Abstract
Deacetylation is the removal of an acetyl group and occurs on a plethora of targets and for a wide range of biological reasons. Several pathogens deacetylate their surface carbohydrates to evade immune response or to support biofilm formation. Furthermore, dynamic acetylation/deacetylation cycles govern processes from chromatin remodeling to posttranslational modifications that compete with phosphorylation. Acetylation usually occurs on nitrogen and oxygen atoms and are referred to as N- and O-acetylation, respectively. This review discusses the structural prerequisites that enzymes must have to catalyze the deacetylation reaction, and how they adapted by formation of specific substrate and metal binding sites.
Subject terms: Pathogens, Carbohydrates, Acetylation, Histone post-translational modifications, Enzyme mechanisms
Marco Bürger and Joanne Chory discuss the structural requirements for enzymes carrying out deacetylation reactions for various functions across phyla. They explore how these enzymes have adapted to and achieved specificity on a large number of target molecules.
Introduction
Intuitively, a deacetylation reaction requires prior acetylation. The introduction of an acetyl group into a molecule results in an amide bond formation upon nitrogen acetylation (N-acetylation) and in the creation of an ester bond if an oxygen is being acetylated (O-acetylation). Historically, N-acetylation of biomolecules has received overwhelming attention over O-acetylation, due to the discovery of outstanding and very general regulatory biological features: In 1976, co-translational N-terminal protein acetylation was first reported1, a modification that has a profound impact on protein stability and localization. The reaction is catalyzed by N-terminal acetyltransferases (NATs), six of which have been identified in humans so far. Another important discovery was the acetylation/deacetylation dynamics that control histone function run by histone acetyltransferase (HAT) and histone deacetylase (HDAC) activities2, and the fact that acetylation is required to activate the tumor-suppressor protein p533. Another prominent example is the acetylation of lysine 40 on α-tubulin, which is a requirement for stable microtubule formation4–6. Compared to these important targets, O-acetylation has remained much less explored. The most prominent example has been the acetylation of serine and threonine residues by the bacterial YopJ effector family. The plague bacterium Yersinia pestis uses acetyltransferases to acetylate phosphosites within the MAPK pathway, thereby cutting off signal transduction in the host cell and sabotaging immune response7. The YopJ superfamily of acetyltransferases is conserved in animal and plant pathogenic bacteria8, and as a matter of fact, the structural details about YopJ activation and mechanism are known from studies about HopZ1a and PopP2, which are produced by the phytopathogens Pseudomonas syringae and Ralstonia solanacearum, respectively9,10. It remains unclear whether protein O-acetylation is exclusively a strategy used by pathogens or whether it is also a common, native reaction, which dynamically regulates the cellular phosphoproteome.
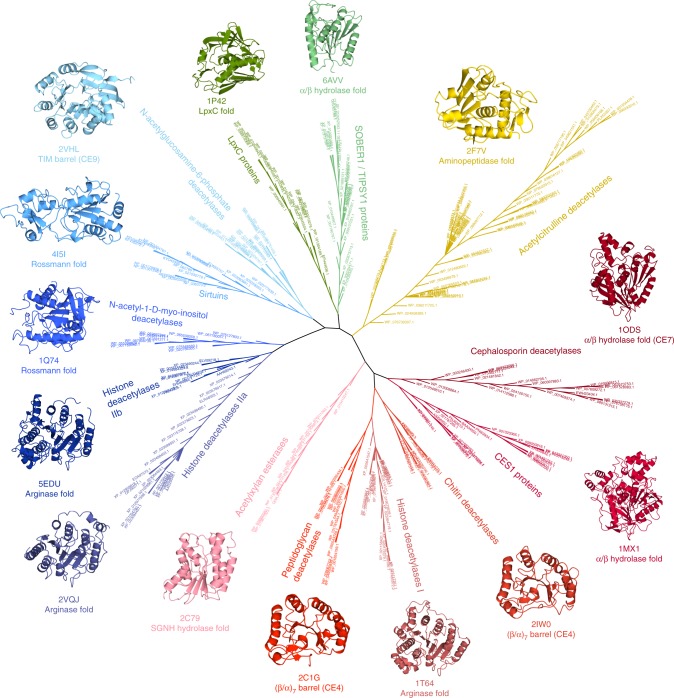
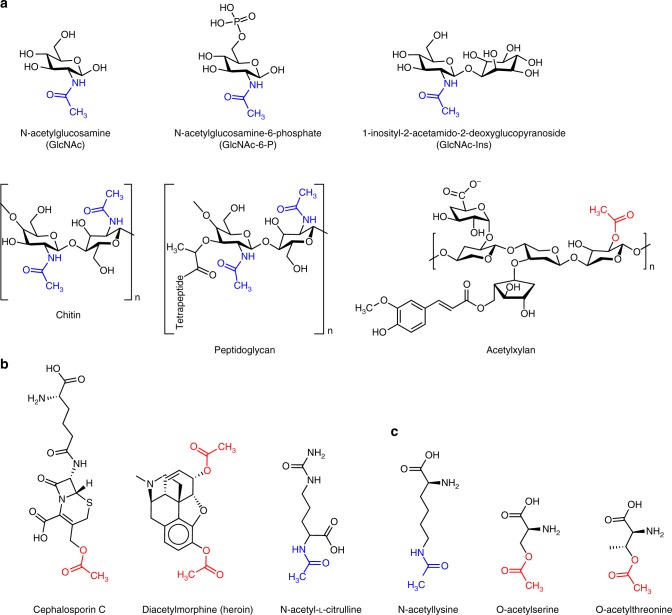
Besides the acetylation and deacetylation of amino acids, we discuss how the high number of acetylated targets and different target molecules has led to the evolution of an evenly high diversity of deacetylating enzymes (Fig. 1). These—while having basic catalytic chemistry in common—are characterized by specific structural features that allows them to recognize a high variety of substrates: For example, the majority of acetylated carbohydrates such as chitin, peptidoglycan, and N-acetylglucosamine are modified with acetyl groups via an amide bond on their nitrogen atoms11, with the exception of acetylxylan, which is O-acetylated via an ester linkage (Fig. 2a). Finally, we outline how small metabolic molecules with O-linked acetyl groups such as the antibiotic cephalosporin C or the opioid heroin (Fig. 2b) usually get deacetylated by promiscuous esterases12,13 and discuss the role of deacetylases in plant immunity.
Fig. 1.
Phylogenetic tree of the deacetylase families discussed in this review, showing NCBI reference sequences (RefSeq)87 accession numbers. Created with SeaView 4.5.488. All protein structures herein were visualized with CCP4mg89. Proteins discussed in this review were taken to represent each family and are displayed showing their folds, PDB codes and annotations from the CAZy database90 (CE4, CE7, and CE9)
Fig. 2.
