Abstract
Cutaneous T-cell lymphoma is a group of incurable extranodal non-Hodgkin lymphomas that develop from the skin-homing CD4+ T cell. Mycosis fungoides and Sézary syndrome are the most common histological subtypes. Although next-generation sequencing data provided significant advances in the comprehension of the genetic basis of this lymphoma, there is not uniform consensus on the identity and prevalence of putative driver genes for this heterogeneous group of tumors. Additional studies may increase the knowledge about the complex genetic etiology characterizing this lymphoma. We used SNP6 arrays and GISTIC algorithm to prioritize a list of focal somatic copy-number alterations in a dataset of multiple sequential samples from 21 Sézary syndrome patients. Our results confirmed a prevalence of significant focal deletions over amplifications: single well-known tumor suppressors, such as TP53, PTEN, and RB1, are targeted by these aberrations. In our cohort, ZEB1 (TCF8, ZFHX1A) spans a deletion having the highest level of significance. In a larger group of 43 patients, we found that ZEB1 is affected by deletions and somatic inactivating mutations in 46.5% of cases; also, we found potentially relevant ZEB1 germline variants. The survival analysis shows a worse clinical course for patients with ZEB1 biallelic inactivation. Multiple abnormal expression signatures were found associated with ZEB1 depletion in Sézary patients we verified that ZEB1 exerts a role in oxidative response of Sézary cells. Our data confirm the importance of deletions in the pathogenesis of cutaneous T-cell lymphoma. The characterization of ZEB1 abnormalities in Sézary syndrome fulfils the criteria of a canonical tumor suppressor gene. Although additional confirmations are needed, our findings suggest, for the first time, that ZEB1 germline variants might contribute to the risk of developing this disease. Also, we provide evidence that ZEB1 activity in Sézary cells, influencing the reactive oxygen species production, affects cell viability and apoptosis.
Introduction
Sézary syndrome (SS) is characterized by erythroderma, lymphadenopathy, and leukemic involvement of the peripheral blood. The circulating atypical lymphocytes usually are CD4+ T helper cells of memory phenotype1. The prevalence of SS is around 0.3 cases per 100,000 people and it accounts for less than 5% of all cutaneous T-cell lymphoma (CTCL). Despite numerous efforts to characterize the events of its pathogenesis, SS remains an incurable and fatal disease. Multiple genomic array-based studies have delineated a complex profile of chromosome aberrations characterizing SS genome with multiple sporadic genetic events of gains/losses in addition to recurrent abnormalities affecting mainly chromosome 8, 9, 10, and 172–5. The mutational landscape of CTCL, compiled with recent reports of next-generation sequencing (NGS) data, identified a broad range of genes variously affected by somatic copy-number alterations (SCNAs) and somatic single-nucleotide variants, involved, predominantly, in the T-cell activation and apoptosis, activation of NF-kB, JAK/STAT signaling, chromatin remodeling, and DNA damage response6–13. Also, NGS data confirmed that a significant proportion of the oncogenic abnormalities identified in CTCL are SCNAs with an average of ~25 SCNAs observed per tumor;6,8,12 of these, focal deletions, resulting in frequent hemi-homozygous loss of target tumor suppressor gene loci, account for a significant proportion of the genetic aberrations, highlighting the significance of these genes as drivers of CTCL pathogenesis6. However, some discrepancies still exist concerning identities and frequencies of alterations found at these putative driver genes across the various studies. Particularly, some investigators reported ZEB1 as variously affected by deletion and somatic mutation at 10p11.22 locus2,6,9,10, whereas others did not7,8. ZEB1 is a zinc-finger-containing transcriptional repressor which regulates interleukin (IL)-stimulated cytokine signaling in normal T cells and it is essential for the correct development of T cells during hematopoiesis14–18. Aberrantly expressed in a variety of human solid cancers, it is assumed to foster migration, invasion, and metastasis, promoting epithelial-to-mesenchymal transition (reviewed in ref. 19). On the contrary, ZEB1 is a candidate tumor suppressor gene in adult T-cell leukemia/lymphoma (ATLL), where it contributes to TGF-β1-mediated growth suppression resistance of malignant CD4+ T cells and ZEB1 mutant mice frequently undergo spontaneous CD4+ T-cell lymphomas20,21. Here, we report ZEB1 as the target of a significant focal deletion with a correspondent transcript downregulation. DNA sequence analysis identified not only somatic inactivating mutations, but also germline variants of potential clinical impact. A tumor suppressor role for ZEB1 in SS is further supported by our survival analysis showing a worse outcome for patients carrying ZEB1 homozygous loss. Gene set enrichment analysis (GSEA) identified, among multiple expression signatures associated with ZEB1 absence in SS patients, the cellular response to oxidative signal. Using ZEB1 knockout (KO) cell lines, we verified that ZEB1 has a role in controlling intracellular reactive oxygen species (ROS) production affecting viability and apoptosis of SS cells. Although our findings require additional functional investigations, they provide the basis to understand the onco-suppressive role of ZEB1 in CTCLs.
Materials and methods
Patients and cell lines
Diagnosis of SS was based on described criteria22. Clinical characteristics of SS patients investigated with SNP6 arrays and GISTIC analysis are shown in Table S1. Hut78 (TIB161), H9 (HTB 176), and HH (CRL2105) cell lines established from peripheral blood of CTCL patients were obtained from American Type Culture Collection (ATCC) and grown in complete RPMI 1640 with 10% FBS (Sigma-Aldrich).
SNP6 DNA array
Tumor CD4+ T lymphocytes and matched granulocyte cells (normal counterpart) were isolated as previously described;3 genomic DNA was extracted using ArchivePure DNA Cell/Tissue Kit (5Prime, Gaithersburg, MD) following the manufacturer instructions. DNA samples were profiled on Affymetrix Genome-Wide Human SNP 6.0 Array as per the manufacturer's protocol (Affymetrix Santa Clara, CA), and the CEL files were generated using the GeneChip Command Console software 4.0 (Affymetrix). Normalization and data modeling have been performed with dChip23, and tumors were analyzed using paired normal samples as the reference. The calculated tumor-to-normal log2 ratio data were then exported and segmented with the Circular Binary Segmentation algorithm (CBS; α = 0.01, permutations = 10,000)24 via GenePattern platform at http://software.broadinstitute.org/cancer/software/genepattern/. Thresholds for copy-number change were fixed at ± 0.3 for gains/losses. We used the same segmentation algorithm applied for SNP6 arrays also for re-analyzing our previous 10 K SNP dataset3 to estimate the regions of copy-number changes, similar to our previous work on PTEN analysis25. With respect to this work, however, we have introduced additional follow-up samples/patients arrays (indicated with N superscript in Table 1) and, for those tumors with enough specimen material for new hybridizations, we have substituted the 10 K with SNP6 platform (S superscript in Table 1); finally, for more consistency of 10 K analysis results, we have eliminated SS patients with neoplastic clonal expansion equal or below 50% (i.e., SS44 and 46 of Table S1 in Cristofoletti et al.25). For the identification of significant regions of SCNAs, we applied the standard GISTIC (Genomic Identification of Significant Targets in Cancer) algorithm26 on the segmented SNP6 data using the GISTIC GenePattern module previous removal of genomic regions of structural copy-number variations identified in the Database of Genomic Variants (DGV, http://dgv.tcag.ca/dgv/app/home).
Table 1.
Status of ZEB1 locus 10p11.22 (chr10:31,648,147–31,856,740)
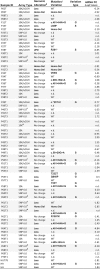
|
FC fold change, UPD uniparental disomy determined by dChip SNP software (Caprini, E et al. 2009), Homo Del homozygous deletion, WT wild type, n.d. not determined, G germline, S somatic
§Determined by Circular Segmentation Algorithm (Olshen et al.24):
Loss = log2 value ≤ −0.3; Homo Del = log2 value ≤ −2.0; no change = −0.3 ≤ log2 value ≤ + 0.3
Referred to the data set of Cristofoletti et al.25: S indicates array sample substitution, N indicates new sample array
Note: light-gray shadow includes multiple samples from the same patient. Bold font indicates ZEB1 DNA mutation/variation
Real-time PCR
Total RNA extraction by TRIzol reagent (Invitrogen) was performed according to the manufacturer’s instruction. Mature miR200c was analyzed using the TaqMan microRNA assay (Applied Biosystems) with RNU48 and miR16 as normalization references. ZEB1 expression analysis was performed with GoScript Reverse Transcriptase and GoTaq qPCR master mix (Promega), using β-actin as normalization reference, on ABI PRISM 7000 SDS (Applied Biosystem). Relative expression was calculated using the comparative Ct method (2–ΔΔCt)27. The following primers were used: ZEB1 forward: 5’-TGCCAACAGACCAGACAGTG-3′ reverse: 5′-CTGTCATCCTCCCAGCAGTT-3′. β-actin forward: 5′-GATGAGATTGGCATGGCTTT-3′ reverse: 5′-GTCACCTTCACCGTTCCAGT-3′.
