Abstract Abstract
Background: When phenotypic characters are described in the literature, they may be constrained or clarified with additional information such as the location or degree of expression, these terms are called “modifiers”. With effort underway to convert narrative character descriptions to computable data, ontologies for such modifiers are needed. Such ontologies can also be used to guide term usage in future publications. Spatial and method modifiers are the subjects of ontologies that already have been developed or are under development. In this work, frequency (e.g., rarely, usually), certainty (e.g., probably, definitely), degree (e.g., slightly, extremely), and coverage modifiers (e.g., sparsely, entirely) are collected, reviewed, and used to create two modifier ontologies with different design considerations. The basic goal is to express the sequential relationships within a type of modifiers, for example, usually is more frequent than rarely, in order to allow data annotated with ontology terms to be classified accordingly.
Method: Two designs are proposed for the ontology, both using the list pattern: a closed ordered list (i.e., five-bin design) and an open ordered list design. The five-bin design puts the modifier terms into a set of 5 fixed bins with interval object properties, for example, one_level_more/less_frequently_than, where new terms can only be added as synonyms to existing classes. The open list approach starts with 5 bins, but supports the extensibility of the list via ordinal properties, for example, more/less_frequently_than, allowing new terms to be inserted as a new class anywhere in the list. The consequences of the different design decisions are discussed in the paper. CharaParser was used to extract modifiers from plant, ant, and other taxonomic descriptions. After a manual screening, 130 modifier words were selected as the candidate terms for the modifier ontologies. Four curators/experts (three biologists and one information scientist specialized in biosemantics) reviewed and categorized the terms into 20 bins using the Ontology Term Organizer (OTO) (http://biosemantics.arizona.edu/OTO). Inter-curator variations were reviewed and expressed in the final ontologies.
Results: Frequency, certainty, degree, and coverage terms with complete agreement among all curators were used as class labels or exact synonyms. Terms with different interpretations were either excluded or included using “broader synonym” or “not recommended” annotation properties. These annotations explicitly allow for the user to be aware of the semantic ambiguity associated with the terms and whether they should be used with caution or avoided. Expert categorization results showed that 16 out of 20 bins contained terms with full agreements, suggesting differentiating the modifiers into 5 levels/bins balances the need to differentiate modifiers and the need for the ontology to reflect user consensus. Two ontologies, developed using the Protege ontology editor, are made available as OWL files and can be downloaded from https://github.com/biosemantics/ontologies.
Contribution: We built the first two modifier ontologies following a consensus-based approach with terms commonly used in taxonomic literature. The five-bin ontology has been used in the Explorer of Taxon Concepts web toolkit to compute the similarity between characters extracted from literature to facilitate taxon concepts alignments. The two ontologies will also be used in an ontology-informed authoring tool for taxonomists to facilitate consistency in modifier term usage.
Keywords: frequency modifiers, certainty modifiers, degree modifiers, coverage modifiers, Modifier Ontology, phenotype modifiers, user warrant, literary warrant, user consensus
Introduction
Despite the development and use of sensor technology in biomedical domains and applications, phenotypic character descriptions published in the literature remain an indispensable resource for ecological and systematics research.
Anatomical and quality ontologies have been developed to support the curation workflows that aim to convert narrative phenotypical characters to ontological statements for cross-taxon inferences and computation. Uber-anatomy Ontology (UBERON), Hymenoptera Anatomy Ontology (HAO), and the Plant Ontology (PO) are some examples of anatomical ontologies that contain anatomical structure terms and their relationships (Cooper et al. 2013, Yoder et al. 2010, Mungall et al. 2016). The Phenotypic Quality Ontology (PATO) is a taxon-neutral quality ontology that treats character and character value terms (Gkoutos et al. 2017, Gkoutos et al. 2004). These ontologies are often used by EQ-based approaches, where Entity and Quality are post-composed to create an ontological statement for a character (Gkoutos et al. 2009, Dahdul et al. 2018a). Other phenotype ontologies, such as the Flora Phenotype Ontology or FLOPO (Hoehndorf et al. 2016), have also been developed to include complete characters.
Modifier terms are used widely in phenotypic character descriptions but have not been treated formally in an ontology. Hagedorn (2007) provided a good definition for phenotype character modifiers:
A modifier is a unit of information that adds detail (or constraints) to the statement to which it is applied. When the modifier information is ignored, the original statement must retain a substantial, albeit more general meaning. A modifier may be applied to statements already modified. Modifiers themselves are constrained by a terminology.
Further, Hagedorn comprehensively summarized the existing studies and arrived at a modifier taxonomy, consisting of 11 groups of modifiers. In this work, we attempt to construct modifier ontologies that treat four groups of the modifiers that have general usage across many characters and share the same characteristics of having implied order among the terms, for example, rarely is less frequent than often, perhaps is less certain than clearly. This sequential relationship is the key semantics we would like to capture in the modifier ontologies because it will be the key for a computer to understand:
How to compare modifiers semantically
When to inherit a character from a family level description to a genus level
How to use them in an identification key application
We propose two alternative approaches to constructing a modifier ontology and discuss the tradeoffs between the two. Both approaches are grounded to a set of modifier words extracted from 30 volumes of Flora of North America (Flora of North America Editorial Committee 1993), the Flora of China (Flora of China Editorial Committee 1994), and a large number of taxonomic publications (ca. 21,000 treatments) on ants, algal fossils, and other taxon groups.
Related work
While a standard formula for building ontologies is yet to be proposed, Z39.19 National Standard for Monolingual Controlled Vocabulary Construction NISO (National Information Standards Organization) (2010) Z39.19-2005 laid out the fundamental principles for controlled vocabularies, which apply equally well to ontology building. These principles are “eliminating ambiguity, controlling synonyms, establishing relationships among terms where appropriate, [and] testing and validation of terms” p. 12 of the NISO (National Information Standards Organization) 2010. In addition, the OBO Foundry Principles provide a set of guidelines that OBO Foundry ontologies are expected to follow, covering aspects ranging from ontology content, from definitions and relations (mostly under-development) to ontology management (Smith et al. 2007).
