Abstract
Perinatal HIV infection is characterized by faster HIV disease progression and higher initial rate of HIV replication compared to adults. While antiretroviral therapy (ART) has greatly reduced HIV replication to undetectable levels, there is persistent elevated inflammation associated with HIV disease progression. Alteration of gut microbiota is associated with increased inflammation in chronic adult HIV infection. Here, we aim to study the gut microbiome and its role in inflammation in treated and untreated HIV-infected children. Examination of fecal microbiota revealed that perinatally infected children living with HIV had significantly higher levels of genus Prevotella that persisted despite ART. These children also had higher levels of soluble CD14 (sCD14), a marker of microbial translocation, and IP-10 despite therapy. The Prevotella positively correlated with IP-10 levels in both treated and untreated HIV-infected children, while genus Prevotella and species Prevotella copri was inversely associated with CD4 count. Relative abundance of genus Prevotella and species Prevotella copri showed positive correlation with sCD14 in ART-suppressed perinatally HIV-infected children. Our study suggests that gut microbiota may serve as one of the driving forces behind the persistent inflammation in children despite ART. Reshaping of microbiota using probiotics may be recommended as an adjunctive therapy along with ART.
Introduction
Due to advent of antiretroviral therapy (ART), HIV infection has now become a chronic disease that can be controlled with the ongoing treatment1. In addition, there has been 70% reduction in perinatal HIV infection between 2000 and 2015 as a result of introduction of ART in HIV-infected pregnant women (Children and HIV, fact sheet, UNAIDS, 2016). While ART substantially reduces HIV viral replication in infected individuals, chronic complications such as cardiovascular disease, metabolic syndrome, liver disease, kidney disease, neurocognitive impairment, and accelerated aging are being reported in higher frequency among horizontally HIV-infected adults2,3. During HIV infection, substantial CD4+T cells depletion occurs in the gastrointestinal tract. This loss is preferentially within Th17 cells that help in mucosal defense and maintain the intestinal epithelial barrier4. The depletion of Th17 cells in intestine has been reported to be associated with the disruption of epithelial barrier in HIV-infected individuals, leading to translocation of microbial products such as bacterial DNA5 and lipopolysaccharides6 into systemic circulation. Microbial translocation leads to immune activation and inflammation7 that persists despite suppressive ART8–11 and may contribute to non-AIDS complications12,13.
Dysbiosis is associated with different inflammatory conditions like inflammatory bowel diseases (IBD)14, irritable bowel syndrome (IBS)15, diabetes16, obesity17,18, rheumatoid arthritis19, allergic disease20 and multiple sclerosis21,22. Alteration in intestinal microbiota has also been reported in HIV-infected individuals23–34. Gori et al. showed increased prevalence of Candida albicans and Pseudomonas aeruginosa with significantly lower abundance of beneficial bacteria like Lactobacilli and Bifidobacteria in the stool samples of treatment naïve adults with acute HIV infection compared to healthy controls24. During early HIV infection, high abundance of Lactobacillales in the gut of untreated HIV-infected subjects is associated with increased CD4+ cell percentage, lower viral load and reduced microbial translocation that continued even after ART35. Later, several authors showed gut microbiota compositional changes during chronic HIV infection. They found significant increase in the relative abundance of genus Prevotella and decreased Bacteroides in the mucosal and fecal samples of adult HIV-infected subjects25–28,32,36. Furthermore, HIV-associated taxa in adults strongly correlated with mucosal and systemic inflammation25,27,31,33.
Most of the previous work on the role of gut microbiota in HIV pathogenesis focused adult HIV-infected subjects. A recent study by Bender et al. reported the gut microbiome differences in HIV exposed uninfected infants (HEU) when compared to HIV unexposed uninfected children (HUU). This difference in the gut microbiome of HEU infants was associated with distinct breast milk oligosaccharides of HIV-infected mothers37. However, there are currently no reports on the role of gut microbiota in intestinal barrier integrity and microbial translocation in the setting of perinatal HIV-infection.
Children with HIV infection experience faster disease progression compared to adults, with death occurring among up to 50% of untreated children within 2 years of life38. This rapid progression is associated with high viral load and increased depletion of CD4+ lymphocytes in children than adults. Perinatally HIV-infected children also showed viral mutation and poor drug adherence39. Unlike adult HIV infection, perinatally HIV-infected children are exposed to HIV either in utero or during birth. This raises a possibility that perinatally HIV-infected children may either be tolerized and have least impact on gut microbiome or have distinct pattern of gut microbiota compared to HIV-infected adults. Given the fact that the development of gut microbiota starts at birth and there is striking differences in HIV pathogenesis between adult and vertical HIV infection, we sought to study the composition and role of gut microbiota among treatment naïve and ART-suppressed perinatally HIV-infected children. In this study, we identified the difference in the fecal microbiota as a substitute to gut microbiota of perinatally HIV-infected children and age-matched uninfected controls. We carried out high throughput 16S rRNA gene sequencing to assess fecal microbiota and explored the relationship of unique fecal microbiota composition of treatment naïve and ART-suppressed perinatally HIV-infected children with systemic inflammation and microbial translocation.
Results
Perinatally HIV-infected children show a distinct pattern of gut microbiota compared to uninfected controls
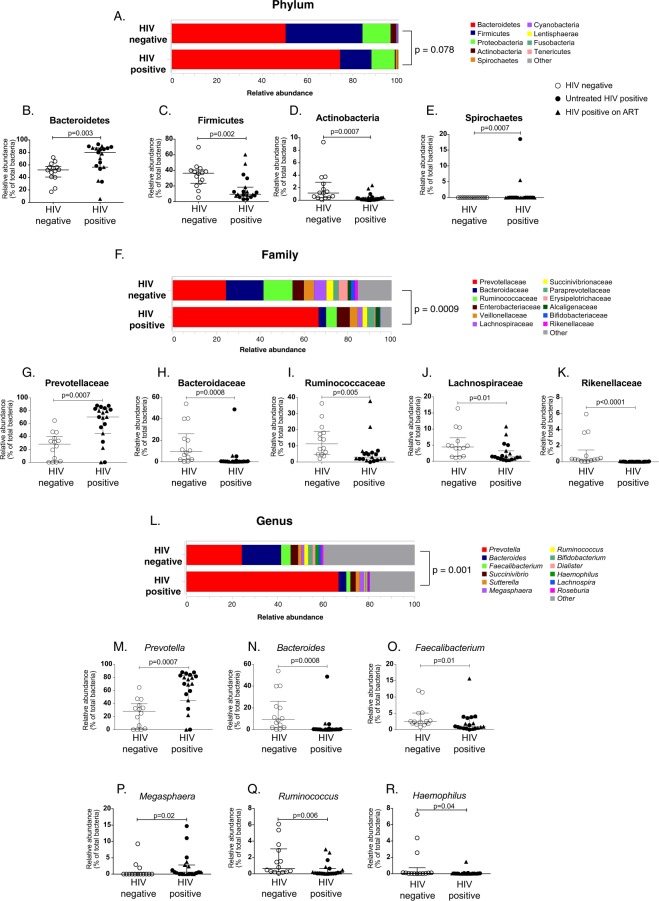
To determine the composition of gut microbiota in the setting of perinatal HIV infection, we assessed fecal microbiota in perinatally HIV-infected children. We determined the relative abundance of fecal microbiota at the phylum, family, and genus level and compared their levels between perinatally HIV-infected children and uninfected controls. Perinatally HIV-infected children showed distinct pattern of fecal microbiota compared to uninfected controls (Fig. 1A–R). The three most predominant phyla accounting for >97% of the overall bacterial abundance in fecal samples of both HIV-infected children and uninfected controls were Bacteroidetes, Firmicutes and Proteobacteria. No significant difference was observed in the overall relative abundances at the phylum level between perinatally HIV-infected children and uninfected controls (p = 0.08) (Fig. 1A). However, when relative abundance of individual phylum was compared between HIV-infected children and controls, Bacteroidetes and Spirochaetes were found to be significantly elevated (p = 0.002 and p = 0.0007, respectively) whereas Firmicutes, Actinobacteria and Cyanobacteria were significantly lower (p = 0.002, p = 0.0007 and p = 0.03, respectively) in HIV-infected children compared to uninfected controls (Figs 1B–E and S1B). The relative abundance of the remaining phyla did not show any significant difference between perinatally HIV-infected children and uninfected controls (Fig. S1A,C–E). On comparing the bacteria at the family and genus level, we observed overall relative abundances to be significantly different between HIV-infected children and uninfected controls (p = 0.0001 and p = 0.002, respectively) (Fig. 1F,L). Then, we individually compared the relative abundance of top abundant bacterial families between perinatally HIV-infected children and uninfected controls and observed that the relative abundance of Prevotellaceae was significantly higher (p = 0.0007), while Bacteroidaceae (p = 0.0008), Ruminococcaceae (p = 0.005), Lachnospiraceae (p = 0.01), and Rikenellaceae (p < 0.0001) were significantly lower in perinatally HIV-infected children compared to uninfected controls (Figs 1G–K, S2A–G). Among the abundant genera, the relative abundance of Prevotella and Megasphaera were significantly increased (p = 0.0007, and p = 0.02, respectively), whereas that of Bacteroides (p = 0.0008), Faecalibacterium (p = 0.01), Ruminococcus (p = 0.006) and Haemophilus (p = 0.04) was significantly lower in HIV-infected children compared to uninfected controls (Fig. 1M–R). The relative abundance of remaining abundant families and genera did not show difference between perinatally HIV-infected children and uninfected controls (Figs S2 and S3). The ratio of abundance of Bacteroides to Prevotella was significantly lower in perinatally HIV-infected children compared to uninfected controls (p = 0.0008, Fig. S5A). Our data shows that perinatally HIV-infected children have distinct pattern of gut microbiota compared to uninfected controls. The median values of relative abundance of significantly distinct phyla, families, and genera between perinatally HIV-infected children and uninfected controls are shown in Table S1.