Overview of the diversity of deacetylase substrates discussed in this review. a Carbohydrates, b small molecules, and c amino acid residues. N-acetyl are highlighted in blue and O-acetyl in red, respectively. Chemical structures were drawn using ChemDraw (Perkin Elmer)
Carbohydrate de-N-acetylases
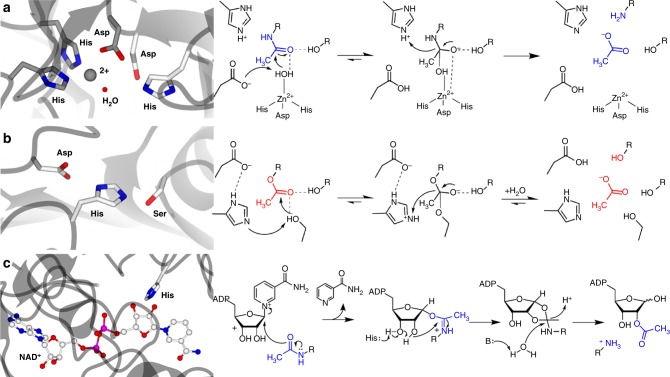
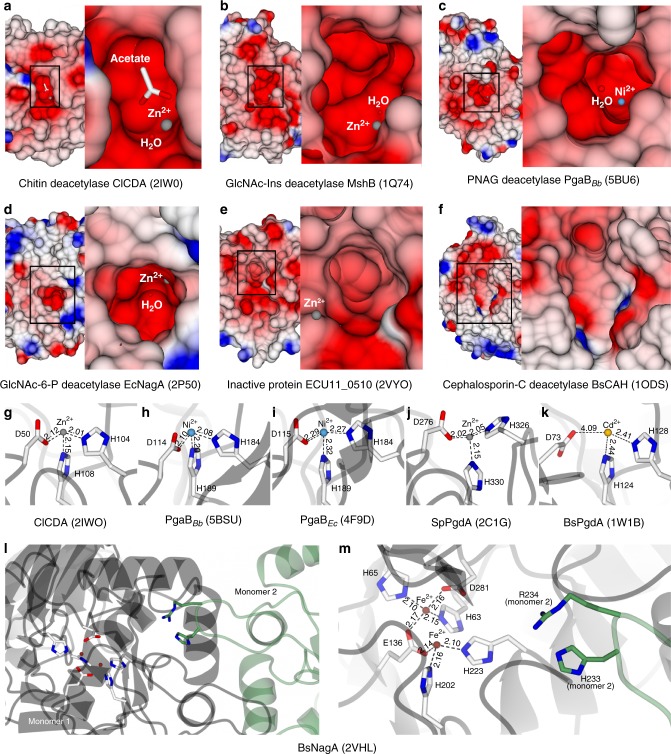
Oligomerized sugars are among the most abundant and most durable building blocks in life14. Their diversity ranges from chitin, which makes up the exoskeleton of insects and the cell walls of fungi to peptidoglycan, the constitutive cell wall component of many bacteria. Remodeling of these oligomers by deacetylation has proven crucial for pathogenesis and host immune evasion. Most of these modifications are N-acetylation events and therefore, despite different folds, one common feature of the deacetylating enzymes can be found in their active sites. To break the amide bond, a His–His–Asp triad that binds a divalent metal cation and a catalytic Asp–His pair with a nearby water molecule are an often-found architecture15,16. The catalytic base aspartate will abstract a proton from the water, making it a nucleophile. This deprotonation is aided by the metal ion, which decreases the pKa of the water. The catalytic acid histidine protonates the nitrogen-bound reaction intermediate, breaking the amide bond and leaving a free amine and an acetate17,18. This mechanism holds true not only for acetylated sugar oligomers but for N-acetylation in general (Fig. 3a). Thus, the main differences between the enzymes lie in their surface topology, accommodating substrates of different shapes and sizes. The crystal structure of the chitin deacetylase ClCDA from the fungal plant pathogen Colletotrichum lindemuthianum revealed a (β/α)(7) barrel fold and a prominent cleft with strong negative charge as binding site for the acetyl group19 (Fig. 4a). Deacetylated chitin (chitosan) is a very poor substrate for chitinases20; therefore deacetylation helps avoiding the creation of chitin breakdown products, which would otherwise be recognized by the plant’s immune system.
Fig. 3.
Deacetylases can be classified in three major groups, according to their catalytic sites. Exemplary active site arrangements and simple catalytic mechanisms of a de-N-acetylases (PDB code 4F9D25), b de-O-acetylases (PDB code 3M8154), and c class III HDACs (PDB code 4I5I65). N-acetyl is highlighted in blue and O-acetyl in red, respectively. Typical catalytic site residues are shown with white carbon atoms and metal binding residues in a are shown with carbon atoms colored in gray
Fig. 4.
Deacetylases feature a signature binding groove with electronegative charge. a–f Protein surface representations of deacetylases with different substrate specificities. Electrostatic surface potentials are contoured from −12.8 kT e−1 (red) to +12.8 kT e−1 (blue). g–k Divalent metal coordination by different deacetylases. Distances are in Å. l Dimeric composition and m metal binding sites (white) and substrate binding residues (green) of the N-acetylglucosamine-6-phosphate deacetylase from Bacillus subtilis
The crystal structure of 1-d-myo-inosityl 2-acetamido-2-deoxy-alpha-d-glucopyranoside deacetylase (MshB) from Mycobacterium tuberculosis revealed a typical Rossman fold but also features a strong electronegative surface cleft, which has a deeper depression compared to the chitin deacetylase (Fig. 4b)21. The substrate GlcNAc-Ins is not as large as the polymeric chitin; therefore, a more profiled binding cleft has evolved, providing a site located deeper in the protein to bind the acetyl group. GlcNAc-Ins is a precursor in the biosynthesis of mycothiol22, an important reducing agent in actinomycetes including mycobacteria, helping to control reactive oxygen species. In contrast, the periplasmatic, metal-dependent poly-acetyl-d-glucosamine (PNAG) deacetylase from Bordetella bronchiseptica (BpsB) displays a (β/α)(7) barrel fold and a surface topology characterized by a more distinct electronegative spot for binding of the acetate, surrounded by less negative charge23. Deacetylation of PNAGs is crucial for Bordetella biofilm stability and colonialization of the respiratory tract. A recent study about PgaB and BpsB has revealed that the proteins’ C-terminal domains, which were previously thought to be catalytically inactive, have glycoside hydrolase activity24. This activity requires deacetylated PNAGs. The current model, therefore, suggests a sequential degradation of PNAG molecules by PgaB proteins, first by deacetylating the substrate and then by hydrolyzing the glycosidic bonds. For reasons of clarity, the authors of the above-mentioned publication have also introduced new names for B. bronchiseptica BpsB and E. coli PgaB, which are now referred to as PgaBBb and PgaBEc, respectively. In the crystal structure of PgaBBb, a nickel ion was found in the catalytic center. Biochemical tests showed the highest enzyme activity with Ni2+ and Co2+, and the Escherichia coli homolog PgaBEc was found with a nickel cation in the metal binding site, too25. Compared to the chitin deacetylase ClCDA, the substrate binding site of PgaBBb is deeper, and the metal ion is located more towards the center of the cavity (Fig. 4c). The distances of the metal ion to the side chains of the His/His/Asp triad are smaller in the zinc-bound structure of ClCDA than in both nickel-bound structures of PgaBBb and PgaBEc (Fig. 4g–i). The higher mass of the atomic nucleus of zinc compared to nickel causes a more contracted electron shell and consequently a smaller atomic van der Waals radius of the zinc atom. The atomic van der Waals radii of zinc and nickel are 1.39 and 1.63 Å, respectively, and generally, the van der Waals radii decrease with an increasing atomic mass within elements 19–30 in period 4 of the periodic table26. Therefore, a slightly larger metal binding site might be needed to accommodate a cobalt or nickel compared to zinc. Although the differences in these distances are rather subtle, they might be the reason why cobalt and nickel increase biochemical activity of PgaB while the smaller zinc does not. A smaller metal might be bound to the metal-binding triad with insufficient affinity or might be inefficiently positioned towards the catalytic water.
Compared to both PgaBBb and PgaBEc, the crystal structure of the N-acetylglucosamine-6-phosphate (GlcNAc-6-P) deacetylase from E. coli folds into a TIM barrel architecture and the protein surface features a much less electronegative charge around the substrate binding site27. Instead, the protein surface is more hydrophobic around a distinct cavity of sharp electronegative charge (Fig. 4d). Next to the entrance to the site, an electropositive spot for the coordination of the phosphate group of the GlcNAc-6-P molecule is located. The catalytic centers of several solved structures of GlcNAc-6-P deacetylases have turned out to be relatively different regarding the side chains involved in metal and substrate binding: The above-mentioned enzyme from E. coli features a His/His/Glu metal binding site and a catalytic center composed of Asp/Asn/Gln plus the nucleophilic water28. However, these asparagine and glutamine residues are substituted by two histidines in the homologous enzyme from Thermotoga maritima (PDB code 1O12). The ortholog from Bacillus subtilis incorporates two divalent iron ions. It is assumed that one of the two metals facilitates the nucleophilic attack activated by the other metal through stabilization of the substrate29. Additionally, the active GlcNAc-6-P deacetylase in B. subtilis is a dimer (Fig. 4l), in which the His/His/Glu and His/His/Asp metal binding sites are being contributed by one monomer and the substrate binding residues by the other monomer. Only upon dimerization do the two parts come close enough to form an active substrate binding site (Fig. 4m).
