Abstract
In this short review, we give an overview of microtubule nucleation within cells. It is nearly 30 years since the discovery of γ-tubulin, a member of the tubulin superfamily essential for proper microtubule nucleation in all eukaryotes. γ-tubulin associates with other proteins to form multiprotein γ-tubulin ring complexes (γ-TuRCs) that template and catalyse the otherwise kinetically unfavourable assembly of microtubule filaments. These filaments can be dynamic or stable and they perform diverse functions, such as chromosome separation during mitosis and intracellular transport in neurons. The field has come a long way in understanding γ-TuRC biology but several important and unanswered questions remain, and we are still far from understanding the regulation of microtubule nucleation in a multicellular context. Here, we review the current literature on γ-TuRC assembly, recruitment, and activation and discuss the potential importance of γ-TuRC heterogeneity, the role of non-γ-TuRC proteins in microtubule nucleation, and whether γ-TuRCs could serve as good drug targets for cancer therapy.
Keywords: centrosome, gamma-tubulin ring complex, g-TuRC, microtubule, MTOC
Introduction
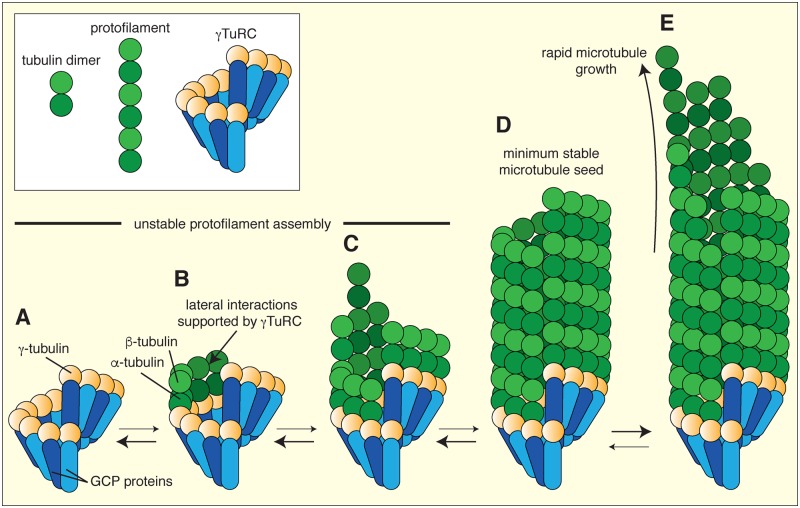
Microtubules are polarised polymers involved in a wide range of cellular processes including chromosome separation, intracellular transport, organelle positioning, cell–cell signalling and cell motility [1]. Tight spatial and temporal regulation of the formation, organisation, and dynamic behaviour of the microtubule cytoskeleton is extremely important. Consequently, cells have developed complex mechanisms to regulate microtubule nucleation, polymerisation and catastrophe, severing, stabilisation and transportation. Collectively, these processes establish diverse microtubule networks with highly specialised functions in different cells or at different times during the cell cycle. Multiprotein γ-tubulin ring complexes (γ-TuRCs) template microtubule nucleation within cells (Figure 1) [2–5]. The γ-tubulin molecules within this complex are positioned in a single-turn helical pattern [6] and the end-on interactions between γ-tubulin and α/β-tubulin dimers most likely help to support the lateral association between α/β-tubulin dimers during their assembly into protofilaments; this is thought to promote the otherwise kinetically unfavourable formation of a short tubular structure (the microtubule seed), which, once beyond a certain size threshold, can rapidly polymerise into a microtubule filament ([3,7]; Figure 1). γ-TuRCs appear to regulate microtubule polarity, as they are always positioned at the α-tubulin-containing ‘minus end’ with the highly dynamic β-tubulin-exposed plus end extending outwards (Figure 1). Microtubule nucleation must be highly regulated in space and time to ensure the correct formation of microtubule networks [8,9]. Thus, γ-TuRCs are normally activated only once recruited to specific sites within the cell known as microtubule organising centres (MTOCs). How γ-TuRCs are assembled, recruited and activated are still key areas of interest in the field.
Figure 1. Templated microtubule nucleation.
(A) γ-tubulin molecules (yellow) within the γ-TuRC are positioned in a single-turn helix via their binding to GCP proteins (blue). (B) γ-tubulin molecules bind to incoming α/β-tubulin dimers from the cytosol and this is thought to promote the lateral interaction between the α/β-tubulin dimers as they grow into protofilaments (a protofilament is a single end-to-end chain of tubulin dimers). (C) Microtubule assembly progresses slowly through an unstable stage where disassembly is more likely than continued assembly (as indicated by the thickness of the two-way arrows). (D) Assembly is thought to reach a stable stage, where a microtubule seed containing sufficient tubulin dimers has formed (although the size of this stable seed remains unclear). (E) Once the stable seed has formed, microtubule polymerisation is favoured and can progress rapidly. Abbreviation: GCP, γ-tubulin complex protein.
γ-TuRC composition
The γ-tubulin small complex
The core subunit of the γ-TuRC is the γ-Tubulin Small Complex (γ-TuSC), a heterotetramer of ~300 kDa containing two molecules of γ-tubulin and one each of γ-tubulin complex protein 2 (GCP2) and GCP3 [10–14] (Figure 2A). In budding yeast, seven γ-TuSCs form a single-turn helix template with approximately the same pitch and diameter as a microtubule [6]. A similar process likely occurs in higher eukaryotes, but other types of GCP molecules are predicted to replace some of the GCP2/3 molecules within the ring [15] (see below). GCP2 and GCP3 are conserved in all eukaryotes and are essential for cell viability in all organisms in which they have been studied [10,12,16–22]. There is even some degree of functional conservation between species, as the homologues of GCP2 and GCP3 in fission yeast can be replaced to some extent by their human or budding yeast counterparts [23].
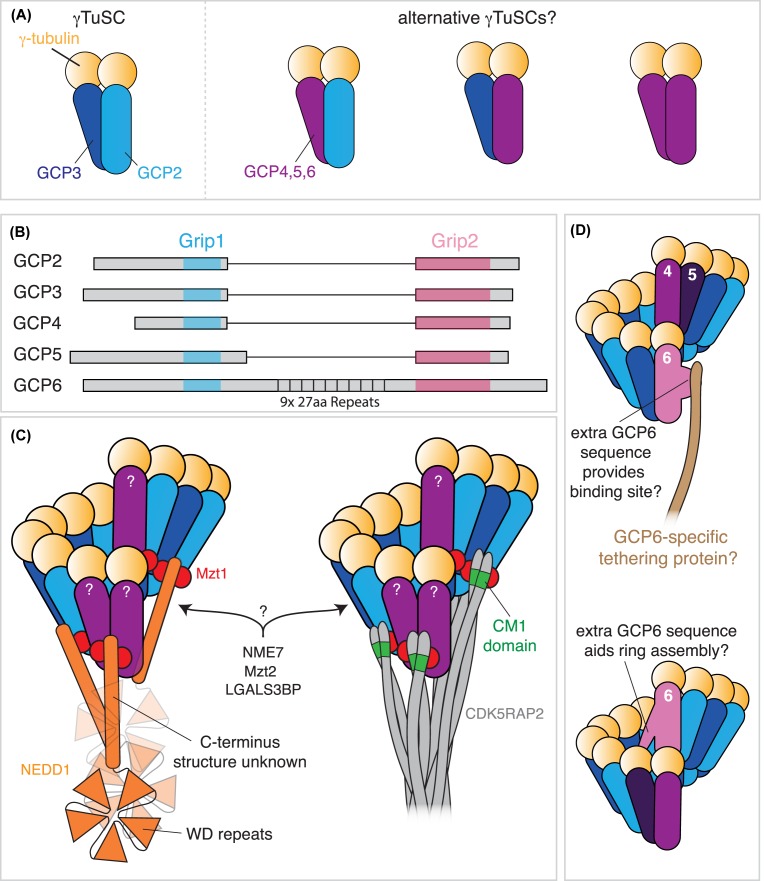
Figure 2. Non-core γ-TuRC components.
(A) The canonical γ-TuSC comprises two molecules of γ-tubulin and one each of GCP2 and GCP3, but alternative γ-TuSCs may exist in which GCP2, GCP3, or both are replaced by either GCP4, 5, or 6. (B) GCP2–6 all contain a Grip1 and a Grip2 domain. The Grip1 domain mediates interactions between GCP proteins, while the Grip2 domain mediates interactions with γ-tubulin. In addition, GCP6 contains an expanded central region that includes nine 27-aa repeats of unknown function. (C) GCP4, 5 and 6 (depicted here in purple) are predicted to replace some of the GCP2 and GCP3 molecules within the ring, but their exact positions remain unknown. MZT1 binds to the N-terminal regions of GCP proteins and acts as an adapter protein for the binding of the tethering protein NEDD1 (left) and tethering proteins that contain an N-terminal CM1-domain, such as CDK5RAP2 (right). NEDD1 contains putative WD40 repeats that form a β-propeller structure known to mediate protein–protein interactions (presumably with proteins at MTOCs). The structure of the C-terminus of NEDD1 is currently unknown but is required for binding to the γ-TuRC. The positions of NME7 kinase, MZT2 and LGALS3BP remain unknown. (D) The function of the extra sequence in GCP6 is unknown, but may provide a binding site for a GCP6-specific tethering protein (top) or may function in ring assembly by forming interactions with other γ-TuRC components (bottom). Abbreviations: CM1, centrosomin motif 1; MZT1, MOZART1; MZT2, MOZART 2.
