SUMMARY
Noncoding Y RNAs are present in both animal cells and many bacteria. In all species examined, Y RNAs tether the Ro60 protein to an effector protein to perform various cellular functions. Recently, a new Y RNA subfamily was identified in bacteria. Bioinformatic analyses of these YrlA (Y RNA-like A) RNAs predict that the effector-binding domain resembles tRNA. We present the structure of this domain, the overall folding of which is strikingly similar to canonical tRNAs. The tertiary interactions that are responsible for stabilizing tRNA are present in YrlA, making it a close tRNA mimic. However, YrlA lacks a free CCA end and contains a kink in the stem corresponding to the anticodon stem. Since nucleotides in the D and T stems are conserved among YrlAs, they may be an interaction site for an unknown factor. Our experiments identify YrlA RNAs as a new class of tRNA mimics.
Keywords: Y RNA, YrlA, tRNA-like element, noncoding RNA
Graphical Abstract
eTOC BLURB
Wei Wang et al. described the crystal structure of the S. Typhimurium YrlA RNA effector binding module, which closely mimics that of tRNA. This module is present in a wide range of bacteria. It may function by tethering an effector molecule to the Ro60/Rsr protein to modulate RNA metabolism.
INTRODUCTION
In addition to the canonical tRNAs that function in protein synthesis, several RNAs depend on structural similarity to tRNA in order to function. These tRNA mimics include the bacterial transfer-messenger RNA (tmRNA) that rescues stalled ribosomes from mRNAs lacking stop codons and the tRNA-like structures that contribute to translation and replication of positive-strand RNA viruses. For tmRNA, the tRNA-like portion undergoes aminoacylation and binds the elongation factor EF-Tu, allowing it to enter the A-site of arrested ribosomes and function as an acceptor for the stalled polypeptide (Keiler, 2015). Although the viral tRNA-like sequences have diverse functions in translation and replication, several are substrates for the CCA-adding enzyme and a tRNA synthetase and interact with the elongation factor EF-1A (Dreher, 2009). Additionally, tRNA-like structures at the 3′ end of several mammalian long noncoding RNAs (lncRNAs) are cleaved by the tRNA 5′ maturation enzyme RNase P, resulting in 3′ end formation of the upstream lncRNA and release of a tRNA-like noncoding RNA (ncRNA) of unknown function (Sunwoo et al., 2009; Wilusz et al., 2008).
Another class of RNAs that are proposed to mimic tRNA consists of bacterial ncRNAs known as YrlA (Y RNA-like A) RNAs (Chen et al., 2014). These ncRNAs are members of the Y RNA family, 80 to ~220 nucleotides (nt) ncRNAs that were initially identified in human cells because they are bound by the Ro60 autoantigen, a major target of autoantibodies in patients with systemic lupus erythematosus (Wolin et al., 2013). Studies in vertebrate cells revealed that Y RNAs regulate the subcellular location of Ro60 and its association with other proteins and RNAs. For example, Y RNA binding masks a nuclear accumulation signal on Ro60, retaining it in the cytoplasm (Sim et al., 2009). Y RNAs also scaffold the association of Ro60 with other proteins, as binding of the zipcode-binding protein ZBP1 to a mouse Y RNA adapts the Ro60/Y RNA complex for nuclear export (Sim et al., 2012). In addition to Y RNAs, the ring-shaped Ro60 binds the 3′ ends of some misfolded ncRNAs in its central cavity and adjacent structured RNA regions on its outer surface (Fuchs et al., 2006; Stein et al., 2005). Because Y RNAs bind overlapping sites on the Ro60 outer surface, Y RNAs may regulate the access of misfolded ncRNAs to the Ro60 cavity (Stein et al., 2005).
Studies of bacterial Y RNAs have revealed that, as in animal cells, their functions are intertwined with that of the Ro60 protein. In Deinococcus radiodurans, the bacterium where Ro60 and Y RNAs have been most extensively characterized, at least two Y RNAs, called Yrn1 (Y RNA 1) and Yrn2, are bound and stabilized by the Ro60 ortholog Rsr (Ro sixty-related) (Chen et al., 2000; Chen et al., 2013; Chen et al., 2007). Consistent with co-regulation, these ncRNAs are encoded upstream of Rsr and on the same DNA strand. One role of Yrn1 is to tether Rsr to the ring-shaped 3′ to 5′ exoribonuclease polynucleotide phosphorylase (PNPase), forming a double-ringed RNA degradation machine called RYPER (Ro60/Y RNA/PNPase Endonuclease RNP) (Chen et al., 2013). In RYPER, single-stranded RNA threads from the Rsr ring into the PNPase cavity for degradation, rendering PNPase more effective in degrading structured RNA (Chen et al., 2013). In addition to its role in RYPER, Rsr assists 23S rRNA maturation by two 3′ to 5′ exoribonucleases, RNase II and RNase PH, during heat stress, where Rsr functions as a free protein, and is inactive when bound to Y RNA (Chen et al., 2007). Thus, in addition to acting as a tether, Yrn1 may function as a gate to block access of other RNAs to Rsr.
Y RNAs are modular, a feature that is critical for carrying out their functions. All characterized Y RNAs contain a long stem, formed by base-pairing the 5′ and 3′ ends of the RNA, that contains the Ro60 binding site. Although in both metazoans and D. radiodurans, the sequences required for Ro60 binding map to a conserved helix (Chen et al., 2013; Green et al., 1998), a structure of a Xenopus laevis (X. laevis) Ro60/Y RNA complex revealed that Ro60 primarily interacts with the 5′ strand (Stein et al., 2005). Consistent with the idea that base-specific interactions with this strand are critical for Ro60 recognition, only the 5′ strand of the helix is conserved across bacterial species (Chen et al., 2014). The other end of all Y RNAs consists of internal loops and stem-loops that interact with other proteins. For example, to form the mammalian Ro60/Y RNA/ZBP1 complex, ZBP1 interacts with the large internal loop of the Y RNA (Köhn et al., 2010; Sim et al., 2012), while in D. radiodurans RYPER, this portion of Yrn1 interacts with the KH and S1 single-stranded RNA-binding domains of PNPase (Chen et al., 2013). Thus, one role of this second Y RNA module is to tether Ro60 to an effector protein.
Remarkably, for many bacterial Y RNAs, the effector-binding module bears a striking resemblance to tRNA. The first member of this Y RNA subfamily was identified in the enteric bacterium Salmonella enterica serovar Typhimurium (Herein referred to as S. Typhimurium), where it and a second Y RNA were bound by Rsr and encoded 3′ to this protein (Chen et al., 2013). Because the two Y RNAs in S. Typhimurium appeared to represent a separate evolutionary lineage from the more metazoan-like Y RNAs characterized in D. radiodurans, these RNAs were designated YrlA and YrlB (Y RNA-like A and B)(Chen et al., 2013). Homology searches revealed that RNAs resembling YrlA were widespread, as they were detected near Rsr in >250 bacterial species representing at least 10 distinct phyla (Chen et al., 2014). Identification of conserved sequences and secondary structures within these RNAs revealed similarities to the D, T and acceptor stem-loops of tRNA (Figure 1) (Chen et al., 2014). Consistent with a tRNA-like fold, S. Typhimurium YrlA is a substrate for two tRNA modification enzymes, TruB and DusA, that modify the T and D loops, respectively (Chen et al., 2014).