Figure 1.
(A) Stacked bar plot depicting mean relative abundance of top nine abundant phyla in the fecal samples of HIV negative and HIV positive children. Dot Plots representing relative abundance of phyla (B) Bacteroidetes (C) Firmicutes, (D) Actinobacteria, and (E) Spirochaetes in HIV negative and HIV positive children. (F) Stacked bar plot showing mean relative abundance of top twelve abundant families in the fecal samples of HIV negative and HIV positive children. Dot Plots representing relative abundance of families (G) Prevotellaceae, (H) Bacteroidaceae (I) Ruminococcaceae (J) Lachnospiraceae and (K) Rikenellaceae in HIV negative and HIV positive children. (L) Stacked bar plot showing mean relative abundance of top twelve abundant genera in the fecal samples of HIV negative and HIV positive children. Dot Plots representing relative abundance of genera (M) Prevotella (N) Bacteroides (O) Faecalibacterium (P) Megasphaera (Q) Ruminococcus and (R) Haemophilus in HIV negative and HIV positive children. The horizontal line and deviation in the dot plot represents median and interquartile range (IQR), respectively. Mann-Whitney-U-test was performed for statistical analysis. p < 0.05 was considered to be significant. Significant differences in the overall relative abundance of fecal microbiota at phylum, family and genus level between HIV negative and HIV positive children were estimated using adonis function of vegan R package.
Impact of ART on the fecal microbiota in perinatally HIV-infected children
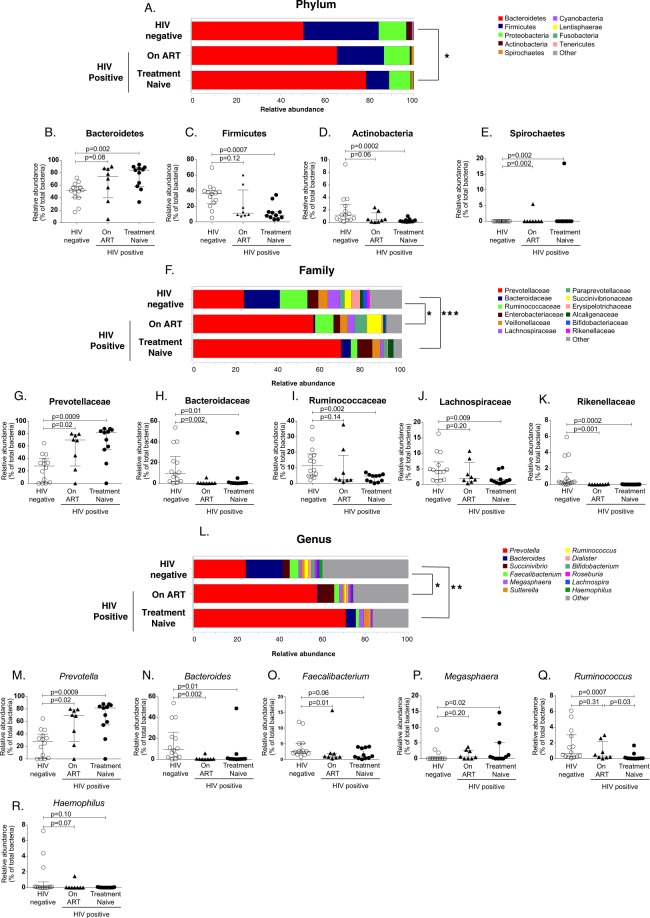
In order to determine the impact of antiretroviral therapy on fecal microbiota of perinatally HIV-infected children, we segregated perinatally HIV-infected children into treatment naïve and ART-experienced group and compared fecal microbiota across three groups. At the level of phylum, we observed overall relative abundance to be significantly different between treatment naïve HIV-infected children and uninfected controls (p = 0.01) but not between ART-suppressed HIV-infected children and uninfected controls (p = 0.27) (Fig. 2A). The relative abundance of Bacteroidetes (p = 0.002) and Spirochaetes (p = 0.002) were found to be significantly higher whereas Firmicutes (p = 0.0007) and Actinobacteria (p = 0.0002) were significantly lower in treatment naïve HIV-infected children compared to uninfected controls (Fig. 2B–E). In ART-suppressed HIV-infected children, the relative abundance of bacterial phyla were normalized to that of uninfected controls except Spirochaetes which remain elevated despite therapy (p = 0.002) (Fig. 2E). Comparisons of bacterial families showed significant difference in the overall relative abundances between uninfected controls and treatment naïve (p = 0.001) or ART-suppressed HIV-infected children (p = 0.02) (Fig. 2F). The relative abundance of Prevotellaceae were significantly increased (p = 0.0009 and p = 0.02 respectively) whereas Bacteroidaceae (p = 0.01 and p = 0.002) and Rikenellaceae (p = 0.0002) were significantly lower in both treatment naïve and ART-suppressed HIV-infected groups compared to uninfected controls (Fig. 2G,H). The relative abundance of other abundant families Ruminococcaceae (p = 0.002) and Lachnospiraceae (p = 0.009) were significantly lower in treatment naïve HIV-infected children whereas no significant difference was found in these families in ART-suppressed HIV-infected children compared to controls. (Fig. 2G–K). We also found overall relative abundance to be significantly different at the genus level between uninfected controls and treatment naïve (p = 0.005) or ART-suppressed HIV-infected children (p = 0.01) (Fig. 2L). ART did not show any effect on the relative abundance of Prevotella and Bacteroides. Compared to uninfected controls, treatment naïve and ART-suppressed HIV-infected children had significantly higher relative abundance of Prevotella (p = 0.0009 and p = 0.02 respectively) and lower abundance of Bacteroides (p = 0.01 and p = 0.002 respectively) (Fig. 2M,N). The relative abundance of Megasphaera was significantly higher (p = 0.02) and Ruminococcus (p = 0.0007) was significantly lower only in treatment naïve HIV-infected children compared to controls (Fig. 2P,Q). However, compared to treatment naïve HIV-infected children, the relative abundance of Ruminococcus was significantly higher in ART-suppressed children, suggesting the complete restoration of this genus by ART (Fig. 2Q). We did not observe difference in the relative abundance of Faecalibacterium (p = 0.06) and Haemophilus (p = 0.1) in treatment naïve HIV-infected children compared to controls (Fig. 2O,R). However, ART-suppressed HIV-infected children had decreased Faecalibacterium compared to controls (Fig. 2O). Our results suggest that the fecal microbiota of HIV-infected children did not completely get restored after ART treatment. The median values of relative abundance of significantly distinct phyla, families, and genera among treatment naïve, ART-suppressed HIV-infected children and uninfected controls are shown in Table S2.
Figure 2.
(A) Stacked bar plot depicting mean relative abundance of top nine abundant phyla n the fecal samples of HIV negative, on ART and treatment naïve perinatally HIV-infected children. Comparison of relative abundance of phyla (B) Bacteroidetes (C) Firmicutes, (D) Actinobacteria and (E) Spirochaetes across HIV negative, on ART and treatment naïve perinatally HIV-infected children. (F) Stacked bar plot showing mean relative abundance of top twelve abundant families in the fecal samples of HIV negative, on ART and treatment naïve perinatally HIV-infected children. Comparison of relative abundance of families (G) Prevotellaceae, (H) Bacteroidaceae (I) Ruminococcaceae (J) Lachnospiraceae and (K) Rikenellaceae across HIV negative, on ART and treatment naïve perinatally HIV-infected children. (L) Stacked bar plot showing mean relative abundance of top twelve abundant genera in the fecal samples of HIV negative, on ART and treatment naïve perinatally HIV-infected children. Comparison of relative abundance of genera (M) Prevotella (N) Bacteroides (O) Faecalibacterium (P) Megasphaera (Q) Ruminococcus and (R) Haemophilus across HIV negative, on ART and treatment naïve perinatally HIV-infected children. The horizontal line and deviation in the dot plot represents median and interquartile range (IQR), respectively. Kruskal-Wallis one-way ANOVA with Dunn’s multiple comparison was performed for statistical analysis. p < 0.05 was considered to be significant. Significant differences in the overall relative abundance of fecal microbiota at phylum, family and genus level between HIV negative controls, On ART and treatment naïve HIV-infected children were estimated using adonis function of vegan R package. *p < 0.05, **p < 0.005, p < 0.0005.
