via the high-mobility group (HMG) domain of SSRP1 to structurally distorted DNA (for example, cis-platinum adducts and thymine dimers) and to abnormal chromosomal structures (for example, four-ways and Holliday junctions) (18, 19), and this shifts the substrate preference of FACT-associated CK2 away from SSRP1 toward p53 (16, 17).
We addressed a role of CK2-FACT in the effect of curaxins on p53. Small interfering RNA (siRNA)–mediated knockdown of either CK2 or SSRP1 resulted in reduced activation of p53 by curaxins (Fig. 3G). Similar results were obtained using lentiviral vector–driven expression of short hairpin RNAs (shRNAs) targeting SSRP1 or SPT16 (fig. S3, D and G). Although the effects of inhibitors of CK2/FACT on curaxin-induced p53 activation were not complete, these data show that curaxins most likely act through FACT to induce CK2-dependent activation of p53.
Curaxins cause chromatin trapping of FACT
The involvement of CK2/FACT in curaxin-induced p53 activation suggested that FACT might be the molecular target of curaxins. This observation was supported by our finding that curaxin, but not inactive, structurally similar compound treatment, caused intranuclear redistribution of FACT subunits (Fig. 4 and fig. S4). Curaxins did not affect the total amount of SSRP1 in cells, but caused SSRP1 to rapidly disappear from the soluble protein fraction (Fig. 4, A and B, and fig. S4A) and become strongly associated with chromatin (binding as tightly as basic histones) (Fig. 4C and fig. S4B). Similar curaxin-induced redistribution was observed for the SPT16 subunit of FACT (Fig. 4C and fig. S4B). This effect of curaxins was specific to FACT because it was not observed for an unrelated HMG domain–containing protein, HMGB1 (Fig. 4, C and D), whereas cisplatin caused weak redistribution of FACT and strong redistribution of HMGB1 (Fig. 4D).
Fig. 4.
Curaxin induction of FACT-chromatin binding. (A) Disappearance of SSRP1 from the soluble protein fraction after curaxin treatment. Western analysis of soluble nuclear extracts from RCC45 cells treated with CBLC137 (top), quinacrine (middle), or CBLC100 (bottom) for different times (left panels) or with different doses for 1 hour (right). (B) Western detection of SSRP1 in total cell lysates and the soluble protein fraction of lysates from HT1080 cells treated with quinacrine (5 μM), CBLC137 (2 μM), or CBLC100 (0.2 μM) for 1 hour. C, untreated. (C) SSRP1 and SPT16 redistribution from the nucleoplasm to chromatin after curaxin treatment. Western blotting (upper four panels) and Coomassie staining (bottom panel) of nuclear extracts from HT1080 cells treated with CBLC100 (CX, 0.2 μM, 1 hour) prepared using a modified high-salt extraction method (54). Anti-p53 staining demonstrates the response of cells to curaxins. Histones H2, H3, and H4 remain associated with chromatin even after extraction with 2.5 M NaCl. (D) Western analysis (anti-HMGB1 and anti-SSRP1) of nuclear extracts prepared as in (C) from HT1080 cells treated with quinacrine (5 μM), CBLC100 (0.2 μM), or cisplatin (100 μg/ml) for 6 hours. (E) Fluorescence imaging of live nonfixed HT1080 cells cotransfected with GFP-tagged SSRP1 and RFP-tagged histone H2B expression constructs and treated with CBLC137 (2 μM, 15 min). (F) Curaxin-induced depletion of soluble SSRP1 and SPT16 in vivo. MMTV-neu transgenic mice with palpable spontaneous mammary tumors were given CBLC137 (100 mg/kg) or vehicle (water) by oral gavage. Western blotting with the indicated antibodies was performed on the soluble fraction of lysates prepared from tumors 24 hours after treatment. (G) Quantification of the Western data in (F) with ImageJ software. Data are mean fold change to control normalized to actin ± SD. *P < 0.05, t test. See also fig. S4.
Curaxin-induced redistribution of SSRP1 was confirmed by fluorescence microscopy of cells expressing green fluorescent protein (GFP)–tagged SSRP1 and red fluorescent protein (RFP)–tagged histone H2B. Without treatment, GFP-SSRP1 fluorescence was evenly distributed throughout the nucleus during interphase and throughout the whole cell during mitosis (Fig. 4E and fig. S4, C and D). Within minutes of adding curaxins to the culture medium, the nuclear pattern of GFP-SSRP1 fluorescence changed to match that of either chromatin (visualized by RFP-H2B) or DNA (visualized by Hoechst staining) (Fig. 4E and fig. S4, C and D). Curaxin-induced “chromatin trapping” of FACT was also observed in vivo: Levels of both SSRP1 and SPT16 were reduced in soluble protein fractions prepared from spontaneous mammary tumors of CBLC137-treated MMTV-neu transgenic mice compared to those from untreated mice (Fig. 4, F and G).
Chromatin trapping of FACT contributes to the cytotoxicity of curaxins
In addition to its role in CK2-mediated p53 activation, FACT is known to affect transcription through its role in nucleosome remodeling (20). We tested whether FACT function is essential for the viability of tumor cells using shRNA knockdown of either FACT subunit and observed that depletion of FACT reduced cell survival independently of their p53 status (Fig. 5A and fig. S5F). This suggests that the toxicity of curaxins may be exerted through their interference with FACT function.
Fig. 5.
Association between FACT levels and cell growth, sensitivity to curaxins, and tumor phenotype. (A) Colony formation of HT1080 cells transduced with the indicated shRNAs. Inset: Western blots show shRNA effects on target protein levels. Data are mean of three replicates ± SD. **P < 0.01; ***P < 0.001, t tests. (B) Increased cell sensitivity to curaxin, but not cisplatin, after FACT knockdown. Colonies formed by cells plated 72 hours after shRNA transduction and treated with CBLC137 (0.5 μM) or cisplatin (1 μg/ml) for 24 hours. The mean number of colonies relative to untreated cells transduced with the same shRNA is shown. **P < 0.01; ***P < 0.001, t tests. (C) Enrichment of cells with higher levels of SSRP1 during CBLC137 exposure. FACS detection of GFP-tagged SSRP1 expression in transduced HT1080 cells cultured in 0.3 μM CBLC137 for different time. (D) Quantification of data shown in (C). **P < 0.01; ***P < 0.001, t tests. (E) Western analysis of FACT subunit expression in total cell extracts from tissues of an MMTV-neu mouse with palpable tumors. The mouse had visible lung metastases (lane indicated “lung with mts”). (F) Western analysis of FACT subunit expression in different cultured human cell lines. See also fig. S5.
However, in testing the effect of curaxins on in vitro RNA polymerase II (RNAPII)–driven transcription of a mononucleosomal DNA template, we observed that curaxin treatment did not interfere with RNAPII-mediated transcription or FACT-induced pausing of RNAPII at the nucleosome (fig. S5, A to D).
