Abstract
Background
The rice blast resistance gene Pi54 was cloned from Oryza sativa ssp. indica cv. Tetep, which conferred broad-spectrum resistance against Magnaporthe oryzae. Pi54 allelic variants have been identified in not only domesticates but also wild rice species, but the majority of japonica and some indica cultivars lost the function.
Results
We here found that Pi54 (Os11g0639100) and its homolog Os11g0640600 (named as #11) were closely located on a 25 kbp region in japonica cv. Sasanishiki compared to a 99 kbp region in japonica cv. Nipponbare. Sasanishiki lost at least six genes containing one other R-gene cluster (Os11g0639600, Os11g0640000, and Os11g0640300). Eight AA-genome species including five wild rice species were classified into either Nipponbare or Sasanishiki type. The BB-genome wild rice species O. punctata was Sasanishiki type. The FF-genome wild rice species O. brachyantha (the basal lineage of Oryza) was neither, because Pi54 was absent and the orientation of the R-gene cluster was reversed in comparison with Nipponbare-type species. The phylogenetic analysis showed that #11gene of O. brachyantha was on the root of both Pi54 and #11 alleles. All Nipponbare-type Pi54 alleles were specifically disrupted by 143 and 37/44 bp insertions compared to Tetep and Sasanishiki type. In addition, Pi54 of japonica cv. Sasanishiki lost nucleotide-binding site and leucine-rich repeat (NBS–LRR) domains owing to additional mutations.
Conclusions
These results suggest that Pi54 might be derived from a tandem duplication of the ancestor #11 gene in progenitor FF-genome species. Two divergent structures of Pi54 locus caused by a mobile unit containing the nearby R-gene cluster could be developed before domestication. This study provides a potential genetic resource of rice breeding for blast resistance in modern cultivars sustainability.
Electronic supplementary material
The online version of this article (10.1186/s12284-018-0256-8) contains supplementary material, which is available to authorized users.
Keywords: Oryza, Pi54 alleles, Resistance gene, Blast disease, Evolution, Rice breeding
Background
Oryza sativa, including two major subspecies japonica and indica, is the staple food for half the world and of pivotal importance in worldwide food production and security (Chang 1976; Lu 1999; Ammiraju et al. 2006). It is estimated that we need to produce 60% more rice between 2010 and 2050 to meet increasing demand (Alexandratos and Bruinsma 2012; Saito et al. 2017). Plant diseases are threatening the crops productions worldwide. For example, rice blast disease, caused by the fungus Magnaporthe oryzae (M. oryzae), is one of the most damaging rice diseases, and rice yield loss was up to 100% (Liu et al. 2014). Plants have evolved sophisticated defense response to resist pathogens infection over the course of evolution. The first layer of plant defense is pathogen-associated molecular patterns triggered immunity, and the second layer is effector-triggered immunity, which is mediated by plant resistance genes (R-genes) (Dangl et al. 2013). Most R-genes encode proteins with nucleotide-binding site and leucine-rich repeats (NBS–LRR) domains (Gay et al. 1991; Hammond-Kosack and Jones 1997; Song et al. 1997; Kobe and Kajava 2001; McHale et al. 2006). Multiple R-genes are located in clusters and all these clusters were abundant on chromosome 11 of rice genome (Zhou et al. 2004; Yang et al. 2006; Zhang et al. 2014; Vasudevan et al. 2015; Ashkani et al. 2016).
In recent rice-breeding programs, pyramiding of R-genes has been an effective strategy for achieving durable resistance in commercial crops (Ashkani et al. 2016; Xiao et al. 2017). Until now, over 100 major blast R-genes against M. oryzae have been identified, but only 30 of them have been cloned and characterized (Wang et al. 2017). Nearly all of the cloned R-genes encodes NBS–LRR proteins except Pid2, which encodes a receptor-like kinase (Chen et al. 2006). Some R-genes against rice blast disease such as Pi1, Piz-5, Pita, and Pi5, have been introgressed into agronomically superior rice cultivars by marker-assisted selection (Hittalmani et al. 2000; Narayanan et al. 2002; Liu et al. 2003; Lee et al. 2009). Marker-assisted backcrossing has been used with the blast resistance genes Piz-5 and Pi54 to develop improved restorers, Pusa1602 (with Piz-5) and Pusa1603 (with Pi54) (Singh et al. 2012).
The blast resistance gene Pi54 (also known as Pi-kh), encoding an NBS–LRR protein, was initially identified and cloned from the indica cv. Tetep; it confers broad-spectrum resistance against Indian rice blast isolates (Rai et al. 2011). Nowadays, many blast resistance alleles of Pi54 were also cloned from various wild rice species providing a high degree of resistance to M. oryzae (Das et al. 2012; Devanna et al. 2014). Moreover, Pi54 allelic variants, including resistant and susceptible genotype, have been detected through sequence-based allele mining. The studies revealed that the numbers of coding DNA sequences (CDSs) at the Pi54 varies from 0 to 3 and the predicted proteins consist of 73 to 486 amino acid (AA) residues (Thakur et al. 2015). Specifically, the LRR domain showed a high level nucleotide variation and the selection pressure was high in this domain (Kumari et al. 2013; Thakur et al. 2015). The NBS domain of Pi54 alleles also revealed diversity with amino acid sequence polymorphism (Kumari et al. 2013). However, it is still unclear how Pi54 and its locus diversity has evolved and spread in genus Oryza.
Recent Oryza genome projects have clarified the domestication history of rice (Zhang et al. 2014; Stein et al. 2018). The 27 Oryza species are divided into 11 genome types, 6 of which are diploid (n = 12: AA, BB, CC, EE, FF and GG) and 5 of which are polyploid (n = 24: BBCC, CCDD, HHJJ, HHKK and KKLL). Two species, O. sativa in Asia ~ 10,000 years ago and O. glaberrima in Africa ~ 3000 years ago, have been independently domesticated as rice. Cultivated rice belongs to the AA genome group. Here, we report two considerably divergent structures of the Pi54 gene and its locus in two O. sativa ssp. japonica domesticated cultivars Nipponbare and Sasanishiki. We compared these structures with those in six AA-genome species (O. nivara, O. glumaepatula, O. barthii, O. glaberrima, O. rufipogon, and O. meridionalis), one BB-genome species O. punctata, and one FF-genome species O. brachyantha including not only domesticated rice but also wild species. Our analysis revealed origin and evolution of the Pi54 gene and its locus in genus Oryza and provided potential genetic resources for breeding for rice blast resistance in modern cultivars.
Methods
Plant materials, growth conditions, DNA extraction, and genomic information
Oryza sativa ssp. japonica cultivars, including Hitomebore, Nipponbare, and Sasanishiki, were grown in pots and kept in a growth chamber at 30 °C during the day and 22 °C at night with a 12-h photoperiod after 3 days germinating at 30 °C. DNA was extracted from fresh leaves using a modified cetyl trimethylammonium bromide (CTAB) protocol (Teranishi et al. 2004). The genomic information of Nipponbare, O. glumaepatula (GEN1233_2), O. nivara (IRGC100897), O. barthii (IRGC105608), O. glaberrima (IRGC:96717), O. rufipogon (W1943), O. meridionalis (W2112), O. punctata (IRGC105690), O. brachyantha (IRGC101232), and O. sativa ssp. indica (9311) was downloaded from EnsemblPlants (http://plants.ensembl.org/index.html). The genomic information of Pi54 reference allele of Tetep is available from GenBank with accession number CCD33085.