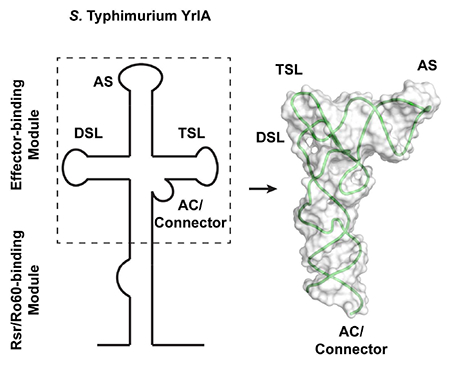
Figure 1.
S. Typhimurium YrlA folds into a structure very similar to that of canonical tRNA. (A) Full-length S. Typhimurium YrlA consists of an Rsr/Ro60-binding module and an effector-binding module (shaded) that resembles tRNA. (B) Secondary structures of the crystallized S. Typhimurium YrlA effector-binding module (nt 16-92) and S. cerevisiae tRNAPhe. Important tertiary interactions are connected by lines and are labeled. Invariant or nearly invariant (>80% conserved) nucleotides in YrlA are in red (Chen et al., 2014). Modified nucleotide sequences to facilitate crystallization are shown as hollow characters. (C) The crystal structure of S. Typhimurium YrlA (green/red, PDB: 6cu1). The conserved nucleotides shown in panel B are in red. (D) Left, the tertiary structure of S. cerevisiae tRNAPhe (orange, PDB: 4tna); right, overlay of the YrlA (green) and tRNAPhe (orange) structures. See also Figure S1 and S2.
To test the hypothesis that the YrlA effector-binding domain folds into a tRNA-like structure, we determined the structure of the tRNA-like domain of S. Typhimurium YrlA by X-ray crystallography. We show that the YrlA effector-binding domain indeed assumes a similar overall fold as tRNA and that the same tertiary interactions that stabilize tRNA are present in YrlA. In support of a critical role for the tRNA-like module, both the ability to fold into a tRNA-like structure and specific sequences within the structure are conserved in YrlA RNAs from a wide range of bacteria.
RESULTS
We determined the crystal structure of the S. Typhimurium YrlA effector-binding module (nucleotides 16-92) at 3 Å resolution (Figure1A, Figure S1). To obtain the high-resolution diffraction data, the 3-nucleotide (nt) loop 56CCG58 of YrlA was changed to a 56GAAA58a tetraloop, where the extra nucleotide was numbered as 58a to maintain the original numbering of the subsequent nucleotides (Figure 1B). 14GG15 and 93C were added to the construct to increase transcription yield and stabilize the stem. The base pair A20-U88 was also mutated to C20-G88 for stem stabilization. The modified YrlA molecules crystallized in the space group C2221 and diffracted to 3.0 Å resolution. A slight variant of this construct, which has an additional cytidine at 3′-end, crystallized in the space group P63 and diffracted to 4.3 Å resolution. The structure was determined by a combination of single-wavelength anomalous dispersion and molecular replacement methods (see Materials and Methods section for more details). The crystals have high solvent content (80.3% for the C2221 crystal) with minor crystal packing contacts between symmetry related molecules (Figure S2). Furthermore, the same YrlA structure was observed in the two different crystal forms. Thus, we believe that the crystal structures represent the native conformation of the S. Typhimurium YrlA effector-binding module. We were able to build 80/81 nucleotides into the electron density map and the refinement statistics are summarized in Table 1.
Table 1.
| Data collection | YrlA Tetraloop | YrlA Tetraloop 3C |
|---|---|---|
| Space group | C2221 | P63 |
| Cell dimensions | ||
| a, b, c (Å) | 59.42, 147.12, 101.58 | 146.46, 146.46, 52.53 |
| α, β, γ(°) | 90, 90, 90 | 90, 90, 120 |
| Resolution (Å)a | 50.0-3.0 (3.05-3.0) | 50.0-4.3 (4.37-4.30) |
| Rmerge (%) | 8.9 (82.7) | 9.6 (>100) |
| <I>/<σ(I)> | 13.5 (2.0) | 14.4 (1.0) |
| Completeness (%) | 98.0 (97.0) | 99.1 (97.4) |
| Redundancy | 3.6 (3.5) | 5.7 (4.8) |
| CC1/2 | 1.00(0.93) | 1.00(0.67) |
| Refinement | ||
| Resolution (Å) | 30.0-3.0 | |
| No. reflections | 8563 (613) | |
| Rwork/ Rfree (%) | 22.3/23.8 (38.1/46.7) | |
| No. atoms | ||
| RNA | 1722 | |
| Ion | 12 | |
| Water | 2 | |
| Mean B-factors (Å2) | ||
| RNA | 140.2 | |
| Ion | 169.7 | |
| Water | 125.6 | |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.009 | |
| Bond angles (°) | 1.04 |
Values in parentheses are for highest-resolution shell.
The overall architecture of S. Typhimurium YrlA resembles that of tRNA
The YrlA RNA folds into a L-shaped structure that is characteristic of tRNAs. As predicted from the secondary structure (Chen et al., 2014), all tRNA equivalent regions are present in the YrlA structure, including a stem-loop region resembling the acceptor stem (AS, nt 50-64), the T stem-loop (TSL, nt 33-49), the D stem-loop (DSL, nt 67-81), and the anticodon stem (AC, nt 24-27 and 83-86). The YrlA structure also contains a Connector region (nt 16-23 and 87-92), linking the tRNA-like module to the Rsr/Ro60 binding module. These structural elements fold into the L-shaped conformation with the two extended stem-loops interacting at an angle very similar to that of tRNAs (Figure 1C). The overall YrlA structure has a backbone RMSD of ~2.3 Å to that of Saccharomyces cerevisiae (S. cerevisiae) tRNAPhe (PDB: 4tna) (Hingerty et al., 1978), based on the alignment of 60 phosphorus atoms (excluding the AS and AC loops and D17 of tRNAPhe). It was shown that YrlA RNAs are substrates for tRNA modification enzymes and contain canonical tRNA modifications such as dihydrouridine in the D loop and pseudouridine in the T loop (Chen et al., 2014). The high degree of structural similarity between the two RNAs provides an explanation for the recognition of YrlAs by tRNA modification enzymes.
Deviating from the canonical tRNA structure, there is a kink between the AC and the Connector regions of YrlA, bending the lower portion of the stem by ~30° (Figure 1C). The Connector region extending from the AC stem is continuous with the Rsr/Ro60-binding module, making YrlA much more elongated than tRNA. Two nucleotides, 22CU23, form a bulge out of the AC/Connector stem. The two bulged nucleotides do not interact with the rest of the molecule and potentially generate a flexible site in the stem for the kink formation. Consistent with this flexible nature they are not well resolved in the electron density map. Interestingly, 23UA24 are conserved among YrlA family members. However, sequence alignment indicates that these two nucleotides are predicted to be located in the variable loop (VL) of most YrlA RNAs, rather than being part of the AC/Connector stem (Chen et al., 2014). Thus, it is unclear whether the kink is a universal feature of YrlA RNAs.