An alternative hypothesis is that depletion of soluble FACT from the nucleoplasm might lead to inhibition of FACT-dependent function. In this case, the sensitivity of cells to curaxins would be dependent on their level of FACT (that is, the less FACT present in cells, the less curaxin required for trapping it and suppressing transcription and vice versa). This was further verified by our finding that shRNA-mediated reduction of FACT increased the sensitivity of cells to treatment with CBLC137 (Fig. 5B). shRNA-transduced cells that survived short-term (24 hours) curaxin treatment resulted in an increased proportion of FACT-positive cells (fig. S5E), which suggests that elevated FACT provided them with a growth advantage in the presence of the drug. Consistent with this, amounts of ectopic GFP-SSRP1 were elevated in cells that survived 96 hours of CBLC137 treatment (Fig. 5, C and D). Thus, depletion of FACT enhanced the cytotoxicity of curaxins, whereas overexpression of FACT counteracted the effect of curaxins to maintain cell viability. These data support the hypothesis that curaxin-induced cytotoxicity is mediated at least in part via depletion of soluble FACT required for transcription of genes within highly structured chromatin (21–23).
To address the mechanism underlying the tumor-specific toxicity of curaxins, we compared FACT subunit expression in tumor and normal cells in vitro and in vivo. Wi38 normal human diploid fibroblasts have undetectable levels of FACT subunits, and immortalized human kidney epithelial cells (NKE-hTERT) have lower levels than several human tumor cell lines (Fig. 5F). Levels of FACT subunits were higher in mammary tumors and in lungs with visible metastases from MMTV-neu mice compared to normal mammary glands, lungs, and other organs from the same animals (except for normal spleen, which contained high levels of FACT) (Fig. 5E). The observed elevation of FACT expression in tumor cells suggests that FACT plays a role in the development, progression, and/or maintenance of tumors and explains why depletion of functional FACT by curaxins is more toxic to tumor cells than to normal cells.
FACT is involved in curaxin-mediated inhibition of NF-κB–driven transcription
The effects of curaxins on NF-κB and FACT suggested involvement of FACT in NF-κB–dependent transcription. Such involvement was supported by our finding that quinacrine and curaxins block NF-κB–dependent transcription downstream of p65 nuclear translocation and DNA binding (7) (Fig. 6, A and B) and confirmed by our demonstration that knockdown of either SSRP1 or SPT16 led to decreased expression of NF-κB–regulated genes, such as IL-8 (interleukin-8), IκBα (inhibitor of NF-κBα), and TNF (tumor necrosis factor) (Fig. 6, C and D). Moreover, chromatin immunoprecipitation (ChIP) experiments showed that curaxin treatment did not alter binding of the p65 subunit of NF-κB, but decreased binding of SSRP1, to the IL-8 promoter in both unstimulated and TNF-stimulated cells (Fig. 6E). Curaxin treatment also inhibited TNF-induced binding of SSRP1 to the TNF promoter (Fig. 6E).
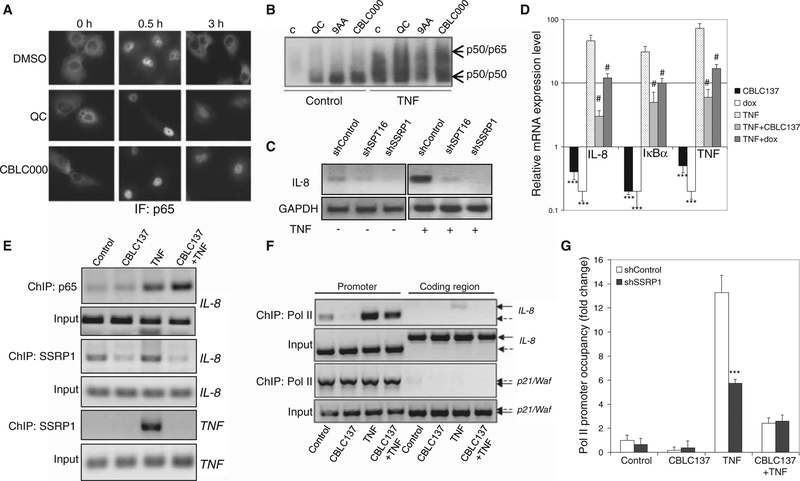
Fig. 6.
Effect of curaxins on NF-κB. (A) Slower reshuttling of NF-κB from the nucleus to the cytoplasm after TNF induction in cells treated with curaxins. Immunofluorescence (IF) staining of p65 in HT1080 cells treated with TNF (10 ng/ml, here and thereafter) in combination with 0.1% DMSO, quinacrine (6 μM), or CBLC000 (1 μM). (B) Effect of curaxins on nuclear accumulation and DNA binding of NF-κB under basal (“control”) and TNF-stimulated conditions. EMSA with 32P-labeled NF-κB consensus binding element and nuclear extracts from H1299 cells left untreated (C) or treated with quinacrine (10 μM), 9-aminoacridine (9AA) (10 μM), or CBLC000 (2 μM) for 2 hours with or without concurrent TNF stimulation. (C and D) Involvement of FACT in TNF-induced NF-κB–dependent transcription. (C) Reverse transcription–PCR (RT-PCR) analysis of IL-8 mRNA expression in shRNA-transduced HT1080 cells left untreated or treated with TNF for 2 hours. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. (D) Real-time quantitative RT-PCR (qPCR) analysis of IL-8, IκBα, and TNF expression in HT1080-tet-ON-shSSRP1 cells treated with CBLC137 (2 μM, 2 hours), doxycycline (dox) (to induce shSSRP1 expression, 5 μg/ml, 48 hours), and/or TNF (2 hours). Mean mRNA levels in treated cells relative to control (±SD) are shown. ***P < 0.001 for comparison to untreated cells; #P < 0.001 (t test) for comparison to TNF-treated cells. The level of SSRP1 mRNA measured by qPCR was reduced fivefold after doxycycline-induced shSSRP1 expression. (E) Reduced presence of SSRP1 on promoters of NF-κB–dependent genes (IL-8 and TNF) after curaxin treatment. ChIP assays using α-SSRP1 or α-p65 were performed on HT1080 cells treated with CBLC137 (1 μM), TNF, or both for 2 hours. (F) Curaxin-mediated inhibition of NF-κB–dependent transcription at the stage of elongation. ChIP using α-RNA Pol II antibody on cells treated as in (E) followed by PCR with primers specific to the promoter or coding region of the IL-8 or p21/Waf genes (used as a control because no induction of p21 is observed after 2 hours of curaxin treatment). Solid and dashed arrows indicate PCR products corresponding to coding and promoter regions, respectively. (G) Reduced RNA Pol II presence on the IL-8 promoter after CBLC137- or shSSRP1-inducing treatment, but no further reduction after combined treatment. ChIP with α-RNA Pol II was performed on HT1080-tet-ON-shSSRP1 (or HT1080-tet-ON-shControl) cells treated with doxycycline for 48 hours and then with CBLC137, TNF, or both as described in (D) for 80 min. ChIP products were assessed by qPCR with IL-8 promoter-specific primers. Data are mean fold change in RNA Pol II binding to the IL-8 promoter relative to untreated shControl-expressing cells ± SD. ***P < 0.001 from shControl (t test). See also fig. S6.