The Basic Formal Ontology (BFO, Arp et al. 2015; https://raw.githubusercontent.com/BFO-ontology/BFO/v2.0/bfo.owl, accessed 4/18/2018) provides a genuine domain independent upper ontology that differentiates a number of fundamental concepts that are useful to guide the development of many ontologies.
Within the BFO framework, character modifiers would fall under the Specifically Dependent Continuant > Quality class. PATO is a taxon-neutral quality ontology (Gkoutos et al. 2004) with the root class “quality” and is tasked to supply quality terms within the BFO framework. Although it is not specified in PATO, the PATO class for quality encompasses terms that would also be subclasses of BFO’s class “specifically dependent continuant”. For consistency with other trait or phenotype ontologies, we place our root class “modifier” as a subclass of PATO quality.
Hagedorn’s dissertation (Hagedorn 2007) comprehensively reviewed then existing data models for descriptive data of organisms, including those used in DELTA and alike, NEXUS, DiversityDescriptions, CBIT Lucid, XPER and alike, Prometheus, and SDD (Lebbe 1984, Maddison et al. 1997, Pullan et al. 2005, Dallwitz et al. 2000, Hagedorn et al. 2006, Hagedorn 2005, CBIT 2007), each has varied support for different types of modifiers. Hagedorn then grouped modifiers into 11 categories:
Spatial modifiers (p. 203, also called “location” or “topological” modifiers). These modifiers indicate a location where a character appear. For example, “at the base”.
Temporal modifiers (p. 204) indicate a time when a character appears. For example, “when old”.
Method modifiers (p. 205) indicate the method that is used to generate or observe a character, for example, “in alcohol”, and “under hand-lens”.
Frequency modifiers (p.206) indicate the probability of observing a true statement, for example, “usually”, “occasionally”, and “rarely”.
Certainty modifiers (p. 207) indicate the probability of a statement being true, for example, “perhaps”, “probably”, “likely”, and “certainly”.
Approximation modifiers (p. 209), a kind of certainty modifier, indicate the degree of inaccuracy of a reported value. For example, “ca.”, “approximately”, “about”, and “roughly”.
Modifiers hinting misinterpretation (p. 209) indicate a stated character is the result of misinterpretation. For example, “by misinterpretation”.
Negation modifiers (p. 211) indicate a negation of a stated character. For example, “not red”.
State modifiers (p. 212) modify the quality, degree, emphasis, or manner, etc. of a state itself. For example, “very”, “weakly”, and “slightly”.
Reliability modifiers (p. 213) indicate the suitability of a character for the purpose of taxon identification.
Other modifiers (p. 214).
The modifier taxonomy proposed in Hagedorn (2007) provides the initial framework for our modifier ontologies.
Over the course of the past ten years, many ontology design patterns have been proposed (e.g., Aranguren et al. 2008, Egaña et al. 2008, Presutti et al. 2012). A design pattern is a general, repeatable solution to a commonly occurring problem. Design patterns have been widely used in software engineering for years to develop reusable and maintainable code bases. The list pattern for ontology development is particularly relevant to modifier ontologies (http://ontologydesignpatterns.org/wiki/Submissions:List accessed 5/27/2018) because the order of the terms is the important semantic relationship that needs to be made explicit to support the applications noted above.
Material and methods
Define the Scope
Ontologies concerning Categories 1-3 in Hagedorn’s taxonomy have been developed or are under development, for example, the Biological Spatial Ontology, (BSPO, Dahdul et al. 2014), the Measurement Method Ontology (Shimoyama et al. 2012), and the Experimental Condition Ontology (Shimoyama et al. 2012). Categories 7 and 10 are defined solely for the purpose of taxon identification and consist of a closed set of system defined terms. These categories are out of scope of the modifier ontology, which focuses on groups of modifiers that have general usage across many characters and are sequentially related to one another. The negation modifiers, or Category 8, was also excluded because negations can be handled with the logical NOT operator. Category 9 derives more specific states from a base state and most of such modifiers are character dependent, for example, “dull” can only modify color characters or sharpness of some edges. However, a subset of the state modifiers, degree modifiers, does have general applicability. Based on this analysis, the scope of our modifier ontologies covers Frequency, Certainty, Degree, and Coverage modifiers (defined below). Coverage modifiers were added after reviewing the candidate terms extracted from a wide range of taxonomic descriptions.
Frequency: the probability of observing a quality
Certainty: the probability of a quality being true
Degree: the measure or intensity of a quality, ranging from the minimal to extremely intense
Coverage: the spatial extent or scope of a quality, ranging from very sparse coverage to complete coverage of an entity.
Data Collection
Following the literary warrant principle ANSI/NISO (National Information Standards Organization) 2010, we intended for the modifier ontology to include modifier terms used in published taxonomic descriptions. CharaParser (Cui 2012), now a part of the Explorer of Taxon Concepts web toolkit (Cui et al. 2016), was used to parse taxonomic descriptions and extract modifiers from a variety of taxonomic publications (https://www.dropbox.com/sh/msnqb0aqjgwlgaw/AAA-jUfSq14vrnM-AgKSjd49a?dl=0), covering ants, diatoms, plants, and fungi. CharaParser markups biological entities, characters, relationships, and modifiers in taxonomic descriptions. A few thousand unique modifier terms/phrases were extracted and after a manual review of these extracted phrases, 130 unique, one-word modifiers within the scope defined above were selected. Multiple-word phrases or expressions were not considered in this work to limit its scope.
Modeling
We observed that the modifier terms were ordinal values. To express the sequential relationships among the terms of each modifier type, two inverse and transitive properties were needed in the ontology: proceeds and follows. Subproperties of proceeds and follows can be defined for each of the modifier types, for example, more_frequently_than and less_frequently_than (Fig. 1). For some applications, there may be a need to treat these ordinal values as interval values. To support this need, further subproperties can be created, for example, one_level_more_frequently_than and one_level_less_frequently_than, making the semantic distance between adjacent nodes equal (i.e., “one level”). The form of this set of property and subproperties is similar to the preceded_by and immediately_preceded_by subproperties of temporally_related_to in the Relations Ontology (RO, http://www.obofoundry.org/ontology/ro.html, accessed 5/27/2018), but the former not only takes out the possibility of inserting an intermediate node between two existing nodes, it further equalizes the distances between any adjacent nodes to “one level”. Consumers of the ontology may define the level based on their specific needs.
