Abstract
The explosion of microbial genome sequences has shown that bacteria harbor an immense, largely untapped potential for the biosynthesis of diverse natural products, which have traditionally served as an important source of pharmaceutical compounds. Most of the biosynthetic genes that can be detected bioinformatically are not, or only weakly, expressed under standard laboratory growth conditions. Herein we review three recent approaches that have been developed for inducing these so-called silent biosynthetic gene cluster: insertion of constitutively active promoters using CRISPR-Cas9, high-throughput elicitor screening for identification of small molecule inducers, and reporter-guided mutant selection for creation of overproducing strains. Together with strategies implemented previously, these approaches promise to unleash the products of silent gene clusters in years to come.
Graphical abstract
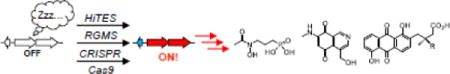
Introduction
Natural products remain a source of inspiration for chemists, biologists, and pharmacologists alike. Extensive analyses by Newman and Cragg have shown just how valuable these metabolites have been, especially in a clinical setting [1,2]: In the past ~35 years, more than half of the drugs approved by the FDA in the United States were based on natural products. The recent explosion in microbial genome sequences shows that this bounty is merely the tip of the iceberg and that there lies a hidden realm of dark microbial matter that we cannot readily access [3–7]. Specifically, genome sequences of prolific secondary metabolite producers routinely reveal many biosynthetic gene clusters (BGCs) – the sets of genes responsible for the synthesis of a natural product – that are not actively expressed, or only lowly-expressed, under standard laboratory growth conditions. These so-called silent or cryptic BGCs outnumber the constitutively active ones by a factor of 5–10 [6–8]. Hence methods that reliably awaken them would dramatically enhance our reservoir of potentially therapeutic small molecules [6–13].
Aside from pinpointing molecules of pharmaceutical relevance, uncovering the hidden metabolomes of bacteria also carries untold lessons in metabolism, chemical ecology, and biosynthesis [13–17]. Although the primary metabolomes of bacteria are generally well-understood, the complete secondary metabolome for any given prolific producer has yet to be determined. Just how many secondary metabolites can an organism with ~30 BGCs produce? Is this metabolome condition-dependent? How common is cross-talk between BGCs and are there genomic rules for identifying it? A global understanding of secondary metabolism would begin to provide answers to these questions.
Perhaps more important than what is produced by the store of silent BGCs is answering the question of why it is produced. That is to say, once the constituents of the secondary metabolome have been established, the biological functions and roles they play in the physiology of the producing host can be explored. Secondary metabolites generally facilitate the interaction of a microbe with its environment and, as such, unveiling the secondary metabolomes of bacteria will shed light onto microbial interactions and chemical ecology [18,19]. This knowledge could in turn permit a bottoms-up reconstruction of the multipartite interactions, mediated by the exchange of secondary metabolites [13–16]. Key to this exchange are the regulatory circuits that control cryptic or induced secondary metabolism, which presently is poorly-understood [13,20–22]. The question of when and how a bacterium ‘chooses’ to activate a given silent BGC is a fascinating and unresolved one, and detailed studies are sure to illuminate mechanisms, perhaps new ones, by which exogenous signals or cues govern this process. Lastly, once new cryptic metabolites have been characterized and linked to their cognate BGCs, biosynthetically-interesting transformations can be explored, thus revealing new biosynthetic routes and enzymatic reactions.
That silent BGCs are an important frontier of natural products research has been recognized by the research community and a number of methods have been developed for activating them. Among the first methods were expression of a BGC in a heterologous host, insertion of constitutive or inducible promoters, co-culture methods, and ribosome engineering. These approaches have been extensively covered in the past [8–13,23]. Herein we review the most recent strategies that have been developed, as well as improvements on previous methodologies (Fig. 1). These include application of CRISPR-Cas9 methods for introducing active promoters, high-throughput elicitor screening (HiTES) to identify small molecule inducers, and reporter-guided random or transposon mutagenesis, all for the purpose of awakening silent BGCs.
Figure 1.
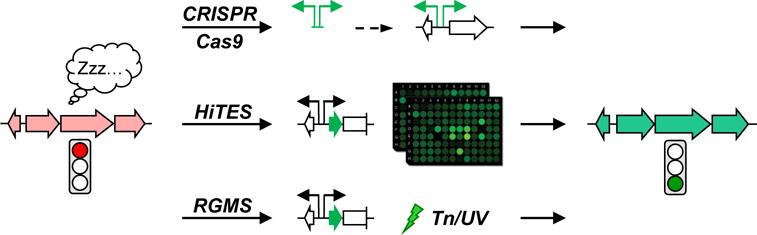
Recent strategies developed for activation of silent BGCs that are discussed in this review. The strategies include insertion of constitutive promoters using CRISPR/Cas9, identification of small molecule elicitors using HiTES, and selection of mutants that induce a given BGC using RGMS. All three approaches have recently been implemented in Streptomyces spp. See text for details.