GCP proteins and the γ-TuRC
Many species possess three additional GCP proteins, GCP4, 5 and 6 (Table 1), which share sequence and structural similarity with GCP2 and GCP3 [5,14,24,25]. GCP proteins have species-specific names, so for simplicity we will use the human nomenclature for all species. The structural similarity of GCP4, 5 and 6 with GCP2/3, together with their low stoichiometry within the γ-TuRC [25–27], suggests that GCP4, 5 and 6 replace some of the GCP2/3 molecules in the γ-TuRC ring ([3,5,24]; Table 1 and Figure 2A). This hypothesis is supported by the co-fractionation of GCP4 and GCP5 with the γ-TuSC [28] and domain swapping and FRET-based interaction experiments [29]. GCP2–6 possess a Grip1 and a Grip2 domain (Figure 2B); the N-terminal Grip1 domain appears to mediate lateral interactions between GCP proteins, while the C-terminal Grip2 domain associates with γ-tubulin [21,22,30,31] and can be swapped between GCP proteins [29]. Although the precise function and position of GCP4, 5 and 6 remain unclear (Figure 2C), their removal in different systems reduces γ-TuRC abundance in cytosolic extracts [19,28,32,33], suggesting a role in γ-TuRC assembly. Charged regions in GCP4 may prevent bidirectional lateral contacts with other GCP proteins, suggesting that GCP4 is involved in ring initiation or termination [15]. Nevertheless, unlike GCP2 and 3, GCP4, 5 and 6 are not essential for viability in Drosophila, fission yeast or Aspergillus [19,32,34,35], showing that proper assembly of γ-TuRCs (at least in the cytosol) is not required for a large proportion of γ-TuRC function. This is most likely because γ-TuRCs can assemble in the absence of GCP4, 5 and 6 at MTOCs (as suggested in [32]), similar to the natural situation in yeast cells (see below). One key function of GCP4, 5 and 6 in higher eukaryotes may, therefore, be to provide greater γ-TuRC diversity in the presence of a wider range of MTOCs (suggested in [5]). For example, GCP6 is involved in the localisation of γ-TuRCs to keratin fibres in epithelial cells [36], and GCP4 and GCP6 are required in both male and female germlines in Drosophila [32,35,37,38]. Very little is known about the additional sequences found in GCP5 and GCP6 (which, in GCP6, contains nine 27-aa repeats) and it is possible that these regions form important interactions either within the γ-TuRC structure or with tethering or modifying proteins at MTOCs (Figure 2D). Elucidating exactly how the extra GCP proteins integrate into γ-TuRCs will shed light on the relevance of these non-essential proteins.
Table 1. Orthologues of proteins involved in microtubule nucleation in selected species.
| Category | Homo sapiens | Drosophila melanogaster | Arabidopsis thaliana | Caenorhabditis elegans | Candida albicans | Aspergillus nidulans | Schizosaccharomyces pombe | Saccharomyces cerevisiae |
|---|---|---|---|---|---|---|---|---|
| γ-TuSC | γ-tubulin | γ-tubulin 23C/37C | TUBG1/2 | Tbg1 | Tub1 | mipA | Tug1/Tubg1 | Tub4 |
| GCP2 | Grip84 | Spc97p/GCP2 | Grip1/Gip1 | Spc97 | GCP2/B | Alp4 | Spc97 | |
| GCP3 | Grip91 | Spc98p/GCP3 | Grip2/Gip2 | Spc98 | GCP3/C | Alp6 | Spc98 | |
| γ-TuRC GCPs | GCP4 | Grip75 | GCP4 | ? | - | GCP4/D | Gfh1 | - |
| GCP5 | Grip128 | GCP5 | ? | - | GCP5/E | Mod21 | - | |
| GCP6 | Grip163 | GCP6 | ? | - | GCP6/F | Alp16 | - | |
| γ-TuRC other | NEDD1/GCP-WD | Grip71 | Nedd1 | ? | - | - | - | - |
| MOZART1 | Mozart1 | GIP1a/b | Mzt1 | Mzt1 | Mzt1/MztA | Mzt1 | - | |
| MOZART2A | - | - | ? | - | - | - | - | |
| MOZART2B | ||||||||
| NME7 | Nmdyn-D7 | ? | - | ? | ? | ? | ? | |
| LGALS3BP | ? | ? | ? | ? | ? | ? | ? | |
| CM1-domain γ-TuRC tethering | CDK5RAP2, Myomegalin, Pericentrin | Cnn, (Plp - no CM1 domain) | ? | ? | Spc110, Spc72 | PcpA, ApsB | Pcp1, Mto1/Mto2 | Spc110, Spc72 |
| γ-TuRC- independent microtubule nucleators | chTog/CKAP5 | Msps | ? | Zyg9 | ? | ? | Alp14 | Stu2 |
| TPX2 | Mei38 | Tpx2 | Tpxl-1 | - | - | - | - |
The table shows orthologues of γ-TuRC proteins, CM1-domain-containing γ-TuRC tethering proteins and γ-TuRC-independent proteins involved in microtubule nucleation across a selection of species, as indicated. ‘-’ refers to cases where attempts in the literature have failed to identify an orthologue and ‘?’ refers to cases where we are unaware of any attempts to identify an orthologue. Abbreviation: CM1, centrosomin motif 1.
Other γ-TuRC proteins
In organisms other than budding yeast, γ-TuRCs contain additional proteins. The first was discovered in Drosophila and named Dgrip71WD [39] (now known simply as Grip71). Despite its name, Grip71 lacks Grip domains and instead contains N-terminal WD40 repeats (predicted to form a β-propeller structure that mediates protein–protein interactions) and a C-terminal domain with unknown structure required for Grip71’s association with the γ-TuRC (Figure 2C) [40–42]. Grip71 and its mammalian homologues (NEDD1, alternatively named as GCP-WD) have been extensively studied [32,39–56], collectively showing that these proteins are dispensable for γ-TuRC assembly and instead function in γ-TuRC recruitment to different MTOCs.
More recently, additional components of the γ-TuRC were identified: MOZART1 (MZT1), MOZART2 (MZT2), LGALS3BP and the kinase NME7 ( [26,27,57–59]; Table 1 and Figure 2C). Of these, MZT1 is the most widely conserved (Table 1) and has been the most extensively studied [28,38,58,60–67]. MZT1 homologues are small (~8.5 kDa) and comprise just three α-helices [65,66]; studies in plants, yeast and cultured human cells have shown that MZT1 interacts directly with the N-terminal region of GCP proteins (i.e. at the base of the γ-TuRC) [28,60–65] and that they regulate the interaction between the γ-TuRC and γ-TuRC-tethering proteins [28,64]. In all three systems, removal or knockdown of MZT1 impairs γ-TuRC recruitment to various MTOCs and cells fail in mitosis [28,58,60–64,66,67]. Surprisingly, however, MZT1 is not essential in Drosophila and is expressed only in the testes, where it is important for γ-TuRC recruitment to basal bodies, but not to mitochondria, in developing sperm cells [38]. It remains to be tested whether MZT1 also defines specific subsets of γ-TuRCs in other multicellular animal systems, and what impact this might have on γ-TuRC recruitment and function. Although MZT1 is clearly involved in γ-TuRC recruitment in cells where it is expressed, there are conflicting reports regarding a possible role for MZT1 in γ-TuRC assembly [28,64].
Less is known about the other newly discovered γ-TuRC proteins. MOZART2A and 2B are found only in deuterostomes; they have no sequence relation to MZT1 but are also small (~16.5 kDa) and are involved in γ-TuRC recruitment, although potentially only during interphase [26]. The kinase NME7 is required for efficient nucleation from centrosomes in human cells and increases the in vitro nucleation activity of purified human γ-TuRCs [68]. This increase, however, is relatively small and so other mechanisms may work synergistically with NME7 phosphorylation. Whether NME7 kinases function at γ-TuRCs in other systems remains to be tested. No study has yet investigated a potential role for LGALS3BP at γ-TuRCs, but its expression levels affect centrosome number and structure [59]. In summary, more work is needed before we have a full understanding of these additional γ-TuRC components.
γ-TuRC assembly, recruitment and activation
Assembly of the γ-TuRC ring
In the past, different models of γ-tubulin complex-mediated microtubule nucleation (template compared with protofilament) had been proposed [69,70]. The consensus now is that the template model is correct and that microtubule nucleation is catalysed after multiple γ-TuSCs assemble, often with other proteins, into ring-like structures. These structures can be regarded as γ-TuRCs, regardless of whether they contain γ-TuRC-specific components. One key question is how γ-TuRCs assemble. This is complex because assembly may occur in a cell- or MTOC-specific manner. In higher eukaryotes, such as humans, Xenopus and Drosophila, γ-TuRCs can assemble in the cytosol before being recruited to an MTOC. This cytosolic assembly depends, to potentially differing degrees, on GCP4, 5 and 6 [28,29,32,33,35] and, in some cell types, on Mzt1 [64]. In fission yeast, despite the presence of a Mzt1 homologue and GCP4, 5 and 6 homologues, γ-TuRCs are largely absent from the cytosol [34] and so presumably form specifically at MTOCs; here, assembly is catalysed by the binding of the Mto1/Mto2 complex (see below) [71]. Similarly, in budding yeast, which contains only γ-tubulin, GCP2 and GCP3, γ-TuRCs are absent from the cytosol and assembly occurs exclusively at Spindle Pole Bodies (SPBs, the yeast equivalent of centrosomes), stimulated by the binding of Spc110 (see below) [6]. Thus, at least two classes of γ-TuRC assembly exist: cytosolic assembly and MTOC-specific assembly, and it remains possible that both classes coexist within the same species or cell type.
Recruitment to MTOCs
γ-TuRCs can be recruited to different MTOCs such as the centrosome, Golgi, nuclear envelope, cell cortex, mitochondria, and even the sides of pre-existing microtubules. This depends on the organism, cell type and cell cycle stage, and needs to be tightly regulated to ensure correct microtubule network formation. Consequently, there are a variety of proteins that can recruit γ-TuRCs to different MTOCs (that we term γ-TuRC-tethering proteins), including yeast Spc110, Spc72, Mto1 and Pcp1, Drosophila Cnn and Grip71 and mammalian CDK5RAP2, Myomegalin, Pericentrin and NEDD1. γ-TuRCs can also be recruited to different MTOCs by the same tethering protein. For example, NEDD1 recruits γ-TuRCs both to centrosomes and to the sides of pre-existing microtubules (via the multiprotein Augmin/HAUS complex) in human cells [40,41,46,72–75], and, while the major isoform of Drosophila Cnn recruits γ-TuRCs to centrosomes in dividing cells [76–80], testes-specific isoforms of Cnn recruit γ-TuRCs to mitochondria in sperm cells [81]. There are also differences between species because the Drosophila homologue of NEDD1, Grip71, is important for γ-TuRC recruitment to spindles (via Augmin) but not to centrosomes [32,47,82]. Most γ-TuRC-tethering proteins (except the Grip71/NEDD1 homologues) contain an N-terminal centrosomin motif 1 (CM1) domain of ~60 amino acids [83,84], which is necessary for binding and recruiting γ-TuRCs to MTOCs [27,64,76,85–87]. Evidence from the structure of Spc110-bound γ-TuRCs suggests the CM1 domain binds directly to the ring of GCP proteins [6], although this remains unproven. The CM1 domain is also necessary for γ-TuRC assembly in yeast [71,87] and for ectopic activation of γ-TuRCs in human and fission yeast cells [27,71]. Thus, the CM1 domain is capable of linking the processes of γ-TuRC recruitment, assembly, and activation.
