Abstract
The ability to activate or repress specific genes in the brain could have a tremendous impact for understanding and treating neurological disorders. Artificial transcription factors based on zinc finger, TALE, and CRISPR/Cas9 programmable DNA-binding platforms have been widely used to regulate the expression of specific genes in cultured cells, but their delivery into the brain represents a critical challenge to apply such tools in live animals. In previous work, we developed a purified, zinc finger-based artificial transcription factor that could be injected systemically, cross the blood-brain barrier, and alter expression of a specific gene in the brain of an adult mouse model of Angelman syndrome. Importantly, our mode of delivery produced widespread distribution throughout the brain. Here we describe our most current methods for the production and purification of the factor, dosage optimization, and use of live animal fluorescence imaging to visualize the kinetics of distribution.
Keywords: Engineered zinc finger protein, Animal models, Preclinical studies, Neurologic disease, Gene therapy
1. Introduction
Artificial transcription factors (ATFs) are based on the attachment of transcriptional effector domains to programmable DNA-binding platforms such as zinc fingers (ZFs), transcription activator-like effectors (TALEs), or catalytically inactive clustered regularly interspaced short palindromic repeats/dead Cas9 (CRISPR/dCas9). These tools are capable of activating or repressing specific genes as has been described extensively [1–4]. Widespread delivery of these gene regulators to the brain remains a significant challenge for the study and treatment of neurologic disorders.
Direct injection of viral vectors into the brain is probably the most common approach for the delivery of protein that can alter genetic (DNA sequence) or epigenetic (gene expression) information in the context of brain. One drawback with this method is that it typically only affects cells in close proximity to the injection site (e.g., [5]). This is primarily due to the fact that most viral vectors are not able to efficiently cross the blood-brain barrier (BBB), a blockade created by the tight junctions of endothelial cells that line the vasculature in the central nervous system to shield the brain from substances in the peripheral circulation. Viral vectors, plasmid DNA, and proteins are often injected directly into the brain, resulting in limited distribution. Recently researchers have developed viral vectors with improved abilities to cross the BBB that can be injected systemically and transduce cells widely throughout the brain [6, 7]. However, the amount of such viral particles required for efficient delivery is quite high, and it remains to be seen if such methods will be effective for the application of ATFs in large animals and humans.
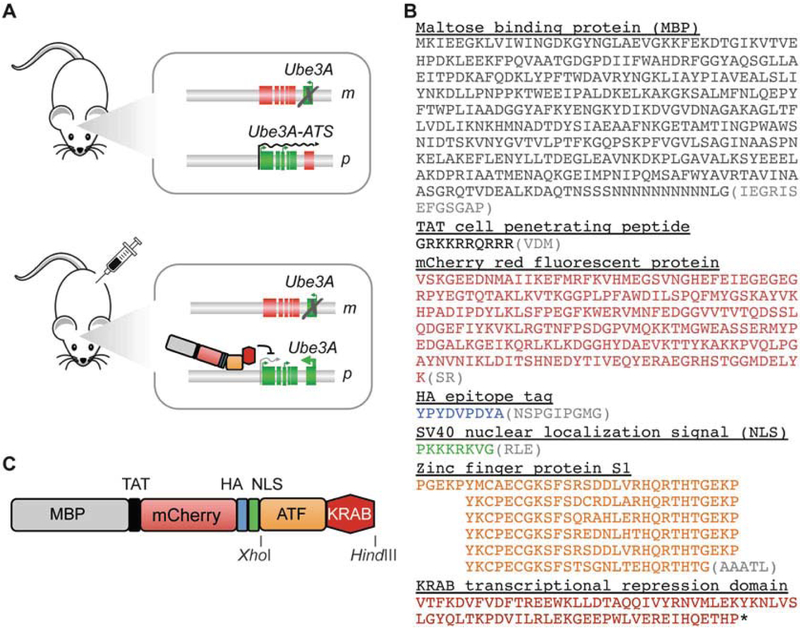
We previously reported the systemic delivery of the comparatively smaller purified ATF protein as a potential therapeutic approach for the treatment of Angelman syndrome (AS) [8]. AS is a rare neurological genetic disorder caused by loss or mutation of the maternal copy of UBE3A in the brain. Due to brain-specific genetic imprinting at this locus, the paternal UBE3A is silenced, resulting in the complete loss of UBE3A expression in brain neurons of patients. Paternal UBE3A silencing is not due to promoter DNA methylation but rather a long antisense transcript, the UBE3A-ATS, that is transcribed across the UBE3A open reading frame in the opposite direction. Inhibition of the UBE3A-ATS transcript could lead to un-silencing of paternal UBE3A, thus providing a therapeutic approach for AS. To inhibit the Ube3a-ATS in a mouse model of AS, we designed a repressive ATF to bind at the transcriptional start site of the transcript (Fig. 1a). The