Using CRISPR-Cas9 to Introduce Constitutive Promoters
Among genetic methods for activation of silent BGCs, perhaps the most widely-used is replacement of the native promoter with known constitutive or inducible ones. Though generally successful, the approach has two key limitations: (1) the organism of choice must be genetically tractable with a sequenced, annotated genome, and (2) the BGC needs to be in a single operon. These limitations make application of the approach challenging in the “talented” Streptomyces genus [7], in which genetic manipulations are slow and multiple genetic insertions very challenging. One solution to this quandary is the application of CRISPR-Cas9 methods, a versatile genome-editing technology that is effective even in many genetically intractable organisms (Fig. 2a) [24–26]. This technology was tailored for application in Streptomyces by several groups simultaneously, who each engineered CRISPR-Cas9-based platforms for the genus and showed their ability to delete genes or gene clusters and perform gene replacements [27–30]. Significantly, these platforms demonstrated increased efficiencies and decreased time investments compared to conventional genetic methods.
Figure 2.
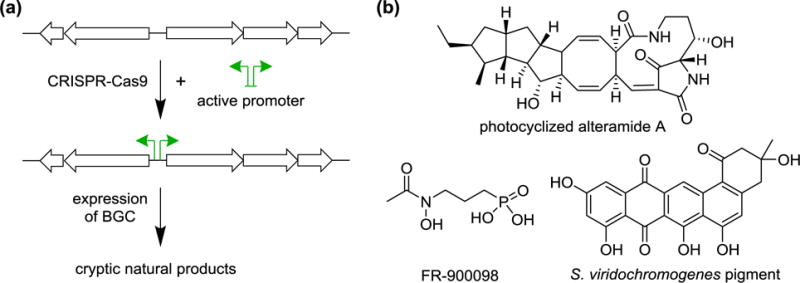
Manipulating expression of silent biosynthetic genes using CRISPR-Cas9. (a) CRISPR-Cas9 methods adapted for use in streptomycetes are used to insert an active uni- or bi-directional promoter to drive expression of silent BGCs. (b) This approach has been used in S. roseosporus and S. viridochromogenes, among other strains, to induce production of alteramide A, FR-9000098, and a type II PKS-derived pigment.
As a proof of concept, Cobb et al. first used CRISPR-Cas9 to knock-in constitutive promoters upstream of previously characterized pigment biosynthetic gene clusters in genetically tractable model Streptomyces strains [27], including the indigoidine cluster in Streptomyces albus and the actinorhodin and undecylprodigiosin clusters in Streptomyces lividans. These efforts resulted in successful activation of pigment production.
The method was further extended to two uncharacterized BGCs in Streptomyces roseosporus with homology to clusters with known products [31]. Knock-in of a constitutive promoter upstream of one cluster induced the production of the polycyclic tetramate macrolactams alteramide A and dihydromaltophilin, both previously reported metabolites (Fig. 2b) [32,33]. The second cluster targeted showed high homology to the known phosphonate FR-900098 [34], and a bidirectional promoter cassette was introduced to drive its expression. The engineered strain showed increased production of FR-900098 (Fig. 2b).
Finally, the strategy was also applied to uncharacterized BGCs in three Streptomyces strains. In S. roseosporus, knock-in of an active promoter upstream of a type I polyketide synthase (PKS) or a separate LuxR-regulated PKS induced production of a new metabolites, as determined by LC-MS analysis. In Streptomyces venezuelae, insertion of a bidirectional promoter to drive a type III PKS resulted in production of a pigment, the structure of which was not reported. Finally, in Streptomyces viridochromogenes, an uncharacterized type II PKS was induced, and the major product of the engineered strain was structurally elucidated. Based on NMR analysis, the novel brown pigment was shown to consist of a dihydrobenzo[α]naphtha-cenequinone core with an unusual cyclohexanone modification. (Fig. 2b) [31].
In an interesting extension of these studies, Kang et al took advantage of CRISPR-Cas9 and transformation-associated recombination (TAR) in a strategy they named mCRISTAR [35]. In this approach, CRISPR-Cas9 was used to induce double-stranded DNA breaks in promoter regions of silent BGCs. The resulting fragments were reassembled via TAR with synthetic promoters replacing the native ones. The refactored clusters were then heterologously expressed. The method was successfully used to activate tetarimycin A production, which had previously been identified by heterologous expression of environmental DNA [36]. Related applications have been reported as well: some Aspergillus fumigatus isolates are known to produce the pigment trypacidin. By comparing sequences of trypacidin-producing and non-producing strains, Brakhage and colleagues identified a single nucleotide insertion in the PKS-encoding gene of the latter group [37]. CRISPR-Cas9 editing was then used to carry out a single-nucleotide deletion, which resulted in restored synthesis of trypacidin in the non-producing strain.