Given the role of the CM1 domain in γ-TuRC activation (see also below), it seems sensible that CM1-domain proteins bind γ-TuRCs only at MTOCs, which presumably explains why endogenous CM1-domain proteins do not readily co-purify with γ-TuRCs from cytosolic extracts [14,25,39,58]. In contrast, Grip71/NEDD1 homologues do co-purify with γ-TuRCs and so do bind γ-TuRCs in the cytosol [39–41]. The reason for this difference is unclear, especially as Grip71 and NEDD1 are not required for cytosolic γ-TuRC assembly [32,40,41]. Intriguingly, the binding of CM1-domain proteins and Grip71/NEDD1 homologues is mutually exclusive, at least in some cell types [27,49], suggesting that they bind to the γ-TuRC in a similar region. It is therefore possible that Grip71/NEDD1 homologues bound to γ-TuRCs mask the binding of CM1-domain proteins in the cytosol in order to prevent premature γ-TuRC activation. Alternatively, the CM1-domain proteins may be unable to bind γ-TuRCs in the cytosol until they undergo MTOC-specific post-translational modifications. Importantly, both possibilities would help avoid the premature activation of γ-TuRCs. Moreover, it may be important to regulate closely which tethering protein binds the γ-TuRC, as NEDD1-bound γ-TuRCs serve to anchor microtubules while CDK5RAP2-bound γ-TuRCs nucleate microtubules in mouse keratinocytes [49]. It will be important in future to understand more about how the binding of different γ-TuRC-tethering proteins can regulate γ-TuRC recruitment and function.
Regulation by phosphorylation and isoform expression
Many phosphorylation sites have been identified in various γ-TuRC components [36,88–95] and γ-TuRC tethering proteins, including Spc110 [84,91,96–100], NEDD1 [41,46,50–54,94,101], Grip71 [95], Pericentrin [102,103], CDK5RAP2 [94,103], Cnn [95,104–107] and Mto2 [108]. Several of these sites have been characterised and have been shown to play roles in γ-TuRC assembly, recruitment or activation, including at specific MTOCs. For example, phosphorylation of Ser405 in NEDD1 is required for chromosomal microtubule nucleation, but not centrosomal nucleation, in Xenopus egg extracts [50], while other phosphorylation sites in NEDD1 regulate its binding to the γ-TuRC [51,54]. Phosphorylation can also negatively regulate γ-TuRC recruitment and activity, as phosphorylation of GCP6 by CDK1 inhibits its binding to intermediate filaments in cultured human epithelial cells [36], and hyperphosphorylation of Mto2 in fission yeast during mitosis leads to the disassembly of the Mto1/Mto2 complex and subsequent inactivation of γ-TuRCs at non-SPB sites [108]. It is now important to try and understand exactly what effects these, and other, phosphorylation events have on the γ-TuRC to induce recruitment, activity or assembly, for example by inducing structural changes in the γ-TuRC. It will also be important to understand how phosphorylation can finely tune nucleation events in a cell- and MTOC-specific manner.
As mentioned above, the γ-TuRC-tethering proteins that contain a CM1 domain typically associate with γ-TuRCs only at MTOCs, and phosphorylation is a plausible mechanism to control this. For example, phosphorylation of Spc110 regulates its binding to γ-TuRCs to some degree [84] (although Spc110 oligomerisation is clearly also important [87]). Whether phosphorylation regulates the binding of other CM1-domain proteins remains unclear, but there is circumstantial evidence that this occurs in Drosophila. Drosophila Cnn is phosphorylated specifically at centrosomes [104] where it is important for γ-TuRC recruitment [76–80], and unphosphorylated Cnn does not associate with γ-TuRCs in in vitro binding assays [38]. How phosphorylation might regulate binding is unclear; it has been suggested that the extreme N-terminal region of Cnn folds back and inhibits the CM1 domain, as this N-terminal region is absent from testes-specific isoforms of Cnn that are able to bind γ-TuRCs in vitro [38]. Thus, the addition of negatively charged phosphate groups may help expose the CM1 domain to allow γ-TuRC binding. A similar region in yeast Spc110 could also be regulatory, as it is absent from the structure of Spc110-bound γ-TuRCs [6] and it is not essential for γ-TuRC binding [87]. This suggests that similar regulatory regions may exist in other CM1-domain proteins, but more work is needed to test these models and reveal any conservation across species. Given the recent findings that different Cnn isoforms bind differently to γ-TuRCs [38] and recruit γ-TuRCs to different MTOCs [81], it will be important to explore potential isoform differences in other γ-TuRC-tethering proteins and in Cnn homologues in different species, particularly as different CDK5RAP2 isoforms do exist [109–111]. It is likely that a combination of phosphorylation and isoform differences contributes to the tight spatiotemporal regulation of γ-TuRC recruitment and activation.
Activation of γ-TuRCs
The assembly of the γ-TuRC alone is thought to be insufficient for it to promote microtubule nucleation; it must also be activated. Purified γ-TuRCs have relatively low nucleating activity, but this increases when they are mixed with fragments of tethering proteins that contain the CM1 domain [27]. When these truncated fragments are expressed in cells, ectopic microtubules are nucleated throughout the cytoplasm [27]. How the binding of CM1-domain proteins induces γ-TuRC activity is not known, but the structure of the budding yeast γ-TuRC suggests that a flexible linker region in GCP3 must move in order to position the γ-tubulin molecules correctly for microtubule assembly [6]. This structure, however, was generated from Spc110-bound γ-TuRCs, suggesting that binding of a CM1-domain protein is not sufficient to move the GCP3 linker, at least in budding yeast. Moreover, artificially inducing structural rearrangements that better positioned the γ-tubulin molecules resulted in only approximately two-fold enhancement of nucleation activity [112]. Thus, other mechanisms must exist, one of which is phosphorylation. Mimicking phosphorylation of Saccharomyces cerevisiae Spc110 increases nucleation activity approximately three-fold [84], and NME7 kinase increases the nucleation activity of human γ-TuRCs by approximately 2.5-fold [68]. Whether these phosphorylation events lead to a conformational change in the γ-TuRC, or bring about some other process, is not known. In summary, it is likely that, in vivo, a combination of allosteric regulation by CM1-domain proteins and phosphorylation helps to activate γ-TuRCs. It is possible that different activation mechanisms function in different cells and at different MTOCs and how this is regulated remains an exciting area for future research.
γ-TuRC heterogeneity
All eukaryotes express the core γ-TuRC components γ-tubulin, GCP2 and GCP3, but not necessarily all of the additional components. For example, the Candida albicans genome contains only MZT1 in addition to the core components [64], while Drosophila additionally contains GCP4, GCP5, GCP6, NEDD1/Grip71, MZT1 and NME7 [5]. The classical view is that all γ-TuRCs within the same organism have the same protein composition, but this has now been disproved. The first evidence came from studies showing that not all mammalian γ-TuRCs contain NEDD1 [27,62]. This led to the suggestion that subpopulations of γ-TuRCs may exist [5]. It was then shown that NEDD1 and CDK5RAP2 bound mutually exclusively to γ-TuRCs in mouse keratinocytes and that NEDD1-bound γ-TuRCs functioned to anchor microtubules while CDK5RAP2-bound γ-TuRCs nucleated microtubules (Figure 3A) [49]. Most recently, it was shown that Drosophila MZT1 is expressed only in the testes and is present in, and required for, the γ-TuRCs that are recruited to basal bodies, but not mitochondria, in sperm cells (Figure 3B) [38]. The differential association of Mzt1 with only a subset of γ-TuRCs may also occur in Arabidopsis thaliana [62], although this analysis is complicated by the presence of two MZT1 genes in plants. Collectively, these studies have now shown beyond doubt that γ-TuRC composition can vary between tissues and even between MTOCs within the same cell, and that this can influence γ-TuRC recruitment and function.
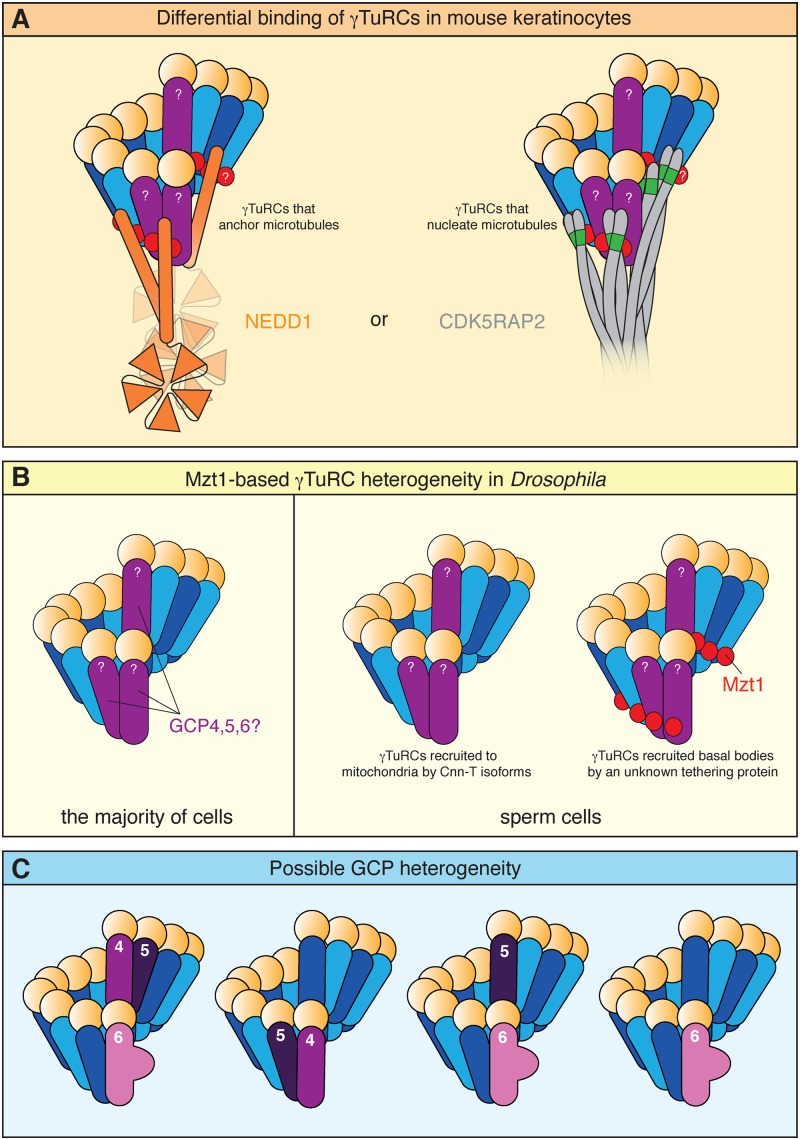
Figure 3. Known and potential forms of γ-TuRC heterogeneity.