Another protein family, peptidoglycan deacetylases, also facilitate bacterial evasion of host immune response and are, therefore, important virulence factors. In particular, deacetylated peptidoglycan is not recognized by mammalian lysozyme, which requires the N-acetyl group for catalysis30. The peptidoglycan GlcNAc deacetylase from Streptococcus pneumoniae displays a prominent electronegative cleft on the protein surface, constituting the substrate binding groove18. The cleft is surrounded by charge as negative except for a few electropositive spots on the side. The homolog from B. subtilis possesses a more hydrophobic surrounding but both proteins have a His/His/Asp metal binding site and a His/Asp/water catalytic center in common31. Both proteins have a (β/α)(7) barrel structure. Interestingly, the Streptococcus enzyme was found with a zinc ion coordinated inside the substrate binding site while attempts to bind zinc to the Bacillus homolog failed. Instead, it was eventually complexed with cadmium. A comparison of the two metal binding sites showed longer distances between metal-coordinating residues in the B. subtilis structure compared to S. pneumoniae (Fig. 4 j–k), therefore a larger ion might be needed for activity (cadmium has a van der Waals radius of 1.58 Å compared to 1.39 Å of zinc). In 2009, the crystal structure of a biochemically uncharacterized, putative amidase from the eukaryotic parasite Encephalitozoon cuniculi was published32. No activity on any tested substrate has been observed and distortions in the protein’s active site arrangements underlined the importance of the correct positioning of the parts that compose substrate binding site and catalytic center. The shape of the substrate binding groove looks dissimilar to the ones found in the previously discussed enzymes, and the active site metal ion is located more than 5 Å away from the position found in other deacetylases (Fig. 4e). In addition, the reaction-activating aspartic acid has shifted away from the active site. Therefore, the enzyme appears either generally catalytically inactive or it binds a molecule structurally much different than previously tested sugar substrates.
Another important enzyme implicated in pathogenicity is UDP-(3-O-(R-3-hydroxymyristoyl))-N-acetylglucosamine deacetylase (LpxC). It catalyzes the first committed step of lipid A biosynthesis33,34. Lipid A is the membrane anchor of lipopolysaccharide (LPS) or endotoxin, the major component of the outer membrane of Gram-negative bacteria35. LpxC has a unique overall fold that does not match any other known architectures. LpxC has a zinc binding site composed of an aspartate and two histidines36. The surface of the protein features a hydrophobic tunnel going through the upper part of the protein to accommodate the lipid moiety and a strong electronegative substrate binding site. It had been unclear whether a bifunctional general acid–base glutamate in the active site would promote the nucleophilic attack of a water, with a histidine stabilizing the oxyanion intermediate37, but without acting as a general acid or if the glutamate and histidine act as a typical general acid–base catalyst pair38. The first mechanism is similar to the one found for carboxypeptidase A39. Here, the glutamate activates the nucleophilic zinc–water complex, but it also facilitates the collapse of the tetrahedral reaction intermediate through partial protonation of the amine leaving group. In the second mechanism, the glutamate also activates the nucleophilic zinc–water complex, but the amine leaving group is protonated by the histidine, leading to the breakdown of the tetrahedral intermediate. More recent crystal structures have supported the latter model40.
Carbohydrate de-O-acetylases / acetylxylan esterases
Acetylxylan esterases or acetylxylan deacetylases are part of a concerted enzymatic action to break down plant cell wall xylan41–43, and they usually target the O-2 and/or O-3 position on the acetylxylan molecule44. A 2006 study revealed the crystal structures of two related metal-dependent xylan esterases from Streptomyces lividans and Clostridium thermocellum45. The S. lividans protein turned out reminiscent of the structural organization known from sugar de-N-acetylases, with a (β/α)(7) barrel fold and a His/His/Asp triad binding a zinc cation. In contrast, the C. thermocellum homolog displays a different loop conformation in the metal binding center plus a histidine to tyrosine substitution, leading to a cobalt ion being coordinated by an aspartate and a histidine. The second histidine is found replaced by four water molecules. In both structures the nucleophilic water is accompanied by a catalytic base aspartate and a catalytic acid histidine. Interestingly, both enzymes displayed the highest activity with Co2+, however, while the S. lividans protein retained a third of its activity when provided Zn2+, the activity of the C. thermocellum homolog was almost completely diminished. Both enzymes were able to tolerate manganese.
Other xylan esterases have a different architecture: The structure of acetylxylan esterase (AXE II) from the fungus Penicillium purpurogenum revealed a canonical SGNH hydrolase fold with a catalytic triad comprising a nucleophilic serine, a catalytic acid histidine and a catalytic base aspartate46. The mechanism of de-O-acetylation is somewhat similar to de-N-acetylation but does not involve a nucleophilic water or a metal ion. The reason for this is the presence of a serine residue. Upon substrate binding, the aspartate in the catalytic triad forms a low-energy barrier with the histidine, increasing the histidine’s pKa on the imidazole ring. The histidine will act as a general base and abstracts a proton from the hydroxyl group on the serine, making it nucleophilic. A tetrahedral reaction intermediate is then formed. The proton is transferred to the ester bond, which breaks upon protonation (Fig. 3b). A later crystal structure of AXE II from the thermophilic soil bacterium Geobacillus stearothermophilus showed the same organization of the catalytic site as the P. purpurogenum enzyme. Interestingly, the G. stearothermophilus protein turned out to be an octamer, which was confirmed in solution47. A possible reason for this quaternary structure could be either specificity or higher catalytic efficiency since this organization moves the active sites of the monomers in close proximity to each other. The hydrogen bonds and π-stacking interactions that stabilize the octamerization are located closely to the active sites, suggesting that the multimeric arrangement might stabilize the catalytic loops. Indeed, a follow-up study has mentioned dimerizing mutations of the protein that lead to a reduction in enzymatic activity48,49. Alternatively, the assembly might simply increase stability, considering that G. stearothermophilus is a thermophilic organism.
Multimeric cephalosporin-C and multi-substrate esterases
Cephalosporin C50 was first isolated from the fungus Acremonium51 and —like most other β-lactam antibiotics such as penicillin52—targets bacterial cell wall biosynthesis53. The acetyl group in cephalosporin C makes it a target for bacterial deacetylases, the activity of which leads to inactivation of the chemical. The structure of a cephalosporin C esterase from Thermotoga maritima has been solved and revealed a classical α/β hydrolase fold and a catalytic Ser/His/Asp triad. The protein features an electronegative binding site for the acetyl group right after the active site serine and an additional, larger cavity immediately adjacent to it54. This second depression most likely serves to accommodate the bulky core structure of the cephalosporin molecule. A homologous structure from Bacillus subtilis showed an almost identical fold but displayed a less electronegative charge on the surface and a smaller binding groove (Fig. 4f), possibly indicating a slightly divergent substrate specificity12. One additional and interesting feature is the higher oligomeric order of the proteins. The enzymes are native hexamers, with their active sites pointing inwards a substrate tunnel, making their assemblies reminiscent of the self-organizing proteasome architecture (Fig. 5). In one of the T. maritima structures, the entrance to this tunnel is covered by several phenylalanine residues, indicating a possible gatekeeper role for these amino acids and a selection towards more hydrophobic molecules. Both surface topology and multimerization appear to contribute to the substrate specificity of these enzymes, which is not limited to cephalosporin C but it is likely that they serve as a general de-O-acetylation conduit for small molecules.
Fig. 5.