Figure 1.
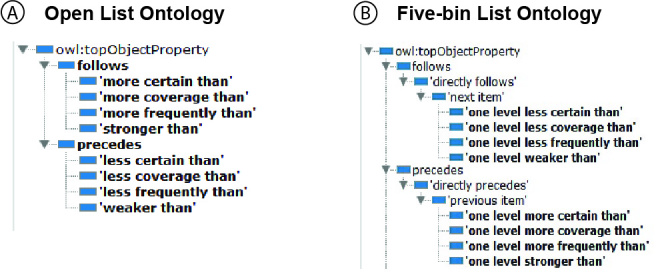
List related object properties in Open List and Five-Bin Ontologies
In applying the list pattern to build the modifier ontologies, we have the choice of keeping the list open or making it closed. An ontology was implemented with each of the two approaches. The open list approach does not limit the size of the list (Fig. 2A). Each modifier type is modeled as an open list, where new modifiers can be inserted to the list as classes as long as the proceeds and follows relationship pairs are established between the new term and their neighboring terms. Fig. 2A shows a conceptual structure of an open list, where a new term (marked as 5) is being inserted into the list.
Figure 2.
Open List vs. Closed List.
Similar to the open list approach, in the closed list approach, each modifier type is modeled as a list. However, a closed list has a fixed size, where new modifier terms can only be added as synonyms to some existing nodes (terms) in the list (Fig. 2B).
Open list allows new nodes (i.e., classes) to be inserted anywhere in the list, causing a shift of relative positions of existing nodes, for example, when node 5 is inserted, the original node 5 becomes node 6 (Fig. 2A). Closed list has fixed number of nodes, and new terms can only be added as synonyms. It’s possible for a term to be a synonym of two different nodes, and such a term is a broader synonym of the relevant nodes. Arrowed lines between nodes represent inverse object properties (proceeds and follows).
Both approaches have desirable and undesirable consequences. An open list is more flexible because not only can new types be easily added as a new list, but new modifier terms can also be added either as a class or a synonym. An open list is not suitable to model interval values because when a new term is added as a class, it changes the positions of all the nodes after the insertion point and therefore the relative positions of affected nodes to all other nodes. This changes the semantic distance between affected nodes. As shown (Fig. 2A), when a new node is added at position 5, the original node 5 becomes node 6, and the distance between this node and node 1 is increased by one. Before the insertion, the similarity between node 5 and node 4 is the same as the similarity between node 4 and node 3. After the insertion, node 5 (now node 6) becomes less similar to node 4 than node 3 is to node 4.
A closed list is a better fit for modeling interval values because the length of the list (the total semantic range) and the position of the nodes in the list are fixed. This fixed structure makes it easy to define the nodes as disjoint classes and to define a list to include only the given classes. This, in effect, creates a “closed world”, making it possible for the machine to classify an unknown entity (i.e., if an unknown entity is one-level preceding node 4 and one-level following node 2, then it must be node 3). Such classification reasoning cannot be done with an open list due to the “open world” assumption of OWL ontologies: the unknown entity may be node 3 but it could also be a node that has not yet been defined.
We also note that open lists allow the ontology to be loaded with more nuanced terms (classes) in a list. Users need to be very cautious when using this feature. Many modifier terms only have subtle differences in meaning and these subtle differences are also quite subjective. This creates two major difficulties in maintaining the ontology’s stability and usability. First, ontology curators and ontology users may not share the same understanding of these terms (and human readable definitions for the terms will not solve this problem). Second, it will be very difficult for different users of the ontologies to use these terms consistently or even for the same users to use these terms consistently over time. The same is true for different curators managing the ontologies.
We implemented two modifier ontologies using the approaches respectively because the need for being flexible and the need for stronger machine reasoning capability seem to be important. Users should decide which implementation better meets their needs.
Term Categorization Consensus
Both open and closed list ontologies need to start by crystallizing the sequential relationships among the available terms for a modifier type. To reveal experts’ shared understanding of modifier terms, five bins were created for each of the four modifier types. For example, for the frequency modifiers, the five bins are frequency_0, frequency_25, frequency_50, frequency_75, and frequency_100. The number five was selected to strike a balance between the need to differentiate a good number of levels in each type of modifiers and the requirement for intuitive and consistent categorization of the terms by the users.
The three leading co-authors and the corresponding author categorized the 130 terms into 20 bins (5 bins for each type of modifier) using OTO (Huang et al. 2015, http://biosemantics.arizona.edu:8080/OTO). Since the terms are on the ordinal scale, the experts were not given numerical ranges for the bins but were instructed to simply categorize the terms based on their intuition: do you feel “sometimes” is more similar to 50% frequency or 75% frequency? OTO supports multi-user categorization of terms and synonyms and records all user decisions and comments. It also allows the user to put the same term into multiple bins (Fig. 3). After independent categorization of the terms, experts met virtually and finalized categorization.
Figure 3.
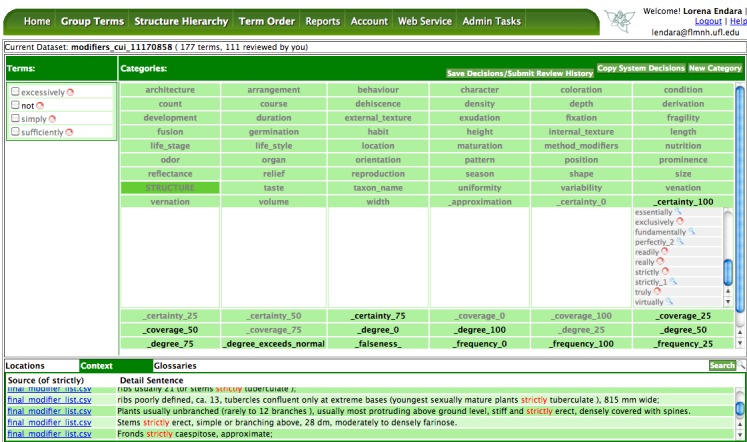
OTO Group Terms User Interface.