In summary, CRISPR-Cas9-mediated gene editing is a generalizable strategy that could lead to additional natural products. A limitation that remains is that genetic procedures, even if simplified, are required. Still, we expect to see further applications of this strategy in the future.
High-throughput Elicitor Screening
The small molecule signals that may induce silent BGCs in the gifted actinomycetes and other bacteria remain poorly understood [6,7,38,39]. To address this aspect, we previously developed a chemogenetic method for identifying elicitors for silent BGCs [13,40]. Referred to as HiTES (high-throughput elicitor screening), the approach consists of insertion of a reporter gene into the BGC of interest to provide a rapid read-out for its expression. Subsequently, small molecule libraries are screened to identify candidate elicitors. We first implemented the approach in the β-Proteobacterium Burkholderia thailandensis [40,41] Recently, it has been extended to streptomycetes, specifically S. albus (Fig. 3) [42,43]. The sur non-ribosomal peptide synthetase (NRPS) gene cluster, unexplored in S. albus, was chosen as a test case. Two different reporter constructs were generated: one containing a promoter-reporter construct, in which the silent sur promoter (Psur) was fused to a triple eGFP cassette (Psur-eGFPx3), and inserted into a neutral site on the S. albus chromosome. A second strain carried a site-specific insertion of eGFPx3 directly downstream of the native Psur promoter. HiTES with both strains using a ~500-member natural product library identified ivermectin and etoposide as the best elicitors of the sur BGC (Fig. 3). Ivermectin is used globally as an antiparasitic agent while etoposide is employed clinically as an anticancer drug. Needless to say, the stimulatory properties of these famous metabolites were not anticipated, demonstrating one of the strengths of the HiTES approach.
Figure 3.
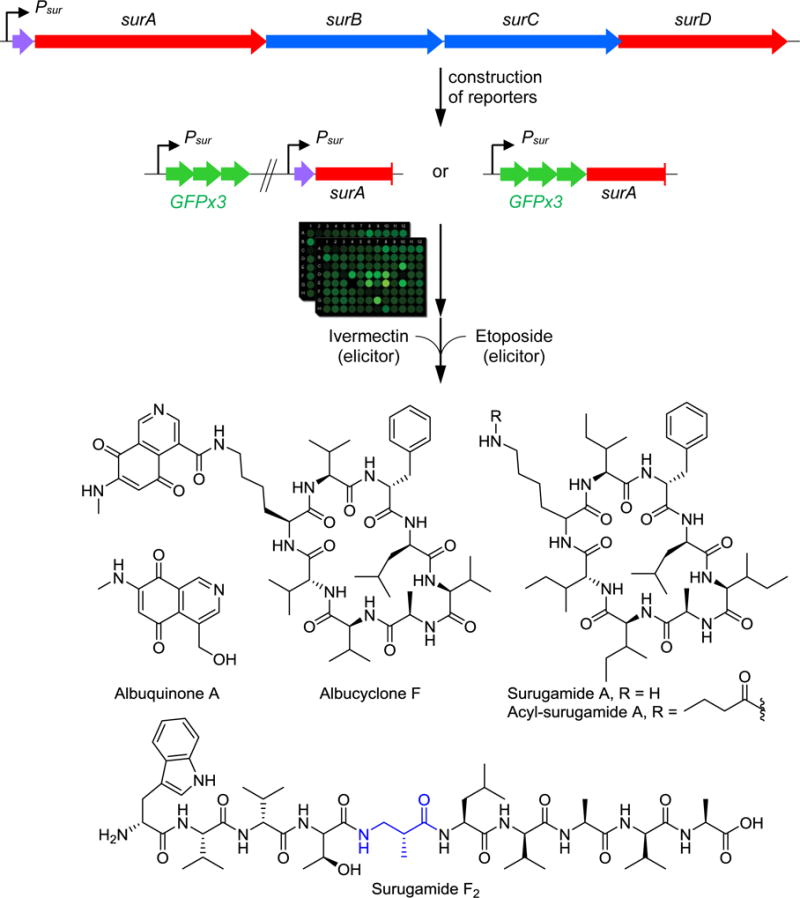
High-throughput elicitor screening in S. albus. The cryptic sur cluster was targeted with two reporter strains, one where the promoter-reporter construct Psur-eGFPx3 was inserted into a neutral chromosomal site and a second, in which eGFPx3 was incorporated site-specifically into the sur cluster downstream of Psur. HiTES with these reporters revealed etoposide and ivermectin as inducers of the sur cluster, which was subsequently shown to produce a bouquet of metabolites, including surugamides, acyl-surugamide A, albucyclones, and surugamide F variants. Both elicitors globally enhanced secondary metabolite synthesis, leading to the identification of a new metabolite, albuquinone A.