(A) In mouse keratinocytes, γ-TuRCs are bound mutually exclusively by either NEDD1 or CDK5RAP2, suggesting that both tethering proteins bind to a similar region of the γ-TuRC. NEDD1-bound γ-TuRCs serve to anchor microtubules while CDK5RAP2-bound γ-TuRCs nucleate microtubules. The position of GCP4, 5 and 6 within γ-TuRCs in these cells remains unknown. (B) In Drosophila, most γ-TuRCs do not contain MZT1 (red), which is expressed only in the testes. Within the testes, MZT1 is predominantly expressed in sperm cells but in early elongating sperm is only present in γ-TuRCs that are recruited to the basal body, and is not present in γ-TuRCs recruited to mitochondria. The position of GCP4, 5 and 6 within Drosophila γ-TuRCs remains unknown. (C) While the position of GCP4, 5 and 6 within γ-TuRCs remains unknown, positive FRET data in HeLa cells suggest that GCP4 and GCP5 are adjacent to each other within the ring, while negative FRET data suggest GCP6 is not adjacent to either GCP4 or GCP5 (left) [29]. Stoichiometry measurements from HEK293T cells and immunoprecipitation experiments from HeLa cells suggest that some complexes do not contain GCP6 (middle left) [27] and that some complexes do not contain GCP4 (middle right) [58] respectively. GCP6 can still associate with γ-TuRCs in the absence of GCP4 or GCP5, suggesting that some complexes can form with only GCP6 (right) [28].
What about other γ-TuRC components? Current data does not rule out that they could also confer γ-TuRC heterogeneity (Figure 3C). This can be difficult to test, as experiments typically assay the whole population of γ-TuRCs. For example, while GCP4, 5 or 6 can co-immunoprecipitate with each other in all pairwise combinations from human cell extracts [25], it remains possible that different subsets coexist, e.g. GCP4/5-only complexes, GCP5/6-only complexes and GCP4/6-only complexes. Recent FRET and cross-linking experiments in HeLa cells detected direct interactions between GCP4 and GCP5, suggesting these proteins are adjacent within γ-TuRCs, but the data cannot rule out that these interactions took place in only a subset of complexes [29]. Indeed, a comparison of protein levels after immunoprecipitating GCP6 or γ-tubulin from HeLa cells shows that similar amounts of GCP5 are co-immunoprecipitated but that much less GCP4 is co-immunoprecipitated with GCP6 [58], suggesting that some complexes contain GCP5 and GCP6 but not GCP4. Moreover, GCP6 can localise to SPBs in the absence of GCP4 and GCP5 in both fission yeast [67] and Aspergillus nidulans [19] and GCP6-containing γ-TuRCs can be detected in extracts that have been depleted of GCP4 or GCP5 [28], suggesting that GCP6-only complexes can exist. Most data highlight GCP6 as being more important for γ-TuRC assembly than GCP4 or GCP5 [19,28,67], but γ-TuRCs purified from human embryonic kidney cells using CDK5RAP2 fragments contain sub-stoichiometric levels of GCP6, i.e. <1 molecule per γ-TuRC [27], suggesting that not all γ-TuRCs contain GCP6. This may reflect differences in the composition of γ-TuRCs between cell types or between γ-TuRCs bound by different tethering proteins or may simply reflect the difficulty of measuring the stoichiometry of the γ-TuRC components. Clearly more work is required to see whether other types of γ-TuRC heterogeneity really do exist and what functional relevance this might have.
Other microtubule nucleation factors
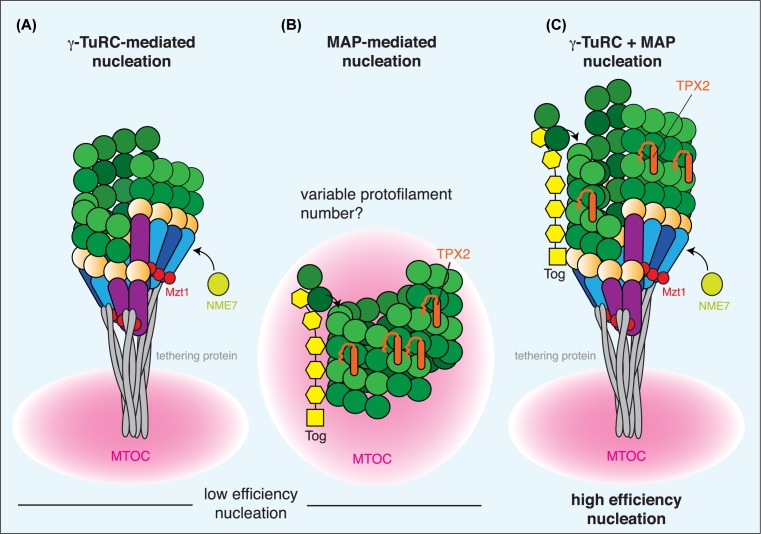
It is now becoming clear that two types of non-γ-TuRC proteins can promote microtubule nucleation: Tog domain proteins, such as XMAP215, and homologues of TPX2 [113–117]. Tog domain proteins are microtubule polymerases that regulate microtubule growth by promoting the longitudinal addition of tubulin dimers via interactions between tubulin and the Tog domains, and it is now thought that this also occurs during the early stages of microtubule nucleation [113–118]. Strong evidence suggests that this involves interactions between Tog domain proteins and γ-TuRCs, as budding yeast Stu2 forms a complex with Spc72-bound γ-TuRCs [119], fission yeast Alp14 co-immunoprecipitates with γ-TuRC components [116], and the C-terminal domain of XMAP215 binds γ-tubulin [117]. An elegant model has been proposed [118] in which the γ-TuRC mediates the lateral interactions between tubulin dimers and sets the 13-protofilament lattice structure, while Tog domain proteins, bound to the γ-TuRC via their C-terminal domain, promote the longitudinal addition of tubulin dimers. TPX2 homologues, however, are also important for microtubule nucleation. It was shown that Xenopus TPX2 promotes the phosphorylation of NEDD1 by Aurora A to stimulate microtubule nucleation [101], but it has recently emerged that TPX2 homologues also help prevent catastrophe of nascent microtubule seeds [113–115,120]. Recent structural data show that TPX2 proteins bind across longitudinal and lateral tubulin interfaces within the microtubule lattice [121]. Thus, microtubule nucleation is likely most robust in the presence of the γ-TuRC, a Tog domain protein and a TPX2 homologue (Figure 4).
Figure 4. Different modes of microtubule nucleation.
(A) The γ-TuRC templates the addition of tubulin dimers to form a microtubule, but is predicted to promote only lateral and not longitudinal interactions between tubulin dimers. (B) Under certain conditions, the combination of tubulin dimers and microtubule-associated proteins (MAPs) is sufficient to promote microtubule nucleation. Tog domain proteins help polymerise the microtubule by promoting the longitudinal addition of tubulin dimers. TPX2 homologues bind across tubulin dimers within the lattice and help prevent catastrophe of the nascent microtubule seed. (C) In vivo, it is likely that a combination of a γ-TuRC and these MAPs drive highly efficient microtubule nucleation. This presumably occurs at centrosomes, where all of these proteins concentrate, but whether other MTOCs (that are less-efficient microtubule nucleators) use specific mechanisms remains unknown.
While XMAP215 and TPX2 homologues appear to work synergistically with a template to promote microtubule nucleation [113,114,116,117], there is strong evidence that microtubules can be nucleated both in vitro and in vivo in the absence of γ-TuRCs [113,115,122–127]. Consistent with this, in vitro studies have shown that XMAP215 and TPX2 homologues are sufficient for microtubule nucleation at relatively low tubulin concentrations [113,115], and the Tog domain protein Stu2, but not the γ-TuRC, is required for kinetochore-driven microtubule formation in budding yeast [127]. In contrast, however, XMAP215-mediated nucleation is inefficient in the absence of γ-TuRCs in Xenopus egg extracts [117], and Zyg9 and Msps are not required for γ-TuRC-independent microtubule nucleation from centrosomes in Caenorhabditis elegans embryos [124] or from acentrosomal sites in cultured Drosophila cells [126] respectively. Thus, mechanisms of microtubule nucleation may vary and might be related to the particular cell type or MTOC; it is also possible that other regulators of microtubule nucleation remain to be discovered.
Given that at least some microtubule populations can be nucleated independently of γ-TuRCs, it is worth contemplating why cells need γ-TuRCs at all. We believe there are at least three reasons: firstly, templated microtubule nucleation is more efficient [13,27,49,69,84,87,112–114], presumably as it promotes the lateral interactions between tubulin dimers. Secondly, γ-TuRCs can define the 13 protofilament arrangement found in most cell types, which may (or may not) allow molecular motors a more efficient route along straight protofilaments [128]. Thirdly, γ-TuRCs can help regulate microtubule polarity by defining the position of the minus end. Collectively, these functions presumably explain why γ-TuRCs are essential for cell and organism viability [10,12,16–22].
The γ-TuRC as a drug target
γ-TuRCs may offer good anticancer targets given their roles during cell division. Currently, some of the most common and most effective chemotherapy agents, such as Taxanes and Vinca Alkaloids, bind microtubules directly. These compounds, however, often lead to a condition known as chemotherapy-induced peripheral neuropathy (CIPN) [129,130]. CIPN presents as numbness, pain, tingling, and heightened sensitivity in the extremities; it limits drug dosage and/or duration and can persist after chemotherapy, and it is a major cause of cancer survivor disability [129,130]. The cellular mechanism by which CIPN occurs is not fully understood, but dying back of axonal projections in the epidermis has been observed in patients and in model systems and this could be caused by axonal transport defects [131–133]; however, alternative mechanisms have been suggested, including mitotoxicity and disruption to calcium homoeostasis [134]. Targeting γ-TuRCs, instead of microtubules directly, may offer a viable alternative [44,135,136] because inhibiting γ-TuRCs would perturb cancer cells [44,135] but may not have a dramatic effect on mature neurons, which would already have generated and stabilised their microtubule networks. For example, axonal transport along stable pre-existing microtubules in neurons might remain unperturbed after γ-TuRC inhibition. Moreover, microtubule severing in neurons may be able to compensate for any reduction in microtubule generation via the γ-TuRC pathway. That said, there is evidence that γ-TuRCs bind to the sides of pre-existing microtubules and regulate microtubule dynamics [125,137], and so it will first be important to assess the role of γ-TuRCs in neurons. The challenge will then be to develop drugs that can inhibit γ-TuRC function in a highly specific manner. Currently, the only γ-TuRC-inhibiting drug is Gatastatin, which was identified by testing derivatives of drugs known to bind α/β-tubulin and was found to bind γ-tubulin with a 12-fold greater affinity than α/β-tubulin [138]. It may also be important, however, to consider γ-TuRC heterogeneity, as this may help increase specificity. Thus, the non-core γ-TuRC components may provide good targets for anticancer drugs, as their inhibition may affect only subsets of γ-TuRCs. Of course, we first need to understand more about γ-TuRC heterogeneity and the role of each γ-TuRC component within the complex.