Terms to be categorized are in the Terms panel on the left, and the bins are shown in the Categories panel on the right. The source sentences where terms were used are shown in the Context tab in the lower panel. The user drags and drops a term into a bin. The red circle next to a term indicates users have different categorization decisions on the term. Click on the red circle, different decisions will be shown in a pop-up window. Synonyms of a term are shown with an indent below their preferred term. If a term is put into multiple bins, a numerical index is attached to the term to create copies of terms. The term set used in this study is "modifiers_cui_11170858" on OTO, accessible to any OTO registered user.
Ontology Construction
After the terms are categorized and categorization reviewed and discussed by the experts, Protege was used to implement the ontologies. Following the user warrant principle (NISO (National Information Standards Organization) 2010), expert consensus on term categorization forms the basis for constructing the ontologies (Tables 1, 2, 3, 4). The following scheme was used to construct a base ontology to which different data properties were then added to create the open list and the five-bin ontologies:
Table 1.
Frequency, certainty, degree, and coverage modifiers with complete consensus among four experts. Proposed labels are in bold. Expert contributed terms are in quotation marks.
| frequency_0 | frequency_25 | frequency_50 | frequency_75 | frequency_100 |
| never | infrequently, occasionally, seldom, uncommonly, rarely | sometimes | frequently, often, regularly, usually | always, consistently |
| certainty_0 | certainty_25 | certainty_50 | certainty_75 | certainty_100 |
| “uncertain” “unclearly” “doubtfully” |
perhaps, possibly | presumably, seemingly | approximately, nearly | decidedly, definitely, distinctly, effectively, essentially, evidentially, evidently, fundamentally,obviously,patently,readily, truly, undoubtedly, virtually |
| degree_0 | degree_25 | degree_50 | degree_75 | degree_100 |
| inconspicuously imperceptibly “unnoticeably” |
barely, faintly, feebly, gently, hardly, lightly, merely, obscurely, scarcely, slightly, subtly | moderately, relatively, modestly | appreciably, considerably, greatly, highly, much, particularly, profoundly, significantly, strongly, very, noticeably, visibly |
boldly, conspicuously, prominently, extremely exceedingly, enormously, exceptionally, extraordinarily, grossly |
| coverage_0 | coverage_25 | coverage_50 | coverage_75 | coverage_100 |
| sparsely, sparingly | “densely” | entirely, throughout, uniformly |
Table 2.
Frequency, certainty, degree, and coverage modifiers with type but not bin consensus among four experts.
| Terms | Suggested bins | |||
| certainty | almost | certainty_100 | certainty_75 | |
| apparently | certainty_100 | certainty_75 | ||
| basically | certainty_100 | certainty_75 | ||
| practically | certainty_100 | certainty_75 | ||
| probably | certainty_75 | certainty_50 | certainty_25 | |
| reportedly | certainty_75 | certainty_50 | ||
| degree | strikingly | degree_100 | degree_75 | |
| notably | degree_50 | degree_75 | ||
| quite | degree_50 | degree_75 | ||
| rather | degree_50 | degree_75 | ||
| fairly | degree_50 | degree_25 | ||
| mildly | degree_50 | degree_25 | ||
| somewhat | degree_50 | degree_25 | ||
| sufficiently | degree_50 | degree_100 | ||
| markedly | degree_100 | degree_75 | ||
Table 3.
Terms that have bin consensus but not type consensus among four experts.
| Term | Frequency | Degree | Certainty | Coverage |
| chiefly | _75 | _75 | ||
| mainly | _75 | _75 | ||
| primarily | _75 | _75 | ||
| strictly | _100 | _100 | ||
| exclusively | _100 | _100 | ||
| extensively | _75 | _75 | ||
| fully | _100 | _100 | ||
| totally | _100 | _100 | ||
| completely | _100 | _100 | ||
| largely | _75 | _75 | ||
| mostly | _75 | _75 | ||
| partly | _50 | _50 | ||
| partially | _50 | _50 | ||
| indistinctly | _25 | _25 | ||
| vaguely | _25 | _25 | ||
| perfectly | _100 | _100 | _100 | |
| predominantly | _75 | _75 | ||
| prevalently | _75 | _75 | _75 | |
| commonly | _75 | _75 | _75 | |
| typically | _75 | _75 | _75 |
Table 4.
Modifier terms with poor consensus on both type and bin, and their treatment in the ontology
| Term | Bins the terms were categorized into by different experts | Treatment of the term for the ontology | ||||
| Frequency | Certainty | Degree | Coverage | Other | ||
| altogether | _100 | yes | Colloquial, excluded from ontology E.g., The black spot altogether absent |
|||
| casually | _25 | State[pattern] modifier, excluded E.g., Veins regularly or casually anastomosing. |
||||
| copiously | _75 | State [quantity], excluded from ontology E.g., Petiole copiously glandular when young |
||||
| dominantly | _75 | _75, _100 | _75 | Included as not Recommended E.g., Cells dominantly solitary, but short chains can be found |
||
| eccentrically | yes | Spatial modifier, excluded E.g., Anthers eccentrically peltate |
||||
| excessively | _75 | yes | Not character modifier, excluded E.g., Females excessively rare |
|||
| generally | _75 | _50, _75 | _75 | Included as not Recommended E.g., head otherwise generally smooth and shining. E.g., branches generally quadrangular |
||
| imperfectly | _75 | _25 | State modifier, excluded E.g., Rays furcate or imperfectly so. Ovary superior, imperfectly 2-loculed |
|||
| incompletely | _75 | State and other modifier, excluded E.g., Legumes incompletely 2-locular. E.g., Lamina incompletely 2-pinnate at base. E.g., Scales incompletely cover underlying leaves. |
||||
| intensely | _75, _100 | yes | State [color] modifier, excluded E.g., Petals intensely violet |
|||
| intermittently | _50 | _25, _50 | Included as notRecommended E.g., Sori spreading intermittently along individual veins almost from midrib to margine. |
|||
| no | _0 | _0 | Negation, excluded | |||
| not | _0 | _0 | Negation, excluded | |||
| powerfully | _100 | State[Size] modifier, excluded E.g., Larvae with mandibles powerfully developed for ant larvae |
||||
| really | _100 | yes | Not modify characters, excluded E.g., Really 3 convexities exist. |
|||
| remarkably | _75 | yes | Included as notRecommended E.g., Style remarkably exserted. |
|||
| richly | _100 | yes | Coverage and state modifiers, excluded. E.g., Vein richly anastomosing Stems richly pubescent. |
|||
| roughly | _50 | _50 | yes | State and other modifiers. Included as notRecommended E.g., Bark roughly furrowed. Stigma roughly rectangular. |
||
| simply | yes | State modifier, excluded. E.g., margin regularly doubly serrate, rarely simply serrate. |
||||
| unusually | _75 | yes | Included as notRecommended E.g., Head unusually small |
|||
| widely | _100 | yes | State modifier, excluded E.g., Stem leaves widely spaced |
|||
- Terms with experts’ full agreement on its type and its bin are considered as class label candidates (Table 1).