PCR analyses
A primer set Pi54 MAS (Additional file 1: Table S1) (Ramkumar et al. 2010) was used to identify the difference of Pi54 allele between Nipponbare, Sasanishiki and Hitomebore. To detect the presence or absence of genes in Pi54 locus, in total 14 genes based on the Os-Nipponbare-Reference-IRGSP-1.0 reference genome (International Rice Genome Sequencing Project, IRGSP), primer sets were prepared and listed in Additional file 1: Table S1. The 14 genes were named as #1 to #14. To identity Nipponbare and Sasanishiki-type Pi54 locus in 24 modern japonica cultivars (including Nipponbare, Sasanishiki, and Hitomebore), specific PCR primer sets were designed with two ways, either according to left-side and right-side border sequences of the divergent region of Pi54 locus or Sasanishiki-specific polymorphisms of Pi54 and #11 genes. The primer sequences were listed in Additional file 1: Table S2. All the primers were designed in Primer3Plus software (Untergasser et al. 2007). PCR condition was using the following temperature profile: initial DNA denaturation, 94 °C for 3 min; followed by 35 cycles of denaturation, 98 °C for 10s; annealing, 68 °C for 15 s; extension, 68 °C for 30s or 1 min (according to the product size); and final extension at 72 °C for 5 min and then hold at 4 °C using PrimerSTAR GXL DNA polymerase (Takara, Japan).
Sequence analysis of the Pi54 locus of Sasanishiki using a BAC clone library
A high-density Sasanishiki bacterial artificial chromosome (BAC) library in the plndigoBAC-5 (Takano et al. 2013) was used. A BAC clone harboring gene loci from Os11g0638700 (named as #1) to Os11g0641300 (named as #14) was obtained and filtered using the primer sets Os11g0639000 (named as #3) and Os11g0640800 (named as #12) (Additional file 1: Table S1). The sequencing primers for confirming exact location of the inserted fragment in BAC clone were pIB FP and pIB RP. The selected BAC clone was used to extract the plasmid in a large-construct DNA purification kit (NucleoBond Xtra BAC). The position of the inserted Sasanishiki fragment is 25,220,854 to 25,448,655 bp on chromosome 11 according to the reference genome; it harbors the genes from #1 (25,237,345 to 25,239,585) to #14 (25,399,957 to 25,400,721). The region containing Pi54 (#4)–#12 was amplified from this clone by long-range PCR. The obtained PCR product was subcloned and sequenced by ABI3130 Genetic Analyzer with a BigDye Terminator Sequence Ready kit (Applied Biosystems, http://www.appliedbiosystems.com). Sequencing primers are listed in Additional file 1: Tables S3 and S4.
Multiple sequence alignments were generated in GenomeMatcher (Ohtsubo et al. 2008) and CLUSTALX2 (Larkin et al. 2007) software. Gene loci of the sequenced fragment was predicted by Augustus software (http://bioinf.uni-greifswald.de/augustus/submission.php). Sequence similarity analysis was performed with the ncbi-blast-2.6.0 tool (ftp://ftp.ncbi.nlm.nih.gov/blast/executables/blast+/LATEST). The threshold expectation value was set to 10− 4, which was determined empirically to filter out most of the spurious hits. The translated AA sequence of each of the predicted genes of Sasanishiki and of each of genes in Nipponbare (Os11g0638700 to Os11g0641300) was used as a query against the genome of the other accessions that was downloaded from EnsemblPlants.
Structural and phylogenetic analyses of the homologous genes Pi54 and #11
The protein sequences encoded by two predicted Sasanishiki genes were acquired in Augustus. The NBS and LRR domains were searched using the CDART (https://www.ncbi.nlm.nih.gov/Structure/lexington/lexington.cgi?cmd=rps) and Pfam (http://pfam.xfam.org/) tools. The coiled-coil (CC) domain was predicted using the COILS server (http://embnet.vital-it.ch/software/COILS_form.html). Repetitive elements were analyzed using the RepeatMasker Web Server (http://www.repeatmasker.org/cgi-bin/WEBRepeatMasker). Information on transposable elements was downloaded from the Repbase database (http://www.girinst.org/repbase/).
To assess the evolutionary relationship of the conserved DNA sequences of alleles Pi54 and #11, molecular phylogenetic analysis was performed by the Maximum Likelihood method (ML) based on the JTT matrix-based model (Jones et al. 1992) in MEGA 7 software (Kumar et al. 2016). DNA sequences (21 in total) from Nipponbare, Sasanishiki, O. glumaepatula, O. nivara, O. barthii, O. glaberrima, O .meridionalis, O. rufipogon, O. punctata, O. brachyantha and O. sativa ssp. indica 9311 and Tetep were used.
Results
Divergent structures of the Pi54 locus in O. sativa ssp. japonica cultivars
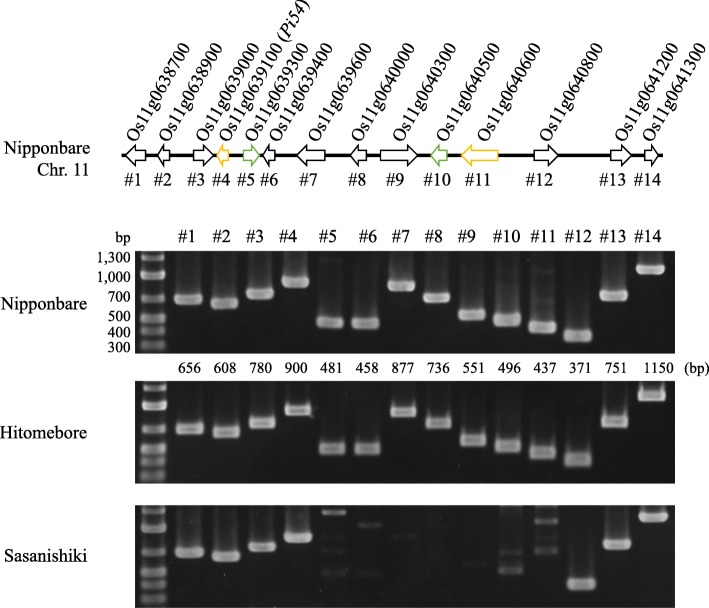
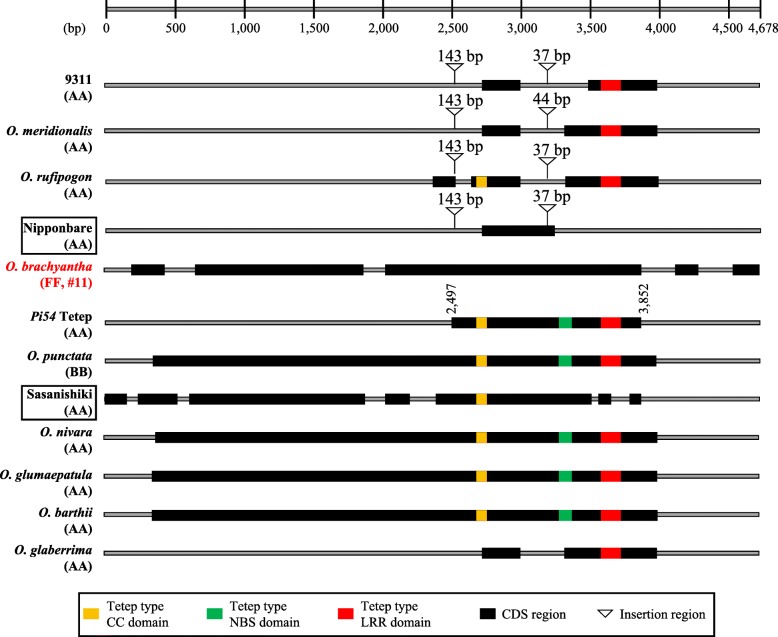
A PCR-based co-dominant molecular marker, Pi54 MAS, has been developed targeting a 144 bp insertion/deletion polymorphism in the exon of Pi54 gene (Ramkumar et al. 2010). Pi54 MAS is able to distinguish resistant (without 144 bp insertion) and susceptible genotype (with 144 bp insertion). To observe the Pi54 alleles (Os11g0639100: named as #4) among O. sativa ssp. japonica cv. Nipponbare, Sasanishiki, and Hitomebore, we performed PCR amplification with Pi54 MAS using genomic DNA. The results clearly indicated that Nipponbare and Hitomebore harbored the insertion but no such insertion was found in Sasanishiki (Additional file 1: Figure S1). To analyze the Pi54 locus in Sasanishiki, we designed 14 primer sets to detect genes Os11g0638700 (#1) to Os11g0641300 (#14) in Nipponbare, Sasanishiki, and Hitomebore genome (Materials and method; Additional file 1: Table S1). All expected fragments were amplified successfully from the Nipponbare and Hitomebore genomes, but specific amplifications of genes from Os11g0639300 (#5) to Os11g0640600 (#11) failed in Sasanishiki genome (Fig. 1). To obtain the genomic fragment of Sasanishiki Pi54 locus, we performed long-range PCR with the forward primer of Pi54 and the reversed primer of #12 (Additional file 1: Table S1), but PCR amplification failed using Sasanishiki genomic DNA directly. Therefore, we screened a 23 K BAC clone library of the Sasanishiki genome to analyze the structure of the Pi54 locus in Sasanishiki (Takano et al. 2013). We finally isolated a positive clone (plate No. 48, P12), sequenced it and found that this 25,061 bp nucleotide fragment contained 4 genes (Pi54, an unknown gene, #11, and #12; Sasanishiki type), while the 98,798 bp Nipponbare fragment included 9 genes (Nipponbare type) (Additional file 1: Figure S2). The genes from #5 to Os11g0640500 (#10) were lost in Sasanishiki. The genes #1, Os11g0638900 (#2), Os11g0639000 (#3), Os11g0641200 (#13), and #14 of Sasanishiki were more than 98% identical to those of Nipponbare, while Pi54, #11, and #12 were less than 93% identical (Table 1). The unknown gene found between Pi54 and #11 in Sasanishiki was absent in the Nipponbare genome.