A major difference between S. Typhimurium YrlA and tRNA is that the AS, which terminates in 3′-CCA in all mature tRNAs, is instead a closed loop in YrlA. (Figure 1A). In addition, YrlA lacks the anticodon loop. The length of the AS of YrlAs varies between species (Chen et al., 2014). For S. Typhimurium YrlA, the stem is only six base pairs, which is one base pair less than that of tRNAs. For other species, such as some cyanobacteria, this YrlA stem is predicted to be much longer (Chen et al., 2014). Interestingly, Mycobacterium smegmatis YrlA, which resembles bona fide tRNAs in containing a seven-base pair AS, is a substrate for RNase P. Following cleavage, the fragment corresponding to a tRNA 3′ end undergoes exonucleolytic nibbling and CCA addition (Chen et al., 2014). However, since most YrlA RNAs contain acceptor stems that are predicted to be poor RNase P substrates, the majority of YrlA RNAs likely resemble circularly permuted tRNAs with closed loop-containing acceptor stems (Chen et al., 2014).
YrlA is stabilized by the same tertiary interactions as tRNAs
In addition to the similarities in overall folding, the tertiary interactions that stabilize the L-shaped structure of YrlA resemble those of tRNA (Figure 2). A major feature of tRNA folding is the interaction between the DSL and the TSL regions to form the tRNA elbow, which serves as a binding site for numerous enzymes that recognize tRNA (reviewed in (Zhang and Ferré-D’Amaré, 2016)). In YrlA, the interaction between nucleotides U38-A42 of the T loop and two guanines in the D loop closely mimics the elbow region of tRNA (Figure 2A, 2B). The first two nucleotides in the YrlA T loop, U38 and U39, stack with nucleotides in the T stem. The third nucleotide, C40, forms a Watson-Crick base pair with G75 in the DSL. There is a gap between the fourth and fifth nucleotides, G41 and A42, in which G74 intercalates to form a continuous stacking interaction.
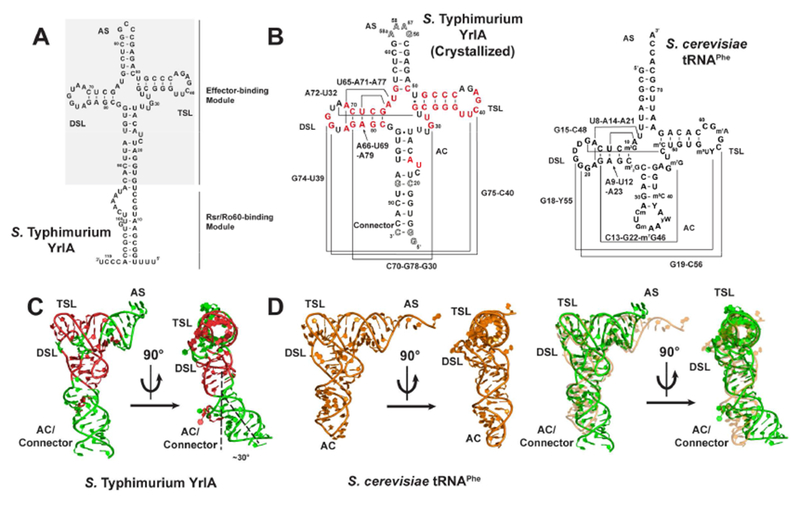
Figure 2.
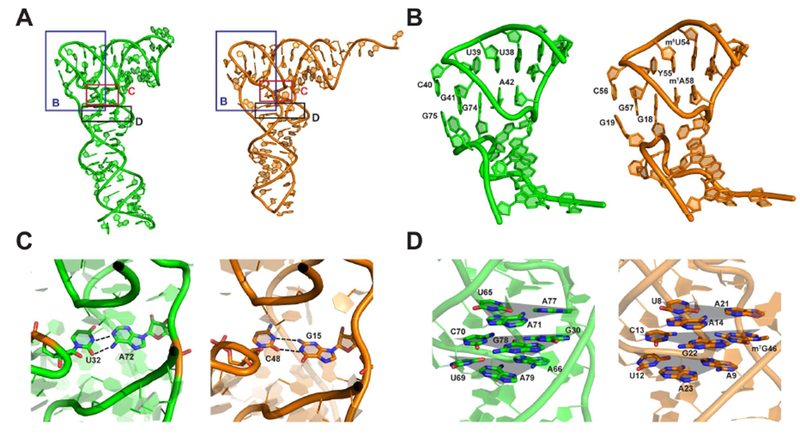
S. Typhimurium YrlA is stabilized by tRNA-like interactions. (A) Overall structures of S. Typhimurium YrlA (Green, PDB: 6cu1) and S. cerevisiae tRNAPhe (Orange, PDB: 4tna). The areas shown in (B), (C) and (D) are boxed. (B) YrlA has DSL-TSL interactions that resemble that of tRNAPhe. (C) YrlA contains a Levitt base pair (A72-U32) similar to that of tRNAPhe (G15-C48). (D) U65-A71-A77, C70-G78-G30 and A66-U69-A79 of YrlA closely resemble the base triples of tRNAPhe, U8-A14-A21, C13-G22-m7G46 and A9-U12-A23. The base triples are highlighted by shaded triangles (gray).
Other tertiary interactions important for stabilizing tRNA structure are also present in YrlA. For example, U32:A72 form a base pair equivalent to the Levitt base pair C48:G15 of tRNAPhe (Figure 2C) (Levitt, 1969). This base pair is evolutionarily conserved in YrlA RNAs (Chen et al., 2014). In addition, in YrlA, U65-A71-A77, C70-G78-G30 and A66-U69-A79 form three base triplets that stack on one another. The equivalent base triplets are also found in tRNAPhe (Figure 2D).
With the S. Typhimurium YrlA structure reported here, the crystal structures of three tRNA-like elements (TLE) have now been determined. The other two TLE structures are Thermus thermophilus (T. thermophilus) tmRNA and the TLS (tRNA-like structure) at the 3′-end of the turnip yellow mosaic virus (TYMV) genome (Bessho et al., 2007; Colussi et al., 2014; Gutmann et al., 2003). These structures share the L-shaped tRNA fold (Figure 3A) but not all tRNA features are present in all three TLEs. Notably, tmRNA and TYMV TLS contain free 3′-CCA ends and anticodon domain mimics (in the case of tmRNA, a portion of its SmpB protein partner substitutes for the anticodon stem-loop) (Bessho et al., 2007; Gutmann et al., 2003) (Figure 3A). The presence of these structural features is consistent with the ability of these two TLEs to be charged with amino acids and with their biological functions (Dreher, 2009; Keiler, 2015).
Figure 3.