Conclusion and perspectives
The combinatorial complexity created by the variety of γ-TuRC components and their various tethering proteins is likely to grant cells the ability to regulate very precisely the assembly, recruitment and activity of their γ-TuRCs. The γ-TuRC has been studied for decades and many key insights have been made. One of the most important developments since the discovery of γ-tubulin [139,140] was the determination of the yeast γ-TuRC structure at near atomic resolution [6]. This finally proved the template model, helped reveal a potentially key step in γ-TuRC activation [112] and, combined with the crystal structure of human GCP4 [15], has changed the way we think about the position and function of the GCP proteins [29]. Clearly, a similar structure of a γ-TuRC from a higher eukaryote would be extremely informative. The more we understand, however, the more questions arise. For example, what causes the flexible region in GCP3 to move in order to position the γ-tubulin molecules correctly? Do all GCP proteins fit into the same γ-TuRC ring? If so, how? If not, how do different GCP proteins affect γ-TuRC behaviour? How do phosphorylation events induce functional changes in the γ-TuRC? How is the binding between γ-TuRCs and γ-TuRC-tethering proteins regulated in a multi-MTOC, multicellular context? These are all exciting questions for the future.
Summary
Structural data from yeast has established that the template model for γ-TuRC-mediated microtubule nucleation is correct. In budding yeast, γ-TuRCs comprise a single-turn helical ring of seven γ-TuSCs. In higher eukaryotes, it is likely that GCP4, 5 and 6 replace some of the GCP2/3 molecules within the helical ring. How this occurs, and the precise function of GCP4, 5 and 6, remains unclear.
Several γ-TuRC components have been discovered only recently. Of these, MOZART1 is the most conserved through evolution and is the best studied, functioning in γ-TuRC recruitment (and possibly γ-TuRC assembly) in several systems.
γ-TuRCs are recruited to various microtubule organising centres (MTOCs) in cells via γ-TuRC-tethering proteins that normally contain an N-terminal CM1 domain. Binding of these CM1 domain proteins to γ-TuRCs is important for γ-TuRC recruitment, but can also influence γ-TuRC assembly and activation.
The composition of γ-TuRCs varies between species, but can also vary within the same species and even within the same cell. More work is needed to understand the extent of this heterogeneity and its functional relevance.
There is an emerging role for non-γ-TuRC proteins in microtubule nucleation. Several recent studies have shown that chTOG domain proteins and TPX2 homologues work synergistically with γ-TuRCs (or artificial templates) for efficient microtubule nucleation.
γ-TuRCs have been identified as potential anti-cancer targets and the first γ-tubulin inhibitor, gatastatin, has recently been developed. γ-TuRC-inhibiting drugs could in theory lead to a reduction in the occurrence of chemotherapy-induced peripheral neuropathy (CIPN), although this remains to be explored.
Abbreviations
- CIPN
chemotherapy-induced peripheral neuropathy
- CM1
centrosomin motif 1
- GCP
γ-tubulin complex protein
- MTOC
microtubule organising centre
- MZT1
MOZART1
- MZT2
MOZART2
- SPB
spindle pole body
- γ-TuRC
γ-tubulin ring complex
- γ-TuSC
γ-tubulin small complex
Competing interests
The authors declare that there are no competing interests associated with the manuscript.
References
- 1.Goodson H.V. and Jonasson E.M. (2018) Microtubules and microtubule-associated proteins. Cold Spring Harb. Perspect. Biol. 10, a022608 10.1101/cshperspect.a022608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sulimenko V., Hájková Z., Klebanovych A. and Dráber P. (2017) Regulation of microtubule nucleation mediated by γ-tubulin complexes. Protoplasma 254, 1–13 10.1007/s00709-016-1070-z [DOI] [PubMed] [Google Scholar]
- 3.Kollman J.M., Merdes A., Mourey L. and Agard D.A. (2011) Microtubule nucleation by γ-tubulin complexes. Nat. Rev. Mol. Cell Biol. 12, 709–721 10.1038/nrm3209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Farache D., Emorine L., Haren L. and Merdes A. (2018) Assembly and regulation of γ-tubulin complexes. Open Biol. 8, 170266 10.1098/rsob.170266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Teixidó-Travesa N., Roig J. and Lüders J. (2012) The where, when and how of microtubule nucleation - one ring to rule them all. J. Cell Sci. 125, 4445–4456 10.1242/jcs.106971 [DOI] [PubMed] [Google Scholar]
- 6.Kollman J.M., Polka J.K., Zelter A., Davis T.N. and Agard D.A. (2010) Microtubule nucleating gamma-TuSC assembles structures with 13-fold microtubule-like symmetry. Nature 466, 879–882 10.1038/nature09207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roostalu J. and Surrey T. (2017) Microtubule nucleation: beyond the template. Nat. Rev. Mol. Cell Biol. 91, 321. [DOI] [PubMed] [Google Scholar]
- 8.Sanchez A.D. and Feldman J.L. (2016) Microtubule-organizing centers: from the centrosome to non-centrosomal sites. Curr. Opin. Cell Biol. 44, 93–101 10.1016/j.ceb.2016.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petry S. and Vale R.D. (2015) Microtubule nucleation at the centrosome and beyond. Nat. Cell Biol. 17, 1089–1093 10.1038/ncb3220 [DOI] [PubMed] [Google Scholar]
- 10.Knop M. and Schiebel E. (1997) Spc98p and Spc97p of the yeast gamma-tubulin complex mediate binding to the spindle pole body via their interaction with Spc110p. EMBO J. 16, 6985–6995 10.1093/emboj/16.23.6985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Murphy S.M., Urbani L. and Stearns T. (1998) The mammalian gamma-tubulin complex contains homologues of the yeast spindle pole body components spc97p and spc98p. J. Cell Biol. 141, 663–674 10.1083/jcb.141.3.663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vardy L. and Toda T. (2000) The fission yeast gamma-tubulin complex is required in G(1) phase and is a component of the spindle assembly checkpoint. EMBO J. 19, 6098–6111 10.1093/emboj/19.22.6098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oegema K., Wiese C., Martin O.C., Milligan R.A., Iwamatsu A., Mitchison T.J.. et al. (1999) Characterization of two related Drosophila gamma-tubulin complexes that differ in their ability to nucleate microtubules. J. Cell Biol. 144, 721–733 10.1083/jcb.144.4.721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gunawardane R.N., Martin O.C., Cao K., Zhang L., Dej K., Iwamatsu A.. et al. (2000) Characterization and reconstitution of Drosophila gamma-tubulin ring complex subunits. J. Cell Biol. 151, 1513–1524 10.1083/jcb.151.7.1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guillet V., Knibiehler M., Gregory-Pauron L., Remy M.-H., Chemin C., Raynaud-Messina B.. et al. (2011) Crystal structure of γ-tubulin complex protein GCP4 provides insight into microtubule nucleation. Nat. Struct. Mol. Biol. 18, 915–919 10.1038/nsmb.2083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martin O.C., Gunawardane R.N., Iwamatsu A. and Zheng Y. (1998) Xgrip109: a gamma tubulin-associated protein with an essential role in gamma tubulin ring complex (gammaTuRC) assembly and centrosome function. J. Cell Biol. 141, 675–687 10.1083/jcb.141.3.675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu J. and Lessman C.A. (2007) Soluble tubulin complexes, gamma-tubulin, and their changing distribution in the zebrafish (Danio rerio) ovary, oocyte and embryo. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 147, 56–73 10.1016/j.cbpb.2006.12.014 [DOI] [PubMed] [Google Scholar]
- 18.Seltzer V., Janski N., Canaday J., Herzog E., Erhardt M., Evrard J.-L.. et al. (2007) Arabidopsis GCP2 and GCP3 are part of a soluble gamma-tubulin complex and have nuclear envelope targeting domains. Plant J. 52, 322–331 10.1111/j.1365-313X.2007.03240.x [DOI] [PubMed] [Google Scholar]
- 19.Xiong Y. and Oakley B.R. (2009) In vivo analysis of the functions of gamma-tubulin-complex proteins. J. Cell Sci. 122, 4218–4227 10.1242/jcs.059196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Colombié N., Vérollet C., Sampaio P., Moisand A., Sunkel C., Bourbon H.-M.. et al. (2006) The Drosophila gamma-tubulin small complex subunit Dgrip84 is required for structural and functional integrity of the spindle apparatus. Mol. Biol. Cell 17, 272–282 10.1091/mbc.e05-08-0722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knop M., Pereira G., Geissler S., Grein K. and Schiebel E. (1997) The spindle pole body component Spc97p interacts with the gamma-tubulin of Saccharomyces cerevisiae and functions in microtubule organization and spindle pole body duplication. EMBO J. 16, 1550–1564 10.1093/emboj/16.7.1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geissler S., Pereira G., Spang A., Knop M., Souès S., Kilmartin J.. et al. (1996) The spindle pole body component Spc98p interacts with the gamma-tubulin-like Tub4p of Saccharomyces cerevisiae at the sites of microtubule attachment. EMBO J. 15, 3899–3911 10.1002/j.1460-2075.1996.tb00764.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Riehlman T.D., Olmsted Z.T., Branca C.N., Winnie A.M., Seo L., Cruz L.O.. et al. (2013) Functional replacement of fission yeast γ-tubulin small complex proteins Alp4 and Alp6 by human GCP2 and GCP3. J. Cell Sci. 126, 4406–4413 10.1242/jcs.128173 [DOI] [PubMed] [Google Scholar]