Fig. 1.
PCR analysis of 14 genes (Os11g0638700–Os11g0641300) in Nipponbare, Hitomebore and Sasanishiki. Marker fragment sizes are shown on the left PCR product sizes are shown at the bottom of the Nipponbare pane
Table 1.
Comparison of the sequences of genes Os11g0638700 to Os11g0641300 between Nipponbare and Sasanishiki
| Sequence alignments | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Nipponbare | Sasanishiki | NT | AA | ||||||
| Gene_list | NTa | AAb | Gene_list | NT | AA | E-values c | Identities | E-values | Identities |
| Os11g0638700 (#1) | 2241 | 746 | #1 | 2260 | 648 | 0 | 2237/2241 (99%) | 0 | 648/712 (91%) |
| Os11g0638900 (#2) | 1835 | 180 | #2 | 1835 | 180 | 0 | 1833/1835 (99%) | 4E-136 | 180/180 (100%) |
| Os11g0639000 (#3) | 4349 | 173 | #3 | 4268 | 222 | 0 | 2882/2933 (98%) | 4E-088 | 124/151 (82%) |
| Os11g0639100 (#4) | 941 | 112 | #4 | 3735 | 920 | 0 | 864/939 (92%) | 1E-049 | 88/100 (88%) |
| – | – | – | Unknown | 2659 | 281 | – | – | – | – |
| Os11g0639300 (#5) | 2276 | 726 | – | – | – | – | – | – | – |
| Os11g0639400 (#6) | 722 | 107 | – | – | – | – | – | – | – |
| Os11g0639600 (#7) | 4823 | 935 | – | – | – | – | – | – | – |
| Os11g0640000 (#8) | 1947 | 648 | – | – | – | – | – | – | – |
| Os11g0640300 (#9) | 4157 | 1118 | – | – | – | – | – | – | – |
| Os11g0640500 (#10) | 2196 | 731 | – | – | – | – | – | – | – |
| Os11g0640600 (#11) | 6464 | 1101 | #11 | 8509 | 924 | 0 | 2593/3179 (82%) | 8E-122 | 224/410 (55%) |
| Os11g0640800 (#12) | 3842 | 620 | #12 | 2086 | 623 | 0 | 1720/1843 (93%) | 0 | 552/620 (89%) |
| Os11g0641200 (#13) | 5361 | 358 | #13 | 5313 | 616 | 0 | 5271/5301 (99%) | 0 | 306/353 (87%) |
| Os11g0641300 (#14) | 765 | 236 | #14 | 913 | 236 | 0 | 764/765 (99%) | 1E-174 | 236/236 (100%) |
aNT, nucleotide sequence
bAA, amino acid sequence
cE-values, expectation values
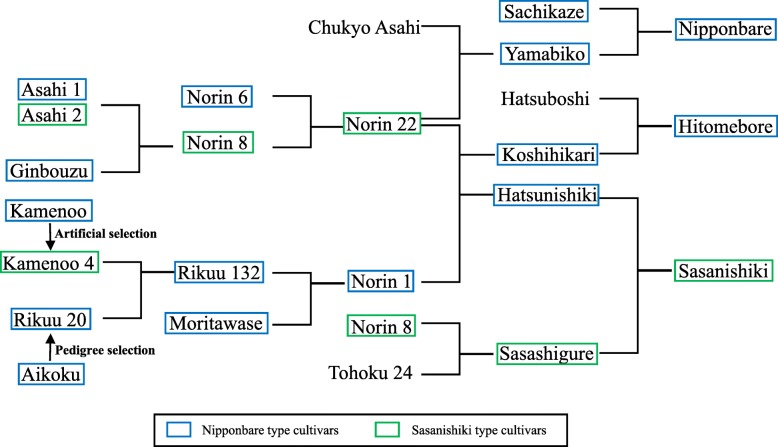
To analyze the breeding history of the Pi54 locus, we designed specific PCR primer sets to amplify the upstream and downstream border sites and found that the divergent structures of the Pi54 locus are randomly distributed in the parental lines of Nipponbare, Sasanishiki, and Hitomebore (Additional file 1: Figure. S3, Fig. 2). The modern rice breeding in Japan was from Norin cultivars. Intriguingly, both types were found in early lines (Asahi 1 and Asahi 2, and Kamenoo, and Kamenoo 4) established before modern breeding more than 100 years ago (Fig. 2). These two types were also found in japonica rice cultivated in China with Hokushitami and Zaijian being the Sasanishiki type and Hexi 23 the Nipponbare type. These results suggest that each type of the Pi54 locus was stably and widely spread among japonica cultivars before modern artificial breeding history.
Fig. 2.