Mutation analysis
The nine coding exons of ZEB1 gene were amplified by genomic PCR using 12 couples of primers. PCR products were then subjected to direct Sanger nucleotide sequencing. Primers’ sequence is reported in Table S2. ZEB1 sequence variants were assessed to determine potential effects on splicing using Human Splicing Finder Version 3.1 (available in the public domain at: http://www.umd.be/HSF3/index.html). The effects of amino acid substitutions on protein function were assessed using the computational prediction scores accessible through VarSome search engine available at: https://varsome.com/.
Survival and statistical analyses
A time-to-event analysis was performed using nonparametric Kaplan–Meier product-limit survival estimates, and differences between Kaplan–Meier survival curves were calculated using the Mantel–Haenszel log-rank test. Analyses were performed using survival curve and survival difference modules at: http://software.broadinstitute.org/cancer/software/genepattern/.
Gene set enrichment analysis
The GSE17601 dataset published in 20093 was processed and analyzed by GenePattern (http://software.broadinstitute.org/cancer/software/genepattern) using GEOImporter module (v5) and GSEA module (v17) with the Molecular Signatures Database “Hallmarks” gene set collection; briefly, this method identifies predefined database gene sets associated with phenotypic differences utilizing t-test statistics28. The default GSEA basic parameters were used with Gene Set as permutation-type option due to the small size of the dataset (<7); to find gene sets that correlate with ZEB1 expression profile (continuous phenotype label), Pearson metric was used for ranking genes.
CRISPR/Cas9 knockout
CRISPR–Cas9 technique was used to knock out ZEB1. All-in-one vectors (pCLIP-ALL-hCMV-Puro) expressing Cas9 nuclease and specific or control gRNA were purchased from Transomic Technologies (Huntsville, AL, USA). For detailed information on the construction of ZEB1 heterozygous and homozygous knockout cell lines, please refer to Supplementary Methods. Briefly, SS cell lines were first characterized for endogenous ZEB1 level (see Figure S1), and H9 cell line was chosen for knockout. Then, Amaxa™ Nucleofector™ Technology (Lonza Cologne GmbH) was used for cell transfection, and selection was performed with 1 µg/mL of puromycin for 72 h. Next, clonal cells were generated by single-cell dilutions and screened by a custom-designed drop-off assay performed with droplet-digital PCR (Biorad) as illustrated in Figure S2. Finally, western blot analysis was used to assess the level of ZEB1 protein as shown in Figure S3.
Western blot
Western blot (WB) analyses were conducted as previously described25. Membranes were probed with primary antibodies for ZEB1 (1:400; Santa Cruz) and β-actin (1:5.000; Santa Cruz) (Santa Cruz Biotechnology, Inc., Dallas, TX, USA). Immunodetection was performed with appropriate horseradish peroxidase-linked secondary antibodies and enhanced chemiluminescence detection reagents (GEHealthcare, Amersham Biosciences, Little Chalfont, UK). The film was scanned on a GS-710 Calibrated Imaging Densitometer and analyzed by means of Quantity One Software Version 4.1.1 (Bio-Rad Laboratories, Hercules, CA, USA) and by Image J open-source software (https://imagej.net).
ROS generation assay
Cells were stained with the oxidation-sensitive dye H2DCFDA (20 μmol/L) (Abcam) for 30 min. Then cells were treated with the indicated concentration of glucose oxidase (from Aspergillus niger, Sigma-Aldrich) for 4 hours, and ROS generation was determined by fluorescence-activated cell sorting (FacsCalibur, BD Biosciences, San Jose, CA, USA).
Cell viability and cell death assays
The cells were treated with increasing concentrations of glucose oxidase, as indicated, for 24 hours and measured by the methyl thiazolyl tetrazolium (MTT) assay using ELISA reader (Biorad). Each experiment was performed three times, and results are shown as means ± SEM. Apoptosis was assessed by AnnexinV–FITC and propidium iodide (Sigma-Aldrich). Samples were acquired on a flow cytometer and analyzed using CellQuest software (BD Biosciences).
Statistical analysis
Where not otherwise specified, statistical analyses were performed with GraphPad PRISM 6 software (GraphPad Software Inc., La Jolla, CA). Differences were evaluated with two-tailed Student’s t test and Pearson correlation test. P ≤ .05 was considered significant.
Results
GISTIC analysis of somatic DNA copy-number alterations in Sézary syndrome identified ZEB1 gene as the target of a highly significant focal deletion of 10p11.22
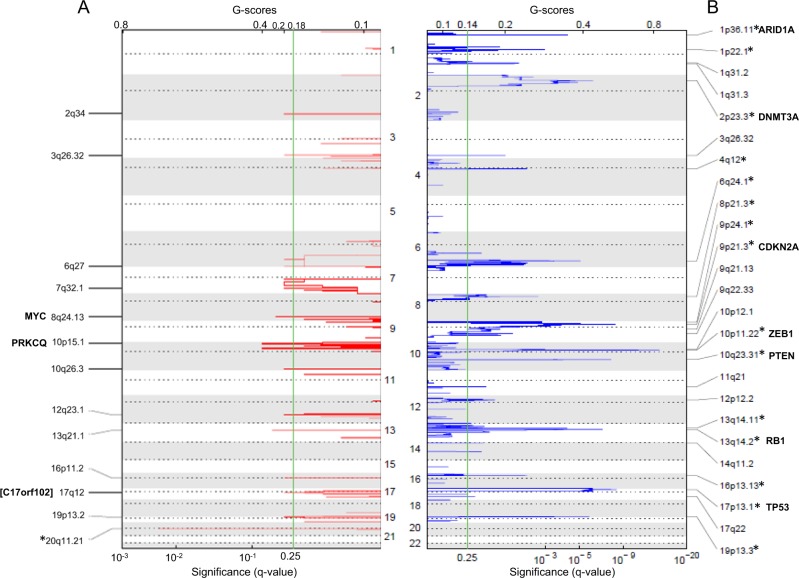
We used SNP 6 arrays and GISTIC2.0 algorithm to identify genes targeted by focal SCNAs, likely representing candidate “drivers” for cancer growth26,29, in a dataset composed of 33 samples, collected at different time points, from 21 SS patients and 3 CTCL cell lines (Hut78, H9, HH). Applying the default threshold for significance (qv.thresh = 0.25), we found 13 regions of gains and 25 genomic losses across our dataset (Fig. 1 and Table S3). Considering as significant all events with false discovery rate (FDR) q-values < 0.01, only 20q11.21 could be taken into account among the focal amplifications; this locus, however, was found only in the Hut78 and H9 cell lines, but in none of the SS samples (data not shown), suggesting that it represents an alteration related to the in vitro replication. If the chromosomes most frequently affected by copy-number gains were considered (i.e., chromosome 10p, 8/8q, and 17q2–5), the following GISTIC segments may assume importance: 10p15.1 with 19% of SS tumors affected (4/21) spanning PRKCQ gene, 8q24.13, altering 62% of cases (13/21 and 3/3 cell lines) encompassing two genes, one of which is MYC, already cited as candidate gene in SS2, and 17q12, involving 52% of tumors (11/21 and 2/3 cell lines), where the GISTIC peak falls in close proximity of an uncharacterized open- reading frame (C17orf102) (Fig. 1). On the other hand, we found 15 focal deletions having FDR q-values < 0.01, four of which identified single consensus cancer drivers: 10p11.22 targeted ZEB1 gene, which showed a deletion frequency of 47.6% (10/21 tumors and 3/3 cell lines); 17p13.1 interval, deleted in 81% of SS (17/21 and 1/3 cell line), identified TP53; 10q23.31 deletion fragment present in 57% of cases (12/21 and 3/3 cell lines) enclosed PTEN; and 13q14.2 deletion, affecting 19% of tumors (4/21 and 3/3 cell lines), involved RB1. The 9p21.3 GISTIC peak encompassed ten genes including CDKN2A as the main candidate cancer driver of this interval deleted in 33% of SS (7/21 and 3/3 cell lines) (Table S3). The high level of significance of ZEB1 deletion in our cohort of SS cases, prompted us to re-examine the 10 K array dataset (see Materials and methods for details) and combine the 10 K and the SNP6 results to investigate the ZEB1 locus alterations in a total of 66 specimens from 43 SS patients and 3 CTCL cell lines (Table 1). We found loss of genetic material (defined as −2.0 < Log2 values ≤ −0.3) for about 35% (15/43) of SS cases and 3 cell lines, homozygous DNA deletions (defined as Log2 value ≤ −2.0) in ~9% (4/43) of SS patients, one case (P28) of uniparental disomy (UPD, i.e., a deletion followed by a re-duplication of the remaining allele3), for a total of about 46.5% (20/43) of patients, and 3 cell lines affected by ZEB1 locus abnormalities (Table 1). Eight of ten cases with one or more follow-ups (P08, P32, P38, P39, P53, P61, P64, and P68) display ZEB1 loss in all the samples examined, whereas in two patients (P23, P63), the deletion appears from the second follow-up (median of the first observation time 4.32 months, median of follow-up observation time 29.47 months). This suggests that ZEB1 aberrations might be related to disease onset rather than to tumor progression. Altogether, our data identified ZEB1 as the target of a highly significant SCNA within 10p11.22 locus affecting almost half of the SS patients and the CTCL cell lines analyzed.
Fig. 1. Gistic plots.