- 24.Guillet V., Knibiehler M., Gregory-Pauron L., Remy M.-H., Chemin C., Raynaud-Messina B.. et al. (2011) Crystal structure of γ-tubulin complex protein GCP4 provides insight into microtubule nucleation. Nat. Struct. Mol. Biol. 18, 915–919 10.1038/nsmb.2083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murphy S.M., Preble A.M., Patel U.K., O’Connell K.L., Dias D.P., Moritz M.. et al. (2001) GCP5 and GCP6: two new members of the human gamma-tubulin complex. Mol. Biol. Cell 12, 3340–3352 10.1091/mbc.12.11.3340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Teixidó-Travesa N., Villén J., Lacasa C., Bertran M.T., Archinti M., Gygi S.P.. et al. (2010) The gammaTuRC revisited: a comparative analysis of interphase and mitotic human gammaTuRC redefines the set of core components and identifies the novel subunit GCP8. Mol. Biol. Cell 21, 3963–3972 10.1091/mbc.e10-05-0408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choi Y.-K., Liu P., Sze S.K., Dai C. and Qi R.Z. (2010) CDK5RAP2 stimulates microtubule nucleation by the gamma-tubulin ring complex. J. Cell Biol. 191, 1089–1095 10.1083/jcb.201007030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cota R.R., Teixidó-Travesa N., Ezquerra A., Eibes S., Lacasa C., Roig J.. et al. (2017) MZT1 regulates microtubule nucleation by linking γTuRC assembly to adapter-mediated targeting and activation. J. Cell Sci. 130, 406–419 10.1242/jcs.195321 [DOI] [PubMed] [Google Scholar]
- 29.Farache D., Jauneau A., Chemin C., Chartrain M., Remy M.-H., Merdes A.. et al. (2016) Functional analysis of gamma-tubulin complex proteins indicates specific lateral association via their N-terminal domains. J. Biol. Chem. 291, 23112–23125, 10.1074/jbc.M116.744862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kollman J.M., Zelter A., Muller EGD, Fox B., Rice L.M., Davis T.N.. et al. (2008) The structure of the gamma-tubulin small complex: implications of its architecture and flexibility for microtubule nucleation. Mol. Biol. Cell 19, 207–215 10.1091/mbc.e07-09-0879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nguyen T., Vinh D.B., Crawford D.K. and Davis T.N. (1998) A genetic analysis of interactions with Spc110p reveals distinct functions of Spc97p and Spc98p, components of the yeast gamma-tubulin complex. Mol. Biol. Cell 9, 2201–2216 10.1091/mbc.9.8.2201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vérollet C., Colombié N., Daubon T., Bourbon H.-M., Wright M. and Raynaud-Messina B. (2006) Drosophila melanogaster gamma-TuRC is dispensable for targeting gamma-tubulin to the centrosome and microtubule nucleation. J. Cell Biol. 172, 517–528 10.1083/jcb.200511071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang L., Keating T.J., Wilde A., Borisy G.G. and Zheng Y. (2000) The role of Xgrip210 in gamma-tubulin ring complex assembly and centrosome recruitment. J. Cell Biol. 151, 1525–1536 10.1083/jcb.151.7.1525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anders A., Lourenço P.C.C. and Sawin K.E. (2006) Noncore components of the fission yeast gamma-tubulin complex. Mol. Biol. Cell 17, 5075–5093 10.1091/mbc.e05-11-1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vogt N., Koch I., Schwarz H., Schnorrer F. and Nüsslein-Volhard C. (2006) The gammaTuRC components Grip75 and Grip128 have an essential microtubule-anchoring function in the Drosophila germline. Development 133, 3963–3972 10.1242/dev.02570 [DOI] [PubMed] [Google Scholar]
- 36.Oriolo A.S., Wald F.A., Canessa G. and Salas P.J.I. (2007) GCP6 binds to intermediate filaments: a novel function of keratins in the organization of microtubules in epithelial cells. Mol. Biol. Cell 18, 781–794 10.1091/mbc.e06-03-0201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schnorrer F., Luschnig S., Koch I. and Nüsslein-Volhard C. (2002) Gamma-tubulin37C and gamma-tubulin ring complex protein 75 are essential for bicoid RNA localization during drosophila oogenesis. Dev. Cell 3, 685–696 10.1016/S1534-5807(02)00301-5 [DOI] [PubMed] [Google Scholar]
- 38.Tovey C.A., Tubman C.E., Hamrud E., Zhu Z., Dyas A.E., Butterfield A.N.. et al. (2018) γ-TuRC heterogeneity revealed by analysis of Mozart1. Curr. Biol., 28, 2314–2323 10.1016/j.cub.2018.05.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gunawardane R.N., Martin O.C. and Zheng Y. (2003) Characterization of a new gammaTuRC subunit with WD repeats. Mol. Biol. Cell 14, 1017–1026 10.1091/mbc.e02-01-0034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Haren L., Remy M.-H., Bazin I., Callebaut I., Wright M. and Merdes A. (2006) NEDD1-dependent recruitment of the gamma-tubulin ring complex to the centrosome is necessary for centriole duplication and spindle assembly. J. Cell Biol. 172, 505–515 10.1083/jcb.200510028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lüders J., Patel U.K. and Stearns T. (2006) GCP-WD is a gamma-tubulin targeting factor required for centrosomal and chromatin-mediated microtubule nucleation. Nat. Cell Biol. 8, 137–147 10.1038/ncb1349 [DOI] [PubMed] [Google Scholar]
- 42.Manning J.A., Shalini S., Risk J.M., Day C.L. and Kumar S. (2010) A direct interaction with NEDD1 regulates gamma-tubulin recruitment to the centrosome. PLoS ONE 5, e9618 10.1371/journal.pone.0009618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu L. and Wiese C. (2008) Xenopus NEDD1 is required for microtubule organization in Xenopus egg extracts. J. Cell Sci. 121, 578–589 10.1242/jcs.018937 [DOI] [PubMed] [Google Scholar]
- 44.Tillement V., Haren L., Roullet N., Etievant C. and Merdes A. (2009) The centrosome protein NEDD1 as a potential pharmacological target to induce cell cycle arrest. Mol. Cancer 8, 10 10.1186/1476-4598-8-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Manning J.A., Lewis M., Koblar S.A. and Kumar S. (2010) An essential function for the centrosomal protein NEDD1 in zebrafish development. Cell Death Differ. 17, 1302–1314 10.1038/cdd.2010.12 [DOI] [PubMed] [Google Scholar]
- 46.Johmura Y., Soung N.-K., Park J.-E., Yu L.-R., Zhou M., Bang J.K.. et al. (2011) Regulation of microtubule-based microtubule nucleation by mammalian polo-like kinase 1. Proc. Natl Acad. Sci. U.S.A. 108, 11446–11451 10.1073/pnas.1106223108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reschen R.F., Colombié N., Wheatley L., Dobbelaere J., St Johnston D., Ohkura H.. et al. (2012) Dgp71WD is required for the assembly of the acentrosomal Meiosis I spindle, and is not a general targeting factor for the γ-TuRC. Biol. Open 1, 422–429 10.1242/bio.2012596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walia A., Nakamura M., Moss D., Kirik V., Hashimoto T. and Ehrhardt D.W. (2014) GCP-WD mediates γ-TuRC recruitment and the geometry of microtubule nucleation in interphase arrays of Arabidopsis. Curr. Biol. 24, 2548–2555 10.1016/j.cub.2014.09.013 [DOI] [PubMed] [Google Scholar]
- 49.Muroyama A., Seldin L. and Lechler T. (2016) Divergent regulation of functionally distinct γ-tubulin complexes during differentiation. J. Cell Biol. 213, 679–692 10.1083/jcb.201601099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pinyol R., Scrofani J. and Vernos I. (2013) The role of NEDD1 phosphorylation by Aurora A in chromosomal microtubule nucleation and spindle function. Curr. Biol. 23, 143–149 10.1016/j.cub.2012.11.046 [DOI] [PubMed] [Google Scholar]
- 51.Gomez-Ferreria M.A., Bashkurov M., Helbig A.O., Larsen B., Pawson T., Gingras A.-C.. et al. (2012) Novel NEDD1 phosphorylation sites regulate γ-tubulin binding and mitotic spindle assembly. J. Cell Sci. 125, 3745–3751 10.1242/jcs.105130 [DOI] [PubMed] [Google Scholar]
- 52.Haren L., Stearns T. and Lüders J. (2009) Plk1-dependent recruitment of gamma-tubulin complexes to mitotic centrosomes involves multiple PCM components. PLoS ONE 4, e5976 10.1371/journal.pone.0005976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sdelci S., Schütz M., Pinyol R., Bertran M.T., Regué L., Caelles C.. et al. (2012) Nek9 phosphorylation of NEDD1/GCP-WD contributes to Plk1 control of γ-tubulin recruitment to the mitotic centrosome. Curr. Biol. 22, 1516–1523 10.1016/j.cub.2012.06.027 [DOI] [PubMed] [Google Scholar]
- 54.Zhang X., Chen Q., Feng J., Hou J., Yang F., Liu J.. et al. (2009) Sequential phosphorylation of Nedd1 by Cdk1 and Plk1 is required for targeting of the gammaTuRC to the centrosome. J. Cell Sci. 122, 2240–2251 10.1242/jcs.042747 [DOI] [PubMed] [Google Scholar]
- 55.Zeng C.J.T., Lee Y.-R.J. and Liu B. (2009) The WD40 repeat protein NEDD1 functions in microtubule organization during cell division in Arabidopsis thaliana. Plant Cell 21, 1129–1140 10.1105/tpc.109.065953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ma W., Baumann C. and Viveiros M.M. (2010) NEDD1 is crucial for meiotic spindle stability and accurate chromosome segregation in mammalian oocytes. Dev. Biol. 339, 439–450 10.1016/j.ydbio.2010.01.009 [DOI] [PubMed] [Google Scholar]
- 57.Janski N., Herzog E. and Schmit A.-C. (2008) Identification of a novel small Arabidopsis protein interacting with gamma-tubulin complex protein 3. Cell Biol. Int. 32, 546–548 10.1016/j.cellbi.2007.11.006 [DOI] [PubMed] [Google Scholar]
- 58.Hutchins J.R.A., Toyoda Y., Hegemann B., Poser I., Hériché J.-K., Sykora M.M.. et al. (2010) Systematic analysis of human protein complexes identifies chromosome segregation proteins. Science 328, 593–599 10.1126/science.1181348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fogeron M.-L., Müller H., Schade S., Dreher F., Lehmann V., Kühnel A.. et al. (2013) LGALS3BP regulates centriole biogenesis and centrosome hypertrophy in cancer cells. Nat. Commun. 4, 1531 10.1038/ncomms2517 [DOI] [PubMed] [Google Scholar]
- 60.Janski N., Masoud K., Batzenschlager M., Herzog E., Evrard J.-L., Houlné G.. et al. (2012) The GCP3-interacting proteins GIP1 and GIP2 are required for γ-tubulin complex protein localization, spindle integrity, and chromosomal stability. Plant Cell 24, 1171–1187 10.1105/tpc.111.094904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Masuda H., Mori R., Yukawa M. and Toda T. (2013) Fission yeast MOZART1/Mzt1 is an essential γ-tubulin complex component required for complex recruitment to the MTOC, but not its assembly. Mol. Biol. Cell 24, 2894–2906 10.1091/mbc.e13-05-0235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nakamura M., Yagi N., Kato T., Fujita S., Kawashima N., Ehrhardt D.W.. et al. (2012) Arabidopsis GCP3-interacting protein 1/MOZART 1 is an integral component of the γ-tubulin-containing microtubule nucleating complex. Plant J. 71, 216–225 10.1111/j.1365-313X.2012.04988.x [DOI] [PubMed] [Google Scholar]