Previous reports show that transcription factors (including NF-κB) can bind to promoters in the presence of nucleosomes (24, 25), whereas binding of the basal transcription machinery (particularly RNAPII) requires assistance from chromatin remodeling factors, including FACT (20, 22). Therefore, lack of functional FACT on NF-κB–dependent promoters in curaxin-treated cells or cells with shRNA-mediated FACT knockdown might lead to reduced RNAPII binding and account for the suppression of NF-κB–dependent transcription observed in curaxin-treated cells. As predicted by this model, curaxin treatment reduced the amount of RNAPII bound to the IL-8 promoter and eliminated RNAPII binding to the coding region of the IL-8 gene, illustrating that IL-8 transcription was blocked predominantly at the stage of elongation (Fig. 6F). There was no additional decrease in RNAPII presence on the IL-8 promoter if shRNA-targeting SSRP1 was combined with curaxin treatment (Fig. 6G), indicating that the effect of curaxins on RNAPII is indeed exerted via FACT.
The effects of curaxins on FACT are different from those of cisplatin and do not involve DNA damage
SSRP1 binds to DNA in the minor groove when its spatial architecture is disturbed by cisplatin- or ultraviolet (UV)–induced covalent modifications (18, 19) or by bending caused by formation of cruciform DNA structures (26). However, quinacrine and curaxins did not induce DNA damage as judged by Comet and γH2AX staining assays, even when used at concentrations up to 10-fold higher than those required to activate p53 (Fig. 7, A and B, and fig. S7, A and B). Furthermore, curaxins did not promote radiation-induced carcinogenesis in cancer-prone p53 heterozygous mice (fig. S6C). Thus, the mechanism underlying FACT modulation by curaxins must be fundamentally different from that of DNA-damaging agents like cisplatin. Indeed, curaxins did not induce binding of recombinant SSRP1 to double-stranded DNA oligonucleotides in vitro, whereas oligonucleotides modified by cisplatin or UV treatment were highly affinitive to SSRP1 (fig. S7D). In contrast, when DNA was presented as native chromatin in a cell-free system, SSRP1 binding to chromatin was stimulated by curaxins (Fig. 7C) but not cisplatin (27). CBLC137 only slightly induced binding of FACT to a reconstituted mononucleosomal template in vitro (Fig. 7D), which suggests that more complex chromatin structures are the target of FACT binding in curaxin-treated cells.
Fig. 7.
Binding of curaxins to DNA without induction of detectable DNA damage. (A) Comet assays were performed on HeLa cells left untreated (control) or treated with DMSO (0.1%), CBLC120 (an inactive curaxin analog, 10 μM), quinacrine (10 μM), CBLC137 (2 μM), CBLC100(0.5 μM), or doxorubicin (0.5 μM) for 6 hours. The amount of DNA breaks is indicated by the mean tail moment of individual cells ± SD (n > 10). *P < 0.05 for comparison to control cells (Kruskal-Wallis one-way ANOVA on ranks). (B) Failure of curaxins to induce histone H2AX phosphorylation. Immunofluorescence staining of HT1080 cells treated with doxorubicin (0.5 μM), CBLC101 (an inactive curaxin analog, 10 μM), CBLC137 (1 μM), or CBLC100 (0.2 μM) for 6 hours. (C) Stimulatory effect of CBLC137 on in vitro binding of SSRP1 to chromatin. Chromatin purified from HeLa cells was incubated for 20 min with FLAG-tagged SSRP1 (100 ng) and/or CBLC137 (2 μM). Reactions were spun down and the soluble and chromatin-bound (pellet) fractions were assessed by anti-FLAG Western blotting. (D) Lack of CBLC137 effect on in vitro binding of FACT to nucleosomal DNA. Autoradiogram of 32P-labeled mononucleosomal DNA (N) incubated with recombinant FACT (10 pM each subunit) and/or CBLC137 (2 μM) for 20 min. N(2) and N(3), di- and trinucleosomes formed upon FACT addition; asterisk, smear induced upon CBLC137 addition. (E) Computer modeling of quinacrine (left panel; electron density model of DNA with stick model of quinacrine) and CBLC137 (right panel; space-filling model) binding to double-stranded DNA. (F) Inverse proportionality of curaxin DNA binding constants and EC50 for p53 reporter activation. See also fig. S7.
The chemical structure of curaxins suggested that they might be DNA intercalators (Fig. 1A). In silico modeling showed that active curaxins bind to DNA with the carbazole core intercalated between DNA bases and a positively charged side chain aligned in the minor groove (Fig. 7E). Quinacrine, which is functionally similar to curaxins, is a known DNA intercalator (28), and DNA binding of two quinacrine enantiomers was shown to correlate with their effects on p53 and NF-κB (3). In silico modeling was substantiated by the experiments showing that all tested active curaxins, but not inactive analogs, can alter the mobility of plasmid DNA in electrophoretic mobility shift assays (EMSAs) (fig. S7E). The potency of p53 and NF-κB modulation by curaxins correlated with their DNA binding affinity (Fig. 7F and fig. S7E). Moreover, curaxins preferentially bind to dAdT-rich sequences (fig. S7F), which are preferred binding sites of HMG domains such as that in SSRP1 (29). We also monitored distribution of several fluorescent curaxins in live cells. In contrast to inactive analogs, active compounds appeared to bind to nuclear structures with highest accumulation in perinuclear membrane and perinucleolar areas, where the heterochromatin zones with high AT content are concentrated (fig. S7G).
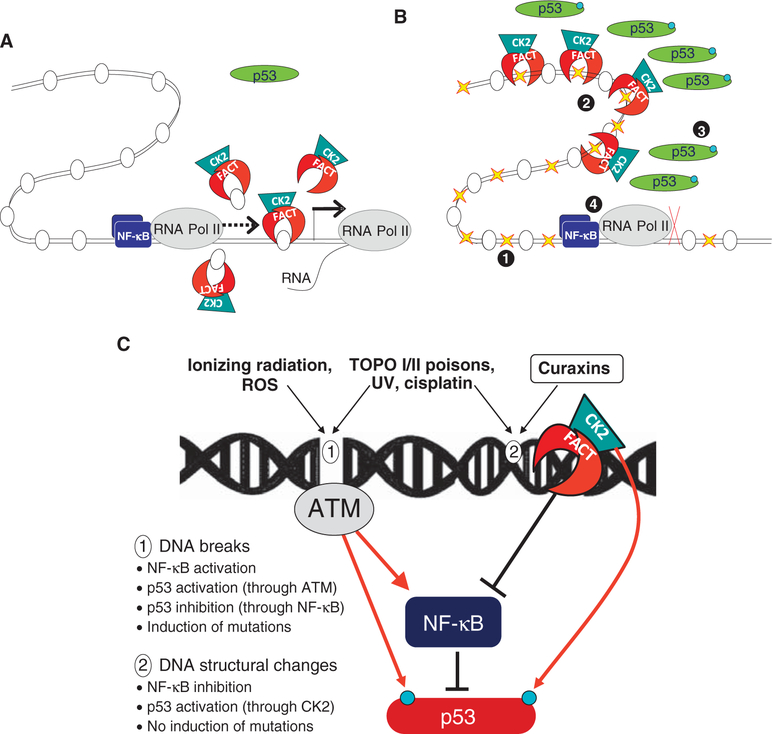
Together, these data support a model in which curaxins bind to DNA and disturb chromatin architecture such that FACT becomes “trapped.” This results in activating phosphorylation of p53 by FACT-associated CK2 and reduced NF-κB–dependent transcription because of depletion of soluble FACT (Fig. 8).