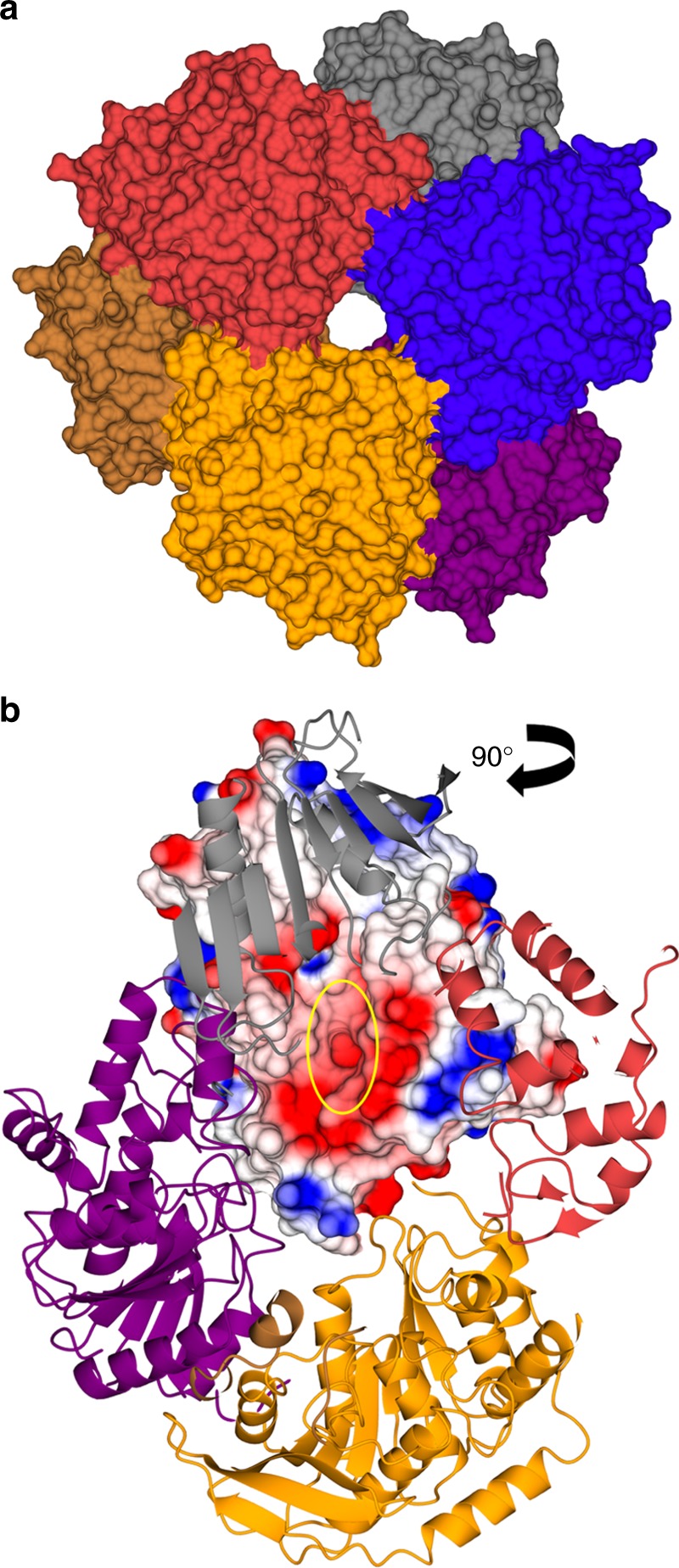
Some deacetylases achieve specificity through oligomerization. a Oligomerization properties of small molecule deacetylases highlighting the narrow tunnel entrance to the hexamer and b the active sites (yellow circle) positioned towards the inside of the substrate conduit (PDB code 1ODS12). The electrostatic surface potential is contoured from −12.8 kT e−1 (red) to +12.8 kT e−1 (blue)
Another example of a hexameric and promiscuous detoxification deacetylase is human carboxylesterase 1 (CES1)13. CES1 also folds into a canonical α/β hydrolase architecture with a classical Ser/His/Asp catalytic triad55. The specificity towards small molecules in likely given by its oligomerization properties, with the enzyme known for deacetylation of cocaine and heroin. CES1 exists in a trimer-hexamer equilibrium, with the hexamer composed of two stacked trimers. Binding of a small molecule to the allosteric Z-sites of the hexamer causes it to separate into two active trimers. Like the active site, the allosteric site is promiscuous too, and only the size of the molecule appears important for binding to the Z-site. The reason for this allosteric regulation is not fully understood but the fact that the inside of the trimer is lined up with hydrophobic side chains (except for the active sites) might require a higher oligomeric organization for protein stability.
Histone / lysine deacetylases
Modification of histones has an important role in regulation of chromatin structure and gene activity56, and are modulated by the activities of histone acetyltransferases (HATs) and histone deacetylases (HDACs)2. The acetylation occurs on lysine residues lying in the N-termini of core histones57 but the activity of HDACs is not limited to histones. HDAC6, for instance, deacetylates α-tubulin58. The enzymes’ cofactors divide them into two major groups: zinc and NAD+ dependent59,60. Whereas zinc-dependent HDACs are characterized by a canonical arginase fold, the NAD+-dependent class displays a typical Rossmann architecture (Table 1, Fig. 1), a structural motif frequently found in proteins that bind nucleotides. Sequence homology classifies them in class I, IIa, IIb, and III HDCAs. Class I enzymes are expressed in all tissues in humans and mice, and they feature a zinc binding site made of an Asp/Asp/His triad and a catalytic site composed of two histidines, one tyrosine, and a nucleophilic water. They also bind a potassium ion about 7 Å away from the catalytic zinc that might have a role in substrate binding61. Besides their expression in a tissue-specific manner, one feature that distinguishes class IIa HDACs from class I enzymes is the presence of an additional non-catalytic but structural zinc ion. This additional metal is coordinated by two histidines and two cysteines, which are strictly conserved within the class IIa family but absent in other HDACs62.
Table 1.
