Abstract
Background
Fluorodeoxyglucose f18 positron emission tomography (18F-FDG PET) is regarded as the only functional neuroimaging biomarker for degeneration which can be used to increase the certainty of Alzheimer’s disease (AD) pathophysiological process in research settings or as an optional clinical tool where available. Although a decline in FDG metabolism was confirmed in some regions known to be associated with AD, there was little known about the genetic association of FDG metabolism in AD cohorts. In this study, we present the first genome-wide association study (GWAS) analysis of brain FDG metabolism.
Methods
A total of 222 individuals were included from the Alzheimer’s Disease Neuroimaging Initiative 1 (ADNI-1) cohort. All subjects were restricted to non-Hispanic Caucasians and met all quality control (QC) criteria. Associations of 18F-FDG with the genetic variants were assessed using PLINK 1.07 under the additive genetic model. Genome-wide associations were visualized using a software program R 3.2.3.
Results
One significant SNP rs12444565 in RNA-binding Fox1 (RBFOX1) was found to have a strong association with 18F-FDG (P=6.06×10–8). Rs235141, rs79037, rs12526331 and rs12529764 were identified as four suggestive loci associated with 18F-FDG.
Conclusions
Our study results suggest that a genome-wide significant SNP (rs12444565) in the RBFOX1, and four suggestive loci (rs235141, rs79037, rs12526331 and rs12529764) are associated with 18F-FDG.
Keywords: Alzheimer’s Disease Neuroimaging Initiative 1 (ADNI-1), Alzheimer’s disease (AD), fluorodeoxyglucose f18 (18F-FDG), RNA-binding Fox1 (RBFOX1)
Introduction
A decline in glucose metabolism in the posterior cingulate cortex (PCC) is one of the earliest biomarkers for AD, which are present years before symptom onset (1). Furthermore, it actually predicts the conversion from healthy aging to mild cognitive impairment (MCI), and from MCI to AD (2,3), signifying its important role in AD progression. Evidences showed that glucose metabolic disorder occurs before the onset of AD and glucose metabolism is a sensitive indicator of cognitive and functional changes in AD and MCI, which may be valuable in predicting cognitive decline in future (4,5). A study demonstrated that enhancing glucose uptake in neurons has a strong neuroprotective effect which can improve proteostasis in the fly model of AD (6). AD patients typically show a very characteristic pattern of hypometabolism in the posterior cingulate, parietotemporal cortices and frontal lobes (the so-called AD metabolic pattern).
Fluorodeoxyglucose f18, also known as fluorodeoxyglucose and abbreviated as 18F-FDG or FDG, is a radioactive drug used in medical imaging of positron emission tomography (PET). The uptake of 18F-FDG by tissues is a marker of glucose uptake which is closely related to tissue metabolism. Currently, 18F-FDG PET is gaining increased acceptance as a valuable tool to investigate pre-symptomatic AD (7-10). 18F-FDG PET, which enables evaluation of cerebral glucose metabolism, is increasingly used to support clinical diagnosis of suspected dementia patients. According to the new recommendations by the Alzheimer’s Association, 18F-FDG PET is regarded as the only functional neuroimaging biomarker for degeneration which can be used to increase the certainty of AD pathophysiological process in research settings (clinical trials, investigational studies) or regarded as an optional clinical tool where available (11-13).
Although a decline in FDG metabolism was confirmed in some regions known to be associated with AD, there was little known about the genetic association of FDG metabolism in AD cohorts. We hypothesized that FDG may constitute a suitable endophenotype for a GWAS designed to identify genetic factors and highlight relevant physiological and pathophysiological processes. In this study, we present the first GWAS analysis of FDG metabolism in Alzheimer’s Disease Neuroimaging Initiative 1 (ADNI-1) database. Through the analysis of AD cases, we hope to identify novel variants specific to FDG metabolism by 18F-FDG PET.
Methods
ADNI dataset
Data used in this study were obtained from the ADNI database (www.loni.ucla.edu/ADNI). The ADNI was launched in 2003 as a public-private partnership, led by Principal Investigator Michael W. Weiner, MD, VA Medical Center and University of California-San Francisco. ADNI was established to test whether serial magnetic resonance imaging (MRI), PET, other biomarkers, and clinical and neuropsychological assessment can be combined to measure the progression of MCI and early AD.