- 63.Dhani D.K., Goult B.T., George G.M., Rogerson D.T., Bitton D.A., Miller C.J.. et al. (2013) Mzt1/Tam4, a fission yeast MOZART1 homologue, is an essential component of the γ-tubulin complex and directly interacts with GCP3Alp6. Mol. Biol. Cell 24, 3337–3349 10.1091/mbc.e13-05-0253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lin T.-C., Neuner A., Flemming D., Liu P., Chinen T., Jäkle U.. et al. (2016) MOZART1 and γ-tubulin complex receptors are both required to turn γ-TuSC into an active microtubule nucleation template. J. Cell Biol. 215, 823–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cukier C.D., Tourdes A., El-Mazouni D., Guillet V., Nomme J., Mourey L.. et al. (2017) NMR secondary structure and interactions of recombinant human MOZART1 protein, a component of the gamma-tubulin complex. Protein Sci. 26, 2240–2248 10.1002/pro.3282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Batzenschlager M., Masoud K., Janski N., Houlné G., Herzog E., Evrard J.-L.. et al. (2013) The GIP gamma-tubulin complex-associated proteins are involved in nuclear architecture in Arabidopsis thaliana. Front. Plant Sci. 4, 480 10.3389/fpls.2013.00480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Masuda H. and Toda T. (2016) Synergistic role of fission yeast Alp16GCP6 and Mzt1MOZART1 in γ-tubulin complex recruitment to mitotic spindle pole bodies and spindle assembly. Mol. Biol. Cell 27, 1753–1763, 10.1091/mbc.e15-08-0577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liu P., Choi Y.-K. and Qi R.Z. (2014) NME7 is a functional component of the γ-tubulin ring complex. Mol. Biol. Cell 25, 2017–2025 10.1091/mbc.e13-06-0339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zheng Y., Wong M.L., Alberts B. and Mitchison T. (1995) Nucleation of microtubule assembly by a gamma-tubulin-containing ring complex. Nature 378, 578–583 10.1038/378578a0 [DOI] [PubMed] [Google Scholar]
- 70.Erickson H.P. and Stoffler D. (1996) Protofilaments and rings, two conformations of the tubulin family conserved from bacterial FtsZ to alpha/beta and gamma tubulin. J. Cell Biol. 135, 5–8 10.1083/jcb.135.1.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lynch E.M., Groocock L.M., Borek W.E. and Sawin K.E. (2014) Activation of the γ-tubulin complex by the Mto1/2 complex. Curr. Biol., 24, 896–903 10.1016/j.cub.2014.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lawo S., Bashkurov M., Mullin M., Ferreria M.G., Kittler R., Habermann B.. et al. (2009) HAUS, the 8-subunit human Augmin complex, regulates centrosome and spindle integrity. Curr. Biol. 19, 816–826 10.1016/j.cub.2009.04.033 [DOI] [PubMed] [Google Scholar]
- 73.Uehara R., Nozawa R.-S., Tomioka A., Petry S., Vale R.D., Obuse C.. et al. (2009) The augmin complex plays a critical role in spindle microtubule generation for mitotic progression and cytokinesis in human cells. Proc. Natl Acad. Sci. U.S.A. 106, 6998–7003 10.1073/pnas.0901587106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhu H., Coppinger J.A., Jang C.-Y., Yates J.R. and Fang G. (2008) FAM29A promotes microtubule amplification via recruitment of the NEDD1-gamma-tubulin complex to the mitotic spindle. J. Cell Biol. 183, 835–848 10.1083/jcb.200807046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhu H., Fang K. and Fang G. (2009) FAM29A, a target of Plk1 regulation, controls the partitioning of NEDD1 between the mitotic spindle and the centrosomes. J. Cell Sci. 122, 2750–2759 10.1242/jcs.048223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhang J. and Megraw T.L. (2007) Proper recruitment of gamma-tubulin and D-TACC/Msps to embryonic Drosophila centrosomes requires Centrosomin Motif 1. Mol. Biol. Cell 18, 4037–4049 10.1091/mbc.e07-05-0474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Conduit P.T., Richens J.H., Wainman A., Holder J., Vicente C.C., Pratt M.B.. et al. (2014) A molecular mechanism of mitotic centrosome assembly in Drosophila. Elife 3, 10.7554/eLife.03399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Vaizel-Ohayon D. and Schejter E.D. (1999) Mutations in centrosomin reveal requirements for centrosomal function during early Drosophila embryogenesis. Curr. Biol. 9, 889–898 10.1016/S0960-9822(99)80393-5 [DOI] [PubMed] [Google Scholar]
- 79.Megraw T.L., Li K., Kao L.R. and Kaufman T.C. (1999) The centrosomin protein is required for centrosome assembly and function during cleavage in Drosophila. Development 126, 2829–2839 [DOI] [PubMed] [Google Scholar]
- 80.Eisman R.C., Phelps M.A.S. and Kaufman T. (2015) An amino-terminal polo kinase interaction motif acts in the regulation of centrosome formation and reveals a novel function for centrosomin (cnn) in Drosophila. Genetics 201, 685–706 10.1534/genetics.115.181842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chen J.V., Buchwalter R.A., Kao L.-R. and Megraw T.L. (2017) A splice variant of centrosomin converts mitochondria to microtubule-organizing centers. Curr. Biol., 27, 1928–1940 10.1016/j.cub.2017.05.090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dobbelaere J., Josué F., Suijkerbuijk S., Baum B., Tapon N. and Raff J. (2008) A genome-wide RNAi screen to dissect centriole duplication and centrosome maturation in Drosophila. PLoS Biol. 6, e224 10.1371/journal.pbio.0060224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sawin K.E., Lourenço P.C.C. and Snaith H.A. (2004) Microtubule nucleation at non-spindle pole body microtubule-organizing centers requires fission yeast centrosomin-related protein mod20p. Curr. Biol. 14, 763–775 10.1016/j.cub.2004.03.042 [DOI] [PubMed] [Google Scholar]
- 84.Lin T.-C., Neuner A., Schlosser Y.T., Schiebel E., Scharf A.N. and Weber L. (2014) Cell-cycle dependent phosphorylation of yeast pericentrin regulates γ-TuSC-mediated microtubule nucleation. Elife 3, e02208 10.7554/eLife.02208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Fong K.-W., Choi Y.-K., Rattner J.B. and Qi R.Z. (2008) CDK5RAP2 is a pericentriolar protein that functions in centrosomal attachment of the gamma-tubulin ring complex. Mol. Biol. Cell 19, 115–125 10.1091/mbc.e07-04-0371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Samejima I., Miller V.J., Groocock L.M. and Sawin K.E. (2008) Two distinct regions of Mto1 are required for normal microtubule nucleation and efficient association with the gamma-tubulin complex in vivo. J. Cell Sci. 121, 3971–3980 10.1242/jcs.038414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lyon A.S., Morin G., Moritz M., Yabut K.C.B., Vojnar T., Zelter A.. et al. (2016) Higher-order oligomerization of Spc110p drives γ-tubulin ring complex assembly. Mol. Biol. Cell 27, 10.1091/mbc.e16-02-0072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Vogel J., Drapkin B., Oomen J., Beach D., Bloom K. and Snyder M. (2001) Phosphorylation of gamma-tubulin regulates microtubule organization in budding yeast. Dev. Cell 1, 621–631 10.1016/S1534-5807(01)00073-9 [DOI] [PubMed] [Google Scholar]
- 89.Keck J.M., Jones M.H., Wong C.C.L., Binkley J., Chen D., Jaspersen S.L.. et al. (2011) A cell cycle phosphoproteome of the yeast centrosome. Science 332, 1557–1561 10.1126/science.1205193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lin T.-C., Gombos L., Neuner A., Sebastian D., Olsen J.V., Hrle A.. et al. (2011) Phosphorylation of the yeast γ-tubulin Tub4 regulates microtubule function. PLoS ONE 6, e19700 10.1371/journal.pone.0019700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fong K.K., Zelter A., Graczyk B., Hoyt J.M., Riffle M., Johnson R.. et al. (2018) Novel phosphorylation states of the yeast spindle pole body. Biol. Open 10.1242/bio.033647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bahtz R., Seidler J., Arnold M., Haselmann-Weiß U., Antony C., Lehmann W.D.. et al. (2012) GCP6 is a substrate of Plk4 and required for centriole duplication. J. Cell Sci. 125, 486–496 10.1242/jcs.093930 [DOI] [PubMed] [Google Scholar]
- 93.Alvarado-Kristensson M., Rodríguez M.J., Silió V., Valpuesta J.M. and Carrera A.C. (2009) SADB phosphorylation of gamma-tubulin regulates centrosome duplication. Nat. Cell Biol. 11, 1081–1092 10.1038/ncb1921 [DOI] [PubMed] [Google Scholar]
- 94.Dephoure N., Zhou C., Villén J., Beausoleil S.A., Bakalarski C.E., Elledge S.J.. et al. (2008) A quantitative atlas of mitotic phosphorylation. Proc. Natl Acad. Sci. U.S.A. 105, 10762–10767 10.1073/pnas.0805139105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhai B., Villén J., Beausoleil S.A., Mintseris J. and Gygi S.P. (2008) Phosphoproteome analysis of Drosophila melanogaster embryos. J. Proteome Res. 7, 1675–1682 10.1021/pr700696a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Friedman D.B., Sundberg H.A., Huang E.Y. and Davis T.N. (1996) The 110-kD spindle pole body component of Saccharomyces cerevisiae is a phosphoprotein that is modified in a cell cycle-dependent manner. J. Cell Biol. 132, 903–914 10.1083/jcb.132.5.903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Friedman D.B., Kern J.W., Huneycutt B.J., Vinh D.B., Crawford D.K., Steiner E.. et al. (2001) Yeast Mps1p phosphorylates the spindle pole component Spc110p in the N-terminal domain. J. Biol. Chem. 276, 17958–17967 10.1074/jbc.M010461200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Huisman S.M., Smeets M.F.M.A. and Segal M. (2007) Phosphorylation of Spc110p by Cdc28p-Clb5p kinase contributes to correct spindle morphogenesis in S. cerevisiae. J. Cell Sci. 120, 435–446 10.1242/jcs.03342 [DOI] [PubMed] [Google Scholar]
- 99.Stirling D.A. and Stark M.J. (1996) The phosphorylation state of the 110 kDa component of the yeast spindle pole body shows cell cycle dependent regulation. Biochem. Biophys. Res. Commun. 222, 236–242 10.1006/bbrc.1996.0728 [DOI] [PubMed] [Google Scholar]
- 100.Liang F., Richmond D. and Wang Y. (2013) Coordination of chromatid separation and spindle elongation by antagonistic activities of mitotic and S-phase CDKs. PLoS Genet. 9, e1003319 10.1371/journal.pgen.1003319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Scrofani J., Sardon T., Meunier S. and Vernos I. (2015) Microtubule nucleation in mitosis by a RanGTP-dependent protein complex. Curr. Biol. 25, 131–140 10.1016/j.cub.2014.11.025 [DOI] [PubMed] [Google Scholar]