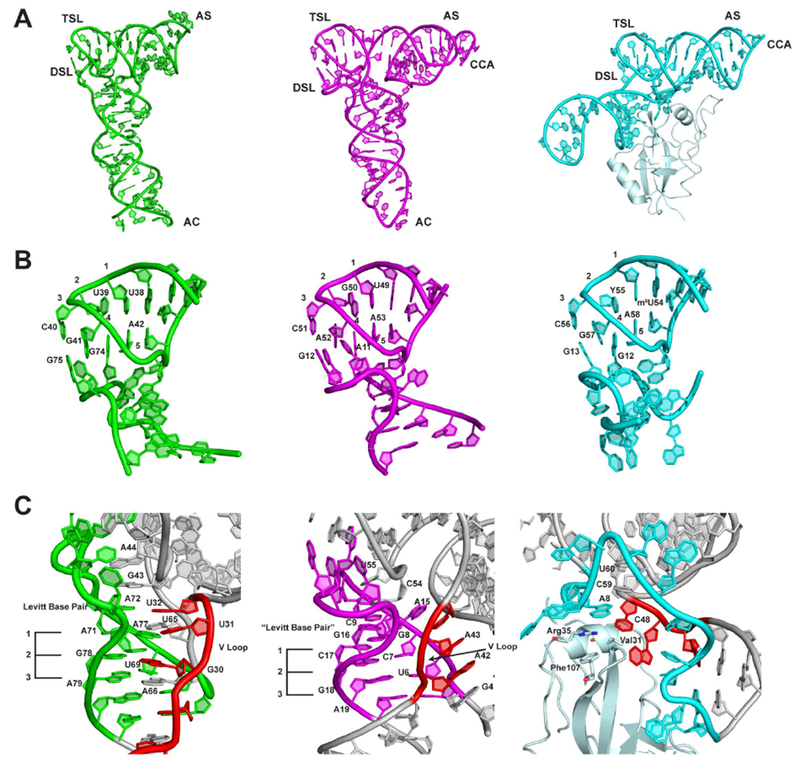
Comparison of the crystal structures of S. Typhimurium YrlA (Green, PDB: 6cu1), TYMV TLS (Magenta, PDB: 4p5j) and T. thermophilus tmRNA-SmpB complex (Cyan, PDB: 2czj). (A) The three TLEs share the same overall L-shaped fold. (B) The interactions between TSL and DSL are very similar between YrlA, TLS and tmRNA-SmpB. (C) The interactions between the VL and DSL show large differences but maintain the same architecture in the three TLEs. The VLs are colored in red. The three stacking layers of base triples or base pairs are labeled with numbers 1-3. [The numbering of TYMV TLS and T. thermophilus tmRNA-SmpB complex are based on previous publications (Bessho et al., 2007; Colussi et al., 2014)]
The DSL-TSL interaction is well conserved among the three TLEs (Figure 3B). This is perhaps not surprising as this is a major interaction defining the tRNA fold. In all cases, two guanine nucleotides in the DSL interact with a T loop through hydrogen bonding and base stacking interactions (Figure 3B). Since these three molecules have completely different evolutionary trajectories, the highly similar DSL-TSL interaction and hence the tRNA-like fold must be a key feature of their function, such that all three TLEs have either retained it (in the case of YrlA and tmRNA, which may have evolved from tRNA), or acquired and maintained it (viral TLEs).
In contrast, the interactions between the DSL and the VL regions are quite different for the three TLEs. In YrlA, the nucleotides in the DSL region interact with the VL region through hydrogen bonding and base stacking interactions and form three base-triplet layers which are very similar to that of canonical tRNAs. These three base layers further stack with the Levitt base pair A72-U32 and with two nucleotides in the T loop, G43 and A44, stabilizing the L-shaped fold (Figure 3C). TYMV TLS uses a different strategy to stabilize the tRNA-like fold. Its VL does not form hydrogen bonds with the DSL. Instead, all three residues in the VL (A42, A43 and U44) flip out to form a continuous stack with A3, G4 and A15. As a consequence, the nucleotides in the D stem form three base pairs instead of base triplets. Nonetheless, the DSL base pairs maintain a stacking interaction with two nucleotides in the T loop (C54 and U55) via a Levitt base pair equivalent (G16-C9) (Figure 3C). Strikingly, these interactions are also mimicked in the case of tmRNA but are conveyed through the SmpB protein, where Val 31, Arg35 and Phe107 bridge the base-pairing and stacking interactions. The completely different strategies used by the three TLEs to stabilize the VL-DSL connection highlight the importance of this region in maintaining the tRNA-like fold. These differences are also consistent with the fact that most tRNA-binding factors do not recognize this portion of tRNAs.
The YrlA effector domain consists of a conserved tRNA core with variable stems and loops
A sequence alignment of YrlAs from various bacteria shows that the nucleotides involved in stabilizing the tertiary interactions are highly conserved (Figure 4A). These nucleotides include those in the D and T loops, the pyrimidine at the last position of the VL and the 65UA66 dinucleotide that connects the AS and the DSL. Most YrlA species also maintain the Levitt base pair (Levitt, 1969) that is important for stabilizing the L-shaped structure (32U and 72A in S. Typhimurium YrlA). Thus, we predict that YrlAs from other species will have the same overall tRNA-like fold as observed in the current structure.
Figure 4.
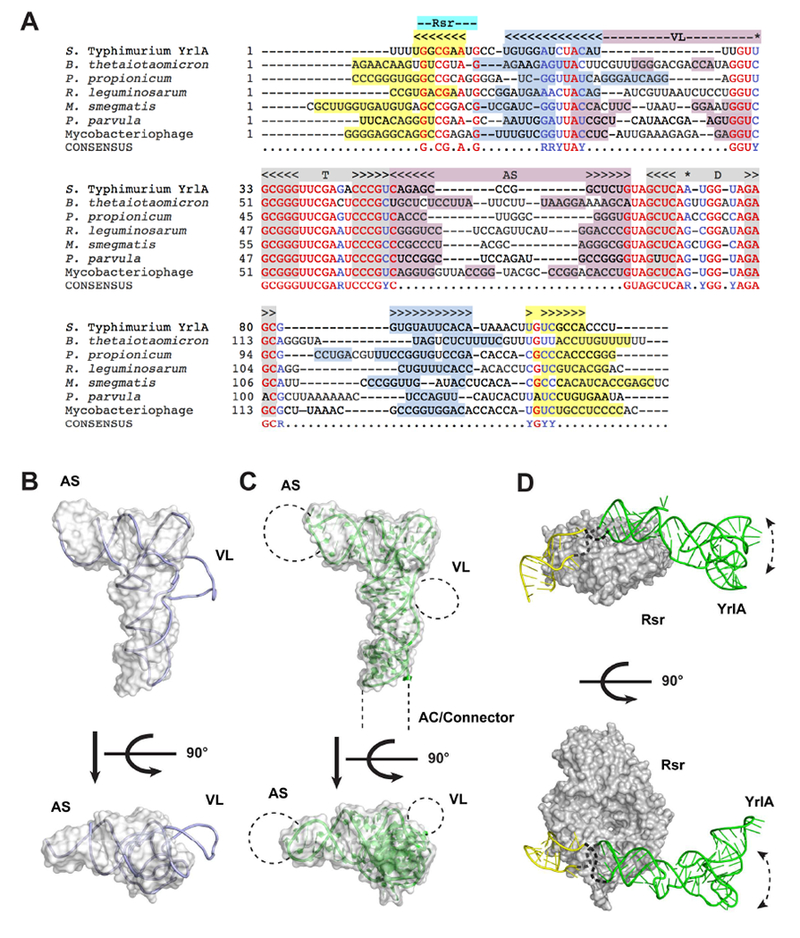
YrlA from various species likely share the same tRNA-like fold. (A) Sequences of representative YrlA RNAs were aligned using Clustal Omega (Goujon et al., 2010; Sievers et al., 2011) and further adjusted manually. The positions of the DSL and TSL are indicated in gray above the alignments and nucleotides that basepair to form the D- and T-stems are shaded gray. The positions of the AS and VL are indicated in pink, the AC and the Connector regions are labeled light blue and the Rsr binding site is indicated with a cyan label. Nucleotides with the potential to form helices within the stem created by basepairing the 5′ and 3′ ends are highlighted light blue and yellow. Nucleotides that are identical in six of the seven YrlA RNA sequences are in red. Nucleotide positions that are conserved as either purines or pyrimidines in six of the seven YrlAs are in blue. The shaded nucleotides in the VL region can potentially form duplexes. Nucleotides forming the Levitt base pair are indicated with a star. (B) The structure of a representative class II tRNA, T. thermophilus tRNATyr (1h3e, chain B, light blue) was superimposed onto the S. Typhimurium YrlA structure (gray surface). (C) Schematic model of a generalized YrlA RNA. The S. Typhimurium YrlA structure is shown as both cartoon (green) and gray surface. The different sizes of the AS, VL and Connector regions are represented with dashed lines. (D) Model of the S. Typhimurium Rsr-YrlA complex. X. laevis Ro in complex with its Y RNA fragment (PDB: 1yvp) was used as a model for Rsr (gray surface) and the YrlA Rsr/Ro60 binding module (yellow). The effector-binding module of YrlA is shown in green.