ATF was composed of an N-terminal maltose-binding protein for purification, a cell-penetrating peptide consisting of the 10-aa transduction domain of the HIV transactivator protein (TAT, residues 48–57), an mCherry red fluorescent protein to aid in protein solubility and visualization, an HA epitope tag for detection, an SV40 nuclear localization signal to ensure nuclear delivery, an engineered zinc finger protein (designated “S1”), and a KRAB transcriptional repression domain that was appended to the C-terminus (Fig. 1b). Encouragingly, we observed that after intraperitoneal (i.p.) or subcutaneous (s.c.) injection at 160–200 mg/kg, the ATF was able to cross the BBB and distribute widely throughout the brain. Due to the mCherry component, we were also able to observe by live animal fluorescence that peak accumulation in the cranium occurred approximately 4–8 h after injection and was essentially cleared by the kidneys by 24 h. Based on the apparent 16-h half-life of the ATF, we performed three injections per week for 4 weeks, after which we observed significant activation of Ube3a expression in the brain of a mouse model of AS, based on immunohistochemistry and Western blot. No overt toxicity or animal distress was noted over the 4-week treatment period [8].
Fig. 1.
Design of ATF S1-KRAB to cross BBB and upregulate Ube3a gene expression. (a) Cartoon of an Angelman syndrome (AS) mouse model. The maternal (m) and paternal (p) alleles of the imprinted locus are shown. Expressed genes are indicated as green boxes; silenced genes are depicted as red boxes. In an untreated mouse (top panel), the brain-specific Ube3a-ATS transcript (indicated by black arrow) silences the paternal copy of Ube3a. Deletion of the maternal copy therefore results in loss of Ube3a expression in the brain. Upon injection with ATF S1-KRAB (bottom panel), the protein crosses the blood-brain barrier, enters the cell nucleus, and binds to its target DNA sequence (Ube3a-ATS) in the mouse brain. The resulting downregulation of Ube3a-ATS allows upregulation of the paternally silenced Ube3a. (b) Diagram of ATF S1-KRAB indicating individual protein domains and restriction sites XhoI and HindIII for subcloning. (c) Protein sequence of ATF S1-KRAB. Individual domains are indicated. Intervening sequences are shown as gray sequences in parenthesis
In principle, this method could be used to deliver similar ATFs designed to target other promoters or DNA elements in the brain by using a different zinc finger protein or other programmable DNA-binding domains such as TALEs or catalytically deactivated CRISPR/Cas9 (dCas9). It should be possible to exchange the KRAB transcriptional repression domain with an activation domain (e.g., VP64 or p300) or writers or erasers of epigenetic information (e.g., DNMT3A or G9A) yielding artificial epigenome editors. The TAT cell-penetrating peptide had been previously shown to deliver proteins across the BBB to the brains of mice following systemic injection [9–11], although its actual role in facilitating both BBB crossing and neuronal cell entry is still under investigation. We have noticed that even seemingly minor changes in the composition of the ATF can have profound effects on the efficiency of protein production and purification. The expression and purification steps described in the protocol that follows have been optimized for the ATF S1-KRAB, with additional modifications adopted since our original study [8]. Modified ATFs will likely require empirical re-optimization of the purification protocol.
2. Materials
2.1. Bacterial Cell Expression and Purification of ZF-ATFs
An artificial transcription factor consisting of an engineered zinc finger protein with an attached effector domain. The zinc finger-based ATF S1-KRAB described in this method can be obtained from the authors upon request (see Notes 1–3).