Overview of the proteins and their molecular features discussed in this review
| Name | PDB code | Substrate | Co-factor | Catalytic domain | Biological function |
|---|---|---|---|---|---|
| Carbohydrate deacetylases | |||||
|
Colletotrichum lindemuthianum CDA |
2IW0 | Chitin | Zn2+ | (β/α)(7) barrel | Fungal chitin deacetylation, evasion of plant host response |
|
Mycobacterium tuberculosis MshB |
1Q74 | GlcNAc-Ins | Zn2+ | Rossmann fold | Biosynthesis of the bacterial reducing agent mycothiol |
|
Bordetella bronchiseptica BpsB |
5BU6 | PNAG oligomers | Ni2+ | (β/α)(7) barrel | Biofilm formation |
|
Escherichia coli PgaB |
4F9D | PNAG oligomers | Ni2+ | (β/α)(7) barrel | Biofilm formation |
|
Escherichia coli NagA |
1YRR, 2P50 | GlcNAc-6-P | Zn2+ | TIM barrel | Murein recycling |
|
Thermotoga maritima NagA |
1O12 | GlcNAc-6-P | Zn2+ | TIM barrel | Murein recycling |
|
Bacillus subtilis NagA |
2VHL | GlcNAc-6-P | 2 Zn2 | TIM barrel | Murein recycling |
|
Streptococcus pneumoniae PgdA |
2C1G | Peptidoglycan | Zn2+ | (β/α)(7) barrel | Bacterial peptidoglycan deacetylation, evasion of immune response |
|
Bacillus subtilis PgdA |
1W1B | Peptidoglycan | Cd2+ | (β/α)(7) barrel | Bacterial peptidoglycan deacetylation, evasion of immune response |
|
Encephalitozoon cuniculi U11_0510 |
2VYO | Unknown | Zn2+ | (β/α)(7) barrel | Unknown, likely inactive protein |
|
Aquifex aeolicus LpxC |
1P42 | UDP-N-acetylglucosamine | Zn2+ | LpxC fold | Lipid A biosynthesis |
|
Streptomyces lividans Axe |
2CC0 | Acetylxylan | Zn2+ | (β/α)(7) barrel | Plant cell wall degradation |
|
Clostridium thermocellum Axe |
2C79 | Acetylxylan | Co2+ | (β/α)(7) barrel | Plant cell wall degradation |
|
Penicillium purpurogenum Axe2 |
1BS9 | Acetylxylan | SGNH hydrolase fold | Plant cell wall degradation | |
|
Geobacillus stearothermophilus Axe2 |
3W7V | Acetylxylan | SGNH hydrolase fold | Plant cell wall degradation | |
| Small molecule deacetylases | |||||
| Thermotoga maritima AxeA | 1VLQ, 3M81 | Cephalosporin C | α/β hydrolase fold | Xylooligosaccharide/Cephalosporin C hydrolysis | |
|
Bacillus subtilis CAH |
1ODS | Cephalosporin C | α/β hydrolase fold | Xylooligosaccharide/Cephalosporin C hydrolysis | |
|
Homo sapiens CES1 |
1MX1, 1MX5 | Small molecules | α/β hydrolase fold | Promiscuous multi-drug degradation | |
| Xanthomonas campestris ACDase | 2F7V | N-acetyl-l-citrulline | Co2+ | Aminopeptidase fold | Arginine biosynthesis |
| Histone deacetylases | |||||
|
Homo sapiens HDAC8 |
1T64 | Acetyllysines on histones | Zn2+ | Arginase fold | Histone deacetylation |
|
Homo sapiens HDAC4 |
2VQJ | Acetyllysines on histones | Zn2+ | Arginase fold | Histone deacetylation |
|
Homo sapiens HDAC6 |
5EDU | Acetyllysines on histones | Zn2+ | Arginase fold | Histone deacetylation |
|
Homo sapiens SIRT1 |
4I5I | Acetyllysines on histones | NAD+ | Rossmann fold | Histone deacetylation |
| Protein deacetylases | |||||
|
Arabidopsis thaliana SOBER1 |
6AVV | Acetylated proteins | α/β hydrolase fold | Hypersensitive response (HR) in plant immunity |
Class IIa HDACs are catalytically weak deacetylases, due to the substitution of the catalytically important tyrosine by a histidine, and histones are not their substrates. The current thinking, therefore, has accepted a non-catalytic role for class IIa HDACs. Only their interaction in a complex with SMRT/N-CoR and HDAC3, a class I HDAC, provides catalytic activity, which entirely comes from the associated HDAC3 protein63. Class IIb members strongly resemble class I enzymes, with slight differences in how the catalytic machinery functions. A study of the class IIb HDAC6 suggests that the tandem histidines in the catalytic site occupy separate roles as general base and general acid64. In HDAC8, a class I HDAC, the second histidine serves as both, general base and acid61.
Class III HDACs, also called sirtuins, have an NAD+-dependent mechanism. The NAD+ forms a ternary complex with the enzyme and the substrate. Nicotinamide is then released from the NAD+, followed by the transfer of the acetyl group from the substrate onto the ADP-ribose, leaving the substrate’s lysine deacetylated. Thus, sirtuins possess both deacetylase and ADP ribosyl transferase activities65. All sirtuins seem to have a conserved histidine residue that acts as the general base, deprotonating one of the ribose oxygens66 (Fig. 3c).
In addition to these groups, plants express another family of HDACs, called type-2 HDACs (HD2s). HD2s were first identified in maize67, and have since been found in almost all land plants. About 60% of HD2s contain a zinc-finger domain and HD2s without this domain are only found in angiosperms68,69. HD2s seem to be involved in leaf development70 and abiotic stress response71, both of which are plant-specific functions. Although there are reports suggesting that HD2s might have deacetylase activity, as they seem to have a role in the deacetylation of nuclear proteins72, and are able to modulate levels of histone acetylation73, direct biochemical or structural evidence proving that they are deacetylases is still missing.
N-acetyl-l-citrulline deacetylases
The first step in arginine biosynthesis is the acetylation of glutamate. This is followed by the so-called acetyl cycle, in which the acetyl group gets passed on from glutamate, finally leading to the formation of N-acetyl-l-ornithine. N-acetyl-l-ornithine will then be deacetylated, recycling the acetyl group onto glutamate, thereby regenerating N-acetyl-l-glutamate and yielding l-ornithine, an arginine precursor74. This last step is catalyzed by the enzyme acetylornithine deacetylase75. However, arginine biosynthesis in Proteobacteria follows a different path and does not involve the direct deacetylation of l-ornithine but rather a transcarbamoylation into N-acetyl-l-citrulline and its deacetylation to l-citrulline76,77. Thus, a different enzyme is needed: N-acetyl-l-citrulline deacetylase (ACDase). In 2007, the crystal structure of ACDase from Xanthomonas campestris was solved78. The protein possesses a metal binding site of His/Asp/Glu that coordinates a cobalt ion within an aminopeptidase architecture. The cobalt is in contact with the nucleophilic water within the catalytic site comprising a glutamate as a single bifunctional general acid–base catalyst. The binding site constitutes a deep electronegative cleft, with a long hydrophobic arm forming the C-terminal domain. The fact that ACDases are Proteobacteria-specific, which involves major pathogens, makes it an interesting drug target.
Plant protein deacetylases
Recently, a family of plant-specific protein deacetylases has been identified that are implicated in suppression of the host’s hypersensitive response (HR) against pathogens. The protein is called SOBER1, an α/β hydrolase highly related to acyl-protein thioesterases79. An insertion in the catalytic lid-loop renders the protein’s specificity into a protein deacetylase by blocking the hydrophobic tunnel that would otherwise accommodate longer substrates. The SOBER1 family can be further broken down into a TIPSY1 group, which possesses deacetylase activity but does not suppress hypersensitive response80. The proteins are highly promiscuous and SOBER1’s confirmed deacetylation substrates so far are the bacterial effector protein AvrBsT and the microtubule-associated ACIP1, which in turn is a substrate of AvrBsT. Another recent story reported that SOBER1 is able to suppress HR elicited by multiple bacterial acetyltransferases81. Though specific deacetylation sites are not known to date, the lack of a metal ion and the presence of a classical Ser/His/Asp catalytic triad suggest that SOBER1 and TIPSY1 are protein de-O-acetylases and are unable to break amide bonds.
Potential human protein de-O-acetylases
Reports about human protein deacetylases have exclusively focused on histone/lysine deacetylases and until today, no protein de-O-acetylases have been identified. The above-mentioned SOBER1/TIPSY1 family is only conserved in plants and the oligomerization properties of the promiscuous human CES1 esterase likely limits its substrate scope to small molecules. However, a protein called LYPLAL1 has been identified as an enzyme with disputed catalytic function. Genetic association studies have suggested its role in fatty acid metabolism and the protein acting as a phospholipase82–84. Like SOBER1, it is structurally related to acyl-protein thioesterases (APTs) and it was long thought to be the third human APT85. However, a structural study concluded that a different loop conformation impairs the substrate binding tunnel, changing LYPLAL1’s substrate specificity towards short acyl groups86. The presence of a Ser/His/Asp catalytic triad and the fact that LYPLAL1 is a monomer suggest a possible function as a protein de-O-acetylase.
Summary and outlook
Deacetylation events occur on a high number of chemically very different biomolecules (Fig. 2). Some reactions manipulate cell surface features to evade host immune response, others are involved in the regulation of basic cellular events and or counter-react prior acetylation events by bacterial effector proteins. Based on their catalyzed reaction, deacetylases can be divided into major groups: Metal-dependent de-N-acetylases and de-O-acetylases featuring a classical catalytic triad (Table 1). All deacetylases possess a distinct binding site for the acetyl group on their protein surface, characterized by a strong electronegative charge. Thus, substrate specificity originates from the immediate surrounding of the acetyl binding pocket. Further specificity appears to be given by oligomerization properties, creating a tunnel-like structure with a molecular weight cutoff selecting for small molecules.
Targeting deacetylases with small molecules might be a promising strategy to improve human health and aid agriculture. Especially bacterial and fungal deacetylases constitute an interesting drug target, since they are involved in pathogenicity or bacteria-specific metabolism, such as evasion of the mammalian immune response20,30, amino acid biosynthesis76 or biofilm formation23. In addition, inhibitors for the SOBER1 family of protein deacetylases might be a strategy to support the plant’s immune response against pathogenic Pseudomonas and Xanthomonas, based on the findings that SOBER1 suppresses hypersensitive response in Brassicaceae79,80.