Fig. 8.
Proposed model of curaxins’ mechanism of activity (see details in the text). (A and B) FACT involved in transcription elongation on normal conditions (A) is trapped in chromatin in curaxin-treated cells (B). 1, curaxin binds DNA and changes chromatin architecture; 2, FACT is trapped in chromatin; 3, p53 is phosphorylated by CK2; 4, NF-κB transcription is blocked. (C) Two types of consequences of small-molecule DNA interactions. 1, DNA breaks result from ionizing radiation and reactive oxygen species (ROS) reaction with DNA; 2, Changes in DNA 3D structure caused by nonreactive intercalators such as curaxins. Inhibitors of topoisomerases or compounds causing covalent modifications of DNA may cause structural changes in DNA and breaks. Red arrow, effects leading to an increase in the activity or the targeted factor; black lines, effect leading to an inhibition of the activity of the targeted factor.
DISCUSSION
This study addresses the efficacy and mechanism of action of curaxins, a newly identified class of anticancer compounds. Curaxins showed broad anticancer activity in mice at doses not causing systemic toxicity or genotoxicity, the major drawback of current cancer treatments with other DNA-targeted chemotherapeutic agents, which promote mutations that can lead to treatment resistance and development of secondary cancers (30–32). Another benefit of curaxins is that they simultaneously activate p53 and suppress NF-κB, which are widely recognized as critical anticancer targets. Specific features of NF-κB–p53 crosstalk may contribute to the efficacy of curaxins. For example, because overactive NF-κB is a frequent cause of p53 suppression in tumors (7, 33–36), inhibition of NF-κB by curaxins is expected to amplify their effect on p53 activation. In contrast, chemotherapeutic agents that cause DNA breaks are known to activate NF-κB in tumor cells through ATM-mediated phosphorylation of IκB kinase γ (IKKγ) (37–39). Because NF-κB negatively regulates p53 (6, 7, 33, 34, 36), activation of NF-κB by these drugs works against effective activation of p53 and thereby reduces their overall anticancer effect (Fig. 8B). Indeed, we demonstrated that this pathway accounts for the failure of DNA-damaging agents to activate p53 in RCC45 cells. siRNA-mediated knockdown of IKKγ prevented DNA damage–dependent activation of NF-κB and made p53 responsive to DNA-damaging agents (fig. S6). Thus, although we initially set out to identify molecules acting directly on p53 (7), our cell-based readout allowed us to identify an important mechanism of p53 deregulation in tumor cells as well as molecules capable of simultaneously modulating the activity of several cellular pathways (8).
Curaxins’ effects on p53 and NF-κB stem from the ability of curaxins to alter functions of FACT. Our demonstration that FACT mediates the antitumor effects of curaxins revealed FACT as a prospective anticancer therapeutic target. The potential importance of FACT as an anticancer target is supported by our finding that tumor cells with reduced levels of FACT are not viable (Fig. 5A), whereas Wi38 normal diploid fibroblasts exist without detectable FACT expression (Fig. 5F). In addition, previous studies found that SSRP1 expression was increased in ovarian cancer cells compared to normal cells (40). Our observation of elevated FACT expression in tumor tissues relative to normal tissues (Fig. 5, E and F) suggests that FACT provides tumor cells with a selective advantage under normal conditions, but also makes them more sensitive to the cytotoxic effects of curaxins. The tumor selectivity of curaxins might be due to differences in chromatin structure/function that leads to a greater requirement for FACT activity in tumor cells than in normal cells. Alternatively, the role of FACT may be similar in normal and tumor tissues, but FACT-dependent processes such as NF-κB–directed transcription may be more important for tumor cells than normal cells.
The normal functions of FACT include binding of histone dimers and tetramers and remodeling of nucleosomes in the vicinity of RNAPs (22). This function is vital for transcription of genes with ordered nucleosome structure (21–23) and presumably requires free soluble FACT. Our data show that curaxins induce changes in FACT localization, leading to depletion of soluble FACT from the nucleoplasm because of its tight association with chromatin (Fig. 4, fig. S4, and model in Fig. 8A). This “trapping” of FACT in chromatin is likely due to its affinity toward altered chromatin architecture caused by DNA intercalation of curaxins. The resulting decrease in free FACT leads to the observed suppression of NF-κB–dependent transcription in curaxin-treated cells and may affect other transcriptional programs as well.
Binding of FACT to curaxin-impregnated chromatin also leads to p53 activation. FACT binds to distorted DNA through the HMG domain of SSRP1 (19). This presumably makes the adjacent intrinsically disordered domain of SSRP1 inaccessible to phosphorylation by CK2 (41). We propose that a similar situation exists when FACT binds to chromatin in curaxin-treated cells such that CK2, lacking SSRP1 as a substrate, “switches attention” and phosphorylates Ser392 of p53.
In addition to its roles in transcription, FACT is also involved in replication (42, 43), mitosis (44), and homologous recombination (45). It remains to be determined whether curaxins affect these processes and, if so, how they contribute to the antitumor effect of the compounds. The requirement for SSRP1 in mitosis (44) might explain the growth-inhibitory effect that curaxins have on normal cells in vitro. In vivo, however, no adverse effects on normal tissues were apparent after curaxin treatment because reduced proliferation of most cell types would not present as evident toxicity (fig. S2).
It remains to be determined exactly how curaxins interact with FACT and/or alter its activities. Although intercalation of curaxins into DNA does not cause DNA damage (Fig. 7 and fig. S7), it likely alters chromatin structure. Curaxins alter the intracellular localization of FACT, which is a chromatin structure–sensitive complex (Fig. 4 and fig. S4). This occurs rapidly after curaxin treatment (Fig. 4A). Moreover, purified SSRP1 binds to chromatin in vitro when it is mixed with chromatin and curaxin at 4°C (Fig. 7C). This suggests that chromatin binding by FACT does not result from curaxin-modified signaling but from rapid changes in chromatin structure caused by curaxin and generation of sites attractive for FACT binding (Fig. 8A). The nature of the proposed curaxin-induced changes in chromatin structure remains unclear; however, it is possible that curaxins induce binding of FACT to protein-DNA complexes that are present in heterochromatin, but not in simple nucleosomal arrays (no significant curaxin-induced binding of FACT to nucleosomes was observed in in vitro assays) (Fig. 7D). Although the DNA crosslinks induced by cisplatin or UV recruit FACT to the distorted DNA, curaxin impregnation is not sufficient to induce FACT binding to linear DNA (fig. S7D). Rather, tight binding of FACT occurs only when curaxins are used in the context of DNA packaged within native chromatin structure (Fig. 7C), presumably due to formation of stronger or different modifications of DNA structure in the context of chromatin. Preliminary experiments demonstrated that curaxins change the superhelicity of DNA, which may lead to the formation of local protrusions and cruciform structures, especially in the context of heterochromatin, and such structures are natural sites of FACT binding (23).