A modified pMAL-c2X prokaryotic expression vector (New England Biolabs, Ipswich, MA) with an expression cassette containing (1) an N-terminal maltose-binding protein (MBP) for purification, (2) a TEV1 protease cleavage site, (3) a cell-penetrating peptide consisting of the 10-aa transduction domain of the HIV-transactivator protein (TAT, residues 48–57), (4) mCherry red fluorescent protein to aid in protein solubility and visualization, (5) an HA epitope tag for detection, and (6) an SV40 nuclear localization signal to ensure nuclear delivery, a XhoI/HindIII cloning site for the zinc finger-KRAB artificial transcription factor. This vector is available from the authors upon request. The complete sequence of the expression cassette for the full-length ATF S1-KRAB protein is provided in Fig. 1c (see Note 4).
Reagents for traditional or Gibson molecular cloning, including restriction enzymes XhoI and HindIII.
Chemically competent NEB5α E. coli bacteria (New England Biolabs). Store at −80 °C. (See Note 5.)
Carbenicillin antibiotic stock at 100 mg/mL in H2O. Keep stock at −20 °C.
10-cm plates of Luria broth (LB) agar (see Note 6) supplemented with carbenicillin at 50 μg/mL. Prepare no more than 1 month in advance and store at 4 °C.
1× Luria broth medium (see Note 6).
Isopropyl β-d-1-thiogalactopyranoside (IPTG) stock at 0.5 M in H2O. Keep stock at −20 °C.
Zinc chloride (ZnCl2) stock at 1 M in H2O. Can be stored at room temperature.
Zinc buffer A (ZBA): 10 mM Tris-base, 90 mM KCl, 1 mM MgCl2, and 100 μM ZnCl2. Adjust pH to 8.5 using HCl (see Note 7). Store at room temperature.
Bacterial culture shakers at both 37 and 4 °C.
Centrifuge for bacterial cultures.
Microfluidizer (microfluidics model M-110Y).
Chromatography columns (~100 mL volume) with valves.
Amylose resin (New England Biolabs, E8021L).
Maltose.
Dithiothreitol (DTT) stock at 1 M in H2O. Aliquot in 0.5–2 mL volumes and store at −20 °C.
Elution Buffer: ZBA, 1 M maltose.
Centricon Plus-70 spin concentrators (Millipore, Billerica, MA, UFC710008).
Refrigerated tabletop centrifuge.
Nalgene Rapid-Flow sterile disposable filter units, 0.2 μm (Thermo Fisher).
HEK-Blue LPS Detection Kit (InvivoGen, San Diego, CA, rep-lps2).
4–20% Mini-PROTEAN TGX Precast Protein Gels (Bio-Rad, Hercules, CA, 4561096).
Apparatus and materials for performing Coomassie-stained SDS PAGE.
Nanodrop UV spectrophotometer or equivalent.
Glycerol, sterilized by autoclave.
2.2. Determining the Maximum Tolerated Dose
Purified ATF protein in elution buffer, 30% glycerol, 5 mM DTT.
C57BL/6 mice of either sex, approximately 8 weeks of age.
1 mL syringe with 25-gauge hypodermic needle.
Isoflurane and appropriate apparatus.
CO2 chamber or other apparatus for humane euthanasia.
2.3. Determining ATF Kinetics Using Live Animal Fluorescent Imaging
-
1–5.
Same as Subheading 2.2 above.
-
6.
Maestro 2 (PerkinElmer, Waltham, MA) live animal fluorescence imager.
-
7.
Shaver and hair removal lotion (Nair).
-
8.
Ophthalmic ointment (Lacri-lube or Puralube).
3. Methods
3.1. Bacterial Cell Expression and Purification of ZF-ATFs
Prepare the prokaryotic expression vector. The ZF-KRAB coding region can be cloned into the modified pMAL-c2X vector between XhoI and HindIII.
Transform the vector into NEB5α E. coli by standard heatshock methods.
Plate the transformed bacteria on LB agar + 50 μg/mL carbenicillin and incubate overnight.
Pick a single colony to inoculate into 5 mL of LB medium + 50 μg/mL carbenicillin. Incubate overnight with shaking at 37 °C.