Regions of gain A and loss B delineated by GISTIC analysis of 33 SS samples. Significance is reported as false discovery rate-corrected q-value. Chromosomes are indicated in the middle and the labeled cytobands correspond to the peak of significance, *regions with q-values < 0.01. Genes of interest are indicated, the gene nearest to the GISTIC peak is in brackets
ZEB1 mRNA expression in SS tumors is dependent on copy-number status
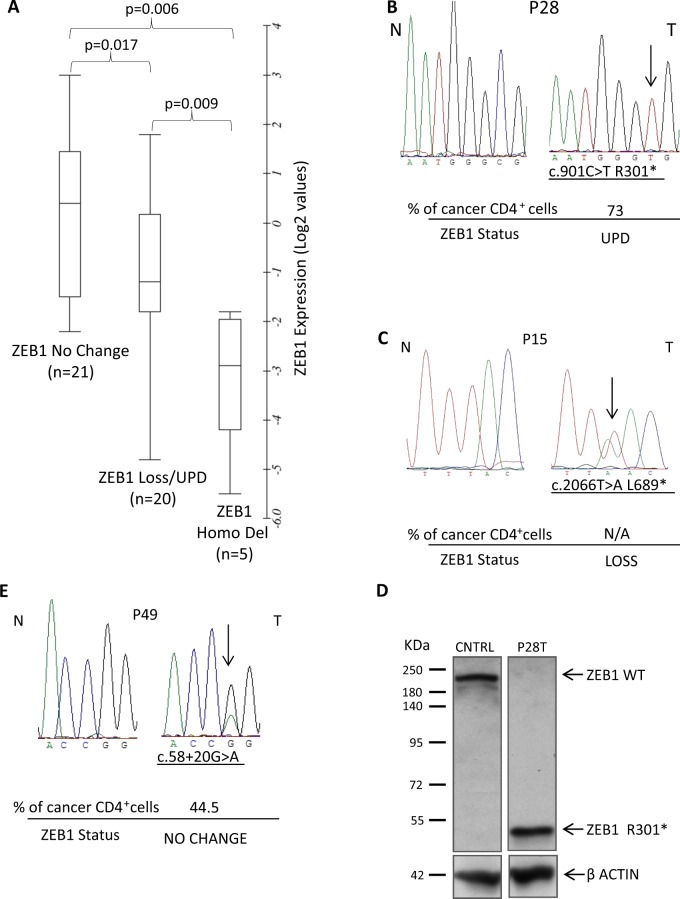
To verify whether the high frequency of ZEB1 SCNA led also to an altered transcript level, we analyzed the mRNA expression in 43 samples, with available RNA, from 35 SS patients and 3 cell lines (Table 1); we used CD4+ T cells from four healthy controls to calibrate the fold-change (FC) values of ZEB1 in quantitative PCR assays. Comparing samples showing loss (20/46) or homozygous deletion (5/46) with samples exhibiting no change of ZEB1 copy number (21/46), we found ZEB1 expression values proportional to copy-number data with significant statistical differences (Fig. 2A), indicating that ZEB1 mRNA level correlates with its genomic locus status.
Fig. 2.
A Correlation analysis of ZEB1 somatic copy-number alterations and gene expression quantitative real-time PCR data, log2 transformed; a box-plot was generated with Online Box Plot Generator (http://www.alcula.com/calculators/statistics/box-plot/) and BoxPlot Grapher (http://www.imathas.com/stattools/boxplot.html); p-values were calculated by two-tailed Student’s t test. B Sequence electropherograms showing the c.901 C > T, R301* somatic mutation identified in the tumor sample (T) of patient P28 compared with the normal matched DNA (N). C Sequence electropherograms of the ZEB1 somatic mutation (c.2066 T > A, L689*) identified in SS patients P15. D Western blot analysis of ZEB1 displaying the R301* truncated protein expression in neoplastic lymphocytes of P28 compared to ZEB1 expression in the CD4 + lymphocytes of a healthy volunteer. E Sequence electropherogram identifying the c.58 + 20 G > A substitution in the tumor DNA (T) of patient P49 compared with the normal matched DNA (N). For each SS patient sequenced, the percentage of CD4 + neoplastic cells and the ZEB1 status is indicated; a cutaneous biopsy of P15 has been used for sequence analysis of tumor DNA (T) since the patient underwent bone marrow transplantation
ZEB1 mutation analysis in SS patients
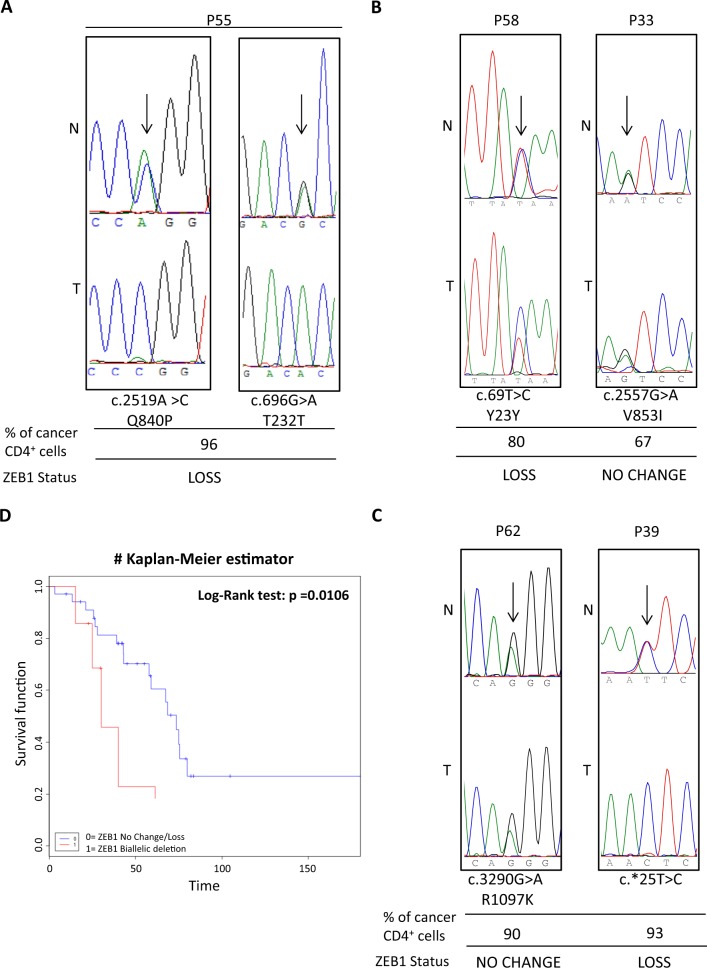
We then performed ZEB1 DNA sequence analysis in 38 SS patients and 3 CTCL cell lines. We detected seven coding nucleotide substitutions in 6/38 cases and four noncoding variations affecting 14 patients and 1 cell line. The available normal matched DNA allowed us to establish that three were somatic and eight were germline substitutions (Table 1). Two somatic mutations were nonsense predicting to generate ZEB1-truncated proteins: c.901 C > T (R301*) in P28 and c.2066 T > A (L689*) in P15 (Fig. 2B, C). These SS cases display also UPD and loss of ZEB1 locus, respectively. Western blot analysis revealed that c.901 C > T mutated gene produces a single fragment of a lower molecular mass compared with the full-length protein (Fig. 2D); if, similarly, the c.2066 T > A mutated gene is translated in a shortened 689 amino acids, then both proteins will likely result in defective molecules (Fig. S4). The third somatic variation, identified in P49 associated with a normal copy number of ZEB1, is a transition (c.58 + 20 G > A) occurring within the first intron of the gene (Fig. 2E). Computational analysis predicted the nucleotide change as probably having no effect on splicing (see Materials and methods for mutation analyses). Since the eight other substitutions were germline, we sought to determine their pathogenicity using data from dbSNP and Genome Aggregation Database (gnomAD)30 (Table 2). The intronic substitutions c.481 + 64 A > G and c.685–15 G > A, identified in 10/38 and 2/38 SS patients, respectively, are genetic variants reported as having minor allele frequencies (MAF) in dbSNP, or allele frequencies of the alternative (non-reference) allele (AF) in gnomAD, above 0.05, thereby likely representing a genetic variant of benign nature. Conversely, we found three rare genetic variations (MAF and AF < 0.05): one missense transversion (c.2519 A > C, Q840P) and two synonymous transitions (c.696 G > A, T232T and c.69 T > C,Y23Y). The missense substitution is reported as having clinical relevance owing to its association with late-onset Fuchs corneal dystrophy (FCD) and shown to cause loss of function (LoF) by an in vivo complementation assay31. Noticeably, the patient (P55) carrying this variant displays also concomitant loss of ZEB1 locus (Fig. 3A, left panel) possibly resulting in the deficiency of a functional allele. The c.696 G > A, T232T was found in the same patient (P55) (Fig. 3A, right panel), and the c.69 T > C,Y23Y in P58 SS case (Fig. 3B, left panel) both combined with ZEB1 deletions; these silent rare variations were tested for their effects on putative exonic splicing enhancers and silencers32, but the analysis did not detect any significant splicing motif alteration; it cannot be excluded, however, that they might have other consequences on mRNA stability and/or translation efficiency33. Finally, we identified the following three germline variants: (1) the c.2557 G > A (V853I) missense substitution in P33 associated with no change of ZEB1 locus (Fig. 3B, right panel), (2) the c.3290 G > A (R1097K), found in P62 carrying a normal ZEB1 copy number, and (3) c.*25 T > C present in P39 with ZEB1 loss (Fig. 3C), none of which was found in the dbSNP and gnomAD (Table 2). It is worth noting that these variations were all absent also from the COSMIC (Catalogue of Somatic Mutations in Cancer) database, suggesting that they do not occur in sporadic cancers. Multiple lines of computational evidence suggest no impact of c.2557 G > A (V853I) and c.3290 G > A (R1097K) on gene or gene product; however, due to the absence from a large general population, the significance of these variant alleles remains uncertain.