Subjects
In this study, a total of 222 individuals (AD =37, MCI =126, healthy controls, HC =59) were included from the ADNI-1 cohort. All subjects were restricted to non-Hispanic Caucasians and met all quality control (QC) criteria. The analysis was restricted to non-Hispanic Caucasians to reduce potential bias from population stratification which might confound GWAS results. The detailed demographic information has been shown in Table 1.
Table 1. Demographic information of sample characteristics.
| ADNI-1 baseline diagnosis | AD (n=37) | MCI (n=126) | HC (n=59) | Total (n=222) |
|---|---|---|---|---|
| Age (years), mean ± SD (range) | 74.44±7.460 (58.2–89.6) | 72.82±7.339 (55.0–87.3) | 74.50±5.550 (62.3–88.1) | 73.54±6.950 (55.0–89.60) |
| Gender, male/female | 19/18 | 68/58 | 30/29 | 117/105 |
| APOE ε4 carrier (%) | 62.16 | 49.21 | 33.90 | 47.30 |
| FDG, mean ± SD | 5.50±0.722 | 6.16±0.636 | 6.53±0.590 | 6.12±0.718 |
AD, Alzheimer’s disease; APOE, Apolipoprotein E; HC, healthy control; MCI, mild cognitive impairment; SD, standard deviation.
Glucose metabolism assessed by 18F-FDG PET and QC
The data for FDG analysis were obtained from UC Berkeley and Lawrence Berkeley National Laboratory (http://adni.loni.usc.edu/data-samples/access-data/). In the analysis, five regions containing the left and right angular gyri, bilateral posterior cingulate, the left and right temporal gyri were analyzed as meta ROIs (regions of interest). The short procedures were as follows. Firstly, we downloaded the PET data from LONI (http://loni.usc.edu/). Then, these images were normalized to the MNI PET templates in SPM. The average numbers of FDG scans for each subject were calculated, and the intensity values were computed using SPM subroutines. Finally, the average value of the top 50% of voxels in a hand-drawn pons/cerebellar vermis region was extracted from the T1 template in the MNI space. The average of each meta ROI was standardized through the pons/vermis reference region average (Landau et al., 2011). 18F-FDG PET measurement 366–399 MBq of 18F-FDG was injected intravenously into all subjects and the follow-up time was 30 min. During this time, the subjects were undisturbed and resting quietly in a dark room with their eyes open. The subjects were then imaged with their eyes open for an 8-min image collection consisting of four 2-min dynamic frames.
Genotyping and QC
The samples were genotyped with the Human 610-Quad BeadChip. Stringent QC assessment was performed using the PLINK software with the following criteria: minimum call rate for SNPs and individuals >98%, minimum minor allele frequencies (MAF) >0.04, and Hardy-Weinberg equilibrium test P>0.001. The restriction to SNPs with a MAF greater than 4% served to reduce the probability of false positive results and enhance statistical power. Rs7412 and rs429358 that define apolipoprotein E (APOE) alleles, were genotyped separately with an APOE genotyping kit.
Statistical analyses
Associations of 18F-FDG with the genetic variants were assessed using PLINK 1.07 under an additive genetic model. The thresholds of P<10–6 and P<10–7 were used for suggestive and genome-wide significant associations respectively. Our analysis included a total of 319,792 genotyped variants. Age, gender and diagnosis were included as covariates. Bonferroni correction of the P values by the total number of acceptable quality SNPs was used for multiple test correction. Genome-wide associations were visualized using a software program R 3.2.3.
Results
The sample contained 222 (AD =37, MCI =126, HC =59) non-Hispanic Caucasians in the ADNI-1 cohort. Table 1 shows the detailed demographic information and data of 18F-FDG in each group.