Detailed secondary metabolite analyses of S. albus as well as a sur-deficient S. albus strain in the presence and absence of the elicitors identified the products of the BGC and led to 14 novel cryptic metabolites in 4 compound families [42] (Fig. 3): These included (i) the surugamides, a family of cyclic octapeptides containing a combination of L- and D-amino acids [44]; (ii) acylated surugamides with a butyryl group modifying the side-chain amine of a Lys residue, (iii) the albucyclones, which contain an unusual isoquinoline quinone that decorates the surugamide scaffold, and (iv) a group of linear decameric peptides with D- and L-amino acids as well as an uncommon β-Ala residue [42,45]. An additional metabolite, albuquinone A, was also identified but did not stem from the sur BGC, indicating that etoposide and ivermectin induced other biosynthetic loci as well (Fig. 3)
Surugamides A, D, and F were previously found from a marine Streptomyces species, Streptomyces sp. JAMM992 [44,45]. Biosynthetic investigations showed that the sur cluster in this strain and in S. albus synthesized two completely disparate products. The two flanking NRPSs (SurA and SurD, Fig. 3) generated the octapeptides, while the two internal NRPSs (SurB and SurC) gave rise to the linear NRPs, providing an unorthodox instance of one cluster producing two disparate compound families [42,45].
HiTES in S. albus not only helped to identify the products of the sur gene cluster, but also aided in uncovering some aspects of its regulation. The expression of surR, a cluster-specific transcriptional regulator was found to be downregulated by both ivermectin and etoposide [42]. The identification of a pathway-specific repressor and modulation of its expression by exogenous molecules provide a starting point for investigating the regulatory circuits that control the sur cluster [42,43]. As this example demonstrates, the advantage of HiTES is that it allows insights into both the product(s) and regulation of a silent BGC. The finding that low doses of antibiotics enhance secondary metabolite biosynthesis will surely motivate further research in the future [40,42]. A current limitation is that genetic manipulations are required; nonetheless, with the unique ability to connect small molecule elicitors to silent BGCs, additional applications and improvements of HiTES are sure to follow.
Reporter-guided Mutant Selection
Reporter-guided mutant selection (RGMS) is a recently developed method for enhancing microbial secondary metabolism. It combines two effective technologies: genome-wide mutagenesis to generate genetic diversity, and a reporter system to facilitate selection of mutants, in which the desired BGCs are induced. Additionally, invaluable mechanistic insights can be gleaned regarding the genes that, directly or indirectly, regulate and silence a given BGC. Perhaps the first use of RGMS applied to natural products was demonstrated by Askenazi et al. in selecting Aspergillus terreus mutants with increased production of lovastatin, a natural product and cholesterol-lowering drug with potent inhibitory activity against 3-hydroxy-3-methylglutaryl-coenzyme A (HMG-CoA) reductase [46]. A promoter-reporter fusion plasmid containing the lovF promoter and the reporter gene ble, coding for phleomycin-resistance, was constructed and transformed into the fungus. LovF expression was monitored through phleomycin-resistance. Transformants were mutagenized with UV light and mutants were selected on agar plates with elevated concentrations of the drug. About 50% of the phleomycin-resistant mutants showed overproduction of lovastatin, thus validating the approach.
The strategy was later adapted by Xiang et al. to improve secondary metabolite production in Streptomyces spp. [47]. In order to reduce the high rates of false positive mutants from single-reporter screens, a double-reporter system was developed, in which xylE, a catecholase that stains colonies brown in the presence of catechol, was fused to neo (coding for kanamycin resistance) to provide faithful read-out of gene expression (Fig. 4). Mutants that lead to induction of the chosen BGC would be both Kan-resistant and catecholase-positive. In Streptomyces clavuligerus, the xylE-neo cassette was fused with the promoter of ccaR gene, a transcriptional activator of clavulanic acid (CA) biosynthesis. Upon UV mutagenesis, 90% of mutants showed improved CA production. Recently, this method was also successfully applied in two other streptomycetes, S. venezuelae and Streptomyces sp. PGA64, leading to the induction of jadomycin biosynthesis and to the discovery of two new gaudimycin derivatives (Fig. 4) [48].
Figure 4.