Inoculate the 5 mL overnight cultures into 800 mL of LB medium + 50 μg/mL carbenicillin. Incubate with shaking overnight at 37 °C.
At optical density ~1.0, induce protein expression by moving the culture to 4 °C (see Note 8) and adding IPTG to 0.75 mM and 1 mL of zinc chloride. Shake gently at 4 °C for 4 days.
To release the protein from the bacteria, pellet and resuspend the culture in 30 mL of cold ZBA (see Note 9). Apply the suspension to a microfluidizer according to the manufacturer’s instructions (see Note 10). Keep the lysate on ice.
Prepare gravity flow amylose resin purification columns by applying enough amylose resin to produce a 30-mL compact bed in the columns (see Note 11). All steps can be performed at room temperature. Wash the columns twice with four column volumes of deionized water and then one column volume of ZBA. Finally, apply the microfluidized lysates to the columns. The initial drip rate should be approximately two drops/s but will decrease as the solution in the column decreases.
Elute the protein in ~150 mL of elution buffer + 5 mM DTT (add DTT just before use). Keep eluted samples on ice.
Concentrate the eluate to 16 mg/mL using a Centricon Plus-70 in a refrigerated centrifuge at 2000 × g for ~60 min (depending on the initial concentration) at 4 °C. For maximum tolerated dose studies, additional higher concentrations may be desired.
Sterilize the protein sample using Nalgene Rapid-Flow sterile disposable filter units to remove any residual bacteria. Purified, sterilized protein samples are routinely checked by an endotoxin kit (e.g., HEK-Blue LPS Detection Kit) to assure no detectable endotoxins are present.
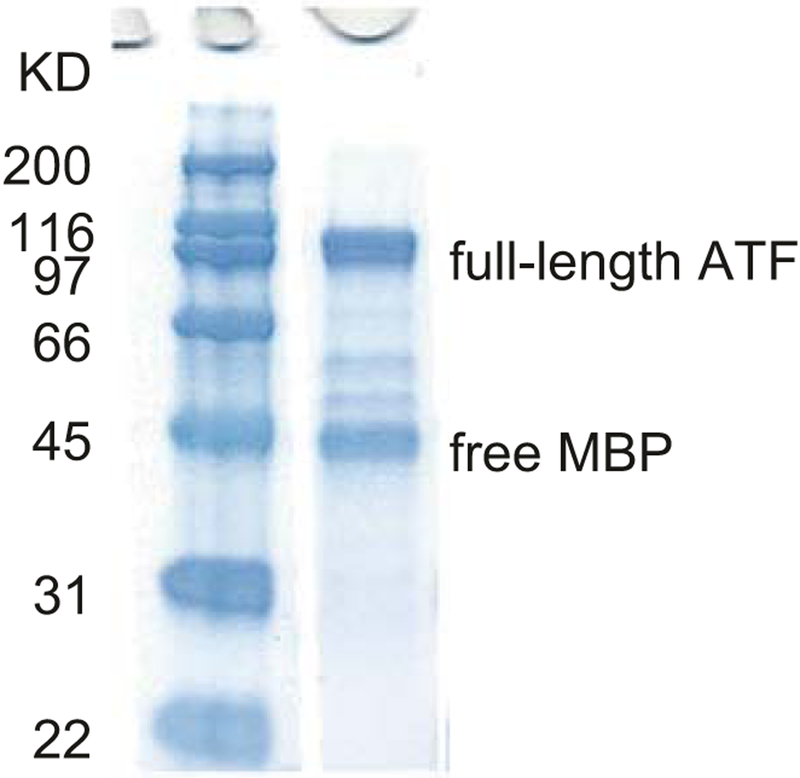
Measure protein concentration using a Nanodrop UV spectrophotometer at A280, blanking with elution buffer. The procedure typically yields 4 g total protein/L of culture for S1-KRAB, but values will likely change for other ATFs. Evaluate protein integrity by SDS polyacrylamide gel electrophoresis using a 4–20% TGX precast protein gel followed by staining with Coomassie blue (see Fig. 2). Usually there is a 44-kDa band corresponding to free MBP. Protein concentrations for injections refer to the full-length + free-MBP band intensities, of which only half was considered to be the 100-kDa full-length protein.