Microbial taxa associated with HIV and ART usage
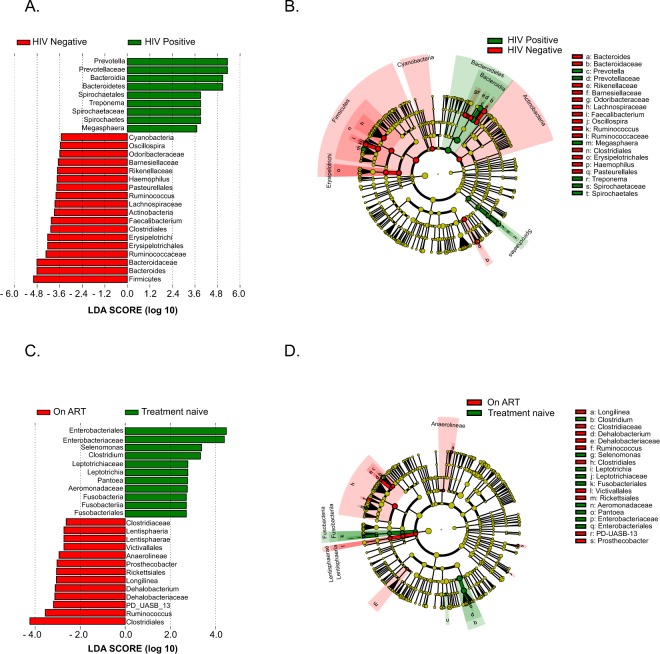
In order to gain insight into the bacterial taxa mostly associated with HIV, we performed linear discriminant analysis effect size analysis (LEfSe) that revealed significant difference in the abundance of 27 taxa between perinatally HIV-infected children and uninfected controls (LDA score >2.5, Fig. 3A,B). In addition to the taxa shown in Fig. 1, LEfSe revealed decreased Odoribacteraceae, Barnesiellaceae and Oscillospira in HIV-infected children compared to controls (Fig. 3A,B). Further, we carried out LEfSe analysis to determine the taxa most associated with ART usage. We found 32 significant differentially abundant taxa between treatment naïve and ART-suppressed children. Of the families, Clostridiaceae and Dehalobacteriaceae were enriched whereas Enterobacteriaceae and Leptotrichiaceae were decreased in ART-suppressed HIV-infected children. Additionally, genera Selenomonas, Clostridium, Leptotrichia and Pantoea were enriched in treatment naïve HIV-infected children whereas Longilinea, Dehalobacterium, PD-UASB-13 were more prevalent in ART-suppressed children (Fig. 3C,D).
Figure 3.
(A) LEfSe analysis showing the bacterial taxa that were significantly different in abundance between HIV positive children and HIV negative controls. Taxa enriched in HIV positive children are shown in green with positive LDA score and HIV negative controls in red with negative LDA score. Taxa passing LDA threshold value of >2.4 are only shown. (B) Cladogram representing the differentially abundant taxa between HIV positive children and HIV negative controls. Bacterial taxa enriched in HIV positive children and HIV negative controls are shown in green and red respectively. Brightness of each dot is proportional to taxon abundance. (C) LEfSe identifies significantly differentially abundant taxa between treatment naïve and On ART HIV-infected children and HIV. Taxa enriched in treatment naive HIV-infected children are shown in green with positive LDA score and On ART HIV-infected children in red with negative LDA score. Taxa passing LDA threshold value of >2.4 are only shown. (D) Taxonomic Cladogram. Red and green shows bacterial taxa enriched in On ART and treatment naïve HIV-infected children and uninfected controls, respectively. Brightness of each dot is proportional to taxon abundance.
Analysis of diversity of microbial community
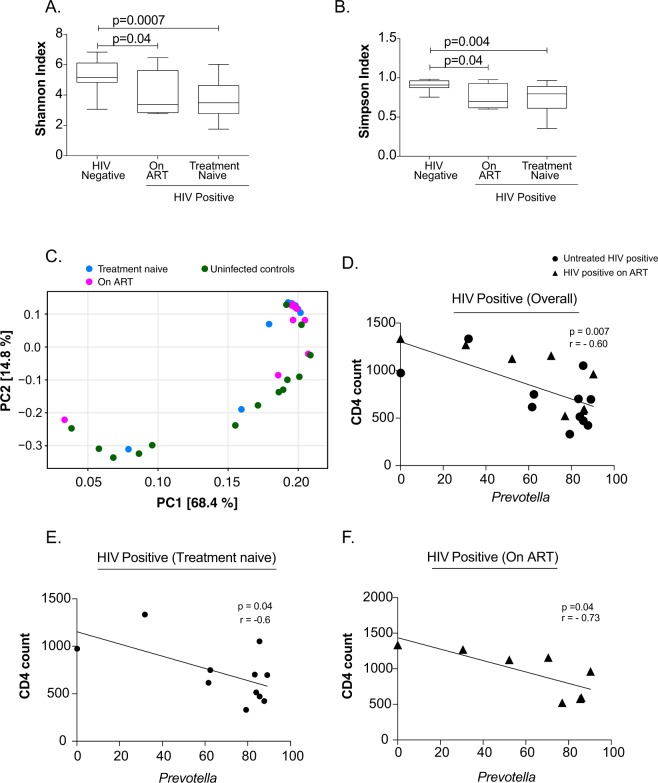
Alpha diversity is the assessment of diversity within a habitat. We analysed the alpha diversity of fecal microbiota using the Shannon and Simpson index. The fecal microbiota diversity of both treatment-naïve and ART-suppressed HIV-infected children was significantly decreased when compared to uninfected controls (Fig. 4A,B). Beta diversity with principal component analysis (PCA) showed that the fecal microbiota composition of most of the treatment-naïve and ART-suppressed HIV-infected children were grouped into cluster and different from uninfected controls (Fig. 4C).
Figure 4.
(A) Shannon and (B) Simpson indices were used to estimate the α-diversity of fecal microbiome of HIV negative controls, ART-suppressed and treatment naïve perinatally HIV infected children. (C) Plot of principle component analysis of fecal microbiome of HIV negative controls (green), On ART (pink) and treatment naïve (blue) perinatally HIV infected subjects. Association between relative abundance of Prevotella and CD4 count in (D) HIV positive children (overall), (E) treatment naïve and (F) On ART perinatally HIV infected children. Statistical analysis was carried out using the Spearman t-test. A linear regression line is included in each plot. p < 0.05 was considered to be significant.
Prevotella is indirectly associated with CD4 T cells in perinatally HIV-infected children
As CD4 depletion is a hallmark of HIV disease progression, we sought to determine association of genera that were significantly differentially abundant in HIV-infected children with CD4 count. The genus Prevotella was only found to be indirectly associated with CD4 count in all HIV-infected children (p = 0.007; r = −0.6) (Figs 4D and 6C). The indirect association between Prevotella and CD4 T cell count remained significant in treatment naïve and ART-suppressed perinatally HIV-infected children (Fig. 4E,F). Prevotella did not show any correlation with CD4 T cell count in uninfected controls (data not shown).
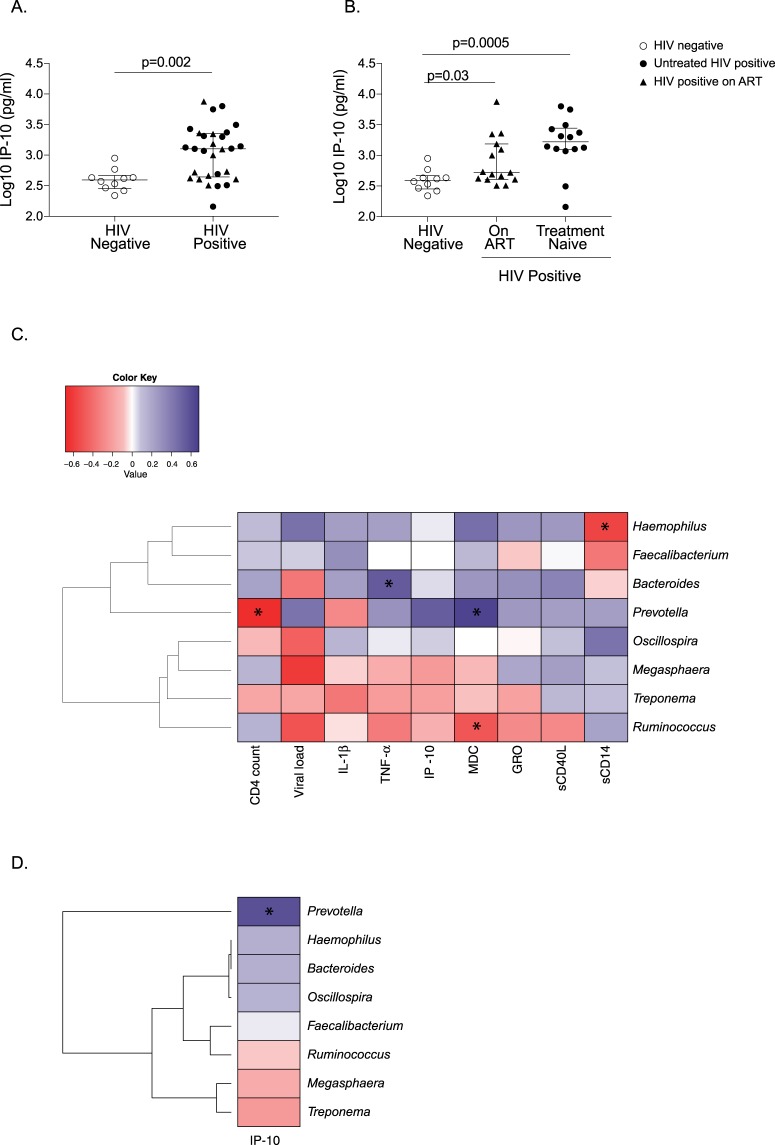
Figure 6.