Table 2.
ZEB1 germline substitutions
| Alternative allele frequencies (AF) | ||||||
|---|---|---|---|---|---|---|
| Nucleotide position$ | Amino acid change | Exon/intron | No. of cases (%) | dbSNP rs* global MAF | gnomAD§ exome | gnomAD§ whole genome |
| c.69 T > C | Y23Y | Exon 2 | 1/38 (2.63) | rs775270565 C = 0.00002/2 | C = 0.00001223 | C = 0.0000323 |
| c.481 + 64 A > G | – | Intron 4 | 10/38 (26.3) | rs2839664 G = 0.0787/394 | – | G = 0.1308 |
| c.685-15 G > A | – | Intron 5 | 2/38 (5.2) | rs220060 G = 0.0787/394 | A = 0.9458 | A = 0.9197 |
| c.696 G > A | T232T | Exon 6 | 1/38 (2.63) | rs149166539 A = 0.0002/1 | A = 0.001227 | A = 0.0009054 |
| c.2519 A > C | Q840P | Exon 7 | 1/38 (2.63) | rs118020901 C = 0.0030/15 | C = 0.007583 | C = 0.009168 |
| c.2557 G > A | V853I | Exon 7 | 1/38 (2.63) | Absent | Allele not found despite > 20X coverage | Allele not found despite > 20X coverage |
| c.3290 G > A | R1097K | Exon 9 | 1/38 (2.63) | Absent | 0.00 | Allele not found despite > 20X coverage |
| c.*25 T > C | – | Exon 9 | 1/38 (2.63) | Absent | Allele not found despite > 20X coverage | Allele not found despite > 20X coverage |
$Ref Seq NM_030751 (CDS), from 5′to 3′ end
* https://www.ncbi.nlm.nih.gov/snp (Build 151). The current default global population is 1000Genome phase 3 genotype data from 2500 worldwide individuals, released in the May 2013 dataset. §Genome aggregation database r2.0.2 accessed through https://varsome.com/. The data set spans 123,136 exome sequencesand 15,496 whole-genome sequences from unrelated individuals
Fig. 3.
A Sequence electropherograms showing the germline heterozygous c.2519 A > C (Q840P) (left) and c.696 G > A (T232T) (right) rare variations identified in the normal DNA of P55 (N) which become hemizygous following the loss of the wild-type allele in the tumor sample (T). B, C Sequence electropherograms of DNA patients displaying rare c.69 T > C, Y23Y, and the newly identified c.2557 G > A, V853I; c.3290 G > A, R1097K; c.*25 T > C germline variants. The percentage of neoplastic CD4 + cells are indicated for each patient as well as the ZEB1 status. D Comparison of survival time (months) between SS patients carrying no change or loss of copy number (n = 34, blue line) vs. biallelic deletion (n = 7, red line) of ZEB1 gene. Patients with truncating (R301*, L689*) or loss-of-function mutations (Q840P) associated with loss were included in the biallelic deleted group. Forty-one out of 43 patients were used for this analysis because one was lost at follow-up and the other had an evolution of SS after a long stable MF disease
Biallelic inactivation of ZEB1 reduces survival in SS patients
We then evaluated if ZEB1 deficiency might affect SS patients survival. In our series, median overall survival was 48 months for ZEB1 no change or loss of copy number (n = 34), compared with 30 months for cases with biallelic loss (n = 7), including patients carrying inactivating (i.e., nonsense mutations) or Q840P LoF variation associated with loss. Overall survival analysis with the Kaplan–Meier estimator revealed reduced survival probability among patients with biallelic loss (log-rank test, P = 0.0106; Fig. 3D). Thus, beyond representing a potential oncogenetic event, lack of ZEB1 function also affects SS prognosis.
Gene sets enriched in ZEB1-depleted SS samples
To gain further insights into ZEB1 role in SS disease, we used our previously published gene expression (GE) dataset (GSE 176013) to explore the transcriptional patterns of ZEB1-deficient and ZEB1-expressing patients. Considering the available 32 arrays in the GE dataset, we had corresponding ZEB1 genetic and expression data for 19 samples (Table 1: P02T, P05T, P08T2, P22T, P23T3, P25T, P28T, P30T1, P32T1, P33T, P34T, P35T, P36T, P37T, P38T1, P39T1, P40T3, P43T2, P45T1). Clustering these SS cases for ZEB1 expression, we had six samples showing a positive log2 FC values (P30T1, P33T, P35T, P36T, P39T1, P45T1) that we labeled as ZEB1 OVER class; in order to maximize the transcriptional differences, we compared this group of cases with a comparable size set of six ZEB1-depleted samples, i.e., exhibiting either homozygous deletion of ZEB1 (P05T, P22T, P32T1), UPD (P28T) or strong mRNA down-regulation with log2 FC expression level ≤ −2.0 (P23T3, P25T); for simplicity, we indicated this class as ZEB1 HD (i.e., HomoDel). We used GSEA and the Molecular Signatures Database “Hallmarks” gene set collection34 to investigate the molecular alterations enriched in the two different groups of patients; The analysis yielded 32 gene sets that were enriched, with FDR < 0.25, in the ZEB1 HD class (Table 3A). To further verify our findings and select the Hallmarks signatures dependant on ZEB1 expression, the same dataset of samples was used to identify lists of enriched gene sets negatively correlated with the expression profile of ZEB1 (Affymetrix ID: 210875_s_at), i.e., gene sets made up of core enriched genes mostly upregulated in ZEB1-deficient samples, but downregulated in ZEB1-expressing cases. A negative association was found for 11 gene sets (FDR < 0.25) all of which were included in the ZEB1 HD class of enriched signatures (Table 3B). Notably, the presence of the Hallmark of IL2_STAT5 signaling either in positive association with ZEB1 HD class or in negative correlation with ZEB1 expression support our analyses since ZEB1 is a known repressor of IL2 transcription in T cells14,15. The results identified the cell response to interferon (IFN)α and IFNγ as highly significant signatures associated with ZEB1 depletion, suggesting it might have a role in the immune-regulatory activities mediated by these cytokines in SS T lymphocytes. Of note also the correlation with the tumor necrosis factor (TNF)α signaling via NF-kB, one of the most important activated pathway conveying persistent survival signals in SS35. Additionally, we found enriched gene sets, such as DNA repair and the P53 pathway, indicating a potential ZEB1 control also in the regulation of genomic DNA integrity. To test this hypothesis, we compared the fraction of tumor genome affected by SCNAs for nine SS samples with ZEB1 homo-deletions or strong mRNA under expression and eight SS tumors with ZEB1-positive expression calculated as the proportion of genome altered (PGA) described by Thu KL et al.36 No significant results were found considering either all lesions (i.e., gains and losses) or losses and gains taken separately (Supplementary Figure S5), thus, ZEB1 depletion does not seem to increase genetic instability in SS. The more widespread changes in the transcriptional program observed comparing ZEB1 HD with ZEB1 OVER classes might be explained, at least in part, as the downstream effects of ZEB1 lack of activities distinguishing the two groups. In support of this data interpretation, we found the upregulation of the oxidative phosphorylation enzymes in both types of analyses (Tables 3A and 3B), and the hallmarks of reactive oxygen species pathways and peroxisome associated with ZEB1 HD class (Table 3A). This is of particular interest as ZEB1 is indeed involved in ROS-induced cellular response as a crucial target of the miR-200 family members that are upregulated under oxidative stress conditions37–41.
Table 3.