Although S. Typhimurium YrlA has a short VL, many YrlA species contain the long VLs (Figure 4A) that are characteristic of class II tRNAs. When a representative class II tRNA structure was superimposed on the structure of S. Typhimurium YrlA, only the VL protrudes from the L-shaped volume defined by this YrlA (Figure 4B). Class II tRNA VLs contain short duplexes, which could also exist in YrlA species with long VLs (Figure 4A). In addition, the sequence alignment indicates that the AS of YrlA RNAs varies in both sequence and length, as does the portion of the YrlA that corresponds to the AC. These comparisons support a model in which the YrlA core folds into a conserved tRNA-like structure, while the sizes of the AS, AC/Connector and VL regions vary (Figure 4C).
Potential interactions of YrlA RNA with cellular factors
Interestingly, although sequences within the D and T stems are not required to maintain the tRNA L-shape, these nucleotides are conserved in YrlA RNAs (Figure 4A). This suggests that the YrlA D and T stems are under evolutionary selection pressure. Most aminoacyl-tRNA synthetases recognize the AC and the AS (Giegé and Eriani, 2014). In addition, the tRNA elbow, where the D and T loops interact, is recognized by the ribosome, ncRNAs and many tRNA-interacting proteins (Zhang and Ferré-D’Amaré, 2016). To our knowledge, no cellular factors have been shown to function by recognizing specific sequences within the T and D stems of tRNAs or TLEs. One hypothesis for the high degree of sequence conservation in the T and D stems of YrlA is that a novel factor may recognize this region with some sequence specificity. An alternative but not exclusive possibility is that a need to maintain very stable D- and T- stems has driven conservation of the many G-C basepairs.
We compared available structural information to gain insight into the possible architecture of the Rsr-YrlA. YrlA RNAs contain a conserved Rsr/Ro60-binding module (Figure 1A); hence the structure of the Rsr-YrlA module is believed to resemble that of the X. laevis Ro60-Y RNA complex (PDB: 1yvp) (Stein et al., 2005). For S. Typhimurium YrlA, the tRNA-like effector-binding module, with structure reported herein, is connected to the Rsr/Ro60-binding module by unpaired short loops of 5 nucleotides. This allows us to model the Rsr-YrlA complex by positioning our structure close to the upper surface of the X. laevis Ro60-Y RNA structure (Figure 4D, upper panel). This surface of Ro60 presents positive charges that were shown to be important for both Y RNA and misfolded 5S rRNA binding (Fuchs et al., 2006; Stein et al., 2005). Thus, as predicted for vertebrate Y RNAs and Ro60, YrlA could serve as a gatekeeper to regulate access of other RNAs to Rsr.
The unknown effector factor that binds YrlA is presumably bound to the elbow region of the YrlA tRNA-like domain. In the presence of misfolded RNA and/or the effector protein, the tRNA-like domain of YrlA may reposition. For instance, the architecture of the D. radiodurans RYPER has been determined by single particle electron microscopy (EM) and the Y RNA is predicted to bend downward from the Rsr surface, allowing the stemloop-containing module to contact one or more S1/KH domains of PNPase (Chen et al., 2013). The architecture of the S. Typhimurium Rsr-YrlA-effector complex remains to be determined.
DISCUSSION
Although the structure of the metazoan Y RNA module that binds the Ro60 autoantigen was elucidated using X-ray crystallography (Stein et al., 2005), high resolution structures of the effector-binding domains of these RNAs have been lacking. Our crystal structure of the tRNA-like module of S. Typhimurium YrlA RNA reveals that this module not only adopts an overall L-shaped structure similar to tRNA, but is also stabilized by the same tertiary interactions. Since all sequences involved in critical tertiary interactions are strongly conserved in YrlA RNAs, we predict that the ability to fold into the canonical tRNA L-shape is a general feature of this Y RNA family. Moreover, the high degree of sequence conservation at the YrlA region corresponding to the tRNA elbow, particularly within the T and D stems, contrasts with the variable sizes of the AS, AC/Connector and VL regions. Based on the extreme conservation of these T and D stem sequences, one hypothesis is that the effector(s) that bind YrlA RNAs will be one or more tRNA-binding protein(s) that recognize the elbow region with some sequence specificity.
The strong resemblance of YrlA RNAs to tRNAs lends support to the proposal that YrlA and other Y RNAs evolved from tRNA (Chen et al., 2014). Consistent with this hypothesis, Y RNAs are encoded adjacent to one or more tRNAs in some bacteria (Chen et al., 2014). Additionally, the finding that YrlA RNAs differ from bona fide tRNAs in that the TSL occurs 5′ to the DSL supports a recent model in which these RNAs originated from dimeric tRNA transcripts (Sim and Wolin, 2018). In this model, the YrlA TSL derived from the first tRNA, while the DSL derived from the second tRNA. Since the YrlA AS would originate from the spacer between the two tRNAs, a model in which YrlA evolved multiple times in distinct bacteria would provide an explanation for the variable length of this stem. Alternatively, if YrlA evolved from a single primordial dimeric tRNA, there may have been less pressure to maintain the length of the AS. In either case, the additional sequences in a dimeric pre-tRNA, such as the DSL and AC of the first tRNA and the AC and TSL of the second tRNA, could potentially basepair to form a stem containing a sequence recognized by Ro60.