Comparison of Plasma IP-10 levels in (A) HIV positive and HIV negative children and (B) HIV negative, on ART and treatment naïve perinatally HIV-infected children. The horizontal line and deviation in the dot plot represents median and interquartile range (IQR), respectively. Mann-Whitney U-test and Kruskal-Wallis one-way ANOVA with Dunn’s multiple comparison were performed for statistical analysis. p < 0.05 was considered to be significant. (C) Heatmap representing the associations of relative abundance of significantly distinct genera with CD4 count, viral load, IL-1β, TNF-α, IP-10, GRO, MCD, sCDl40L and microbial translocation marker, sCD14. Blue and red shading represents positive and negative association respectively. Clustering was carried out based on the association of genera with clinical and immune parameters. (D) Heatmap representing the associations of absolute abundance of significantly distinct genera with IP-10. Blue shading indicates positive association and red shading indicates negative association. Clustering was carried out based on the association of genera with IP-10. Statistical analysis was carried out using Spearman t-test. p < 0.05 was considered to be significant. *p < 0.05.
Increased abundance of Prevotella copri in the gut of perinatally HIV-infected children despite ART
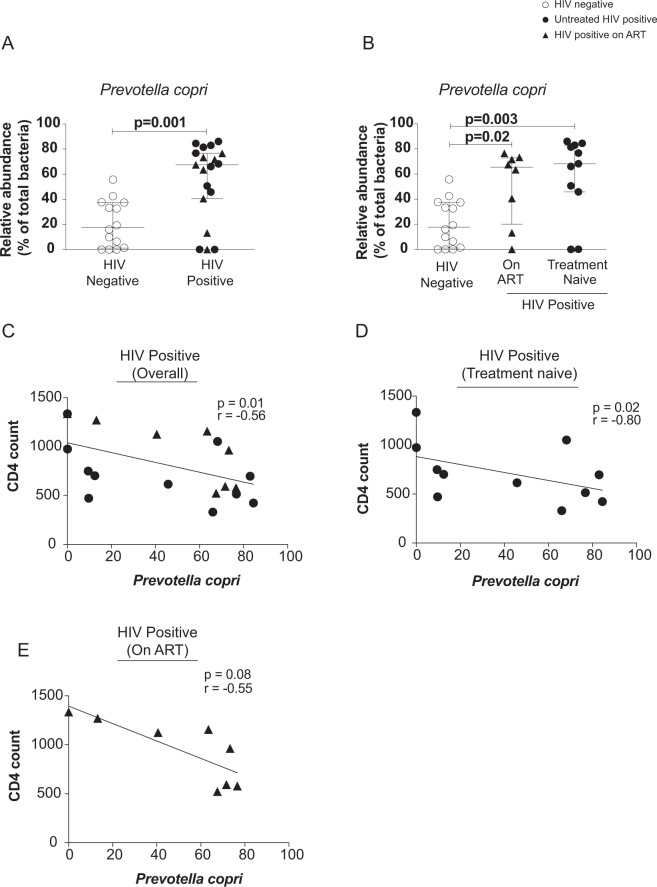
We were intrigued by the increased relative abundance of Prevotella in perinatally HIV-infected children despite suppressive therapy and wanted to further characterize the genus Prevotella to species level. At species level, we identified Prevotella copri and Prevotella stercorea. However, only the relative abundance of Prevotella copri was found to be elevated in HIV-infected children compared to controls (p = 0.001) (Fig. 5A). The relative abundance of Prevotella copri was also significantly elevated in treatment naïve (p = 0.003) and ART-suppressed HIV-infected children (p = 0.02) compared to uninfected controls, suggesting that ART does not normalize the levels of Prevotella copri in HIV-infected children (Fig. 5B). Additionally, the relative abundance of other abundant species Bacteroides plebieus, Bacteroides uniformis and Faecalibacterium prausnitzii in the fecal sample of HIV-infected children was significantly decreased compared to controls (Fig. S4) and remained lowered regardless of treatment. Like genus Prevotella, the relative abundance of P. copri also showed indirect association with CD4 T cells in overall and treatment naïve perinatally HIV-infected children (Fig. 5C,D) however it failed to achieve level of significance in ART-suppressed HIV-infected children (p = 0.08; Fig. 5E). The median values for the relative abundance of Prevotella copri in three groups are shown in Table S2.
Figure 5.
Dot plots representing relative abundance of Prevotella copri in the fecal sample of (A) HIV positive and HIV negative children and (B) HIV negative, On ART and treatment naïve perinatally HIV-infected children. The horizontal line and deviation in the dot plot represents median and interquartile range (IQR), respectively. Mann-Whitney U-test and Kruskal-Wallis one-way ANOVA with Dunn’s multiple comparison were performed for statistical analysis. The p < 0.05 was considered to be significant. Correlation between relative abundance of Prevotella copri and CD4 count in (C) HIV positive children (overall), (D) treatment naïve. (E) On ART perinatally HIV infected children. Statistical analysis was carried out using Spearman t-test. A linear regression line is included in each plot. p < 0.05 was considered to be significant.
Soluble IP-10 is elevated in HIV-infected children and are associated with Prevotella despite ART
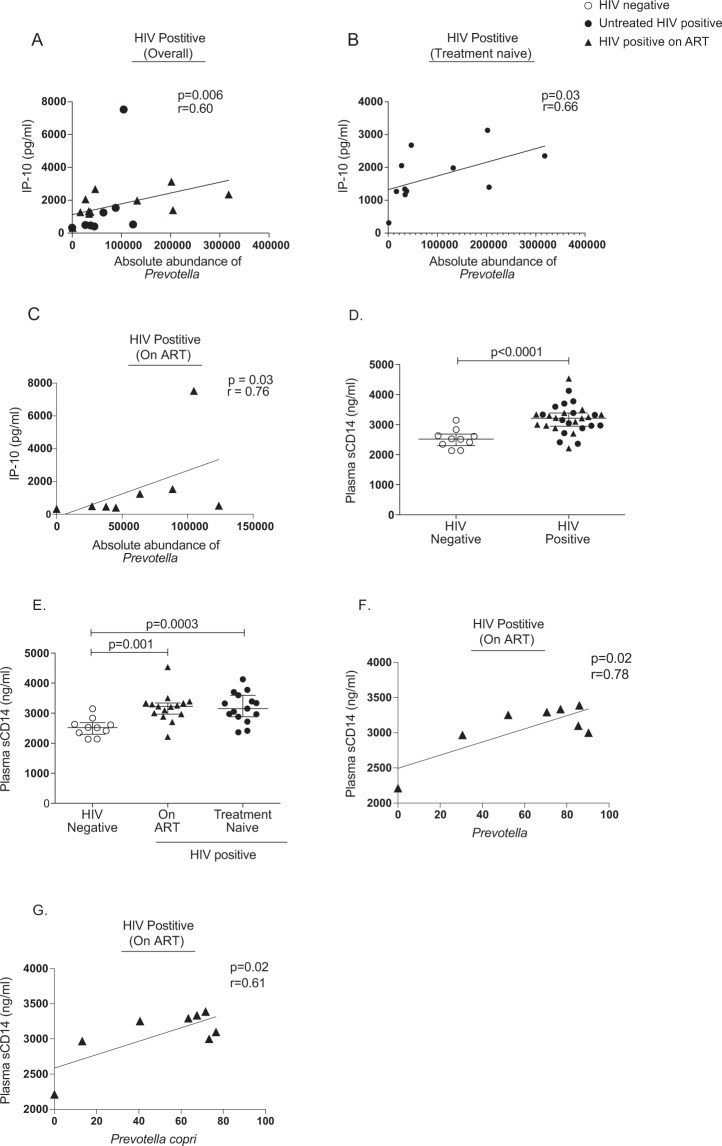
Multiplex analysis revealed significantly high levels of IL-1β, TNF-α, IP-10 (Interferon-gamma inducible protein 10), MDC (Macrophage-derived chemokine), GRO (Growth regulated oncogene chemokine), and sCD40L (Soluble CD40-ligand) in treatment naïve HIV-infected children compared to uninfected controls. Treatment appeared to restore the levels of these cytokines except IP-10 (Table 1). The IP-10 level was significantly elevated in perinatally HIV-infected children (p = 0.002) and it remained elevated in treated group (p = 0.03) in comparison to uninfected controls (Fig. 6A,B). This suggests a potential role of IP-10 in maintaining the inflammatory environment in HIV-infected children on treatment. Next, we performed multivariate analysis to evaluate the association of significantly distinct genera with cytokines that were significantly elevated in HIV-infected children. Notably, Prevotella was directly correlated (r = 0.65, p = 0.005) and Ruminococcus was indirectly correlated (r = −0.49, p = 0.048) with MDC levels. Bacteroides showed direct association with TNF- α (r = 0.54, p = 0.02). Since, IP-10 was the only cytokine to be elevated despite therapy in HIV-infected children, we also evaluated the association of absolute abundance of significantly differentially abundant genera with IP-10 level. We found IP-10 levels directly correlated only with the absolute abundance of Prevotella in perinatally HIV-infected children with or without treatment (Figs 6D and 7A–C). The levels of IL-2, IL-3, IL-4, IL-5, IL-8, IL-9, IL-1RA, IL-13, IL-12p40, TNF-β, MIP-1β, Flt-3L, and MCP-3 were undetectable.
Table 1.