- 102.Lee K. and Rhee K. (2011) PLK1 phospγ-tubulin ring complexhorylation of pericentrin initiates centrosome maturation at the onset of mitosis. J. Cell Biol., 195, 1093–1101 10.1083/jcb.201106093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Santamaria A., Wang B., Elowe S., Malik R., Zhang F., Bauer M.. et al. (2011) The Plk1-dependent phosphoproteome of the early mitotic spindle. Mol. Cell. Proteomics 10, 10.1074/mcp.M110.004457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Conduit P.T., Feng Z., Richens J.H., Baumbach J., Wainman A., Bakshi S.D.. et al. (2014) The centrosome-specific phosphorylation of Cnn by Polo/Plk1 drives Cnn scaffold assembly and centrosome maturation. Dev. Cell 28, 659–669 10.1016/j.devcel.2014.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bodenmiller B., Malmstrom J., Gerrits B., Campbell D., Lam H., Schmidt A.. et al. (2007) PhosphoPep-a phosphoproteome resource for systems biology research in Drosophila Kc167 cells. Mol. Syst. Biol. 3, 139 10.1038/msb4100182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bodenmiller B., Campbell D., Gerrits B., Lam H., Jovanovic M., Picotti P.. et al. (2008) PhosphoPep-a database of protein phosphorylation sites in model organisms. Nat. Biotechnol. 26, 1339–1340 10.1038/nbt1208-1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Eisman R.C., Phelps M.A.S. and Kaufman T.C. (2009) Centrosomin: a complex mix of long and short isoforms is required for centrosome function during early development in Drosophila melanogaster. Genetics 182, 979–997 10.1534/genetics.109.103887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Borek W.E., Groocock L.M., Samejima I., Zou J., de Lima Alves F., Rappsilber J.. et al. (2015) Mto2 multisite phosphorylation inactivates non-spindle microtubule nucleation complexes during mitosis. Nat. Commun. 6, 7929 10.1038/ncomms8929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kraemer N., Issa-Jahns L., Neubert G., Ravindran E., Mani S., Ninnemann O.. et al. (2015) Novel alternative splice variants of mouse Cdk5rap2. PLoS ONE 10, e0136684 10.1371/journal.pone.0136684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wang Z., Zhang C. and Qi R.Z. (2014) A novel myomegalin isoform functions in Golgi microtubule organization and ER-Golgi transport. J. Cell Sci. 127, 4904–4917 10.1242/jcs.155408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Park J.S.Y., Lee M.-K., Kang S., Jin Y., Fu S., Rosales J.L.. et al. (2015) Species-specific expression of full-length and alternatively spliced variant forms of CDK5RAP2. PLoS ONE 10, e0142577 10.1371/journal.pone.0142577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Kollman J.M., Greenberg C.H., Li S., Moritz M., Zelter A., Fong K.K.. et al. (2015) Ring closure activates yeast γTuRC for species-specific microtubule nucleation. Nat. Struct. Mol. Biol. 10.1038/nsmb.2953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Roostalu J., Cade N.I. and Surrey T. (2015) Complementary activities of TPX2 and chTOG constitute an efficient importin-regulated microtubule nucleation module. Nat. Cell Biol. 17, 1422–1434 10.1038/ncb3241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wieczorek M., Bechstedt S., Chaaban S. and Brouhard G.J. (2015) Microtubule-associated proteins control the kinetics of microtubule nucleation. Nat. Cell Biol. 10.1038/ncb3188 [DOI] [PubMed] [Google Scholar]
- 115.Woodruff J.B., Ferreira Gomes B., Widlund P.O., Mahamid J., Honigmann A. and Hyman A.A. (2017) The centrosome is a selective condensate that nucleates microtubules by concentrating tubulin. Cell 169, 1066–1077 10.1016/j.cell.2017.05.028 [DOI] [PubMed] [Google Scholar]
- 116.Flor-Parra I., Iglesias-Romero A.B. and Chang F. (2018) The XMAP215 ortholog Alp14 promotes microtubule nucleation in fission yeast. Curr. Biol., 28, 1681–1691 10.1016/j.cub.2018.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Thawani A., Kadzik R.S. and Petry S. (2018) XMAP215 is a microtubule nucleation factor that functions synergistically with the γ-tubulin ring complex. Nat. Cell Biol. 20, 1–18 10.1038/s41556-018-0091-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Lüders J. (2018) XMAP215 joins microtubule nucleation team. Nat. Cell Biol. 20, 508–510 10.1038/s41556-018-0100-9 [DOI] [PubMed] [Google Scholar]
- 119.Usui T., Maekawa H., Pereira G. and Schiebel E. (2003) The XMAP215 homologue Stu2 at yeast spindle pole bodies regulates microtubule dynamics and anchorage. EMBO J. 22, 4779–4793 10.1093/emboj/cdg459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Reid T.A., Schuster B.M., Mann B.J., Balchand S.K., Plooster M., McClellan M.. et al. (2016) Suppression of microtubule assembly kinetics by the mitotic protein TPX2. J. Cell Sci. 129, 1319–1328 10.1242/jcs.178806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zhang R., Roostalu J., Surrey T. and Nogales E. (2017) Structural insight into TPX2-stimulated microtubule assembly. Elife 6, 1518 10.7554/eLife.30959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sampaio P., Rebollo E., Varmark H., Sunkel C.E. and González C. (2001) Organized microtubule arrays in gamma-tubulin-depleted Drosophila spermatocytes. Curr. Biol. 11, 1788–1793 10.1016/S0960-9822(01)00561-9 [DOI] [PubMed] [Google Scholar]
- 123.Llamazares S., Tavosanis G. and González C. (1999) Cytological characterisation of the mutant phenotypes produced during early embryogenesis by null and loss-of-function alleles of the gammaTub37C gene in Drosophila. J. Cell Sci. 112, 659–667 [DOI] [PubMed] [Google Scholar]
- 124.Hannak E., Oegema K., Kirkham M., Gönczy P., Habermann B. and Hyman A.A. (2002) The kinetically dominant assembly pathway for centrosomal asters in Caenorhabditis elegans is gamma-tubulin dependent. J. Cell Biol. 157, 591–602 10.1083/jcb.200202047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bouissou A., Vérollet C., Sousa A., Sampaio P., Wright M., Sunkel C.E.. et al. (2009) {gamma}-Tubulin ring complexes regulate microtubule plus end dynamics. J. Cell Biol. 187, 327–334 10.1083/jcb.200905060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Rogers G.C., Rusan N.M., Peifer M. and Rogers S.L. (2008) A multicomponent assembly pathway contributes to the formation of acentrosomal microtubule arrays in interphase Drosophila cells. Mol. Biol. Cell 19, 3163–3178 10.1091/mbc.e07-10-1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kitamura E., Tanaka K., Komoto S., Kitamura Y., Antony C. and Tanaka T.U. (2010) Kinetochores generate microtubules with distal plus ends: their roles and limited lifetime in mitosis. Dev. Cell 18, 248–259 10.1016/j.devcel.2009.12.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Chaaban S. and Brouhard G.J. (2017) A microtubule bestiary: structural diversity in tubulin polymers. Mol. Biol. Cell 28, 2924–2931 10.1091/mbc.e16-05-0271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Argyriou A.A., Kyritsis A.P., Makatsoris T. and Kalofonos H.P. (2014) Chemotherapy-induced peripheral neuropathy in adults: a comprehensive update of the literature. Cancer Manag. Res. 6, 135–147 10.2147/CMAR.S44261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Fukuda Y., Li Y. and Segal R.A. (2017) A mechanistic understanding of axon degeneration in chemotherapy-induced peripheral neuropathy. Front. Neurosci. 11, 481 10.3389/fnins.2017.00481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.LaPointe N.E., Morfini G., Brady S.T., Feinstein S.C., Wilson L. and Jordan M.A. (2013) Effects of eribulin, vincristine, paclitaxel and ixabepilone on fast axonal transport and kinesin-1 driven microtubule gliding: implications for chemotherapy-induced peripheral neuropathy. Neurotoxicology 37, 231–239 10.1016/j.neuro.2013.05.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Theiss C. and Meller K. (2000) Taxol impairs anterograde axonal transport of microinjected horseradish peroxidase in dorsal root ganglia neurons in vitro. Cell Tissue Res. 299, 213–224 10.1007/s004410050019 [DOI] [PubMed] [Google Scholar]
- 133.Shemesh O.A. and Spira M.E. (2010) Paclitaxel induces axonal microtubules polar reconfiguration and impaired organelle transport: implications for the pathogenesis of paclitaxel-induced polyneuropathy. Acta Neuropathol. 119, 235–248 10.1007/s00401-009-0586-0 [DOI] [PubMed] [Google Scholar]
- 134.Gornstein E.L. and Schwarz T.L. (2017) Neurotoxic mechanisms of paclitaxel are local to the distal axon and independent of transport defects. Exp. Neurol. 288, 153–166 10.1016/j.expneurol.2016.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Whitehurst A.W., Bodemann B.O., Cardenas J., Ferguson D., Girard L., Peyton M.. et al. (2007) Synthetic lethal screen identification of chemosensitizer loci in cancer cells. Nature 446, 815–819 10.1038/nature05697 [DOI] [PubMed] [Google Scholar]
- 136.Cala O., Remy M.-H., Guillet V., Merdes A., Mourey L., Milon A.. et al. (2013) Virtual and biophysical screening targeting the γ-tubulin complex - a new target for the inhibition of microtubule nucleation. PLoS ONE 8, e63908 10.1371/journal.pone.0063908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Bouissou A., Vérollet C., de Forges H., Haren L., Bellaiche Y., Perez F.. et al. (2014) γ-Tubulin Ring Complexes and EB1 play antagonistic roles in microtubule dynamics and spindle positioning. EMBO J. 33, 114–128 10.1002/embj.201385967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Chinen T., Liu P., Shioda S., Pagel J., Cerikan B., Lin T.-C.. et al. (2015) The γ-tubulin-specific inhibitor gatastatin reveals temporal requirements of microtubule nucleation during the cell cycle. Nat. Commun. 6, 8722 10.1038/ncomms9722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Weil C.F., Oakley C.E. and Oakley B.R. (1986) Isolation of mip (microtubule-interacting protein) mutations of Aspergillus nidulans. Mol. Cell. Biol. 6, 2963–2968 10.1128/MCB.6.8.2963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Oakley C.E. and Oakley B.R. (1989) Identification of gamma-tubulin, a new member of the tubulin superfamily encoded by mipA gene of Aspergillus nidulans. Nature 338, 662–664 10.1038/338662a0 [DOI] [PubMed] [Google Scholar]