Gene sets enriched in ZEB1 HD class (A). Gene sets enriched in negative correlation with ZEB1 profile (B)
| NAME | SIZE | ES | NES | NOM p-val | FDR q-val | |
|---|---|---|---|---|---|---|
| (A) | ||||||
| 1 | HALLMARK_OXIDATIVE_PHOSPHORYLATION | 188 | 0.67 | 2.67 | 0 | 0 |
| 2 | HALLMARK_INTERFERON_ALPHA_RESPONSE | 77 | 0.74 | 2.62 | 0 | 0 |
| 3 | HALLMARK_INTERFERON_GAMMA_RESPONSE | 171 | 0.65 | 2.58 | 0 | 0 |
| 4 | HALLMARK_COMPLEMENT | 181 | 0.58 | 2.31 | 0 | 0 |
| 5 | HALLMARK_MYC_TARGETS_V1 | 171 | 0.57 | 2.29 | 0 | 0 |
| 6 | HALLMARK_ADIPOGENESIS | 169 | 0.53 | 2.08 | 0 | 0 |
| 7 | HALLMARK_ALLOGRAFT_REJECTION | 191 | 0.52 | 2.07 | 0 | 0 |
| 8 | HALLMARK_REACTIVE_OXIGEN_SPECIES_PATHWAY | 43 | 0.58 | 1.89 | 0 | 0.001 |
| 9 | HALLMARK_APOPTOSIS | 157 | 0.48 | 1.88 | 0 | 0.001 |
| 10 | HALLMARK_IL6_JAK_STAT3_SIGNALING | 85 | 0.51 | 1.85 | 0 | 0.001 |
| 11 | HALLMARK_PROTEIN_SECRETION | 88 | 0.5 | 1.83 | 0 | 0.001 |
| 12 | HALLMARK_IL2_STAT5_SIGNALING | 169 | 0.45 | 1.78 | 0 | 0.003 |
| 13 | HALLMARK_MTORC1_SIGNALING | 182 | 0.45 | 1.75 | 0 | 0.003 |
| 14 | HALLMARK_FATTY_ACID_METABOLISM | 140 | 0.45 | 1.73 | 0.001 | 0.004 |
| 15 | HALLMARK_DNA_REPAIR | 135 | 0.45 | 1.71 | 0.001 | 0.005 |
| 16 | HALLMARK_KRAS_SIGNALING_UP | 191 | 0.42 | 1.69 | 0 | 0.006 |
| 17 | HALLMARK_XENOBIOTIC_METABOLISM | 185 | 0.42 | 1.67 | 0.001 | 0.008 |
| 18 | HALLMARK_INFLAMMATORY_RESPONSE | 185 | 0.41 | 1.64 | 0 | 0.01 |
| 19 | HALLMARK_HEME_METABOLISM | 183 | 0.41 | 1.63 | 0.001 | 0.011 |
| 20 | HALLMARK_TNFA_SIGNALING_VIA_NFKB | 185 | 0.41 | 1.63 | 0.002 | 0.01 |
| 21 | HALLMARK_G2M_CHECKPOINT | 176 | 0.41 | 1.62 | 0.001 | 0.01 |
| 22 | HALLMARK_UNFOLDED_PROTEIN_RESPONSE | 104 | 0.42 | 1.55 | 0.011 | 0.018 |
| 23 | HALLMARK_E2F_TARGETS | 167 | 0.39 | 1.53 | 0.003 | 0.02 |
| 24 | HALLMARK_COAGULATION | 132 | 0.39 | 1.51 | 0.011 | 0.024 |
| 25 | HALLMARK_CHOLESTEROL_HOMEOSTASIS | 60 | 0.44 | 1.51 | 0.031 | 0.023 |
| 26 | HALLMARK_PEROXISOME | 94 | 0.4 | 1.45 | 0.041 | 0.04 |
| 27 | HALLMARK_TGF_BETA_SIGNALING | 49 | 0.43 | 1.45 | 0.043 | 0.041 |
| 28 | HALLMARK_P53_PATHWAY | 182 | 0.36 | 1.4 | 0.024 | 0.059 |
| 29 | HALLMARK_ANDROGEN_RESPONSE | 94 | 0.36 | 1.34 | 0.077 | 0.098 |
| 30 | HALLMARK_UV_RESPONSE_UP | 150 | 0.34 | 1.33 | 0.051 | 0.099 |
| 31 | HALLMARK_ESTROGEN_RESPONSE_LATE | 193 | 0.33 | 1.31 | 0.061 | 0.115 |
| 32 | HALLMARK_PANCREAS_BETA_CELLS | 36 | 0.42 | 1.3 | 0.129 | 0.114 |
| (B) | ||||||
| 1 | HALLMARK_INTERFERON_ALPHA_RESPONSE | 77 | −0.57 | −2.86 | 0 | 0 |
| 2 | HALLMARK_INTERFERON_GAMMA_RESPONSE | 171 | −0.47 | −2.68 | 0 | 0 |
| 3 | HALLMARK_TNFA_SIGNALING_VIA_NFKB | 185 | −0.28 | −1.65 | 0 | 0.029 |
| 4 | HALLMARK_IL2_STAT5_SIGNALING | 169 | −0.28 | −1.6 | 0 | 0.034 |
| 5 | HALLMARK_APOPTOSIS | 157 | −0.28 | −1.58 | 0 | 0.031 |
| 6 | HALLMARK_DNA_REPAIR | 135 | −0.28 | −1.56 | 0.008 | 0.029 |
| 7 | HALLMARK_P53_PATHWAY | 182 | −0.25 | −1.45 | 0.005 | 0.059 |
| 8 | HALLMARK_ADIPOGENESIS | 169 | −0.24 | −1.39 | 0.014 | 0.081 |
| 9 | HALLMARK_OXIDATIVE_PHOSPHORYLATION | 188 | −0.24 | −1.37 | 0.02 | 0.089 |
| 10 | HALLMARK_UV_RESPONSE_UP | 150 | −0.23 | −1.28 | 0.059 | 0.153 |
| 11 | HALLMARK_COMPLEMENT | 181 | −0.21 | −1.21 | 0.073 | 0.239 |
SIZE size of gene set, ES enriched score, NES normalized enriched score, NOM p-val nominal p-value, FDR q-val, false discovery rate q-value
Bold font denotes common gene sets enriched in both analyses
ZEB1 is not regulated by miR200c in SS
We tried to assess if any relationship existed between ZEB1 and miR200 expression in SS. From microarray data of our previous study on miRNA transcription profiling in SS42, we identify miR200c as the family member having the highest variance in expression between patients and controls; thus we performed a real time quantitative assay to estimate miR200c FC values in 18 SS patients/cell lines (P25T, P30T1, P37T, P39T1, P39T2, P40T3, P43T3, P45T1, P48T1, P51T1, P62T1, P63T1, P67T1, P68T1, P69T, HUT78, H9, and HH cell lines) using the same set of healthy donors employed for ZEB1 expression analysis. No significant correlation emerged between the log2 FC values of ZEB1 and miR200c in a Pearson’s correlation analysis (R = 0.0144, p-value = 0.956). Also, dividing the same group of samples for ZEB1 expression level, we calibrated the miR200c FC values of the under-expressing cases (P25T, P37T, P40T3, P48T1, P62T1, P67T1, P68T1, P69T, HUT78) on the average value of the over-expressing tumors (P30T1, P39T1, P39T2, P43T3, P45T1, P51T1, P63T1, H9, HH); in this case a weak negative correlation emerged that, however, did not reach the significance (Pearson’s correlation R = −0.426, p-value = 0.25). Our results suggest that ZEB1 role in oxidative phosphorylation response of SS cells is independent from miR200c expression.
Oxidative stress and ZEB1 depletion in SS cells
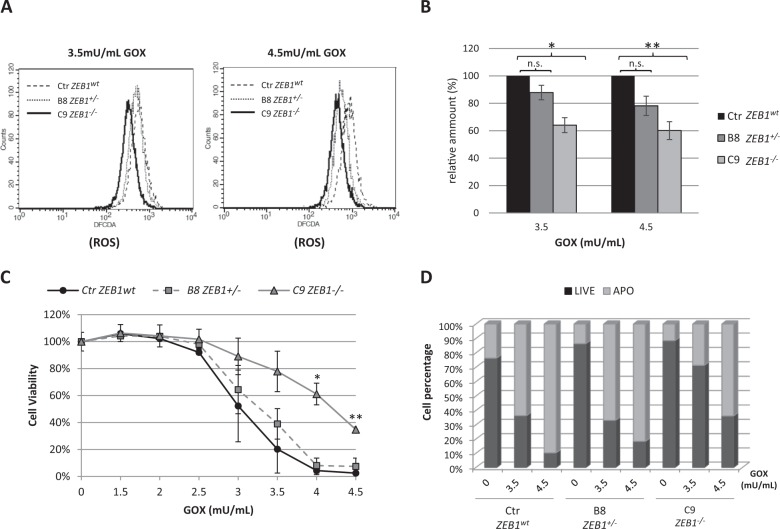
The stimulation of T-cell receptor (TCR) generates an oxidative signal that is crucial to trigger the activation-induced cell death (AICD), a key process required to keep in balance the immune system43,44. This prompted us to study the effects of ZEB1 deficiency in SS context of oxidative condition. To this end, we used heterozygous and homozygous CRISPR/Cas9-generated ZEB1 KO cell clones, derived from H9 SS cell line (see Supplementary Methods), treated with glucose oxidase (GOX) which reacts with glucose and oxygen for a continuous enzymatic generation of H2O2. This reagent, used at relatively low concentration (2–5 mU/mL), it is used to reproduce a physiological condition of constant low level H2O2-mediated oxidative signal45. Flow cytometry analysis of cells cultured with increasing concentrations of GOX for 4 hours showed a sustained increase in intracellular ROS production, however, if compared with the WT CRISPR/Cas9 control cells (Ctr ZEB1wt), ZEB1−/− cell clone (C9 ZEB1−/−) showed an appreciable lower amount of ROS at 3.5 and 4.5 mU/mL of GOX concentration, while the fluorescence intensity of ZEB1+/− cells (B8 ZEB1+/−) overlapped the WT control at 3.5 mU/mL and exhibited an intermediate profile between the two genotypes at 4.5 mU/mL of GOX (Fig. 4A). When ROS levels were calculated relatively to ZEB1wt control cells, significant statistical decreases were measured for ZEB1−/− cells at both GOX concentrations, whereas no significant differences emerged in ZEB1+/− clone (Fig. 4B). These results suggest that ZEB1-deficient SS cells are more competent to respond to H2O2-induced oxidative stress counterbalancing increasing intracellular ROS production. Cell viability assays of ZEB1wt and KO clones showed that ZEB1−/− cells exhibited higher cell viability with respect to ZEB1wt and ZEB1+/– following the exposure to increase GOX-induced oxidative stress with significant differences observed at 4.0 and 4.5 mU/mL (Fig. 4C). Flow cytometry, used to quantify cellular apoptosis in response to 3.5 and 4.5 mU/mL of GOX, revealed that ZEB1−/− KO showed 29 and 64% of apoptotic cells, respectively; conversely, ZEB1wt exhibited 64 and 94% of apoptosis at the corresponding GOX concentrations, comparable with the 67 and 82% values observed for ZEB1+/− KO (Fig. 4D). Collectively, our data show that ZEB1-devoid SS cells are able to respond to oxidative stress counteracting ROS imbalance, this ability influences cell survival and apoptosis and it requires the absence of both ZEB1 alleles.