Cytokine levels in perinatally HIV-infected children and HIV negative controls.
| Nature | Analyte (pg/ml) | HIV-infected children | HIV negative control | |
|---|---|---|---|---|
| Treatment naïve Median (IQR) | On ART Median (IQR) | Median (IQR) | ||
| Proinflammatory | IL-1α | 214.7 (113.6–334.4) | 156.2 (81.44–266.8) | 215.6 (59.57–326.4) |
| IL-1β | 10.12 (0.0–12.03) | 5.345 (0.7575–12.30) | 17.60 (4.888–30.79) | |
| IL-6 | 19.27 (10.54–40.71) | 21.31 (0.0–32.84) | 46.78 (5.043–63.64) | |
| IL-7 | 58.33 (41.91–69.07) | 44.48 (37.38–52.47) | 58.75 (35.05–76.12) | |
| IL-12p70 | 12.48 (0.7100–22.64) | 8.625 (4.180–12.48) | 27.60 (2.390–51.71) | |
| IL-15 | 9.810 (0.0–15.90) | 7.055 (2.295–14.35) | 19.12 (2.860–33.05) | |
| IL-17A | 7.870 (4.070–13.29) | 6.425 (2.080–11.26) | 15.98 (4.440–23.43) | |
| INF-γ | 23.15 (14.44–31.37) | 19.37 (9.908–34.73) | 34.06 (18.03–50.19) | |
| TNF-α | 103.9 (91.28–128.9) | 57.9 (47.76–75.61) | 67.26 (45.94–77.95) | |
| GM-CSF | 87.53 (52.17–104.5) | 54.9 (31.34–90.03) | 124.2 (57.50–167.2) | |
| Anti-inflammatory | IL-10 | 28.85 (8.430–44.44) | 11.60 (0.0–23.26) | 33.35 (0.2075–47.67) |
| Chemoattractants | IP-10 | 2582 (1739–4812) | 884.7 (580–1936) | 569 (435.5–825.4) |
| MCP-1 | 634 (591–746.3) | 473.7 (382.1–543) | 650.6 (482.3–806) | |
| MDC | 770.6 (546.3–883.2) | 466.5 (394.9–522.8) | 573.4 (469.2–636.3) | |
| MIP-1α | 39.42 (21.87–50.21) | 36.93 (0.0–50.56) | 39.13 (8.218–56.40) | |
| EOTAXIN | 10.48 (8.040–12.66) | 7.860 (0–13.89) | 10.59 (3.463–15.64) | |
| Fractalkine | 186.5 (59.9–227.2) | 149 (118.6–211) | 270.9 (95.77–416.2) | |
| Growth Factors | VEGF | 206.7 (117.7–329) | 138.5 (86.69–252.2) | 315.5 (119.7–481.2) |
| EGF | 20.82 (4.340–28.56) | 20.70 (14.33–22.61) | 15.25 (8.505–25.95) | |
| FGF-2 | 31.12 (17.25–36.75) | 21.28 (16.16–30.61) | 40.34 (22.49–57.55) | |
| TGF-α | 0.1500 (0.0–0.5400) | 0.2050 (0.0–0.5850) | 0.7850 (0.1150–1.533) | |
| GRO | 2563 (1271–3056) | 582.2 (491.3–788.2) | 581.9 (491.3–772.1) | |
| G-CSF | 21.33 (17.14–27.44) | 28.81 (22.48–38.60) | 22.39 (15.23–30.01) | |
| INFα2 | 221.3 (123.8–275.4) | 190.5 (132.8–231.5) | 312.2 (160.8–432.1) | |
| sCD40L | 7247 (5101–8489) | 2023 (1549–4095) | 3904 (2762–4629) | |
Bold numbers denote values different from HIV NEG, p < 0.05.
Figure 7.
Association between absolute abundance of Prevotella and plasma IP-10 level (pg/ml) in (A) HIV positive children, (B) on ART and (C) treatment naïve perinatally HIV infected children. Statistical analysis was performed using Spearman t-test. A linear regression line is included in each plot. p < 0.05 was considered to be significant. Comparison of Plasma sCD14 levels in (D) HIV positive and HIV negative children and (E) HIV negative, on ART and treatment naïve perinatally HIV-infected children. The horizontal line and deviation in the dot plot represents median and interquartile range (IQR), respectively. Mann-Whitney U-test and Kruskal-Wallis one-way ANOVA with Dunn’s multiple comparison were performed for statistical analysis. p < 0.05 was considered to be significant. Association between (F) relative abundance of Prevotella and plasma sCD14 level (ng/ml) (G) relative abundance of Prevotella copri and plasma sCD14 level (ng/ml) in ART-suppressed perinatally HIV infected children. Statistical analysis was performed using Spearman t-test. A linear regression line is included in each plot. The p < 0.05 was considered to be significant.
Increased levels of plasma sCD14 in perinatally HIV-infected children despite ART
We evaluated the level of microbial translocation marker, sCD14 in the plasma of perinatally HIV-infected children. Plasma level of sCD14 was significantly elevated in perinatally HIV-infected children compared to uninfected controls (Fig. 7D). ART could not normalize the level of sCD14 in the plasma of treated HIV-infected children (Fig. 7E). Given that gut microbiota maintains gut epithelial barrier, we then evaluated whether significantly differentially abundant genera in HIV-infected children was associated with plasma level of sCD14. We found Haemophilus to be negatively associated with sCD14 (r = −0.56, p = 0.01) (Fig. 6C). Relative abundance of Prevotella and also Prevotella copri directly correlated with plasma sCD14 level in treated HIV-infected children (Fig. 7F,G).
Discussion
In this paper, we studied the fecal microbiota in treatment-naïve and ART-suppressed perinatally HIV-infected children, and compared them with that of uninfected controls. Our study showed a unique pattern of fecal microbiota with elevated relative abundance of the genus Prevotella and species Prevotella copri in HIV-infected children that persisted during suppressive ART. The genus Prevotella was directly associated with soluble IP-10 and inversely correlated with CD4 count despite ART. The genus Prevotella and species Prevotella copri positively correlated with sCD14, a marker of microbial translocation, in ART-suppressed perinatally HIV-infected children implying that Prevotella may potentially be involved in persistent inflammation in perinatally HIV-infected children during therapy.
Alteration in the overall gut microbial diversity during HIV infection in adults currently remains controversial. Some studies reported gut microbiota to be less diverse in untreated HIV-infected adults28,29,40,41 or the diversity to be decreased after ART28,36, while others found no alteration in the diversity of gut microbiota26,27,31,33,34. In our study, we found decreased α-diversity in the fecal microbiota of HIV-infected children despite therapy. This discrepancy among previous studies could be due to different ethnicity, age, different stage of HIV infection of enrolled HIV-infected subjects, selection of sample type, use of antibiotics by study participants or different technologies used to study gut microbiota composition. Perinatally HIV-infected children and uninfected controls participated in our study had preserved CD4 count and shared similar geographical region and dietary habits. As antibiotics alter gut microbiota composition profoundly42, we only included children who were not on antibiotics within last one month of sample collection. Similar to studies on HIV-infected adults, beta diversity indices such as PCA showed composition of fecal microbiota in most of the perinatally HIV-infected children to be different from uninfected controls. Moreover, most of the treated HIV-infected children were clustered with treatment naïve HIV-infected children suggesting similar pattern of gut microbiota in treated and untreated perinatally HIV-infected children. Thus, despite successful viral suppression, ART could not completely restore fecal microbiota in children with HIV.
Untreated HIV-infected children had increased relative abundance of Bacteroidetes and decreased Firmicutes whereas studies on HIV-infected adults reported decreased Firmicutes with no difference in Bacteroidetes abundance compared to controls25,29,30,43. We did not observe any difference in the relative abundance of phylum Proteobacteria in perinatally HIV-infected children compared to uninfected controls, which is in contrast to a recent study by Dinh et al. who showed significantly enriched relative abundance of Proteobacteria in the fecal samples of adults living with HIV31. The variations in the relative abundance of Proteobacteria in HIV-infected subjects have been observed across studies. Some previous studies that evaluated intestinal mucosal biopsies showed increased relative abundance of Proteobacteria in HIV-positive patients25,28, whereas other failed to observe any difference in the fecal and rectal sponge samples of HIV-positive patients compared to healthy controls26,29.
Our observations of increased relative abundance of Prevotellaceae and decreased relative abundance of Bacteroidaceae and Rikenellaceae in untreated HIV-infected children are in agreement with the previous findings in HIV-infected adults25,27,36. Similar to HIV-infected adults25–28,32, we also observed significantly increased relative abundance of Prevotella with decreased abundance of Bacteroides in perinatally HIV-infected children. Increase in gut Prevotella has been reported in chronic inflammatory diseases such as Rheumatoid arthritis44,45 and metabolic syndrome46. Recent studies have shown that the high abundance of Prevotella in HIV-infected individuals is attributed to their sexual behavior rather than to the disease itself32,47. However, in our study prior sexual behavior does not have any relevance because the study is focussed on perinatally HIV-infected children who have either not reached or just reached an early stage of puberty. A study by Dillon et al. showed positive correlation of relative abundance of Prevotella with mucosal CD4+ and CD8+ T cell activation, and pro-inflammatory cytokine-producing mucosal CD4+ and CD8+ T cells25 however, Ling et al. did not find any significant correlation between Prevotella abundance and systemic inflammatory cytokines27 in HIV-infected adults. This suggests that the potential inflammatory role of Prevotella is only confined to the local gut immune system. In contrast to the previous study in adults25,27, we found that relative abundance of Prevotella was indirectly associated with peripheral CD4 count and directly associated with MDC in perinatally HIV-infected children. MDC has emerged as novel modulator of lung inflammation in mice48 and an inflammatory marker to assess the severity of atopic dermatitis in children and infants49. More recently, the increased levels of MDC in HIV-infected adults have also been reported50. This suggests the potential inflammatory role of MDC in HIV-infected children, however the level of MDC normalizes after initiation of ART. Interestingly, absolute abundance of Prevotella was found to be directly associated with soluble IP-10 in untreated and ART-experienced perinatally HIV-infected children. Our results indicate that Prevotella outgrowth (represented by increase in the absolute number of Prevotella), rather than Prevotella evenness may drive the production of IP-10.