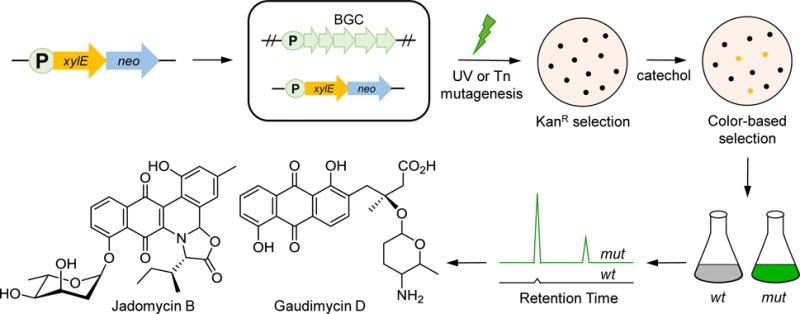
Reporter-guided mutant selection in Streptomyces spp. A double reporter construct, xylE-neo, is placed under the control of a desired silent promoter and the strain is then mutagenized using UV, chemical or transposon mutagenesis. Desired mutants are selected on Kan plates and further by screening for catecholase-positive colonies. The metabolome of the mutant and wt strain are then compared by HPLC-MS to identify the products of the chosen silent cluster. This approach led to overproduction of jadomycin B and gaudimycin D, among other products.
The disadvantage of UV mutagenesis is that the gene(s) implicated cannot be easily mapped. Moreover, multiple mutations may be induced thus complicating downstream analyses. Transposon (Tn) mutagenesis, on the other hand, is more amenable to identifying the genes, whose mutations alter secondary metabolism. Along these lines, Xu et al. recently performed Tn mutagenesis in Streptomyces coelicolor to screen for mutants with altered levels of production of the tripyrrole antibiotic undecylprodigiosin (RED), the bright red color of which provides a tractable read-out [49]. The approach identified hundreds of genes that influenced RED biosynthesis, including those in DNA metabolism, protein modification, branched-chain amino acid biosynthesis, and genes involved in signaling, stress, and transcriptional regulation. These results underline the complexity of biological pathways that affect natural product biosynthesis.
Ahmed and coworkers combined the advantages of the experiments above, that is mutagenesis by Tn insertion (rather than by UV) in conjunction with genetic reporters (rather than compound-specific colorimetric read-out), to investigate candicidin production in S. albus [50]. This cluster is not silent, and hence the authors searched for genes that affect its expression. A Tn insertion into a transcriptional regulator resulted in production of the chemical trigger, butenolide 4, which then led to overproduction of previously-known candicidins and antimycins. The authors suggested that the transcriptional regulator acts as a new regulatory element in the butenolide system to control secondary metabolism in S. albus. Accordingly, RGMS, like HiTES, can report on the product of a given BGC and its underlying regulation.
Perspective
The development of an arsenal of approaches for inducing silent BGCs in a short period of time has been impressive [8,13,51]. Though largely complementary, each of the approaches old and new still has disadvantages that may prevent it from broad application. In our estimation, the ideal method is one that avoids challenging cloning and genetic procedures, uses mono-culture methods, and side-steps heterologous expression. Genetic procedures slow down the pace of discovery and reduce the throughput of any method. Co-culture, while in some cases successful, sometimes suffers from reproducibility and, when conducted on agar surfaces, scalability. Heterologous expression has been very useful but is generally-speaking not well suited to compounds that arise from crosstalk of multiple genetic loci and to the large BGCs typically expressed in actinomycetes. Methods that alter the regulation of a given BGC, i.e. promoter insertion and heterologous expression, cannot provide insights into the regulatory networks that underlie cryptic metabolism. As such, the search for the ideal, high-throughput method for activating silent BGCs continues. But the success with the approaches developed so far clearly illustrates that the pursuit of such a method will be worth-while.
Highlights.
Silent biosynthetic gene clusters are a treasure trove of new secondary metabolites
CRISPR-Cas9 methods allow for site-specific insertion of active promoters
HiTES enables identification of small molecule inducers for any silent gene cluster
RGMS combines promoter-reporters and mutagenesis to activate silent pathways
These and other approaches promise to unlock the biosynthetic potential of bacteria
Acknowledgments
Research in our group on the activation of silent BGCs was supported by the National Institutes of Health (grant DP2-AI-124786), the Searle Scholars Program, and the Pew Biomedical Scholars Program.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1•.Newman DJ, Cragg GM. Natural products as sources of new drugs from 1981 to 2014. J Nat Prod. 2016;79:629–661. doi: 10.1021/acs.jnatprod.5b01055. A comprehensive treatment of the importance of natural products as clinical agents in the past ~34 years. [DOI] [PubMed] [Google Scholar]