Fig. 4.
A Flow cytometry analysis showing the levels of ROS produced in ZEB1wt control cells and ZEB1 KO clones following exposure to 3.5 mU/mL and 4.5 mU/mL of GOX concentration; ROS are detected as H2DCFDA fluorogenic dye intensity. B The amount of ROS produced in ZEB1 KO clones, relative to the WT control, following 3.5 and 4.5 mU/mL of GOX treatment for 4 h. Data are expressed as means ± SEM (n = 3), *p < 0.05 **p < 0.01 vs. control. C MTT assay showing the differences in cell viability between the ZEB1wt control cells and ZEB1 KO clones treated with increasing concentration of GOX; *p < 0.05, **p < 0.001 vs. control. D Bar graph quantifying the percentage of dead and living cell sub-populations in ZEB1wt control and KO clones according to the indicated treatments. Data are expressed as percentage of total cell numbers. Light-gray bars indicate Annexin V-positive apoptotic cells
Discussion
We have generated a high-resolution SNP6 data set of SS tumors; in the absence of hallmarks of disease progression, we used multiple follow-up samples to increase the detection of positively selected clonal DNA aberration in the disease course. The GISTIC algorithm delineated a profile of significant regions of gain and loss, with a large predominance of focal deletions over amplifications. This is in line with previously published studies and support the hypothesis that deletion events, targeting critical tumor suppressors, represent a mechanism of CTCL pathogenesis5,6. We narrowed some of the most significant focal deletions to critical tumor suppressor targets, such as TP53, PTEN, and RB1 (Fig. 1), thereby involving their loss as driver events in SS tumorigenesis. Recent studies, integrating copy-number and single-nucleotide variant analyses, identified these genes with variable frequencies in CTCL cohorts: TP53 was affected in 21–92.5% of cases, PTEN ranged from 20 to 30%, and RB1 and CDKN2A were targeted in 16–39% and 32.5–58% of tumors, respectively6,8,9,11,46. Our data also confirmed targets such as PRKCQ found altered in 20–30% of CTCLs6,11, and support other candidates like the chromatin-modifying genes ARID1A, implicated in 33–62.5% of CTCLs, and DNMT3A involved in 9–42.5% of cases6,8,9,11,12. This great variability in the alteration frequencies might depend on sample sizes/compositions and/or diversities of technical/analytical approaches. In our cohort, the GISTIC analysis identified 10p11.22 as the region most significantly affected by loss of genetic material and ZEB1 as the target of this region; also, we found the presence of nonsense somatic mutations, predicting to generate defective proteins coupled with ZEB1 deletion. A combined burden of deletions and somatic mutations, affecting ZEB1, has been reported with frequencies ranging from 45 to 65% of CTCLs2,6,9,10. Thus, our and previous findings support ZEB1 as a strong candidate tumor suppressor gene in CTCL as already described in ATLL;20 differently from canonical cancer genes like TP53, PTEN, RB1, and CDKN2A recurrently inactivated also in other cancer types, ZEB1 seems to be a lineage-specific tumor suppressor. Here, we show, for the first time, that SS patients harbor not only ZEB1 somatic mutations, but also germline variations; we found that 8/11 allelic variants were of germline origin (Table 1 and Table 2). We found substitutions reported as either rare variants (MAF/AF < 5%) or totally absent from dbSNPs or gnomAD, representing, thereby, novel sequence variations (Table 2). Importantly, the Q840P (c.2519 A > C) is a ZEB1 LoF variation associated with a rare inherited disorder of the corneal endothelium31. Interestingly, Hidaka et al.20, reported the identification of ZEB1 c.232 A > C (N78T) variation in the Hut102 CTCL cell line, which, also, is recurrent in FCD patients and causes a protein impairment31. Although the germline origin of the N78T variation cannot be proved for Hut102, it is worth noting that it is absent from the COSMIC database, whereas it is cataloged as a variation with a global MAF < 5% in the dbSNP (rs80194531) and gnomAD, further supporting our findings. Rare genetic variations play an important role in human disorders, as they may cause many Mendelian and rare forms of common diseases47. Evolutionary theory predicts that disease alleles are likely to be rare as a result of purifying selection47,48 and indeed, LoF variants are especially rare49,50. Also, empirical evidences have shown that low frequency and rare variants are associated with complex diseases, including cancer51–53. Moreover, emerging evidence suggests that synonymous SNPs could affect, besides mRNA splicing, also mRNA stability, protein expression, and enzymatic activity contributing to human disease risk and other complex traits (reviewed in ref. 54). Although further observations and experimental validations are necessary to establish the functional consequences of ZEB1 germline variations in SS, this is the first report to suggest a genetic risk for this rare neoplasm. Overall survival analysis, in our SS series, highlighted an association of poorer outcome with biallelic loss of ZEB1, which supports its recessive nature of tumor suppressor and underlines its potential role as a risk-stratification and prognostic marker. In CTCL, ZEB1 acts as a transcriptional repressor contributing to an oncogenic regulatory mechanism involving IL15, HDAC1, HDAC6, and miR-2116. Here, we found that ZEB1 absence is associated with multiple dysregulated pathways (Table 3), among which, the upregulation of IL2 signaling and the oxidative stress cell response were already shown to depend on ZEB1 expression. In oxidative stress condition, ZEB1 downregulation by miR-200c has been described; we could not confirm this relationship in SS, rather ZEB1 expression is mainly dependant on the integrity of its genomic locus (Fig. 2A). We verified that ZEB1−/− SS cells respond to oxidative stress with significant lower intracellular ROS levels, higher cell viability, and resistance to apoptosis. ROS mediate apoptosis in a number of cell types; in T cells, re-stimulation of previously activated T cells via the TCR increases ROS concentrations leading to AICD, an important regulatory mechanism of T-cell homeostasis55. Indeed, CTCLs have been shown to be AICD-resistant, while inhibition of the NF-kB pathway induces CTCL cell death via free intracellular iron and massive ROS production56,57. NF-kB exerts a negative control on ROS formation, which is central for the suppression of TNFα-induced apoptosis58–61. Interestingly, we found that upregulation of TNFα signaling via NF-kB is significantly correlated with ZEB1 absence, suggesting a control of this pathway in SS disease. Recently, Morel et al. reported that ZEB1 reduces cellular ROS in mammary stem cells following an oncogenic stimulus, while the expression of several detoxifying enzymes was increased; among them, they identified MSRB3 as a transcriptional target of ZEB1, implying its direct role in the control of ROS production62. We did not find MSRB3, but HIF1A, another antioxidant gene found by Morel et al., positively correlated with ZEB1 depletion. It would be interesting to further investigate if ZEB1 is capable, not only to induce ROS-scavenging factors, but also to repress them in a cell-type-specific manner. Although we found molecular pathways of genomic instability associated with ZEB1 absence, no significant differences emerged in our PGA analysis between ZEB1-depleted and ZEB1-expressing tumors; this might be due to additional independent factors contributing to DNA damage such as defects in genes deputed to genome maintenance11. Also, it has been shown very recently that ZEB1 mediates the epigenetic silencing of protective type III IFN response in airway mucosal epithelium63. Here, we found ZEB1 depletion highly correlated with IFNα and IFNγ response (Table 3). IFNs have been proposed to play an important role in regulating the abundance and persistence of memory T cell;64,65 particularly, previous studies showed that IFNα/β inhibited activated T-cell apoptosis directly, without induction of secondary mediators, promoting, among the diverse survival pathways, also the upregulation of intracellular antioxidant molecule glutathione66–68. Although our observations await further confirmations and mechanistic experimental validations, they provide evidence for uncharacterized ZEB1 roles in regulating T-cell homeostasis, paving the way for future developments and treatment opportunities.
Ethics approval and consent to participate
This study was conducted in accordance with Good Clinical Practice Guidelines and the Declaration of Helsinki and approved by the Ethical Committee of the Istituto Dermopatico dell’Immacolata (ID n. 4/CE/2015). A written consent was obtained from all patients.
