Abstract
Canalization, or phenotypic robustness in the face of environmental and genetic perturbation, is an emergent property of living systems. Although this phenomenon has long been recognized, its molecular underpinnings have remained enigmatic until recently. Here, we review the contributions of the molecular chaperone Hsp90, a protein that facilitates the folding of many key regulators of growth and development, to canalization of phenotype – and de-canalization in times of stress – drawing on studies in eukaryotes as diverse as baker’s yeast, mouse ear cress, and blind Mexican cavefish. Hsp90 is a hub of hubs that interacts with many so-called ‘client proteins’ that affect virtually every aspect of cell signaling and physiology. As Hsp90 facilitates client folding and stability, it can epistatically suppress or enable the expression of genetic variants in its clients and other proteins that acquire client status through mutation. Hsp90’s vast interaction network explains the breadth of its phenotypic reach, including Hsp90-dependent de novo mutations and epigenetic effects on gene regulation. Intrinsic links between environmental stress and Hsp90 function thus endow living systems with phenotypic plasticity in fluctuating environments. As environmental perturbations alter Hsp90 function, they also alter Hsp90’s interaction with its client proteins, thereby re-wiring networks that determine the genotype-to-phenotype map. Ensuing de-canalization of phenotype creates phenotypic diversity that is not simply stochastic, but often has an underlying genetic basis. Thus, extreme phenotypes can be selected, and assimilated so that they no longer require environmental stress to manifest. In addition to acting on standing genetic variation, Hsp90 perturbation has also been linked to increased frequency of de novo variation and several epigenetic phenomena, all with the potential to generate heritable phenotypic change. Here, we aim to clarify and discuss the multiple means by which Hsp90 can affect phenotype and possibly evolutionary change, and identify their underlying common feature: at its core, Hsp90 interacts epistatically through its chaperone function with many other genes and their gene products. Its influence on phenotypic diversification is thus not magic but rather a fundamental property of genetics.
Keywords: Canalization, buffering, capacitor, potentiate, robustness, Hsp90
1 Introduction
1.1 The concept of canalization from theory to experiment
From bacteria to humankind, a multitude of mechanisms ensure the fidelity of information transfer and its phenotypic manifestation. Indeed, when faced with ever changing environments, organisms must be robust to survive. Yet evolutionary success also requires adaptability, whether through phenotypic plasticity or the generation and selection of heritable biological novelty. Although genotype inarguably influences the development and expression of phenotype, selective forces ultimately operate on phenotypes. Mechanisms that influence the capacity of genetic variants to alter traits can thus have a fundamental impact on the adaptive landscape.
Some traits are more stable across environments and genetic backgrounds than others. A canalized trait is one that is stable in the face of genetic or environmental perturbation (Figure 1A). For example, many aspects of gross morphology (e.g. body plan and organ systems in animals) are reproducible across a wide variety of environments and genetic backgrounds. Other traits are less canalized. In Caenorhabditis elegans, for example, genetic and environmental perturbation readily gives rise to differences in gender distribution, fecundity, and fat storage, but has less impact on cell number and lineage or movement and development of the pharynx, vulva, and other organs [1]. In contrast to animals, plant body plans and morphologies are far more responsive to environmental change due to continuous development; nevertheless, organ identity is largely preserved even in the face of severe stress [2].
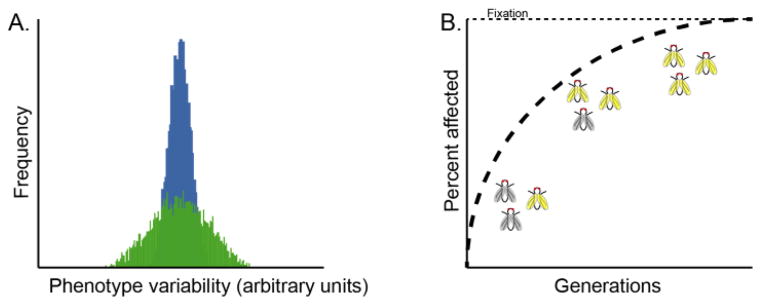
Figure 1. Canalization minimizes phenotypic variation.
A. Quantitative traits exhibit some degree of variation, represented here by a distribution. A canalized trait shows tight distributions (blue) regardless of genetic or environmental perturbation. Some traits can be de-canalized by environmental or genetic perturbations which increases the degree of phenotypic variation (green). B. A possible mechanism of assimilation of a new phenotype occurs via de-canalization. Over several generations of selection for a crossveinless wing phenotype in Drosophila, the rare phenotype was assimilated to a large fraction of the population.
As outlined in several previous reviews, the remarkable phenotypic robustness of wild-type organisms is commonly attributed to features of the underlying genetic networks, such as genetic redundancy, network connectivity, feedback loops, modularity, and the presence of microRNAs [1, 3–27]. In model organisms, perturbation of any of these mechanisms, or environmental change, can decrease phenotypic robustness and release cryptic genetic variation [3, 10–12, 20, 28–33].
Canalization can be advantageous in constant environments, or even in the presence of modest fluctuations, maintaining a mean distribution of phenotypes that hews closely to what has been adaptive to an ancestral population [34]. Yet extreme canalization can create a phenotypic ‘lock-in’ that could be disadvantageous if the environment shifts such that the prior distribution of phenotypes is maladaptive [35]. In this review, we focus on the stress-regulated molecular chaperone Hsp90 as a mechanism for canalization that offers some resolutions to this paradox, linking the degree of trait canalization to the severity of environmental change.
1.2 Hsp90 provides a mechanism for de-canalization mediated by stress
Seminal studies investigating the concept of canalization were carried out more than half a century ago by the ‘father of epigenetics,’ Conrad Hal Waddington. While conducting heat shock experiments with Drosophila pupae, Waddington noted that a small fraction of the resulting adult flies developed crossveinless wings [36]. Selection enriched this phenotype to near fixation in the population, suggesting a stable epigenetic or genetic basis. Remarkably, after a few generations of selection, the crossveinless phenotype was evident even in the absence of stress [36] (Figure 1B); that is, this once rare and environmentally contingent phenotype was readily assimilated into the population as a stable trait. This observation laid the groundwork for many future studies of canalization (and de-canalization) and its possible contribution to evolutionary change. In the intervening decades, these concepts have been applied to many other systems, ranging from heritability of tadpole body size mediated by dietary changes [37] to caterpillar body color driven by temperature shifts [38].
Working half a century after Waddington, Rutherford and Lindquist observed a strikingly similar set of phenomena linked to the activity of the molecular chaperone Hsp90 when investigating Drosophila melanogaster harboring heterozygous mutants of the HSP83 gene (for simplicity we will hereafter use “HSP90” when referring to the gene and “Hsp90” when referring to the protein in all organisms). They noticed that mutant flies occasionally developed crossveinless wings, among other morphological abnormalities [39]. The specific kind of Hsp90-dependent phenotype observed strongly depended on the genetic background examined, consistent with an underlying genetic basis. Thermal stress produced the same phenotypes in the same backgrounds. Just as in Waddington’s studies, selection over several generations enriched these phenotypes in the population, consistent with a stable epigenetic or genetic basis. Selection on an eye and a wing trait also rendered these phenotypes independent of perturbation by either direct interference with Hsp90 function or environmental stress. The data conform to one view of the definition of assimilation, as originally articulated by Waddington, and implicated Hsp90 as a possible molecular mechanism contributing to Waddington’s seminal observations.
Subsequent studies, highlighted in later sections, have extended the hypothesis that Hsp90 broadly influences the phenotypic manifestation of genetic diversity beyond D. melanogaster [18, 40, 41]. Hsp90 influences the phenotypic outcomes of diverse genetic variants in plants (Arabidopsis thaliana) [42], zebrafish [43], fungi [44, 45], worms [46], Mexican cavefish [47], and even humans [48–50] (Figure 2A–D). The broad conservation of Hsp90’s role in shaping phenotype highlights the importance of canalization in understanding the trajectory from genotype to phenotype, in particular with regard to understanding complex human diseases.
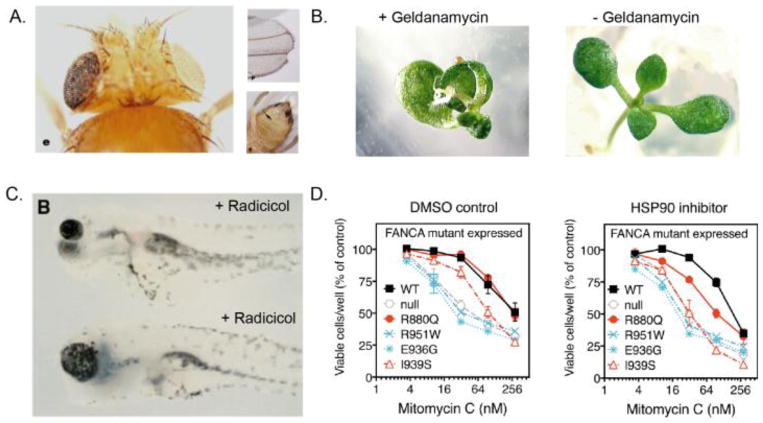
Figure 2. Phenotypic variability revealed by inhibition of Hsp90.
A. Inhibition of Hsp90 in Drosophila reveals phenotypes including black facets in one eye, notched wings, and extraneous tissue (reprint from Rutherford and Lindquist, 1998). B. Inhibition of Hsp90 by geldanamycin in A. thaliana reveals phenotypes including disruption of typical symmetry and oval shaped, flat leaves (reprint from Queitsch et al., 2002). C. Inhibition of Hsp90 in Mexican cavefish, A. mexicanus, results in variable eye size of larval fish (reprint from Rohner et al., 2013). D. Human cells expressing the FANCA mutant allele R880Q exhibit increased sensitivity to Hsp90 inhibitor ganetespib but a wild-type allele or non-buffered alleles do not. In this case, the detrimental growth phenotype revealed upon Hsp90 inhibition is only observed in a specific FANCA mutant background (reprint from Karras et al., 2017).
Hsp90 is conserved in many but not all bacteria and only a few Archaea, suggesting it may have been gained and lost several times outside of eukarya [51]. Although the bacterial Hsp90 homolog HtpG does not seem to exhibit the same buffering functions of eukaryotic Hsp90, other chaperones such as GroEL and DnaK have been shown to buffer deleterious effects of genetic variation and exert similar effects on the evolution of their client proteins [52–55].
1.3 Hsp90 – a special chaperone
Hsp90 is one of the most abundant proteins in eukaryotes [56]. Hsp90 was first discovered and named for its thermal induction as a heat shock protein response. Expression of Hsp90 and other heat shock proteins increases dramatically upon exposure to heat through a combination of transcriptional and translational mechanisms [57]. Along with other heat shock proteins (e.g. the chaperones Hsp70 and Hsp40) Hsp90 acts to help unfolded proteins achieve their proper conformation in the crowded intracellular milieu [58]. Several features distinguish Hsp90 from other chaperones. It is normally expressed at relatively high levels – beyond what is required for growth in organisms where this has been tested [59] – and it is induced by myriad environmental stresses. In some instances, however, even this increased expression of Hsp90 cannot contend with the increasing number of unfolded proteins under stress, resulting in a reduction of net chaperone activity [60, 61]. It has been hypothesized that the basal reservoir of Hsp90 activity in unstressed conditions allows cells to rapidly respond to modest levels of environmental fluctuations.
Over one thousand proteins have been identified in physical interaction screens using Hsp90 as bait [62–64]; however, Hsp90 clients are not a random sampling of all proteins, but a select cohort enriched in kinases and transcription factors [63] that are known to be conformationally dynamic. These clients reside in nearly every developmental and signaling pathway in eukaryotes [58, 65–67], providing a plausible explanation for Hsp90’s broad influence on the relationship between genotype and phenotype. In contrast, generalist chaperones such as Hsp70 have many more clients that are a more random sampling of the proteome. They are also thought to act earlier in folding pathways than Hsp90 [58].
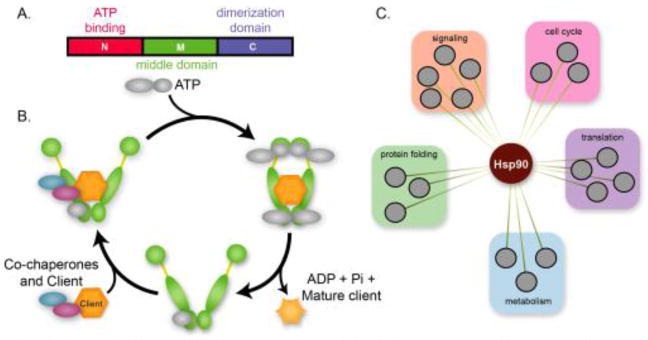
Hsp90’s N-terminus contains a conserved ATPase domain that powers conformational dynamics essential for client folding [68]; its C-terminus is critical for dimerization (Figure 3A). The basis of client recognition is complex. Structural studies with a range of client proteins suggest that interactions with the middle region are important for protein-protein interactions [69–71], but the interfaces identified also include some portions of the N- and C-termini [72–74]. Regardless of the precise protein binding domain location, it is clear that protein interactions and ATP hydrolysis are required for client folding. Combinatorial recognition with co-chaperones also plays an important role in this process (see below). Subdomain-FRET-analyses and crystal structures have illuminated the dynamic cycles that fuel the ‘molecular clamp’ mechanism of Hsp90 binding and release driven by ATP hydrolysis (Figure 3B) [75, 76]. Figure 3B illustrates the chaperone cycle that has been well defined for the progesterone receptor, a model client of Hsp90 [77]. However, our mechanistic knowledge is incomplete; e.g. we are unable to predict whether a particular protein is an Hsp90 client or whether a mutation will confer or revoke client status. Clients are often directed to Hsp90 via other (co)chaperones, dozens of which are known. For instance, Hsp90’s capacity to catalyze the folding of many of its kinase client proteins is dependent on the Cdc37 co-chaperone [73, 78–83]. Like other Hsp90 clients, these kinases are thought to be initially metastable, but they are stabilized through interaction with Hsp90 and become functional. Other client proteins rely on other co-chaperones, such as SQUINT in A. thaliana [84]. We point the reader to recent reviews detailing the extended family of Hsp90 proteins, diverse co-chaperones, post-translational control, and mechanisms of action of all chaperone proteins [58, 78, 83, 85–95]. Our knowledge of the multitude of Hsp90 client proteins continues to expand with the utilization of new techniques [63]. Collectively, these studies demonstrate that Hsp90 is a hub-of-hubs, linked to nearly every process within the cell (Figure 3C).