The breeding history of modern O. sativa ssp. japonica cultivars Nipponbare, Hitomebore, and Sasanishiki was obtained from Agriknowledge database (https://agriknowledge.affrc.go.jp/). The modern rice breeding in Japan was from Norin cultivars
Pi54 locus in the genus Oryza
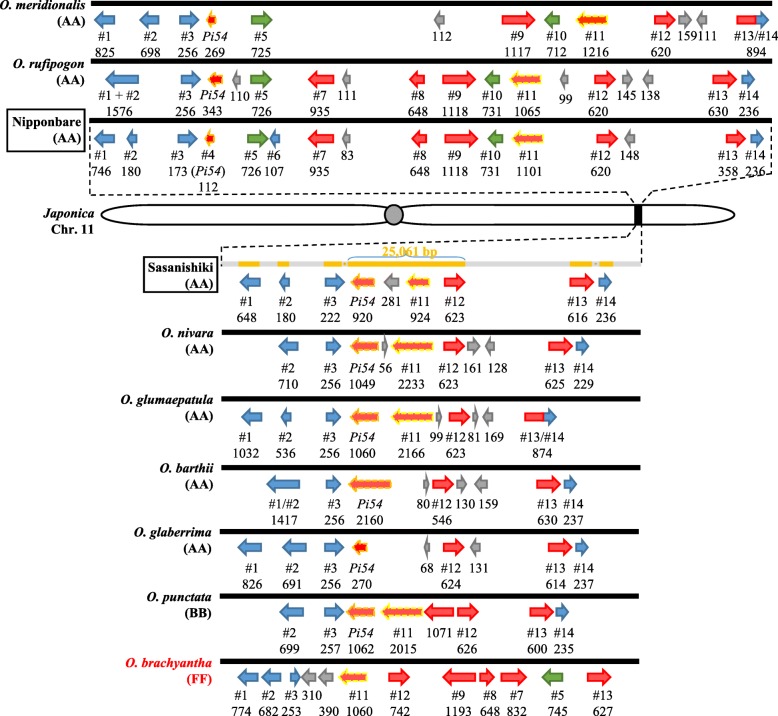
Next, we investigated whether the two types of the Pi54 locus are conserved. The Pi54 locus of six AA-genome species was Nipponbare type in O. rufipogon and O. meridionalis and Sasanishiki type in O. nivara, O. glumaepatula, O. barthi, and O. glaberrima (Fig. 3). The synteny from Pi54 to #12 was conserved within the types and the synteny of both flanking regions (#1 to Pi54 on the left side and #12 to #14 on the right side) was highly conserved in all AA-genome species (Fig. 3). A multiple alignment analysis of DNA sequences from Pi54 to #12 showed more than 90% identity within each type but little identity between the Nipponbare and Sasanishiki types (Fig. 4). The Nipponbare type had a long insertion containing genes #5 to #10 in comparison with the Sasanishiki type. In the African wild rice O. barthii and O. glaberrima, as well as in Nipponbare type species O. rufipogon (Asian) and O. meridionalis (Australian), different insertions and deletions were found (Fig. 4). Altogether, these results indicate that the two divergent Pi54 loci developed before the establishment of AA genome species.
Fig. 3.
Genes homologous to #1 to #14. Nipponbare: the region corresponding to 25,237,345–25,400,721 bp on chromosome 11 of Os-Nipponbare-Reference-IRGSP-1.0. Arrows: red, NBS–LRR family-like genes; blue, miscellaneous genes; gray, presumed genes; green, translocated genes. Arrows with orange and/or yellow dashed (alleles of the Pi54 and #11, respectively) line show genes that were used to conduct phylogenetic tree. Protein size (AA) is indicated under each gene ID. Gray bars in Sasanishiki correspond to unsequenced regions
Fig. 4.
Multiple sequence alignment of divergent structures in the Pi54 locus. DNA sequences from Pi54 to #12 in 10 species were extracted and analyzed in GenomeMatcher software (Ohtsubo et al. 2008). The corresponding region in the reference genome Os-Nipponbare-Reference-IRGSP-1.0 is 25,263,336–25,362,133 bp on chromosome 11. Red arrows represent gene loci
The structure of the Pi54 locus in O. punctata (BB-genome) lacked the region from genes #5 to #10, and thus was Sasanishiki type (Figs. 3, and 4), whereas that in O. brachyantha (FF-genome) consisted of genes #1, #2, #3, #11, #12, #9, #8, #7, #5, and #13, but Pi54 and #10 were absent. In comparison with other Nipponbare type species, the orientation of #9, #8, #7, and #5 in O. brachyantha was reversed (Figs. 3, and 4). Thus, in FF-genome species, the Pi54 locus had characters of both the Sasanishiki (close location of #3 and #11) and Nipponbare type (presence of #5, #7, #8 and #9).
Diversity of the homologous genes Pi54 and #11
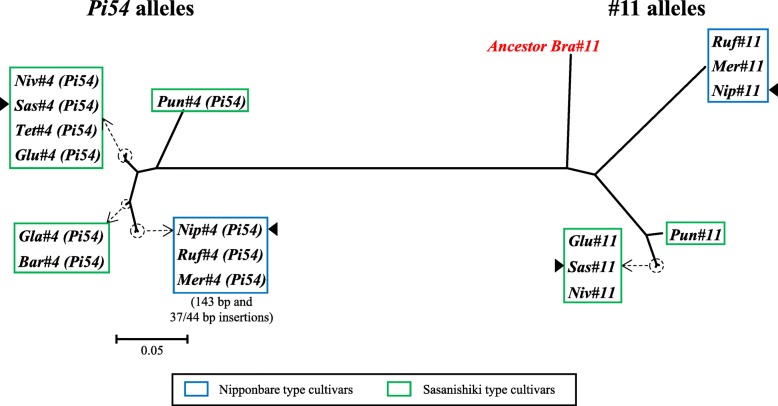
In Nipponbare, in comparison with a 112 AA protein of gene Pi54, its homolog #11 (EnsemblPlants database) is a 1101 AA protein. Genome-wide blast analysis showed that all six AA-genome wild species analyzed carry orthologues of both Pi54 and #11. Phylogenetic analysis showed that Pi54 and #11 of Nipponbare were most closely related to those of Nipponbare type species O. rufipogon (Asian) and O. meridionalis (Australian) (Fig. 5). In contrast, Pi54 and #11 of Sasanishiki were very similar to those of Sasanishiki type species, such as O. nivara (Asian) and O. glumaepatula (South American). The Pi54 alleles of African O. barthii and O. glaberrima (Sasanishiki type species) was the sister group only of Nipponbare type species, and the lowest common ancestor of them was close to other Sasanishiki type species (such as O. nivara and O. glumaepatula).
Fig. 5.
A maximum-likelihood phylogenetic tree showing relationships among the alleles of the Pi54 and #11 genes. The tree contains 18 genomic DNA sequences both coding and non-coding. Sequences of 10 Pi54 alleles were extracted according to the functional Pi54 sequence in Tetep. Sequences of 8 alleles of #11 are complete coding DNA sequence (CDS). Nip, Nipponbare; Sas, Sasanishiki; Tet, Tetep; Glu, O. glumaepatula; Niv, O. nivara; Bar, O. barthii; Gla, O. glaberrima; Ruf, O. rufipogon; Mer, O. meridionalis; Pun, O. punctata; Bra, O. brachyantha. Locations of Pi54 alleles on chromosome 11: Nip, 25,263,336–25,264,503; Ruf, 25,053,651–25,054,821; Mer, 21,747,946–21,749,123; Glu, 24,126,550–24,127,540; Niv, 22,154,478–22,155,468; Bar, 20,525,896–20,526,886; Gla, 18,706,523–18,707,513; Pun, 25,829,456–25,830,446. Locations of #11 alleles on chromosome 11: Nip, 25,338,884–25,345,347; Ruf, 25,125,041–25,131,339; Mer, 21,841,707–21,846,125; Glu, 24,139,300–24,147,809; Niv, 22,167,498–22,176,039; Bra, 14,455,855–14,459,981
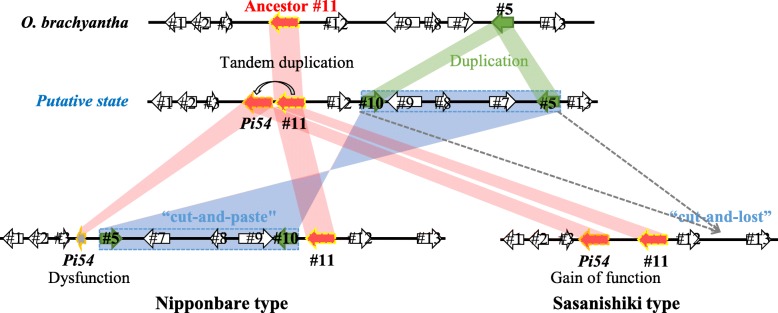
We also performed phylogenetic analysis of Pi54 and its homolog #11 with O. punctata (BB-genome) and O. brachyantha (FF-genome) that is placed in the basal lineage in Oryza (Chen et al. 2013) (Fig. 5). The divergence time between AA- and BB-genome species is estimated as more than 6 million years ago (Mya) and that between FF-and AA-BB genome species as 15 Mya (Stein et al. 2018). We found that #11 gene of O. brachyantha was on the root of both Pi54 alleles and #11 alleles, while #11 of O. punctata was relatively close to Sasanishiki type species. In O. brachyantha (FF), Pi54 was not detected in the expected region. In contrast, in O. punctata (BB), which was close to the root of all Pi54 alleles, Pi54 and an additional gene between #11 and #12 were found (Fig. 4). To sum up, we suggested that #11 of the FF-genome species was duplicated and Pi54 originated from the duplicated before the divergence of FF- and AA-BB- genome species, and that this duplication was caused by natural rather than artificial selection.