For storage of the proteins, add glycerol to 30% and DTT to 5 mM. This typically decreases the concentration from 16 mg/mL to 12 mg/mL total protein. Store at −20 °C (see Note 12). Note that it is difficult to measure the protein concentration after addition of glycerol, so concentration is measured in the previous step.
Fig. 2.
SDS-PAGE analysis of purified ATF S1-KRAB. The gel is stained with Coomassie blue. Full-length protein is visible at 100 kDa, while free MBP is visible at 45 kDa. Full-length is generally considered to represent 50% of the total protein
3.2. Determining the Maximum Tolerated Dose
Obtain prior approval from the Institutional Animal Care and Use Committee of the investigator’s institution before any work on animals is performed.
Mice should be anesthetized with 4% isoflurane before injection (see Notes 13 and 14).
Administer the protein systemically by i.p. or s.c. injection over a series of concentrations representing twofold increments (inject 300 μL at an initial concentration of ~1.5 mg/mL ATF protein, using elution buffer to make the dilutions). Use three mice per dose. A volume of 300 μL of 1.5 mg/mL purified ATF protein corresponds to 0.45 mg of total protein (~0.23 mg of full-length ATF), corresponding to a dose of 18–22.5 mg/kg total protein in a 20–25 g mouse.
Mice should be monitored every hour for the first 8 h and again the following day. Generally, mice will succumb to a lethal dose in the first hour.
All remaining mice are humanly euthanized.
By this approach, the acute exposure threshold for full-length ATF S1-KRAB was achieved at a one-time dose of 740 mg/kg.
A functional dose approximately fourfold less than the maximum tolerated dose, 160–200 mg/kg, was chosen for subsequent studies of ATF S1-KRAB.
3.3. Determining ATF Kinetics Using Live Animal Fluorescent Imaging
One day before injection and imaging, the mice are shaved and additional hair removed using Nair from the injection site, head and back. This is performed early to avoid background signals in the imaging. It is important to be thorough, as residual hair in these areas will block the fluorescent signal (see Note 15).
Mice should be anesthetized with 4% isoflurane before injection. Inject three mice with ATF S1-KRAB at a functional dose (160–200 mg/kg in our experiments) using the preferred route of injection (s.c. in our experiments) (see Note 16). Also inject three mice with a version of the ATF vector that has no ZF or KRAB domains as a negative control. Maintain the mice on 2.5% Isoflurane until the first imaging time point. Ophthalmic ointment should be used to keep the eyes lubricated. The Maestro 2 imager has a built-in heating surface for the mice during the procedures.
Time points for imaging are 15 min, 4 h, 8 h, and 24 h post injection. For mCherry, the green filter of the Maestro 2 imager is used with acquisition settings of 550–800 nm in 10-nm steps. At each time point, separate fluorescent images are taken of the injection site, the head and the back. Optimal exposure times for these regions may vary and should be empirically determined. We typically use exposure times of 229, 1959, and 1986 ms for the injection site, the head and the back, respectively. Up to three mice can be imaged in one exposure, which should consist of at least one treated and one control mouse. The positions of the treated and control mice should be rotated to avoid any biases in exposure.
Mice are revived to ambulation between time points, maintaining heating as required. After the final imaging, all mice are humanly euthanized. Organs (e.g., brain, heart, liver, and kidneys) should be harvested at this point and imaged.