Level of IP-10 has been shown to be elevated in many viral infections, including acute West Nile virus51, acute and chronic HCV52,53 and severe influenza infection54,55 suggesting the role of this chemokine in immune response to viral infection. Elevated IP-10 has been reported during acute and chronic HIV infection52,56–59. Consistent with previous studies on HIV-infected adults60,61, ART could not normalise the plasma level of IP-10 in perinatally HIV-infected children to that of uninfected controls. Previous studies on acute and chronic HIV infection in adults showed positive correlation of IP-10 with viral load57,59 and negatively with CD4 count56, thus proposed IP-10 to be the marker of disease progression. However, we failed to find direct correlation between IP-10 and HIV viral load in perinatally HIV-infected children, possibly because of limited sample size.
Infection of monocyte-derived macrophages (MDM) with HIV-1 has been shown to stimulate the production of GRO-α chemokine in-vitro. This chemokine in turn prompt HIV replication in macrophages and lymphocytes, thereby contributing to HIV pathogenesis62. Our observation of elevated level of GRO in untreated HIV-infected children might be due to viral replication.
Elevated sCD14, a marker of microbial translocation is associated with increased mortality among HIV-infected individuals63 and also sCD14 level in HIV-infected mothers have reported to be directly associated with risk of mother to child transmission64. Consistent with prior findings65–68, perinatally HIV-infected children had increased plasma level of sCD14 compared to uninfected children, showing gut epithelial barrier disruption and microbial translocation that persists even after suppressive therapy. Our study also showed positive correlation of sCD14 with abundance of genus Prevotella and Prevotella copri in treated HIV-infected children. However the presence of virus appears to disrupt this correlation as seen in untreated HIV-infected children. This suggests that either HIV or other unknown factors driven by HIV viral load contribute to microbial translocation in untreated subjects. Additional contributory factors to increased microbial translocation in untreated children are the decreased relative abundance of Lachnospiracea and Ruminococcacea families that include many butyrate-producing bacterial species that are essential for preserving the gut mucosal integrity69.
Ours is the first study to investigate the fecal microbiota composition of treatment naïve and ART-suppressed perinatally HIV-infected children that showed association of genus Prevotella with IP-10 and sCD14, a microbial translocation marker. These results suggest that Prevotella is proinflammatory in nature that may augment the inflammation in perinatally HIV–infected children despite ART, and may potentially promote chronic complications. Our study has few limitations. First, in our cross sectional study fecal microbiota has been used as a proxy for gut microbiota. Fecal microbiota only represents gut lumen microbiota and does not reflect mucosal-associated microbiota, which is important to consider as microbes on the gut mucosa can potentially interact with gut associated lymphoid tissue (GALT). However fecal sample was only the realistic sample for non-invasive study. Second, though we showed impact of ART on the fecal microbiota of HIV-infected children, the effect of individual antiretroviral drugs on fecal microbiota is not investigated in our study.
In summary, our study found distinct gut microbiota composition in untreated and ART-suppressed perinatally HIV-infected children compared to uninfected controls. They had higher abundance of Prevotella and decreased Bacteroides, and elevated levels of IP-10 that persists despite ART. The higher abundance of Prevotella may promote HIV pathogenesis as suggested by the negative association of Prevotella with CD4 count. Interestingly, the direct correlation between Prevotella and IP-10 suggests that IP-10 may be regulated by gut microbiota and could potentially contribute to ongoing inflammation and T cell depletion. Future studies to understand the mechanism of Prevotella-driven inflammation may aid the development of therapeutic interventions that can reconstitute healthy gut microbiota and mitigate the level of inflammation in individuals living with HIV.
Methods
Study Population
Study population included 29 perinatally HIV-1-infected children and 14 uninfected age matched controls. Perinatal infection in the child was confirmed by the documentation of HIV infection within 1 year of life. Among HIV-infected children, 15 children were on ART for more than 6 months, whereas the other 14 children were not on ART (treatment naïve). To study the gut microbiota, 11-treatment naïve and 8 ART-suppressed perinatally HIV-infected children were included together with 14 uninfected controls for the high throughput sequencing of DNA extracted from fecal sample. All treated HIV-infected children had undetectable plasma viral load (VL < 50 copies/ml). Uninfected controls were matched for sex and age to HIV-infected children. The following criteria were used to exclude children: age more than 15 years old, use of antibiotics or probiotics within one month of sample collection, any medical history of intestinal inflammatory disorders, evidence of hepatitis B and C infection, and vaccination within 1 week of sample collection. All study participants belonged to same ethnicity, geographical area and had similar dietary habits. The study was approved by Institutional Review Board (IRB) of St. Johns Medical College & Hospital at Bangalore, Jawaharlal Nehru University (JNU) at New Delhi and King George’s Medical University (KGMU) Lucknow, and was carried out in accordance with the approved guidelines. Informed written consent was obtained from parents/guardian of all the children who participated in our study. The characteristics of subjects are listed in the Table 2.
Table 2.
Characteristics of subjects.
| Parameters | HIV-infected children | HIV negative control | |
|---|---|---|---|
| Treatment naïve | ART-suppressed | ||
| Number | 14 | 15 | 14 |
| Age (years) | 10 (8–12) | 11 (7–12) | 10 (7–15) |
| Male % (n) | 78.5 (11) | 40 (6) | 60 (7) |
| Female % (n) | 21.5 (3) | 60 (9) | 40 (7) |
| Male/Female ratio | 3.6 | 0.6 | 1 |
| CD4 count (cells per µl) | 644 (330–1334) | 1141.5 (523–2406) | 1343 (863–1884) |
| Plasma viral load (HIV-1 RNA copies/ml) | 33,779 (1,461–4,31,237) | <50 | NA |
Values are shown as median (IQR), NA: Not applicable.
Sample collection
Approximately 2–3 g of fresh fecal sample was collected in a sterile plastic container from each child and stored at −80 °C within 30 minutes. Peripheral blood was collected into vacutainer blood collection tubes containing EDTA or Heparin and whole blood was centrifuged to isolate plasma. Plasma was stored at −80 °C for measuring levels of cytokines.
Plasma viral load and CD4 count
Plasma HIV-1 viral load (VL) was measured using Abbott Real Time HIV-1 assay with a lower limit of detection of 50 copies of RNA/ml (Abbott Molecular Inc., Des Plaines, IL, USA. CD4 T cell count was measured using FC500™ flow cytometer (Beckman Coulter, Fullerton, California, USA).
Fecal DNA extraction
Fecal sample stored at −80 °C was thawed on ice and approximately 220 mg of fecal sample was taken in a microcentrifuge tube for bacterial DNA isolation. Microbial DNA was isolated from fecal samples by commercially available QIAamp Fast DNA Stool Mini kit, as per manufacturer’s recommended protocol. The amount and purity of DNA were estimated by absorbance at 260 nm and 280 nm using nanodrop spectrophotometer.
Amplicon Library preparations and Sequencing
Identification of bacteria was carried out by amplifying and sequencing bacterial 16S rRNA gene at Eurofins Genomics India Pvt Ltd (Bangalore, India). Oligonucleotide primers (Forward - 5′ GCCTACGGGNGGCWGCAG 3′ and Reverse-5′ ACTACHVGGGTATCTAATCC 3′) that target V3-V4 region of 16S rRNA gene were used to generate amplicons. The amplicon libraries were prepared using Nextera XT Index Kit (Illumina inc.) as per the 16S Metagenomic Sequencing Library preparation protocol and also include multiplexing index sequences and illumina adapters. The amplicon libraries were purified by 1X AMpureXP beads, quantified using Qubit fluorometer and analyzed in 4200 Tape Station system (Agilent Technologies) using D1000 Screen tape as per manufacturer instructions. Sequencing of the amplicons was performed on Miseq Platform using 2 × 300 v3 Miseq Kit.
Luminex cytokine assays
A Luminex cytokine/chemokine assay (EMD Millipore) was used to examine the plasma samples of 14-treatment naïve, 15 ART-suppressed HIV-infected children and 10 uninfected controls. The assay measured the following serum cytokine/chemokine levels: IL-1α, IL-1β, IL-2, IL-3, IL-4, IL-5, IL-6, IL-7, IL-8, IL-9, IL-10, IL-12p40, IL-12p70, IL-13, IL-15, IL-17A, INF-γ, TNF-α, TNF-β, GM-CSF, IL-1RA, IP-10, MCP-1, MDC, MIP-1β, MIP-1β, EOTAXIN, Fractalkine, VEGF, EGF, FGF-2, TGF-β, GRO, G-CSF, Flt-3L, INF-α2, MCP-3 and sCD40L. Cytokine standards and each sample were tested in duplicate. Data was acquired on Bio-Plex 200 system using Bio-Plex manager software, v4.1 (Bio-Rad).
sCD14 detection
Level of sCD14 in plasma was quantified by commercially available Human sCD14 Elisa kit according to the manufacturers’ protocols (R&D Systems, Minneapolis, MN). Each sample was analysed in duplicate and sCD14 level was expressed in ng/ml.