Previously known nongenotoxic DNA intercalators (46, 47) have not been extensively investigated as possible anticancer agents because it was assumed that induction of DNA damage was an essential aspect of the anticancer mechanism of other DNA intercalators. The strong biological effects caused by nongenotoxic DNA binding of curaxins demonstrate that this is not a valid assumption, thereby opening up a new area for anticancer drug discovery (3).
In conclusion, we have defined curaxins as a new class of small molecules with broad and potent anticancer activity and a mechanism of action that is fundamentally different from current chemotherapeutic drugs. By binding DNA within chromatin and altering FACT activities, curaxins modulate p53 and NF-κB in the directions desired for cancer therapy. Curaxins do not cause DNA damage or affect general transcription and are therefore expected to be well tolerated and safe for use in humans, as we have shown in mice. In addition to building a foundation for possible clinical development of curaxins, this study allowed us to define FACT as a promising anticancer target involved in regulation of multiple cellular pathways that are frequently deregulated in cancer.
MATERIALS AND METHODS
Standard experiments (for example, immunofluorescence, EMSA, Western blotting, Comet assay, in vitro p53 phosphorylation assay, and ChIP) are described in the Supplementary Material.
Cells and reagents
The cell lines used are described in the Supplementary Material. Cell lines with p53- and NF-κB–dependent reporters were generated by lentiviral transduction and have been described (7). Commercially available chemicals are listed in the Supplementary Material. Compounds for screening and hit-to-lead optimization were provided by ChemBridge Inc. siGENOMESMART pool reagents and control ON-TARGETplus Non-Targeting siRNAs were from Dharmacon Inc. Mission lentiviral shRNA vectors were purchased from Sigma-Aldrich Inc. pTRIPZ inducible lentiviral vector or miRNA expression was purchased from Open Biosystem Inc. Cell Lights Histone H2B-RFP-BacMam is from Invitrogen. M30 Apoptosome Elisa kit is from Pevia.
Screening for p53 activators and NF-κB inhibitors
RCC45-p53-Luc and H1299-κB-Luc cells were treated in 96-well plates with 0.08 to 20 μM test compounds in duplicate. TNF (10 ng/ml) was added to H1299-κB-Luc cells to induce NF-κB activity. Luciferase activity was measured 16 hours later (Bright-Glo Luciferase Assay system, Promega). Quinacrine (6 μM) was used as a positive control. Assays were run three times, and the average 50% effective concentration for p53 induction or NF-κB suppression (EC50) was calculated by the sigmoid approximation method.
Cytotoxicity and colony assays
Chemicals were tested by treating cells in 96-well plates for 1 or 24 hours with 0.08 to 20 μM test compounds in triplicate. Cell survival was assessed 72 hours later by methylene blue staining. To test the effect of shRNAs, we infected cells with lentiviral shRNAs at ~80 to 90% multiplicity of infection (MOI), selected them in puromycin (72 hours), and plated them for colony assays or collected them for protein or RNA extraction. Colony number was assessed at different times depending on when different cell types transduced with control shRNA formed visible colonies.
DNA binding assay
The ability of compounds to alter the mobility of plasmid DNA was tested by incubating plasmid DNA in tris-EDTA buffer (pH 8.0) with 0.1 to 10 μM chemicals at room temperature for 20 min followed by electrophoresis (0.8% agarose gel, 1.5 V/cm constant for 16 hours). Gels were stained with ethidium bromide (0.5 μg/ml) and visualized with short-wavelength UV light.
Competition dialysis was performed as described previously (48). Details are provided in the Supplementary Material.
Assembly and transcription of mononucleosomal template was done as described (49). Details are provided in the Supplementary Material. Briefly, a 199–base pair (bp) DNA fragment carrying the 603 nucleosomal positioning sequence (NPS) was isolated by polymerase chain reaction (PCR) from a pUC18-based plasmid with a 5′-biotin–labeled primer. Nucleosomes were assembled on this fragment with salt-denatured histones by gradually decreasing the NaCl concentration from 2 to 0.1 M (overnight dialysis against assembly buffer). The efficiency of mononucleosome assembly was evaluated by 4% native polyacrylamide gel electrophoresis and by testing protection of the NPS against restriction endonuclease digestion.
Transcription of the mononucleosomal templates was performed as in (50) and is described in the Supplementary Material. Recombinant FACT was purified from SF9 cells with constructs provided by D. Reinberg (New York University School of Medicine, New York, NY) and a previously described method (51). Transcripts were purified by phenol-chloroform extraction and ethanol precipitation, resolved by denaturing gel electrophoresis, and visualized/quantified with a PhosphorImager (Molecular Dynamics) and ImageQuant software (GE Healthcare).
Binding of SSRP1 to DNA and chromatin in vitro
Two types of 5′-end 32P-labeled templates were used: a 120-bp double-stranded DNA probe pML20–42 (52) and mononucleosomal DNA prepared as described above. Binding reactions were performed in 20 μl for 45 min at room temperature in 20 mM tris-HCl (pH 7.6); 150 mM NaCl; 10 mM MgCl2; 5% glycerol; 0.5 mM dithiothreitol (DTT); 0.25 mM EDTA with 30 fmol (2 × 103 cpm) DNA probe; and 2, 4, 8, or 16 pmol FLAG-tagged SSRP. Probes were treated with curaxins for 10 min or with cisplatin as described previously (53) before addition of proteins. Binding reactions were resolved on 4.5% native polyacrylamide gels (29% acrylamide/1% bisacrylamide). Gels were dried and visualized with PhosphorImager (Molecular Dynamics). For each chromatin-binding reaction, nuclei from 106 HeLa cells were lysed in no-salt buffer (54) and centrifuged (6500g, 5 min, 4°C). Chromatin pellets were washed twice with 150 mM NaCl buffer and then resuspended in 10 mM tris (pH 7.5), 50 mM NaCl, 10 mM MgCl2, 0.5 mM EDTA, 5% glycerol, and 1 mM DTT and mixed with 100 ng of purified SSRP1 and 5 μM CBLC137 for 2 hours. Reactions were then centrifuged (6500g, 15 min, +4°C). Supernatants were loaded directly onto gels; pellets were sonicated and boiled in Laemmli buffer for 5 min before loading. Electrophoresis and anti-SSRP1 Western blotting were performed as described in the Supplementary Material.