Figure 3. Hsp90 structure and function.
A. The N-terminus of Hsp90 contains a conserved ATP binding domain (lime green), a middle domain that may bind client proteins and co-chaperones (green), and a C-terminal domain responsible for dimerization (blue). B. The Hsp90 chaperone cycle begins with the binding of co-chaperones and clients. Here co-chaperones are colored blue. All other co-chaperones and ATP are drawn in gray. The progesterone receptor is one of the best understood clients. It binds one co-chaperone, then recruits another. After ATP binding, the Hsp90 dimer clamps together. This final hydrolysis step includes binding of another co-chaperone as well as ATP. Upon ATP hydrolysis, the clamp opens, releasing a mature client protein. C. Protein interaction studies have defined the vast network of Hsp90 interactors. Hsp90 interacts with hundreds of proteins of diverse functions including protein folding, signaling, cell cycle, translation and metabolism.
1.4 Missing heritability, canalization, and the evolving genotype-to-phenotype map
Decoding how genetic variation gives rise to phenotypic diversity is the central challenge of genetics and genomics. Modern genomic technologies have enabled unprecedented strides toward cataloging genetic variation across many individuals and mapping functional genomic regions; nevertheless, the complexity of most biological traits, including many diseases, poses enormous challenges for predicting phenotype from sequence. For example, genome wide association studies have been employed in large cohorts of individuals to identify genetic variants that contribute to complex human diseases. These studies have successfully associated thousands of genetic variants with various quantitative phenotypes. However, most of these variants contribute little to disease risk and explain only a small proportion of the observed heritability in families. This “missing heritability” has been attributed to several factors including unidentified sequence variation (e.g. copy number variation in repetitive DNA, large genomic rearrangements) [96], rare alleles of large effect [97, 98], inflated heritability values in families [97], the failure to account for epistasis (i.e. genetic interactions) [97, 98], epigenetic variation [97, 98], variable levels of robustness [99, 100], and cryptic genetic variants. Hsp90 and hubs like it [30, 99] are likely to play a major role in shaping complex traits through their role as strong genetic modifiers.
2 Principles and theory
2.1 Epistasis with environmental contingency
Traditionally, epistasis refers to a non-reciprocal interaction between two alleles in which the phenotypic effect of one allele is dependent upon the presence of the other. The extent of epistasis in genetic networks is vast. For example, systematic studies in yeast have tested all pairs of viable gene deletion alleles, and identified genetic interactions between them [101]. Even in this compact genome, and examining only a single growth condition, these >23 million double mutants revealed nearly one million instances of epistasis. Notably, systematic studies like this one reveal that not all genes exhibit the same degree of epistasis, allowing us to understand and build genetic networks [102]. Genes at the hub of such networks – Hsp90 key among them – show epistatic interactions with a large number of other genes [63]. For example, studies of ~65,000 pairs of genes in C. elegans identified 350 genetic interactions; over a quarter of these involved six hub genes, all encoding chromatin regulators [30]. In other words, network hubs like Hsp90 and chromatin regulators represent a special case of epistasis in which one gene interacts epistatically with many others.
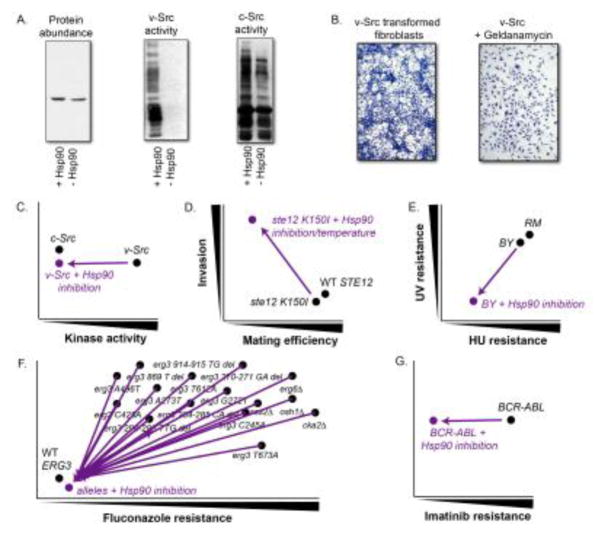
The direct link between Hsp90 activity and environmental conditions make this relationship even more remarkable, and important for linking phenotypic diversification to environmental change. Indeed, because Hsp90 activity itself serves as a ‘stress sensor’ [103, 104], this system ensures that phenotypic diversity can increase in the face of environmental change across entire populations, in contrast to mutations that affect only a few individuals. Although other hubs have been shown to act as strong genetic modifiers [30], there is less evidence that their canalizing or ‘buffering’ function is environmentally responsive. Without plausible paths for de-canalization across many individuals in a population, genetic variation will remain cryptic, and unable to contribute to phenotype or be acted upon by selection. Nevertheless, one can imagine mutations that disrupt such hub genes in certain individuals, a scenario that has been invoked as contributing to complex human diseases [30, 99]. With regard to Hsp90, common environmental stresses and changes in cell physiology during malignant transformation can modulate its activity and hence its interactions with many other genes. The classic example of Hsp90s’ effect on malignant transformation is the well-studied interaction between Hsp90 and v-Src kinase, without which the kinase is not functional (Figure 4A–C) [105, 106]. Thus, changes in the intra-and extracellular environment can readily lead to the release (and masking) of genetic variation, de-canalization of phenotype, and ultimately the assimilation of such traits via selection.
Figure 4. Mechanistic examples of buffering and potentiating.
A. Activity of v-Src but not c-Src is dependent on Hsp90 (reprint from Xu and Lindquist, 1993). B. Likewise, malignant transformation with v-Src is also dependent on Hsp90. Upon treatment with the Hsp90 inhibitor geldanamycin, the normal contact inhibition of growth is restored (reprint from Whitesell et al., 1994). C. Graphic illustration of Hsp90 potentiating oncogenic v-Src constitutively active kinase activity. The original observed phenotype is graphed on a phenotypic scale in black. The phenotype dependent on Hsp90 is graphed in purple with a vector designating the phenotypic difference of buffered alleles. D. Ste12 contributes to both mating and invasion. A wild-type STE12 allele results in normal mating efficiency and invasion. A mutant ste12 K150I allele results in decreased mating efficiency only at high temperature or upon inhibition of Hsp90. The same allele results in increased invasion only at high temperature. E. In the RM background, the MEC1 allele is not dependent on Hsp90. In the BY background, the MEC1 allele is dependent on Hsp90; HU-resistance and UV-resistance decreases when Hsp90 in inhibited. F. Hsp90 potentiates BCR-ABL imatinib resistance. G. Hsp90 potentiates fluconazole resistance in several erg3 alleles and other gene deletions.
Although environmental stress provides a common trigger for changes in Hsp90 activity, genetic and even epigenetic mechanisms may also affect its function. Hsp90 and associated co-chaperones are highly conserved in eukarya, but variation has been observed in some organisms. For example, some Drosophila populations harbor HSP90 variants that can affect phenotypic robustness [107, 108]. In C. elegans, the penetrance of several mutant alleles in developmental pathways can be predicted by endogenous Hsp90 levels in juvenile animals [109]. Hsp90 polymorphisms are also surprisingly common in humans, an exciting topic for future study.
2.2 Genetic assimilation of environmentally sensitive traits
One of the most striking – and thus far least experimentally dissected – aspects of Hsp90-dependent capacitance is the apparent genetic assimilation of previously environmentally and Hsp90-dependent traits (see also accompanying perspectives by L. Loison and G. Geiler-Samerotte in this issue). Empirically, such genetic assimilation has been observed in the laboratory. In Waddington’s original experiments, fixation of the crossveinless wing trait in D. melanogaster was rapid. After several generations of selective breeding the trait was stabilized in the population even in the absence of heat stress, a process that Waddington defined as genetic assimilation [36] (Figure 1B). Likewise, Rutherford and Lindquist observed rapid assimilation of analogous, Hsp90-dependent traits in D. melanogaster [32] (Figure 2A). Laboratory studies with different organisms and employing other environmental triggers have also concluded that the environment can exert a large influence on heritability, presumably by altering the impact of cryptic genetic variation. For example, changes in the broad-sense heritability of tadpole body size, developmental stage, and gut length after shifting to a rich shrimp diet or exposure to a hormone induced by such dietary changes [37].
Although Hsp90 activity can be affected by all environmental stimuli that compromise protein folding, it has long been debated whether Hsp90-dependent variation and its release upon stress can contribute to evolutionary trajectories in nature. Rohner et al. investigated this question in the context of the Mexican cavefish Astyanax mexicanus [47]. Recent geological events have repeatedly isolated sub-populations of this river fish in different cave environments. These isolated populations have experienced a series of highly reproducible changes, specifically loss of pigmentation and eyes. Multiple lines of evidence suggest that these phenotypes are adaptive in the cave environment [47]. Rohner et al. exposed developing river fish with normal eyes to low concentrations of Hsp90 inhibitor, noticing a significant de-canalization of orbit size. Selective breeding of fish with small orbit size led to rapid assimilation of the trait. That is, progeny from such crosses showed small orbits even without any Hsp90 inhibition (Figure 2C). Most remarkably, exposure to cave water conditions, an ecologically relevant stress, also decanalized orbit size in river fish, analogous to the treatment with the Hsp90 inhibitor. Although determining whether Hsp90-dependent de-canalization has contributed to the cave-specific phenotypes in nature stands as a goalpost for future work, this study establishes the expression of Hsp90-dependent, formerly cryptic variation as a plausible explanation for this rapid and recurring adaptation.
3 Evidence for the evolutionary importance of Hsp90-dependent variation
Although it is challenging to devise experiments to conclusively prove that Hsp90-dependent variation contributed to eye loss or other traits, the existence and relative importance of Hsp90-dependent evolution can be detected in genomes, in particular by comparing the evolutionary rates of client and non-client proteins. The capacitor hypothesis posits that clients should be allowed to acquire more and potentially more harmful mutations than non-clients. Evolutionary rate is influenced by many factors, including protein stability, protein interactions, and gene expression. These factors will confound any effect that interaction with Hsp90 may have, making it imperative to conduct analyses of evolutionary rate among genes encoding comparable client and non-client proteins. One solution has been to compare pairs of recent gene duplicates that differ little in sequence and expression. In A. thaliana, the Hsp90-client BES1, a transcription factor in the brassinosteroid pathway, shows relaxed selection compared to its closest paralog the non-client BZR1 [110]. BES1 bears hallmarks of neo- and subfunctionalization, and shows dynamic Hsp90 client status across independent evolutionary paths. In yeast, Hsp90 clients also evolve faster than their non-client paralogs [110].
The availability of large-scale data on Hsp90 clients, in particular on human kinases [63], has finally enabled the systematic exploration of Hsp90’s role in protein evolution across the mammalian lineage. Kinases are monophyletic, akin to the prior studies of gene duplicates. Kinases identified as clients in humans show significantly higher evolutionary rates than non-client kinases across the mammalian lineage; this effect is independent of gene expression and protein interaction, the two major factors influencing evolutionary rate, and comparable in effect size [111]. Across several thousand sequenced humans, genes encoding client kinases showed greater nucleotide diversity than those encoding non-clients. Moreover, the genetic variants in client kinases were predicted to be more damaging to protein function than those in non-clients, consistent with Hsp90’s hypothesized capacitance function. If so, one would predict that once a kinase acquires Hsp90 client status, it would be unlikely to lose it again. Indeed, this is the case. Kinases are of outsized importance in shaping development, physiology, and evolutionary trajectories; they also play a major role in many human diseases, including various cancers. In this context, Hsp90’s role in promoting their divergence and in maintaining their function in the face of accumulating genetic variation is particularly noteworthy.