Sequence data and Statistical analysis
Sequencing data was analysed using QIIME 1.9.170. Raw reads were quality filtered using Trimmomatic v0.38. Quality control involved the removal of reads with errors in barcode or primer sequences, reads containing ambiguous bases and low quality sequences (reads with more than 10% quality threshold (QV) <20 phred score) along with a sliding window of 20 bp and a minimum length of 100 bp. According to barcodes, the high quality reads obtained were then assigned to samples. Using UCLUST in QIIME the high quality sequences were clustered into operational taxonomic units (OTUs) with 97% sequence similarity. OTUs was used to analyze alpha diversity indices (Shannon and Simpson) using QIIME. For PCA, distance matrices were calculated using R. Using Greengenes reference databases, the taxonomies are assigned to the OTUs by UCLUST with a threshold of 90% sequence similarity. Explicet software v2.8.5 was used to analyze OTUs and create figures71. Significant differences in the overall relative abundance of fecal microbiota at phylum, family and genus level between treatment naïve, ART-suppressed HIV-infected children and uninfected controls were estimated using adonis function of vegan R package72. Statistical analysis of alpha diversity indices, the relative abundance of taxa, plasma cytokines, and soluble sCD14 levels between two and three groups was performed using non-parametric Mann Whitney U test and Kruskal-Wallis 1-way ANOVA with Dunn’s multiple comparison respectively in GraphPad Prism v. 6. LEfSe analysis (http://huttenhower.sph.harvard.edu/lefse/) was used to identify differentially abundant bacterial taxa across three groups with alpha value of 0.05 and LDA score of >2.473. Association between two variables was carried out by Spearman rank correlation test. Correlation coefficient r = 1, describes a positive association and r = −1, describes a negative correlation. A p value below 0.05 was considered significant. Heatmap was generated to visualize association between two variables using heatmap.2 function of gplots R package (https://CRAN.R-project.org/package=gplots).
Electronic supplementary material
Acknowledgements
We would like to acknowledge Science and Engineering Research Board-Department of Science and Technology (SERB-DST) grant ECR/2015/000304 (R.T.), UPOE-II Project ID: 15 (R.T.), and DST PURSE II (DST/SR/PURSE Phase II/I) for providing financial support to carry out research work. RT is thankful to UGC-Faculty Recharge Programme (FRP).
Author Contributions
U.K.S., A.S., R.T. designed the experiment. U.K.S., N.R., B.P.G., P.M., performed the experiment. R.T., A.S., H.D., B.P.S., and R.C. contributed reagents/materials. U.K.S., R.T., H.D., B.P.S., and R.C. analyzed the data. U.K.S., R.T., and A.S. wrote the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-35877-4.
References
- 1.Pasternak AO, et al. Cellular levels of HIV unspliced RNA from patients on combination antiretroviral therapy with undetectable plasma viremia predict the therapy outcome. PLoS One. 2009;4:e8490. doi: 10.1371/journal.pone.0008490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marin B, et al. Non-AIDS-defining deaths and immunodeficiency in the era of combination antiretroviral therapy. AIDS. 2009;23:1743–1753. doi: 10.1097/QAD.0b013e32832e9b78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guaraldi G, et al. Premature age-related comorbidities among HIV-infected persons compared with the general population. Clin Infect Dis. 2011;53:1120–1126. doi: 10.1093/cid/cir627. [DOI] [PubMed] [Google Scholar]
- 4.Brenchley JM, et al. Differential Th17 CD4 T-cell depletion in pathogenic and nonpathogenic lentiviral infections. Blood. 2008;112:2826–2835. doi: 10.1182/blood-2008-05-159301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Favre D, et al. Tryptophan catabolism by indoleamine 2,3-dioxygenase 1 alters the balance of TH17 to regulatory T cells in HIV disease. Sci Transl Med. 2010;2:32ra36. doi: 10.1126/scitranslmed.3000632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chege D, et al. Sigmoid Th17 populations, the HIV latent reservoir, and microbial translocation in men on long-term antiretroviral therapy. AIDS. 2011;25:741–749. doi: 10.1097/QAD.0b013e328344cefb. [DOI] [PubMed] [Google Scholar]
- 7.Brenchley JM, et al. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat Med. 2006;12:1365–1371. doi: 10.1038/nm1511. [DOI] [PubMed] [Google Scholar]
- 8.French MA, King MS, Tschampa JM, Da Silva BA, Landay AL. Serum immune activation markers are persistently increased in patients with HIV infection after 6 years of antiretroviral therapy despite suppression of viral replication and reconstitution of CD4+ T cells. J Infect Dis. 2009;200:1212–1215. doi: 10.1086/605890. [DOI] [PubMed] [Google Scholar]
- 9.Neuhaus J, et al. Markers of inflammation, coagulation, and renal function are elevated in adults with HIV infection. J Infect Dis. 2010;201:1788–1795. doi: 10.1086/652749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hunt PW, et al. T cell activation is associated with lower CD4+ T cell gains in human immunodeficiency virus-infected patients with sustained viral suppression during antiretroviral therapy. J Infect Dis. 2003;187:1534–1543. doi: 10.1086/374786. [DOI] [PubMed] [Google Scholar]
- 11.Wada NI, et al. The effect of HAART-induced HIV suppression on circulating markers of inflammation and immune activation. AIDS. 2015;29:463–471. doi: 10.1097/QAD.0000000000000545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Utay NS, Hunt PW. Role of immune activation in progression to AIDS. Curr Opin HIV AIDS. 2016;11:131–137. doi: 10.1097/COH.0000000000000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsu DC, Sereti I. Serious Non-AIDS Events: Therapeutic Targets of Immune Activation and Chronic Inflammation in HIV Infection. Drugs. 2016;76:533–549. doi: 10.1007/s40265-016-0546-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ferreira CM, et al. The central role of the gut microbiota in chronic inflammatory diseases. J Immunol Res. 2014;2014:689492. doi: 10.1155/2014/689492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kennedy PJ, Cryan JF, Dinan TG, Clarke G. Irritable bowel syndrome: a microbiome-gut-brain axis disorder? World J Gastroenterol. 2014;20:14105–14125. doi: 10.3748/wjg.v20.i39.14105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karlsson FH, et al. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature. 2013;498:99–103. doi: 10.1038/nature12198. [DOI] [PubMed] [Google Scholar]
- 17.Damms-Machado A, et al. Effects of surgical and dietary weight loss therapy for obesity on gut microbiota composition and nutrient absorption. Biomed Res Int. 2015;2015:806248. doi: 10.1155/2015/806248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Riva A, et al. Pediatric obesity is associated with an altered gut microbiota and discordant shifts in Firmicutes populations. Environ Microbiol. 2017;19:95–105. doi: 10.1111/1462-2920.13463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang X, et al. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat Med. 2015;21:895–905. doi: 10.1038/nm.3914. [DOI] [PubMed] [Google Scholar]
- 20.Bisgaard, H. et al. Reduced diversity of the intestinal microbiota during infancy is associated with increased risk of allergic disease at school age. J Allergy Clin Immunol128, 646–652 e641-645, 10.1016/j.jaci.2011.04.060 (2011). [DOI] [PubMed]
- 21.Chen J, et al. Multiple sclerosis patients have a distinct gut microbiota compared to healthy controls. Sci Rep. 2016;6:28484. doi: 10.1038/srep28484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miyake S, et al. Dysbiosis in the Gut Microbiota of Patients with Multiple Sclerosis, with a Striking Depletion of Species Belonging to Clostridia XIVa and IV Clusters. PLoS One. 2015;10:e0137429. doi: 10.1371/journal.pone.0137429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Williams B, Landay A, Presti RM. Microbiome alterations in HIV infection a review. Cell Microbiol. 2016;18:645–651. doi: 10.1111/cmi.12588. [DOI] [PubMed] [Google Scholar]
- 24.Gori A, et al. Early impairment of gut function and gut flora supporting a role for alteration of gastrointestinal mucosa in human immunodeficiency virus pathogenesis. J Clin Microbiol. 2008;46:757–758. doi: 10.1128/JCM.01729-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dillon SM, et al. An altered intestinal mucosal microbiome in HIV-1 infection is associated with mucosal and systemic immune activation and endotoxemia. Mucosal Immunol. 2014;7:983–994. doi: 10.1038/mi.2013.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lozupone CA, et al. Alterations in the gut microbiota associated with HIV-1 infection. Cell Host Microbe. 2013;14:329–339. doi: 10.1016/j.chom.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ling Z, et al. Alterations in the Fecal Microbiota of Patients with HIV-1 Infection: An Observational Study in A Chinese Population. Sci Rep. 2016;6:30673. doi: 10.1038/srep30673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mutlu EA, et al. A compositional look at the human gastrointestinal microbiome and immune activation parameters in HIV infected subjects. PLoS Pathog. 2014;10:e1003829. doi: 10.1371/journal.ppat.1003829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McHardy IH, et al. HIV Infection is associated with compositional and functional shifts in the rectal mucosal microbiota. Microbiome. 2013;1:26. doi: 10.1186/2049-2618-1-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu G, Fadrosh D, Ma B, Ravel J, Goedert JJ. Anal microbiota profiles in HIV-positive and HIV-negative MSM. AIDS. 2014;28:753–760. doi: 10.1097/QAD.0000000000000154. [DOI] [PubMed] [Google Scholar]