Extraction of soluble and chromatin-bound proteins from cells
Total cell extracts were prepared by boiling cells in Laemmli buffer for 15 min. Soluble proteins were obtained by lysing cells in Cell Culture Lysis Reagent (Promega) for 10 min on ice followed by spinning down of the insoluble fraction. Extraction of chromatin-bound proteins followed the protocol of Shechter et al. (54) with minor modifications. Instead of one high-salt extraction, serial extractions with 0.15, 1.0, 1.5, 2.5, or 3 M NaCl were done. The final pellet was sonicated and boiled in Laemmli buffer for 5 min. Samples were dialyzed as described (54) and loaded onto 10.5 to 14% tris-HCl gels (Bio-Rad). Gels were either stained with Coomassie blue or blotted onto polyvinylidene difluoride membrane for immunoblotting.
Fluorescent microscopy
Fluorescent images of live cells were obtained with a Zeiss Axio Observer A1 inverted microscope with N-Achroplan 100×/1.25 oil lens, Zeiss MRC5 camera, and AxioVision Rel.4.8 software.
Computer modeling
SAR study and computer modeling of curaxin-DNA binding used several dozen active curaxins and structurally similar molecules unable to activate p53. Computer modeling analyzed superimposition of three-dimensional (3D) conformers of active and inactive molecules docking on DNA with MOLOC (Gerber Molecular Design) molecular mechanics and GOLD programs (Cambridge Crystallographic Data Center).
In vivo evaluation of curaxins in mouse tumor models
All procedures involving mice were approved by the Institutional Animal Care and Use Committee of Roswell Park Cancer Institute (RPCI, Buffalo, NY). The maximum tolerated doses (MTDs) for single and repetitive oral administration of curaxins were defined as the maximum doses not associated with mortality or persistent morbidity. Details are provided in the Supplementary Material.
Curaxins were tested for efficacy against human tumor xenografts established by inoculating (subcutaneously) 1 × 106 to 3 × 106 cultured tumor cells into each rear flank of athymic nude mice (n = 10 per treatment group). For the pancreatic ductal adenocarcinoma model, a severe combined immunodeficient (SCID)/non-obese diabetic (NOD) mouse with a single tumor was generated by subcutaneous transplantation of a tissue piece from a pancreatic cancer patient (provided by E. Repasky, RPCI, Buffalo, NY). This tumor was grown in the donor mouse to ~500 mm3, then excised from the anesthetized mouse, washed with phosphate-buffered saline (PBS), and cut into 8- to 10-mm3 pieces. A single tumor piece was transplanted under the skin of each anesthetized (isoflurane/oxygen gas mixture through mouth mask) recipient mouse. Upon visible/palpable presence of a tumor, tumors were measured with digital vernier calipers and tumor volume was calculated as volume = length × width2/2, where length is the largest diameter and width is the largest diameter perpendicular to the length. Tumors were measured every other day before the start of treatment and twice weekly thereafter. Treatment was started when tumors reached ~50 mm3. Curaxins were administered by oral gavage at rMTD after a 5 days on/2 days off schedule for up to 4 weeks. Mice were euthanized after 4 weeks of treatment or when at least one tumor reached a volume of 1000 mm3. Statistically significant differences in tumor volumes of curaxin- and vehicle-treated groups were determined with Student’s t test (P < 0.05 = significant).
p53 heterozygous mice were obtained as described (55). Forty males and 40 females were exposed to 4-gray total body γ-irradiation (137Cs source) and then separated into two groups. Mice received vehicle (water) or CBLC137 (30 mg/kg, dissolved in water) by gavage once daily for 7 consecutive days of every month of their life span. Animals were observed and weighed daily and were killed in cases of >15% weight loss, development of visible tumors, or other signs suggesting imminent death. Euthanized animals were subjected to gross pathology examination, including collection of tumors for histopathology examination.
Female MMTV-neu mice [Tg(MMTVneu)202Mul/J; Jackson Laboratory] with spontaneous palpable mammary tumors were used in the study.
Supplementary Material
www.sciencetranslationalmedicine.org/cgi/content/full/3/95/95ra74/DC1
Materials and Methods
Fig. S1. Structure and activity of curaxins.
Fig. S2. Antitumor effect of curaxin CBLC137 in xenograft mouse models of cancer.
Fig. S3. Dependence of p53 activation by curaxin on CK2 and FACT.
Fig. S4. Curaxin induction of FACT-chromatin binding.
Fig. S5. Association between FACT levels and cell growth, sensitivity to curaxins, and tumor phenotype.
Fig. S6. Suppression of IKKγ expression makes the p53 present in RCC45 cells responsive to DNA-damaging agents.
Fig. S7. Binding of curaxins to DNA without induction of detectable DNA damage.
Table S1. Physicochemical and pharmacological properties of curaxins described in the paper.
References
Acknowledgments:
We are grateful to E. Feinstein for her contribution to the curaxin development program and for critical reading of the manuscript. We also thank F. Bone, E. Kononov, and A. Cheney for help with animal experiments; E. Repasky and B. Hylander for providing human pancreatic surgical samples; D. Luse for providing reagents and technical advice on the experiments with mononucleosomal templates; D. Reinberg for providing baculovirus vectors for expression of FACT components; S. Lowe for providing C8 cells; B. Vogelstein for the syngenic pair of HCT116 p53 plus and minus cells; L. Korochkina for help with FACT purification; H. Garcia for technical assistance; and P. Stanhope Baker for help with manuscript preparation.
Funding: This work was supported in part by grants from NIH (CA75179) and from Cleveland BioLabs Inc. to A.V.G. and from Incuron Inc. to K.V.G.
Footnotes
Competing interests: K.V.G., A.A.P., C.A.B., and A. V. Gudkov are co-inventors on a patent application that covers composition of matter and use of curaxins, #61/102,913, “Carbazole compounds and therapeutic uses of the compounds.” A. V. Gudkov is a paid consultant and has equity interest in Cleveland BioLabs Inc., which holds patents on and develops curaxins. O.A.G. has been a paid consultant for RPCI, and K.V.G. has received an honorarium from Incuron Inc. C.A.B. has stock options in Cleveland BioLabs Inc. The others authors declare no competing interests.