Electronic supplementary material
Acknowledgements
The authors wish to thank the patients of I Clinical Dermatology Unit of the IDI-IRCCS, G. Alvetreti and E. Ferranti for nursing assistance and collaboration, G. Costanzi for computer technology assistance, and Dr. A. Magenta and Dr. S. Pastore for scientific support and helpful discussion. The authors wish to remember Dr. F. Carlomosti who generously gave initial contribution and prematurely passed away. This study was supported by Associazione Italiana Ricerca sul Cancro IG-15828 (GR), Associazione Italiana Ricerca sul Cancro IG-17748 (MGN), Fondi Ricerca Corrente from Italian Ministry of Health RC2016-17 (EC).
Authors contribution
E.C. conceived and supervised the study, performed sequence, bioinformatics and survival analyses, interpreted and analyzed data, and wrote the paper, G.R. co-designed the study, interpreted data, and revised the paper. A.B. and C.C. collected samples, acquired SNP array data, performed expression and biochemical experiments, cell biology and flow cytometry assays, KO cell lines generation, data acquisition, and interpretation; M.H.C. and V.T. collected samples, processed DNA for SNP array and sequencing, and performed real-time and droplet digital PCR assays; E.S. performed cytofluorimetric analyses of patients samples; A.M. and R.B. recruited eligible patients and collected clinical information; M.G.N. interpreted data and critically revised the paper. All authors read and approved the final paper.
Data availability
The datasets 10 K SNP Arrays (GSE17595) and Gene Expression Arrays (GSE17601) supporting the conclusions of this article are available in GEO public data repository under the super series GSE17602 at the following link: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc = GSE17602, SNP6 Arrays dataset is under ethical evaluation for submission in GEO public data repository.
Conflict of interest
The authors declare that they have no conflict of interest.
Footnotes
Edited by G. Melino
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Antonella Bresin, Cristina Cristofoletti
Contributor Information
Maria Grazia Narducci, Phone: +39 06 66464798, Email: m.narducci@idi.it.
Giandomenico Russo, Phone: +39 06 66462432, Email: g.russo@idi.it.
Electronic supplementary material
Supplementary Information accompanies this paper at (10.1038/s41419-018-1212-7).
References
- 1.Fierro MT, et al. Heterogeneity of circulating CD4+memory T-cell subsets in erythrodermic patients: CD27 analysis can help to distinguish cutaneous T-cell lymphomas from inflammatory erythroderma. Dermatology. 2008;216:213–221. doi: 10.1159/000112928. [DOI] [PubMed] [Google Scholar]
- 2.Vermeer MH, et al. Novel and highly recurrent chromosomal alterations in Sézary syndrome. Cancer Res. 2008;68:2689–2698. doi: 10.1158/0008-5472.CAN-07-6398. [DOI] [PubMed] [Google Scholar]
- 3.Caprini E, et al. Identification of key regions and genes important in the pathogenesis of Sézary syndrome by combining genomic and expression microarrays. Cancer Res. 2009;69:8438–8446. doi: 10.1158/0008-5472.CAN-09-2367. [DOI] [PubMed] [Google Scholar]
- 4.Laharanne Elodie, Oumouhou Naïma, Bonnet Françoise, Carlotti Martina, Gentil Catherine, Chevret Edith, Jouary Thomas, Longy Michel, Vergier Béatrice, Beylot-Barry Marie, Merlio Jean-Philippe. Genome-Wide Analysis of Cutaneous T-Cell Lymphomas Identifies Three Clinically Relevant Classes. Journal of Investigative Dermatology. 2010;130(6):1707–1718. doi: 10.1038/jid.2010.8. [DOI] [PubMed] [Google Scholar]
- 5.Lin WM, et al. Characterization of the DNA copy-number genome in the blood of cutaneous T-cell lymphoma patients. J. Invest. Dermatol. 2012;132:188–197. doi: 10.1038/jid.2011.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi J, et al. Genomic landscape of cutaneous T cell lymphoma. Nat. Genet. 2015;47:1–11. doi: 10.1038/ng.3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ungewickell A, et al. Genomic analysis of mycosis fungoides and Sézary syndrome identifies recurrent alterations in TNFR2. Nat. Genet. 2015;47:1056–1060. doi: 10.1038/ng.3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.da Silva Almeida AC, et al. The mutational landscape of cutaneous T cell lymphoma and Sézary syndrome. Nat. Genet. 2015;47:1465–1470. doi: 10.1038/ng.3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang L, et al. Genomic profiling of Sézary syndrome identifies alterations of key T cell signaling and differentiation genes. Nat. Genet. 2015;47:1426–1434. doi: 10.1038/ng.3444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McGirt LY, et al. Whole-genome sequencing reveals oncogenic mutations in mycosis fungoides. Blood. 2015;126:508–519. doi: 10.1182/blood-2014-11-611194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Woollard WJ, et al. Candidate driver genes involved in genome maintenance and DNA repair in Sézary syndrome. Blood. 2016;127:3387–3397. doi: 10.1182/blood-2016-02-699843. [DOI] [PubMed] [Google Scholar]
- 12.Kiel MJ, et al. Genomic analyses reveal recurrent mutations in epigenetic modifiers and the JAK-STAT pathway in Sézary syndrome. Nat. Commun. 2015;6:8470. doi: 10.1038/ncomms9470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park J, et al. Genomic analysis of 220 CTCLs identifies a novel recurrent gain-of-function alteration in RLTPR (p.Q575E. Blood. 2017;130:1430–1440. doi: 10.1182/blood-2017-02-768234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Williams TM, et al. Identification of a zinc finger protein that inhibits IL-2 gene expression. Sci. (80-). 1991;254:1791–1794. doi: 10.1126/science.1840704. [DOI] [PubMed] [Google Scholar]
- 15.Wang J, et al. The transcription repressor, ZEB1, cooperates with CtBP2 and HDAC1 to suppress IL-2 gene activation in T cells. Int. Immunol. 2009;21:227–235. doi: 10.1093/intimm/dxn143. [DOI] [PubMed] [Google Scholar]
- 16.Mishra A, et al. Mechanism, consequences, and therapeutic targeting of abnormal IL15 signaling in cutaneous T-cell lymphoma. Cancer Discov. 2016;6:986–1005. doi: 10.1158/2159-8290.CD-15-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Higashi Y, et al. Impairment of T cell development in deltaEF1 mutant mice. J. Exp. Med. 1997;185:1467–1479. doi: 10.1084/jem.185.8.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brabletz T, et al. Negative regulation of CD4 expression in T cells by the transcriptional repressor ZEB. Int. Immunol. 1999;11:1701–1708. doi: 10.1093/intimm/11.10.1701. [DOI] [PubMed] [Google Scholar]
- 19.Caramel J, Ligier M, Puisieux A. Pleiotropic roles for ZEB1 in cancer. Cancer Res. 2018;78:30–35. doi: 10.1158/0008-5472.CAN-17-2476. [DOI] [PubMed] [Google Scholar]
- 20.Hidaka T, et al. Down-regulation of TCF8 is involved in the leukemogenesis of adult T-cell leukemia/lymphoma. Blood. 2008;112:383–393. doi: 10.1182/blood-2008-01-131185. [DOI] [PubMed] [Google Scholar]
- 21.Nakahata S, Yamazaki S, Nakauchi H, Morishita K. Downregulation of ZEB1 and overexpression of Smad7 contribute to resistance to TGF-Β1-mediated growth suppression in adult T-cell leukemia/lymphoma. Oncogene. 2010;29:4157–4169. doi: 10.1038/onc.2010.172. [DOI] [PubMed] [Google Scholar]
- 22.Olsen EA, et al. Clinical end points and response criteria in mycosis fungoides and Sézary syndrome: a consensus statement of the international society for cutaneous lymphomas, the United States cutaneous lymphoma consortium, and the cutaneous lymphoma task force of the E. J. Clin. Oncol. 2011;29:2598–2607. doi: 10.1200/JCO.2010.32.0630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin M, et al. dChipSNP: significance curve and clustering of SNP-array-based loss-of-heterozygosity data. Bioinformatics. 2004;20:1233–1240. doi: 10.1093/bioinformatics/bth069. [DOI] [PubMed] [Google Scholar]
- 24.Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–572. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 25.Cristofoletti C., Picchio M. C., Lazzeri C., Tocco V., Pagani E., Bresin A., Mancini B., Passarelli F., Facchiano A., Scala E., Lombardo G. A., Cantonetti M., Caprini E., Russo G., Narducci M. G. Comprehensive analysis of PTEN status in Sezary syndrome. Blood. 2013;122(20):3511–3520. doi: 10.1182/blood-2013-06-510578. [DOI] [PubMed] [Google Scholar]
- 26.Beroukhim R, et al. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc. Natl. Acad. Sci. 2007;104:20007–20012. doi: 10.1073/pnas.0710052104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 28.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mermel CH, et al. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12:R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lek M, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Riazuddin SA, et al. Missense mutations in TCF8 cause late-onset fuchs corneal dystrophy and interact with FCD4 on chromosome 9p. Am. J. Hum. Genet. 2010;86:45–53. doi: 10.1016/j.ajhg.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cartegni L, Chew SL, Krainer AR. Listening to silence and understanding nonsense: Exonic mutations that affect splicing. Nat. Rev. Genet. 2002;3:285–298. doi: 10.1038/nrg775. [DOI] [PubMed] [Google Scholar]