- 31.Dinh DM, et al. Intestinal microbiota, microbial translocation, and systemic inflammation in chronic HIV infection. J Infect Dis. 2015;211:19–27. doi: 10.1093/infdis/jiu409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Noguera-Julian M, et al. Gut Microbiota Linked to Sexual Preference and HIV Infection. EBioMedicine. 2016;5:135–146. doi: 10.1016/j.ebiom.2016.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vujkovic-Cvijin I, et al. Dysbiosis of the gut microbiota is associated with HIV disease progression and tryptophan catabolism. Sci Transl Med. 2013;5:193ra191. doi: 10.1126/scitranslmed.3006438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu W, et al. Association Between Gut Microbiota and CD4 Recovery in HIV-1 Infected Patients. Front Microbiol. 2018;9:1451. doi: 10.3389/fmicb.2018.01451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Perez-Santiago J, et al. Gut Lactobacillales are associated with higher CD4 and less microbial translocation during HIV infection. AIDS. 2013;27:1921–1931. doi: 10.1097/QAD.0b013e3283611816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lozupone CA, et al. HIV-induced alteration in gut microbiota: driving factors, consequences, and effects of antiretroviral therapy. Gut Microbes. 2014;5:562–570. doi: 10.4161/gmic.32132. [DOI] [PubMed] [Google Scholar]
- 37.Bender JM, et al. Maternal HIV infection influences the microbiome of HIV-uninfected infants. Sci Transl Med. 2016;8:349ra100. doi: 10.1126/scitranslmed.aaf5103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shapiro RL, Lockman S. Mortality among HIV-exposed infants: the first and final frontier. Clin Infect Dis. 2010;50:445–447. doi: 10.1086/649887. [DOI] [PubMed] [Google Scholar]
- 39.Allison SM, et al. Assessing medication adherence of perinatally HIV-infected children using caregiver interviews. J Assoc Nurses AIDS Care. 2010;21:478–488. doi: 10.1016/j.jana.2010.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nowak P, et al. Gut microbiota diversity predicts immune status in HIV-1 infection. AIDS. 2015;29:2409–2418. doi: 10.1097/QAD.0000000000000869. [DOI] [PubMed] [Google Scholar]
- 41.Dubourg G, et al. Gut microbiota associated with HIV infection is significantly enriched in bacteria tolerant to oxygen. BMJ Open Gastroenterol. 2016;3:e000080. doi: 10.1136/bmjgast-2016-000080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lange K, Buerger M, Stallmach A, Bruns T. Effects of Antibiotics on Gut Microbiota. Dig Dis. 2016;34:260–268. doi: 10.1159/000443360. [DOI] [PubMed] [Google Scholar]
- 43.Yang L, et al. HIV-induced immunosuppression is associated with colonization of the proximal gut by environmental bacteria. AIDS. 2016;30:19–29. doi: 10.1097/QAD.0000000000000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scher JU, et al. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. Elife. 2013;2:e01202. doi: 10.7554/eLife.01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maeda Y, et al. Dysbiosis Contributes to Arthritis Development via Activation of Autoreactive T Cells in the Intestine. Arthritis Rheumatol. 2016;68:2646–2661. doi: 10.1002/art.39783. [DOI] [PubMed] [Google Scholar]
- 46.Pedersen HK, et al. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature. 2016;535:376–381. doi: 10.1038/nature18646. [DOI] [PubMed] [Google Scholar]
- 47.Kelley CF, et al. The rectal mucosa and condomless receptive anal intercourse in HIV-negative MSM: implications for HIV transmission and prevention. Mucosal Immunol. 2017;10:996–1007. doi: 10.1038/mi.2016.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Richter JR, et al. Macrophage-derived chemokine (CCL22) is a novel mediator of lung inflammation following hemorrhage and resuscitation. Shock. 2014;42:525–531. doi: 10.1097/SHK.0000000000000253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leung TF, et al. Serum concentration of macrophage-derived chemokine may be a useful inflammatory marker for assessing severity of atopic dermatitis in infants and young children. Pediatr Allergy Immunol. 2003;14:296–301. doi: 10.1034/j.1399-3038.2003.00052.x. [DOI] [PubMed] [Google Scholar]
- 50.Ji Y, et al. Changes in intestinal microbiota in HIV-1-infected subjects following cART initiation: influence of CD4+ T cell count. Emerg Microbes Infect. 2018;7:113. doi: 10.1038/s41426-018-0117-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tobler LH, et al. Interferon and interferon-induced chemokine expression is associated with control of acute viremia in West Nile virus-infected blood donors. J Infect Dis. 2008;198:979–983. doi: 10.1086/591466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stacey AR, et al. Induction of a striking systemic cytokine cascade prior to peak viremia in acute human immunodeficiency virus type 1 infection, in contrast to more modest and delayed responses in acute hepatitis B and C virus infections. J Virol. 2009;83:3719–3733. doi: 10.1128/JVI.01844-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Crisan D, Grigorescu MD, Radu C, Suciu A, Grigorescu M. Interferon-gamma-inducible protein-10 in chronic hepatitis C: Correlations with insulin resistance, histological features & sustained virological response. Indian J Med Res. 2017;145:543–550. doi: 10.4103/ijmr.IJMR_1410_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hagau N, et al. Clinical aspects and cytokine response in severe H1N1 influenza A virus infection. Crit Care. 2010;14:R203. doi: 10.1186/cc9324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.de Jong MD, et al. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat Med. 2006;12:1203–1207. doi: 10.1038/nm1477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mhandire K, et al. Plasma IP-10 Concentrations Correlate Positively with Viraemia and Inversely with CD4 Counts in Untreated HIV Infection. Open AIDS J. 2017;11:24–31. doi: 10.2174/1874613601711010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee S, et al. Interferon-inducible protein 10 (IP-10) is associated with viremia of early HIV-1 infection in Korean patients. J Med Virol. 2015;87:782–789. doi: 10.1002/jmv.24026. [DOI] [PubMed] [Google Scholar]
- 58.Roberts L, et al. Plasma cytokine levels during acute HIV-1 infection predict HIV disease progression. AIDS. 2010;24:819–831. doi: 10.1097/QAD.0b013e3283367836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Keating SM, et al. The effect of HIV infection and HAART on inflammatory biomarkers in a population-based cohort of women. AIDS. 2011;25:1823–1832. doi: 10.1097/QAD.0b013e3283489d1f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kamat A, et al. A plasma biomarker signature of immune activation in HIV patients on antiretroviral therapy. PLoS One. 2012;7:e30881. doi: 10.1371/journal.pone.0030881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stylianou E, Aukrust P, Bendtzen K, Muller F, Froland SS. Interferons and interferon (IFN)-inducible protein 10 during highly active anti-retroviral therapy (HAART)-possible immunosuppressive role of IFN-alpha in HIV infection. Clin Exp Immunol. 2000;119:479–485. doi: 10.1046/j.1365-2249.2000.01144.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lane BR, Strieter RM, Coffey MJ, Markovitz DM. Human immunodeficiency virus type 1 (HIV-1)-induced GRO-alpha production stimulates HIV-1 replication in macrophages and T lymphocytes. J Virol. 2001;75:5812–5822. doi: 10.1128/JVI.75.13.5812-5822.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sandler NG, et al. Plasma levels of soluble CD14 independently predict mortality in HIV infection. J Infect Dis. 2011;203:780–790. doi: 10.1093/infdis/jiq118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shivakoti R, et al. Soluble CD14: An Independent Biomarker for the Risk of Mother-to-Child Transmission of HIV in a Setting of Preexposure and Postexposure Antiretroviral Prophylaxis. J Infect Dis. 2016;213:762–765. doi: 10.1093/infdis/jiv479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bi, X. et al. Impact of HIV Infection and Anti-Retroviral Therapy on the Immune Profile of and Microbial Translocation in HIV-Infected Children in Vietnam. Int J Mol Sci17, 10.3390/ijms17081245 (2016). [DOI] [PMC free article] [PubMed]
- 66.Wallet MA, et al. Microbial translocation induces persistent macrophage activation unrelated to HIV-1 levels or T-cell activation following therapy. AIDS. 2010;24:1281–1290. doi: 10.1097/QAD.0b013e328339e228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Alvarez P, et al. Immune activation despite preserved CD4 T cells in perinatally HIV-infected children and adolescents. PLoS One. 2017;12:e0190332. doi: 10.1371/journal.pone.0190332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pilakka-Kanthikeel S, et al. Increased gut microbial translocation in HIV-infected children persists in virologic responders and virologic failures after antiretroviral therapy. Pediatr Infect Dis J. 2012;31:583–591. doi: 10.1097/INF.0b013e31824da0f5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hamer HM, et al. Butyrate modulates oxidative stress in the colonic mucosa of healthy humans. Clin Nutr. 2009;28:88–93. doi: 10.1016/j.clnu.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 70.Kuczynski, J. et al. Using QIIME to analyze 16S rRNA gene sequences from microbial communities. Curr Protoc Bioinformatics Chapter 10, Unit10 17, 10.1002/0471250953.bi1007s36 (2011). [DOI] [PMC free article] [PubMed]
- 71.Robertson CE, et al. Explicet: graphical user interface software for metadata-driven management, analysis and visualization of microbiome data. Bioinformatics. 2013;29:3100–3101. doi: 10.1093/bioinformatics/btt526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jari Oksanen, F. G. B., Michael Friendly, Roeland Kindt,, Pierre Legendre, D. M., Peter R. Minchin, R. B. O’Hara,, Gavin L. Simpson, P. S., M. Henry H. Stevens, Eduard Szoecs, & Wagner, H. Vegan: Community Ecology Package. R Package Version. 2.0-10, https://cran.r-project.org, https://github.com/vegandevs/vegan (2016).
- 73.Segata N, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011;12:R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.