- Within the group of terms for each type and bin (e.g., frequency_75, see Table 1), experts selected one term that best represents the class and this term becomes the class label. This label has the least chance for end users to confuse it with other class labels.
- The rest of the terms become the exact synonyms of the class (oboInOWL#hasExactSynonym).
- Two exceptions are “throughout” and “uniformly” categorized under coverage_100. This will be discussed in the Discussion section.
Terms with experts’ full agreement on its type, but not on its bin are included in the ontology but annotated as “not recommended” (a new annotation), because there is a good chance for the terms to confuse the end users of the ontology. These terms should be included in the ontology as “not recommended” to discourage the continued usage in scientific publications (Table 2).
Terms with experts’ full agreement on its bin, but not on its type (Table 3) are included in the ontology as broader synonyms (oboInOWL#hasBroaderSynonym). We follow the best practice of the Plant Ontology Consortium and use broader synonym annotations to indicate if the term is considered a synonym of two or more different classes (Cooper et al. 2013).
- Terms without full agreement on its type nor its bin are either included as “not-recommended” or excluded from the ontology (Table 4).
- Informal terms (colloquial terms) are excluded from the ontology.
- If an ambiguous modifier is deemed to have a high probability of being used, it is included in the ontology as a not recommended term.
- State modifiers that fell into Category 9 in Hagedorn (2007) were excluded from the ontology as explained in the “Define the Scope” section.
For bins where no terms with full agreement is found, experts contributed terms from their vocabulary. Descriptive sentences using these terms were then checked in other sources and terms with full expert agreement were included in the ontology. In Table 1, expert-contributed terms are enclosed with quotation marks.
Classes were given a human readable definition based on their type definition. For example:
Frequently (the class label for Frequency_75) is a frequency modifier that indicates around 75% probability of observing a quality.
For the open list ontology, ordinal properties such as more_frequently_than and less_frequently_than were used to indicate the order of the classes in a list. The five-bin implementation of the ontology uses interval properties such as one_level_more_frequently_than and one_level_less_frequently_than. In addition, five-bin version also uses only (opposed to some) existence indicators, disjoint statements, and logical OR operators to make the lists “closed” worlds.
Results
Term Categorization Result
Modifier terms categorized with full agreement on both modifier type and bin accounted for 57.7% of all categorized terms (Table 1). 11.5% terms had agreement on the type, but not on the bin (Table 2), while another 15.4% had agreement on the bin, but not on the type (Table 3). The remainder 16.2% of modifier terms had no agreement on the bin nor the type (Table 4). Four of the twenty bins did not have any terms with full agreement on both type and bin, and three of which are related to coverage. To make the ontology more complete, experts contributed four terms (shown in quotation marks in (Table 1) that filled two of the four empty bins.
Ontology Result
Phenotype Modifier Ontology (open list) and Phenotype Modifier Ontology (5-bin) were created, each contains 44 classes and 128 terms. The ontologies can be accessed at https://github.com/biosemantics/ontologies (Fig. 4).
Figure 4.

Classes in the modifier ontologies.
In the current modifier ontologies, a set of inverse object properties are defined for each type of modifier (e.g., more_frequently_than, less_frequently_than in the open list version, and one_level_ more_frequently_than, one_level_less_frequently_than in the five-bin version), as opposed to using one generic object property for all types of modifiers (Fig. 1). We believe this treatment better models reality because one level of frequency can be semantically different from one level of certainty. These object properties are subproperties of follows/precedes or next item/previous item properties imported from the list pattern.
Discussion
An ontology is a conceptual representation of the consensus of a domain. In the modifier domain, we show that there is a level of consensus among the experts: 16 of 20 bins end up holding terms with full agreement. We acknowledge stronger/weaker consensus can be obtained if we had used smaller/larger number of bins. This result suggests that five bins capture a good amount of consensus and a reasonable number of levels most applications need to distinguish within a modifier type. Since the two ontologies share the same set of terms, the consensus gathered from the experts are presented in both. We would like users to decide which ontology works better for their application and it would be interesting to see how the open list ontology evolves with use over time.
In the process of categorizing the terms, Certainty and Degree modifiers were the most difficult to separate among the four types of modifiers. We note that characters that are intense or with great measurements may imply a high certainty of the observation of the character. However, a high certainty does not always correlate with a stronger degree. Based on this observation, terms primarily describing a degree should be categorized as degree and not extended automatically to certainty. For example, authors may have used the words “visibly” and “noticeable” to indicate certainty on characters, however, knowing the ambiguity associated with certainty and degree terms, we need to alert future authors to the difference.
Relatively fewer terms were consistently categorized into Coverage (Table 1). The vast majority (90%) of the terms that had only type disagreement were categorized as Coverage by at least one expert (Table 3). Terms such “mostly” and “generally” are used frequently in phenotype descriptions, but it was not easy to ascertain what the authors tried to express with the term. For example, “leaves mostly short-petiolate”, was the author trying to say “leaves clearly short-petiolate”(degree), “most leaves short-petiolate” (coverage), or even “leaves usually short-petiolate” (frequency)? Such terms are included in the ontology with an annotation (broader synonym or not recommended) to alert future authors of the ambiguity with hope that these terms will not be used. We also considered the term “intermittently” as a potential coverage_50 modifier to fill the empty bin in Table 1, but there was only one usage of the term (Table 4) in over 21,000 descriptions included in this exercise, and the experts could not agree on its meaning. We decided to leave the empty bins for future work.