In contrast to eukarya, Hsp90 is not essential in bacteria (where it is known as HtpG), and a broad characterization of its molecular function and possible role in capacitance is lacking in this domain of life. A recent study used genome-scale phylogenetic analysis to identify genes that co-evolve with bacterial Hsp90 [51]. Genes whose gains and losses are coordinated with Hsp90 throughout bacterial evolution tended to function in large protein complexes associated with motility and secretion, suggesting that Hsp90 may aid the assembly of protein complexes. Indeed, experimental validation identified Hsp90 clients such as the flagellar protein FliN and the chemotaxis kinase CheA; Escherichia coli HSP90 mutants showed impaired motility and chemotaxis. The presence of bacterial HSP90 across all sequenced species is associated with a preference for multiple habitats. Taken together, bacterial Hsp90 appears to aid the assembly of membrane protein complexes and facilitates adaptation to novel environments, both functions that seem to preface its much larger and essential role in protein folding and evolution in eukarya.
To summarize, these studies confirm one of the two predictions of the capacitor hypothesis: genetic variation indeed accumulates at higher rates in genes encoding client proteins. The other prediction of the capacitor hypothesis posits that intermittent stress leads to expression of this accumulated variation followed by selection. This prediction has also been addressed in recent studies, as we will discuss in more detail below (see 5.3).
4 Protein folding as a general mechanism
4.1 Open questions
Studies from yeast to human have established Hsp90’s conserved capacity to buffer genetic variation. Although several Hsp90-dependent loci have been mapped in natural populations [44, 112, 113] it has largely remained unresolved how precisely Hsp90 perturbation enables cryptic genetic variation to contribute to phenotype. Does this variation occur in client proteins, or can it occur in non-clients, or even regulatory regions? Could any variant in a client protein be Hsp90-responsive? In short, what variation can the chaperone buffer? Below we synthesize results from experiments seeking to answer these questions.
4.2 Hsp90 reveals cryptic genetic variants responsible for many traits
Our understanding of how Hsp90 affects cryptic genetic variation comes from a series of genetic studies across model organisms. In flies, plants, yeast, fish, and other model systems, Hsp90 perturbation generates background-specific traits [39, 42, 47, 48] (Figure 2), allowing Hsp90-dependent loci to be mapped in some systems [44, 112, 113]. Thus far, however, few Hsp90-dependent variants have been identified at the gene level [44, 114]. As a result, our mechanistic understanding of Hsp90-mediated capacitance is limited, while biochemical studies of Hsp90 function have flourished [67]. In the absence of systematic studies of Hsp90-dependent variation in a wide array of client and non-client proteins, it has been impossible to rigorously determine features that render a given variant Hsp90-responsive. The handful of known examples suggest that Hsp90-dependent variants can occur in clients, non-clients that interact with client proteins, and even in regulatory regions that are bound by clients. We discuss specific examples in section 5. Recent studies suggest that disease-associated variants in various proteins often show Hsp90-dependence [48]; early work found this to be true for mutations in oncogenic proteins [106, 115].
5 Specific examples across eukarya
5.1 Alleles that are buffered
A capacitor stores electrical energy for later rapid release. Hsp90 has been proposed to function analogously for genetic variants, allowing them to accumulate (or be ‘stored’) as phenotypically silent mutations, only to be expressed after a shift in environment. That is, ensuing reduction of Hsp90 function may unmask the phenotypic consequences of genetic variants that were previously cryptic. As discussed above our understanding of specific variants that are influenced by this mechanism is incomplete, although there is convincing evidence that they are common in natural populations [29, 112, 113].
Greater detail on buffered variants is emerging from saturating mutagenesis studies of individual genes. For example, “deep mutational scanning” has identified Hsp90-buffered variants of the Ste12 transcription factor in Saccharomyces cerevisiae [114] (Figure 4D). The wild-type Ste12 protein is not a client of Hsp90. At standard growth conditions, the Hsp90-dependent Ste12 variants were competent for both mating and invasion. Ste12 drives the expression program for both traits, which are mutually exclusive, yet share many signaling components, including Ste12. Upon Hsp90 inhibition or thermal stress, cells harboring the Hsp90-dependent variants lose the capacity to mate, but become dominantly hyperinvasive even in the absence of the known invasion cofactor. Indeed, temperature-regulated invasiveness is a common phenotype of fungal pathogens [116–119]. The Ste12 example is striking because loss of Hsp90 does not simply lead to degradation of the respective protein and loss of function; rather, loss of Hsp90 shifts the trait preference of Ste12 towards invasion by altering its binding to DNA. Although Hsp90-dependent Ste12 variants are rare, they are accessible through a single amino acid change, suggesting that the chaperone could facilitate a mutational path toward a pathogenic fungal lifestyle by minimizing mating costs at normal temperature and enhancing invasion at the higher host temperature.
In yeast, fine mapping studies have identified several natural genetic variants that are affected by chaperone activity [44]. Two arise in Hsp90 clients: Mec1, the sentinel DNA damage kinase, and Nfs1, an essential sulfur donor in FeS cluster biogenesis and tRNA thiolation. Under normal growth conditions, natural genetic variation in these genes is phenotypically silent. But upon pharmacological or environmental inhibition of Hsp90 activity, vineyard alleles of these genes give rise to new phenotypes. The vineyard allele of Mec1 produces resistance to DNA damage induced by UV-irradiation (Figure 4E), and the vineyard allele of Nfs1 produces resistance to rapamycin. Each of these phenotypes is explained by the known function of the polymorphic genes and their well-characterized interaction with the chaperone. Hsp90 also buffers resistance to the oxidative stressor 1-chloro-2,4-nitrobenzene (CDNB) derived from vineyard alleles of the Ndi1 gene. Remarkably, this trait arises from mutations in the 3′-untranslated sequence of the gene. Upon Hsp90 inhibition, levels of NDI1 mRNA increase by nearly 100-fold in strains harboring the vineyard allele. Forced overexpression of Ndi1 also produces CDNB resistance. Although the NDI1 mRNA transcript itself is of course not a client of Hsp90, several Hsp90 clients are known to bind to its 3′ untranslated region, providing a plausable rationale for this Hsp90-responsive phenotype.
Genotypes and phenotypes have been measured extensively across many dozens of yeast isolates from diverse ecological niches. This approach has made it feasible to assess the global relationship among Hsp90, genotype and phenotype in this organism. Across >100 growth conditions and without Hsp90 perturbation, the correlation between genotype and phenotype is statistically significant, but surprisingly weak. This correlation increases considerably in response to Hsp90 inhibition, suggesting that a considerable fraction of the ‘silent’ genetic variation in this organism has phenotypic consequences in the presence of Hsp90 inhibition [44].
Another striking example comes from mice, where Hsp90 buffers the regulatory influence of certain endogenous retroviruses on neighboring developmental genes [120]. This effect on cis-regulatory variation arises because Hsp90 chaperone activity is required for TRIM28/KAP1-mediated epigenetic silencing of endogenous retroviral elements. This example provides yet another mechanism by which Hsp90 can buffer natural genetic variation, and raises the question of whether the capacitor function of Hsp90 may have facilitated exaptation of endogenous retroviruses as modifiers of gene expression.
As discussed earlier, as the number of sequenced human genomes expands, so does our ability to detect the broad effects of Hsp90 and protein folding in general on human phenotypes. Across >1,500 disease alleles, a recent study found that their relative association with Hsp90 was highly predictive of disease severity, especially when compared to association with Hsp70, another chaperone (Figure 2D) [48]. This finding, which is remarkable given the diversity of genetic causes of disease, illustrates how even specific aspects of chaperone function are fundamentally integrated into the trajectory between genotype and phenotype. A more detailed examination of mutations associated with the cancer-prone syndrome Fanconi Anemia (FA) revealed that Hsp90 perturbation amplified FA-related sensitivities to chemotherapeutics [48]. It is important to reiterate that not all variants are buffered by Hsp90. Many are potentiated, and most are unaffected [121, 122]. This has also been borne out at the phenotypic level. For example in Drosophila some phenotypes are strongly affected by Hsp90 inhibition whereas others are not [39]. Although an integrated understanding of how chaperone activity influences specific disease alleles will require many more both detailed and systematic studies, it is clear that these effects are likely highly significant and widespread.
5.2 Alleles that are potentiated
Hsp90 can also potentiate mutations. That is, when Hsp90 activity is reduced, the capacity of these variants to produce new traits is eliminated. Although we use a different term for this effect (potentiation), it is driven by the same phenomenon that promotes buffering: Hsp90’s function as a chaperone of its client proteins. Like buffering, potentiation reflects Hsp90’s widespread epistasis with regulatory genes. Indeed, the first studies of buffering in A. thaliana also observed instances of potentiation [42], and subsequent studies in S. cereviasiae further defined specific examples of such an effect [44]. However, one of the first oncogenes discovered, v-Src, provided a particularly striking example nearly 30 years ago. v-Src arose from mutations in the c-Src progenitor kinase that lead to its constitutive activation. However, these same mutations render v-Src thermodynamically unstable, making its folding and function dependent on Hsp90 [106, 123] (Figure 4A–C). Many other oncogenes have since been shown to require Hsp90 for the potentiation of their function. Another fascinating example is the BCR-ABL gene fusion that encodes a constitutively active tyrosine kinase. The BCR-ABL inhibitor Imatinib, known commercially as Gleevec, was one of the earliest and most successful targeted therapies for chronic myelogenous leukemia. However, the emergence of resistance has been a common failure mode of this and most other oncogene-directed therapies. A majority of patients treated with Imatinib at an advanced stage will relapse due to a point mutation in BCR-ABL that drives resistance [124]. Treatment with an Hsp90 inhibitor leads to degradation of the Hsp90-client BCR-ABL and loss of imatinib resistance (Figure 4F) [49, 50], highlighting the need to understand the role of Hsp90 and other factors in enabling the accumulation of mutations favorable to cancer cells.
Like buffering, potentiation can also occur indirectly. A striking example is resistance to fluconazole, a main line antifungal drug, in the fungal pathogen Candida albicans. Mutations in ERG3 enable resistance to fluconazole [45], but also result in the production of a toxic sterol. Such variants depend upon Hsp90, which mediates the function of its client Calcineurin and other proteins integral to stress response circuitry. Activation of this stress response alleviates toxicity that would otherwise be induced by the alternate sterol [125]. By chaperoning Calcineurin, Hsp90 therefore allows the ERG3 variants to persist in the population and contribute to the evolution of drug resistance. Mutations in several other genes also potentiated fluconazole resistance dependent on Hsp90 including erg6Δ, osh1Δ, scs2Δ, and cka2Δ (Figure 4G). Genetic assimilation of initially Hsp90-dependent antifungal drug resistance has been observed in multiple clinical isolates evolving in human hosts, suggesting that this process may have a strong influence on human health. Remarkably, dependence on Hsp90 was abrogated by high temperatures, pointing to a potential clinical benefit of fever.
Because the effects of both buffering and potentiation are examples of epistasis, Hsp90 has been referred to as a “global modifier” of genetic variation. Phenotypes arising from buffered and potentiated variants can be influenced by selection, which might in principle affect the distribution of buffered vs. potentiated variants over time and across changing environments. A recent effort attempted to quantify the frequency of buffered and potentiated mutations in yeast cells. Genetic backgrounds that have been subject to minimal selective pressure showed more frequent potentiation, whereas backgrounds that have been subject to more significant selection showed more frequent buffering [126].
5.3 Buffering and potentiation arise naturally from Hsp90’s chaperone function
Although Hsp90’s relationship with phenotype is remarkable, it is decidedly not magic. Hsp90 does not have the capacity to buffer or potentiate every mutation in every gene; rather, its effects on phenotype are a consequence of its biochemical function. Hsp90’s client proteins – largely kinases and transcription factors – occupy key nodes in signaling cascades that govern a multitude of cellular and developmental pathways [58, 63]. Relative to other chaperones, Hsp90 catalyzes folding steps that occur late in the maturation of its client proteins [58]. A case in point are steroid hormone receptors, a class of closely related transcription factors that were among the earliest Hsp90 clients identified and studied. These proteins bind to Hsp90 in an immature but mostly folded state [127]. In the presence of hormone, the protein acquires a fully folded and functional conformation, leading to concomitant release from the chaperone. Other clients have different trajectories; they require phosphorylation or other modifications to reach their stable state or reach their final cellular destination. Loss of Hsp90 activity severely disrupts client protein function, often through degradation or failure to signal or productively interact in protein complexes [18].
As discussed, Hsp90’s biochemical activity and regulation is tightly coupled to fluctuations in the environment. Together with Hsp70, Hsp90 interacts and suppresses Hsf1 activity, the major regulator of the conserved heat shock response that ensues upon proteotoxic stress. It has been appreciated for decades that upon stress Hsf1 trimerizes, releasing Hsp90 and Hsp70 to act on unfolding proteins; as soon as protein folding has recovered, Hsp90 and Hsp70 are once more available to suppress Hsf1 [128, 129]. Active, trimeric Hsf1 activates expression of many chaperones, including Hsp90. There are also many post-transcriptional and post-translational mechanisms that regulate Hsp90 activity – and these too are linked to environmental inputs – including phosphorylation, acetylation, and nitrosylation [58, 130]. Hsp90 also changes its interaction with co-chaperones in different environments, lineages, and disease states which affects its substrate specificity. Together, these regulatory inputs generally up-or down-regulate Hsp90 activity, or direct its activity toward particular cohorts of clients.