- 2.Newman DJ, Cragg GM. Natural products as sources of new drugs over the 30 years from 1981 to 2010. J Nat Prod. 2012;75:311–335. doi: 10.1021/np200906s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bentley SD, Chater KF, Cerdeño-Tárraga A-M, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D, et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2) Nature. 2002;417:141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- 4.Ikeda H, Ishikawa J, Hanamoto A, Shinose M, Kikuchi H, Shiba T, Sakaki Y, Hattori M, Omura S. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat Biotechnol. 2003;21:526–531. doi: 10.1038/nbt820. [DOI] [PubMed] [Google Scholar]
- 5.Oliynyk M, Samborskyy M, Lester JB, Mironenko T, Scott N, Dickens S, Haydock SF, Leadlay PF. Complete genome sequence of the erythromycin-producing bacterium Saccharopolyspora erythraea NRRL23338. Nat Biotechnol. 2007;25:447–453. doi: 10.1038/nbt1297. [DOI] [PubMed] [Google Scholar]
- 6•.Nett M, Ikeda H, Moore BS. Genomic basis for natural product biosynthetic diversity in the actinomycetes. Nat Prod Rep. 2009;26:1362–1384. doi: 10.1039/b817069j. A catalog of the abundance of unassigned biosynthetic gene clusters in well-studied model streptomycetes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7•.Baltz RH. Gifted microbes for genome mining and natural product discovery. J Ind Microbiol Biotechnol. 2017;44:573–588. doi: 10.1007/s10295-016-1815-x. Description of the correlation between genome size and number of biosynthetic gene clusters to assign ‘gifted’ secondary metabolite producing microbial phyla. [DOI] [PubMed] [Google Scholar]
- 8.Rutledge PJ, Challis GL. Discovery of microbial natural products by activation of silent biosynthetic gene clusters. Nat Rev Microbiol. 2015;13:509–523. doi: 10.1038/nrmicro3496. [DOI] [PubMed] [Google Scholar]
- 9.Zhu H, Sandiford SK, van Wezel GP. Triggers and cues that activate antibiotic production by actinomycetes. J Ind Microbiol Biotechnol. 2014;41:371–386. doi: 10.1007/s10295-013-1309-z. [DOI] [PubMed] [Google Scholar]
- 10.Ren H, Wang B, Zhao H. Breaking the silence: new strategies for discovering novel natural products. Curr Opin Biotechnol. 2017;48:21–27. doi: 10.1016/j.copbio.2017.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pettit RK. Mixed fermentation for natural product drug discovery. Appl Microbiol Biotechnol. 2009;83:19–25. doi: 10.1007/s00253-009-1916-9. [DOI] [PubMed] [Google Scholar]
- 12.Wright GD. Opportunities for natural products in 21st century antibiotic discovery. Nat Prod Rep. 2017;34:694–701. doi: 10.1039/c7np00019g. [DOI] [PubMed] [Google Scholar]
- 13.Okada BK, Seyedsayamdost MR. Antibiotic dialogues: induction of silent biosynthetic gene clusters by exogenous small molecules. FEMS Microbiol Rev. 2017;41:19–33. doi: 10.1093/femsre/fuw035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meinwald J. Natural products as molecular messengers. J Nat Prod. 2011;74:305–309. doi: 10.1021/np100754j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van der Meij A, Worsley SF, Hutchings MI, van Wezel GP. Chemical ecology of antibiotic production by actinomycetes. FEMS Microbiol Rev. 2017;41:392–416. doi: 10.1093/femsre/fux005. [DOI] [PubMed] [Google Scholar]
- 16.Flórez LV, Biedermann PH, Engl T, Kaltenpoth M. Defensive symbioses of animals with prokaryotic and eukaryotic microorganisms. Nat Prod Rep. 2015;32:904–936. doi: 10.1039/c5np00010f. [DOI] [PubMed] [Google Scholar]
- 17.Tang MC, Zou Y, Watanabe K, Walsh CT, Tang Y. Oxidative cyclization in natural product biosynthesis. Chem Rev. 2017;117:5226–5333. doi: 10.1021/acs.chemrev.6b00478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Meinwald J, Eisner T. The chemistry of phyletic dominance. Proc Natl Acad Sci USA. 1995;92:14–18. doi: 10.1073/pnas.92.1.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang R, Seyedsayamdost MR. Hijacking exogenous signals to generate new secondary metabolites during symbiotic interactions. Nat Rev Chem. 2017;1:0021. [Google Scholar]