While histone acetylation/deacetylation is well researched, and studies have been carried out to map acetylation sites cell-wide, these attempts have exclusively focused on lysine acetylation. Future studies are, therefore, likely to address O-acetylation and de-O-acetylation events in a global manner. It is still unknown as to how serine and threonine acetylation is used to compete with and regulate the phosphoproteome. A direct competition has only been shown in a handful of cases and each time, a pathogen was involved. In addition, no animal protein de-O-acetylase has been confirmed yet and their prediction from the protein sequence is rather difficult since minor changes to the fold or surface topology can result in a fundamentally altered substrate specificity. Therefore, more data obtained from mass spectrometry and structural studies will be necessary until acetylomic studies catch up with the amount of attention that other posttranslational modifications are receiving.
Acknowledgements
Apologies are offered to those colleagues whose work was not cited due to space constrains. We thank the following members of the Plant Biology Laboratory at the Salk Institute for Biological Studies: Hyun Jee Lee for discussions and help with chemical structures and Björn C. Willige for critical reading of this review.
Author contributions
M.B. and J.C. wrote the review.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Change history
1/3/2019
In the original published version of the article, the protein name for the carbohydrate deacetylase in Geobacillus stearothermophilus was missing from Table 1. The protein name, Axe2 (corresponding to PDB code 3W7V), should have appeared immediately following the species name. In addition, a space was inadvertently omitted between Xanthomonas campestris and ACDase in Table 1. The errors have been corrected in the HTML and PDF versions.
References
- 1.Brown JL, Roberts WK. Evidence that approximately eighty per cent of the soluble proteins from Ehrlich ascites cells are Nalpha-acetylated. J. Biol. Chem. 1976;251:1009–1014. [PubMed] [Google Scholar]
- 2.Inoue A, Fujimoto D. Enzymatic deacetylation of histone. Biochem. Biophys. Res. Commun. 1969;36:146–150. doi: 10.1016/0006-291x(69)90661-5. [DOI] [PubMed] [Google Scholar]
- 3.Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 4.L’Hernault SW, Rosenbaum JL. Chlamydomonas alpha-tubulin is posttranslationally modified by acetylation on the epsilon-amino group of a lysine. Biochemistry. 1985;24:473–478. doi: 10.1021/bi00323a034. [DOI] [PubMed] [Google Scholar]
- 5.Maruta H, Greer K, Rosenbaum JL. The acetylation of alpha-tubulin and its relationship to the assembly and disassembly of microtubules. J. Cell Biol. 1986;103:571–579. doi: 10.1083/jcb.103.2.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Piperno G, LeDizet M, Chang XJ. Microtubules containing acetylated alpha-tubulin in mammalian cells in culture. J. Cell Biol. 1987;104:289–302. doi: 10.1083/jcb.104.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mukherjee S, et al. Yersinia YopJ acetylates and inhibits kinase activation by blocking phosphorylation. Science. 2006;312:1211–1214. doi: 10.1126/science.1126867. [DOI] [PubMed] [Google Scholar]
- 8.Ma KW, Ma W. YopJ family effectors promote bacterial infection through a unique acetyltransferase activity. Microbiol. Mol. Biol. Rev. 2016;80:1011–1027. doi: 10.1128/MMBR.00032-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang ZM, et al. Structure of a pathogen effector reveals the enzymatic mechanism of a novel acetyltransferase family. Nat. Struct. Mol. Biol. 2016;23:847–852. doi: 10.1038/nsmb.3279. [DOI] [PubMed] [Google Scholar]
- 10.Zhang ZM, et al. Mechanism of host substrate acetylation by a YopJ family effector. Nat. Plants. 2017;3:17115. doi: 10.1038/nplants.2017.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aragunde H, Biarnes X, Planas A. Substrate recognition and specificity of chitin deacetylases and related family 4 carbohydrate esterases. Int. J. Mol. Sci. 2018;19:2. doi: 10.3390/ijms19020412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vincent F, et al. Multifunctional xylooligosaccharide/cephalosporin C deacetylase revealed by the hexameric structure of the Bacillus subtilis enzyme at 1.9A resolution. J. Mol. Biol. 2003;330:593–606. doi: 10.1016/s0022-2836(03)00632-6. [DOI] [PubMed] [Google Scholar]
- 13.Munger JS, et al. A serine esterase released by human alveolar macrophages is closely related to liver microsomal carboxylesterases. J. Biol. Chem. 1991;266:18832–18838. [PubMed] [Google Scholar]
- 14.Karrer P, Hofmann A. Polysaccharide XXXIX. Über den enzymatischen Abbau von Chitin und Chitosan I. Helv. Chim. Acta. 1929;12:616–637. [Google Scholar]
- 15.Lombardi PM, Cole KE, Dowling DP, Christianson DW. Structure, mechanism, and inhibition of histone deacetylases and related metalloenzymes. Curr. Opin. Struct. Biol. 2011;21:735–743. doi: 10.1016/j.sbi.2011.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoshida M, Kudo N, Kosono S, Ito A. Chemical and structural biology of protein lysine deacetylases. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2017;93:297–321. doi: 10.2183/pjab.93.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hernick M, Fierke CA. Zinc hydrolases: the mechanisms of zinc-dependent deacetylases. Arch. Biochem. Biophys. 2005;433:71–84. doi: 10.1016/j.abb.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 18.Blair DE, Schuttelkopf AW, MacRae JI, van Aalten DM. Structure and metal-dependent mechanism of peptidoglycan deacetylase, a streptococcal virulence factor. Proc. Natl Acad. Sci. USA. 2005;102:15429–15434. doi: 10.1073/pnas.0504339102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blair DE, et al. Structure and mechanism of chitin deacetylase from the fungal pathogen Colletotrichum lindemuthianum. Biochemistry. 2006;45:9416–9426. doi: 10.1021/bi0606694. [DOI] [PubMed] [Google Scholar]
- 20.Ride JP, Barber MS. Purification and characterization of multiple forms of endochitinase from wheat leaves. Plant Sci. 1990;71:185–197. [Google Scholar]
- 21.Maynes JT, et al. The crystal structure of 1-d-myo-inosityl 2-acetamido-2-deoxy-alpha-d-glucopyranoside deacetylase (MshB) from Mycobacterium tuberculosis reveals a zinc hydrolase with a lactate dehydrogenase fold. J. Biol. Chem. 2003;278:47166–47170. doi: 10.1074/jbc.M308914200. [DOI] [PubMed] [Google Scholar]