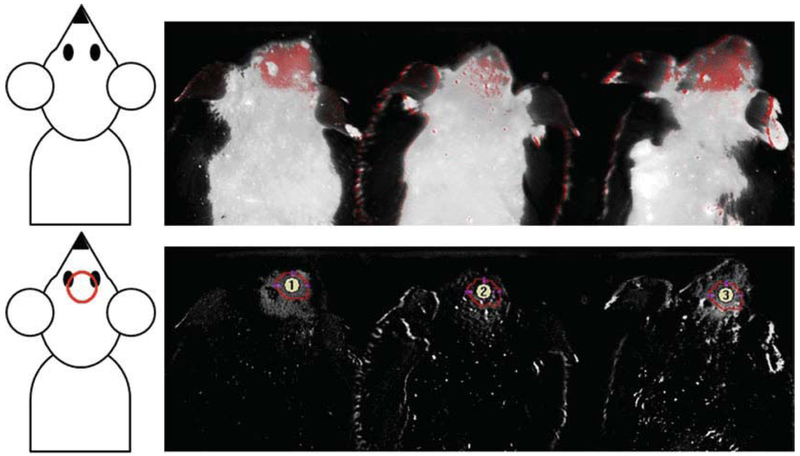
For analysis, all images are based on the same raw inputs for mCherry signal and background control mouse signal. The mCherry signal is usually baselined to a point just outside the direct injection site. Analysis of mCherry signal is performed by measuring the mCherry by using a consistent-sized region of interest (ROI) in the brain region using Maestro software (see Fig. 3). The measurement area will be the same for each mouse across all treatments and time points.
From this analysis in our previous work, it was found that the peak of ATF protein in the brain was found 4–6 h post injection and was largely cleared by 24 h [8]. The estimated half-life of the protein in the brain was ~16 h. The ultimate site of accumulation of the ATF was the kidneys. This information led us to perform subsequent experiments for functional effects by injecting the mice three times per week (Monday, Wednesday, and Friday) for the duration of the treatment period (typically 4 weeks). The imaging data also suggested that we consider renal impairments among the potential side effects of the ATF treatment.
Functional effects of the ATF treatment were typically determined by assays of behavior (e.g., locomotor, anxiety, social, seizure) and molecular events (e.g., immunohistochemistry, RT-qPCR, Western blot, chromatin immunoprecipitation analyzed by PCR (ChIP-PCR)). Such assays are widely used in the study of rodent animal models and will thus not be described here.
Fig. 3.
Maestro 2 live animal florescence imaging and analysis. Three shaved mice were imaged 8 h after injection with an mCherry-containing ATF (left and right mice) or just elution buffer, 30% glycerol, and 5 mM DTT that contains no ATF as a negative control (center mouse). Top panel, combined fluorescence and brightfield. Bottom panel, the florescence signal only with regions of interest for analysis shown as numbered circles
4. Notes
The activity of the ATF to regulate expression of its specific target gene has to be validated first in a mammalian cell culture assay before injection into mice. This protocol assumes such validation experiments will have already been performed.
Design strategies for other ATFs based on zinc fingers, TALEs, and dCas9 have been described [1–4, 12–14]. It is typically convenient to have the coding region commercially synthesized. Anecdotal evidence suggests that injection of purified TALE or Cas9 protein at the doses used for zinc finger-based artificial transcription factors evokes a rapid and strong immune response that is toxic to mice. Also, Cas9 and dCas9 require a guide RNA to recognize its target DNA sequence, in contrast to zinc fingers and TALE proteins. Inclusion of a guide RNA would represent a significant complication for the methods described here.
The expression and purification methods described here will likely require significant optimization for any new type of zinc finger protein or effector domain used.
At the time of this writing, it is not clear if protein domains such as MBP and mCherry are required for full function of the ATF. Similarly it is not clear that the TAT cell-penetrating peptide is required, since it has been shown that the zinc fingers themselves can act as protein transduction domains [16].
BL21 Star cells could also be used for protein expression. The ATF S1-KRAB seems to express equally well in both BL21 Star and NEB5a cells. However, others, such as TALE proteins, express much better in BL21.
Any source of LB medium is usually acceptable. However, we have found for some ATFs (not S1-KRAB) that LB from some vendors produced a dramatic reduction in yield, which was restored by using LB from VWR.
A good guide to the appropriate pH for the ZBA is the protein’s isoelectric point, which can be calculated on the ExPASy server using the known amino acid sequence (http://web.expasy.org/protparam/). Some optimization may be necessary.
The cold temperature induction was critical to obtaining high yields. Induction at room temperature or 37 °C was far less efficient. This unusual requirement was fortuitously observed when several standard induction conditions were tried and proved unsatisfactory. Concurrently, a culture that had been accidentally left at 4 °C for several days turned noticeably pink. This was an indication that the protein was being expressed, at least the mCherry domain. Experimental refinement of methods resulted in the reported production protocol.