As a result of its complex regulation, Hsp90’s epistatic relationship with genetic variants is not static, but highly dependent on specific environmental circumstances, developmental stages, and even diurnal cycles (Figure 5). Thus, organisms have likely experienced environmentally-driven changes in these epistatic relationships during their evolutionary histories. This predictability of Hsp90 activity changes may expose Hsp90-dependent genetic variation to significant selection, purging deleterious alleles. Indeed, the distribution of fitness effects revealed upon Hsp90 inhibition in wild isolates of S. cerevisiae is heavily skewed toward adaptive phenotypes (~50%) relative to the distribution of fitness effects that would be expected for spontaneous mutations (where adaptive effects are rare) [131, 132], arguing for intermittent release of and selection on Hsp90-dependent variation. In A. thaliana plants, deep mutagenesis revealed significantly greater penetrance of EMS mutations upon Hsp90 inhibition; however, many of the Hsp90-dependent phenotypes (but not all) are also observed in wildtype plants treated with Hsp90 inhibitors (Figure 6). These observations in yeast and plants are consistent with two key points we wish to make: first, not all loci are Hsp90-responsive, and second, Hsp90-dependent variants in these loci in natural populations likely experience intermittent release and selection, purging those that are deleterious [133]. Deeper investigation of these observations stands as a goalpost for future studies.
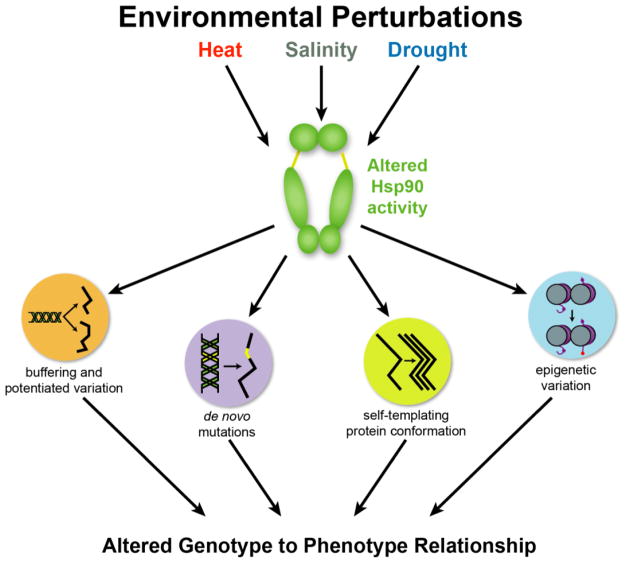
Figure 5. Environmental perturbations regulate Hsp90, a central node that integrates stress sensing with the manifestation and generation of de novo variants.
Many environmental perturbations including heat, salinity, and drought have the potential to alter Hsp90 activity. This altered activity affects cryptic genetic variation, buffered and potentiated variants, de novo mutations, self-templating protein conformations, and epigenetic variation. All of these will in turn alter the relationship between genotype and phenotype.
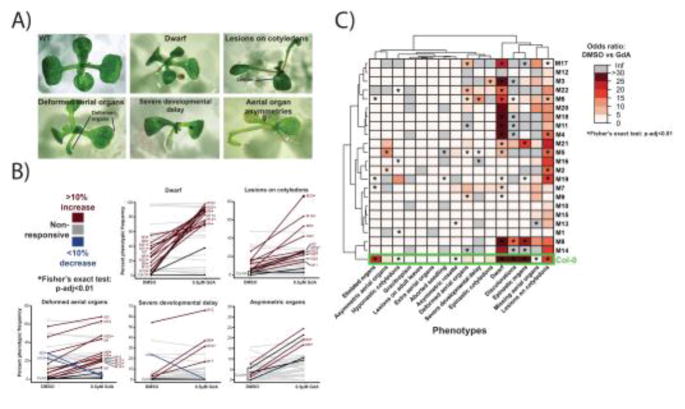
Figure 6. Hsp90-responsive phenotypes in deeply mutagenized Col-0 seedlings often resemble those commonly arising in the wild-type Col-0 background but their frequency in the population and severity is significantly increased.
This observation is consistent with mutations affecting genes encoding clients that are already susceptible to Hsp90 perturbation in the wild-type through standing variation. Novel Hsp90-dependent phenotypes are also observed; genetic variation in the genes underlying these phenotypes has likely been purged in the wild-type Col-0 population. A. Phenotype examples of EMS mutagenized seedlings in the Col-0 background. B. Response to Hsp90 perturbation (geldanamycin: GdA) in Col-0 and mutagenized lines (M3 generation). Dark red color denotes 10% increase in frequency of phenotypes under Hsp90-reduced conditions. Dark blue color denotes 10% decrease in frequency of phenotypes under Hsp90-reduced conditions. Black lines represent phenotypic frequency in the Col-0 background. An asterisk (*) denotes a significant p-value for Fisher’s Exact test (p-adj < 0.05). C. Odds ratios of seedling phenotypes in M3 lines derived from EMS mutagenized Col-0. Seedlings were assayed for 16 early-seedling phenotypes under geldanamycin (GdA) and mock (DMSO) treatment. Increasing red intensity reflects higher Odds Ratios, with grey color reflecting an infinite Odds Ratio for that comparison. An asterisk (*) denotes a significant p-value for Fisher’s Exact test (p-adj < 0.05).
6 Links between Hsp90 and de novo variation
6.1 Hsp90 impacts genome stability
Genome instability encompasses an increased genome-wide frequency of point mutations, insertions/deletions, microsatellite slippage events, somatic homologous recombination, and transposon activity. In human cells, Hsp90 perturbation increases mutation rates of microsatellites [134], and decreases resistance to ionizing radiation [135]. In yeast, strong Hsp90 inhibition can increase rates of aneuploidy [136] and overexpression of HSP90 results in reduced efficiency of DNA repair [137]. In metazoans, Hsp90 inhibition increases transposon transcription and mobility through the disruption of PIWI-protein function (D. melanogaster, C. elegans, mice, and human cells) [95, 138–144]. In the plant A. thaliana, perturbation of HSP90 increases somatic homologous recombination [99] and susceptibility to ionizing radiation [145]. HSP90 perturbation is therefore correlated with, and in several instances causative of, genome instability in eukaryotes (Figure 5). Hsp90’s wide-reaching effects on genome stability is readily explained by its role in chaperoning many proteins functioning in the various DNA maintenance and repair pathways. In addition to the above discussed PIWI-proteins, these include the previously mentioned Mec1 in yeast, telomerase, FANCA in the Fanconi anemia pathway [119], DNA Polymerase subunits, BRCA proteins, and Rad proteins to name a few examples (See reference [132] for database of Hsp90 interactors).
However, given the breadth of standing genetic variation that is responsive to Hsp90 perturbation [29, 39, 42, 48, 60, 120, 146], mutations that newly arise in response to Hsp90 inhibition likely play only a minor role in most Hsp90-dependent phenotypes. Most previous studies validate their genotype-specific Hsp90-dependent phenotypes through treatment of embryos with highly specific Hsp90 inhibitors and moderate temperature stress. Drug and temperature treatments produce the same genotype-specific phenotypes, which is inconsistent with major contributions of de novo mutations. De novo mutations due to these treatments would be somatic mutations, and these most certainly occur at an increased rate in response to Hsp90 inhibition as we and others have shown [99]. However, somatic mutations occurring in individual embryos are extremely unlikely to affect many embryos of a given genotype in the same way; they are also extremely unlikely to be transmitted to the next generation. As the issue of Hsp90-dependent de novo mutations has caused some controversy in the past, we would like to state clearly that although Hsp90 perturbation increases genome instability, the majority of Hsp90-dependent phenotypes described to date are highly unlikely a consequence of de novo mutations, particularly in cases where the high frequency and independent reproducibility of these phenotypes have been validated through drug or environmental treatments of embryos.
The observation that perturbation of a hub like Hsp90 is associated with genomic instability may have broader implications. Increased genome instability via Hsp90 or via perturbation of other hubs may be a general hallmark of de-canalization, which in turn may affect penetrance of genetic variation. In support of this hypothesis, yeast studies identified hub genes in which deletion decreased phenotypic robustness; one-fourth of ~300 identified hubs were genes with critical roles in genome stability [10]. Similarly, environmental stress is known to decrease phenotypic robustness; in many cases, it is also associated with increased genome instability [147, 148]. As genome instability and canalization levels appear to be associated, it has been previously proposed that levels of genome instability could be used as a marker for canalization levels in humans and non-model organisms (and hence as markers for penetrance levels of genetic variation) [99].
6.2 Chromatin-based epigenetic variation
Hsp90 binds to several chromatin regulators, including Trithorax proteins, the INO80 complex, and histone modifying enzymes [149, 150]. These interactions make it likely that Hsp90 can facilitate the generation and phenotypic consequences of epigenetic variation. We discuss two studies from D. melanogaster here. Examining a sensitized Krüppel mutant, Sollars et al. found that Hsp90 inhibition produces heritable phenotypic variation [151]. Krüppel encodes a zinc-finger transcription factor required for abdominal segment patterning; the mutation causes the protein to be ectopically expressed in the eye, altering the pattern of its facets. A genetic enhancer screen for ectopic eye outgrowth yielded loci encoding trithorax group members and five alleles of Hsp90. Strikingly, treating a highly inbred Krüppel line with a specific inhibitor of Hsp90 also produced the ectopic outgrowth near the eye. Despite the line’s nearly isogenic background, selection increased the penetrance of this trait and rendered it independent of Hsp90. One of us has previously argued that the near fixation of this trait may be due to spreading heterochromatin, consistent with the finding that mutations in Trithorax proteins also produce the ectopic outgrowth phenotype in the Krüppel background [152]. This study demonstrated that Hsp90 can affect phenotypes due to epigenetic variation in addition to those due to genetic variation. Although this study expanded the capacitor concept to include epigenetic phenomena, we note that this is still a clear consequence of Hsp90 acting as a strong epistatic modifier. The chaperone acts upon multiple chromatin regulators, hence affecting chromatin states.
Using high-resolution ChIP-seq in D. melanogaster, Sawarkar and colleagues demonstrated that Hsp90 localizes near promoters of many coding and non-coding genes including microRNAS [153]. Hsp90’s interaction with chromatin is indirect, and arises because Hsp90 maintains and optimizes RNA polymerase II pausing by stabilizing the negative elongation factor complex (NELF). Consistent with this model, Hsp90 inhibition leads to upregulation of these genes near its binding sites, and the chaperone is required for maximal activation of paused genes in D. melanogaster and mammalian cells in response to stimuli. Taken together, Hsp90’s effects on chromatin and gene expression are likely complex and locus-specific due to the complex functions of its client proteins. For example, Trithorax proteins are typically associated with euchromatic, transcriptionally active genomic regions; these regions should experience downregulation of expression in response to Hsp90 perturbation [150]. Conversely, paused genes are upregulated in response to Hsp90 inhibition.
6.3 Protein-based epigenetic variation
Originally seen as a rare biological oddity, emerging evidence suggests that self-templating protein conformations – prions – can also commonly serve as epigenetic elements of inheritance [154–156]. Because their propagation is highly dependent on protein conformation, the traits that prions encode are very sensitive to perturbation of molecular chaperones. Most data to date come from a series of studies of amyloid prions in yeast, which have established a critical dependence on the Hsp70 chaperone and the Hsp104 disaggregase for propagation and maintenance [157].
The influence of Hsp90 on prion propagation and maintenance has been more recently examined. For example, propagation of the [URE3] prion requires Hsp90’s interaction with the Cpr7 co-chaperone [158]. However, Hsp90 has no impact on another well-characterized yeast prion, [PSI+], suggesting that this machinery has selective effects on protein-based epigenetic elements [158]. The influence of Hsp90 co-chaperones on spatial quality control of prion-like proteins has also been the subject to recent investigations [159]. Sequestration of amyloid proteins, e.g. the expanded polyglutamine repeats of Huntingtin (Htt103Q), can be protective for the cell. Loss of the Hsp90 co-chaperone Sti1 exacerbates Htt toxicity and blocks the formation of large aggregated protein assemblies. Increased expression of Sti1 has the opposite effect. Thus, this co-chaperone has the capacity to ‘buffer’ the phenotypic impact of protein-based epigenetic variants.
Hsp90 chaperone activity itself appears to matter for the propagation of some protein-based epigenetic elements that do not arise from amyloid fibers. Chakrabortee et al. examined the breadth of protein-based inheritance across the yeast proteome [156]. Transient overexpression of nearly 50 proteins (of 5,300 examined) created traits that remained heritable long after their expression returned to normal. These traits had prion-like patterns of inheritance, were common in wild yeasts, and could be transmitted to naive cells with protein alone. Most inducing proteins were not known prions and did not form amyloid fibers. Instead, they were highly enriched in nucleic-acid-binding proteins with large intrinsically disordered domains that have been widely conserved across evolution. Several required the chaperone activity of Hsp90 to propagate from one generation to the next. Although mechanistic knowledge of these protein-based epigenetic elements is nascent, these observations suggest that Hsp90 can exert a strong impact on the propagation of protein-based epigenetic elements.
Finally, the activity of Hsp90 has recently been linked to the propagation of a protein-based epigenetic trait in mammals [160, 161]. In response to viral infection, RIG-I-like RNA helicases activate the mitochondrial protein MAVS to induce a protective response. It has recently become clear that the mechanism of MAVS activation involves the ubiquitin-mediated formation of large aggregates. These assemblies propagate in a prion-like manner that depends on the Hsp90 chaperone to convert endogenous native MAVS protein into an infectious active form. In the light of new discoveries such as functionally important membrane-free protein assemblies and nuclear foci associated with transcriptional activation or silencing, we speculate that Hsp90’s role in shaping phenotype will only grow in the near future.