PATO has a frequency class and also treats degree terms to an extent, but they both are different from the modifier ontologies. PATO:frequency (PATO_0000044) is a physical quality of a process, “which inheres in a bearer by virtue of the number of the bearer’s repetitive actions in a particular time”. Based on this definition, PATO:frequency is a quality itself and not a modifier to a quality. Using one example to differentiate the two concepts: a PATO:frequency can be rate of heart beat, say 70 times/min, in contrast, our frequency modifiers describe how often we observe a heart beat of 70 times/min. Hence, frequency modifiers are different from PATO:frequency, conceptually. In our ontologies, we used label “frequency_modifier” to make the difference clear.
PATO employs a consistent pattern of representing the extent of measurable qualities as “decreased”, “increased”, or “normal”, for example, increased degree of illumination, decreased length. This is one way to bring out the degree semantics of a quality by referring to an implied normal value. The treatment of degree modifiers in the modifier ontologies is ignorant of any norm, and only attempts to represent the ranges of the degree for a quality.
The concept of modifiers is also used in the Human Phenotype Ontology (Köhler et al. 2016) as reflected in "Clinical modifier" and "Frequency" classes. HPO:Frequency class is similar to our Frequency modifiers in that it bins freqency into a number of ranges: Excluded (0% of the cases), Very rare (1-4%), Occasional (5-29%), Frequent (30-79%), Very frequent (80-99%) and Obligate(100%). HPO:Frequency class is not applicable to our application for several reasons: (1) The class labels (e.g., excluded, obligate) are not terms used by the majority of taxonomists. We believe meaninful class lables are critical to the usability of an ontology. (2) Due to the broad range of various taxon groups we need to cover, precise ranges of percentages of the cases are not going to be applicable to all groups. (3) It is very unlikely for various taxon groups to record and compute the percentage of cases for an undefined number of characters they may care. HPO:Clinical modifier class holds subclasses "Agravated by", "Ameliorated by", "Pain characteristic", "Phenotpic variability", "Position", "Refractory", "Severity", and "Triggered by". All but "Severity" is disjoint from the types of modifiers that we treat in the modifier ontologies. HPO:Severity overlaps with the Degree modifiers, but it holds subclasses that are applicable to clinical settings: Boderline, Mild, Moderate, Severe, and Profound.
While these ontologies recognize the need to treat modifiers seperately and observed sequential relations among the terms, another key difference between the treatment of modifiers in HPO, as compared to our ontology construct, is that the two Modifier Ontologies we created have clear logic definitions order the terms that form a range, while HPO only has human readable definition.
The five-bin ontology is currently being used for comparing taxon concepts in the ETC project (Cui et al. 2016). The Taxonomy Comparison tool of the ETC project uses the morphological characters extracted from taxonomic descriptions to facilitate taxon concept resolution tasks. The intuition is that character evidence documented should correlate well with expert asserted relationships between two taxon concepts: if an expert asserts that one taxon concept is congruent with another, then the characters described for the two concepts should be very similar. ETC Text Capture tools extract characters from text for the Taxonomy Comparison tool to compute the similarity between two characters. For example, are “leaves usually toothed” “leaves often toothed”, and “leaves rarely toothed” essentially the same or somewhat different? With an interval list that has a fixed number of elements, as implemented in the five-bin ontology, the software can be configured to reliably compute the similarity score without being affected by ontology updates.
The two ontologies are being applied in another project entitled “Authors in the driver's seat: fast, consistent, computable phenotype data and ontology production”, recently funded by the US National Science Foundation (Cui et al. 2017). Recognizing that the semantic ambiguity in vocabulary usage by the authors at the time of writing results in inconsistent interpretations of documented characters at the time of use (Cui et al. 2015, Endara et al. 2017, Dahdul et al. 2018), the project aims to investigate effective ways to help phenotype authors converge on their term usage and to produce ontology-informed characters for computer algorithms to harvest. These two modifier ontologies will be compared in empirical studies to evaluate their effectiveness for this purpose. For example, the need of authors to add a term as a class vs. a synonym will be examined, in addition to the frequency of authors adopting a modifier from the given classes and exact synonyms.
Conclusions
The two modifier ontologies were created by following the literary warrant and user warrant principles of the national standard on constructing controlled vocabularies, using the list ontology pattern. The ontologies address four types of modifier terms (frequency, certainty, degree, and coverage) that are used widely in describing phenotype characters but have not been treated by existing ontologies. We have made the ontologies public accessible on GitHub. These ontologies can be used to support machine-based character similarity calculations and to increase author’s awareness of the ambiguities in modifier terms.
Data resources
Included or linked to within the manuscript
Acknowledgements
GVG acknowledges support from H2020-EINFRA (731075) and the US National Science Foundation (IOS:1340112) as well as support from the NIHR Birmingham ECMC, NIHR Birmingham SRMRC and the NIHR Birmingham Biomedical Research Centre and the MRC HDR UK. HC acknowledges support from US National Science Foundation through DBI-1661485, DBI-1147266, DEB-1541506. The views expressed in this publication are those of the authors and not necessarily those of the funding agencies.
Funding program
H2020-EINFRA (731075)
US National Science Foundation IOS:1340112
US National Science Foundation DBI-1661485
US National Science Foundation DBI-1147266
US National Science Foundation DEB-1541506
Contributor Information
Lorena Endara, Email: clendara@gmail.com.
Hong Cui, Email: hongcui@email.arizona.edu.