Protease inhibitors are not used when purifying the ATF S1-KRAB. They are sometimes used with other factors, especially if degradation appears as a significant issue. However, we have generally been cautious out of concern for undesired effects of residual protease inhibitors in animals.
The use of the microfluidizer was found to be critical for obtaining full-length protein. Sonication and freeze/thaw techniques produced fragmented proteins.
The amylose resin can be reused up to 14 times by washing. In some cases, yield seemed to increase using resin that had been used and washed.
Proteins were originally stored at −80 °C, but later studies showed less protein fragmentation due to freeze-thaw when the ATF was stored at −20 °C.
It is often difficult to initially assess the minimum dose required to see a phenotypic response, because the changes in phenotype may require an unknown time to manifest, and the behavioral assays often require many mice to achieve statistical significance. A more pragmatic approach is therefore to determine a maximum tolerated dose and assume that treatments below but near this dose will produce the most dramatic phenotypic effects possible. A maximum tolerated dose for the full-length ATF S1-KRAB in mice was established using the procedure that follows.
In addition to avoiding accidental autoinoculation by trying to inject a moving mouse, anesthesia before injection also provides a transient immunosuppression that prevents an acute immune response to the protein when the bolus is injected. Anesthesia is thus highly recommended.
Measuring how quickly the ATF appears in the brain and how long it remains can provide information for determining if and how often repeat dosing is necessary. This is particularly important for effectors such as KRAB, which we and others have shown produces only a transient effect on gene expression [15]. That is to say, in order for a KRAB ATF to keep its target gene repressed, the protein needs to be physically present. Repeated injection is one way to keep a continuous, or at least periodic, presence of the ATF at the target site. Because the ATF contains an mCherry domain, the fluorescence of the protein can be detected in real-time using the Maestro 2 live animal fluorescence imager. At the University of California, Davis, a Maestro 2 imager is available through the Center for Molecular and Genomic Imaging. The Center maintains the device and provides the required training for all users. The Center also hosts the software for analyzing the native data, which are ultimately output as numerical spreadsheets and TIFF image files.
Ideally, a power calculation would be performed first to determine how many mice would be required to observe the molecular phenotype (i.e., the change in Ube3a expression). This calculation requires an estimation of the variance in the live fluorescence assay. Since the variance may differ for different proteins, testing three mice here can provide information on the variance that can be used to make a more informed calculation for a sufficiently powered experiment. However, in our experience, three mice per group are typically sufficient. More mice would be required to investigate behavioral phenotypes, which would be the final and relevant readouts of this procedure but are beyond the scope of this methods description.
Acknowledgment
We thank Alexa Adams, Jennifer Trang Nguyen, Victoria Le, Anvita Komarla, Joanna Watterson, Andy Tran, Joshua Mandella, Ruth Le, and Michelle McAllister for their assistance with the experiments. We thank Enoch Baldwin and Sarah Lockwood for expert advice and discussions in developing these methods. Imaging work was performed at the Center for Molecular and Genomic Imaging (CMGI), University of California, Davis. We would like to acknowledge Michelle Connell for her help with Maestro 2 imaging. This work was supported by the NIH (NS071028), the Angelman Syndrome Foundation, and the Foundation for Angelman Syndrome Therapeutics. B.J.B. was also funded by an NSF fellowship (0707429) and a grant to UC Davis from the Howard Hughes Medical Institute through the Med into Grad Initiative (56005706) and a CTSC pilot study (TR000002). Maestro imaging was supported by a CMGI pilot grant.