- 22.Newton GL, et al. The glycosyltransferase gene encoding the enzyme catalyzing the first step of mycothiol biosynthesis (mshA) J. Bacteriol. 2003;185:3476–3479. doi: 10.1128/JB.185.11.3476-3479.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Little DJ, et al. The protein BpsB is a poly-beta-1,6-N-acetyl-d-glucosamine deacetylase required for biofilm formation in Bordetella bronchiseptica. J. Biol. Chem. 2015;290:22827–22840. doi: 10.1074/jbc.M115.672469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Little DJ, et al. PgaB orthologues contain a glycoside hydrolase domain that cleaves deacetylated poly-beta(1,6)-N-acetylglucosamine and can disrupt bacterial biofilms. PLoS Pathog. 2018;14:e1006998. doi: 10.1371/journal.ppat.1006998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Little DJ, et al. The structure- and metal-dependent activity of Escherichia coli PgaB provides insight into the partial de-N-acetylation of poly-beta-1,6-N-acetyl-d-glucosamine. J. Biol. Chem. 2012;287:31126–31137. doi: 10.1074/jbc.M112.390005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bondi A. van der Waals volumes and radii. J. Phys. Chem. 1964;68:441–451. [Google Scholar]
- 27.Ferreira FM, et al. Structural analysis of N-acetylglucosamine-6-phosphate deacetylase apoenzyme from Escherichia coli. J. Mol. Biol. 2006;359:308–321. doi: 10.1016/j.jmb.2006.03.024. [DOI] [PubMed] [Google Scholar]
- 28.Hall RS, et al. Structural diversity within the mononuclear and binuclear active sites of N-acetyl-d-glucosamine-6-phosphate deacetylase. Biochemistry. 2007;46:7953–7962. doi: 10.1021/bi700544c. [DOI] [PubMed] [Google Scholar]
- 29.Vincent F, Yates D, Garman E, Davies GJ, Brannigan JA. The three-dimensional structure of the N-acetylglucosamine-6-phosphate deacetylase, NagA, from Bacillus subtilis: a member of the urease superfamily. J. Biol. Chem. 2004;279:2809–2816. doi: 10.1074/jbc.M310165200. [DOI] [PubMed] [Google Scholar]
- 30.Amano K, Hayashi H, Araki Y, Ito E. The action of lysozyme on peptidoglycan with N-unsubstituted glucosamine residues. Isolation of glycan fragments and their susceptibility to lysozyme. Eur. J. Biochem. 1977;76:299–307. doi: 10.1111/j.1432-1033.1977.tb11596.x. [DOI] [PubMed] [Google Scholar]
- 31.Blair DE, van Aalten DM. Structures of Bacillus subtilis PdaA, a family 4 carbohydrate esterase, and a complex with N-acetyl-glucosamine. FEBS Lett. 2004;570:13–19. doi: 10.1016/j.febslet.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 32.Urch JE, et al. Structural and functional characterization of a putative polysaccharide deacetylase of the human parasite Encephalitozoon cuniculi. Protein Sci. 2009;18:1197–1209. doi: 10.1002/pro.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Anderson MS, Raetz CR. Biosynthesis of lipid A precursors in Escherichia coli. A cytoplasmic acyltransferase that converts UDP-N-acetylglucosamine to UDP-3-O-(R-3-hydroxymyristoyl)-N-acetylglucosamine. J. Biol. Chem. 1987;262:5159–5169. [PubMed] [Google Scholar]
- 34.Anderson MS, Robertson AD, Macher I, Raetz CR. Biosynthesis of lipid A in Escherichia coli: identification of UDP-3-O-[(R)-3-hydroxymyristoyl]-alpha-d-glucosamine as a precursor of UDP-N2,O3-bis[(R)-3-hydroxymyristoyl]-alpha-d-glucosamine. Biochemistry. 1988;27:1908–1917. doi: 10.1021/bi00406a017. [DOI] [PubMed] [Google Scholar]
- 35.Raetz CR. Biochemistry of endotoxins. Annu. Rev. Biochem. 1990;59:129–170. doi: 10.1146/annurev.bi.59.070190.001021. [DOI] [PubMed] [Google Scholar]
- 36.Whittington DA, Rusche KM, Shin H, Fierke CA, Christianson DW. Crystal structure of LpxC, a zinc-dependent deacetylase essential for endotoxin biosynthesis. Proc. Natl Acad. Sci. USA. 2003;100:8146–8150. doi: 10.1073/pnas.1432990100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McClerren AL, Zhou P, Guan Z, Raetz CR, Rudolph J. Kinetic analysis of the zinc-dependent deacetylase in the lipid A biosynthetic pathway. Biochemistry. 2005;44:1106–1113. doi: 10.1021/bi048001h. [DOI] [PubMed] [Google Scholar]
- 38.Hernick M, et al. UDP-3-O-((R)-3-hydroxymyristoyl)-N-acetylglucosamine deacetylase functions through a general acid-base catalyst pair mechanism. J. Biol. Chem. 2005;280:16969–16978. doi: 10.1074/jbc.M413560200. [DOI] [PubMed] [Google Scholar]
- 39.Christianson DW, Lipscomb WN. Carboxypeptidase a. Acc. Chem. Res. 1989;22:62–69. [Google Scholar]
- 40.Clayton GM, et al. Structure of the bacterial deacetylase LpxC bound to the nucleotide reaction product reveals mechanisms of oxyanion stabilization and proton transfer. J. Biol. Chem. 2013;288:34073–34080. doi: 10.1074/jbc.M113.513028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bacon JS, Gordon AH, Morris EJ, Farmer VC. Acetyl groups in cell-wall preparations from higher plants. Biochem. J. 1975;149:485–487. doi: 10.1042/bj1490485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Biely P, MacKenzie C, Puls J, Schneider H. Cooperativity of esterases and xylanases in the enzymatic degradation of acetyl xylan. Nat. Biotechnol. 1986;4:731. [Google Scholar]
- 43.Poutanen K, Sundberg M, Korte H, Puls J. Deacetylation of xylans by acetyl esterases of Trichoderma reesei. Appl. Microbiol. Biotechnol. 1990;33:506–510. [Google Scholar]
- 44.Chesson A, Gordon AH, Lomax JA. Substituent groups linked by alkali‐labile bonds to arabinose and xylose residues of legume, grass and cereal straw cell walls and their fate during digestion by rumen microorganisms. J. Sci. Food Agric. 1983;34:1330–1340. [Google Scholar]
- 45.Taylor EJ, et al. Structure and activity of two metal ion-dependent acetylxylan esterases involved in plant cell wall degradation reveals a close similarity to peptidoglycan deacetylases. J. Biol. Chem. 2006;281:10968–10975. doi: 10.1074/jbc.M513066200. [DOI] [PubMed] [Google Scholar]
- 46.Ghosh D, et al. Determination of a protein structure by iodination: the structure of iodinated acetylxylan esterase. Acta Crystallogr. D. Biol. Crystallogr. 1999;55:779–784. doi: 10.1107/s0907444999000244. [DOI] [PubMed] [Google Scholar]
- 47.Lansky S, et al. A unique octameric structure of Axe2, an intracellular acetyl-xylooligosaccharide esterase from Geobacillus stearothermophilus. Acta Crystallogr. D. Biol. Crystallogr. 2014;70:261–278. doi: 10.1107/S139900471302840X. [DOI] [PubMed] [Google Scholar]
- 48.Lansky S, et al. Preliminary crystallographic analysis of a double mutant of the acetyl xylo-oligosaccharide esterase Axe2 in its dimeric form. Acta Crystallogr. F. Struct. Biol. Commun. 2014;70:476–481. doi: 10.1107/S2053230X14004129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alalouf, O. Biochemical characterization and structure-function studies of acetylxylan esterases from Geobacillus stearothermophilus. PhD thesis, Technion - Israel Institute of Technology (2014).