In certain algae and at least one archaeal species, some tRNAs are transcribed as circularly permuted variants that are processed to mature tRNAs (Chan et al., 2011; Maruyama et al., 2010; Soma et al., 2007). Some of these pre-tRNAs resemble YrlA in that the AS is initially a closed loop and the AC stem is initially formed by base pairing the 5′ and 3′ ends of the newly made RNA. These unusual pre-tRNAs are processed to canonical tRNAs by excising and ligating the extended AC stem to form a circular intermediate, followed by opening of the AS by endonucleases such as RNase P and/or RNase Z (Soma et al., 2007). Although enzymes equivalent to the eukaryotic and archaeal splicing endonucleases have not been reported in bacteria, the resemblance of YrlA RNAs to circularly permuted tRNAs raises the possibility that some YrlA RNAs could undergo processing to more closely resemble canonical tRNAs.
The tRNA resemblance is less evident for Yrn1 RNAs: however, these ncRNAs also contain some tRNA-like features. Yrn1 can be folded to contain a TSL that conserves the T stem sequences of YrlA RNAs (Chen et al., 2014). This TSL likely forms in vivo, as it contains pseudouridine at the position corresponding to the pseudouridine in TSLs of all canonical tRNAs (Chen et al., 2014). Since structures have not been reported for the effector-binding domain of Yrn1 or any metazoan Y RNAs, it remains possible that the three stem loops in the Yrn1 effector-binding domain fold in three-dimensions to mimic tRNA.
Although the exact role of YrlA RNA is unknown, it is likely that it functions in RNA degradation and/or repair. Consistent with a role in RNA degradation, some YrlA and Rsr co-purify with PNPase in S. Typhimurium (Chen et al., 2013). If, as described for Yrn1 (Chen et al., 2013), the YrlA effector-binding domain interacts with the S1 and KH domains of PNPase, the highly folded tRNA domain could serve to protect the RNA from endonucleolytic nicks that would render it a substrate for PNPase or other exoribonucleases.
Rsr, YrlB and YrlA have also been proposed to function in RNA repair, since they are encoded adjacent to the RtcB RNA ligase in many bacteria (Burroughs and Aravind, 2016; Chen et al., 2013; Das and Shuman, 2013). In some bacteria, including S. Typhimurium, this operon (rsr-yrlBA-rtcBA) encodes both RtcB, the ligase that joins pre-tRNA halves following intron excision in Archaea and metazoans (Englert et al., 2011; Popow et al., 2011; Tanaka et al., 2011; Tanaka and Shuman, 2011), and RtcA, an RNA terminal phosphate cyclase (Das and Shuman, 2013; Filipowicz et al., 1985). Although the substrates of RtcB in bacteria are largely unknown, E. coli RtcB repairs 16S rRNA following cleavage by the MazF toxin (Temmel et al., 2017). Because Rsr and Y RNAs are encoded adjacent to RtcB in bacteria from multiple phyla, it was proposed that Rsr and one or more Y RNAs function as cofactors to enhance RtcB activity (Burroughs and Aravind, 2016). Consistent with a more general role in RNA ligation, Rsr and YrlA are occasionally encoded adjacent to members of other RNA ligase families (Burroughs and Aravind, 2016).
Interestingly, in certain other bacteria, RtcB is encoded adjacent to a protein containing a Band-7 domain and a predicted ncRNA, called band 7-associated tRNA (b7a-tRNA), that strongly resembles an authentic tRNA (Burroughs and Aravind, 2016). Consistent with functional redundancy between b7a-tRNA and YrlA, these bioinformatics searches predict that occasionally b7a-tRNA is encoded adjacent to Rsr and YrlA is adjacent to the Band-7 domain protein. Although the existence of the putative b7a-tRNA has not been validated experimentally, these predictions, together with our finding that the YrlA effector-binding domain folds similarly to tRNA, support the hypothesis that tRNA-like molecule(s) contribute, directly or indirectly, to RNA ligation.
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dr. Yong Xiong (yong.xiong@yale.edu).
EXPERIMENTAL MODEL DETAILS
In this study, plasmid used for in vitro transcription of S. Typhimurium YrlA was extracted from E. coli strain DH5α, which was cultured by shaking at 37°C in Luria Broth (0.5% w/v Yeast Extract, 1% w/v Tryptone, 1% NaCl). T7 RNA polymerase was purified from E. coli strain BL21(DE3} which was cultured by shaking at 16°C in Terrific Broth (2.4% w/v Yeast Extract, 1.2% w/v Tryptone, 0.4% v/v glycerol, 0.017 M KH2PO4, 0.072 M K2HPO4).
METHOD DETAILS
Plasmid construction and RNA purification
The tRNA-like domain of S. Typhimurium YrlA was cloned into the EcoRI and Nhel sites of plasmid pHDV4 (Walker et al., 2003), such that the YrlA coding sequence was followed by the HDV ribozyme. S. Typhimurium YrlA was transcribed from HindIII-linearized plasmid using T7 RNA polymerase (Milligan et al., 1987). The transcription reaction was mixed with an equal volume of ribozyme denaturing buffer (8M Urea and 0.5M MgCl2) and incubated at 37 °C for 2h to increase HDV ribozyme cleavage efficiency (Rosenstein and Been, 1990). Following T7 transcription and HDV cleavage, the sequence of the resulting RNA is: GGGUGGCUCUACAUUUGUUGCGGGUUCGAGACCCGUCAGAGCCCGGCUCUGUAGCUCAAUGGUAGAGCGGUGUAGUCACC. The underlined nucleotides indicate modifications to the original YrlA sequence in order to stabilize the AC/Connector stem. The YrlA product was precipitated with ethanol and purified by polyacrylamide-urea gel electrophoresis. YrlA variants were made using QuikChange Site-Directed Mutagenesis (Agilent).
Crystallization and data collection
The purified YrlA RNAs were folded by heating to 95 °C for 2 min, transferring to 60 °C and incubating for 2 min. MgCl2 was added at 60°C to a final concentration of 10 mM and the reaction was quenched on ice for 30 min before use. The folded RNA was then buffer exchanged to RNA crystallization buffer (50 mM sodium cacodylate, 50 mM KCl, 1 mM MgCl2 and 0.1 mM EDTA) using Amicon Ultra Centrifugal Unit (Merck Millipore, Billerica, MA). The RNA was concentrated to a final concentration of 1.2-1.5 mg/ml and screened for crystals using the microbatch under oil method using the Nucleix Suite (Qiagen, Germantown, MD). Crystals were readily formed under numerous conditions overnight.
Three YrlA variants were crystallized, namely YrlA WT, YrlA Tetraloop and YrlA Tetraloop 3C, in which an additional cytosine was added to the 3′-end of YrlA Tetraloop. The best crystallization conditions for each YrlA constructs were summarized in Table S1. For experimental phase using the single-wavelength anomalous dispersion (SAD) method, YrlA-Tetraloop-3C crystals were soaked with 1mM Iridium (III) hexamine trichloride solution at room temperature for 30min before freezing. Crystals were cryoprotected by Paratone oil (Hampton Research, Aliso Viejo, CA) or 30% glycerol. Diffraction data were collected at the Advanced Photon Source beamlines 24ID-C and 24ID-E. The YrlA-Tetraloop-3C derivative data diffracted to 4.3 Å resolution. The YrlA-Tetraloop crystal showed anisotropic diffraction with good data to 3.0 Å resolution. The data statistics are summarized in Table 1.