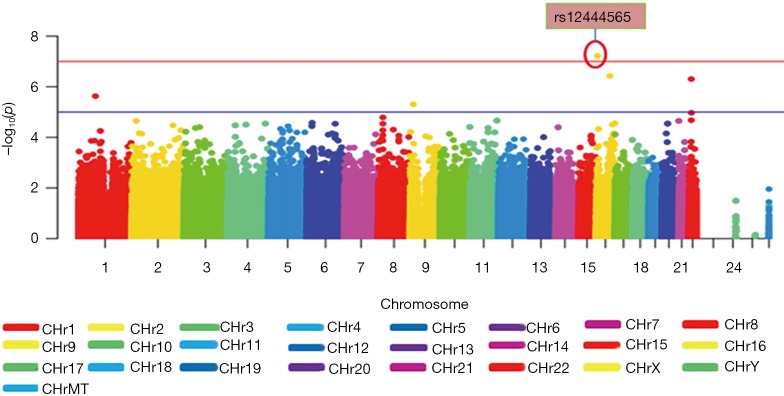
Figure 1 displays the Manhattan plot of 18F-FDG in the ADNI-1 cohort. After adjustment for age, gender and diagnosis, a genome-wide significant association of rs12444565 with 18F-FDG was detected. The analysis identified four additional SNPs with suggestive association (rs235141, rs79037, rs12526331 and rs12529764).
Figure 1.
Genome-wide association results for 18F-FDG in ADNI-1. The y-axis shows the P values (on the –log10 scale) for each association test. The x-axis is the chromosomal position of each SNP. The gold horizontal line at 10−7 indicates genome-wide significance. A genome-wide significant association of rs12444565 with 18F-FDG was detected. Four additional SNPs with suggestive association (rs235141, rs79037, rs12526331 and rs12529764) were identified. 18F-FDG, fluorodeoxyglucose f18; ADNI-1, Alzheimer’s Disease Neuroimaging Initiative 1.
One significant SNP rs12444565 in RNA-binding Fox1 (RBFOX1) was found to have a strong association with 18F-FDG (P=6.06×10–8) (in Table 2). This locus survived after multiple testing based on both permutation and Bonferroni corrections [empirical P (EMP1) =0.0002; permutation-based corrected empirical P (EMP2) =0.005; Bonferroni corrected P=0.005]. SNPs which did not reach genome-wide significance and whose P values were between 10–7 and 10–6 including rs235141, rs79037, rs12526331 and rs12529764 were also listed in Table 2.
Table 2. SNPs associated with 18F-FDG.
| SNP | CHR | Position | Associated area | 18F-FDG P value |
|---|---|---|---|---|
| rs12444565 | 16 | 7347272 | Left angular | 5.89×10–8 |
| rs235141 | 16 | 66209852 | Left angular | 3.80×10–7 |
| rs79037 | 22 | 27037216 | Left angular | 4.95×10–7 |
| rs12526331 | 6 | 80086431 | Cigulumpost | 1.57×10–7 |
| rs12529764 | 6 | 80086864 | Cigulumpost | 1.57×10–7 |
18F-FDG, fluorodeoxyglucose f18; CHR, chromosome.
Discussion
A GWAS was performed in ANDI-1 to study the effect of genetic variation on the cerebral glucose metabolism. The use of quantitative traits in GWAS has been shown to have an increased statistical power and a decreased requirement of sample size, so this study has more advantages than traditional case-control designs (14-16). We identified a genome-wide significant association of a SNP in the regions of RBFOX1 with glucose metabolism and four additional SNPs with suggestive association (rs235141, rs79037, rs12526331 and rs12529764) by using 18F-FDG as quantitative traits.
RBFOX1, a neuronal RNA binding protein, also known as Ataxin-2-binding protein 1 (A2BP1) or FOX1, was initially identified as an interacting partner of ATAXIN-2 (17) and was expressed in neuronal tissues, muscle and heart (18,19). The RBFOX1 gene locus is located on chromosome region 16p13.3 (20). Through the combination with (U) of GCAUG element in mRNA precursor, RBFOX1 has been reported to play a key role in alternative splicing in the key genes of neuronal development, such as calcitonin/calcitonin gene related peptide, CaV1.2 voltage-controlled calcium channel and N-methyl-D-aspartate (NMDA) receptor 1 (18,19,21-23). Defects in these key processes may lead to the structural and functional defects of cortical neurons, leading to the pathophysiology of neurodevelopmental disorders with RBFOX1 abnormalities (24). Genetic studies, including GWAS, have proven that RBFOX1 is associated with many neuropsychiatric disorders including autism spectrum disorder (ASD), epilepsy, attention deficit hyperactivity disorder (ADHD), generalised anxiety disorder (GAD), intellectual disabilities (IDs), and schizophrenia (25-30), which strongly suggest that partial RBFOX1 deficiency is a risk factor for the recurrence of human neurodevelopmental defects.