Insertions (143 bp and 37/44 bp) were found to be conserved in the Pi54 alleles of 9311, O. rufipogon and O. meridionalis (Nipponbare type), but not in Tetep, O. nivara, O. glumaepatula, O. barthii, or O. glaberrima (Sasanishiki type) (Fig. 6, Additional file 1: Figure S4). A 143 bp insertion is associated with susceptibility to blast disease (e.g., Nipponbare and Swarna), and resistant genotypes have no such insertion (e.g., Tetep and Suraksha) (Ramkumar et al. 2010). We found that the sequence of this insertion has high similarity to the non-autonomous DNA transposon Helitron-N91 (Additional file 1: Figure S5). The 37 or 44 bp insertion results in a loss of the NBS domain (Fig. 6). Sasanishiki lacks these insertions but has other mutations in the NBS and LRR domains, which may result in loss of Pi54 function (Fig. 6).
Fig. 6.
Schematic representation of DNA sequences of Pi54 alleles. The sequences of Nipponbare, O. rufipogon, O. meridionalis, O. sativa ssp. indica (9311 and Tetep), and O. glaberrima include the gene sequences and 800 bp upstream sequences, while the other 5 sequences are gene sequences only (starting from ATG). CC, coiled coil
Interspersed repeats of the Pi54 locus in Oryza species
To better understand the divergent structures of the Pi54 locus, we analyzed interspersed repeat sequence in the region from Pi54 to #12 (exception: from #11 to #5 of in O. brachyantha) (Table 2). All AA-genome species harbored a high level of interspersed repeats. For example, interspersed repeats occupied 55.64% of 99 kbp in Nipponbare and 33.43% of 25 kbp in Sasanishiki. In comparison with AA-genome species, the percentages of repetitive sequence of the ancestral species O. brachyantha (FF-genome, #11 to #5, 32 kbp) and O. punctata (BB-genome, Pi54 to #12, 22 kbp) were significantly low, which occupied 10.90% and 3.17%, respectively. This high level of repetitive sequence in the Pi54 locus of AA-genome species potentially lead to considerable diversity between Nipponbare and Sasanishiki type species. To further understand evolution of duplicated genes (#4 and #11; #5 and #10) in the Pi54 locus, we analyzed transposons in the flanking regions of these duplicated genes in O. brachyantha, Nipponbare type species Nipponbare, O. rufipogon, and O. meridionalis, and Sasanishiki type species O. nivara. 26 non-autonomous transposons of both retroelements and DNA transposons were identified in total (Additional file 1: Table S5). In particular, the number of transposon that located on upstream of #5 is significantly higher than the other flanking regions of duplicated genes. Thus, the high level of interspersed repeats and a great number of transposons in the Pi54 locus of AA-genome species may reflect underlying mechanisms of establishment of divergent structures and also increase genome instability.
Table 2.
Interspersed repeat sequence analysis of divergent structures in the Pi54 locus
| Species | Length (bp) | Percentage of sequence | |||
|---|---|---|---|---|---|
| Retroelements | DNA transposons | Unclassified | Total interspersed repeats | ||
| O. meridionalis (AA) | 112,929 | 9.34% | 21.09% | 6.81% | 37.24% |
| O. rufipogon (AA) | 92,702 | 13.45% | 21.07% | 18.34% | 52.86% |
| Nipponbare (AA) | 98,798 | 12.73% | 21.35% | 21.56% | 55.64% |
| O. brachyantha (FF) | 32,280 | 7.59% | 2.37% | 0.94% | 10.90% |
| O. punctata (BB) | 22,183 | 1.58% | 1.59% | 0.00% | 3.17% |
| Sasanishiki (AA) | 25,061 | 27.36% | 5.99% | 0.09% | 33.43% |
| O. nivara (AA) | 25,651 | 25.92% | 5.11% | 0.63% | 31.66% |
| O. glumaepatula (AA) | 22,674 | 18.87% | 5.45% | 0.10% | 24.42% |
| O. barthii (AA) | 16,743 | 0.00% | 19.76% | 1.84% | 21.60% |
| O. glaberrima (AA) | 26,686 | 6.76% | 39.67% | 2.13% | 48.56% |
Discussion
The evolutionary mechanisms of R-genes
The mechanism of interaction between host plants with R-genes encoding NBS–LRR protein and pathogen is explained by gene-for-gene resistance theory (Van Der Biezen and Jones 1998; McHale et al. 2006). R and Avr genes have co-evolved with natural selection in host–pathogen interactions (Sharma et al. 2012). Rapid evolution of the fungus M. oryzae occurs through non-synonymous variations, which frequently result in gain or loss of function of the Avr genes (Xue et al. 2012; Huang et al. 2014; Zhang et al. 2015). Resistant plants have acquired high levels of allelic diversity, new R-genes or copy number variations (CNVs) thereby increasing the durability of resistance (Yu et al. 2011; Jacob et al. 2013; Wang et al. 2014). In Oryza, the copy number of R-genes is markedly higher in both the indica and japonica cultivars than in wild rice, probably because of artificial selection for increased R-gene diversity and CNVs (Zhang et al. 2014; Stein et al. 2018). For example, 631 R-genes are predicted in the Nipponbare genome but only 307 in the genome of wild species O. brachyantha (Zhang et al. 2014; Zhu et al. 2014).
In this study, we found that an R-gene cluster located on the Pi54 locus. In terms of Pi54 gene of this locus, the family size is 2.8 ± 1.1 genes per species in 13 Oryza species, considerably smaller than the sizes of the RGA4 (9.8 ± 3.1), RGA5 (22.5 ± 5.2), Pik1 (5.6 ± 1.6), and Pik2 (18.7 ± 3.5) families (Stein et al. 2018). Suggested that Pi54 family was conserved between modern and ancestral Oryza species with a small and stable gene family size. Therefore, to enhance the durability of rice blast resistance, Pi54 may evolve through high levels of allelic diversity rather than CNVs. In terms of R-genes #7 to #9 that encode NBS–LRR proteins, the copy number of these three genes in Nipponbare is 36, 8, and 3, respectively, in contrast to 26, 5, and 4 in the progenitor O. brachyantha (EnsemblPlants database). Suggested that R-genes #7 to #9 might evolve with either high levels of allelic diversity or CNVs. To sum up, many R-genes could evolve with high levels of allelic diversity as Pi54. Thus, for Pi54-like R-genes, the study of exploring allelic diversity would be essential.