- 33.Tennessen JA, et al. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Sci. (80-). 2012;336:64–69. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liberzon A, et al. The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sors A. Down-regulating constitutive activation of the NF- B canonical pathway overcomes the resistance of cutaneous T-cell lymphoma to apoptosis. Blood. 2006;107(6):2354–2363. doi: 10.1182/blood-2005-06-2536. [DOI] [PubMed] [Google Scholar]
- 36.Thu KL, et al. Lung adenocarcinoma of never smokers and smokers harbor differential regions of genetic alteration and exhibit different levels of genomic instability. PLoS. One. 2012;7:e33003. doi: 10.1371/journal.pone.0033003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Magenta A, et al. MiR-200c is upregulated by oxidative stress and induces endothelial cell apoptosis and senescence via ZEB1 inhibition. Cell Death Differ. 2011;18:1628–1639. doi: 10.1038/cdd.2011.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cortez MA, et al. Therapeutic delivery of miR-200c enhances radiosensitivity in lung cancer. Mol. Ther. 2014;22:1494–1503. doi: 10.1038/mt.2014.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wu Y, Gu C, Huang X. Sevoflurane protects against hepatic ischemia/reperfusion injury by modulating microRNA-200c regulation in mice. Biomed. Pharmacother. 2016;84:1126–1136. doi: 10.1016/j.biopha.2016.10.024. [DOI] [PubMed] [Google Scholar]
- 40.D’Agostino M, et al. Circulating miR-200c is up-regulated in paediatric patients with familial hypercholesterolaemia and correlates with miR-33a/b levels: implication of a ZEB1-dependent mechanism. Clin. Sci. (Lond.) 2017;131:2397–2408. doi: 10.1042/CS20171121. [DOI] [PubMed] [Google Scholar]
- 41.Yi Y, et al. Ultraviolet A irradiation induces senescence in human dermal fibroblasts by down-regulating DNMT1 via ZEB1. Aging. 2018;10:212–228. doi: 10.18632/aging.101383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Narducci MG, et al. MicroRNA profiling reveals that miR-21, miR486 and miR-214 are upregulated and involved in cell survival in Sézary syndrome. Cell Death Dis. 2011;2:e151. doi: 10.1038/cddis.2011.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Devadas S, Zaritskaya L, Rhee SG, Oberley L, Williams MS. Discrete generation of superoxide and hydrogen peroxide by T cell receptor stimulation. J. Exp. Med. 2002;195:59–70. doi: 10.1084/jem.20010659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gulow K, et al. HIV-1 trans-activator of transcription substitutes for oxidative signaling in activation-induced T cell death. J. Immunol. 2005;174:5249–5260. doi: 10.4049/jimmunol.174.9.5249. [DOI] [PubMed] [Google Scholar]
- 45.Kaczara P, Sarna T, Burke JM. Dynamics of H2O2 availability to ARPE-19 cultures in models of oxidative stress. Free Radic. Biol. Med. 2010;48:1064–1070. doi: 10.1016/j.freeradbiomed.2010.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prasad Aparna, Rabionet Raquel, Espinet Blanca, Zapata Luis, Puiggros Anna, Melero Carme, Puig Anna, Sarria-Trujillo Yaris, Ossowski Stephan, Garcia-Muret Maria P., Estrach Teresa, Servitje Octavio, Lopez-Lerma Ingrid, Gallardo Fernando, Pujol Ramon M., Estivill Xavier. Identification of Gene Mutations and Fusion Genes in Patients with Sézary Syndrome. Journal of Investigative Dermatology. 2016;136(7):1490–1499. doi: 10.1016/j.jid.2016.03.024. [DOI] [PubMed] [Google Scholar]
- 47.Gibson G. Rare and common variants: twenty arguments. Nat. Rev. Genet. 2012;13:135–145. doi: 10.1038/nrg3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kryukov GV, Shpunt A, Stamatoyannopoulos JA, Sunyaev SR. Power of deep, all-exon resequencing for discovery of human trait genes. Proc. Natl. Acad. Sci. U. S. A. 2009;106:3871–3876. doi: 10.1073/pnas.0812824106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.MacArthur DG, et al. A systematic survey of loss-of-function variants in human protein-coding genes. Science. 2012;335:823–828. doi: 10.1126/science.1215040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.1000 Genomes Project Consortium. et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rivas MA, et al. Deep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel disease. Nat. Genet. 2011;43:1066–1073. doi: 10.1038/ng.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gudmundsson J, et al. A study based on whole-genome sequencing yields a rare variant at 8q24 associated with prostate cancer. Nat. Genet. 2012;44:1326–1329. doi: 10.1038/ng.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jonsson T, et al. A mutation in APP protects against Alzheimer’s disease and age-related cognitive decline. Nature. 2012;488:96–99. doi: 10.1038/nature11283. [DOI] [PubMed] [Google Scholar]
- 54.Sauna ZE, Kimchi-Sarfaty C. Understanding the contribution of synonymous mutations to human disease. Nat. Rev. Genet. 2011;12:683–691. doi: 10.1038/nrg3051. [DOI] [PubMed] [Google Scholar]
- 55.Hildeman Da, Mitchell T, Kappler J, Marrack P. T cell apoptosis and reactive oxygen species. J. Clin. Invest. 2003;111:575–581. doi: 10.1172/JCI200318007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Klemke CD, et al. Lack of T-cell receptor-induced signaling is crucial for CD95 ligand up-regulation and protects cutaneous T-cell lymphoma cells from activation-induced cell death. Cancer Res. 2009;69:4175–4183. doi: 10.1158/0008-5472.CAN-08-4631. [DOI] [PubMed] [Google Scholar]
- 57.Kiessling MK, et al. Inhibition of constitutively activated nuclear factor-K,B induces reactive oxygen species- and iron-dependent cell death in cutaneous T-cell lymphoma. Cancer Res. 2009;69:2365–2374. doi: 10.1158/0008-5472.CAN-08-3221. [DOI] [PubMed] [Google Scholar]
- 58.Sakon S, et al. NF- B inhibits TNF-induced accumulation of ROS that mediate prolonged MAPK activation and necrotic cell death. EMBO J. 2003;22:3898–3909. doi: 10.1093/emboj/cdg379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pham CG, et al. Ferritin heavy chain upregulation by NF-kappaB inhibits TNFalpha-induced apoptosis by suppressing reactive oxygen species. Cell. 2004;119:529–542. doi: 10.1016/j.cell.2004.10.017. [DOI] [PubMed] [Google Scholar]
- 60.Kamata H, et al. Reactive oxygen species promote TNFalpha-induced death and sustained JNK activation by inhibiting MAP kinase phosphatases. Cell. 2005;120:649–661. doi: 10.1016/j.cell.2004.12.041. [DOI] [PubMed] [Google Scholar]
- 61.Papa S, et al. The NF-kappaB-mediated control of the JNK cascade in the antagonism of programmed cell death in health and disease. Cell Death Differ. 2006;13:712–729. doi: 10.1038/sj.cdd.4401865. [DOI] [PubMed] [Google Scholar]
- 62.Morel AP, et al. A stemness-related ZEB1-MSRB3 axis governs cellular pliancy and breast cancer genome stability. Nat. Med. 2017;23:568–578. doi: 10.1038/nm.4323. [DOI] [PubMed] [Google Scholar]
- 63.Yang J, Tian B, Sun H, Garofalo RP, Brasier AR. Epigenetic silencing of IRF1 dysregulates type III interferon responses to respiratory virus infection in epithelial to mesenchymal transition. Nat. Microbiol. 2017;2:17086. doi: 10.1038/nmicrobiol.2017.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Akbar, A. N., Lord, J. M. & Salmon, M. IFN-alpha and IFN-beta: a link between immune memory and chronic inflammation. Immunol. Today21, 337–342 (2000). [DOI] [PubMed]
- 65.Whitmire JK, Eam B, Benning N, Whitton JL. Direct interferon-gamma signaling dramatically enhances CD4+and CD8+T cell memory. J. Immunol. 2007;179:1190–1197. doi: 10.4049/jimmunol.179.2.1190. [DOI] [PubMed] [Google Scholar]
- 66.Marrack P, Kappler J, Mitchell T. Type I interferons keep activated T cells alive. J. Exp. Med. 1999;189:521–529. doi: 10.1084/jem.189.3.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pilling D, et al. Interferon-beta mediates stromal cell rescue of T cells from apoptosis. Eur. J. Immunol. 1999;29:1041–1050. doi: 10.1002/(SICI)1521-4141(199903)29:03<1041::AID-IMMU1041>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 68.Hyde H, Borthwick NJ, Janossy G, Salmon M, Akbar AN. Upregulation of intracellular glutathione by fibroblast-derived factor(s): enhanced survival of activated T cells in the presence of low Bcl-2. Blood. 1997;89:2453–2460. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets 10 K SNP Arrays (GSE17595) and Gene Expression Arrays (GSE17601) supporting the conclusions of this article are available in GEO public data repository under the super series GSE17602 at the following link: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc = GSE17602, SNP6 Arrays dataset is under ethical evaluation for submission in GEO public data repository.