- 20•.Rigali S, Titgemeyer F, Barends S, Mulder S, Thomae AW, Hopwood DA, van Wezel GP. Feast or famine: the global regulator DasR links nutrient stress to antibiotic production by Streptomyces. EMBO Rep. 2008;9:670–675. doi: 10.1038/embor.2008.83. The first report of the connection between nutrient sensing, DasR, and secondary metabolism, leading to induction of silent BGCs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoon V, Nodwell JR. Activating secondary metabolism with stress and chemicals. J Ind Microbiol Biotechnol. 2014;4:415–424. doi: 10.1007/s10295-013-1387-y. [DOI] [PubMed] [Google Scholar]
- 22.Bibb MJ. Regulation of secondary metabolism in streptomycetes. Curr Opin Microbiol. 2005;8:208–215. doi: 10.1016/j.mib.2005.02.016. [DOI] [PubMed] [Google Scholar]
- 23.Ochi K, Hosaka T. New strategies for drug discovery: activation of silent or weakly expressed microbial gene clusters. Appl Microbiol Biotechnol. 2013;97:87–98. doi: 10.1007/s00253-012-4551-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wright AV, Nuñez JK, Doudna JA. Biology and applications of CRISPR systems: Harnessing nature’s toolbox for genome engineering. Cell. 2016;164:29–44. doi: 10.1016/j.cell.2015.12.035. [DOI] [PubMed] [Google Scholar]
- 25.Doudna JA, Charpentier E. The new frontier of genome engineering with CRISPR-Cas9. Science. 2014;346:1258096. doi: 10.1126/science.1258096. [DOI] [PubMed] [Google Scholar]
- 26.Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32:347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27•.Cobb RE, Wang Y, Zhao H. High-efficiency multiplex genome editing of Streptomyces species using an engineered CRISPR/Cas system. ACS Synth Biol. 2015;4:723–728. doi: 10.1021/sb500351f. Development of an engineered CRISPR/Cas9 system for multiplexed genome editing of Streptomyces spp. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang H, Zheng G, Jiang W, Hu H, Lu Y. One-step high-efficiency CRISPR/Cas9-mediated genome editing in Streptomyces. Acta Biochim Biophys Sin (Shanghai) 2015;47:231–243. doi: 10.1093/abbs/gmv007. [DOI] [PubMed] [Google Scholar]
- 29.Tong Y, Charusanti P, Zhang L, Weber T, Lee SY. CRISPR-Cas9 based engineering of actinomycetal genomes. ACS Synth Biol. 2015;4:1020–1029. doi: 10.1021/acssynbio.5b00038. [DOI] [PubMed] [Google Scholar]
- 30.Zeng H, Wen S, Xu W, He Z, Zhai G, Liu Y, Deng Z, Sun Y. Highly efficient editing of the actinorhodin polyketide chain length factor gene in Streptomyces coelicolor M145 using CRISPR/Cas9-CodA(sm) combined system. Appl Microbiol Biotechnol. 2015;99:10575–10585. doi: 10.1007/s00253-015-6931-4. [DOI] [PubMed] [Google Scholar]
- 31••.Zhang MM, Wong FT, Wang Y, Luo S, Lim YH, Heng E, Yeo WL, Cobb RE, Enghiad B, Ang EL, Zhao H. CRISPR-Cas9 strategy for activation of silent Streptomyces biosynthetic gene clusters. Nat Chem Biol. 2017;13:607–609. doi: 10.1038/nchembio.2341. Activation of silent BGCs via CRISPR-Cas9-mediated insertion of active promoters in streptomycetes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shigemori H, Bae MA, Yazawa K, Sasaki T, Kobayashi J. Alteramide A, a new tetracyclic alkaloid from a bacterium Alteromonas sp. associated with the marine sponge Halichondria okadai. J Org Chem. 1992;57:4317–4320. [Google Scholar]
- 33.Graupner PR, Thornburgh S, Mathieson JT, Chapin EL, Kemmitt GM, Brown JM, Snipes CE. Dihydromaltophilin; a novel fungicidal tetramic acid containing metabolite from Streptomyces sp. J Antibiot. 1997;50:1014–1019. doi: 10.7164/antibiotics.50.1014. [DOI] [PubMed] [Google Scholar]
- 34.Eliot AC, Griffin BM, Thomas PM, Johannes TW, Kelleher NL, Zhao H, Metcalf WW. Cloning, expression, and biochemical characterization of Streptomyces rubellomurinus genes required for biosynthesis of antimalarial compound FR900098. Chem Biol. 2008;15:765–770. doi: 10.1016/j.chembiol.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kang HS, Charlop-Powers Z, Brady SF. Multiplexed CRISPR/Cas9- and TAR-mediated promoter engineering of natural product biosynthetic gene clusters in yeast. ACS Synth Biol. 2016;5:1002–1010. doi: 10.1021/acssynbio.6b00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kallifidas D, Kang HS, Brady SF. Tetarimycin A, an MRSA-active antibiotic identified through induced expression of environmental DNA gene clusters. J Am Chem Soc. 2012;134:19552–19555. doi: 10.1021/ja3093828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weber J, Valiante V, Nødvig CS, Mattern DJ, Slotkowski RA, Mortensen UH, Brakhage AA. Functional reconstitution of a fungal natural product gene cluster by advanced genome editing. ACS Synth Biol. 2017;6:62–68. doi: 10.1021/acssynbio.6b00203. [DOI] [PubMed] [Google Scholar]