It has been reported that RBFOX1 plays a pivotal role in neuronal excitation regulation, which affects susceptibility to epilepsy (23,31). Many studies have suggested that a lot of RBFOX1 target transcripts play a role in epileptogenesis (32-34), while the rare external absence of RBFOX1 gene increases the risk of idiopathic generalized epilepsy (35). In mice, the brain’s specific homozygous and heterozygous RBFOX1 knockouts showed spontaneous epileptic seizures and dramatic epileptic responses without altering the morphology of the brain (23).
Human genetic studies have identified RBFOX1 as a candidate gene for ASD. Studies have shown RBFOX1 target transcripts are overlapping significantly in ASD (31). In addition, RBFOX1 was found to act as a “hub” in the gene transcription group network in the brain of ASD patients (31). Abnormal RBFOX1 gene encoding an mRNA-splicing factor, has been proved to cause ASD. In ASD patients, the expression of RBFOX1 was reduced in relation to the splicing of the target exon (36). A large number of splicing changes were detected in 48 genes in the brain of a group of ASD patients, whose RBFOX1 was estimated to be down-regulated (36).
Rs235141, rs79037, rs12526331 and rs12529764 (located on chromosome region 16:66209852, 22:27037216, 6:80086431 and 6:80086864 respectively) were four suggestive loci associated with glucose metabolism. There were no relevant studies available to study their roles and relationships with neuropsychiatric disorders.
The limitation of this study was the moderate sample size of GWAS. In addition, ADNI data was restricted to non-Hispanic Caucasians to avoid genetic stratification due to race. Therefore, our results can not represent other races.
To conclude, we detected a genome-wide significant SNP (rs12444565) in RBFOX1 and four suggestive loci (rs235141, rs79037, rs12526331 and rs12529764) associated with 18F-FDG. Accumulating evidence suggested that aberrations in RBFOX1-regulated circuitry were risk factors for multiple neuropsychiatric disorders, especially epilepsy and ASD. Therefore, 18F-FDG PET may be applied to some neuropsychiatric disorders especially epilepsy and ASD to predict the occurrence and development as well as therapy of the diseases through the genotyping in the future. Independent, larger datasets will be needed to replicate research to confirm this result.
Acknowledgements
Data collection and sharing was funded by ADNI. ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Alzheimers Association; Alzheimers Drug Discovery Foundation; BioClinica, Inc.; Biogen Idec Inc.; Bristol-Myers Squibb Company; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; GE Healthcare; Innogenetics, N.V.; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Medpace, Inc.; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Synarc Inc.; and Takeda Pharmaceutical Company. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
Footnotes
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- 1.Minoshima S, Giordani B, Berent S, et al. Metabolic reduction in the posterior cingulate cortex in very early Alzheimer's disease. Ann Neurol 1997;42:85-94. 10.1002/ana.410420114 [DOI] [PubMed] [Google Scholar]
- 2.de Leon MJ, Convit A, Wolf OT, et al. Prediction of cognitive decline in normal elderly subjects with 2-[(18)F]fluoro-2-deoxy-D-glucose/poitron-emission tomography (FDG/PET). Proc Natl Acad Sci U S A 2001;98:10966-71. 10.1073/pnas.191044198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chételat G, Desgranges B, Sayette VD, et al. Mild cognitive impairment Can FDG-PET predict who is to rapidly convert to Alzheimer’s disease? Neurology 2003;60:1374. 10.1212/01.WNL.0000055847.17752.E6 [DOI] [PubMed] [Google Scholar]