An evolutionary model of two divergent structures of the Pi54 locus
To gain insights into evolution of the two divergent structures of the Pi54 locus, we conducted genome-wide blast analysis (Fig. 3) and a phylogenetic analysis (Fig. 5) and deduced evolutionary history (Fig. 7). In the FF-genome species O. brachyantha, only #11 gene was localized in the Pi54 locus but its homolog Pi54 gene was undetectable in the whole genome. In addition, the phylogenetic analysis revealed that #11 gene of O. brachyantha was on the root of both Pi54 and #11 alleles in other Oryza species. Taken together, we assumed that a tandem duplication of the ancestral #11 gene in progenitor FF-genome species led to the emergence of Pi54. Gene #5 with a domain of unknown function DUF594 was also duplicated, leading to the emergence of #10, which was then inserted in inverted orientation downstream of #9. The genes surrounded by homologous genes #5 and #10 might have become a mobile unit. In the Nipponbare type, this unit was integrated between Pi54 and #11 in inverted orientation by “cut-and-paste” and might result in a shortening of the N-terminal portion of Pi54 gene. As with the opposite orientation of the region from #5 to #10 (~ 60 kbp) in O. brachyantha, some other paracentric inversions have been reported between AA- and FF-genome species either (Stein et al. 2018). In the Sasanishiki type, this unit was “cut-and-lost” (Fig. 7). Besides, the Pi54 locus of Nipponbare and Sasanishiki type were firstly observed in wild species O. meridionalis (AA-genome) and O. punctata (BB-genome), respectively. The mean AA-BB divergence time was 6.76 Mya, while the split time between O. meridionalis and other AA-genome species was 2.41 Mya (Stein et al. 2018). Along with domestication of O. sativa and O. glaberrima starting from ~ 10,000 and ~ 3000 years ago separately, these findings revealed that these two divergent Pi54 loci had been developed before domestication. Taken together, in the evolutionary model, the duplication events are important factors in the generation of gain-of-function genes, including R genes (Hulbert et al. 2001; Leister 2004; Guo et al. 2011). The evolution of the two divergent structures of the Pi54 locus revealed an ongoing birth and death process in R-genes.
Fig. 7.
Model of the evolution of the two types of Pi54 locus. In particular, the Pi54 allele in Sasanishiki has lost its function because of several SNPs and small insertions and deletions. Dashed rectangle represents the mobile unit
Intriguingly, a similar unit (the same pattern as #5 and #10 with the orientation of genes toward each other and several genes between them) surrounded by OPUNC01G04010 and OPUNC01G04050 of the #5 gene family exists on chromosome 1 in the BB-genome species O. punctata (Additional file 1: Table S6). This unit is absent in several AA-genome species (O. sativa cv. Nipponbare, O. rufipogon, O. meridionalis, O. nivara, and O. glumaepatula), while the flanking sequences of this unit are well conserved. In the FF-genome species O. brachyantha, linkage of a single gene of the #5 gene family (OB01G13970) to the flanking sequences strongly supports our evolutionary model of the two divergent structures of the Pi54 locus. In Nipponbare genome, the #5 gene family is expanded and 35 homologous genes are identified (EnsemblPlants database). It may contribute instability of rice genomes.
Surviving genus Oryza with the divergent Pi54 loci
Pi54 alleles with high levels of tolerance to rice blast disease were identified and clone from not only domesticated but also wild rice species (Das et al. 2012; Devanna et al. 2014). In case of domesticated rice, it has been reported that the frequency of the functional Pi54 genotype is higher in the indica subspecies (up to 87.9%) than in javanica (8.6%, now known as tropical japonica) and japonica (3.5%) (Vasudevan et al. 2015). Even in japonica cv. Sasanishiki that the nucleotide sequence of Pi54 allele is similarity to indica cv. Tetep, a dysfunctional Pi54 genotype appeared because of accumulation of polymorphisms and small insertions or deletions (Fig. 6). In case of wild rice species, we found that Pi54 alleles in O. rufipogon and O. meridionalis (distributed in East Asia and Oceania (Stein et al. 2018)) were dysfunctional because of small (143 bp and 37/44 bp) insertions or a mobile unit surrounded by the homologous genes #5 and #10.
Recently, Zhong et al. (2018) found that the blast fungi M. oryzae were grouped into three major globally distributed clades. In the clade 2, most pathogens were isolated from indica rice-growing area. In the clade 3, most were isolated from japonica rice-growing area. Each clade shows different mating type (Zhong et al. 2018). Therefore, Pi54 function may be essential for indica cultivars to resist the clade 2 pathogens but not for most japonica cultivars and several wild rice species, such as Nipponbare, O. rufipogon, and O. meridionalis. Taken together, we came up with a hypothesis of strategies of the divergent Pi54 loci. Sasanishiki type species might evolve with the functional Pi54 genotype at the cost of the loss of several NBS–LRR proteins (#7 to #9). In the Nipponbare type species, Pi54 alleles became a pseudogene, and three R-genes (#7 to #9) accumulated in the adjacent region as compensation. Both type Pi54 loci could confer genus Oryza with advantages surviving diverse pathogens distributed in different areas.
However, under global warming, pyramiding of the blast resistance gene Pi54 into japonica commercial cultivars would be urgent for rice breeding programs, because the geographic range of pathogens interacted with Pi54 alleles will expand owing to increasing temperatures (Stein et al. 2018). Our study of the Pi54 locus in Nipponbare and Sasanishiki type species provides evolutionary insights into the generation of diversity and a potential genetic resource of rice breeding for blast resistance in modern cultivars sustainability.
Conclusions
We found two divergent structures of Pi54 locus that one carried dysfunctional Pi54 gene but an R-gene cluster accumulated in the Pi54 locus as compensation in an approximately 99 kbp region and was defined as Nipponbare type. Another one only harbored functional Pi54 gene (except Sasanishiki) but at the cost of loss of the R-gene cluster in an around 25 kbp region and was considered as Sasanishiki type. Both of the divergent Pi54 loci are widely distributed in modern japonica cultivars and wild rice species, including AA-, BB-, and FF-genome species. Furthermore, these two divergent loci had been developed before domestication and thus was caused by natural selection rather than artificial selection. Together with phylogenetic analysis, we came up with an evolutionary model of the two divergent structures of Pi54 locus. This study could contribute to rice breeding programs and pave a novel way to understanding genetic evolution.
Additional file
Figure S1. Selection and identification of Pi54 alleles using primer set Pi54 MAS. Figure S2. Alignment of the divergent regions from Sasanishiki and Nipponbare. Figure S3. Detection of divergent structures at the Pi54 locus in modern O. sativa cultivars. Figure S4. DNA sequences of Pi54 alleles. Figure S5. Alignment of Helitron-N91 and the 143 bp insertion in Nipponbare. Table S1. Primer sets for PCR amplification. Table S2. Primer sets for identification of Nipponbare and Sasanishiki type species. Table S3 Primers for sequencing. Table S4. Primers for sequencing the genes flanking the 25,061 bp region in Sasanishiki. Accession codes. Sequence data of Sasanishiki the Pi54 locus. Table S5. Transposable elements in the flanking regions of Pi54, #5, #10 and #11. Table S6. Genes Orthologous to a mobile unit on chromosome 1 in Oryza species. (PDF 1280 kb)
Acknowledgements
We greatly appreciate Dr. Yoshiyuki Ohtsubo for the contributing to the data analysis by GenomeMatcher software. Rice seeds used in this study were provided by the Genebank Project, NARO (National Agriculture and Food Research Organization, Japan).