Funding program
H2020-EINFRA (731075)
US National Science Foundation IOS:1340112
US National Science Foundation DBI-1661485
US National Science Foundation DBI-1147266
US National Science Foundation DEB-1541506
References
- Aranguren Mikel Egaña, Antezana Erick, Kuiper Martin, Stevens Robert. Ontology Design Patterns for bio-ontologies: a case study on the Cell Cycle Ontology. http://dx.doi.org/10.1186/1471-2105-9-s5-s1. BMC Bioinformatics. 2008;9:S1. doi: 10.1186/1471-2105-9-s5-s1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arp R., Smith B., Spear A. D. Building Ontologies with Basic Formal Ontology. The MIT Press; Cambridge, Massachusetts; London, England: 2015. [Google Scholar]
- CBIT Lucid. http://www.lucidcentral.com/en-us/home/aboutlucid.aspx. 2007 1.
- Cooper L. D., Walls R. L., Elser J., Gandolfo M. A., Stevenson D. W., Smith B., Preece J., Athreya B., Mungall C. J., Rensing S., Hiss M., Lang D. The Plant Ontology as a tool for comparative plant anatomy and genomic analyses. Plant Cell Physiology. 2013;54(e1):1–23. doi: 10.1093/pcp/pcs163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui H. CharaParser for fine-grained semantic annotation of organism morphological descriptions. Journal of the Association for Information Science and Technology. 2012;63:738–754. doi: 10.1002/asi.22618. [DOI] [Google Scholar]
- Cui Hong, Dahdul Wasila, Dececchi A. T., Ibrahim Nizar, Mabee Paula, Balhoff J. P., Gopalakrishnan Hariharan. CharaParser+EQ: Performance evaluation without gold standard. http://dx.doi.org/10.1002/pra2.2015.145052010020. Proceedings of the Association for Information Science and Technology. 2015;52(1):1–10. doi: 10.1002/pra2.2015.145052010020. [DOI] [Google Scholar]
- Cui H., Dongfang X., Chong S. S., Ramirez M., Rodenhausen T., Macklin J., Ludascher B., Morris P. J., Soto E. M., N Mongiardino Koch. Introducing Explorer of Taxon Concepts with a case study on spider measurement matrix building. BMC Bioinformatics. 2016;17:471. doi: 10.1186/s12859-016-1352-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Hong, Reznicek Anton, Macklin James, Sachs Joel. Authors in the driver's seat: fast, consistent, computable phenotype data and ontology production. https://www.nsf.gov/awardsearch/showAward?AWD_ID=1661485&HistoricalAwards=false. [2018-05-21T00:00:00+03:00];
- Dahdul W., Cui H., Mabee P. M., Mungall C. J., Osumi-Sutherland D., Walls R. L., Haendel M. A. Nose to tail, roots to shoots: spatial descriptors for phenotypic diversity in the Biological Spatial Ontology. Journal of Biomedical Semantics. 2014;5(34):1–13. doi: 10.1186/2041-1480-5-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahdul Wasila, Dececchi T. Alexander, Ibrahim Nizar, Cui Hong, Balhoff James P., Mabee Paula, Vision Todd, Lapp Hilmar. Gold Standard Corpus, Ontologies, And Entity-Quality Ontology Annotations For Evolutionary Phenotypes. http://dx.doi.org/10.5281/ZENODO.1345307. Zenodo. 2018 doi: 10.5281/ZENODO.1345307. [DOI]
- Dahdul Wasila, Manda Prashanti, Cui Hong, Balhoff J. P, Dececchi Alexander, Ibrahim Nizar, Lapp Hilmar, Vision Todd, Mabee P. M. Annotation of phenotypes using ontologies: a Gold Standard for the training and evaluation of natural language processing systems. http://dx.doi.org/10.1101/322156. BioRxiv. 2018:322156. doi: 10.1101/322156. [DOI] [PMC free article] [PubMed]
- Dallwitz M. J., Paine T. A., Zurcher E. J. Edition 4.12. [Included in the CSIRO DELTA program package] CSIRO Division of Entomology; Canberra: 2000. User’s guide to the DELTA system. A general system for processing taxonomic descriptions.156 [Google Scholar]
- Egaña M., Rector A., Stevens R., Antezana E. Applying ontology design patterns in bio-ontologies. In: Gangemi A., Euzenat J., editors. Knowledge Engineering: Practice and Patterns. Springer; Berlin, Heidelberg: 2008. 7-16 [Google Scholar]
- Endara Lorena, Cole Heather A., Burleigh J. Gordon, Nagalingum Nathalie S., Macklin James A., Liu Jing, Ranade Sonali, Cui Hong. Building the "Plant Glossary"—A controlled botanical vocabulary using terms extracted from the Floras of North America and China. http://dx.doi.org/10.12705/664.9. Taxon. 2017;66(4):953–966. doi: 10.12705/664.9. [DOI] [Google Scholar]
- Committee Flora of China Editorial. Flora of China. Science Press & Missouri Botanical Garden Press; Beijing & St. Louis: 1994. [Google Scholar]
- Committee Flora of North America Editorial. Flora of North America North of Mexico. Oxford University Press; New York and Oxford: 1993. [Google Scholar]
- Gkoutos Georgios V, Green Eain CJ, Mallon Ann-Marie, Hancock John M, Davidson Duncan. Using ontologies to describe mouse phenotypes. http://dx.doi.org/10.1186/gb-2004-6-1-r8. Genome Biology. 2004;6(1):R8. doi: 10.1186/gb-2004-6-1-r8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gkoutos G. V., Mungall C., Dolken S., Ashburner M., Lewis S., Hancock J., Schofield P., Kohler S., Robinson P. N. Entity/Quality-Based Logical Definitions for the Human Skeletal Phenome using PATO. 2009 Annual International Conference of the IEEE Engineering in Medicine and Biology Society; 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gkoutos G. V., Schofield P. N., Hoehndorf Robert. The anatomy of phenotype ontologies: principles, properties and applications. http://dx.doi.org/10.1093/bib/bbx035. Briefings in Bioinformatics. 2017:bbx035. doi: 10.1093/bib/bbx035. [DOI] [PMC free article] [PubMed]