Structure determination and refinement
The structure of YrlA RNA was determined by a combination of SAD and molecular replacement methods. The initial phase information was obtained by SAD phasing using SHELX C/D/E (Sheldrick, 2008) and SOLVE (Terwilliger, 2004) from the Iridium hexamine derivative data set. Nine Iridium sites were found. Clear electron density for the tRNA-shaped molecule was visible after solvent flattening using RESOLVE (Terwilliger, 2004) (figure of merit 0.66) (Figure S1). The anticodon loop and the CCA end of S. cerevisiae tRNAPhe structure (PDB: 4tna) (Hingerty et al., 1978) were truncated and the resulting model was placed in the SAD map by phased molecular replacement using Molrep (Vagin and Teplyakov, 1997). The relative orientation of the truncated tRNAPhe arms was adjusted manually according to the SAD map and the resulting model was used as the search model for the native YrlA-Tetraloop data set using Phaser (McCoy et al., 2007). There is one RNA molecule in the asymmetric unit of the crystal. The initial S. Typhimurium YrlA model was rebuilt manually using the tRNAPhe structure as guidance. The initial model was refined by iterative rounds of restrained refinement using refmac5 (Vagin et al., 2004) followed by manual rebuilding with Coot (Emsley and Cowtan, 2004). Jelly body restraint with a sigma of 0.02 was used and RNA secondary structure restrains were also applied. Overall B-factor refinement was carried out initially and individual B-factor refinement was performed in the later stage of refinement. In the last steps, TLS and restrained refinement were performed. B-factor sharpening was carried out to facilitate model building (Liu and Xiong, 2014). Iterative cycles of automatic rebuilding using Phenix.ERRASER (Chou et al., 2012), manual rebuilding, and refmac5 refinement were further preformed to improve the model geometry. The structure was refined to final R/R-free of 22.5%/24.3% with good geometry and excellent electron density, as shown in both the 2Fo-Fc map and simulated annealing omit map (Figure S1). Refinement statistics are summarized in Table 1.
QUANTIFICATION AND STATISTICAL ANALYSIS
Multiple sequence alignment of bacteria Y RNAs was performed using the Clustal Omega Program (www.ebi.ac.uk/Tools/msa/clustalo/) (Sievers et al., 2011). Diffraction data processing was performed using HKL2000 (www.hkl-xray.com/hkl-2000) (Otwinowski and Minor, 1997). Iridium atom positions were determined by SHELX C/D/E (shelx.uni-goettingen.de/) (Sheldrick, 2008). SOLVE, RESOLVE (Terwilliger, 2004) and Phenix.ERRASER (Chou et al., 2012) were part of the Phenix program (www.phenix-online.org/) (Adams et al., 2010). Molrep (Vagin and Teplyakov, 1997), Phaser (McCoy et al., 2007) and refmac5 (Vagin et al., 2004) were part of the CCP4 suite (www.ccp4.ac.uk/) (Winn et al., 2011). Model building was performed using COOT (www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/) (Emsley and Cowtan, 2004).
DATA AND SOFTWARE AVAILABILITY
The crystal structure of the S. Typhimurium YrlA effector-binding module (nucleotide 16-92) is deposited in the Protein Data Bank (www.rcsb.org) under the PDB ID code 6cu1.
Supplementary Material
HIGHLIGHTS.
S. Typhimurium YrlA effector binding module closely mimics tRNA in structure
Nucleotides stabilizing the tRNA-like fold are conserved in bacteria and phages
The results shed light onto how an effector could be tethered to Ro60/Rsr by YrlA
YrlA RNAs are a new class of tRNA mimics
ACKNOWLEDGEMENTS
The authors would like to thank the staff at the Advanced Photon Source beamline 24-ID for assistance in data collection. This work was supported in part by National Institutes of Health Grants R01AI116313 (Y.X.) and R01GM073863 (to S.L.W.) and by the Intramural Research Program, Center for Cancer Research, National Cancer Institute, National Institutes of Health (X.C. and S.L.W.).
The authors declare no conflicts of interest.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, et al. (2010). PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessho Y, Shibata R, Sekine S.-i., Murayama K, Higashijima K, Hori-Takemoto C, Shirouzu M, Kuramitsu S, and Yokoyama S (2007). Structural basis for functional mimicry of long-variable-arm tRNA by transfer-messenger RNA. Proc. Natl. Acad. Sci. 104, 8293–8298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burroughs AM, and Aravind L (2016). RNA damage in biological conflicts and the diversity of responding RNA repair systems. Nucleic Acids Res. 44, 8525–8555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan PP, Cozen AE, and Lowe TM (2011). Discovery of permuted and recently split transfer RNAs in Archaea. Genome Biology 12, R38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Quinn AM, and Wolin SL (2000). Ro ribonucleoproteins contribute to the resistance of Deinococcus radiodurans to ultraviolet irradiation. Genes Dev. 14, 777–782. [PMC free article] [PubMed] [Google Scholar]
- Chen X, Sim S, Wurtmann EJ, Feke A, and Wolin SL (2014). Bacterial noncoding Y RNAs are widespread and mimic tRNAs. RNA 20, 1715–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Taylor David W., Fowler Casey C., Galan Jorge E., Wang H-W, and Wolin Sandra L. (2013). An RNA Degradation Machine Sculpted by Ro Autoantigen and Noncoding RNA. Cell 153, 166–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Wurtmann EJ, Van Batavia J, Zybailov B, Washburn MP, and Wolin SL (2007). An ortholog of the Ro autoantigen functions in 23S rRNA maturation in D. radiodurans. Genes Dev. 21, 1328–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou F-C, Sripakdeevong P, Dibrov SM, Hermann T, and Das R (2012). Correcting pervasive errors in RNA crystallography through enumerative structure prediction. Nat. Methods 10, 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colussi TM, Costantino DA, Hammond JA, Ruehle GM, Nix JC, and Kieft JS (2014). The structural basis of transfer RNA mimicry and conformational plasticity by a viral RNA. Nature 511, 366–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das U, and Shuman S (2013), 2′ -Phosphate cyclase activity of RtcA: a potential rationale for the operon organization of RtcA with an RNA repair ligase RtcB in Escherichia coli and other bacterial taxa. RNA 19, 1355–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreher TW (2009). Role of tRNA-like structures in controlling plant virus replication. Virus Res. 139, 217–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, and Cowtan K (2004). Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- Englert M, Sheppard K, Aslanian A, Yates JR, and Soil D (2011). Archaeal 3′ -phosphate RNA splicing ligase characterization identifies the missing component in tRNA maturation. Proc. Natl. Acad. Sci. 108, 1290–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipowicz W, Strugala K, Konarska M, and Shatkin AJ (1985). Cyclization of RNA 3′-terminal phosphate by cyclase from HeLa cells proceeds via formation of N(3′)pp(5′)A activated intermediate. Proc Natl Acad Sci U S A 82, 1316–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs G, Stein AJ, Fu C, Reinisch KM, and Wolin SL (2006). Structural and biochemical basis for misfolded RNA recognition by the Ro autoantigen. Nat. Struct. Mol. Biol. 13, 1002–1009. [DOI] [PubMed] [Google Scholar]