- 50.Abraham EP, Newton GG. The structure of cephalesporin C. Biochem. J. 1961;79:377–393. doi: 10.1042/bj0790377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brotzu G. Ricerche su di un nuovo antibiotico. Lav. dell’Instituto d’Igiene di Cagliari. 1948;1948:1–11. [Google Scholar]
- 52.Fleming A. On the antibacterial action of cultures of a penicillium, with special reference to their use in the isolation of B. influenzæ. Br. J. Exp. Pathol. 1929;10:226–236. [Google Scholar]
- 53.Park JT, Strominger JL. Mode of action of penicillin. Science. 1957;125:99–101. doi: 10.1126/science.125.3238.99. [DOI] [PubMed] [Google Scholar]
- 54.Levisson M, et al. Functional and structural characterization of a thermostable acetyl esterase from Thermotoga maritima. Proteins. 2012;80:1545–1559. doi: 10.1002/prot.24041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bencharit S, Morton CL, Xue Y, Potter PM, Redinbo MR. Structural basis of heroin and cocaine metabolism by a promiscuous human drug-processing enzyme. Nat. Struct. Biol. 2003;10:349–356. doi: 10.1038/nsb919. [DOI] [PubMed] [Google Scholar]
- 56.Allfrey VG, Faulkner R, Mirsky AE. Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc. Natl Acad. Sci. USA. 1964;51:786–794. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wade PA, Pruss D, Wolffe AP. Histone acetylation: chromatin in action. Trends Biochem. Sci. 1997;22:128–132. doi: 10.1016/s0968-0004(97)01016-5. [DOI] [PubMed] [Google Scholar]
- 58.Hubbert C, et al. HDAC6 is a microtubule-associated deacetylase. Nature. 2002;417:455–458. doi: 10.1038/417455a. [DOI] [PubMed] [Google Scholar]
- 59.Khochbin S, Verdel A, Lemercier C, Seigneurin-Berny D. Functional significance of histone deacetylase diversity. Curr. Opin. Genet. Dev. 2001;11:162–166. doi: 10.1016/s0959-437x(00)00174-x. [DOI] [PubMed] [Google Scholar]
- 60.Seto E, Yoshida M. Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harb. Perspect. Biol. 2014;6:a018713. doi: 10.1101/cshperspect.a018713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Somoza JR, et al. Structural snapshots of human HDAC8 provide insights into the class I histone deacetylases. Structure. 2004;12:1325–1334. doi: 10.1016/j.str.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 62.Bottomley MJ, et al. Structural and functional analysis of the human HDAC4 catalytic domain reveals a regulatory structural zinc-binding domain. J. Biol. Chem. 2008;283:26694–26704. doi: 10.1074/jbc.M803514200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fischle W, et al. Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Mol. Cell. 2002;9:45–57. doi: 10.1016/s1097-2765(01)00429-4. [DOI] [PubMed] [Google Scholar]
- 64.Hai Y, Christianson DW. Histone deacetylase 6 structure and molecular basis of catalysis and inhibition. Nat. Chem. Biol. 2016;12:741–747. doi: 10.1038/nchembio.2134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhao X, et al. The 2.5 A crystal structure of the SIRT1 catalytic domain bound to nicotinamide adenine dinucleotide (NAD+) and an indole (EX527 analogue) reveals a novel mechanism of histone deacetylase inhibition. J. Med. Chem. 2013;56:963–969. doi: 10.1021/jm301431y. [DOI] [PubMed] [Google Scholar]
- 66.Yuan H, Marmorstein R. Structural basis for sirtuin activity and inhibition. J. Biol. Chem. 2012;287:42428–42435. doi: 10.1074/jbc.R112.372300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lusser A, Brosch G, Loidl A, Haas H, Loidl P. Identification of maize histone deacetylase HD2 as an acidic nucleolar phosphoprotein. Science. 1997;277:88–91. doi: 10.1126/science.277.5322.88. [DOI] [PubMed] [Google Scholar]
- 68.Grandperret V, Nicolas-Frances V, Wendehenne D, Bourque S. Type-II histone deacetylases: elusive plant nuclear signal transducers. Plant Cell Environ. 2014;37:1259–1269. doi: 10.1111/pce.12236. [DOI] [PubMed] [Google Scholar]
- 69.Bourque S, et al. The evolution of HD2 proteins in green plants. Trends Plant. Sci. 2016;21:1008–1016. doi: 10.1016/j.tplants.2016.10.001. [DOI] [PubMed] [Google Scholar]
- 70.Zhou C, et al. Expression and function of HD2-type histone deacetylases in Arabidopsis development. Plant J. 2004;38:715–724. doi: 10.1111/j.1365-313X.2004.02083.x. [DOI] [PubMed] [Google Scholar]
- 71.Luo M, et al. HD2C interacts with HDA6 and is involved in ABA and salt stress response in Arabidopsis. J. Exp. Bot. 2012;63:3297–3306. doi: 10.1093/jxb/ers059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bourque S, et al. Type-2 histone deacetylases as new regulators of elicitor-induced cell death in plants. New Phytol. 2011;192:127–139. doi: 10.1111/j.1469-8137.2011.03788.x. [DOI] [PubMed] [Google Scholar]
- 73.Ding B, Bellizzi Mdel R, Ning Y, Meyers BC, Wang GL. HDT701, a histone H4 deacetylase, negatively regulates plant innate immunity by modulating histone H4 acetylation of defense-related genes in rice. Plant Cell. 2012;24:3783–3794. doi: 10.1105/tpc.112.101972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cunin R, Glansdorff N, Pierard A, Stalon V. Biosynthesis and metabolism of arginine in bacteria. Microbiol. Rev. 1986;50:314–352. doi: 10.1128/mr.50.3.314-352.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Meinnel T, Schmitt E, Mechulam Y, Blanquet S. Structural and biochemical characterization of the Escherichia coli argE gene product. J. Bacteriol. 1992;174:2323–2331. doi: 10.1128/jb.174.7.2323-2331.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shi D, et al. Crystal structure of N-acetylornithine transcarbamylase from Xanthomonas campestris: a novel enzyme in a new arginine biosynthetic pathway found in several eubacteria. J. Biol. Chem. 2005;280:14366–14369. doi: 10.1074/jbc.C500005200. [DOI] [PubMed] [Google Scholar]
- 77.Morizono H, et al. Acetylornithine transcarbamylase: a novel enzyme in arginine biosynthesis. J. Bacteriol. 2006;188:2974–2982. doi: 10.1128/JB.188.8.2974-2982.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Shi D, Yu X, Roth L, Tuchman M, Allewell NM. Structure of a novel N-acetyl-L-citrulline deacetylase from Xanthomonas campestris. Biophys. Chem. 2007;126:86–93. doi: 10.1016/j.bpc.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 79.Cunnac S, et al. A conserved carboxylesterase is a SUPPRESSOR OF AVRBST-elicited resistance in Arabidopsis. Plant Cell. 2007;19:688–705. doi: 10.1105/tpc.106.048710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Burger M, Willige BC, Chory J. A hydrophobic anchor mechanism defines a deacetylase family that suppresses host response against YopJ effectors. Nat. Commun. 2017;8:2201. doi: 10.1038/s41467-017-02347-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Choi, S., Jayaraman, J., & Sohn, K. H. Arabidopsis thaliana SOBER1 (SUPPRESSOR OF AVRBST-ELICITED RESISTANCE 1) suppresses plant immunity triggered by multiple bacterial acetyltransferase effectors. New Phytol.219, 324–335 (2018). [DOI] [PubMed]
- 82.Steinberg GR, Kemp BE, Watt MJ. Adipocyte triglyceride lipase expression in human obesity. Am. J. Physiol. Endocrinol. Metab. 2007;293:E958–E964. doi: 10.1152/ajpendo.00235.2007. [DOI] [PubMed] [Google Scholar]
- 83.Heid IM, et al. Meta-analysis identifies 13 new loci associated with waist-hip ratio and reveals sexual dimorphism in the genetic basis of fat distribution. Nat. Genet. 2010;42:949–960. doi: 10.1038/ng.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Benjamin AM, et al. Gene by sex interaction for measures of obesity in the framingham heart study. J. Obes. 2011;2011:329038. doi: 10.1155/2011/329038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zeidman R, Jackson CS, Magee AI. Protein acyl thioesterases (review) Mol. Membr. Biol. 2009;26:32–41. doi: 10.1080/09687680802629329. [DOI] [PubMed] [Google Scholar]
- 86.Burger M, et al. Crystal structure of the predicted phospholipase LYPLAL1 reveals unexpected functional plasticity despite close relationship to acyl protein thioesterases. J. Lipid Res. 2012;53:43–50. doi: 10.1194/jlr.M019851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35(Database issue):D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gouy M, Guindon S, Gascuel O. SeaView version 4: A multiplatform graphical user interface for sequence alignment and phylogenetic tree building. Mol. Biol. Evol. 2010;27:221–224. doi: 10.1093/molbev/msp259. [DOI] [PubMed] [Google Scholar]
- 89.McNicholas S, Potterton E, Wilson KS, Noble ME. Presenting your structures: the CCP4mg molecular-graphics software. Acta Crystallogr. D. Biol. Crystallogr. 2011;67:386–394. doi: 10.1107/S0907444911007281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lombard V, Golaconda Ramulu H, Drula E, Coutinho PM, Henrissat B. The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 2014;42(Database issue):D490–D495. doi: 10.1093/nar/gkt1178. [DOI] [PMC free article] [PubMed] [Google Scholar]