7 Conclusions and future directions
It has been nearly two decades since Rutherford and Lindquist proposed that Hsp90 might impact the relationship between genotype and phenotype, and suggested that this might provide a link between environmental change and evolutionary processes [32]. Their work attracted a great deal of interest; yet, it was also highly controversial. And, it must be said, for good reasons. As exciting as the initial findings in D. melanogaster were, the adaptive value of the traits exposed upon Hsp90 inhibition, and the nature of their underlying genetics, was unclear.
Since then, many studies have moved the capacitance model from a compelling hypothesis to a plausible mechanism that has contributed to the evolution of genomes. Work in plants, fungi, and human cancers have identified many genetic variants and loci that are affected by Hsp90 activity. Indeed, the pervasive influence of Hsp90 on mutations linked to cancer has sparked great interest in the therapeutic value of chaperone inhibitors [115, 162–164]. In a wide variety of fungal pathogens, Hsp90 fuels the rapid acquisition of mutations that confer resistance to environmental stressors, including antifungal drugs [165–169], further contributing to the interest in Hsp90 as a therapeutic target.
Perhaps the strongest evidence for Hsp90’s influence on evolution comes from systematic studies of genotype-to-phenotype relationships. In S. cerevisiae strains from diverse ecotypes, inhibiting Hsp90 leads to a far more adaptive distribution of fitness effects than would be expected from random mutations, suggesting that selection has previously acted on the genetic variation responsible for these traits. Indeed, modest reduction in Hsp90 function (elicited either phamacologically or by a moderate environmental stress) improved the correlation between genotype and phenotype across more than 100,000 polymorphisms in sequenced yeast strains [44]. Even more compelling evidence has come with systematic annotation of human kinase clients of Hsp90 function. A key tenet of the original capacitor hypothesis is that Hsp90 client proteins should be able to accumulate mutations at a higher rate than non-clients. Indeed, across the human protein kinase superfamily – and mammals more generally – Hsp90 client status promotes increased evolutionary rate [111]. Collectively, these studies provide strong evidence that Hsp90 has been playing an important role in shaping the evolution of current genomes.
Although Hsp90 exerts a large influence on the phenotypic manifestation of genetic and epigenetic variation, it is not magic and clearly does not universally act on any type of variation. In fact, the most universal mechanism for buffering the effects of genetic variation is being diploid, which prevents most mutations from having a phenotypic impact (because they are recessive). Yet even diploidy fails at buffering dominant mutations, which are estimated to constitute ca. 10–20% of variants from studies in D. melanogaster and S. cerevisiae [170, 171]. Likewise, many human diseases, including various cancers, arise from haploinsufficiency or other dominant mutations [172]. Experiments in plants and yeast suggest that Hsp90 activity can influence many natural genetic variants [29, 31], and the polymorphisms that have been fine-mapped to date are plausibly linked to Hsp90 chaperone function [29]. This is a natural consequence of Hsp90’s interaction with large swaths of cellular circuitry integral to growth and development. Theory holds that in any sexually reproducing organism the costs of maintaining a system for increasing variation will eventually be separated from beneficial variants via meiotic recombination [173]. Yet because Hsp90’s chaperone activity is required for nearly all aspects of eukaryotic biology, such separation is likely impossible. Furthermore, the large number of Hsp90 clients and their distribution throughout the genome makes it likely that combinatorial gain (and loss) of chaperone-dependent variants can occur at each generation.
Hsp90’s influence on evolutionary processes is now indisputable – we can detect the chaperone’s impact in genomes. Yet a better understanding of the genotype-to-phenotype map across multiple environments will improve our understanding of how this chaperone alters adaptive landscapes (Figure 7). Fortunately, we have new tools in hand to achieve this goal. Using deep mutational scanning, it is now possible to systemically interrogate how Hsp90 affects variants at every position within a protein. Likewise, new crossing strategies in model organisms [174] and advances in human functional genomics [48] will enable identification of Hsp90-dependent genetic variants at single nucleotide resolution and on an unprecedented scale. These powerful new approaches promise to transform our mechanistic understanding of how the Hsp90 chaperone acts as a global regulator of the evolving genotype-to-phenotype map.
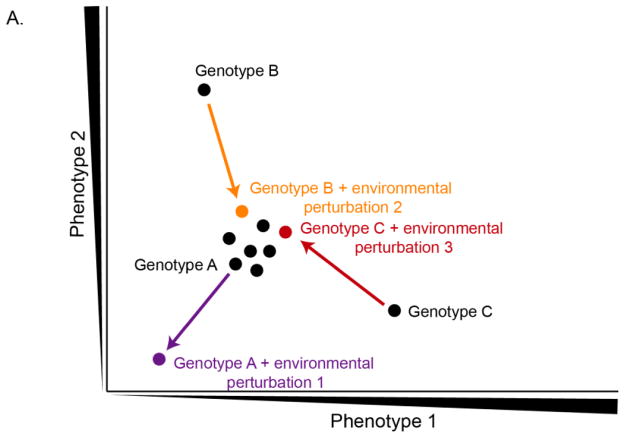
Figure 7. Phenotypic neighborhoods are rewired by changes in environment.
In this cartoon example, two phenotypes are graphed in two dimensional phenotypic spaces (axes). Each point represents a different genotype. The same genotype is graphed twice when it presents a different phenotype in a different environmental condition (colored points). Phenotypes dependent on the three different environmental perturbations are graphed (purple, orange, red) at the end of vectors representing the difference due to buffering or potentiation. Research illuminating examples such as these will greatly advance our understanding of how Hsp90 and environmental perturbations alter phenotypic landscapes.
Acknowledgments
Funding sources
This work was supported by a National Institutes of Health New Innovator Award (NIH-DP2-GM119140), a Searle Scholar Award (14-SSP-210), a Kimmel Scholar Award (SKF-15-154), and by a Science and Engineering Fellowship from the David and Lucile Packard Foundation to D.F.J. Further, this work was supported by National Institute of Health awards (NIH-R01-GM114166, NIH -R21-AG0520203, NIH-R01-GM122088) and a National Science Foundation award (NSF MCB-1516701) to CQ. G.A.M. is supported on the Genetic Approaches to Aging Training Grant (NIH-T32-AG00057). R.A.Z. is supported by a Stanford Dean’s Postdoctoral Fellowship.
We would like acknowledge our late mentor, Susan Lindquist, a fearless scientist who together with her postdoctoral fellow Suzannah Rutherford discovered that perturbation of Hsp90 in flies produces rare genotype-specific phenotypes that respond to selection. Sue recognized the enormous importance of this unexpected chance observation, and connected it to Waddington’s seminal work on canalization decades earlier. In doing so, she laid the groundwork for a myriad of subsequent studies by her group and many others that demonstrated the crucial role of protein folding in both evolution and disease, opening new therapeutic avenues for disorders like ALS, Parkinson’s, and Alzheimer’s. We mentees are immensely grateful for the opportunity to have worked with her and to have taken part in some of these studies. Her scientific genius has inspired our work and immeasurably enriched the scientific community. This is a review she wanted to write herself; we humbly apologize for our imperfect attempt at doing her fine mind and writing justice. However, as her generation’s most eminent geneticist, Sue gave this piece its title (or nearly so): “It’s not magic, it’s genetics”.
Footnotes
Competing interest statement
The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Felix MA, Wagner A. Robustness and evolution: concepts, insights and challenges from a developmental model system. Heredity (Edinb) 2008;100(2):132–40. doi: 10.1038/sj.hdy.6800915. [DOI] [PubMed] [Google Scholar]
- 2.Lachowiec J, Queitsch C, Kliebenstein DJ. Molecular mechanisms governing differential robustness of development and environmental responses in plants. Ann Bot. 2016;117(5):795–809. doi: 10.1093/aob/mcv151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergman A, Siegal ML. Evolutionary capacitance as a general feature of complex gene networks. Nature. 2003;424(6948):549–52. doi: 10.1038/nature01765. [DOI] [PubMed] [Google Scholar]
- 4.Ciliberti S, Martin OC, Wagner A. Innovation and robustness in complex regulatory gene networks. Proc Natl Acad Sci U S A. 2007;104(34):13591–6. doi: 10.1073/pnas.0705396104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Visser JA, Hermisson J, Wagner GP, Ancel Meyers L, Bagheri-Chaichian H, Blanchard JL, Chao L, Cheverud JM, Elena SF, Fontana W, Gibson G, Hansen TF, Krakauer D, Lewontin RC, Ofria C, Rice SH, von Dassow G, Wagner A, Whitlock MC. Perspective: Evolution and detection of genetic robustness. Evolution Int J Org Evolution. 2003;57(9):1959–72. doi: 10.1111/j.0014-3820.2003.tb00377.x. [DOI] [PubMed] [Google Scholar]
- 6.Hornstein E, Shomron N. Canalization of development by microRNAs. Nat Genet. 2006;38(Suppl):S20–4. doi: 10.1038/ng1803. [DOI] [PubMed] [Google Scholar]
- 7.Jarosz DF, Taipale M, Lindquist S. Protein homeostasis and the phenotypic manifestation of genetic diversity: principles and mechanisms. Annu Rev Genet. 2010;44:189–216. doi: 10.1146/annurev.genet.40.110405.090412. [DOI] [PubMed] [Google Scholar]
- 8.Leclerc RD. Survival of the sparsest: robust gene networks are parsimonious. Mol Syst Biol. 2008;4:213. doi: 10.1038/msb.2008.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lehner B. Selection to minimise noise in living systems and its implications for the evolution of gene expression. Mol Syst Biol. 2008;4:170. doi: 10.1038/msb.2008.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levy SF, Siegal ML. Network hubs buffer environmental variation in Saccharomyces cerevisiae. PLoS Biol. 2008;6(11):e264. doi: 10.1371/journal.pbio.0060264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li X, Cassidy JJ, Reinke CA, Fischboeck S, Carthew RW. A microRNA imparts robustness against environmental fluctuation during development. Cell. 2009;137(2):273–82. doi: 10.1016/j.cell.2009.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Manu, Surkova S, Spirov AV, Gursky VV, Janssens H, Kim AR, Radulescu O, Vanario-Alonso CE, Sharp DH, Samsonova M, Reinitz J. Canalization of gene expression and domain shifts in the Drosophila blastoderm by dynamical attractors. PLoS Comput Biol. 2009;5(3):e1000303. doi: 10.1371/journal.pcbi.1000303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Masel J, Siegal ML. Robustness: mechanisms and consequences. Trends Genet. 2009;25(9):395–403. doi: 10.1016/j.tig.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raser JM, O’Shea EK. Noise in gene expression: origins, consequences, and control. Science. 2005;309(5743):2010–3. doi: 10.1126/science.1105891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wagner A. Robustness against mutations in genetic networks of yeast. Nat Genet. 2000;24(4):355–61. doi: 10.1038/74174. [DOI] [PubMed] [Google Scholar]
- 16.Wagner A. Gene duplications, robustness and evolutionary innovations. Bioessays. 2008;30(4):367–73. doi: 10.1002/bies.20728. [DOI] [PubMed] [Google Scholar]
- 17.Salathia N, Queitsch C. Molecular mechanisms of canalization: Hsp90 and beyond. J Biosci. 2007;32(3):457–63. doi: 10.1007/s12038-007-0045-9. [DOI] [PubMed] [Google Scholar]
- 18.Sangster TA, Lindquist S, Queitsch C. Under cover: causes, effects and implications of Hsp90-mediated genetic capacitance. Bioessays. 2004;26(4):348–62. doi: 10.1002/bies.20020. [DOI] [PubMed] [Google Scholar]
- 19.Rutherford S, Hirate Y, Swalla BJ. The Hsp90 capacitor, developmental remodeling, and evolution: the robustness of gene networks and the curious evolvability of metamorphosis. Crit Rev Biochem Mol Biol. 2007;42(5):355–72. doi: 10.1080/10409230701597782. [DOI] [PubMed] [Google Scholar]