Funding
This work was funded in part by the Ministry of Education, Culture, Sports, Science and Technology (MEXT KAKENHI grant number 15H04616 and 18H03947).
Availability of data and materials
The datasets supporting the conclusions of this article are provided within the article and its additional files.
Abbreviations
- AA
Amino acid
- BAC
Bacterial artificial chromosome
- CC
Coiled-coil domain
- CDS
Coding DNA sequence
- CNV
Copy number variation
- CTAB
Cetyl trimethylammonium bromide
- LRR
Leucine-rich repeats domain
- NBS
Nucleotide-binding site domain
- R-gene
Resistance gene
Authors’ contributions
LZ performed the experiments and data analysis and co-wrote the paper. YN, TE, JH, and MT contributed to selecting rice materials and data analysis. SS performed data analysis and co-wrote the paper. AH designed the experiments and co-wrote the paper. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Alexandratos N, Bruinsma J. World agriculture towards 2030/2050: the 2012 revision. Rome: ESA Working paper FAO; 2012. [Google Scholar]
- Ammiraju JSS, Luo MZ, Goicoechea JL, et al. The Oryza bacterial artificial chromosome library resource: construction and analysis of 12 deep-coverage large-insert BAC libraries that represent the 10 genome types of the genus Oryza. Genome Res. 2006;16:140–147. doi: 10.1101/gr.3766306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashkani S, Rafii MY, Shabanimofrad M, et al. Molecular progress on the mapping and cloning of functional genes for blast disease in rice (Oryza sativa L.): current status and future considerations. Crit Rev Biotechnol. 2016;36:353–367. doi: 10.3109/07388551.2014.961403. [DOI] [PubMed] [Google Scholar]
- Chang T-T. The origin, evolution, cultivation, dissemination, and diversification of Asian and African Rices. Euphytica. 1976;25:425–441. doi: 10.1007/BF00041576. [DOI] [Google Scholar]
- Chen JF, Huang QF, Gao DY et al (2013) Whole-genome sequencing of Oryza brachyantha reveals mechanisms underlying Oryza genome evolution. Nat Commun 4. 10.1038/ncomms2596 [DOI] [PMC free article] [PubMed]
- Chen X, Shang J, Chen D, et al. A B-lectin receptor kinase gene conferring rice blast resistance. Plant J. 2006;46:794–804. doi: 10.1111/j.1365-313X.2006.02739.x. [DOI] [PubMed] [Google Scholar]
- Dangl J. L., Horvath D. M., Staskawicz B. J. Pivoting the Plant Immune System from Dissection to Deployment. Science. 2013;341(6147):746–751. doi: 10.1126/science.1236011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A, Soubam D, Singh PK, et al. A novel blast resistance gene, Pi54rh cloned from wild species of rice, Oryza rhizomatis confers broad spectrum resistance to Magnaporthe oryzae. Funct Integr Genomics. 2012;12:215–228. doi: 10.1007/s10142-012-0284-1. [DOI] [PubMed] [Google Scholar]
- Devanna Navadagi B., Vijayan Joshitha, Sharma Tilak R. The Blast Resistance Gene Pi54of Cloned from Oryza officinalis Interacts with Avr-Pi54 through Its Novel Non-LRR Domains. PLoS ONE. 2014;9(8):e104840. doi: 10.1371/journal.pone.0104840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay NJ, Packman LC, Weldon MA, Barna JCJ. A leucine-rich repeat peptide derived from the Drosophila toll receptor forms extended filaments with a β-sheet structure. FEBS Lett. 1991;291:87–91. doi: 10.1016/0014-5793(91)81110-T. [DOI] [PubMed] [Google Scholar]
- Guo Y-L, Fitz J, Schneeberger K, et al. Genome-wide comparison of nucleotide-binding site-leucine-rich repeat-encoding genes in Arabidopsis. Plant Physiol. 2011;157:757–769. doi: 10.1104/pp.111.181990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond-Kosack KE, Jones JDG. Plant disease resistance genes. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:575–607. doi: 10.1146/annurev.arplant.48.1.575. [DOI] [PubMed] [Google Scholar]
- Hittalmani S, Parco A, Mew TV, et al. Fine mapping and DNA marker-assisted pyramiding of the three major genes for blast resistance in rice. Theor Appl Genet. 2000;100:1121–1128. doi: 10.1007/s001220051395. [DOI] [Google Scholar]
- Huang Ju, Si Weina, Deng Qiming, Li Ping, Yang Sihai. Rapid evolution of avirulence genes in rice blast fungus Magnaporthe oryzae. BMC Genetics. 2014;15(1):45. doi: 10.1186/1471-2156-15-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulbert SH, Webb CA, Smith SM, Sun Q. Resistance gene complexes: evolution and utilization. Annu Rev Phytopathol. 2001;39:285–312. doi: 10.1146/annurev.phyto.39.1.285. [DOI] [PubMed] [Google Scholar]
- Jacob F, Vernaldi S, Maekawa T. Evolution and conservation of plant NLR functions. Front Immunol. 2013;4:1–16. doi: 10.3389/fimmu.2013.00297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Kobe B, Kajava AV. The leucine-rich repeat as a protein recognition motif. Curr Opin Struct Biol. 2001;11:725–732. doi: 10.1016/s0959-440x(01)00266-4. [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari A, Das A, Devanna BN, et al. Mining of rice blast resistance gene Pi54 shows effect of single nucleotide polymorphisms on phenotypic expression of the alleles. Eur J Plant Pathol. 2013;137:55–65. doi: 10.1007/s10658-013-0216-5. [DOI] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, et al. Clustal W and clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lee S.-K., Song M.-Y., Seo Y.-S., Kim H.-K., Ko S., Cao P.-J., Suh J.-P., Yi G., Roh J.-H., Lee S., An G., Hahn T.-R., Wang G.-L., Ronald P., Jeon J.-S. Rice Pi5-Mediated Resistance to Magnaporthe oryzae Requires the Presence of Two Coiled-Coil-Nucleotide-Binding-Leucine-Rich Repeat Genes. Genetics. 2009;181(4):1627–1638. doi: 10.1534/genetics.108.099226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leister D. Tandem and segmental gene duplication and recombination in the evolution of plant disease resistance genes. Trends Genet. 2004;20:116–122. doi: 10.1016/j.tig.2004.01.007. [DOI] [PubMed] [Google Scholar]
- Liu SP, Li X, Wang CY, et al. Improvement of resistance to rice blast in Zhenshan 97 by molecular marker-aided selection. Acta Bot Sin. 2003;45:1346–1350. [Google Scholar]
- Liu W, Liu J, Triplett L, et al. Novel insights into Rice innate immunity against bacterial and fungal pathogens. Annu Rev Phytopathol. 2014;52:213–241. doi: 10.1146/annurev-phyto-102313-045926. [DOI] [PubMed] [Google Scholar]
- Lu BR. Taxonomy of the genus Oryza (Poaceae): historical perspective and current status. Int Rice Res Notes. 1999;24:4–8. [Google Scholar]
- McHale L, Tan X, Koehl P, Michelmore RW. Plant NBS-LRR proteins: adaptable guards. Genome Biol. 2006;7:212. doi: 10.1186/gb-2006-7-4-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narayanan NN, Baisakh N, Vera Cruz CM, et al. Molecular breeding for the development of blast and bacterial blight resistance in Rice cv. IR50. Crop Sci. 2002;42:2072–2079. doi: 10.2135/cropsci2002.2072. [DOI] [Google Scholar]