REFERENCES AND NOTES
- 1.Burdak-Rothkamm S, Prise KM, New molecular targets in radiotherapy: DNA damage signalling and repair in targeted and non-targeted cells. Eur. J. Pharmacol 625, 151–155 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Darzynkiewicz Z, Traganos F, Wlodkowic D, Impaired DNA damage response—An Achilles’ heel sensitizing cancer to chemotherapy and radiotherapy. Eur. J. Pharmacol 625, 143–150 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gurova K, New hopes from old drugs: Revisiting DNA-binding small molecules as anticancer agents. Future Oncol. 5, 1685–1704 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kawanishi S, Hiraku Y, Amplification of anticancer drug-induced DNA damage and apoptosis by DNA-binding compounds. Curr. Med. Chem. Anticancer Agents 4, 415–419 (2004). [DOI] [PubMed] [Google Scholar]
- 5.Gudkov AV, Gurova KV, Komarova EA, Inflammation and p53: A tale of two stresses. Genes Cancer 2, 503–516 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dey A, Tergaonkar V, Lane DP, Double-edged swords as cancer therapeutics: Simultaneously targeting p53 and NF-κB pathways. Nat. Rev. Drug Discov 7, 1031–1040 (2008). [DOI] [PubMed] [Google Scholar]
- 7.Gurova KV, Hill JE, Guo C, Prokvolit A, Burdelya LG, Samoylova E, Khodyakova AV, Ganapathi R, Ganapathi M, Tararova ND, Bosykh D, Lvovskiy D, Webb TR, Stark GR, Gudkov AV, Small molecules that reactivate p53 in renal cell carcinoma reveal a NF-κB-dependent mechanism of p53 suppression in tumors. Proc. Natl. Acad. Sci. U.S.A 102, 17448–17453 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gurova KV, Hill JE, Razorenova OV, Chumakov PM, Gudkov AV, p53 pathway in renal cell carcinoma is repressed by a dominant mechanism. Cancer Res. 64, 1951–1958 (2004). [DOI] [PubMed] [Google Scholar]
- 9.Buschmann T, Adler V, Matusevich E, Fuchs SY, Ronai Z, p53 phosphorylation and association with murine double minute 2, c-Jun NH2-terminal kinase, p14ARF, and p300/CBP during the cell cycle and after exposure to ultraviolet irradiation. Cancer Res. 60, 896–900 (2000). [PubMed] [Google Scholar]
- 10.Ito A, Lai CH, Zhao X, Saito S, Hamilton MH, Appella E, Yao TP, p300/CBP-mediated p53 acetylation is commonly induced by p53-activating agents and inhibited by MDM2. EMBO J. 20, 1331–1340 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wadgaonkar R, Collins T, Murine double minute (MDM2) blocks p53-coactivator interaction, a new mechanism for inhibition of p53-dependent gene expression. J. Biol. Chem 274, 13760–13767 (1999). [DOI] [PubMed] [Google Scholar]
- 12.Blattner C, Tobiasch E, Litfen M, Rahmsdorf HJ, Herrlich P, DNA damage induced p53 stabilization: No indication for an involvement of p53 phosphorylation. Oncogene 18, 1723–1732 (1999). [DOI] [PubMed] [Google Scholar]
- 13.Kulkarni A, Das KC, Differential roles of ATR and ATM in p53, Chk1, and histone H2AX phosphorylation in response to hyperoxia: ATR-dependent ATM activation. Am. J. Physiol. Lung Cell. Mol. Physiol 294, L998–L1006 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kapoor M, Hamm R, Yan W, Taya Y, Lozano G, Cooperative phosphorylation at multiple sites is required to activate p53 in response to UV radiation. Oncogene 19, 358–364 (2000). [DOI] [PubMed] [Google Scholar]
- 15.Cuddihy AR, Wong AH, Tam NW, Li S, Koromilas AE, The double-stranded RNA activated protein kinase PKR physically associates with the tumor suppressor p53 protein and phosphorylates human p53 on serine 392 in vitro. Oncogene 18, 2690–2702 (1999). [DOI] [PubMed] [Google Scholar]
- 16.Keller DM, Lu H, p53 serine 392 phosphorylation increases after UV through induction of the assembly of the CK2·hSPT16·SSRP1 complex. J. Biol. Chem 277, 50206–50213 (2002). [DOI] [PubMed] [Google Scholar]
- 17.Keller DM, Zeng X, Wang Y, Zhang QH, Kapoor M, Shu H, Goodman R, Lozano G, Zhao Y, Lu H, A DNA damage–induced p53 serine 392 kinase complex contains CK2, hSpt16, and SSRP1. Mol. Cell 7, 283–292 (2001). [DOI] [PubMed] [Google Scholar]
- 18.Bruhn SL, Pil PM, Essigmann JM, Housman DE, Lippard SJ, Isolation and characterization of human cDNA clones encoding a high mobility group box protein that recognizes structural distortions to DNA caused by binding of the anticancer agent cisplatin. Proc. Natl. Acad. Sci. U.S.A 89, 2307–2311 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yarnell AT, Oh S, Reinberg D, Lippard SJ, Interaction of FACT, SSRP1, and the high mobility group (HMG) domain of SSRP1 with DNA damaged by the anticancer drug cisplatin. J. Biol. Chem 276, 25736–25741 (2001). [DOI] [PubMed] [Google Scholar]
- 20.Belotserkovskaya R, Oh S, Bondarenko VA, Orphanides G, Studitsky VM, Reinberg D, FACT facilitates transcription-dependent nucleosome alteration. Science 301, 1090–1093 (2003). [DOI] [PubMed] [Google Scholar]
- 21.Jimeno-González S, Gómez-Herreros F, Alepuz PM, Chávez S, A gene-specific requirement for FACT during transcription is related to the chromatin organization of the transcribed region. Mol. Cell. Biol 26, 8710–8721 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reinberg D, Sims RJ III, de FACTo nucleosome dynamics. J. Biol. Chem 281, 23297–23301 (2006). [DOI] [PubMed] [Google Scholar]
- 23.Singer RA, Johnston GC, The FACT chromatin modulator: Genetic and structure/function relationships. Biochem. Cell Biol 82, 419–427 (2004). [DOI] [PubMed] [Google Scholar]
- 24.Pazin MJ, Sheridan PL, Cannon K, Cao Z, Keck JG, Kadonaga JT, Jones KA, NF-κB-mediated chromatin reconfiguration and transcriptional activation of the HIV-1 enhancer in vitro. Genes Dev. 10, 37–49 (1996). [DOI] [PubMed] [Google Scholar]
- 25.Steger DJ, Workman JL, Stable co-occupancy of transcription factors and histones at the HIV-1 enhancer. EMBO J. 16, 2463–2472 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gariglio M, Ying GG, Hertel L, Gaboli M, Clerc RG, Landolfo S, The high-mobility group protein T160 binds to both linear and cruciform DNA and mediates DNA bending as determined by ring closure. Exp. Cell Res 236, 472–481 (1997). [DOI] [PubMed] [Google Scholar]
- 27.Dejmek J, Iglehart JD, Lazaro JB, DNA-dependent protein kinase (DNA-PK)–dependent cisplatin-induced loss of nucleolar facilitator of chromatin transcription (FACT) and regulation of cisplatin sensitivity by DNA-PK and FACT. Mol. Cancer Res 7, 581–591 (2009). [DOI] [PubMed] [Google Scholar]
- 28.Jones RL, Davidson MW, Wilson WD, Comparative viscometric analysis of the interaction of chloroquine and quinacrine with superhelical and sonicated DNA. Biochim. Biophys. Acta 561, 77–84 (1979). [DOI] [PubMed] [Google Scholar]