- Giegé R, and Eriani G (2014). Transfer RNA Recognition and Aminoacylation by Synthetases In eLS (John Wiley & Sons, Ltd; ). [Google Scholar]
- Goujon M, McWilliam H, Li W, Valentin F, Squizzato S, Paern J, and Lopez R (2010). A new bioinformatics analysis tools framework at EMBL–EBI. Nucleic Acids Res. 38, W695–W699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green CD, Long KS, Shi H, and Wolin SL (1998). Binding of the 60-kDa Ro autoantigen to Y RNAs: evidence for recognition in the major groove of a conserved helix. RNA 4, 750–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutmann S, Haebel PW, Metzinger L, Sutter M, Felden B, and Ban N (2003). Crystal structure of the transfer-RNA domain of transfer-messenger RNA in complex with SmpB. Nature 424, 699–703. [DOI] [PubMed] [Google Scholar]
- Hingerty B, Brown RS, and Jack A (1978). Further refinement of the structure of yeast tRNAPhe. J. Mol. Biol. 124, 523–534. [DOI] [PubMed] [Google Scholar]
- Keiler KC (2015). Mechanisms of ribosome rescue in bacteria. Nat Rev Micro 13, 285–297. [DOI] [PubMed] [Google Scholar]
- Köhn M, Lederer M, Wachter K, and Huttelmaier S (2010). Near-infrared (NIR) dye-labeled RNAs identify binding of ZBP1 to the noncoding Y3-RNA. RNA 16, 1420–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt M (1969). Detailed Molecular Model for Transfer Ribonucleic Acid. Nature 224, 759–763. [DOI] [PubMed] [Google Scholar]
- Liu C, and Xiong Y (2014). Electron Density Sharpening as a General Technique in Crystallographic Studies. J. Mol. Biol. 426, 980–993. [DOI] [PubMed] [Google Scholar]
- Maruyama S, Sugahara J, Kanai A, and Nozaki H (2010). Permuted tRNA Genes in the Nuclear and Nucleomorph Genomes of Photosynthetic Eukaryotes. Mol. Biol. Evol. 27, 1070–1076. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, and Read RJ (2007). Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan JF, Groebe DR, Witherell GW, and Uhlenbeck OC (1987). Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 15, 8783–8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, and Minor W (1997). Processing of X-ray diffraction data collected in oscillation mode In Methods Enzymol. (Academic Press; ), pp. 307–326. [DOI] [PubMed] [Google Scholar]
- Popow J, Englert M, Weitzer S, Schleiffer A, Mierzwa B, Mechtler K, Trowitzsch S, Will CL, Lührmann R, Söil D, et al. (2011). HSPC117 Is the Essential Subunit of a Human tRNA Splicing Ligase Complex. Science 331, 760. [DOI] [PubMed] [Google Scholar]
- Rosenstein SP, and Been MD (1990). Self-cleavage of hepatitis delta virus genomic strand RNA is enhanced under partially denaturing conditions. Biochemistry 29, 8011–8016. [DOI] [PubMed] [Google Scholar]
- Sheldrick G (2008). A short history of SHELX. Acta Crystallogr. A 64, 112–122. [DOI] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, et al. (2011). Fast, scalable generation of high - quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim S, Weinberg DE, Fuchs G, Choi K, Chung J, and Wolin SL (2009). The Subcellular Distribution of an RNA Quality Control Protein, the Ro Autoantigen, Is Regulated by Noncoding Y RNA Binding. Molecular Biology of the Cell 20, 1555–1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim S, and Wolin SL (2018). Bacterial Y RNAs: Gates, Tethers and tRNA Mimics. Microbiology Spectrum 6, doi:10.1128/microbiolspec.RWR-0023-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim S, Yao J, Weinberg DE, Niessen S, Yates JR, and Wolin SL (2012). The zipcode-binding protein ZBP1 influences the subcellular location of the Ro 60-kDa autoantigen and the noncoding Y3 RNA. RNA 18, 100–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soma A, Onodera A, Sugahara J, Kanai A, Yachie N, Tomita M, Kawamura F, and Sekine Y (2007). Permuted tRNA Genes Expressed via a Circular RNA Intermediate in Cyanidioschyzon merolae. Science 318, 450. [DOI] [PubMed] [Google Scholar]
- Stein AJ, Fuchs G, Fu C, Wolin SL, and Reinisch KM (2005). Structural Insights into RNA Quality Control: The Ro Autoantigen Binds Misfolded RNAs via Its Central Cavity. Cell 121, 529–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunwoo H, Dinger ME, Wilusz JE, Amaral PP, Mattick JS, and Spector DL (2009). MEN ε/β nuclear-retained non-coding RNAs are up-regulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 19, 347–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka N, Meineke B, and Shuman S (2011). RtcB, a Novel RNA Ligase, Can Catalyze tRNA Splicing and HAC1 mRNA Splicing in Vivo. J. Biol. Chem. 286, 30253–30257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka N, and Shuman S (2011). RtcB Is the RNA Ligase Component of an Escherichia coli RNA Repair Operon. J. Biol. Chem. 286, 7727–7731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temmel H, Müller C, Sauert M, Vesper O, Reiss A, Popow J, Martinez J, and Moll I (2017). The RNA ligase RtcB reverses MazF-induced ribosome heterogeneity in Escherichia coli. Nucleic Acids Res. 45, 4708–4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger T (2004). SOLVE and RESOLVE: automated structure solution, density modification, and model building. Journal of Synchrotron Radiation 11, 49–52. [DOI] [PubMed] [Google Scholar]
- Vagin A, and Teplyakov A (1997). MOLREP: an Automated Program for Molecular Replacement. J. Appl. Crystallogr. 30, 1022–1025. [Google Scholar]
- Vagin AA, Steiner RA, Lebedev AA, Potterton L, McNicholas S, Long F, and Murshudov GN (2004). REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr. D 60, 2184–2195. [DOI] [PubMed] [Google Scholar]
- Walker SC, Avis JM, and Conn GL (2003). General plasmids for producing RNA in vitro transcripts with homogeneous ends. Nucleic Acids Res. 31, e82–e82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE, Freier SM, and Spector DL (2008), 3′ End Processing of a Long Nuclear-Retained Noncoding RNA Yields a tRNA-like Cytoplasmic RNA. Cell 135, 919–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AGW, McCoy A, et al. (2011). Overview of the CCP4 suite and current developments. Acta Crystallogr. D 67, 235–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolin SL, Belair C, Boccitto M, Chen X, Sim S, Taylor DW, and Wang HW (2013). Non-coding Y RNAs as tethers and gates. RNA Biology 10, 1602–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, and Ferré-D’Amaré RA (2016). The tRNA Elbow in Structure, Recognition and Evolution. Life 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