- 4.Sun H, Hu B, Yao Z, et al., editors. A PET study of discrimination of cerebral glucose metabolism in Alzheimer's disease and mild cognitive impairment. Int Conf Biomed Health Inform 2014. doi: 10.1109/BMEI.2013.6746897. [DOI] [Google Scholar]
- 5.Li XY, Men WW, Zhu H, et al. Age- and Brain Region-Specific Changes of Glucose Metabolic Disorder, Learning, and Memory Dysfunction in Early Alzheimer’s Disease Assessed in APP/PS1 Transgenic Mice Using 18F-FDG-PET. Int J Mol Sci 2016;17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duran-Aniotz C, Hetz C. Glucose Metabolism: A Sweet Relief of Alzheimer’s Disease. Curr Biol 2016;26:R806-9. 10.1016/j.cub.2016.07.060 [DOI] [PubMed] [Google Scholar]
- 7.Illán IA, Górriz JM, Ramírez J, et al. 18F-FDG PET imaging analysis for computer aided Alzheimer’s diagnosis. Inform Sci 2011;181:903-16. 10.1016/j.ins.2010.10.027 [DOI] [Google Scholar]
- 8.Gallivanone F, Rosa PA, Castiglioni I. Statistical Voxel-Based Methods and [18F]FDG PET Brain Imaging: Frontiers for the Diagnosis of AD. Curr Alzheimer Res 2016;13:682-94. 10.2174/1567205013666151116142039 [DOI] [PubMed] [Google Scholar]
- 9.Herholz K, Westwood S, Haense C, et al. Evaluation of a calibrated (18)F-FDG PET score as a biomarker for progression in Alzheimer disease and mild cognitive impairment. J Nucl Med 2011;52:1218-26. 10.2967/jnumed.111.090902 [DOI] [PubMed] [Google Scholar]
- 10.Prestia A, Muscio C, Caroli A, et al. Computer-aided diagnostic reporting of FDG PET for the diagnosis of Alzheimer’s disease. Clin Transl Imaging 2013;1:279-88. 10.1007/s40336-013-0031-1 [DOI] [Google Scholar]
- 11.Cabral C, Morgado PM, Campos Costa D, et al. Predicting conversion from MCI to AD with FDG-PET brain images at different prodromal stages. Comput Biol Med 2015;58:101-9. 10.1016/j.compbiomed.2015.01.003 [DOI] [PubMed] [Google Scholar]
- 12.Herholz K. Use of FDG PET as an imaging biomarker in clinical trials of Alzheimer's disease. Biomark Med 2012;6:431-9. 10.2217/bmm.12.51 [DOI] [PubMed] [Google Scholar]
- 13.Arbizu J, Prieto E, Martínez-Lage P, et al. Automated analysis of FDG PET as a tool for single-subject probabilistic prediction and detection of Alzheimer's disease dementia. Eur J Nucl Med Mol Imaging 2013;40:1394-405. 10.1007/s00259-013-2458-z [DOI] [PubMed] [Google Scholar]
- 14.Meyer-Lindenberg A, Weinberger DR. Intermediate phenotypes and genetic mechanisms of psychiatric disorders. Nat Rev Neurosci 2006;7:818-27. 10.1038/nrn1993 [DOI] [PubMed] [Google Scholar]
- 15.Potkin SG, Turner JA, Guffanti G, et al. Genome-wide strategies for discovering genetic influences on cognition and cognitive disorders: methodological considerations. Cogn Neuropsychiatry 2009;14:391-418. 10.1080/13546800903059829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Allen A, Epstein MP, Satten GA. Score-based adjustment for confounding by population stratification in genetic association studies. Genet Epidemiol 2010;34:383-5. 10.1002/gepi.20487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shibata H, Huynh DP, Pulst SM. A novel protein with RNA-binding motifs interacts with ataxin-2. Hum Mol Genet 2000;9:1303-13. 10.1093/hmg/9.9.1303 [DOI] [PubMed] [Google Scholar]
- 18.Jin Y, Suzuki H, Maegawa S, et al. A vertebrate RNA-binding protein Fox-1 regulates tissue-specific splicing via the pentanucleotide GCAUG. EMBO J 2003;22:905-12. 10.1093/emboj/cdg089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Underwood JG, Boutz PL, Dougherty JD, et al. Homologues of the Caenorhabditis elegans Fox-1 protein are neuronal splicing regulators in mammals. Mol Cell Biol 2005;25:10005-16. 10.1128/MCB.25.22.10005-10016.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lucarelli P, Palminiello S, Saccucci P, et al. Association study of autistic disorder and chromosome 16p. Am J Med Genet A 2003;119A:242-6. 10.1002/ajmg.a.10187 [DOI] [PubMed] [Google Scholar]