- Ohtsubo Yoshiyuki, Ikeda-Ohtsubo Wakako, Nagata Yuji, Tsuda Masataka. GenomeMatcher: A graphical user interface for DNA sequence comparison. BMC Bioinformatics. 2008;9(1):376. doi: 10.1186/1471-2105-9-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai AK, Kumar SP, Gupta SK, et al. Functional complementation of rice blast resistance gene Pi-k(h)(Pi54) conferring resistance to diverse strains of Magnaporthe oryzae. J Plant Biochem Biotechnol. 2011;20:55–65. doi: 10.1007/s13562-010-0026-1. [DOI] [Google Scholar]
- Ramkumar G, Srinivasarao K, Mohan KM, et al. Development and validation of functional marker targeting an InDel in the major rice blast disease resistance gene Pi54 (Pik h ) Mol Breed. 2010;27:129–135. doi: 10.1007/s11032-010-9538-6. [DOI] [Google Scholar]
- Saito K, van Oort P, Dieng I, et al (2017) Yield gap analysis towards meeting future rice demand Faso; Ibrahim Bassoro and Delphine Mapiemfu Lamare, IRAD, Cameroon; Côte d’Ivoire; belay a. Bayuh, EIAR, Ethiopia; Famara Jaiteh, NARI, the Sékou, IRAG, Guinea; Raymond Rabeson, FOFIFA, Madagascar; Nia. In: achieving sustainable cultivation of rice volume 2. Burleigh Dodds science publishing, pp 189–214
- Sharma TR, Rai AK, Gupta SK, et al. Rice blast management through host-plant resistance: retrospect and prospects. Agric Res. 2012;1:37–52. doi: 10.1007/s40003-011-0003-5. [DOI] [Google Scholar]
- Singh VK, Singh A, Singh SP, et al. Incorporation of blast resistance into “PRR78”, an elite basmati rice restorer line, through marker assisted backcross breeding. F Crop Res. 2012;128:8–16. doi: 10.1016/j.fcr.2011.12.003. [DOI] [Google Scholar]
- Song WY, Pi LY, Wang GL, et al. Evolution of the rice Xa21 disease resistance gene family. Plant Cell. 1997;9:1279. doi: 10.1105/tpc.9.8.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein JC, Yu Y, Copetti D, et al. Genomes of 13 domesticated and wild rice relatives highlight genetic conservation, turnover and innovation across the genus Oryza. Nat Genet. 2018;50:285–296. doi: 10.1038/s41588-018-0040-0. [DOI] [PubMed] [Google Scholar]
- Takano N, Takahashi Y, Yamamoto M, et al. Isolation of a novel UVB-tolerant rice mutant obtained by exposure to carbon-ion beams. J Radiat Res. 2013;54:637–648. doi: 10.1093/jrr/rrt007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teranishi M, Iwamatsu Y, Kumagai T. Ultraviolet-B sensitivities in japanese lowland rice cultivars: Cyclobutane pyrimidine dimer photolyase activity and gene mutation. Plant Cell Physiol. 2004;45:1848–1856. doi: 10.1093/pcp/pch215. [DOI] [PubMed] [Google Scholar]
- Thakur S, Singh PK, Das A, et al. Extensive sequence variation in rice blast resistance gene Pi54 makes it broad spectrum in nature. Front Plant Sci. 2015;6:345. doi: 10.3389/fpls.2015.00345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Untergasser A, Nijveen H, Rao X, et al. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 2007;35:W71–W74. doi: 10.1093/nar/gkm306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Biezen EA, Jones JDG. Plant disease-resistance proteins and the gene-for-gene concept. Trends Biochem Sci. 1998;23:454–456. doi: 10.1016/S0968-0004(98)01311-5. [DOI] [PubMed] [Google Scholar]
- Vasudevan K, Gruissem W, Bhullar NK. Identification of novel alleles of the rice blast resistance gene Pi54. Sci Rep. 2015;5:15678. doi: 10.1038/srep15678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WANG Bao-hua, Ebbole Daniel J., WANG Zong-hua. The arms race between Magnaporthe oryzae and rice: Diversity and interaction of Avr and R genes. Journal of Integrative Agriculture. 2017;16(12):2746–2760. doi: 10.1016/S2095-3119(17)61746-5. [DOI] [Google Scholar]
- Wang M, Yu Y, Haberer G, et al. The genome sequence of African rice (Oryza glaberrima) and evidence for independent domestication. Nat Genet. 2014;46:982. doi: 10.1038/ng.3044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao N, Wu YY, Pan CH et al (2017) Improving of Rice blast resistances in japonica by pyramiding major R genes. Front Plant Sci 7. 10.3389/fpls.2016.01918 [DOI] [PMC free article] [PubMed]
- Xue M, Yang J, Li Z, et al. Comparative analysis of the genomes of two field isolates of the Rice blast fungus Magnaporthe oryzae. PLoS Genet. 2012;8:e1002869. doi: 10.1371/journal.pgen.1002869.t001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SH, Feng ZM, Zhang XY, et al. Genome-wide investigation on the genetic variations of rice disease resistance genes. Plant Mol Biol. 2006;62:181–193. doi: 10.1007/s11103-006-9012-3. [DOI] [PubMed] [Google Scholar]
- Yu P, Wang C, Xu Q, et al. Detection of copy number variations in rice using array-based comparative genomic hybridization. BMC Genomics. 2011;12:372. doi: 10.1186/1471-2164-12-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang QJ, Zhu T, Xia EH, et al. Rapid diversification of five Oryza AA genomes associated with rice adaptation. Proc Natl Acad Sci U S A. 2014;111:E4954–E4962. doi: 10.1073/pnas.1418307111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Wang L, Wu W, et al. Function and evolution of Magnaporthe oryzae avirulence gene AvrPib responding to the rice blast resistance gene Pib. Sci Rep. 2015;5:11642. doi: 10.1038/srep11642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Z, Chen M, Lin L, et al. Population genomic analysis of the rice blast fungus reveals specific events associated with expansion of three main clades. ISME J. 2018;12:1867–1878. doi: 10.1038/s41396-018-0100-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou T, Wang Y, Chen JQ, et al. Genome-wide identification of NBS genes in japonica rice reveals significant expansion of divergent non-TIR NBS-LRR genes. Mol Gen Genomics. 2004;271:402–415. doi: 10.1007/s00438-004-0990-z. [DOI] [PubMed] [Google Scholar]
- Zhu T, Xu PZ, Liu JP, et al. Phylogenetic relationships and genome divergence among the AA- genome species of the genus Oryza as revealed by 53 nuclear genes and 16 intergenic regions. Mol Phylogenet Evol. 2014;70:348–361. doi: 10.1016/j.ympev.2013.10.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Selection and identification of Pi54 alleles using primer set Pi54 MAS. Figure S2. Alignment of the divergent regions from Sasanishiki and Nipponbare. Figure S3. Detection of divergent structures at the Pi54 locus in modern O. sativa cultivars. Figure S4. DNA sequences of Pi54 alleles. Figure S5. Alignment of Helitron-N91 and the 143 bp insertion in Nipponbare. Table S1. Primer sets for PCR amplification. Table S2. Primer sets for identification of Nipponbare and Sasanishiki type species. Table S3 Primers for sequencing. Table S4. Primers for sequencing the genes flanking the 25,061 bp region in Sasanishiki. Accession codes. Sequence data of Sasanishiki the Pi54 locus. Table S5. Transposable elements in the flanking regions of Pi54, #5, #10 and #11. Table S6. Genes Orthologous to a mobile unit on chromosome 1 in Oryza species. (PDF 1280 kb)
Data Availability Statement
The datasets supporting the conclusions of this article are provided within the article and its additional files.