- Hagedorn Gregor. DiversityDescriptions (DeltaAccess) (version 1.9). Documentation of the information model. http://diversityworkbench.net/OldModels/Descriptions/Model/2005-03-30/DModelDD19.html
- Hagedorn Gregor, Thiele Kevin, Morris Robert, Heidorn P. B. The “Structured Descriptiv http://diversityworkbench.net/OldModels/Descriptions/Model/2005-03-30/DModelDD19.htmle Data (SDD)” w3c-xml-schema, version 1.1. http://rs.tdwg.org/UBIF/2006/rddl.html
- Hagedorn G. Structuring Descriptive Data of Organisms — Requirement Analysis and Information Models (Strukturierung organismischer Beschreibungsdaten – Anforderungsanalyse und Informationsmodelle) Institute for Plant Virology, Microbiology and Biosafety, Federal Biological Research Center for Agriculture and Forestry, Universität Bayreuth; Berlin: 2007. [Google Scholar]
- Hoehndorf Robert, Alshahrani Mona, Gkoutos G. V., Gosline George, Groom Quentin, Hamann Thomas, Kattge Jens, de Oliveira S. M., Schmidt Marco, Sierra Soraya, Smets Erik, Vos R. A., Weiland Claus. The Flora Phenotype Ontology (FLOPO): tool for integrating morphological traits and phenotypes of vascular plants. http://dx.doi.org/10.1186/s13326-016-0107-8. Journal of Biomedical Semantics. 2016;7(1):65. doi: 10.1186/s13326-016-0107-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Fengqiong, Macklin J. A., Cui Hong, Cole H. A., Endara Lorena. OTO: Ontology Term Organizer. http://dx.doi.org/10.1186/s12859-015-0488-1. BMC Bioinformatics. 2015;16:47. doi: 10.1186/s12859-015-0488-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler Sebastian, Vasilevsky Nicole A., Engelstad Mark, Foster Erin, McMurry Julie, Aymé Ségolène, Baynam Gareth, Bello Susan M., Boerkoel Cornelius F., Boycott Kym M., Brudno Michael, Buske Orion J., Chinnery Patrick F., Cipriani Valentina, Connell Laureen E., Dawkins Hugh J. S., DeMare Laura E., Devereau Andrew D., de Vries Bert B. A., Firth Helen V., Freson Kathleen, Greene Daniel, Hamosh Ada, Helbig Ingo, Hum Courtney, Jähn Johanna A., James Roger, Krause Roland, Laulederkind Stanley J. F., Lochmüller Hanns, Lyon Gholson J., Ogishima Soichi, Olry Annie, Ouwehand Willem H., Pontikos Nikolas, Rath Ana, Schaefer Franz, Scott Richard H., Segal Michael, Sergouniotis Panagiotis I., Sever Richard, Smith Cynthia L., Straub Volker, Thompson Rachel, Turner Catherine, Turro Ernest, Veltman Marijcke W. M., Vulliamy Tom, Yu Jing, Ziegenweidt Julie von, Zankl Andreas, Züchner Stephan, Zemojtel Tomasz, Jacobsen Julius O. B., Groza Tudor, Smedley Damian, Mungall Christopher J., Haendel Melissa, Robinson Peter N. The Human Phenotype Ontology in 2017. http://dx.doi.org/10.1093/nar/gkw1039. Nucleic Acids Research. 2016;45:D865–D876. doi: 10.1093/nar/gkw1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebbe J. Manuel d’utilisation du logiciel XPER. Micro Application; Paris: 1984. 148 [Google Scholar]
- Maddison David R., Swofford David L., Maddison Wayne P. NEXUS: An Extensible File Format for Systematic Information. http://dx.doi.org/10.2307/2413497. Systematic Biology. 1997;46(4):590. doi: 10.2307/2413497. [DOI] [PubMed] [Google Scholar]
- Mungall C. J., Haendel M., Dahdul W., Ibrahim N., Segerdell E., Blackburn D. C., Comte A., Niknejad A., Dececchi T. A. UBERON Ontology. http://purl.obolibrary.org/obo/uberon/releases/2016-01-26/uberon.owl. [2018-05-27T00:00:00+03:00];
- Organization) NISO (National Information Standards. Guidelines for the construction, format, and management of monolingual controlled vocabularies. Baltimore: National Information Standards Organization. http://www.niso.org/apps/group_public/ download.php/12591/z39-19-2005r2010.pdf
- Presutti V., Blomqvist E., Daga E., Gangemi A. Pattern-Based Ontology Design. In: Suárez-Figueroa M. C., Gómez-Pérez A., Motta E., Gangemi A., editors. Ontology Engineering in a Networked World. Springer; New York: 2012. 35-64 [Google Scholar]
- Pullan Martin R., Armstrong Kate E., Paterson Trevor, Cannon Alan, Kennedy Jessie B., Watson Mark F., McDonald Sarah, Raguenaud Cédric. The Prometheus Description Model: an examination of the taxonomic description-building process and its representation. Taxon. 2005;54(3):751–765. doi: 10.2307/25065431. [DOI] [Google Scholar]
- Shimoyama Mary, Nigam Rajni, McIntosh Leslie Sanders, Nagarajan Rakesh, Rice Treva, Rao D. C., Dwinell Melinda R. Three ontologies to define phenotype measurement data. http://dx.doi.org/10.3389/fgene.2012.00087. Frontiers in Genetics. 2012;3 doi: 10.3389/fgene.2012.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith Barry, Ashburner Michael, Rosse Cornelius, Bard Jonathan, Bug William, Ceusters Werner, Goldberg Louis J, Eilbeck Karen, Ireland Amelia, Mungall Christopher J, Leontis Neocles, Rocca-Serra Philippe, Ruttenberg Alan, Sansone Susanna-Assunta, Scheuermann Richard H, Shah Nigam, Whetzel Patricia L, Lewis Suzanna. The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. http://dx.doi.org/10.1038/nbt1346. Nature Biotechnology. 2007;25(11):1251–1255. doi: 10.1038/nbt1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder Matthew J., Mikó István, Seltmann Katja C., Bertone Matthew A., Deans Andrew R. A gross anatomy ontology for Hymenoptera. http://dx.doi.org/10.1371/journal.pone.0015991. PLoS ONE. 2010;5(12):e15991. doi: 10.1371/journal.pone.0015991. [DOI] [PMC free article] [PubMed] [Google Scholar]