References
- 1.Blancafort P, Segal DJ, Barbas CF 3rd (2004) Designing transcription factor architectures for drug discovery. Mol Pharmacol 66(6):1361–1371 [DOI] [PubMed] [Google Scholar]
- 2.Polstein LR, Perez-Pinera P, Kocak DD, Vockley CM, Bledsoe P, Song L, Safi A, Crawford GE, Reddy TE, Gersbach CA (2015) Genome-wide specificity of DNA binding, gene regulation, and chromatin remodeling by TALE- and CRISPR/Cas9-based transcriptional activators. Genome Res 25(8):1158–1169. https://doi.org/10.1101/gr.179044.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thakore PI, Black JB, Hilton IB, Gersbach CA (2016) Editing the epigenome: technologies for programmable transcription and epigenetic modulation. Nat Methods 13(2):127–137. https://doi.org/10.1038/nmeth.3733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thakore PI, Gersbach CA (2016) Design, assembly, and characterization of TALE-based transcriptional activators and repressors. Methods Mol Biol 1338:71–88. https://doi.org/10.1007/978-1-4939-2932-0_7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cearley CN, Wolfe JH (2006) Transduction characteristics of adeno-associated virus vectors expressing cap serotypes 7, 8, 9, and Rh10 in the mouse brain. Mol Ther 13(3):528–537 [DOI] [PubMed] [Google Scholar]
- 6.Deverman BE, Pravdo PL, Simpson BP, Kumar SR, Chan KY, Banerjee A, Wu WL, Yang B, Huber N, Pasca SP, Gradinaru V (2016) Credependent selection yields AAV variants for widespread gene transfer to the adult brain. Nat Biotechnol 34(2):204–209. https://doi.org/10.1038/nbt.3440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gray SJ, Matagne V, Bachaboina L, Yadav S, Ojeda SR, Samulski RJ (2011) Preclinical differences of intravascular AAV9 delivery to neurons and glia: a comparative study of adult mice and nonhuman primates. Mol Ther 19(6):1058–1069. https://doi.org/10.1038/mt.2011.72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bailus BJ, Pyles B, McAlister MM, O’Geen H, Lockwood SH, Adams AN, Nguyen JT, Yu A, Berman RF, Segal DJ (2016) Protein delivery of an artificial transcription factor restores widespread Ube3a expression in an Angelman syndrome mouse brain. Mol Ther 24(3):548–555. https://doi.org/10.1038/mt.2015.236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gong B, Cao Z, Zheng P, Vitolo OV, Liu S, Staniszewski A, Moolman D, Zhang H, Shelanski M, Arancio O (2006) Ubiquitin hydrolase Uch-L1 rescues beta-amyloidinduced decreases in synaptic function and contextual memory. Cell 126(4):775–788. https://doi.org/10.1016/j.cell.2006.06.046 [DOI] [PubMed] [Google Scholar]
- 10.Wagstaff KM, Jans DA (2006) Protein transduction: cell penetrating peptides and their therapeutic applications. Curr Med Chem 13(12):1371–1387 [DOI] [PubMed] [Google Scholar]
- 11.Mae M, Langel U (2006) Cell-penetrating peptides as vectors for peptide, protein and oli-gonucleotide delivery. Curr Opin Pharmacol 6(5):509–514 [DOI] [PubMed] [Google Scholar]
- 12.Bhakta MS, Henry IM, Ousterout DG, Das KT, Lockwood SH, Meckler JF, Wallen MC, Zykovich A, Yu Y, Leo H, Xu L, Gersbach CA, Segal DJ (2013) Highly active zinc-finger nucleases by extended modular assembly. Genome Res 23(3):530–538. gr.143693.112 [pii]. https://doi.org/10.1101/gr.143693.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bhakta MS, Segal DJ (2010) The generation of zinc finger proteins by modular assembly. Methods Mol Biol 649:3–30. https://doi.org/10.1007/978-1-60761-753-2_1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee CM, Cradick TJ, Fine EJ, Bao G (2016) Nuclease target site selection for maximizing on-target activity and minimizing off-target effects in genome editing. Mol Ther 24(3):475–487. https://doi.org/10.1038/mt.2016.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O’Geen H, Ren C, Nicolet CM, Perez AA, Halmai J, Le VM, Mackay JP, Farnham PJ, Segal DJ (2017) dCas9-based epigenome editing suggests acquisition of histone meth-ylation is not sufficient for target gene repression. Nucleic Acids Res 45(17):9901–9916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaj T, Liu J, Anderson KE, Sirk SJ, Barbas CF 3rd (2014) Protein delivery using Cys2-His2 zinc-finger domains. ACS Chem Biol 9(8):1662–1667. https://doi.org/10.1021/cb500282g [DOI] [PMC free article] [PubMed] [Google Scholar]