- 29.Bewley CA, Gronenborn AM, Clore GM, Minor groove-binding architectural proteins: Structure, function, and DNA recognition. Annu. Rev. Biophys. Biomol. Struct 27, 105–131 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fonseca FL, Sant Ana AV, Bendit I, Arias V, Costa LJ, Pinhal AA, del Giglio A, Systemic chemotherapy induces microsatellite instability in the peripheral blood mononuclear cells of breast cancer patients. Breast Cancer Res. 7, R28–R32 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gonzalez-Angulo AM, Morales-Vasquez F, Hortobagyi GN, Overview of resistance to systemic therapy in patients with breast cancer. Adv. Exp. Med. Biol 608, 1–22 (2007). [DOI] [PubMed] [Google Scholar]
- 32.Sakai W, Swisher EM, Karlan BY, Agarwal MK, Higgins J, Friedman C, Villegas E, Jacquemont C, Farrugia DJ, Couch FJ, Urban N, Taniguchi T, Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers. Nature 451, 1116–1120 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo C, Gasparian AV, Zhuang Z, Bosykh DA, Komar AA, Gudkov AV, Gurova KV, 9-Aminoacridine-based anticancer drugs target the PI3K/AKT/mTOR, NF-κB and p53 pathways. Oncogene 28, 1151–1161 (2009). [DOI] [PubMed] [Google Scholar]
- 34.Jung KJ, Dasgupta A, Huang K, Jeong SJ, Pise-Masison C, Gurova KV, Brady JN, Small-molecule inhibitor which reactivates p53 in human T-cell leukemia virus type 1-transformed cells. J. Virol 82, 8537–8547 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shao J, Fujiwara T, Kadowaki Y, Fukazawa T, Waku T, Itoshima T, Yamatsuji T, Nishizaki M, Roth JA, Tanaka N, Overexpression of the wild-type p53 gene inhibits NF-κB activity and synergizes with aspirin to induce apoptosis in human colon cancer cells. Oncogene 19, 726–736 (2000). [DOI] [PubMed] [Google Scholar]
- 36.Tergaonkar V, Perkins ND, p53 and NF-κB crosstalk: IKKα tips the balance. Mol. Cell 26, 158–159 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Arlt A, Schäfer H, NFκB-dependent chemoresistance in solid tumors. Int. J. Clin. Pharmacol. Ther 40, 336–347 (2002). [DOI] [PubMed] [Google Scholar]
- 38.Pham CG, Bubici C, Zazzeroni F, Knabb JR, Papa S, Kuntzen C, Franzoso G, Upregulation of Twist-1 by NF-κB blocks cytotoxicity induced by chemotherapeutic drugs. Mol. Cell. Biol 27, 3920–3935 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shen HM, Tergaonkar V, NFκB signaling in carcinogenesis and as a potential molecular target for cancer therapy. Apoptosis 14, 348–363 (2009). [DOI] [PubMed] [Google Scholar]
- 40.Hudson ME, Pozdnyakova I, Haines K, Mor G, Snyder M, Identification of differentially expressed proteins in ovarian cancer using high-density protein microarrays. Proc. Natl. Acad. Sci. U.S.A 104, 17494–17499 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsunaka Y, Toga J, Yamaguchi H, Tate S, Hirose S, Morikawa K, Phosphorylated intrinsically disordered region of FACT masks its nucleosomal DNA binding elements. J. Biol. Chem 284, 24610–24621 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tan BC, Liu H, Lin CL, Lee SC, Functional cooperation between FACT and MCM is coordinated with cell cycle and differential complex formation. J. Biomed. Sci 17, 11 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tan BC, Chien CT, Hirose S, Lee SC, Functional cooperation between FACT and MCM helicase facilitates initiation of chromatin DNA replication. EMBO J. 25, 3975–3985 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zeng SX, Li Y, Jin Y, Zhang Q, Keller DM, McQuaw CM, Barklis E, Stone S, Hoatlin M, Zhao Y, Lu H, Structure-specific recognition protein 1 facilitates microtubule growth and bundling required for mitosis. Mol. Cell. Biol 30, 935–947 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kumari A, Mazina OM, Shinde U, Mazin AV, Lu H, A role for SSRP1 in recombination-mediated DNA damage response. J. Cell. Biochem 108, 508–518 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Snyder RD, Assessment of atypical DNA intercalating agents in biological and in silico systems. Mutat. Res 623, 72–82 (2007). [DOI] [PubMed] [Google Scholar]
- 47.Snyder RD, Hendry LB, Toward a greater appreciation of noncovalent chemical/DNA interactions: Application of biological and computational approaches. Environ. Mol. Mutagen 45, 100–105 (2005). [DOI] [PubMed] [Google Scholar]
- 48.Ren J, Chaires JB, Rapid screening of structurally selective ligand binding to nucleic acids. Methods Enzymol. 340, 99–108 (2001). [DOI] [PubMed] [Google Scholar]
- 49.Lowary PT, Widom J, New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol 276, 19–42 (1998). [DOI] [PubMed] [Google Scholar]
- 50.Pal M, Luse DS, The initiation–elongation transition: Lateral mobility of RNA in RNA polymerase II complexes is greatly reduced at +8/+9 and absent by +23. Proc. Natl. Acad. Sci. U.S.A 100, 5700–5705 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Loyola A, He S, Oh S, McCafferty DG, Reinberg D, Techniques used to study transcription on chromatin templates. Methods Enzymol. 377, 474–499 (2004). [DOI] [PubMed] [Google Scholar]
- 52.Pal M, McKean D, Luse DS, Promoter clearance by RNA polymerase II is an extended, multistep process strongly affected by sequence. Mol. Cell. Biol 21, 5815–5825 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jin Y, Lipscomb JD, Probing the mechanism of C–H activation: Oxidation of methylcubane by soluble methane monooxygenase from Methylosinus trichosporium OB3b. Biochemistry 38, 6178–6186 (1999). [DOI] [PubMed] [Google Scholar]
- 54.Shechter D, Dormann HL, Allis CD, Hake SB, Extraction, purification and analysis of histones. Nat. Protoc 2, 1445–1457 (2007). [DOI] [PubMed] [Google Scholar]
- 55.Leonova KI, Shneyder J, Antoch MP, Toshkov IA, Novototskaya LR, Komarov PG, Komarova EA, Gudkov AV, A small molecule inhibitor of p53 stimulates amplification of hematopoietic stem cells but does not promote tumor development in mice. Cell Cycle 9, 1434–1443 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lowe SW, Ruley HE, Jacks T, Housman DE, p53-dependent apoptosis modulates the cytotoxicity of anticancer agents. Cell 74, 957–967 (1993). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
www.sciencetranslationalmedicine.org/cgi/content/full/3/95/95ra74/DC1
Materials and Methods
Fig. S1. Structure and activity of curaxins.
Fig. S2. Antitumor effect of curaxin CBLC137 in xenograft mouse models of cancer.
Fig. S3. Dependence of p53 activation by curaxin on CK2 and FACT.
Fig. S4. Curaxin induction of FACT-chromatin binding.
Fig. S5. Association between FACT levels and cell growth, sensitivity to curaxins, and tumor phenotype.
Fig. S6. Suppression of IKKγ expression makes the p53 present in RCC45 cells responsive to DNA-damaging agents.
Fig. S7. Binding of curaxins to DNA without induction of detectable DNA damage.
Table S1. Physicochemical and pharmacological properties of curaxins described in the paper.
References