- 38.Wright GD. Opportunities for natural products in 21st century antibiotic discovery. Nat Prod Rep. 2017;34:694–701. doi: 10.1039/c7np00019g. [DOI] [PubMed] [Google Scholar]
- 39.Bachmann BO, Van Lanen SG, Baltz RH. Microbial genome mining for accelerated natural products discovery: is a renaissance in the making? J Ind Microbiol Biotechnol. 2014;41:175–184. doi: 10.1007/s10295-013-1389-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40••.Seyedsayamdost MR. High-throughput platform for the discovery of elicitors of silent bacterial gene clusters. Proc Natl Acad Sci USA. 2014;111:7266–7271. doi: 10.1073/pnas.1400019111. Development of HiTES in B. thailandensis and discovery of low doses of antibiotics as cell-wide inducers of silent BGCs. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Okada BK, Wu Y, Mao D, Bushin LB, Seyedsayamdost MR. Mapping the trimethoprim-induced secondary metabolome of Burkholderia thailandensis. ACS Chem Biol. 2016;11:2124–2130. doi: 10.1021/acschembio.6b00447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42•.Xu F, Nazari B, Moon K, Bushin LB, Seyedsayamdost MR. Discovery of a cryptic antifungal compound from Streptomyces albus J1074 using high-throughput elicitor screens. J Am Chem Soc. 2017;139:9203–9212. doi: 10.1021/jacs.7b02716. Implementation of HiTES in S. albus and discovery of ivermectin/etoposide as inducers of novel cryptic metabolites. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rosen PC, Seyedsayamdost MR. Though much is taken, much abides: finding new antibiotics using old ones. Biochemistry. 2017;56:4925–4926. doi: 10.1021/acs.biochem.7b00782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takada K, Ninomiya A, Naruse M, Sun Y, Miyazaki M, Nogi Y, Okada S, Matsunaga S. Surugamides A-E, cyclic octapeptides with four D-amino acid residues, from a marine Streptomyces sp.: LC-MS-aided inspection of partial hydrolysates for the distinction of D- and L-amino acid residues in the sequence. J Org Chem. 2013;78:6746–6750. doi: 10.1021/jo400708u. [DOI] [PubMed] [Google Scholar]
- 45•.Ninomiya A, Katsuyama Y, Kuranaga T, Miyazaki M, Nogi Y, Okada S, Wakimoto T, Ohnishi Y, Matsunaga S, Takada K. Biosynthetic gene cluster for surugamide A encompasses an unrelated decapeptide, surugamide F. Chembiochem. 2016;17:1709–1712. doi: 10.1002/cbic.201600350. Discovery of two unrelated surugamides from the same BGC in a marine Streptomyces strain. [DOI] [PubMed] [Google Scholar]
- 46.Askenazi M, Driggers EM, Holtzman DA, Norman TC, Iverson S, Zimmer DP, Boers ME, Blomquist PR, Martinez EJ, Monreal AW, et al. Integrating transcriptional and metabolite profiles to direct the engineering of lovastatin-producing fungal strains. Nat Biotechnol. 2003;21:150–156. doi: 10.1038/nbt781. [DOI] [PubMed] [Google Scholar]
- 47.Xiang SH, Li J, Yin H, Zheng JT, Yang X, Wang HB, Luo JL, Bai H, Yang KQ. Application of a double-reporter-guided mutant selection method to improve clavulanic acid production in Streptomyces clavuligerus. Metab Eng. 2009;11:310–318. doi: 10.1016/j.ymben.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 48••.Guo F, Xiang S, Li L, Wang B, Rajasärkkä J, Gröndahl-Yli-Hannuksela K, Ai G, Metsä-Ketelä M, Yang K. Targeted activation of silent natural product biosynthesis pathways by reporter-guided mutant selection. Metab Eng. 2015;28:134–142. doi: 10.1016/j.ymben.2014.12.006. Induction of silent BGCs using double reporter-guided mutant selection in streptomycetes. [DOI] [PubMed] [Google Scholar]
- 49•.Xu Z, Wang Y, Chater KF, Ou HY, Xu HH, Deng Z, Tao M. Large-scale transposition mutagenesis of Streptomyces coelicolor identifies hundreds of genes influencing antibiotic biosynthesis. Appl Environ Microbiol. 2017;83:e02889–16. doi: 10.1128/AEM.02889-16. Saturating Tn mutagenesis in S. coelicolor that implicates hundreds of genes in the production of secondary metabolites. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ahmed Y, Rebets Y, Tokovenko B, Brötz E, Luzhetskyy A. Identification of butenolide regulatory system controlling secondary metabolism in Streptomyces albus J1074. Sci Rep. 2017;7:9784. doi: 10.1038/s41598-017-10316-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rigali S, Anderssen S, Naômé A, van Wezel GP. Cracking the regulatory code of biosynthetic gene clusters as a strategy for natural product discovery. Biochem Pharmacol. 2018 doi: 10.1016/j.bcp.2018.01.007. In Press. [DOI] [PubMed] [Google Scholar]