- 21.Zhou HL, Baraniak AP, Lou H. Role for Fox-1/Fox-2 in mediating the neuronal pathway of calcitonin/calcitonin gene-related peptide alternative RNA processing. Mol Cell Biol 2007;27:830-41. 10.1128/MCB.01015-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee JA, Tang ZZ, Black DL. An inducible change in Fox-1/A2BP1 splicing modulates the alternative splicing of downstream neuronal target exons. Genes Dev 2009;23:2284-93. 10.1101/gad.1837009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gehman LT, Stoilov P, Maguire J, et al. The splicing regulator Rbfox1 (A2BP1) controls neuronal excitation in the mammalian brain. Nat Genet 2011;43:706-11. 10.1038/ng.841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hamada N, Ito H, Nishijo T, et al. Essential role of the nuclear isoform of RBFOX1, a candidate gene for autism spectrum disorders, in the brain development. Sci Rep 2016;6:30805. 10.1038/srep30805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Griswold AJ, Dueker ND, Van Booven D, et al. Targeted massively parallel sequencing of autism spectrum disorder-associated genes in a case control cohort reveals rare loss-of-function risk variants. Mol Autism 2015;6:43. 10.1186/s13229-015-0034-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bhalla K, Phillips HA, Crawford J, et al. The de novo chromosome 16 translocations of two patients with abnormal phenotypes (mental retardation and epilepsy) disrupt the A2BP1 gene. J Hum Genet 2004;49:308-11. 10.1007/s10038-004-0145-4 [DOI] [PubMed] [Google Scholar]
- 27.Elia J, Glessner JT, Wang K, et al. Genome-wide copy number variation study associates metabotropic glutamate receptor gene networks with attention deficit hyperactivity disorder. Nat Genet 2011;44:78-84. 10.1038/ng.1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davis LK, Maltman N, Mosconi MW, et al. Rare Inherited A2BP1 Deletion In A Proband With Autism And Developmental Hemiparesis. Am J Med Genet A 2012;158A:1654-61. 10.1002/ajmg.a.35396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davies MN, Verdi S, Burri A, et al. Generalised Anxiety Disorder--A Twin Study of Genetic Architecture, Genome-Wide Association and Differential Gene Expression. PLoS One 2015;10:e0134865. 10.1371/journal.pone.0134865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kohannim O, Hibar DP, Jahanshad N, et al. Predicting temporal lobe volume on mri from genotypes using L(1)-L(2) regularized regression. Proc IEEE Int Symp Biomed Imaging 2012:1160-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Voineagu I, Wang X, Johnston P, et al. Transcriptomic Analysis of Autistic Brain Reveals Convergent Molecular Pathology. Nature 2011;474:380-4. 10.1038/nature10110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pandolfo M, editor. Genetics of Epilepsy. Seminars in Neurology; 2011. [DOI] [PubMed] [Google Scholar]
- 33.Fogel BL, Wexler E, Wahnich A, et al. RBFOX1 regulates both splicing and transcriptional networks in human neuronal development. Hum Mol Genet 2012;21:4171-86. 10.1093/hmg/dds240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Veeramah KR, O'Brien JE, Meisler MH, et al. De Novo Pathogenic SCN8A Mutation Identified by Whole-Genome Sequencing of a Family Quartet Affected by Infantile Epileptic Encephalopathy and SUDEP. Am J Hum Genet 2012;90:502-10. 10.1016/j.ajhg.2012.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lal D, Trucks H, Møller RS, et al. Rare exonic deletions of the RBFOX1 gene increase risk of idiopathic generalized epilepsy. Epilepsia 2013;54:265-71. 10.1111/epi.12084 [DOI] [PubMed] [Google Scholar]
- 36.Weyn-Vanhentenryck SM, Mele A, Yan Q, et al. HITS-CLIP and integrative modeling define the Rbfox splicing-regulatory network linked to brain development and autism. Cell Rep 2014;6:1139-52. 10.1016/j.celrep.2014.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]