- 20.Baggs JE, Price TS, DiTacchio L, Panda S, Fitzgerald GA, Hogenesch JB. Network features of the mammalian circadian clock. PLoS Biol. 2009;7(3):e52. doi: 10.1371/journal.pbio.1000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Whitacre JM. Biological robustness: paradigms, mechanisms, and systems principles. Front Genet. 2012;3:67. doi: 10.3389/fgene.2012.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gibson G, Wagner G Department of Genetics GHNCSURNC, Gardner Hall NCSURNC, E. Department of OMLYUNHC S. Evolutionary Biology, Canalization in evolutionary genetics: a stabilizing theory? BioEssays. 2017;22(4):372–380. doi: 10.1002/(SICI)1521-1878(200004)22:4<372::AID-BIES7>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 23.Debat V, David P. Mapping phenotypes: canalization, plasticity and developmental stability. Trends in Ecology & Evolution. 2001;16(10):555–561. [Google Scholar]
- 24.Wagner A. Circuit topology and the evolution of robustness in two-gene circadian oscillators. 2005 doi: 10.1073/pnas.0501094102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Flatt T. The evolutionary genetics of canalization. Q Rev Biol. 2005;80(3):287–316. doi: 10.1086/432265. [DOI] [PubMed] [Google Scholar]
- 26.Dworkin I. In: Canalization, Cryptic Variation, and Developmental Buffering: A Critical Examination and Analytical Perspective, Variation—a central concept in biology. Hallgrímsson B, Hall BK, editors. Chapter 8. 2005. pp. 131–158. [Google Scholar]
- 27.Gibson G, Dworkin I. Uncovering cryptic genetic variation. Nat Rev Genet. 2004;5(9):681–90. doi: 10.1038/nrg1426. [DOI] [PubMed] [Google Scholar]
- 28.Burga A, Casanueva MO, Lehner B. Predicting mutation outcome from early stochastic variation in genetic interaction partners. Nature. 2011;480(7376):250–3. doi: 10.1038/nature10665. [DOI] [PubMed] [Google Scholar]
- 29.Jarosz DF, Lindquist S. Hsp90 and environmental stress transform the adaptive value of natural genetic variation. Science. 2010;330(6012):1820–4. doi: 10.1126/science.1195487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lehner B, Crombie C, Tischler J, Fortunato A, Fraser AG. Systematic mapping of genetic interactions in Caenorhabditis elegans identifies common modifiers of diverse signaling pathways. Nat Genet. 2006;38(8):896–903. doi: 10.1038/ng1844. [DOI] [PubMed] [Google Scholar]
- 31.Queitsch C, Sangster TA, Lindquist SL. Hsp90 as a capacitor of phenotypic variation. Nature. 2002;417(6889):618–24. doi: 10.1038/nature749. [DOI] [PubMed] [Google Scholar]
- 32.Rutherford SL, Lindquist S. Hsp90 as a capacitor for morphological evolution. Nature. 1998;396(6709):336–42. doi: 10.1038/24550. [DOI] [PubMed] [Google Scholar]
- 33.Yeyati PL, Bancewicz RM, Maule J, van Heyningen V. Hsp90 selectively modulates phenotype in vertebrate development. PLoS Genet. 2007;3(3):e43. doi: 10.1371/journal.pgen.0030043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cohen D. Optimizing reproduction in a randomly varying environment. J Theor Biol. 1966;12(1):119–29. doi: 10.1016/0022-5193(66)90188-3. [DOI] [PubMed] [Google Scholar]
- 35.Lenski RE, Barrick JE, Ofria C. Balancing robustness and evolvability. PLoS Biol. 2006;4(12):e428. doi: 10.1371/journal.pbio.0040428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Waddington CH. Genetic Assimilation of an Acquired Character. Evolution. 1953;7(2):118–126. [Google Scholar]
- 37.Ledon-Rettig CC, Pfennig DW, Crespi EJ. Diet and hormonal manipulation reveal cryptic genetic variation: implications for the evolution of novel feeding strategies. Proceedings of the royal society. 2010 doi: 10.1098/rspb.2010.0877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suzuki Y, Nijhout HF. Evolution of a polyphenism by genetic accommodation. Science. 2006;311(5761):650–2. doi: 10.1126/science.1118888. [DOI] [PubMed] [Google Scholar]
- 39.Rutherford SL, Lindquist S. Hsp90 as a capacitor for morphological evolution. Nature. 1998 doi: 10.1038/24550. [DOI] [PubMed] [Google Scholar]
- 40.Milton CC, Ulane CM, Rutherford S. Control of Canalization and Evolvability by Hsp90. PLoS ONE. 2006 doi: 10.1371/journal.pone.0000075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salathia N, Queitsch C. Molecular mechanisms of canalization: Hsp90 and beyond. 2007 doi: 10.1007/s12038-007-0045-9. [DOI] [PubMed] [Google Scholar]
- 42.Queitsch C, Sangster TA, Lindquist S. Hsp90 as a capacitor of phenotypic variation. Nature. 2002 doi: 10.1038/nature749. [DOI] [PubMed] [Google Scholar]
- 43.Yeyati PL, Bancewicz RM, Maule J, van Heyningen V. Hsp90 Selectively Modulates Phenotype in Vertebrate Development. PLoS Genetics. 2009 doi: 10.1371/journal.pgen.0030043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jarosz DF, Lindquist S. Hsp90 and Environmental Stress Transform the Adaptive Value of Natural Genetic Variation. Science. 2010 doi: 10.1126/science.1195487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cowen LE, Lindquist S. Hsp90 Potentiates the Rapid Evolution of New Traits: Drug Resistance in Diverse Fungi. 2005 doi: 10.1126/science.1118370. [DOI] [PubMed] [Google Scholar]
- 46.Casanueva MO, Burga A, Lehner B. Fitness Trade-Offs and Environmentally Induced Mutation Buffering in Isogenic C. elegans. Science. 2012 doi: 10.1126/science.1213491. [DOI] [PubMed] [Google Scholar]
- 47.Rohner N, Jarosz DF, Kowalko JE, Yoshizawa M, Jeffery WR, Borowsky RL, Lindquist S, Tabin CJ. Cryptic Variation in Morphological Evolution: HSP90 as a Capacitor for Loss of Eyes in Cavefish. Science. 2013 doi: 10.1126/science.1240276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Karras GI, Yi S, Sahni N, Fischer M, Xie J, Vidal M, D’Andrea AD, Whitesell L, Lindquist S. HSP90 Shapes the Consequences of Human Genetic Variation. Cell. 2017;168(5):856–866 e12. doi: 10.1016/j.cell.2017.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gorre ME, Ellwood-Yen K, Chiosis G, Rosen N, Sawyers CL. BCR-ABL point mutants isolated from patients with imatinib mesylate-resistant chronic myeloid leukemia remain sensitive to inhibitors of the BCR-ABL chaperone heat shock protein 90. Blood. 2002;100(8):3041–4. doi: 10.1182/blood-2002-05-1361. [DOI] [PubMed] [Google Scholar]
- 50.Nimmanapalli R, O’Bryan E, Huang M, Bali P, Burnette PK, Loughran T, Tepperberg J, Jove R, Bhalla K. Molecular Characterization and Sensitivity of STI-571 (Imatinib Mesylate, Gleevec)- resistant. Bcr-Abl-positive, Human Acute Leukemia Cells to SRC Kinase Inhibitor PD180970 and 17-Allylamino-17-demethoxygeldanamycin. 2016 [PubMed] [Google Scholar]
- 51.Press MO, Li H, Creanza N, Kramer G, Queitsch C, Sourjik V, Borenstein E. Genome-scale co-evolutionary inference identifies functions and clients of bacterial Hsp90. PLoS Genet. 2013;9(7):e1003631. doi: 10.1371/journal.pgen.1003631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maisnier-Patin S, Roth JR, Fredriksson A, Nystrom T, Berg OG, Andersson DI. Genomic buffering mitigates the effects of deleterious mutations in bacteria. Nat Genet. 2005;37(12):1376–9. doi: 10.1038/ng1676. [DOI] [PubMed] [Google Scholar]
- 53.Aguilar-Rodriguez J, Sabater-Munoz B, Montagud-Martinez R, Berlanga V, Alvarez-Ponce D, Wagner A, Fares MA. The Molecular Chaperone DnaK Is a Source of Mutational Robustness. Genome Biol Evol. 2016;8(9):2979–2991. doi: 10.1093/gbe/evw176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bogumil D, Dagan T. Chaperonin-dependent accelerated substitution rates in prokaryotes. Genome Biol Evol. 2010;2:602–8. doi: 10.1093/gbe/evq044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kadibalban AS, Bogumil D, Landan G, Dagan T. DnaK-Dependent Accelerated Evolutionary Rate in Prokaryotes. Genome Biol Evol. 2016;8(5):1590–9. doi: 10.1093/gbe/evw102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.PaxDb: Protein Abundance Database. 2017 https://pax-db.org/
- 57.Lindquist S. Regulation of protein synthesis during heat shock. Nature. 1981;293(5830):311–4. doi: 10.1038/293311a0. [DOI] [PubMed] [Google Scholar]
- 58.Taipale M, Jarosz DF, Lindquist S. HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nat Rev Mol Cell Biol. 2010;11(7):515–28. doi: 10.1038/nrm2918. [DOI] [PubMed] [Google Scholar]
- 59.Lindquist KAB, Farrelly FW, Finkelstein DB, Taulien J. hsp82 is an essential protein that is required in higher concentrations for growth of cells at higher temperatures. 1989 doi: 10.1128/mcb.9.9.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cowen LE, Lindquist S. Hsp90 potentiates the rapid evolution of new traits: drug resistance in diverse fungi. Science. 2005;309(5744):2185–9. doi: 10.1126/science.1118370. [DOI] [PubMed] [Google Scholar]
- 61.Queitsch C, Hong SW, Vierling E, Lindquist S. Heat shock protein 101 plays a crucial role in thermotolerance in Arabidopsis. Plant Cell. 2000;12(4):479–92. doi: 10.1105/tpc.12.4.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhao R, Davey M, Hsu Y-C, Kaplanek P, Tong A, Parsons AB, Krogan N, Cagney G, Mai D, Greenblatt J, Boone C, Emili A, Houry WA. Navigating the Chaperone Network: An Integrative Map of Physical and Genetic Interactions Mediated by the Hsp90 Chaperone. Cell. 2005;120(5):715–727. doi: 10.1016/j.cell.2004.12.024. [DOI] [PubMed] [Google Scholar]
- 63.Taipale M, Tucker G, Peng J, Krykbaeva I, Lin ZY, Larsen B, Choi H, Berger B, Gingras AC, Lindquist S. A quantitative chaperone interaction network reveals the architecture of cellular protein homeostasis pathways. Cell. 2014;158(2):434–448. doi: 10.1016/j.cell.2014.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sahni N, Yi S, Taipale M, Fuxman Bass JI, Coulombe-Huntington J, Yang F, Peng J, Weile J, Karras GI, Wang Y, Kovacs IA, Kamburov A, Krykbaeva I, Lam MH, Tucker G, Khurana V, Sharma A, Liu YY, Yachie N, Zhong Q, Shen Y, Palagi A, San-Miguel A, Fan C, Balcha D, Dricot A, Jordan DM, Walsh JM, Shah AA, Yang X, Stoyanova AK, Leighton A, Calderwood MA, Jacob Y, Cusick ME, Salehi-Ashtiani K, Whitesell LJ, Sunyaev S, Berger B, Barabasi AL, Charloteaux B, Hill DE, Hao T, Roth FP, Xia Y, Walhout AJM, Lindquist S, Vidal M. Widespread macromolecular interaction perturbations in human genetic disorders. Cell. 2015;161(3):647–660. doi: 10.1016/j.cell.2015.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sangster TA, Queitsch C. The HSP90 chaperone complex, an emerging force in plant development and phenotypic plasticity. Curr Opin Plant Biol. 2005;8(1):86–92. doi: 10.1016/j.pbi.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 66.Biebl FHS, Johannes B, Maximilian M. The HSP90 chaperone machinery. Nature Reviews Molecular Cell Biology. 2017;18(6):345. doi: 10.1038/nrm.2017.20. [DOI] [PubMed] [Google Scholar]
- 67.Schopf FH, Biebl MM, Buchner J. The HSP90 chaperone machinery. Nat Rev Mol Cell Biol. 2017;18(6):345–360. doi: 10.1038/nrm.2017.20. [DOI] [PubMed] [Google Scholar]
- 68.Krukenberg KA, Street TO, Lavery LA, Agard DA. Conformational dynamics of the molecular chaperone Hsp90. Quarterly reviews of biophysics. 2011;44(2):229–55. doi: 10.1017/S0033583510000314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Meyer P, Prodromou C, Liao C, Hu B, Roe SM, Vaughan CK, Vlasic I, Panaretou B, Piper PW, Pearl LH. Structural basis for recruitment of the ATPase activator Aha1 to the Hsp90 chaperone machinery. EMBO J. 2004;23(6):1402–10. doi: 10.1038/sj.emboj.7600141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Meyer P, Prodromou C, Hu B, Vaughan C, Roe SM, Panaretou B, Piper PW, Pearl LH. Structural and functional analysis of the middle segment of hsp90: implications for ATP hydrolysis and client protein and cochaperone interactions. Mol Cell. 2003;11(3):647–58. doi: 10.1016/s1097-2765(03)00065-0. [DOI] [PubMed] [Google Scholar]
- 71.Sato S, Fujita N, Tsuruo T. Modulation of Akt kinase activity by binding to Hsp90. Proc Natl Acad Sci U S A. 2000;97(20):10832–7. doi: 10.1073/pnas.170276797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Karagoz GE, Duarte AM, Akoury E, Ippel H, Biernat J, Moran Luengo T, Radli M, Didenko T, Nordhues BA, Veprintsev DB, Dickey CA, Mandelkow E, Zweckstetter M, Boelens R, Madl T, Rudiger SG. Hsp90-Tau complex reveals molecular basis for specificity in chaperone action. Cell. 2014;156(5):963–74. doi: 10.1016/j.cell.2014.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Vaughan CK, Gohlke U, Sobott F, Good VM, Ali MM, Prodromou C, Robinson CV, Saibil HR, Pearl LH. Structure of an Hsp90-Cdc37-Cdk4 complex. Mol Cell. 2006;23(5):697–707. doi: 10.1016/j.molcel.2006.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Street TO, Lavery LA, Agard DA. Substrate binding drives large-scale conformational changes in the Hsp90 molecular chaperone. Mol Cell. 2011;42(1):96–105. doi: 10.1016/j.molcel.2011.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hugel CR, Johannes B, Martin H, Moritz M, Thorsten The large conformational changes of Hsp90 are only weakly coupled to ATP hydrolysis. Nature Structural and Molecular Biology. 2009;16(3):281. doi: 10.1038/nsmb.1557. [DOI] [PubMed] [Google Scholar]
- 76.Hugel BH, Florian K, Martin Z, Philipp W, Thorsten Multidomain structure and correlated dynamics determined by self-consistent FRET networks. Nature Methods. 2016;14(2):174. doi: 10.1038/nmeth.4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cintron NS, Toft D. Defining the requirements for Hsp40 and Hsp70 in the Hsp90 chaperone pathway. J Biol Chem. 2006;281(36):26235–44. doi: 10.1074/jbc.M605417200. [DOI] [PubMed] [Google Scholar]
- 78.Pearl LH. Hsp90 and Cdc37 – a chaperone cancer conspiracy. Current Opinion in Genetics & Development. 2005;15(1):55–61. doi: 10.1016/j.gde.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 79.Verba KA, Agard DA. How Hsp90 and Cdc37 Lubricate Kinase Molecular Switches. Trends in biochemical sciences. 2017;42(10):799–811. doi: 10.1016/j.tibs.2017.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cutforth T, Rubin GM. Mutations in Hsp83 and cdc37 impair signaling by the sevenless receptor tyrosine kinase in Drosophila. Cell. 1994;77(7):1027–36. doi: 10.1016/0092-8674(94)90442-1. [DOI] [PubMed] [Google Scholar]
- 81.Stepanova L, Leng X, Parker SB, Harper JW. Mammalian p50Cdc37 is a protein kinase-targeting subunit of Hsp90 that binds and stabilizes Cdk4. Genes Dev. 1996;10(12):1491–502. doi: 10.1101/gad.10.12.1491. [DOI] [PubMed] [Google Scholar]
- 82.Vaughan CK, Mollapour M, Smith JR, Truman A, Hu B, Good VM, Panaretou B, Neckers L, Clarke PA, Workman P, Piper PW, Prodromou C, Pearl LH. Hsp90-dependent activation of protein kinases is regulated by chaperone-targeted dephosphorylation of Cdc37. Molecular cell. 2008;31(6):886–95. doi: 10.1016/j.molcel.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Riggs DL, Cox MB, Cheung-Flynn J, Prapapanich V, Carrigan PE, Smith DF. Functional specificity of co-chaperone interactions with Hsp90 client proteins. Crit Rev Biochem Mol Biol. 2004;39(5–6):279–95. doi: 10.1080/10409230490892513. [DOI] [PubMed] [Google Scholar]
- 84.Earley KW, Poethig RS. Binding of the cyclophilin 40 ortholog SQUINT to Hsp90 protein is required for SQUINT function in Arabidopsis. J Biol Chem. 2011;286(44):38184–9. doi: 10.1074/jbc.M111.290130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Breiman A. Plant Hsp90 and its Co-Chaperones. Current protein & peptide science. 2017;15(3):232–244. doi: 10.2174/1389203715666140331115603. [DOI] [PubMed] [Google Scholar]
- 86.Smith DLR, Marc BC, Joyce C-F, Viravan P, Patricia EC, David F. Functional Specificity of Co-Chaperone Interactions with Hsp90 Client Proteins. 2010 doi: 10.1080/10409230490892513. [DOI] [PubMed]
- 87.Zuehlke A, Johnson JL. Hsp90 and co-chaperones twist the functions of diverse client proteins. Biopolymers. 2010;93(3):211–7. doi: 10.1002/bip.21292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Salminen A, Ojala J, Kaarniranta K, Hiltunen M, Soininen H. Hsp90 regulates tau pathology through co-chaperone complexes in Alzheimer’s disease. Progress in neurobiology. 2011;93(1):99–110. doi: 10.1016/j.pneurobio.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 89.Fan AC, Young JC. Function of cytosolic chaperones in Tom70-mediated mitochondrial import. Protein and peptide letters. 2011;18(2):122–31. doi: 10.2174/092986611794475020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhang XC, Millet YA, Cheng Z, Bush J, Ausubel FM. Jasmonate signalling in Arabidopsis involves SGT1b-HSP70-HSP90 chaperone complexes. Nat Plants 1. 2015 doi: 10.1038/nplants.2015.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Edkins AL. CHIP: a co-chaperone for degradation by the proteasome. Sub-cellular biochemistry. 2015;78:219–42. doi: 10.1007/978-3-319-11731-7_11. [DOI] [PubMed] [Google Scholar]
- 92.Calderwood SK. Cdc37 as a co-chaperone to Hsp90. Sub-cellular biochemistry. 2015;78:103–12. doi: 10.1007/978-3-319-11731-7_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Baindur-Hudson S, Edkins AL, Blatch GL. Hsp70/Hsp90 organising protein (hop): beyond interactions with chaperones and prion proteins. Sub-cellular biochemistry. 2015;78:69–90. doi: 10.1007/978-3-319-11731-7_3. [DOI] [PubMed] [Google Scholar]
- 94.Guy NC, Garcia YA, Sivils JC, Galigniana MD, Cox MB. Functions of the Hsp90-binding FKBP immunophilins. Sub-cellular biochemistry. 2015;78:35–68. doi: 10.1007/978-3-319-11731-7_2. [DOI] [PubMed] [Google Scholar]
- 95.Karam JA, Parikh RY, Nayak D, Rosenkranz D, Gangaraju VK. Co-chaperone Hsp70/Hsp90-organizing protein (Hop) is required for transposon silencing and Piwi-interacting RNA (piRNA) biogenesis. J Biol Chem. 2017;292(15):6039–6046. doi: 10.1074/jbc.C117.777730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Eichler EE, Flint J, Gibson G, Kong A, Leal SM, Moore JH, Nadeau JH. Missing heritability and strategies for finding the underlying causes of complex disease. Nat Rev Genet. 2010;11(6):446–50. doi: 10.1038/nrg2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Gibson G. Rare and common variants: twenty arguments. Nat Rev Genet. 2012;13(2):135–45. doi: 10.1038/nrg3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, Cho JH, Guttmacher AE, Kong A, Kruglyak L, Mardis E, Rotimi CN, Slatkin M, Valle D, Whittemore AS, Boehnke M, Clark AG, Eichler EE, Gibson G, Haines JL, Mackay TF, McCarroll SA, Visscher PM. Finding the missing heritability of complex diseases. Nature. 2009;461(7265):747–53. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Queitsch C, Carlson KD, Girirajan S. Lessons from model organisms: phenotypic robustness and missing heritability in complex disease. PLoS Genet. 2012;8(11):e1003041. doi: 10.1371/journal.pgen.1003041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li J, Browning S, Mahal SP, Oelschlegel AM, Weissmann C. Darwinian evolution of prions in cell culture. Science. 2010;327(5967):869–72. doi: 10.1126/science.1183218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.van Leeuwen J, Pons C, Mellor JC, Yamaguchi TN, Friesen H, et al. Exploring genetic suppression interactions on a global scale. 2016 doi: 10.1126/science.aag0839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Costanzo M, VanderSluis B, Koch EN, Baryshnikova A, Pons C, Tan G, Wang W, Usaj M, Hanchard J, Lee SD, Pelechano V, Styles EB, Billmann M, van Leeuwen J, van Dyk N, Lin ZY, Kuzmin E, Nelson J, Piotrowski JS, Srikumar T, Bahr S, Chen Y, Deshpande R, Kurat CF, Li SC, Li Z, Usaj MM, Okada H, Pascoe N, San Luis BJ, Sharifpoor S, Shuteriqi E, Simpkins SW, Snider J, Suresh HG, Tan Y, Zhu H, Malod-Dognin N, Janjic V, Przulj N, Troyanskaya OG, Stagljar I, Xia T, Ohya Y, Gingras AC, Raught B, Boutros M, Steinmetz LM, Moore CL, Rosebrock AP, Caudy AA, Myers CL, Andrews B, Boone C. A global genetic interaction network maps a wiring diagram of cellular function. Science. 2016;353(6306) doi: 10.1126/science.aaf1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Borkovich KA, Farrelly FW, Finkelstein DB, Taulien J, Lindquist S. hsp82 is an essential protein that is required in higher concentrations for growth of cells at higher temperatures. Mol Cell Biol. 1989;9(9):3919–30. doi: 10.1128/mcb.9.9.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lindquist S, Craig EA. The heat-shock proteins. Annu Rev Genet. 1988;22:631–77. doi: 10.1146/annurev.ge.22.120188.003215. [DOI] [PubMed] [Google Scholar]
- 105.Whitesell L, Mimnaugh EG, De Costa B, Myers CE, Neckers LM. Inhibition of heat shock protein HSP90-pp60v-src heteroprotein complex formation by benzoquinone ansamycins: essential role for stress proteins in oncogenic transformation. Proc Natl Acad Sci U S A. 1994;91(18):8324–8. doi: 10.1073/pnas.91.18.8324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Xu Y, Lindquist S. Heat-shock protein hsp9O governs the activity of pp6Ovsrc kinase. PNAS. 1993;90:7074–7078. doi: 10.1073/pnas.90.15.7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chen B, Wagner A. Hsp90 is important for fecundity, longevity, and buffering of cryptic deleterious variation in wild fly populations. BMC Evol Biol. 2012 doi: 10.1186/1471-2148-12-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Sgro CM, Wegener B, Hoffmann AA. A naturally occurring variant of Hsp90 that is associated with decanalization. Proc Biol Sci. 2010;277(1690):2049–57. doi: 10.1098/rspb.2010.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Casanueva MO, Burga A, Lehner B. Fitness trade-offs and environmentally induced mutation buffering in isogenic C. elegans. Science. 2012;335(6064):82–5. doi: 10.1126/science.1213491. [DOI] [PubMed] [Google Scholar]
- 110.Lachowiec J, Lemus T, Thomas JH, Murphy PJ, Nemhauser JL, Queitsch C. The protein chaperone HSP90 can facilitate the divergence of gene duplicates. Genetics. 2013;193(4):1269–77. doi: 10.1534/genetics.112.148098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lachowiec J, Lemus T, Borenstein E, Queitsch C. Hsp90 promotes kinase evolution. Mol Biol Evol. 2015;32(1):91–9. doi: 10.1093/molbev/msu270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sangster TA, Salathia N, Lee HN, Watanabe E, Schellenberg K, Morneau K, Wang H, Undurraga S, Queitsch C, Lindquist S. HSP90-buffered genetic variation is common in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2008;105(8):2969–74. doi: 10.1073/pnas.0712210105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sangster TA, Salathia N, Undurraga S, Milo R, Schellenberg K, Lindquist S, Queitsch C. HSP90 affects the expression of genetic variation and developmental stability in quantitative traits. Proc Natl Acad Sci U S A. 2008;105(8):2963–8. doi: 10.1073/pnas.0712200105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Dorrity MW, Cuperus JT, Carlisle JA, Fields S, Queitsch C. Trait specificity mediated by alternative DNA-binding preferences of a single transcription factor in yeast. 2017 [Google Scholar]
- 115.Whitesell L, Lindquist SL. HSP90 and the chaperoning of cancer. Nat Rev Cancer. 2005;5(10):761–72. doi: 10.1038/nrc1716. [DOI] [PubMed] [Google Scholar]
- 116.Perfect JR. Cryptococcus neoformans: the yeast that likes it hot. FEMS yeast research. 2006;6(4):463–8. doi: 10.1111/j.1567-1364.2006.00051.x. [DOI] [PubMed] [Google Scholar]
- 117.Johnson A. The biology of mating in Candida albicans. Nature reviews. Microbiology. 2003;1(2):106–16. doi: 10.1038/nrmicro752. [DOI] [PubMed] [Google Scholar]
- 118.Dong H, Courchesne W. A novel quantitative mating assay for the fungal pathogen Cryptococcus neoformans provides insight into signalling pathways responding to nutrients and temperature. Microbiology. 1998;144(Pt 6):1691–7. doi: 10.1099/00221287-144-6-1691. [DOI] [PubMed] [Google Scholar]
- 119.Shapiro RS, Cowen LE. Thermal control of microbial development and virulence: molecular mechanisms of microbial temperature sensing. mBio. 2012;3(5) doi: 10.1128/mBio.00238-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hummel B, Hansen EC, Yoveva A, Aprile-Garcia F, Hussong R, Sawarkar R. The evolutionary capacitor HSP90 buffers the regulatory effects of mammalian endogenous retroviruses. Nat Struct Mol Biol. 2017;24(3):234–242. doi: 10.1038/nsmb.3368. [DOI] [PubMed] [Google Scholar]
- 121.Debat V, Milton CC, Rutherford S, Klingenberg CP, Hoffmann AA. Hsp90 and the quantitative variation of wing shape in Drosophila melanogaster. Evolution. 2006;60(12):2529–38. [PubMed] [Google Scholar]
- 122.Takahashi KH. Little effect of HSP90 inhibition on the quantitative wing traits variation in Drosophila melanogaster. Genetica. 2017;145(1):9–18. doi: 10.1007/s10709-016-9940-z. [DOI] [PubMed] [Google Scholar]
- 123.Xu Y, Singer MA, Lindquist S. Maturation of the tyrosine kinase c-src as a kinase and as a substrate depends on the molecular chaperone Hsp90. PNAS. 1999;96:109–114. doi: 10.1073/pnas.96.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Neckers L. Heat shock protein 90: the cancer chaperone. 2007 doi: 10.1007/s12038-007-0051-y. [DOI] [PubMed] [Google Scholar]
- 125.Cowen LE, Steinbach WJ. Stress, drugs, and evolution: the role of cellular signaling in fungal drug resistance. Eukaryotic cell. 2008;7(5):747–64. doi: 10.1128/EC.00041-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Geiler-Samerotte KA, Zhu YO, Goulet BE, Hall DW, Siegal ML. Selection Transforms the Landscape of Genetic Variation Interacting with Hsp90. PLoS Biol. 2016;14(10):e2000465. doi: 10.1371/journal.pbio.2000465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Pratt WB. The role of heat shock proteins in regulating the function, folding, and trafficking of the glucocorticoid receptor. J Biol Chem. 1993;268(29):21455–8. [PubMed] [Google Scholar]
- 128.Voellmy R. On mechanisms that control heat shock transcription factor activity in metazoan cells. Cell Stress Chaperones. 2004;9(2):122–33. doi: 10.1379/CSC-14R.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Voellmy R, Boellmann F. Chaperone regulation of the heat shock protein response. Adv Exp Med Biol. 2007;594:89–99. doi: 10.1007/978-0-387-39975-1_9. [DOI] [PubMed] [Google Scholar]
- 130.Prodromou C. Mechanisms of Hsp90 regulation. The Biochemical journal. 2016;473(16):2439–52. doi: 10.1042/BCJ20160005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Zeyl C, DeVisser JA. Estimates of the rate and distribution of fitness effects of spontaneous mutation in Saccharomyces cerevisiae. Genetics. 2001;157(1):53–61. doi: 10.1093/genetics/157.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Eyre-Walker A, Keightley PD. The distribution of fitness effects of new mutations. Nat Rev Genet. 2007;8(8):610–8. doi: 10.1038/nrg2146. [DOI] [PubMed] [Google Scholar]
- 133.Masel J, Trotter MV. Robustness and evolvability. Trends Genet. 2010;26(9):406–14. doi: 10.1016/j.tig.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Mittelman D, Sykoudis K, Hersh M, Lin Y, Wilson JH. Hsp90 modulates CAG repeat instability in human cells. Cell stress & chaperones. 2010;15(5):753–9. doi: 10.1007/s12192-010-0191-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Dote H, Burgan WE, Camphausen K, Tofilon PJ. Inhibition of hsp90 compromises the DNA damage response to radiation. Cancer research. 2006;66(18):9211–20. doi: 10.1158/0008-5472.CAN-06-2181. [DOI] [PubMed] [Google Scholar]
- 136.Chen G, Bradford WD, Seidel CW, Li R. Hsp90 stress potentiates rapid cellular adaptation through induction of aneuploidy. Nature. 2012;482(7384):246–50. doi: 10.1038/nature10795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Khurana N, Laskar S, Bhattacharyya MK, Bhattacharyya S. Hsp90 induces increased genomic instability toward DNA-damaging agents by tuning down RAD53 transcription. Molecular biology of the cell. 2016;27(15):2463–78. doi: 10.1091/mbc.E15-12-0867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Specchia V, Piacentini L, Tritto P, Fanti L, D’Alessandro R, Palumbo G, Pimpinelli S, Bozzetti MP. Hsp90 prevents phenotypic variation by suppressing the mutagenic activity of transposons. Nature. 2010;463(7281):662–5. doi: 10.1038/nature08739. [DOI] [PubMed] [Google Scholar]
- 139.Piacentini L, Fanti L, Specchia V, Bozzetti MP, Berloco M, Palumbo G, Pimpinelli S. Transposons, environmental changes, and heritable induced phenotypic variability. Chromosoma. 2014;123(4):345–54. doi: 10.1007/s00412-014-0464-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Gangaraju VK, Yin H, Weiner MM, Wang J, Huang XA, Lin H. Drosophila Piwi functions in Hsp90-mediated suppression of phenotypic variation. Nat Genet. 2011;43(2):153–8. doi: 10.1038/ng.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Izumi N, Kawaoka S, Yasuhara S, Suzuki Y, Sugano S, Katsuma S, Tomari Y. Hsp90 facilitates accurate loading of precursor piRNAs into PIWI proteins. RNA. 2013;19(7):896–901. doi: 10.1261/rna.037200.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ryan CP, Brownlie JC, Whyard S. Hsp90 and Physiological Stress Are Linked to Autonomous Transposon Mobility and Heritable Genetic Change in Nematodes. Genome Biol Evol. 2016;8(12):3794–3805. doi: 10.1093/gbe/evw284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Xiol J, Cora E, Koglgruber R, Chuma S, Subramanian S, Hosokawa M, Reuter M, Yang Z, Berninger P, Palencia A, Benes V, Penninger J, Sachidanandam R, Pillai RS. A role for Fkbp6 and the chaperone machinery in piRNA amplification and transposon silencing. Mol Cell. 2012;47(6):970–9. doi: 10.1016/j.molcel.2012.07.019. [DOI] [PubMed] [Google Scholar]
- 144.Ichiyanagi T, Ichiyanagi K, Ogawa A, Kuramochi-Miyagawa S, Nakano T, Chuma S, Sasaki H, Udono H. HSP90alpha plays an important role in piRNA biogenesis and retrotransposon repression in mouse. Nucleic Acids Res. 2014;42(19):11903–11. doi: 10.1093/nar/gku881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kozeko L, Talalaiev O, Neimash V, Povarchuk V. A protective role of HSP90 chaperone in gamma-irradiated Arabidopsis thaliana seeds. Life sciences in space research. 2015;6:51–8. doi: 10.1016/j.lssr.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 146.Rohner N, Jarosz DF, Kowalko JE, Yoshizawa M, Jeffery WR, Borowsky RL, Lindquist S, Tabin CJ. Cryptic variation in morphological evolution: HSP90 as a capacitor for loss of eyes in cavefish. Science. 2013;342(6164):1372–5. doi: 10.1126/science.1240276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Migicovsky Z, Kovalchuk I. Changes to DNA methylation and homologous recombination frequency in the progeny of stressed plants. Biochemistry and cell biology = Biochimie et biologie cellulaire. 2013;91(1):1–5. doi: 10.1139/bcb-2012-0046. [DOI] [PubMed] [Google Scholar]
- 148.Galhardo RS, Hastings PJ, Rosenberg SM. Mutation as a stress response and the regulation of evolvability. Critical reviews in biochemistry and molecular biology. 2007;42(5):399–435. doi: 10.1080/10409230701648502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Sawarkar R, Paro R. Hsp90@chromatin.nucleus: an emerging hub of a networker. Trends in cell biology. 2013;23(4):193–201. doi: 10.1016/j.tcb.2012.11.007. [DOI] [PubMed] [Google Scholar]
- 150.Tariq M, Nussbaumer U, Chen Y, Beisel C, Paro R. Trithorax requires Hsp90 for maintenance of active chromatin at sites of gene expression. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(4):1157–62. doi: 10.1073/pnas.0809669106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Sollars V, Lu X, Xiao L, Wang X, Garfinkel MD, Ruden DM. Evidence for an epigenetic mechanism by which Hsp90 acts as a capacitor for morphological evolution. Nat Genet. 2003;33(1):70–4. doi: 10.1038/ng1067. [DOI] [PubMed] [Google Scholar]
- 152.Sangster TA, Queitsch C, Lindquist SL. Hsp90 and chromatin: where is the link? Cell Cycle. 2003;2(3):166–8. [PubMed] [Google Scholar]
- 153.Sawarkar R, Sievers C, Paro R. Hsp90 globally targets paused RNA polymerase to regulate gene expression in response to environmental stimuli. Cell. 2012;149(4):807–18. doi: 10.1016/j.cell.2012.02.061. [DOI] [PubMed] [Google Scholar]
- 154.Alberti S, Halfmann R, King O, Kapila A, Lindquist S. A systematic survey identifies prions and illuminates sequence features of prionogenic proteins. Cell. 2009;137(1):146–58. doi: 10.1016/j.cell.2009.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Halfmann R, Jarosz DF, Jones SK, Chang A, Lancaster AK, Lindquist S. Prions are a common mechanism for phenotypic inheritance in wild yeasts. Nature. 2012;482(7385):363–8. doi: 10.1038/nature10875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Chakrabortee S, Byers JS, Jones S, Garcia DM, Bhullar B, Chang A, She R, Lee L, Fremin B, Lindquist S, Jarosz DF. Intrinsically Disordered Proteins Drive Emergence and Inheritance of Biological Traits. Cell. 2016;167(2):369–381 e12. doi: 10.1016/j.cell.2016.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Shorter J, Lindquist S. Hsp104, Hsp70 and Hsp40 interplay regulates formation, growth and elimination of Sup35 prions. Embo J. 2008;27(20):2712–24. doi: 10.1038/emboj.2008.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kumar N, Gaur D, Gupta A, Puri A, Sharma D. Hsp90-Associated Immunophilin Homolog Cpr7 Is Required for the Mitotic Stability of [URE3] Prion in Saccharomyces cerevisiae. PLoS Genet. 2015;11(10):e1005567. doi: 10.1371/journal.pgen.1005567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Wolfe KJ, Ren HY, Trepte P, Cyr DM. The Hsp70/90 cochaperone, Sti1, suppresses proteotoxicity by regulating spatial quality control of amyloid-like proteins. Molecular biology of the cell. 2013;24(23):3588–602. doi: 10.1091/mbc.E13-06-0315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Hou F, Sun L, Zheng H, Skaug B, Jiang QX, Chen ZJ. MAVS forms functional prion-like aggregates to activate and propagate antiviral innate immune response. Cell. 2011;146(3):448–61. doi: 10.1016/j.cell.2011.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Xu H, He X, Zheng H, Huang LJ, Hou F, Yu Z, de la Cruz MJ, Borkowski B, Zhang X, Chen ZJ, Jiang QX. Structural basis for the prion-like MAVS filaments in antiviral innate immunity. eLife. 2014;3:e01489. doi: 10.7554/eLife.01489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Dai C, Whitesell L. HSP90: a rising star on the horizon of anticancer targets. Future Oncol. 2005;1(4):529–40. doi: 10.2217/14796694.1.4.529. [DOI] [PubMed] [Google Scholar]
- 163.Cullinan SB, Whitesell L. Heat shock protein 90: a unique chemotherapeutic target. Semin Oncol. 2006;33(4):457–65. doi: 10.1053/j.seminoncol.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 164.Rosenberg SM, Queitsch C. Medicine. Combating evolution to fight disease. Science. 2014;343(6175):1088–9. doi: 10.1126/science.1247472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Cowen LE, Carpenter AE, Matangkasombut O, Fink GR, Lindquist SL. Genetic architecture of Hsp90-dependent drug resistance. Eukaryot Cell. 2006;5(12):2184–8. doi: 10.1128/EC.00274-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Cowen LE. Hsp90 orchestrates stress response signaling governing fungal drug resistance. PLoS pathogens. 2009;5(8):e1000471. doi: 10.1371/journal.ppat.1000471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Singh SD, Robbins N, Zaas AK, Schell WA, Perfect JR, Cowen LE. Hsp90 governs echinocandin resistance in the pathogenic yeast Candida albicans via calcineurin. PLoS pathogens. 2009;5(7):e1000532. doi: 10.1371/journal.ppat.1000532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Shapiro RS, Uppuluri P, Zaas AK, Collins C, Senn H, Perfect JR, Heitman J, Cowen LE. Hsp90 orchestrates temperature-dependent Candida albicans morphogenesis via Ras1-PKA signaling. Current biology: CB. 2009;19(8):621–9. doi: 10.1016/j.cub.2009.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Cowen LE, Singh SD, Kohler JR, Collins C, Zaas AK, Schell WA, Aziz H, Mylonakis E, Perfect JR, Whitesell L, Lindquist S. Harnessing Hsp90 function as a powerful, broadly effective therapeutic strategy for fungal infectious disease. Proc Natl Acad Sci U S A. 2009;106(8):2818–23. doi: 10.1073/pnas.0813394106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Garcia-Dorado A, Caballero A. On the average coefficient of dominance of deleterious spontaneous mutations. Genetics. 2000;155(4):1991–2001. doi: 10.1093/genetics/155.4.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Agrawal AF, Whitlock MC. Inferences about the distribution of dominance drawn from yeast gene knockout data. Genetics. 2011;187(2):553–66. doi: 10.1534/genetics.110.124560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Huang N, Lee I, Marcotte EM, Hurles ME. Characterising and predicting haploinsufficiency in the human genome. PLoS Genet. 2010;6(10):e1001154. doi: 10.1371/journal.pgen.1001154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Raynes Y, Gazzara MR, Sniegowski PD. Mutator dynamics in sexual and asexual experimental populations of yeast. BMC evolutionary biology. 2011;11:158. doi: 10.1186/1471-2148-11-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.She R, Jarosz DF. Mapping Causal Variants with Single-Nucleotide Resolution Reveals Biochemical Drivers of Phenotypic Change. Cell. 2018;172(3):478–490 e15. doi: 10.1016/j.cell.2017.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]